Abstract
Cervical cancer is a common malignant tumor worldwide ranking fourth in incidence and mortality among females, which was reduced significantly by cytology screening and human papilloma virus (HPV) DNA testing. The specificity of cytology is high; however, the sensitivity is low, in contrast to the HPV DNA testing. Despite the success of these measures, new biomarkers are still considered to aim increasing sensitivity and specificity of screening and diagnosis. Significant alterations in microRNA (miRNA) expression have been detected in several cancers with variable consistency. To investigate the stratification role of miRNAs between normal epithelium and cervical intraepithelial neoplasia (CIN2–3), we screened the expression of 667 miRNAs to identify significant markers (n = 10), out of them 9 miRNAs were applied in the study (miR-20b, −24, −26a, −29b, −99a, −100, −147, −212, −515-3p) along with RNU48 and U6 as the references. To benchmark the miRNAs, 22 paired (tumor-free and tumor tissue pairs) laser microdissection-obtained cervical formalin fixed, paraffin embedded tissue samples were assayed. The expression of miR-20b was 2.4 times higher in CIN2–3 samples as compared to normal tissues (p < 0.0001). In the HPV16-positive subsets of the samples (n = 13), miR-20b showed 2.9-times elevation (p < 0.001), whereas miR-515 was 1.15-times downregulated (p < 0.05) in CIN2–3 as compared to normal tissue. These results suggest the potential value of miR-20b as a statification biomarker in order to differentiate neoplastic and non-tumorous cases.
Keywords: Cervical cancer, Cervical intraepithelial neoplasia (CIN), microRNA, Human papilloma virus
Introduction
Cervical cancer is a common malignant tumor worldwide among females regarding both incidence and mortality, ranking fourth after breast, lung, and colorectal cancer with an estimated 570,000 new cases yearly and 311,000 cancer related deaths in 2018 according to GLOBOCAN sources [1]. There is, however, great variability across the geographic regions of the world, with higher numbers occurring in several developing countries, where cervical cancer ranks second for both incidence and mortality [1]. The effect of population-based screening programs using the Papanicolaou („Pap”) test for cervical cancer resulted a significant decrease both in incidence and mortality [1–3]. Introduction of world-wide accepted guidelines for new evaluation methods, including the one formulated at the Bethesda Conference in 2001 [4], led to further decline in cervical cancer incidence. Discovery of the etiological role of the human papilloma virus (HPV) in cervical carcinogenesis [5] and the introduction of population-based vaccination programs were further steps towards the prevention of cervical cancer [6]. It became clear, however, that while being highly specific, the sensitivity of cytology is relatively low regarding the detection of high-grade squamous intraepithelial lesions (HSIL) or cervical intraepithelial neoplasia (CIN) [7]. Recently, more sensitive though less specific HPV DNA molecular detection tests have been introduced into the screening system effectively reducing cervical cancer incidence better than cytology screening alone. The demonstration of high-risk HPV (hrHPV) infection however does not prove whether the infection is transient or of the transforming type. [6, 8–10]
Several new biomarkers have been applied for detection of transformed cells aiming to increase the sensitivity and/or specificity of the screening tests and/or in combination with cytology and/or with HPV DNA testing [8, 11]. More recently, several groups have demonstrated aberrant expression of microRNAs (miRNAs) in premalignant and malignant cervical lesions, suggesting that this might be used as a more specific biomarker [3, 12–19]. The majority of these works analyzed cervical cancer and/or premalignant lesions, with no surrounding tumor-free tissues coming from the same patient. For this reason, we performed a study comparing the miRNA expressions of HSIL/CIN2–3 lesions and histologically non-altered surrounding cervical epithelium in pairs deriving from the same patients.
Materials and Methods
Patients and Specimens
In total, 10 paired (normal/diseased) formalin fixed paraffin embedded (FFPE) histological samples with CIN1, 2, 3 and in situ carcinoma (CIS) were applied for miRNA screening and further 44 FFPE tissues (22 pairs of CIN2–3 and surrounding normal tissue) were chosen for validation of the results. The samples were selected from the archives of the 2nd Department of Pathology of Semmelweis University. The study was performed with the permission of the National Ethical Committee (V-R-021/04346–4/2013), according to the principles of the Declaration of Helsinki and with written consent from the patients. The samples derived form patients between 22 and 45 years of age at the time of obtaining tissue material with an average of 32.9 years (Table 1). Laser captured microdissection was used for selection and isolation of morphologically altered, dysplastic and non-tumorous foci performed by experienced pathologists. The 4–5 μm-thick FFPE sections were cut on FrameSlides 1.4 μm Pet Membrane (MicroDissect GmbH, Herborn, Germany). Leica AS LMD (Meyer Instruments, Houston, Texas, USA) instrument was used for microdissection of the areas of HSIL and non-altered epithelium, which were collected in BRAND®PCR-tubes (0,5 ml, BrandTech® Scientific, Merck KGaA, Darmstadt, Germany). A total of 10–12 microdissected sections were collected from the selected areas for the subsequent RNA isolation.
Table 1.
List of patients
| No | Age | HPV type |
|---|---|---|
| 1 | 31 | 16+ |
| 2 | 30 | 16+ |
| 3 | 38 | Negative |
| 4 | 25 | 16+ |
| 5 | 44 | 16+ |
| 6 | 32 | Negative |
| 7 | 34 | 33+ |
| 8 | 27 | 16+ |
| 9 | 30 | Negative |
| 10 | 32 | 58+ |
| 11 | 23 | 16+ |
| 12 | 25 | 16+; 31+; 33+ |
| 13 | 35 | 18+ |
| 14 | 28 | 16+, 66+, 45+, 59+ |
| 15 | 35 | 16+ |
| 16 | 34 | Negative |
| 17 | 43 | 16+,18+ |
| 18 | 36 | 16+ |
| 19 | 43 | Negative |
| 20 | 22 | 16+ |
| 21 | 35 | LR+ and 56+ |
| 22 | 42 | 16+ |
| Average: 32.9 | ||
HR: high risk HPV types; LR: low risk HPV types
HPV Genotyping Test
The Human papilloma virus (HPV) viral DNA detection was perfomed by CONFIDENCE HPV™ test (GenoID, Budapest, Hungary), which detects HPV16, HPV18 separately, and the other high-risk HPV-types (31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66 and 68) in group using multiplex real-time polymerase chain reaction technology [20].
RNA Isolation
Total RNA including the miRNA fraction was extracted from the removed tissue areas by RecoverAll Total Nucleic Acid Isolation Kit (Ambion brand by Life Technologies, Carlsbad, California, USA) according to the manufacturer’s instructions. The purified RNA was kept until use at −80 °C after the determination of the RNA concentration by a NanoDrop 1000 spectrophotometer (Life Technologies of Thermo Fischer Scientific, Waltham, MA, USA).
miRNA Screening
A preliminary miRNA expression screening was perfomed in 10 paired (normal/tumor) samples using TaqMan Array (TaqMan Array Human MicroRNA Cards Set v2.0, Panel A and B, Life Technologies, Foster City, CA, USA) according to the manufacturer’s instructions (Table 2). The two-card sets contained 667 specific assays to human miRNAs and four control assays (three endogenous control and one negative control assays). In order to increase sensitivity of the analysis for miRNAs being present in lower concentration, a preamplification step was included using Megaplex PreAmp Primers (Applies Biosystems by Life Technologies Foster City, CA, USA). The miRNA measurements were performed in 384-plate format in a 7900HT Fast Real-Time PCR System (Applies Biosystems by Life Technologies Foster City, CA, USA). Ct values were calculated using the SDS software v.2.1 using automatic baseline settings.
Table 2.
Kits used for preliminary miRNA expression screening
| Name of the kit | Catalogue Number |
|---|---|
| TaqMan MicroRNA Reverse Transcription Kit | 4,366,596 |
| Megaplex RT Primers Human Pool A | 4,399,966 |
| Megaplex RT Primers Human Pool B | 4,399,968 |
| TaqMan PreAmp Master Mix Kit | 4,384,267 |
| Megaplex PreAmp Primers Human Pool A | 4,399,233 |
| Megaplex PreAmp Primers Human Pool B | 4,399,201 |
| TaqMan 2X Universal PCR Master Mix, No AmpErase UNG (1 × 5 ml) | 4,324,018 |
| Human Array A and B | 4,400,238 |
Determination of the Expression Levels of miRNAs Identified in the Screening
The expression of the selected miRNAs listed in Table 3 was determined using TaqMan MicroRNA Assays (Life Technologies brand of Thermo Fisher Scientific Inc). Reverse transcription (RT) and quantitative polymerase chain reaction (qPCR) were performed according to manufacturer’s instructions. RT reaction was carried out using TaqMan MicroRNA Reverse Transcription Kit containing 10 ng total RNA. The qPCR was performed by applying TaqMan Universal Master Mix II, no UNG and 0.65 μL of the RT product. Owing to low copy numbers of miRNAs initially present in the samples, two miRNAs (miR-147 and miR-515) were preamplified just before qPCR for 12 cycles including the assays for reference as well. The amplification reaction was run in triplicates on a LightCycler 480 Instrument II (Roche Diagnostics, Indianapolis, IN, USA). Relative expression (2∆Cq, where ΔCq = CqRef-CqmiR) was calculated applying the average of RNU48 and U6 as the most stable reference determined by the NormFinder application [21].
Table 3.
The list of selected miRNAs
| Assay ID | miRNA |
|---|---|
| 001014 | hsa-miR-20b |
| 000402 | hsa-miR-24 |
| 000405 | hsa-miR-26a |
| 000413 | hsa-miR-29b |
| 000435 | hsa-miR-99a |
| 000437 | hsa-miR-100 |
| 000469 | hsa-miR-147 |
| 000515 | hsa-miR-212 |
| 002369 | hsa-miR-515-3p |
| For reference: | |
| 001006 | RNU48 |
| 001973 | U6 |
Statistical Analysis
The differences between normal and HSIL/CIN2–3 lesions were analyzed by means of non-parametric Wilcoxon Matched Pairs Test using STATISTICA software, version 12 (StatSoft Inc., Tulsa, OK, USA). A p value of 0.05 was set as the threshold for statistical significance.
Results
To investigate whether miRNAs could predict the difference between normal and HSIL/CIN2–3 tissue samples, we analyzed the expression of 667 miRNAs in a preliminary set of samples (n = 10). The paired healthy and diseased samples were evaluated according to the average down-regulation / up-regulation and their respective standard deviation and miRNAs with significant values (compared to the zero change) were selected for the study. It is noteworthy that there were significant average fold changes in the data sets. A total of 52 dysplasia upregulated and 27 dowregulated miRNAs with more than 3-fold changes were detected, but the majority of these changes was also highly variable, which renders them statistically insignificant. The final selection of the miRNAs was based on significantly different expression in diseased as compared to healthy tissue pairs (miR-20b, −24, −26a, −100) and additional miRNAs were selected based on a literature review (miR-29b, −99a, −147, −212, −515-3p).
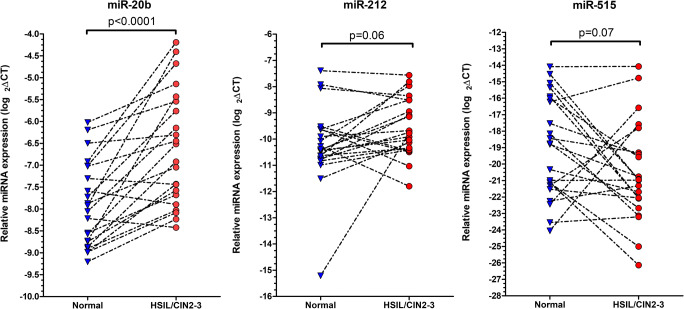
In order to assess the expression of these selected 9 miRNAs in HSIL/CIN2–3 and surrounding morphologically non-altered (“normal”) epithelium, further 22 paired FFPE cervical samples were used from the patient cohort. Statistical analysis of the miRNA expressions in the sample pairs (normal contra HSIL) showed a 2.4 fold overexpression of miR-20b in HSIL/CIN2–3 samples compared to normal tissues with high statistical significance (p < 0.0001) (Fig. 1). Additionally, miR-212 was also elevated with 1.6 fold increase (p < 0.06) and miR-515 showed 4 fold downregulation (p < 0.07) in dysplasia tissue compared to normal; however, these differencies were not statistically significant (Table 4). The other miRNAs analyzed did not show any difference between dysplasia and normal areas.
Fig. 1.
Relative miRNA expression in HSIL/CIN2–3 and corresponding normal cervical samples. Black interrupted lines represent the pairs of samples and indicate the direction of alteration of miRNA expression in dysplastic samples as compared to normal tissue
Table 4.
miRNAs showing statistical differences between HSIL/CIN2–3 and corresponding normal tissue
| miRNA | p value |
|---|---|
| In the 22 pairs | |
| miR-20b | <0.0001 |
| miR-212 | 0.06 |
| miR-515 | 0.07 |
| In HPV16+ samples | |
| miR-20b | <0.001 |
| miR-515 | <0.05 |
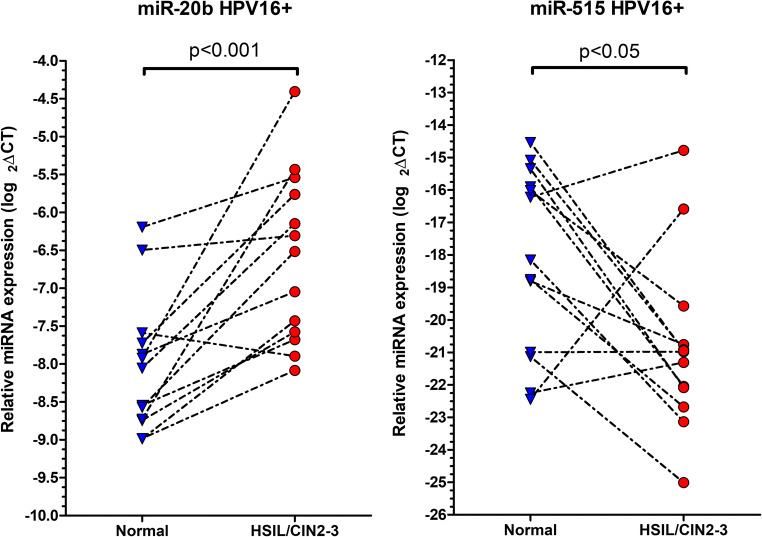
When investigating miRNA expression in the samples tested to be HPV16-positive (n = 13), miR-20b showed 2.9 fold upregulation (p < 0.001) and miR-515 was 1.15 fold downregulated (p < 0.05) in HSIL/CIN2–3 compared to normal tissue, which were statistically significant (Fig. 2).
Fig. 2.
Relative miRNA expression in HSIL/CIN2–3 samples known to be HPV16 positive and in corresponding normal cervical samples (n = 13). Black interrupted lines represent the pairs of samples and indicate the direction of alteration of miRNA expression in dysplastic samples as compared to normal tissue
Discussion
The novelty of our study is the pairs of HSIL/CIN2–3 and morphologically non-altered tissues, derived from the same patients as the source of miRNAs, precisely removed by laser microdissection, which has been rarely done in previous reports [22]. Our results demonstrated significantly increased expression of miR-20b in HSIL/CIN2–3 lesions in comparison to the surrounding morphologically non-altered cervical epithelium; while increased miR-212 and decreased miR-515 expression were also detected, however, these expressional differences were not statistically significant regarding the whole study population.
miRNAs are a family of small (18–25 nucleotides in length) single-stranded non-coding RNA molecules, which regulate gene expression at post-transcriptional level. miRNAs affect messenger RNAs (mRNAs) by degradation or by repression of translation via binding to the 3′ untranslated region of the targeted mRNAs [11, 17, 23]. Until now, approximately 2500 human miRNAs have been described in the miRBase database (www.mirbase.org), consisting of tissue-, organ-, cell- and „differentiation”- specific miRNAs. It has been shown that dysregulated miRNA-expression is involved in various biological processes including carcinogenesis. miRNAs may act as „oncomirs” functioning as either oncogenes or tumor suppressors depending on the type of tissue and stage of tumor development. miRNAs may be up- or downregulated in cancer tissue and thus these molecules have been suggested to be potential novel tumor markers [3, 13, 14, 17, 19, 24–28].
Accordig to Pardini et al., 24 studies targeting miRNA expression in cervical cancer show „great variability”. The variability, however, can be attributed to the differing numbers of samples (and consequently the varied statistical power) and the studied different histological backgrounds (premalignant or malignant lesions). Nevertheless, the analyzed samples (liquid, solid, FFPE or frozen) and the RNA isolation methods are also varied. Despite the mixed backgrounds, miR-29a and miR-21 were the most often found down- and upregulated miRNAs in invasive cervical cancer, while the altered expression of other miRNAs, especially miR-20b, was found to be associated with a range of patological lesions including from premalignant to malignant stages [17]. Cheung et al., by comparing normal epithelium, CIN and carcinoma samples, found 10 upregulated and 2 downregulated miRNAs in CIN lesions. The upregulation of miR-20b was found to be of the highest fold changes (2.89) in CIN2–3 lesions compared to normal tissue. As miR-20b was shown to be upregulated from the stage of premalignancies to invasive cancer, our results confirm the earlier findings regarding HSIL/CIN2–3 samples [29].
miR20b is encoded by the miR-106a-363 cluster located on human chromosome X and an autoregulatory feedback loop exists between E2F1 and miR-20b-5p, which was shown to be involved in proliferation and differentiation [30]. In a breast cancer study, miR-20b was shown to act as a tumor suppressor by inhibiting the migration and invasion of breast cancer cells [30] . Others demonstrated that HPV E6-regulates miR-20b and promotes invasion in cervical cancer by targeting the tissue inhibitor metalloproteinase 2 [31].
Regarding other dysregulated miRNAs in cervical cancer development, Pardini et al. and He et al. reviewed the available studies, with listing miR-26a, −29a, −99a, −100, and − 212 reported to be downregulated in CIN2–3 and/or cervical cancer in comparison to normal cervical epithelium [17, 32]. In addition, aberrant expression of miR-24 and -29b has been observed in cervical cancer [33–35] and downregulated miR-147 and upregulated miR-515-3p have been found in several solid cancers [36, 37]. However, we did not find statistically significant differences in the expression of these miRNAs when all of the 22 paires of HSIL/CIN2–3 samples were analyzed.
When examinig the miRNA expression in the 10 pairs of preliminary samples, we observed highly variable fold changes with relative large standard deviation, suggesting that the samples were rather heterogenous. This means that samples of different molecular background might have been put into the same group of HSIL/CIN2–3, which signifies the finding of a more efficient way for the stratification of samples with the aim to reduce the heterogeneity between the samples even more. For this reason, we chose a subgroup of samples tested positive for HVP16 and found a more upregulated miR-20b in HPV16-positive HSIL/CIN2–3 samples as compared to normal cervical epithelium. Association between miRNA expression and hrHPV infection has been reported [16, 32, 38]. For example, Wang et al. (2014) detected 13 up- and downregulated miRNAs, which were associated with hrHPV 16 and 18 [39].
In association with CIN2–3 and cervical carcinoma, several further miRNAs showing dysregulated expression have been reported, such as miR-218, 29a, −101, −140-5p [40–44]. In the study of Gocze et al. (2015), progressively increased expression of miR-27a was demonstrated in CIN2–3 as compared to CIN1 and in squamous cell carcinoma (SCC) as compared to CIN2–3, while the levels of miR-34a were found to be lower in CIN2–3 when compared to CIN1 and in SCC in comparison to CIN2–3. These data were found to be correlated with HPV16 positivity [14]. Differences were also detected in miRNA expression between SCC and adenocarcinoma [13]. Overexpression of several miRNAs was shown in SCC (miR-21, −27a, −34a, −155, −196, −203, −221) regardless of HPV status, however miR-21, −27a, −34a, −196a, and − 221 characterized HPV positive SCC, in contrast to adenocarcinoma having the same HPV status [13].
More recently, altered miRNA expression has been reported in non-solid tissues, such as biofluids (blood, vaginal mucus, saliva, urine etc.) [18, 45]. Four miRNAs were upregulated in CIN2–3, SCC and adenocarcinoma, showing a correlaion with hrHPV. The data were confirmed in frozen tissues coming from the same patients and miR-20b was among the upregulated miRNAs, as found in our HSIL/CIN2–3 cases. miRNAs may be associated with „Argonaute (AGO) proteins” or located in extracellular vesicles implying that miRNAs are „diversely distributed” across tissues and extracellular fluids, meaning that miRNAs are present intracellularly and extracellularly as well [45].
To summarize, our study revealed overexpression of miR-20b in HSIL/CIN2–3 tissue lesions as compared to the morphologically non-dysplastic adjacent epithelium. This might suggest the use of miR-20b as a potential biomarker in order to differentiate neoplastic from normal tissues. Further studies may examine the use of miR-20b as a marker in vaginal fluid/mucous and/or in liquid based cytology samples.
Acknowledgements
The authors thank Mrs. Elvira Kálé Rigóné for the English proofreading.
Authors Contribution
TS, NV and GL performed the RNA isolation and microRNA evaluation, ÁG and KS provided and selected the cervical samples and performed statistics, MB and AK performed the microRNA screening and HPV testing, NÁ, CJ, ZS and GS designed and coordinated the study. ZS and GS accept full responsibility for the work. All authors read and approved the final manuscript.
Funding Information
Open access funding provided by Semmelweis University. The work was supported by grant #OTKA PD105019 by the Hungarian National Research Foundation and #KMR-12-A FP7-HEALTH-2012-INNOVATION-1-Grant Agreement Number 306037 from the Hungarian Ministry of National Development.
Compliance with Ethical Standards
Conflict of Interest
The authors declare that they have no conflict of interest with respect to the research, authorship, and/or publication of this article.
MB is an employee of Synlab Hungary Kft. and NEUMANN Diagnostics Ltd., AK is of the NEUMANN Diagnostics Ltd. CJ and NV were former employees of NEUMANN Diagnostics Ltd.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Tímea Szekerczés, Ádám Galamb, Gábor Lendvai and Gábor Sobel contributed equally to this work.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Safaeian M, Sherman ME. From papanicolaou to papillomaviruses: evolving challenges in cervical cancer screening in the era of human papillomavirus vaccination. J Natl Cancer Inst. 2013;105:1524–1526. doi: 10.1093/jnci/djt267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Laengsri V, Kerdpin U, Plabplueng C, Treeratanapiboon L, Nuchnoi P. Cervical Cancer markers: epigenetics and microRNAs. Lab Med. 2018;49:97–111. doi: 10.1093/labmed/lmx080. [DOI] [PubMed] [Google Scholar]
- 4.Solomon D, Davey D, Kurman R, Moriarty A, O'Connor D, Prey M, Raab S, Sherman M, Wilbur D, Wright T Jr, Young N, Forum Group Members. Bethesda 2001 Workshop The 2001 Bethesda system: terminology for reporting results of cervical cytology. JAMA. 2002;287:2114–2119. doi: 10.1001/jama.287.16.2114. [DOI] [PubMed] [Google Scholar]
- 5.Crosbie E, Einstein M, et al. Human papillomavirus and cervical cancer. Lancet. 2013;382:889–899. doi: 10.1016/S0140-6736(13)60022-7. [DOI] [PubMed] [Google Scholar]
- 6.Ronco G, Dillner J, Elfström KM, Tunesi S, Snijders PJF, Arbyn M, Kitchener H, Segnan N, Gilham C, Giorgi-Rossi P, Berkhof J, Peto J, Meijer CJLM. Efficacy of HPV-based screening for prevention of invasive cervical cancer: follow-up of four European randomised controlled trials. Lancet. 2014;383:524–532. doi: 10.1016/S0140-6736(13)62218-7. [DOI] [PubMed] [Google Scholar]
- 7.Isidean SD, Franco EL. Embracing a new era in cervical cancer screening. Lancet. 2014;383:493–494. doi: 10.1016/S0140-6736(13)62028-0. [DOI] [PubMed] [Google Scholar]
- 8.Arbyn M, Ronco G, Anttila A, Meijer CJLM, Poljak M, Ogilvie G, Koliopoulos G, Naucler P, Sankaranarayanan R, Peto J. Evidence regarding human papillomavirus testing in secondary prevention of cervical cancer. Vaccine. 2012;30:F88–F99. doi: 10.1016/j.vaccine.2012.06.095. [DOI] [PubMed] [Google Scholar]
- 9.Tornesello ML, Buonaguro L, Giorgi-Rossi P, Buonaguro FM. Viral and cellular biomarkers in the diagnosis of cervical intraepithelial neoplasia and cancer. Biomed Res Int. 2013;2013:519619–519610. doi: 10.1155/2013/519619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nicolás I, Saco A, Barnadas E, Marimon L, Rakislova N, Fusté P, Rovirosa A, Gaba L, Buñesch L, Gil-Ibañez B, Pahisa J, Díaz-Feijoo B, Torne A, Ordi J, del Pino M. Prognostic implications of genotyping and p16 immunostaining in HPV-positive tumors of the uterine cervix. Mod Pathol. 2020;33:128–137. doi: 10.1038/s41379-019-0360-3. [DOI] [PubMed] [Google Scholar]
- 11.Galamb Á, Benczik M, Zinner B, Vígh E, Baghy K, Jeney C, Kiss A, Lendvai G, Sobel G. Dysregulation of microRNA expression in human cervical preneoplastic and neoplastic lesions. Pathol Oncol Res Oncol Ogy Res. 2015;21:503–508. doi: 10.1007/s12253-014-9871-x. [DOI] [PubMed] [Google Scholar]
- 12.Pereira PM, Marques JP, Soares AR, Carreto L, Santos MAS. Microrna expression variability in human cervical tissues. PLoS One. 2010;5:e11780. doi: 10.1371/journal.pone.0011780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gocze K, Gombos K, et al. Unique microRNA expression profiles in cervical cancer. Anticancer Res. 2013;33:2561–2568. [PubMed] [Google Scholar]
- 14.Gocze K, Gombos K, Kovacs K, Juhasz K, Gocze P, Kiss I. MicroRNA expressions in HPV-induced cervical dysplasia and cancer. Anticancer Res. 2015;35:523–530. [PubMed] [Google Scholar]
- 15.Pedroza-Torres A, Fernández-Retana J, Peralta-Zaragoza O, Jacobo-Herrera N, Cantú de Leon D, Cerna-Cortés JF, Lopez-Camarillo C, Pérez-Plasencia C. A microRNA expression signature for clinical response in locally advanced cervical cancer. Gynecol Oncol. 2016;142:557–565. doi: 10.1016/j.ygyno.2016.07.093. [DOI] [PubMed] [Google Scholar]
- 16.Liu SS, Chan KKL, Chu DKH, Wei TN, Lau LSK, Ngu SF, Chu MMY, Tse KY, Ip PPC, Ng EKO, Cheung ANY, Ngan HYS. Oncogenic microRNA signature for early diagnosis of cervical intraepithelial neoplasia and cancer. Mol Oncol. 2018;12:2009–2022. doi: 10.1002/1878-0261.12383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pardini B, De Maria D, et al. MicroRNAs as markers of progression in cervical cancer: a systematic review. BMC Cancer. 2018;18:1–17. doi: 10.1186/s12885-018-4590-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawai S, Fujii T, Kukimoto I, Yamada H, Yamamoto N, Kuroda M, Otani S, Ichikawa R, Nishio E, Torii Y, Iwata A. Identification of miRNAs in cervical mucus as a novel diagnostic marker for cervical neoplasia. Sci Rep. 2018;8:1–11. doi: 10.1038/s41598-018-25310-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang S, Chen X. Identification of potential biomarkers in cervical cancer with combined public mRNA and miRNA expression microarray data analysis. Oncol Lett. 2018;16:5200–5208. doi: 10.3892/ol.2018.9323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kocsis A, Takács T, Jeney C, Schaff Z, Koiss R, Járay B, Sobel G, Pap K, Székely I, Ferenci T, Lai HC, Nyíri M, Benczik M. Performance of a new HPV and biomarker assay in the management of hrHPV positive women: subanalysis of the ongoing multicenter TRACE clinical trial (n > 6,000) to evaluate POU4F3 methylation as a potential biomarker of cervical precancer and cancer. Int J Cancer. 2017;140:1119–1133. doi: 10.1002/ijc.30534. [DOI] [PubMed] [Google Scholar]
- 21.Andersen CL, Jensen JL, Ørntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and Colon Cancer data sets. Cancer Res. 2004;64:5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- 22.Yang Y, Liu Y, Li G, Li L, Geng P, Song H. MicroRNA-214 suppresses the growth of cervical cancer cells by targeting EZH2. Oncol Lett. 2018;16:5679–5686. doi: 10.3892/ol.2018.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen R, Shi YH, et al. Systematic prediction of target genes and pathways in cervical cancer from microRNA expression data. Oncol Lett. 2018;15:9994–10000. doi: 10.3892/ol.2018.8566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dweep H, Sticht C, Pandey P, Gretz N. miRWalk--database: prediction of possible miRNA binding sites by “walking” the genes of three genomes. J Biomed Inform. 2011;44:839–847. doi: 10.1016/j.jbi.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 25.Iorio MV, Croce CM. Causes and consequences of MicroRNA dysregulation. Cancer J (United States) 2012;18:215–222. doi: 10.1097/PPO.0b013e318250c001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles G, Clark NR, Ma’ayan A. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.González-Quintana V, Palma-Berré L, et al. MicroRNAs are involved in cervical cancer development, progression, clinical outcome and improvement treatment response (review) Oncol Rep. 2016;35:3–12. doi: 10.3892/or.2015.4369. [DOI] [PubMed] [Google Scholar]
- 28.Kori M, Arga KY. Potential biomarkers and therapeutic targets in cervical cancer: insights from the meta-analysis of transcriptomics data within network biomedicine perspective. PLoS One. 2018;13:1–27. doi: 10.1371/journal.pone.0200717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheung TH, Man KNM, Yu MY, Yim SF, Siu NSS, Lo KWK, Doran G, Wong RRY, Wang VW, Smith DI, Worley Jr MJ, Berkowitz RS, Chung TKH, Wong YF. Dysregulated microRNAs in the pathogenesis and progression of cervical neoplasm. Cell Cycle. 2012;11:2876–2884. doi: 10.4161/cc.21278. [DOI] [PubMed] [Google Scholar]
- 30.Xu H, Li W, Luo S, Yuan J, Hao L. Adipose derived stem cells promote tumor metastasis in breast Cancer cells by stem cell factor inhibition of miR20b. Cell Signal. 2019;62:109350. doi: 10.1016/j.cellsig.2019.109350. [DOI] [PubMed] [Google Scholar]
- 31.Cheng Y, Geng L, Zhao L, Zuo P, Wang J. Human papillomavirus E6-regulated microRNA-20b promotes invasion in cervical cancer by targeting tissue inhibitor of metalloproteinase 2. Mol Med Rep. 2017;16:5464–5470. doi: 10.3892/mmr.2017.7231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.He Y, Lin J, Ding Y, Liu G, Luo Y, Huang M, Xu C, Kim TK, Etheridge A, Lin M, Kong D, Wang K. A systematic study on dysregulated microRNAs in cervical cancer development. Int J Cancer. 2016;138:1312–1327. doi: 10.1002/ijc.29618. [DOI] [PubMed] [Google Scholar]
- 33.Wang X., Tang S., le S.Y., Lu R., Rader J.S., Meyers C., Zheng Z.M. (2008) Aberrant expression of oncogenic and tumor-suppressive microRNAs in cervical cancer is required for cancer cell growth PLoS One 3 10.1371/journal.pone.0002557 [DOI] [PMC free article] [PubMed]
- 34.Li Y, Zhang Z, Xiao Z, Lin Y, Luo T, Zhou Q, Zhang X. Chemotherapy-mediated miR-29b expression inhibits the invasion and angiogenesis of cervical cancer. Oncotarget. 2017;8:14655–14665. doi: 10.18632/oncotarget.14738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Servín-González LS, Granados-López AJ, et al. Families of microRNAs expressed in clusters regulate cell signaling in cervical cancer. Int J Mol Sci. 2015;16:12773–12790. doi: 10.3390/ijms160612773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y, Zhang H, et al. MicroRNA-147 suppresses proliferation, invasion and migration through the AKT/mTOR signaling pathway in breast cancer. Oncol Lett. 2016;11:405–410. doi: 10.3892/ol.2015.3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Han X, Li X, Zhao H, Zhou D, Sun B, Liu A, Zhang J, Cui Z, Ma X, Yuan L. Serum miR-515-3p, a potential new RNA biomarker, is involved in gastric carcinoma. J Cell Biochem. 2019;120:15834–15843. doi: 10.1002/jcb.28854. [DOI] [PubMed] [Google Scholar]
- 38.Virtanen E., Pietilä T., Nieminen P., Qian K., Auvinen E. (2016) Low expression levels of putative HPV encoded microRNAs in cervical samples. Springerplus 5 10.1186/s40064-016-3524-3 [DOI] [PMC free article] [PubMed]
- 39.Wang X, Wang HK, Li Y, Hafner M, Banerjee NS, Tang S, Briskin D, Meyers C, Chow LT, Xie X, Tuschl T, Zheng ZM. MicroRNAs are biomarkers of oncogenic human papillomavirus infections. Proc Natl Acad Sci U S A. 2014;111:4262–4267. doi: 10.1073/pnas.1401430111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu Z, Mao L, Wang L, Zhang H, Hu X. MiR-218 functions as a tumor suppressor gene in cervical cancer. Mol Med Rep. 2020;21:209–219. doi: 10.3892/mmr.2019.10809. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 41.Nan P, Niu Y, Wang X, Li Q. MiR-29a function as tumor suppressor in cervical cancer by targeting sirt1 and predict patient prognosis. Onco Targets Ther. 2019;12:6917–6925. doi: 10.2147/OTT.S218043. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 42.Jiang W, Pan J-J, Deng YH, Liang MR, Yao LH. Down-regulated serum microRNA-101 is associated with aggressive progression and poor prognosis of cervical cancer. J Gynecol Oncol. 2017;28:e75. doi: 10.3802/jgo.2017.28.e75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Su Y, Xiong J, Hu J, Wei X, Zhang X, Rao L. MicroRNA-140-5p targets insulin like growth factor 2 mRNA binding protein 1 (IGF2BP1) to suppress cervical cancer growth and metastasis. Oncotarget. 2016;7:68397–68411. doi: 10.18632/ONCOTARGET.11722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Varghese VK, Shukla V, Jishnu PV, Kabekkodu SP, Pandey D, Sharan K, Satyamoorthy K. Characterizing methylation regulated miRNA in carcinoma of the human uterine cervix. Life Sci. 2019;232:116668. doi: 10.1016/j.lfs.2019.116668. [DOI] [PubMed] [Google Scholar]
- 45.Berti F, Salviano-Silva A, et al. From squamous intraepithelial lesions to cervical cancer: circulating microRNAs as potential biomarkers in cervical carcinogenesis. Biochim Biophys Acta Rev Cancer. 2019;1872:188306. doi: 10.1016/j.bbcan.2019.08.001. [DOI] [PubMed] [Google Scholar]