Abstract
The development of colorectal cancer is a complex and multistep process mediated by a variety of factors including the dysregulation of genetic and epigenetic under the influence of microenvironment. It is evident that epigenetics that affects gene activity and expression has been recognized as a critical role in the carcinogenesis. Aside from DNA methylation, miRNA level, and genomic imprinting, histone modification is increasingly recognized as an essential mechanism underlying the occurrence and development of colorectal cancer. Aberrant regulation of histone modification like acetylation, methylation and phosphorylation levels on specific residues is implicated in a wide spectrum of cancers, including colorectal cancer. In addition, as this process is reversible and accompanied by a plethora of deregulated enzymes, inhibiting those histone-modifying enzymes activity and regulating its level has been thought of as a potential path for tumor therapy. This review provides insight into the basic information of histone modification and its application in the colorectal cancer treatment, thereby offering new potential targets for treatment of colorectal cancer.
Keywords: Histone modification, Colorectal cancer, Histone acetylation, Histone methylation, Histone phosphorylation
Introduction
The accumulation of genetic and epigenetic dysregulation are two kinds of separate mechanisms which resulted in tumorigenesis. Genetic variations have traditionally been regarded as an important player in the occurrence and development of the tumor. Nevertheless, current data have been accumulating concerning the epigenetic change which is responsible for the genesis and progression of cancer [1]. epigenetics is defined as the study of changes in the expression and regulation of genes does not involved in changes in the DNA sequence which can be classified into DNA methylation, histone modification, miRNA, genomic imprinting and chromosome remodeling [2, 3]. Up to now, DNA methylation and histone modification are the most intensively studied epigenetics; Moreover, Histone modification is one of the important regulatory mechanisms of epigenetics, which is detected primarily in the amino- and carboxy-terminal histone tails. It is also an important mechanism in cancer progression [4]. In this setting, a variety of cancer such as gastric cancer, prostate cancer, lung cancer, et al. have been extensively reported to be correlated with this change [5–7]. Colorectal cancer (CRC) is a multi-factorial disease and retains its status as the most common cause of cancer-related death, with more than 6000,000 estimated death across the globe every year [8]. The etiology of colorectal cancer is not completely established, accordingly, it is critical to probe into the molecular bases of colorectal cancer and markers for the early diagnosis and treatment of this cancer. Aberration in histone modification may become an early diagnosis of colorectal cancer. This review focus on understanding the role of histone modifications in the progression of colorectal cancer and their application in colorectal cancer may provide a new direction for diagnosis, treatment,and prognosis of colorectal cancer.
Histone Modification
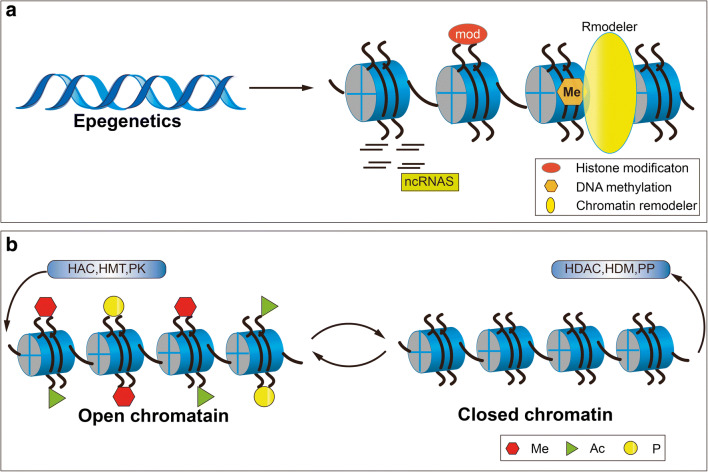
Chromatin is a dynamic molecule with multiple structures and there are two basic forms: heterochromatin and euchromatin. Epigenetic events are essential to regulate the condensation state of chromatin and hence to regulate the function of genome by means of histone modifications, chromatin remodeling, DNA methylation and no-coding RNA [9] Fig. 1. the fundamental unit of chromatin is nucleosome, which is composed of two copies of histones H2A, H2B, H3, and H4, wrapping with approximately 147 base pairs of DNA [10]. Histone, one of the major components of chromatin, has long tails protruding away from the nucleosome, which is susceptible to covalently modified at several places [11]. It includes many kinds of covalent modifications, such as acetylation, phosphorylation, methylation, ubiquitylation, sumoylation and so on [12]. At present, more efforts have been put into two aspects of histone modification namely, acetylation, methylation in terms of the orchestration chromatin structure and gene expression [13, 14]. The combination of single or multi-histone modification can interact with each other which forms “histone modification code” [15]. Covalent modification of histones can change the nucleosomal conformation in such a way that modulate the chromatin structure and expression of genes [16]. currently, an emerging role of aberrant histone modifications have been found in a wide spectrum of cancer, giving rise to the role of histone modification is under extensively investigation [13]. The reason why these aberrant modifications of histone lie in inappropriate targeting of histone-modifying enzymes, including HATs, HDACs, HMTs, and HDMTS, locally at gene promoters, resulting in perturbations or mutations in genes [17].
Fig. 1.
Epigenetics regulate the gene expression without alteration in DNA sequence. It mainly consists of DNA methylation, histone modification, non-coding RNA, chromatin remodeling. a The protruding of amino tails of histone modification can undergo several post-translational modifications that affect the expression of genes. Illustrated is the major histone modifications that have been studied in the setting of colorectal cancer such as acetylation, methylation, and phosphorylation. This process is mediated by HATs, HDACs, HMTs, HDMs and protein kinases (PKs). In open chromatin state, histone tail recruits HATs, HMTs, and PKs which can promote gene transcription. In closed chromatin state, histone tail removes these histone-modifying enzymes which can inhibit the gene transcription. (b)
Histone Acetylation
Reversible histone acetylation is a dynamic process that is achieved by the addition or removal of histone acetyltransferases (HATs) and deacetylases (HDACs) [18]. In recent years, the HATs that has been identified mainly includes P300/CBP, GNAT, MYST, P160, PCAF, TAFII230 families [19]. In the higher eukaryotes, HDACs can be classified into four groups on the basis of their homology with the original yeast enzyme sequence. Class I HDACs, including HDAC1, 2, 3, 8; Class II HDACs composed of HDAC4, 5, 6, 7, 9, 10; Class III HDACs, also known as Sirtuins, comprised of SIRT1–7; Class IV HDACs, including HDAC11 [20]. Among these HDACs Class I, II and IV are zinc-dependent, while Class III are NAD+-dependent. HATs transfer the acetyl group of acetyl coenzyme A to the terminal of histone amino acid and relax the structure of chromatin under the action of electric charge, which is helpful to transcription by means of increasing the accessibility of DNA. On the contrary, HDAC removes the terminal acetyl group of histone lysine making the structure of chromatin is compact which result in the inhibition of transcription [21]. In generally, hyperacetylation leads to the increased expression of the gene which is related to the activation of gene transcription, while hypoacetylation means repression of gene expression [22].
Histone Methylation
Histone methylation refers to the transfer of methyl group to the residues of histone arginine or lysine by taking S-adenosylmethionine as methyl donor under the action of histone methyltransferases (HMTs). Similar to histone acetylation, histone methylation is also a dynamic event which is regulated by different types of HMTs and histone demethytransferases (HDMTs). Histone methylation predominantly occurs in arginine and lysine residues of histone tails and is catalyzed by HMTs. Lysine can be monomethylated, dimethylated or trimethylated, respectively, arginine is either monomethylated or dimethylated. Furthermore, Lysine methylation is a more stable and complex modification of gene expression regulation, which occurs mainly on histone H3 and H4. There are six sites of lysine methylation that have been extensively studied such as H3K9, H3K27, H3K36, H3K79, and H4K20 [23] (Table 1). Protein arginine methylation is chiefly catalyzed by some members of the protein arginine methyltransferase family including PRMT1, PRMT3, PRMT1/HMTA, PRMT4/CARMA, PRMT5 [24]. HMTs comprised of histone arginine methyltransferase (PRMTs) and histone lysine methyltransferase (HKMTs). PRMTs can be classified as two types: Type I catalyzes the formation of mono-methylarginine and asymmetric di-methylarginine; Type II catalyzes the formation of monomethyl arginine and symmetric di-methylarginine. HKMTs mainly include Suv39h1, Suv39h2, G9a, EZH2, SET1, SET2, SET9 which contains the SET domain. DOT1L is the only lysine methyltransferase without the SET domain, which can specifically catalyze the methylation of H3K79 [25]. In addition to the histone methyltransferase, the finding of HDMTs by Shi et al., making the process of histone methylation more dynamic [26]. From then on, a variety of HDMTs has been observed, such as LSD1, UTX, JMJD3, JMJD1A. In contrast to histone acetylation that promotes gene expression, histone lysine methylation has a more complicated effect on gene expression which is depending on the location of modified residues being methylated lead to either transcriptionally-active or -repressive. For instance, methylation at H3K4 and H3K36 sites activate gene transcription. Conversely, methylation at H3K9, H3K27, H3K79, H4K20 sites inhibits gene transcription [27] .
Table 1.
Classification of histone methyltransferases and demethyltransferases
| Class | Members | ||
|---|---|---|---|
| HMTs | PRMTs | Type I | PRM1, PRM3, PMT1/HMT, PRMT4/CAMR1 |
| Type II | PRMT5 | ||
| HKMTs | Type I | G9a, EZH2, Suv39h1, Suv39h2, SET1, SET2, KMT2A | |
| Type II | DOTL1 | ||
| HDMs | LSD1, UTX, JMJD3, JMJD1A | ||
Adapted from Ref. [23]
PRMTs type I: mono-methylarginine and asymmetric dimethylargine; PRMTs type II: mono-methylarginine and symmeric dimethylarginine; HKMTs type I: lysine-specific SET histone transferases; HMTs type II: without lysine SET histone transferases
Histone Phosphorylation
Histone phosphorylation, taking place mainly at serines (S), threonines (T) and tyrosine (Y) residues of histone tails, is one of the histone modifications. Phosphorylation disrupts the interaction between histones and DNA attributed to the instability of chromatin structure which is requirement for the structural recombination of chromatin agglutination into homologous chromosomes during mitosis. It matters that different types of the phosphorylated site within histone is intimately linked to different chromatin functions. Moreover, Histone phosphorylation together with other modifications such as histone acetylation is involved in gene transcription, DNA repair, apoptosis and chromosome condensation. For instance, ChIP sequencing data manifested that histone H3 phosphorylated at tyrosine 41 (H3Y41) presents in transcriptional start sites (TSS) function together with H3K4me3 which implicated in transcriptional activation [28]. Furthermore, the combination of H3Y41 and H3K56 function together can significantly increase the accessibility of DNA by more than an order of magnitude [29]. Histone H3 phosphorylated at threonines45 (H3T45) takes part in apoptosis and DNA replication and can promote the acetylation of H3K56 [30, 31]. There is various kinase responsible for phosphorylation such as Aurora kinase(AKs), protein kinase B (PKB/Akt), cyclin-dependent kinases (CDKs), protein kinase C (PKC), casein kinase 2 (Ck2) and Rad3 related kinase ATR [32], and so on. H3Y41 is catalyzed by JAK2 tyrosine kinase (JAK2) [33]. Bub1 is the kinase responsible for phosphorylation of H2AT120 and H3T3 phosphorylated by Haspin kinase [34, 35].
Histone Modification and Colorectal Cancer
Given that previous research on histone modification in the development of colorectal cancer, the relationship between histone modification and colorectal cancer have been evolving understanding. Dysfunction of histone modification patterns involved in the activation of oncogenes and silence tumor suppressor genes have been verified correlated with the etiology of a variety of human diseases including allergic diseases, multiple sclerosis as well as gastrointestinal cancer [36–38]. Moreover, the alteration of histone modification patterns led to the deregulation of gene expression which plays pivotal roles in the formation of colorectal cancer (Table 2). Therefore, It’s important to investigate the mechanism and biological function of histone modification in CRC, thereby improving the clinical diagnosis and therapy of colorectal cancer.
Table 2.
Histone acetylation/methylation/phosphory marks in CRC
| Modification and sites | Method | Impaired function | References |
|---|---|---|---|
| Histone acetylation marks | |||
| Global H3ac | CHIP, WB | Hyperacetylation (CRC tissues) | [39] |
| Global H4ac | IHC | Hypoacetylayion (CRC cell lines) induced by CPERT | [40] |
| H3K9ac | IHC | Hypoacetylation (CRC liver metastasis) | [41] |
| H3K18ac | IHC | Hypoactylation (CRC cell lines) | [42] |
| H3K27ac | MS,WB | Hyperacetylation (CRC tissues) | [43] |
| H3K56ac |
WB, CHIP RT-qPCR |
Hypoacetylation (CRC cell lines) through RAS-PI3K signal pathway. | [44] |
| H4K12ac | IHC | Hypoacetylation (CRC cell lines) | [42] |
| H4K16ac |
LC-ES/MS IHC |
Hypoacetylation (CRC cell lines, CRC primary tumors) | [45, 46] |
| Histone methylation marks | |||
| H3K4me2 | CHIP, WB | Hypermethylation (CRC tissues) | [39] |
| H3K4me3 | IHC | Hypomethylation (CRC tissues) | [46] |
| H3K9me2 | IHC, WB | Hypermethylation (CRC cells line, CRC liver metastasis) | [41, 47] |
| H3K27me2 | IHC | Hypermethylation (CRC tissues) | [48] |
| H3K27me3 | IHC | Hypermethylation (CRC tissues) | [49] |
| H3K36me2 | IHC | Hypomethylation (CRC liver metastasis) | [48] |
| H3K79me2 | IHC | Hypermethylation (Patient with CRC) | [50] |
| H4K20me2 | LC-ES/MS | Hypomethylation (CRC cell lines) | [45] |
| H4K20me3 | CHIP, PCR | Hypomethylation (CRC patient’s plasma) | [51] |
| Histone phosphorylation marks | |||
| H3S10ph | IHC | Hypophosphorylation (CRC cell lines) | [52] |
| H2AX | IHC | Hyperphosphorylation (CRC patients) | [53] |
Histone Acetylation and Colorectal Cancer
Up to now, it has been the overwhelming accumulation of evidence indicated that histone acetylation finely regulates a wide range of cell functions, such as cell differentiation, assembly of the nucleosome, the change of chromatin structure and stability of gene expression [43]. it’s not surprising that abnormal regulation of histone acetylation is relevance to the predisposition to developing colorectal cancer and this process is modulated by a plethora of deregulated enzymes including HATs, HDATs. Karczmarski et al. used both mass spectrometry (MS) and western blot showed that acetylation of H3K27 was significantly increased in CRC samples compared with normal tissue [42]. Ashktorab et al. reported that acetylation of H3K12ac and H3K18ac was significantly increased in moderate to well differentiated colonic cancer, whereas decreased in poorly differentiated colonic cancer. They also observed that high level of HDAC2 in adenocarcinoma compared with those in adenoma, suggesting that the expression of HDAC2 is closely related to the progression from adenoma to adenocarcinoma [45]. Fraga et al. demonstrated that colorectal cancer is accompanied by reduced histone acetylation on H4K16 in CRC cell lines used both LC-ES-MS and western blot [54]. furthermore, several reports has indicated that global histone acetylation was positively correlated with tumor stage, lymph metastasis, poor survival, poor prognosis, histological subtype and cancer recurrence [17, 39]. Hashimoto et al. uesd multivariate analysis found that up-regulated global expression level of acetylated histone H3 (H3Ac) in colorectal cancer tissues was linked to poor overall survival. in addition, high AS (Allred scoring system) score of H3Ac predicted poor prognosis [41]. Tanagawa et al. demonstrated that global hypoacetylation of H3K9 was significantly associated with the histological type of colorectal cancer using immunohistochemistry [46]. Benard et al. found that increased nuclear expression of H3K56ac and H4K16ac was highly correlations with better survival of CRC patients and a lower chance of tumor recurrence [55].
Recent intensively investigations in several of cancers focus on altering expression of HATs or HDACs uncovered that they contribute to tumorigenesis. Research data have indicated that low expression of males absent on the first (MOF) appeared in colorectal cancer and it mainly correlated with lymph node metastasis and tumor stage in patients with CRC [56, 57]. Previous studies indicated that CLASS I HDACs are generally up-regulated in normal colon tissues and colon cancer cell lines [58, 59]. HDAC1 was shown to higher in CRC tissues than in normal tissues and low expression of it indicated better overall survival (OS) [60]. HDAC2 has been found up-regulated in CRC cell lines as compared with their corresponding normal colonic epithelial cells [61]. Nemati et al. used RT-PCR observed that an increased level of HDAC3 in CRC specimens in relation to poor tumor differentiation [62]. In addition, there is also some study indicated that certain histone acetylation can be targeted via specific signaling pathway [44]. For example, Liu et al. discovered that RAS-PI3K signaling down-regulates the level of H3K56ac which is related to transcription, proliferation, and migration of cancer cells [40]. The study by Zhang et al. uncovered that cell-cycle related and expression-elevated protein (CREPT) cooperated with acetyltransferase P300 stimulates the Wnt/−catenin signaling to promote the expression of H4Ac and H3K27ac [47].
Histone Methylation and Colorectal Cancer
Histone methylation is involved in diverse biological functions including the formation of heterochromatin, inactivation of the X chromosome, DNA damage response and transcriptional regulation. The abnormal biological function of histone methylation regulates pathogenesis of various diseases including tumors. To date, ectopic expression of histone methylation and demethylases have been widely described in several cancers including colorectal cancer. For instance, Nakazawa et al. Used both immunohistochemistry and western blot analysis have revealed that increased expression of global level of dimethylated lysine 9 on histone H3 (H3K9me2) in the nuclei of adenocarcinoma more than that in adenoma, suggesting that hyperacetylation of H3K9m2 might be relevant to the adenoma transition to adenocarcinoma [63]. Yokoyama et al. reported that the methylation level of trimethylated lysine 9 on histone 3 (H3K9me3) was especially up-regulated in invasive regions of colorectal cancer tissues and H3K9 trimethylation was positively related to lymph node metastasis. in addition, elevated expression of H3K9 methyltransferase SUV39H1 was facilitated the development of CRC which resulted in a poor survival rate in mouse [51]. while Gezer.et al. uncovered that histone methylation marks H3K9me3 and H4K20me3 was significantly decreased in plasma of the patient with CRC [48]. High AS score of H3K4me2 is dramatically associated with colorectal cancer clinicopathological factors including deeper tumor invasion and advanced pathological stage [41]. What is more, Multivariate survival analysis revealed that the low expression of H3K4me2 could be served as an independent prognostic factor in CRC patients with metachronous liver metastasis [46]. The low expression level of H4K20me2 was a common hallmark in CRC cell lines [54]. Tamagawa et al. showed that decrease methylation of H3K27me2 in liver metastasis in comparison with primary tumors, whereas the expression of H3K36me2 was reversed. They also demonstrated that the expression level of H3K37me2 is positively associated with tumor size and poorer survival rates and it could be served as an independent prognostic factor for CRC patients with metachronous liver metastasis [49]. Benard et al. found that an up-regulated level of H3K27me3 compared to normal counterparts that were detected by immunohistochemically stained (IHC). it closely linked to better patient survival and longer recurrence-free periods [64]. They also observed that increase expression of H3K4me3 and decrease expression of H3K9me3 and H4K20me3 were associated with shorter survival and higher chances of tumor recurrence in the early stage of colon cancer [50]. Additionally, high levels expression of H3K79me2 was suggested to be a predictor of poor CRC patient survival [65].
Global histone methylation is controlled by histone methyltransferase (HMTS) and demethyltransferase (HDMTS) plays an essential role in the regulation of chromatin structure and function. Many recent studies have discovered that the alteration of HATs and HDMTs was documented in different types of cancers [66]. Kornbluht described that reduction of histone methyltransferase SEDT2 facilitated the CRC development by affecting alternative splicing [67]. The study by Qin et al. showed that the expression of G9A was dramatically increased in CRC tumor tissues and overexpression of G9A was mainly correlated with American Joint Committee on Cancer staging (AJCC), tumor differentiation and tumor relapse of CRC [68]. The histone H3K27 methyltransferase EZH2 expression was up-regulated in CRC, which was predicted shorter survival and advanced stage implying that it could use to an indicator of clinical outcome in CRC patients [69, 70]. Low nuclear expression of demethylase JMJD3 was shown in normal colorectal tissues as compared with CRC tissues and low expression of JMJD3 could serve as an independent predictor of poor prognosis in patients with CRC [71]. Elevate expression of Lysine-specific demethylase (LSD1) observed in colon cancer tissues, and high expression level of LSD1 was strongly correlated with advanced TNM stages and distant metastasis [72].
Histone Phosphorylation and Colorectal Cancer
Histone phosphorylation is essential for maintaining the equilibrium of kinase-phosphatase at kinetochore to refrain from chromosomal instability and cancer. Accordingly, there is an increasing body of investigation evaluating the impact of dysregulated phosphorylation on the development of many human diseases, including colon cancer [73]. As yet, there are few researches address the relationship between histone phosphorylation and colorectal cancer. Several studies have shown that aberrant of phosphorylation histone has been verified to correlated with the pathogenesis of colorectal cancer. For example, downregulation of dual specificity phosphatase 22 (DUSP22) expression was observed in colorectal cancer specimens and reduced DUSP22 expression in stage IV patients was mainly exhibited poor survival outcome [53]. Lee et al. revealed that phosphorylation of the H2AX histone (p-H2AX) have been found elevated in CRC tissues and have been corrected with a more aggressive type of tumor behavior, as well as poor CRC patient survival [74]. Chen et al. found that PKCƐ modulated MllP-S303 phosphorylation and its expression level was associated with metastasis and prognosis of colorectal cancer [52]. Xiao et al. identified that a reduced level of Histone H3 at Ser10 (H3S10) was observed in colon cancer. Meanwhile, the phosphorylation of T-LAK cell-originated protein kinase (TOPK) at Y74 and Y272 facilitated the carcinogenesis of colon cancer [75].
Application of Histone Modification in Colorectal Cancer
Conventional CRC therapies generally including chemotherapy, surgery and radiation therapy. However, clinic efficacy of those treatments is limited. Recent investigation has revealed that the process of histone modification is reversible and their aberrations can be restored to nearly normal status through epigenetic therapy. Thus, histone modification serves as a promising therapeutic target in treating various cancers in combination with conventional treatment. Histone deacetylation and methylation inhibitors are the most widely applied to colorectal cancer.
Histone Methyltransferase Inhibitors and Treatment of Colorectal Cancer
Previous studies have emphasized the significance of histone methylation in the regulation of gene and other physiological processes. Furthermore, aberrant histone methylation as a result of gene mutation is frequently associated with the occurrence and development of cancer. In order to provide a broader platform for cancer treatment, the study communities further identify small molecule inhibitors targeting either histone methyltransferases or demethylases for the therapy of CRC, therefore most of the histone modifying enzymes serve as a drug target has been widely reported [76]. The study by Hsu et al. was reported that LSD1 inhibitors CBB1003 suppress CRC cell growth through down-regulating LGR5 levels and inactivates the Wnt/β-catenin pathway [77]. Enhancer of zeste homolog 2 (EZH2) is a subunit of the polycomb repressive complex 2 (PCR2) and high level of EZH2 has been observed in different cancers including bladder cancer, non-small-cell lung cancer as well as colorectal cancer [78]. Therefore, EZH2-specific inhibitors have been regarded as an appealing target as a result of its oncogenic activities. EZH2 inhibitor GSK346 enjoys good anti-tumor efficacy for it can suppress migration, invasion, and proliferation of CRC cells [79]. Likewise, in vitro investigations have shown that UCN1999 and GSK 343 are two S-adenosyl-L-methionine (SAM) -competitive inhibitors which promoted autophagy through upregulated the expression of LC3 gene resulted in colorectal cancer cell death [80]. Verticillin A, a selective histone methyltransferase inhibitor, not only effectively inhibited the metastatic CRC cell growth but also enhanced the efficacy of CTL immunotherapy to block the progression and metastasis of CRC [81]. Nowadays, in vitro study shows that JIB-04, a novel histone demethylase inhibitors targets colorectal cancer stem cells (CSC), was able to repress CSC growth, invasion, and migration to fight against colorectal cancer [82].
Histone Deacetylase Inhibitors and the Treatment of Colorectal Cancer
According to HDACis’s molecular function, they can be divided into four major groups: the first group is short-chain fatty acids mainly including phenylbutyrate (PB), valproic acid (VPA) and carboxylic acids NaB. The second group is hydroxamates consisting of TSA and SAHA. The third group is benzamides containing MS-275 and MGCD-0103. the fourth group is cyclic peptides. As an emerging sort of anti-cancer drug, increasing evidence has demonstrated that HDACis exert their anti-tumor effects is conveyed by regulating multiple approaches including induce tumor cells cycle arrest, inhibit tumor cell growth, differentiation apoptosis. Furthermore, they facilitate the acetylation of histone and nonhistone protein resulted in the alteration of their transcriptional activity [83]. they inhibit angiogenesis and modulate the miRNA expression in tumor progression [84]. Major clinic HDACis for the treatment of CRC is summarized in Table 2.
The current investigation indicated that deregulation of HATs and histone HDACs is engaged in the progression of a range of cancers, making them spur the considerable interest of the research community [22, 85]. Thus, HDACis become appealed target in attempts to attenuate many human cancers including colorectal cancer. Therefore, various histone deacetylase inhibitors (HDIs) become the favored target in attempts to attenuate much human cancer including colorectal cancer. So far, there are four HDACis have been approved by Food and Drug Administration (FAD) for the treatment of patients with cutaneous T cell lymphoma and peripheral T cell lymphoma [86, 87]. Even if there is a growing list of HDACis applied to colorectal cancer, a wealth of candidates are ongoing intensively study and clinical trials. For example, Trichostatin A (TSA) suppress the growth of CRC cells in vivo by inducing cell cycle arrest and apoptosis through the modulation of JAK2/STAT3 signaling [88]. SAHA known as suberoylanilide hydroxamic acid inhibits colon tumor growth via decreasing the expression of histone deacetylases, cyclin D1 and survivin [89]. in addition, SAHA exerts their anti-proliferative effects in CRC cells through reducing expression of oncogenic miR17–92 cluster miRNAs [90]. Treating colorectal cancer with Valproate (VPA) could depress tumor growth with cell cycle through alteration of H3 and H4 acetylation [91]. Romidepsin, one of the new class of histone deacetylase inhibitors, exhibits its anti-neoplastic effect in colorectal cancer cell lines via induced alteration in protein modification including acetylation and phosphorylation [92]. In vitro data indicated that it was able to induce apoptosis by the generation of reactive oxygen species (ROS) [93]. Butyrate is a kind of short fatty acid which has been shown to significantly effective against the migration and invasion of CRC cell lines via activating the A kt1 and ERK1/2 signaling [94]. Entinostat (MS-275) is a member of benzamides, in various CRC lines and xenograft models it has been shown potent anti-proliferative effects and reduce tumor volume [95] and it facilitates Natural killer (NK) cell killing of tumor cells by regulation both the NKG2D receptor and its ligand, implying that augment NK cell immunotherapy may be a potential approach for solid tumors [96]. Furthermore, It is recently shown that Dihydroxybenzoic acid (DHBA), a kind of benzoic acid derivatives, can suppress HDAC activity resulted in cancer cell growth inhibition via the induction of ROS and cellular apoptosis regulated by Caspase-3 [97]. In CRC cell lines, Belinostat induces apoptosis and inhibit colon cancer cell proliferation by the regulation of proteins including p53, AP1 [98]. In HCC and CRC cell lines treated with panobinostat (LBH589) was able to reduce proliferation and vascularization lead to a suppressed tumor growth [99], this observation is in accordance with previously published results [100] (Table 3).
Table 3.
The implication of histone acetylase inhibitors(HDACis) in CRC treatment
| Class | Specificity | HDACis | Experiment design | Effect | Reference |
|---|---|---|---|---|---|
| Hydroxamic acids | Class I, II | Trichostatin A (TSA) | CRC cell lines | Inhibit CRC cells growth | [87] |
| Vorinostat (SAHA) | Mouse model | Inhibit the growth of colon tumors in nude mice | [88] | ||
| CRC cell lines | Anti-proliferative in CRC cell lines | [89] | |||
| Panobistat | CRC cell lines HCC cell lines | Anti-angiogenic Reduce proliferation | [98] | ||
| Belinostat | CRC cell lines | Induce CRC cell apoptosis | [97] | ||
| Short-chain fatty acids | Class I | Butyrate | CRC cell lines | Inhibit CRC cell migration and invasion | [93] |
| Valproic acid (VPA) | CRC cell lines | Inhibit the growth of CRC cell lines with cell cycle arrest | [90] | ||
| Benzamides | Class I | Entinostat (MS-275) | CRC cell lines mouse model | Anti-proliferative effects | [94] |
| Benzoic acid | Dihydroxy benzoic acid | Colon cell lines | Induce cancer cell death | [96] | |
| Cyclic peptides | Class I, II | Romidepsin (FK228) | CRC cell lines | Anti-tumor activity | [91] |
It has been noted HDAC inhibitors as monotherapy were initially incorporated into the clinical exhibits limited effectiveness, acquisition of drug resistance as well as adverse effect in the treatment of cancer [101, 102]. Hence, the tendency to apply them in corporation with different types of anti-cancer drugs is increasing. Several studies show the combined therapies may amplify the anti-tumor effect to suppress refractory tumors [103]. The plethora of investigation involved in anti-tumor agents and HDAC inhibitors synergistically used. For example, A452, an HDAC6-selective inhibitor, in combination with SAHA enhanced anti-proliferation effects on CRC cell lines compared with single-agent therapy [104]. EZH2 inhibitors and EGFR inhibitors synergisticll induced autophagy and apoptosis, resulting in the inhibition of colon cancer cells [105]. Combination treatment with vorinostat and bortezomib has better suppress proliferation and induce CRC cell cycle arrest than treatment with single-agent therapy [106]. The combined use of Butyrate and irinotecan exhibit potentiate the antineoplastic effects in CRC cell lines, resulting in tumor cell death [107]. while combining entinostat with demethylating agent 5-azacitidine applied with metastatic CRC patients have no activity in colorectal cancer [108]. Moreover, a variety of HDAC inhibitors are lack of specificity, thus, it is more critical to devote to developing effective and specific epigenetic targets against colorectal cancer and provide an optimized therapeutic regimen for CRC patients. Accordingly, in future, the current study is primarily concentrates on synergistic effects achieved by the combination use these agents which will represent great promise for further development in human cancer therapy.
Conclusion and Future Perspectives
In summary, there are plenty of publications with respect to histone modification in colorectal cancer, however, its complete picture remains unclear. This article attempts to comprehensively elucidate the essential role of histone modification in CRC. we are now clear that histone modification is involved in the pathogenesis of CRC. Therefore, it is important for us to apply this new understanding to develop novel therapeutic approaches for cancer. However, there are still some problems. First, the contribution of histone modification dysregulation in colorectal cancer isn’t completely known, hindering the discovery of emerging target for cancer therapy. Second, the different subtypes of CRC caused by emerging abnormal expression pattern of histone modification on specific residues in the progression of CRC that requires specificity of epigenetic drugs to provide individual treatment for CRC patients. Third, considering monotherapy have limited anti-tumor efficacy and lead to adverse effects, combining with other anticancer drugs for the treatment of this malignant neoplasm is absolutely necessary. In the future, with great advances made in this evolving area, the future of epigenetic drug is bright, despite the fact that we have faced these problems. Furthermore, as researcher communities delicate to exploring more advances effective combination inhibitors of epigenetic drugs to alter the course of colorecter cancer. It’s firmly believed that these will bring the benefit to colorectal cancer patients.
Funding
This work was supported by the National Natural Science Foudation of China (Grant No. 81673944).
Compliance with Ethical Standards
Conflict of Interest
The author declare that there are no conflicts of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Okugawa Y, Grady WM, Goel A. Epigenetic alterations in colorectal cancer: emerging biomarkers. Gastroenterology. 2015;149(5):1204–1225.e12. doi: 10.1053/j.gastro.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gieni RS, Hendzel MJ. Polycomb group protein gene silencing, non-coding RNA, stem cells, and cancer. Biochem Cell Biol. 2009;87(5):711–746. doi: 10.1139/O09-057. [DOI] [PubMed] [Google Scholar]
- 3.Lawrence M, Daujat S, Schneider R. Lateral thinking: how histone modifications regulate gene expression. Trends Genet. 2016;32(1):42–56. doi: 10.1016/j.tig.2015.10.007. [DOI] [PubMed] [Google Scholar]
- 4.Werner RJ, Kelly AD, Issa JJ. Epigenetics and precision oncology. Cancer J. 2017;23(5):262–269. doi: 10.1097/PPO.0000000000000281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang WY, Gu JL, Zhen TM. Recent advances of histone modification in gastric cancer. J Cancer Res Ther. 2014;10(Suppl):240–245. doi: 10.4103/0973-1482.151450. [DOI] [PubMed] [Google Scholar]
- 6.Nowacka-Zawisza M, Wiśnik E. DNA methylation and histone modifications as epigenetic regulation in prostate cancer. Oncol Rep. 2017;38(5):2587–2596. doi: 10.3892/or.2017.5972. [DOI] [PubMed] [Google Scholar]
- 7.Mehta A, Dobersch S, Romero-Olmedo AJ, Barreto G. Epigenetics in lung cancer diagnosis and therapy. Cancer Metastasis Rev. 2015;34(2):229–241. doi: 10.1007/s10555-015-9563-3. [DOI] [PubMed] [Google Scholar]
- 8.Ferlay J, Steliarova-Foucher E, Lortet-Tieulent J, Rosso S, Coebergh JWW, Comber H, Forman D, Bray F. Cancer incidence and mortality patterns in Europe: estimates for 40 countries in 2012. Eur J Cancer. 2013;49(6):1374–1403. doi: 10.1016/j.ejca.2012.12.027. [DOI] [PubMed] [Google Scholar]
- 9.De Majo F, Calore M. Chromatin remodelling and epigenetic state regulation by non-coding RNAs in the diseased heart. Non-coding RNA Research. 2018;3(1):20–28. doi: 10.1016/j.ncrna.2018.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coppedè F. Genetic and epigenetic biomarkers for diagnosis, prognosis and treatment of colorectal cancer. World J Gastroenterol. 2014;20(4):943–947. doi: 10.3748/wjg.v20.i4.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293(5532):1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 12.Rothbart SB, Strahl BD. Interpreting the language of histone and DNA modifications. Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms. 2014;1839(8):627–643. doi: 10.1016/j.bbagrm.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fu LN, Tan J, Chen YX, Fang JY. Genetic variants in the histone methylation and acetylation pathway and their risks in eight types of cancers. J Dig Dis. 2018;19(2):102–111. doi: 10.1111/1751-2980.12574. [DOI] [PubMed] [Google Scholar]
- 14.Azieva AM, Sheinov AA, Galkin FA, Georgieva SG, Soshnikova NV. Stability of chromatin remodeling complex subunits is determined by their phosphorylation status. Dokl Biochem Biophys. 2018;479(1):66–68. doi: 10.1134/S1607672918020035. [DOI] [PubMed] [Google Scholar]
- 15.Stral BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403(6):41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 16.Gurard-Levin ZA, Almouzni G. Histone modifications and a choice of variant: a language that helps the genome express itself. F1000Prime Rep. 2014;6:76–86. doi: 10.12703/P6-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seligson DB, Horvath S, McBrian MA, Mah V, Yu H, Tze S, Wang Q, Chia D, Goodglick L, Kurdistani SK. Global levels of histone modifications predict prognosis in different cancers. Am J Pathol. 2009;174(5):1619–1628. doi: 10.2353/ajpath.2009.080874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O'Hagan HM. Chromatin modifications during repair of environmental exposure-induced DNA damage: a potential mechanism for stable epigenetic alterations. Environ Mol Mutagen. 2014;55(3):278–291. doi: 10.1002/em.21830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berndsen CE, Denu JM. Catalysis and substrate selection by histone/protein lysine acetyltransferases. Curr Opin Struct Biol. 2008;18(6):682–689. doi: 10.1016/j.sbi.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Ruijter AJ, van Gennip AH, Caron HN, Kemp S, van Kuilenburg AB. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem J. 2003;370(Pt 3):737–749. doi: 10.1042/BJ20021321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mehrotra S, Galdieri L, Zhang T, Zhang M, Pemberton LF, Vancura A. Histone hypoacetylation-activated genes are repressed by acetyl-CoA- and chromatin-mediated mechanism. Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms. 2014;1839(9):751–763. doi: 10.1016/j.bbagrm.2014.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gräff J, Tsai L. Histone acetylation: molecular mnemonics on the chromatin. Nat Rev Neurosci. 2013;14(2):97–111. doi: 10.1038/nrn3427. [DOI] [PubMed] [Google Scholar]
- 23.Wei S, Li C, Yin Z, Wen J, Meng H, Xue L, Wang J. Histone methylation in DNA repair and clinical practice: new findings during the past 5-years. J Cancer. 2018;9(12):2072–2081. doi: 10.7150/jca.23427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Di Lorenzo A, Bedford MT. Histone arginine methylation. FEBS Lett. 2011;585(13):2024–2031. doi: 10.1016/j.febslet.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wood K, Tellier M, Murphy S. DOT1L and H3K79 methylation in transcription and genomic stability. Biomolecules. 2018;8(1):11. doi: 10.3390/biom8010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119(7):941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 27.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447(7143):407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 28.Dawson MA, Foster SD, Bannister AJ, Robson SC, Hannah R, Wang X, Xhemalce B, Wood AD, Green AR, Gottgens B, et al. Three distinct patterns of histone H3Y41 phosphorylation mark active genes. Cell Rep. 2012;2(3):470–477. doi: 10.1016/j.celrep.2012.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brehove M, Wang T, North J, Luo Y, Dreher SJ, Shimko JC, Ottesen JJ, Luger K, Poirier MG. Histone core phosphorylation regulates DNA accessibility. J Biol Chem. 2015;290(37):22612–22621. doi: 10.1074/jbc.M115.661363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hurd PJ, Bannister AJ, Halls K, Dawson MA, Vermeulen M, Olsen JV, Ismail H, Somers J, Mann M, Owen-Hughes T, et al. Phosphorylation of histone H3 Thr-45 is linked to apoptosis. J Biol Chem. 2009;284(24):16575–16583. doi: 10.1074/jbc.M109.005421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Darieva Z, Webber A, Warwood S, Sharrocks AD. Protein kinase C coordinates histone H3 phosphorylation and acetylation. Elife. 2015;4:e09886. doi: 10.7554/eLife.09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Treviño LS, Wang Q, Walker CL. Phosphorylation of epigenetic “readers, writers and erasers”: implications for developmental reprogramming and the epigenetic basis for health and disease. Prog Biophys Mol Biol. 2015;118(1–2):8–13. doi: 10.1016/j.pbiomolbio.2015.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dawson MA, Bannister AJ, Göttgens B, Foster SD, Bartke T, Green AR, Kouzarides T. JAK2 phosphorylates histone H3Y41 and excludes HP1α from chromatin. Nature. 2009;461(7265):819–822. doi: 10.1038/nature08448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang F, Dai J, Daum JR, Niedzialkowska E, Banerjee B, Stukenberg PT, Gorbsky GJ, Higgins JMG. Histone H3 Thr-3 phosphorylation by Haspin positions Aurora B at centromeres in mitosis. Science. 2010;330(6001):231–235. doi: 10.1126/science.1189435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kawashima SA, Yamagishi Y, Honda T, Ishiguro K, Watanabe Y. Phosphorylation of H2A by Bub1 prevents chromosomal instability through localizing shugoshin. Science. 2010;327(5962):172–177. doi: 10.1126/science.1180189. [DOI] [PubMed] [Google Scholar]
- 36.Alaskhar Alhamwe B, Khalaila R, Wolf J, von Bülow V, Harb H, Alhamdan F, Hii CS, Prescott SL, Ferrante A, Renz H et al (2018) Histone modifications and their role in epigenetics of atopy and allergic diseases. Allergy, Asthma Clin Immunol 14(1) [DOI] [PMC free article] [PubMed]
- 37.He H, Hu Z, Xiao H, Zhou F, Yang B (2018) The tale of histone modifications and its role in multiple sclerosis. Human Genomics 12(1) [DOI] [PMC free article] [PubMed]
- 38.Biswas S, Rao CM. Epigenetics in cancer: fundamentals and beyond. Pharmacol Ther. 2017;173:118–134. doi: 10.1016/j.pharmthera.2017.02.011. [DOI] [PubMed] [Google Scholar]
- 39.Elsheikh SE, Green AR, Rakha EA, Powe DG, Ahmed RA, Collins HM, Soria D, Garibaldi JM, Paish CE, Ammar AA, et al. Global histone modifications in breast Cancer correlate with tumor phenotypes, prognostic factors, and patient outcome. Cancer Res. 2009;69(9):3802–3809. doi: 10.1158/0008-5472.CAN-08-3907. [DOI] [PubMed] [Google Scholar]
- 40.Liu Y, Wang D, Chen S, Zhao L, Sun F. Oncogene Ras/phosphatidylinositol 3-kinase signaling targets histone H3 acetylation at lysine 56. J Biol Chem. 2012;287(49):41469–41480. doi: 10.1074/jbc.M112.367847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hashimoto T, Yamakawa M, Kimura S, Usuba O, Toyono M. Expression of acetylated and Dimethylated histone H3 in colorectal cancer. Dig Surg. 2013;30(3):249–258. doi: 10.1159/000351444. [DOI] [PubMed] [Google Scholar]
- 42.Karczmarski J, Rubel T, Paziewska A, Mikula M, Bujko M, Kober P, Dadlez M, Ostrowski J. Histone H3 lysine 27 acetylation is altered in colon cancer. Clin Proteomics. 2014;11(1):24. doi: 10.1186/1559-0275-11-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun W, Zhou X, Zheng J, Lu M, Nie J, Yang X, Zheng Z. Histone acetyltransferases and deacetylases: molecular and clinical implications to gastrointestinal carcinogenesis: figure 1. Acta Biochim Biophys Sin. 2011;44(1):80–91. doi: 10.1093/abbs/gmr113. [DOI] [PubMed] [Google Scholar]
- 44.Bardhan K, Paschall AV, Yang D, Chen MR, Simon PS, Bhutia YD, Martin PM, Thangaraju M, Browning DD, Ganapathy V, et al. IFN induces DNA methylation-silenced GPR109A expression via pSTAT1/p300 and H3K18 acetylation in colon cancer. Cancer Immunol Res. 2015;3(7):795–805. doi: 10.1158/2326-6066.CIR-14-0164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ashktorab H, Belgrave K, Hosseinkhah F, Brim H, Nouraie M, Takkikto M, Hewitt S, Lee EL, Dashwood RH, Smoot D. Global histone H4 acetylation and HDAC2 expression in Colon adenoma and carcinoma. Dig Dis Sci. 2009;54(10):2109–2117. doi: 10.1007/s10620-008-0601-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tamagawa H, Oshima T, Shiozawa M, Morinaga S, Nakamura Y, Yoshihara M, Sakuma Y, Kameda Y, Akaike M, Masuda M, et al. The global histone modification pattern correlates with overall survival in metachronous liver metastasis of colorectal cancer. Oncol Rep. 2012;27(3):637. doi: 10.3892/or.2011.1547. [DOI] [PubMed] [Google Scholar]
- 47.Zhang Y, Wang S, Kang W, Liu C, Dong Y, Ren F, Wang Y, Zhang J, Wang G, To KF, et al. CREPT facilitates colorectal cancer growth through inducing Wnt/β-catenin pathway by enhancing p300-mediated β-catenin acetylation. Oncogene. 2018;37(26):3485–3500. doi: 10.1038/s41388-018-0161-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gezer U, Üstek D, Yörüker EE, Cakiris A, Abaci N, Leszinski G, Dalay N, Holdenrieder S. Characterization of H3K9me3- and H4K20me3-associated circulating nucleosomal DNA by high-throughput sequencing in colorectal cancer. Tumor Biol. 2013;34(1):329–336. doi: 10.1007/s13277-012-0554-5. [DOI] [PubMed] [Google Scholar]
- 49.Tamagawa H, Oshima T, Numata M, Yamamoto N, Shiozawa M, Morinaga S, Nakamura Y, Yoshihara M, Sakuma Y, Kameda Y, et al. Global histone modification of H3K27 correlates with the outcomes in patients with metachronous liver metastasis of colorectal cancer. Eur J Surg Oncol. 2013;39(6):655–661. doi: 10.1016/j.ejso.2013.02.023. [DOI] [PubMed] [Google Scholar]
- 50.Benard A, Goossens-Beumer IJ, van Hoesel AQ, de Graaf W, Horati H, Putter H, ECM Z, van de Velde CJH, Kuppen PJK. Histone trimethylation at H3K4, H3K9 and H4K20 correlates with patient survival and tumor recurrence in early-stage colon cancer. BMC Cancer. 2014;14(1):531. doi: 10.1186/1471-2407-14-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yokoyama Y, Hieda M, Nishioka Y, Matsumoto A, Higashi S, Kimura H, Yamamoto H, Mori M, Matsuura S, Matsuura N. Cancer-associated upregulation of histone H3 lysine 9 trimethylation promotes cell motilityin vitro and drives tumor formation in vivo. Cancer Sci. 2013;104(7):889–895. doi: 10.1111/cas.12166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen T, Li J, Xu M, Zhao Q, Hou Y, Yao L, Zhong Y, Chou P, Zhang W, Zhou P et al (2017) PKCε phosphorylates MIIP and promotes colorectal cancer metastasis through inhibition of RelA deacetylation. Nat Commun:8(1) [DOI] [PMC free article] [PubMed]
- 53.Yu D, Li Z, Gan M, Zhang H, Yin X, Tang S, Wan L, Tian Y, Zhang S, Zhu Y, et al. Decreased expression of dual specificity phosphatase 22 in colorectal cancer and its potential prognostic relevance for stage IV CRC patients. Tumor Biol. 2015;36(11):8531–8535. doi: 10.1007/s13277-015-3588-7. [DOI] [PubMed] [Google Scholar]
- 54.Fraga MF, Ballestar E, Villar-Garea A, Boix-Chornet M, Espada J, Schotta G, Bonaldi T, Haydon C, Ropero S, Petrie K, et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat Genet. 2005;37(4):391–400. doi: 10.1038/ng1531. [DOI] [PubMed] [Google Scholar]
- 55.Benard A, Goossens-Beumer IJ, van Hoesel AQ, Horati H, de Graaf W, Putter H, Zeestraten ECM, Liefers G, van de Velde CJH, Kuppen PJK. Nuclear expression of histone deacetylases and their histone modifications predicts clinical outcome in colorectal cancer. Histopathology. 2015;66(2):270–282. doi: 10.1111/his.12534. [DOI] [PubMed] [Google Scholar]
- 56.Cao L, Zhu L, Yang J, Su J, Ni J, Du Y, Liu D, Wang Y, Wang F, Jin J, et al. Correlation of low expression of hMOF with clinicopathological features of colorectal carcinoma, gastric cancer and renal cell carcinoma. Int J Oncol. 2014;44(4):1207–1214. doi: 10.3892/ijo.2014.2266. [DOI] [PubMed] [Google Scholar]
- 57.Su J, Wang F, Cai Y, Jin J. The functional analysis of histone acetyltransferase MOF in tumorigenesis. Int J Mol Sci. 2016;17(1):99. doi: 10.3390/ijms17010099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang H, Salz T, Zajac-Kaye M, Liao D, Huang S, Qiu Y. Overexpression of histone deacetylases in cancer cells is controlled by interplay of transcription factors and epigenetic modulators. FASEB J. 2014;28(10):4265–4279. doi: 10.1096/fj.14-250654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lutz L, Fitzner IC, Ahrens T, Geissler AL, Makowiec F, Hopt UT, Bogatyreva L, Hauschke D, Werner M, Lassmann S. Histone modifiers and marks define heterogeneous groups of colorectal carcinomas and affect responses to HDAC inhibitors in vitro. Am J Cancer Res. 2016;6(3):664–676. [PMC free article] [PubMed] [Google Scholar]
- 60.Cao LL, Yue Z, Liu L, Pei L, Yin Y, Qin L, Zhao J, Liu H, Wang H, Jia M. The expression of histone deacetylase HDAC1 correlates with the progression and prognosis of gastrointestinal malignancy. Oncotarget. 2017;8(24):39241–39253. doi: 10.18632/oncotarget.16843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ye P, Xing H, Lou F, Wang K, Pan Q, Zhou X, Gong L, Li D. Histone deacetylase 2 regulates doxorubicin (Dox) sensitivity of colorectal cancer cells by targeting ABCB1 transcription. Cancer Chemother Pharmacol. 2016;77(3):613–621. doi: 10.1007/s00280-016-2979-9. [DOI] [PubMed] [Google Scholar]
- 62.Nemati M, Ajami N, Estiar MA, Rezapour S, Ravanbakhsh Gavgani R, Hashemzadeh S, Samadi Kafil H, Sakhinia E. Deregulated expression of HDAC3 in colorectal cancer and its clinical significance. Adv Clin Exp Med. 2018;27(3):305–311. doi: 10.17219/acem/66207. [DOI] [PubMed] [Google Scholar]
- 63.Nakazawa T, Kondo T, Ma D, Niu D, Mochizuki K, Kawasaki T, Yamane T, Iino H, Fujii H, Katoh R. Global histone modification of histone H3 in colorectal cancer and its precursor lesions. Hum Pathol. 2012;43(6):834–842. doi: 10.1016/j.humpath.2011.07.009. [DOI] [PubMed] [Google Scholar]
- 64.Benard A, Goossens-Beumer IJ, van Hoesel AQ, Horati H, Putter H, Zeestraten EC, van de Velde CJ, Kuppen PJ. Prognostic value of polycomb proteins EZH2, BMI1 and SUZ12 and histone modification H3K27me3 in colorectal cancer. PLoS One. 2014;9(9):e108265. doi: 10.1371/journal.pone.0108265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kryczek I, Lin Y, Nagarsheth N, Peng D, Zhao L, Zhao E, Vatan L, Szeliga W, Dou Y, Owens S, et al. IL-22(+)CD4(+) T cells promote colorectal cancer stemness via STAT3 transcription factor activation and induction of the methyltransferase DOT1L. Immunity. 2014;40(5):772–784. doi: 10.1016/j.immuni.2014.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mu Z, Li H, Fernandez SV, Alpaugh KR, Zhang R, Cristofanilli M. EZH2 knockdown suppresses the growth and invasion of human inflammatory breast cancer cells. J Exp Clin Cancer Res. 2013;32:70. doi: 10.1186/1756-9966-32-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kornblihtt AR. Epigenetics at the base of alternative splicing changes that promote colorectal cancer. J Clin Invest. 2017;127(9):3281–3283. doi: 10.1172/JCI96497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Qin J, Zeng Z, Luo T, Li Q, Hao Y, Chen L. Clinicopathological significance of G9A expression in colorectal carcinoma. Oncol Lett. 2018;15(6):8611–8619. doi: 10.3892/ol.2018.8446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang C. EZH2 and STAT6 expression profiles are correlated with colorectal cancer stage and prognosis. World J Gastroenterol. 2010;16(19):2421–2427. doi: 10.3748/wjg.v16.i19.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen Z, Yang P, Li W, He F, Wei J, Zhang T, Zhong J, Chen H, Cao J. Expression of EZH2 is associated with poor outcome in colorectal cancer. Oncol Lett. 2018;15(3):2953–2961. doi: 10.3892/ol.2017.7647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tokunaga R, Sakamoto Y, Nakagawa S, Miyake K, Izumi D, Kosumi K, Taki K, Higashi T, Imamura Y, Ishimoto T, et al. The prognostic significance of histone lysine demethylase JMJD3/KDM6B in colorectal cancer. Ann Surg Oncol. 2016;23(2):678–685. doi: 10.1245/s10434-015-4879-3. [DOI] [PubMed] [Google Scholar]
- 72.Ding J, Zhang Z, Xia Y, Liao G, Pan Y, Liu S, Zhang Y, Yan Z. LSD1-mediated epigenetic modification contributes to proliferation and metastasis of colon cancer. Br J Cancer. 2013;109(4):994–1003. doi: 10.1038/bjc.2013.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cordeiro MH, Smith RJ, Saurin AT. A fine balancing act: a delicate kinase-phosphatase equilibrium that protects against chromosomal instability and cancer. Int J Biochem Cell Biol. 2018;96:148–156. doi: 10.1016/j.biocel.2017.10.017. [DOI] [PubMed] [Google Scholar]
- 74.Lee YC, Yin TC, Chen YT, Chai CY, Wang JY, Liu MC, Lin YC, Kan JY. High expression of phospho-H2AX predicts a poor prognosis in colorectal cancer. Anticancer Res. 2015;35(4):2447–2453. [PubMed] [Google Scholar]
- 75.Xiao J, Duan Q, Wang Z, Yan W, Sun H, Xue P, Fan X, Zeng X, Chen J, Shao C, et al. Phosphorylation of TOPK at Y74, Y272 by Src increases the stability of TOPK and promotes tumorigenesis of colon. Oncotarget. 2016;7(17):24483. doi: 10.18632/oncotarget.8231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Song Y, Wu F, Wu J (2016) Targeting histone methylation for cancer therapy: enzymes, inhibitors, biological activity and perspectives. J Hematol Oncol 9(1) [DOI] [PMC free article] [PubMed]
- 77.Hsu H, Liu Y, Tseng K, Yang T, Yeh C, You J, Hung H, Chen S, Chen H. CBB1003, a lysine-specific demethylase 1 inhibitor, suppresses colorectal cancer cells growth through down-regulation of leucine-rich repeat-containing G-protein-coupled receptor 5 expression. J Cancer Res Clin Oncol. 2015;141(1):11–21. doi: 10.1007/s00432-014-1782-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Takawa M, Masuda K, Kunizaki M, Daigo Y, Takagi K, Iwai Y, Cho H, Toyokawa G, Yamane Y, Maejima K, et al. Validation of the histone methyltransferase EZH2 as a therapeutic target for various types of human cancer and as a prognostic marker. Cancer Sci. 2011;102(7):1298–1305. doi: 10.1111/j.1349-7006.2011.01958.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ying L, Yan F, Williams BR, Xu P, Li X, Zhao Y, Hu Y, Wang Y, Xu D, Dai J. (−)-Epigallocatechin-3-gallate and EZH2 inhibitor GSK343 have similar inhibitory effects and mechanisms of action on colorectal cancer cells. Clin Exp Pharmacol Physiol. 2018;45(1):58–67. doi: 10.1111/1440-1681.12854. [DOI] [PubMed] [Google Scholar]
- 80.Hsieh YY, Lo HL, Yang PM. EZH2 inhibitors transcriptionally upregulate cytotoxic autophagy and cytoprotective unfolded protein response in human colorectal cancer cells. Am J Cancer Res. 2016;6(8):1661–1680. [PMC free article] [PubMed] [Google Scholar]
- 81.Paschall AV, Yang D, Lu C, Choi JH, Li X, Liu F, Figueroa M, Oberlies NH, Pearce C, Bollag WB, et al. H3K9 Trimethylation silences Fas expression to confer Colon carcinoma immune escape and 5-fluorouracil Chemoresistance. J Immunol. 2015;195(4):1868–1882. doi: 10.4049/jimmunol.1402243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kim MS, Cho HI, Yoon HJ, Ahn Y, Park EJ, Jin YH, Jang YK (2018) JIB-04, a small molecule histone demethylase inhibitor, selectively targets colorectal cancer stem cells by inhibiting the Wnt/β-catenin signaling pathway. Sci Rep 8(1) [DOI] [PMC free article] [PubMed]
- 83.Sonnemann J, Marx C, Becker S, Wittig S, Palani CD, Krämer OH, Beck JF. p53-dependent and p53-independent anticancer effects of different histone deacetylase inhibitors. Br J Cancer. 2014;110(3):656–667. doi: 10.1038/bjc.2013.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gargalionis AN, Piperi C, Adamopoulos C, Papavassiliou AG. Histone modifications as a pathogenic mechanism of colorectal tumorigenesis. Int J Biochem Cell Biol. 2012;44(8):1276–1289. doi: 10.1016/j.biocel.2012.05.002. [DOI] [PubMed] [Google Scholar]
- 85.Liu K, Wang L, Hsu S. Modification of epigenetic histone acetylation in hepatocellular carcinoma. Cancers. 2018;10(1):8. doi: 10.3390/cancers10010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Iyer SP, Foss FF. Romidepsin for the treatment of peripheral T-cell lymphoma. Oncologist. 2015;20(9):1084–1091. doi: 10.1634/theoncologist.2015-0043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Foss F, Advani R, Duvic M, Hymes KB, Intragumtornchai T, Lekhakula A, Shpilberg O, Lerner A, Belt RJ, Jacobsen ED, et al. A phase II trial of Belinostat (PXD101) in patients with relapsed or refractory peripheral or cutaneous T-cell lymphoma. Br J Haematol. 2015;168(6):811–819. doi: 10.1111/bjh.13222. [DOI] [PubMed] [Google Scholar]
- 88.Xiong H, Du W, Zhang Y, Hong J, Su W, Tang J, Wang Y, Lu R, Fang J. Trichostatin A, a histone deacetylase inhibitor, suppresses JAK2/STAT3 signaling via inducing the promoter-associated histone acetylation of SOCS1 and SOCS3 in human colorectal cancer cells. Mol Carcinog. 2012;51(2):174–184. doi: 10.1002/mc.20777. [DOI] [PubMed] [Google Scholar]
- 89.Jin J, Tsao T, Sun P, Yu C, Tzao C. SAHA inhibits the growth of Colon tumors by decreasing histone deacetylase and the expression of cyclin D1 and Survivin. Pathol Oncol Res. 2012;18(3):713–720. doi: 10.1007/s12253-012-9499-7. [DOI] [PubMed] [Google Scholar]
- 90.Humphreys KJ, Cobiac L, Le Leu RK, Van der Hoek MB, Michael MZ. Histone deacetylase inhibition in colorectal cancer cells reveals competing roles for members of the oncogenic miR-17-92 cluster. Mol Carcinog. 2013;52(6):459–474. doi: 10.1002/mc.21879. [DOI] [PubMed] [Google Scholar]
- 91.Strey CW, Schamell L, Oppermann E, Haferkamp A, Bechstein WO, Blaheta RA. Valproate inhibits colon cancer growth through cell cycle modification in vivo and in vitro. Exp Ther Med. 2011;2(2):301–307. doi: 10.3892/etm.2011.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang TY, Chai YR, Jia YL, Gao JH, Peng XJ, Han HF. Crosstalk among the proteome, lysine phosphorylation, and acetylation in romidepsin-treated colon cancer cells. Oncotarget. 2016;7(33):53471–53501. doi: 10.18632/oncotarget.10840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mizutani H, Hiraku Y, Tada-Oikawa S, Murata M, Ikemura K, Iwamoto T, Kagawa Y, Okuda M, Kawanishi S. Romidepsin (FK228), a potent histone deacetylase inhibitor, induces apoptosis through the generation of hydrogen peroxide. Cancer Sci. 2010;101(10):2214–2219. doi: 10.1111/j.1349-7006.2010.01645.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Li Q, Ding C, Meng T, Lu W, Liu W, Hao H, Cao L. Butyrate suppresses motility of colorectal cancer cells via deactivating Akt/ERK signaling in histone deacetylase dependent manner. J Pharmacol Sci. 2017;135(4):148–155. doi: 10.1016/j.jphs.2017.11.004. [DOI] [PubMed] [Google Scholar]
- 95.Bracker TU, Sommer A, Fichtner I, Faus H, Haendler B, Hess-Stumpp H. Efficacy of MS-275, a selective inhibitor of class I histone deacetylases, in human colon cancer models. Int J Oncol. 2009;35(4):909–920. doi: 10.3892/ijo_00000406. [DOI] [PubMed] [Google Scholar]
- 96.Zhu S, Denman CJ, Cobanoglu ZS, Kiany S, Lau CC, Gottschalk SM, Hughes DPM, Kleinerman ES, Lee DA. The narrow-Spectrum HDAC inhibitor Entinostat enhances NKG2D expression without NK cell toxicity, leading to enhanced recognition of cancer cells. Pharm Res. 2015;32(3):779–792. doi: 10.1007/s11095-013-1231-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Anantharaju PG, Reddy BD, Padukudru MA, Kumari Chitturi CM, Vimalambike MG, Madhunapantula SV. Naturally occurring benzoic acid derivatives retard cancer cell growth by inhibiting histone deacetylases (HDAC) Cancer Biol Ther. 2017;18(7):492–504. doi: 10.1080/15384047.2017.1324374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Beck HC, Petersen J, Nielsen SJ, Morsczeck C, Jensen PB, Sehested M, Grauslund M. Proteomic profiling of human colon cancer cells treated with the histone deacetylase inhibitor belinostat. Electrophoresis. 2010;31(16):2714–2721. doi: 10.1002/elps.201000033. [DOI] [PubMed] [Google Scholar]
- 99.Maschauer S, Gahr S, Gandesiri M, Tripal P, Schneider-Stock R, Kuwert T, Ocker M, Prante O. In vivo monitoring of the anti-angiogenic therapeutic effect of the pan-deacetylase inhibitor panobinostat by small animal PET in a mouse model of gastrointestinal cancers. Nucl Med Biol. 2016;43(1):27–34. doi: 10.1016/j.nucmedbio.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 100.LaBonte MJ, Wilson PM, Fazzone W, Groshen S, Lenz HJ, Ladner RD. DNA microarray profiling of genes differentially regulated by the histone deacetylase inhibitors vorinostat and LBH589 in colon cancer cell lines. BMC Med Genet. 2009;2:67. doi: 10.1186/1755-8794-2-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Iwamoto M, Friedman EJ, Sandhu P, Agrawal NGB, Rubin EH, Wagner JA. Clinical pharmacology profile of vorinostat, a histone deacetylase inhibitor. Cancer Chemother Pharmacol. 2013;72(3):493–508. doi: 10.1007/s00280-013-2220-z. [DOI] [PubMed] [Google Scholar]
- 102.Hu Q, Baeg GH. Role of epigenome in tumorigenesis and drug resistance. Food Chem Toxicol. 2017;109(Pt 1):663–668. doi: 10.1016/j.fct.2017.07.022. [DOI] [PubMed] [Google Scholar]
- 103.Papavassiliou KA, Papavassiliou AG. Histone deacetylases inhibitors: conjugation to other anti-tumour pharmacophores provides novel tools for cancer treatment. Expert Opin Investig Drugs. 2013;23(3):291–294. doi: 10.1517/13543784.2014.857401. [DOI] [PubMed] [Google Scholar]
- 104.Won H, Ryu H, Shin D, Yeon S, Lee DH, Kwon SH. A452, an HDAC6-selective inhibitor, synergistically enhances the anticancer activity of chemotherapeutic agents in colorectal cancer cells. Mol Carcinog. 2018;57:1383–1395. doi: 10.1002/mc.22852. [DOI] [PubMed] [Google Scholar]
- 105.Katona BW, Liu Y, Ma A, Jin J, Hua X. EZH2 inhibition enhances the efficacy of an EGFR inhibitor in suppressing colon cancer cells. Cancer Biol Ther. 2014;15(12):1677–1687. doi: 10.4161/15384047.2014.972776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pitts TM, Morrow M, Kaufman SA, Tentler JJ, Eckhardt SG. Vorinostat and bortezomib exert synergistic antiproliferative and proapoptotic effects in colon cancer cell models. Mol Cancer Ther. 2009;8(2):342–349. doi: 10.1158/1535-7163.MCT-08-0534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Encarnacao JC, Pires AS, Amaral RA, Goncalves TJ, Laranjo M, Casalta-Lopes JE, Goncalves AC, Sarmento-Ribeiro AB, Abrantes AM, Botelho MF. Butyrate, a dietary fiber derivative that improves irinotecan effect in colon cancer cells. J Nutr Biochem. 2018;56:183–192. doi: 10.1016/j.jnutbio.2018.02.018. [DOI] [PubMed] [Google Scholar]
- 108.Azad NS, El-Khoueiry A, Yin J, Oberg AL, Flynn P, Adkins D, Sharma A, Weisenberger DJ, Brown T, Medvari P, et al. Combination epigenetic therapy in metastatic colorectal cancer (mCRC) with subcutaneous 5-azacitidine and entinostat: a phase 2 consortium/stand up 2 cancer study. Oncotarget. 2017;8(21):35326. doi: 10.18632/oncotarget.15108. [DOI] [PMC free article] [PubMed] [Google Scholar]