Abstract
Pathological complete response (pCR) after neoadjuvant chemotherapy (NAC) has been proposed as a surrogate endpoint for the prediction of long-term survival in breast cancer (BC); however, an increased pCR rate has not clearly correlated with improved survival. We hypothesized that some transcriptomic and functional pathway features correlate with survival after pCR in BC. We utilized 2 published NAC cohorts, 105 women with gene expression data before, “Baseline”, and that changed during NAC, “Delta”, and TCGA database with 1068 BC patients to investigate the relationship between the efficacy of NAC and survival utilizing differentially expressed-mRNAs, construction and analysis of the mRNA-hub gene network, and functional pathway analysis. In mRNA expression profiling, S100A8 was a gene involved in survival after pCR in Baseline and NDP was a gene involved in recurrence after pCR in Delta. In functional pathway analysis, we found multiple pathways involved in survival after pCR. In mRNA-hub gene analysis, HSP90AA1, EEF1A1, APP, and HSPA4 were related to recurrence in BC patients with pCR due to NAC. TP53, EGFR, CTNNB1, ERBB2, and HSPB1 may play a significant role in survival for patients with pCR. Interestingly, high HSP90AA1, HSPA4, S100A8, and TP53, and low EEF1A1, EGFR, and CTNNB1 expressing tumors have significantly worse overall survival in TCGA BC cohort. We demonstrated the genes and functional pathway features associated with pCR and survival utilizing the bioinformatics approach to public BC cohorts. Some genes involved in recurrence after pCR due to NAC also served as prognostic factors in primary BC.
Keywords: Breast cancer, pCR and survival, cancer genomics, functional pathway, bioinformatics
Introduction
Breast cancer (BC) is the most commonly diagnosed cancer and the second leading cause of cancer deaths among American women, and thus has been identified as a public health priority in the United States. The lifetime risk of developing BC today is one in every eight women [1]. Currently, surgery, radiotherapy, and chemo-/endocrine-therapy are the major treatment options for BC. Neoadjuvant chemotherapy (NAC), which is systemic therapy delivered before definitive BC surgery, has been widely applied for the following three reasons. First, NAC reduces the size and extent of locally advanced tumors, which allows for breast conserving surgery [2]. Second, NAC allows for early identification of unresponsive tumors and provides an opportunity to terminate the ineffective therapy and/or to switch to an alternative regimen [3]. Indeed, NAC trials have been used for the rapid assessment of drug efficacy that sped up the development and approval of drugs for early BC during the last two decades [4]. For example, the GeparTrio study showed that the regimen based upon NAC response was significantly better in prolonging disease free survival (DFS) and overall survival (OS) than a non-individualized approach with a fixed number of cycles, especially among patients with hormone receptor (HR)-positive tumors [5-7]. Third, response to NAC is used as an early predictive indicator of the prognosis of BC patients.
In general, pathological complete response (pCR) has been used as a surrogate endpoint for the prediction of long-term survival such as DFS and OS [2]; however, this notion has recently been challenged. First, responses to conventional NAC differ by the BC subtypes, complicating the investigation of the predictive value of biomarkers. Thus, pCR is currently utilized as a “surrogate marker” for accelerated drug registration only in aggressive BC subtypes such as triple negative (TN) or human epidermal growth factor receptor 2 (HER2)-positive cases [8-10]. Second, BC cells may remain in dormancy and survive in patients that achieved pCR to NAC. Multiple mechanisms have been proposed to explain how cancer cells become dormant, and how they become reactivated and exit dormancy [11].
To this end, further elucidation of the relationship between pCR and survival should enhance the role of pCR after NAC as a surrogate marker for survival for the BC patients. One of the approaches to exploit the full potential of NAC is to identify the key genes that are expressed prior to and that changed during the treatment and correlate them with survival. We hypothesized that some transcriptomic and functional pathway features correlate with pCR to NAC and survival in BC cohorts. To test this hypothesis we utilized mRNA expression profiles, construction and analysis of the mRNA-hub gene network, and functional and pathway enrichment analysis.
Materials and methods
Neoadjuvant chemotherapy cohorts
Two Gene Expression Omnibus (GEO) datasets, GSE32603 and GSE87455, were used to examine the association between response to anthracycline-based chemotherapy and survival in patients who underwent NAC (Figure S1). Microarray gene expression data in GEO datasets (http://www.ncbi.nlm.nih.gov/geo) were queried from the National Center for Biotechnology Information. In the GSE32603 cohort, out of 46 primary BC patients treated with anthracycline based chemotherapy (AC) followed by optimal taxane based chemotherapy (OTC), 36 women who had both gene expression data for before (T1) and during (T2) AC were analyzed [12]. In the GSE87455 cohort, out of 150 primary BC patients treated with epirubicin + docetaxel + bevacizumab (EDB), 69 women who had both gene expression data for T1 and T2 were analyzed [13]. Both cohorts were used to support the authenticity of the association between the effect of NAC and clinical outcomes. We defined the gene expression profile in T1 as “Baseline”, and the change from T1 to T2 as “Delta”.
Screening for differentially expressed mRNAs
The Student’s t-test was used to compare the difference between binary variables. Top 10 differentially expressed-mRNAs were selected based on |log2[fold change (FC)]|. P-value <0.05 and |log2FC| >0.17 were set as the thresholds for screening differentially expressed-mRNAs. This screening method was referred to as “Previous analysis” in prior publication [14].
Gene Ontology (GO) annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis
GO annotation and KEGG pathway enrichment analyses for the predicted target genes of the top 20 differentially expressed-mRNAs were conducted by using the R package “clusterProfiler”.
Construction and analysis of mRNA hub gene network
To assess the interactive relationship among the predicted target genes in each category, the STRING database (http://string-db.org) was used to construct the mRNA-hub gene network as follows; first, we chose 10 genes with the largest difference in expression using mRNA data. Next, we built the network using these top 10 genes and their interactors (genes that interact with them). Finally, we screened out the top 25 hub genes through the resulting network.
mRNA expression data from the cancer genome atlas (TCGA)
TCGA was supervised by the National Cancer Institute (NCI) and the National Human Genome Research Institute [15]. The gene expression levels (mRNA expression z-score from RNA-sequence) from Genomic Identification of Significant Targets in Cancer for TCGA cohort was downloaded through cBioportal (TCGA, PanCancer Atlas) [16,17] as described before [18,19]. The expression levels of potential mRNAs were extracted from the downloaded mRNA information.
Statistical analysis
All statistical analyses were performed using R software (http://www.r-project.org/) and Bioconductor (http://bioconductor.org/). The student-t test was used to assess baseline differences between binary variables. In the analysis of OS, the Kaplan-Meier method was used to estimate survival rates, and differences between survival curves were evaluated by the log-rank test. Differences were considered significant with a P-value <0.05.
Results
Identification of mRNA expression profile involved in recurrence after pCR in NAC cohorts
To clarify the relationship between pCR and survival, we divided BC patients into two groups in two ways in each cohort (Figure S2): Category 1, we divided BC patients into two groups with clinical integrity, clinical concordance versus discordance. Clinical concordance was defined as a group of patients who achieved pCR/not relapsed for 5 years, and non-pCR/relapsed within 5 years, after NAC. On the other hand, clinical discordance was defined as a group of patients who achieved pCR/relapsed and non-pCR/not relapsed, after NAC (Figure S2A). Category 2, we divided NAC treated BC patients according to whether they relapsed in the group that achieved pCR (Figure S2B). By using Category 1 as supporting data for Category 2, the genes associated with recurrence in pCR could be determined more accurately (Figure S2). Two gene expression microarray data sets GSE32603 and GSE87455 were downloaded from the GEO database [12,13]. The Student’s t-test was used to compare the difference between binary variables (Category 1: clinical concordance to discordance; Category 2: recurrence to no recurrence among pCR patients). The data were processed by unpaired t-test (P<0.05, |log2FC| >0.17). Here, we explored what kind of genes were up- or down-regulated in the course of treatment with chemotherapy. In Baseline analysis (analysis of T1 in Figure S1), the top 10 mRNAs with more or less mRNA expression at baseline are listed in Table 1. There was no common gene as Category 1 between GSE32603 and GSE87455. In Category 2, S100A8 was the common gene between GSE32603 and GSE87455. Since S100A8 decreased (no recurrence after pCR) in GSE32603 and increased (recurred after pCR) in GSE87455, it was suggested that S100A8 may behave differently in each regimen. In Delta analysis (analysis of the gene expression difference between T1 and T2 in Figure S1), in Category 1, 140 mRNAs (62 up-regulated and 78 down-regulated mRNAs) were screened out in the GSE32603 and 9 mRNAs (2 up-regulated and 7 down-regulated mRNAs) were screened out in the GSE87455. In Category 2, 1302 mRNAs (429 up-regulated and 873 down-regulated mRNAs) were screened out in the GSE32603 and 279 mRNAs (139 up-regulated and 140 down-regulated mRNAs) were screened out in the GSE87455. The top 10 mRNAs with a remarkable difference between T1 and T2 are listed in Table 2. NDP was recognized in Category 1 and 2 of GSE87455. Since NDP was decreased (clinical discordance) in Category 1 and increased (recurrence after pCR) in Category 2, it was speculated to be a specific gene involved in recurrence after pCR.
Table 1.
Identification of top 10 differentially expressed mRNAs in each category of each NAC cohort in Baseline
| Category 1 | Category 2 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
||||||||||
| GSE32603 | GSE87455 | GSE32603 | GSE87455 | ||||||||
|
|
|
|
|
||||||||
| mRNA | logFC | P-Value | mRNA | logFC | P-Value | mRNA | logFC | P-Value | mRNA | logFC | P-Value |
| C4A | -1.574 | 0.002 | ANKRD30A | -2.154 | 0.001 | LITAF | 3.450 | 0.011 | MUCL1 | 3.597 | 0.022 |
| GTF2IRD1 | 1.436 | 0.005 | AGR2 | -2.043 | 0.000 | LTF | -3.277 | 0.006 | S100P | 3.176 | 0.015 |
| NTN4 | -1.404 | 0.020 | FOXA1 | -2.023 | 0.000 | UBD | -3.025 | 0.010 | MUC1 | 2.742 | 0.004 |
| C1orf35 | 1.218 | 0.003 | TFF3 | -1.992 | 0.002 | CDH2 | -2.887 | 0.006 | S100A7 | 2.720 | 0.000 |
| F2RL2 | -1.145 | 0.025 | CEACAM6 | -1.877 | 0.004 | LYZ | -2.695 | 0.007 | S100A9 | 2.708 | 0.006 |
| RARRES3 | -1.127 | 0.035 | GABRP | 1.581 | 0.003 | MAOB | -2.650 | 0.008 | S100A8 | 2.459 | 0.044 |
| CSK | 1.119 | 0.029 | PIP | -1.533 | 0.030 | S100A8 | -2.559 | 0.019 | HBA2 | -2.249 | 0.031 |
| TRIM29 | -1.082 | 0.048 | MLPH | -1.491 | 0.001 | EFHD1 | 2.511 | 0.037 | HBB | -2.243 | 0.030 |
| KRT19 | -1.076 | 0.038 | TFF1 | -1.447 | 0.022 | RMND5B | 2.354 | 0.002 | VTCN1 | 2.213 | 0.000 |
| SLC40A1 | -1.054 | 0.001 | SRARP | -1.409 | 0.004 | KRT18 | -2.342 | 0.021 | HBA1 | -2.175 | 0.033 |
Abbreviations: NAC, neoadjuvant chemotherapy; FC, fold change.
Table 2.
Identification of top 10 differentially expressed mRNAs in each category of each NAC cohort in Delta
| Category 1 | Category 2 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
||||||||||
| GSE32603 | GSE87455 | GSE32603 | GSE87455 | ||||||||
|
|
|
|
|
||||||||
| mRNA | logFC | P-Value | mRNA | logFC | P-Value | mRNA | logFC | P-Value | mRNA | logFC | P-Value |
| CYC1 | 1.558 | 0.031 | CPB1 | -1.106 | 0.020 | LITAF | -4.238 | 0.009 | ALB | -2.651 | 0.028 |
| C1orf35 | -1.557 | 0.009 | NDP | -0.924 | 0.009 | ACRV1 | -3.879 | 0.044 | NDP | 2.247 | 0.032 |
| CXCL9 | 1.505 | 0.020 | SCGB2A1 | -0.897 | 0.018 | DNAH14 | 3.681 | 0.002 | GJA1 | 2.157 | 0.030 |
| SLC4A4 | 1.398 | 0.025 | WFDC2 | -0.865 | 0.010 | CPB1 | 3.526 | 0.002 | MOXD1 | 2.131 | 0.013 |
| MMP7 | 1.350 | 0.016 | CRABP1 | -0.827 | 0.003 | JAM2 | 3.173 | 0.011 | CRYAB | -1.939 | 0.006 |
| CXCL10 | 1.349 | 0.026 | APOC1 | 0.719 | 0.016 | PVALB | 3.016 | 0.009 | CTXN1 | 1.925 | 0.002 |
| FGFR1 | -1.318 | 0.048 | MSLN | -0.677 | 0.021 | CEP55 | -2.902 | 0.010 | C1orf115 | -1.823 | 0.0002 |
| SIVA1 | 1.270 | 0.018 | HDC | -0.670 | 0.021 | ISG15 | 2.838 | 0.002 | THBS4 | -1.771 | 0.036 |
| CNN3 | 1.223 | 0.006 | DCD | 0.646 | 0.015 | ZCCHC9 | -2.800 | 0.011 | VASH2 | 1.638 | 0.049 |
| FAM26F | 1.203 | 0.003 | GRIA2 | -0.573 | 0.010 | LRRC2 | 2.799 | 0.005 | ANGPTL8 | -1.632 | 0.017 |
Abbreviations: NAC, neoadjuvant chemotherapy; FC, fold change.
Construction and analysis of mRNA hub gene network
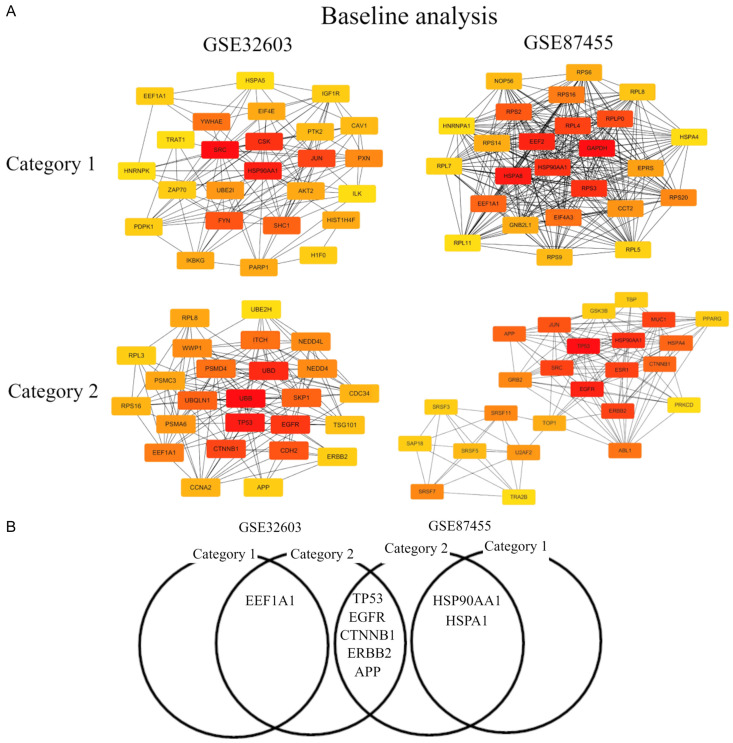
The construction of the mRNA-hub gene network is considerably helpful for us to identify the most potentially functional mRNAs [20]. Here, we explored the relationship between mRNAs in each category of each GEO. After analyzing the data from STRING using Cytoscape software, we first screened out the top 25 hub nodes according to degree related with the top 10 mRNA with more changes in each NAC cohort in Category 1 and 2, in the same way as published in [14] (Tables 3 and 4). For better visualization of the interactions of these hub genes, additionally, we constructed networks based on the screened top 25 hub genes related with the top 10 mRNAs with more changes, as presented in Figures 1 and 2. In Baseline analysis, the top 10 hub genes were SRC, HSP90AA1, CSK, JUN, FYN, SHC1, YWHAE, PXN, UBE2I, and HIST1H4F, among which SRC showed the highest node degree (degree = 43) in Category 1 of GSE32603. In Category 1 of GSE87455, the top 10 hub genes were GAPDH, HSPA8, EEF2, HSP90AA1, RPS3, RPL4, RPLP0, RPS2, EEF1A1, and EIF4A3, among which GAPDH showed the highest node degree (degree = 305). In Category 2 of GSE32603, the top 10 hub genes were TP53, EGFR, HSP90AA1, MUC1, ERBB2, SRC, ESR1, JUN, HSPA4, and CTNNB1, among which TP53 showed the highest node degree (degree = 37). In GSE87455, the top 10 hub genes were UBB, TP53, UBD, EGFR, CTNNB1, CDH2, SKP1, UBQLN1, ITCH, and PSMD4, among which UBB showed the highest node degree (degree = 50) (Table 3). Of note, EEF1A1 in GSE32603 and HSP90AA1 and HSPA4 in GSE87455 were genes common to Category 1 and 2. It was suggested that EEF1A1 may have a role in BC recurrence after pCR due to AC followed by OTC. On the other hand, HSP90AA1 and HSPA4 may have a role in BC recurrence after pCR due to EDB. TP53, EGFR, CTNNB1, ERBB2, and APP were genes common to GSE32603 and GSE87455 in Category 2, suggesting that these mRNAs may play a significant role in survival for patients with pCR (Figure 1).
Table 3.
Identification of top 25 hub nodes according to degree related with the top 10 mRNA with more changes in each category of each NAC cohort in Baseline
| Category 1 | Category 2 | ||||||
|---|---|---|---|---|---|---|---|
|
|
|
||||||
| GSE32603 | GSE87455 | GSE32603 | GSE87455 | ||||
|
|
|
|
|
||||
| Gene Symbol | Degree | Gene Symbol | Degree | Gene Symbol | Degree | Gene Symbol | Degree |
| SRC | 43 | GAPDH | 305 | UBB | 50 | TP53 | 37 |
| HSP90AA1 | 41 | HSPA8 | 260 | TP53 | 44 | EGFR | 31 |
| CSK | 33 | EEF2 | 236 | UBD | 37 | HSP90AA1 | 30 |
| JUN | 29 | HSP90AA1 | 225 | EGFR | 36 | MUC1 | 25 |
| FYN | 26 | RPS3 | 225 | CTNNB1 | 31 | ERBB2 | 25 |
| SHC1 | 24 | RPL4 | 222 | CDH2 | 30 | SRC | 25 |
| YWHAE | 23 | RPLP0 | 215 | SKP1 | 26 | ESR1 | 25 |
| PXN | 22 | RPS2 | 214 | UBQLN1 | 26 | JUN | 23 |
| UBE2I | 19 | EEF1A1 | 212 | ITCH | 25 | HSPA4 | 21 |
| HIST1H4F | 18 | EIF4A3 | 211 | PSMD4 | 25 | CTNNB1 | 21 |
| AKT2 | 18 | RPS16 | 208 | EEF1A1 | 24 | APP | 21 |
| IKBKG | 18 | RPS20 | 207 | WWP1 | 22 | ABL1 | 20 |
| EIF4E | 18 | CCT2 | 205 | NEDD4 | 22 | GRB2 | 19 |
| PARP1 | 18 | EPRS | 202 | NEDD4L | 22 | SRSF7 | 19 |
| PTK2 | 17 | RPS6 | 201 | PSMA6 | 21 | SRSF11 | 19 |
| CAV1 | 17 | GNB2L1 | 201 | RPL8 | 20 | U2AF2 | 18 |
| EEF1A1 | 16 | RPS14 | 201 | PSMC3 | 20 | TOP1 | 17 |
| PDPK1 | 16 | RPS9 | 200 | CDC34 | 19 | SRSF5 | 16 |
| ZAP70 | 16 | NOP56 | 200 | RPS16 | 19 | SRSF3 | 15 |
| IGF1R | 16 | RPL8 | 198 | CCNA2 | 19 | GSK3B | 15 |
| H1F0 | 16 | RPL5 | 196 | TSG101 | 18 | SAP18 | 15 |
| HNRNPK | 15 | RPL7 | 196 | RPL3 | 17 | TBP | 15 |
| ILK | 15 | HSPA4 | 194 | ERBB2 | 17 | PPARG | 15 |
| HSPA5 | 15 | HNRNPA1 | 194 | APP | 17 | TRA2B | 14 |
| TRAT1 | 15 | RPL11 | 194 | HGS | 16 | PRKCD | 14 |
Abbreviations: NAC, neoadjuvant chemotherapy.
Table 4.
Identification of top 25 hub nodes according to degree related with the top 10 mRNA with more changes in each category of each NAC cohort in Delta
| Category 1 | Category 2 | ||||||
|---|---|---|---|---|---|---|---|
|
|
|
||||||
| GSE32603 | GSE87455 | GSE32603 | GSE87455 | ||||
|
|
|
|
|
||||
| Gene symbol | Degree | Gene symbol | Degree | Gene symbol | Degree | Gene symbol | Degree |
| AKT1 | 82 | GRIA2 | 11 | GAPDH | 87 | ALB | 76 |
| TP53 | 70 | GRIA1 | 8 | HSPA8 | 74 | TP53 | 68 |
| SRC | 67 | MSLN | 8 | HSP90AA1 | 72 | INS | 60 |
| FGFR1 | 58 | APP | 7 | RPS27A | 63 | AKT1 | 59 |
| PIK3R1 | 57 | GRIP1 | 6 | HSPA4 | 61 | FN1 | 51 |
| CTNNB1 | 51 | RAB11A | 6 | UBB | 57 | MAPK1 | 49 |
| PDGFRB | 49 | NDUFB5 | 5 | ENO1 | 52 | APP | 47 |
| HSP90AA1 | 47 | MRPL28 | 4 | EEF2 | 47 | CASP3 | 46 |
| FGF2 | 47 | PICK1 | 4 | VCP | 45 | VEGFA | 44 |
| PTPN11 | 46 | ATP5F1 | 4 | ISG15 | 44 | CTNNB1 | 41 |
| SOS1 | 44 | MYO5A | 4 | HNRNPA2B1 | 44 | CCND1 | 36 |
| CYC1 | 41 | GAPDH | 4 | GNB2L1 | 42 | CDH1 | 36 |
| CDH1 | 40 | NDUFB9 | 4 | HSPD1 | 41 | CRYAB | 35 |
| STAT3 | 40 | MRPL16 | 4 | HSP90AB1 | 41 | APOE | 35 |
| PIK3R2 | 39 | NDUFV3 | 3 | CCT2 | 39 | UBC | 35 |
| ERBB3 | 38 | RALA | 3 | PKM | 38 | GJA1 | 31 |
| UBC | 37 | SDCBP | 3 | HNRNPK | 37 | F2 | 30 |
| PLCG1 | 35 | GRN | 3 | PGK1 | 36 | SNCA | 29 |
| GAB1 | 35 | MRPL1 | 3 | ELAVL1 | 36 | APOA1 | 29 |
| JAK2 | 35 | EIF1AX | 2 | EEF1G | 35 | FGF2 | 28 |
| SHC1 | 35 | RNMTL1 | 2 | TSG101 | 35 | FGA | 28 |
| MDM2 | 31 | GRID2 | 2 | HSPB1 | 34 | HSPB1 | 27 |
| CRK | 31 | PTPRF | 2 | CCT4 | 34 | HP | 27 |
| VDAC1 | 31 | AGAP2 | 2 | ANXA2 | 33 | TJP1 | 26 |
| VAV1 | 31 | TRMT6 | 1 | EIF4G1 | 33 | BCL2L1 | 26 |
Abbreviations: NAC, neoadjuvant chemotherapy.
Figure 1.
The top 25 mRNA-hub genes in baseline analysis. (A) mRNA hub gene network and (B) Venn diagram. Category 1, we divided BC patients into two groups with clinical integrity, clinical concordance versus discordance. Clinical concordance was defined as a group of patients who were pCR/not relapsed and non-pCR/relapsed, according to NAC. On the other hand, clinical discordance was defined as a group of patients who were pCR/relapsed and non-pCR/not relapsed, according to NAC. Category 2, we divided NAC treated BC patients according to whether they relapsed in the group having pCR. Two microarray data sets GSE32603 and GSE87455 were downloaded from the GEO database. The data were processed by unpaired t-test (P<0.05, |log2FC| >0.17). Abbreviations: BC, breast cancer; pCR, pathological complete response; NAC, neoadjuvant chemotherapy; GEO, Gene Expression Omnibus.
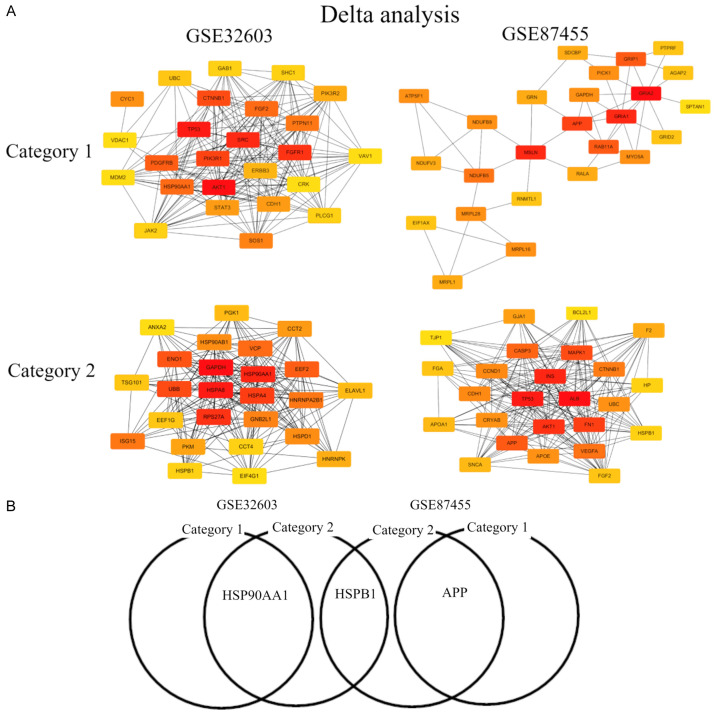
Figure 2.
The top 25 mRNA-hub genes in delta analysis. (A) mRNA hub gene network and (B) Venn diagram. Category 1, we divided BC patients into two groups with clinical integrity, clinical concordance versus discordance. Clinical concordance was defined as a group of patients who were pCR/not relapsed and non-pCR/relapsed, according to NAC. On the other hand, clinical discordance was defined as a group of patients who were pCR/relapsed and non-pCR/not relapsed, according to NAC. Category 2, we divided NAC treated BC patients according to whether they relapsed in the group having pCR. Two microarray data sets GSE32603 and GSE87455 were downloaded from the GEO database. The data were processed by unpaired t-test (P<0.05, |log2FC| >0.17). Abbreviations: BC, breast cancer; pCR, pathological complete response; NAC, neoadjuvant chemotherapy; GEO, Gene Expression Omnibus.
In Delta analysis, the top 10 hub genes were AKT1, TP53, SRC, FGFR1, PIK3R1, CTNNB1, PDGFRB, HSP90AA1, FGF2, and PTPN11, among which AKT1 showed the highest node degree (degree = 82) in Category 1 of GSE32603. In Category 1 of GSE87455, the top 10 hub genes were GRIA2, GRIA1, MSLN, APP, GRIP1, RAB11A, NDUFB5, MRPL28, PICK1, and ATP5F1, among which GRIA2 showed the highest node degree (degree = 11). In Category 2 of GSE32603 the top 10 hub genes were GAPDH, HSPA8, HSP90AA1, RPS27A, HSPA4, UBB, ENO1, EEF2, VCP, and ISG15, among which GAPDH showed the highest node degree (degree = 87). In GSE87455 the top 10 hub genes were ALB, TP53, INS, AKT1, FN1, MAPK1, APP, CASP3, VEGFA, and CTNNB1, among which ALB showed the highest node degree (degree = 76) (Table 4). Of note, HSP90AA1 in GSE32603 and APP in GSE87455 were genes common to Category 1 and 2. It was suggested that HSP90AA1 may have a role in BC recurrence of BC after pCR due to AC followed by OTC. On the other hand, APP may have a role in BC recurrence after pCR due to EDB. HSPB1 was the gene common to GSE32603 and GSE87455 in Category 2, suggesting that these mRNAs may play a significant role in BC patient survival after pCR. In summary, HSP90AA1 was identified by both analyses as a common gene for Category 1 and 2 and is suggested to strongly correlate with BC recurrence after NAC.
Functional and pathway enrichment analyses
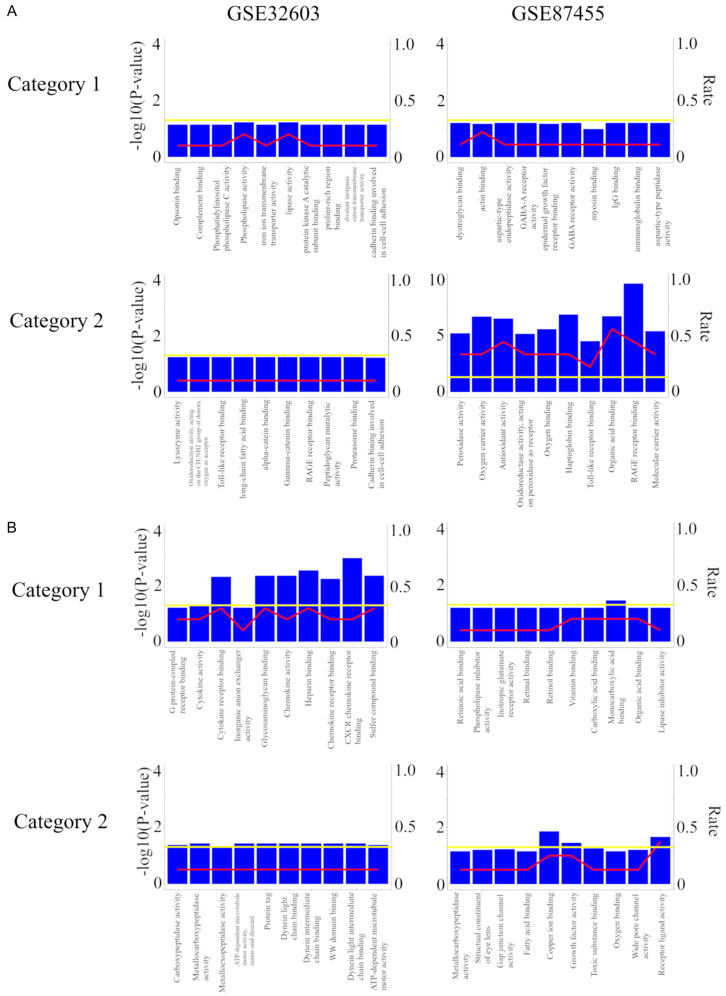
To further explore the systematic features and biological functions of the identified genes, GO functional annotation and KEGG pathway enrichment analyses were performed by R package, “clusterProfiler”. In Baseline analysis, the GO terms of the identified genes of the top 10 mRNAs with more changes are shown in Figure 3A. In Category 1, there was no significant pathway in the GSE32603 and GSE87455. In Category 2, there was no significant pathway in the GSE32603, but the GSE87455 shows RAGE receptor binding, Haptoglobin binding, Organic acid binding, Oxygen carrier activity, Antioxidant activity, Oxygen binding, Molecular carrier activity, Peroxidase activity, Oxidoreductase activity, acting on peroxide as receptor, and Toll-like receptor binding.
Figure 3.
GO annotation analysis for the target genes of the top 10 most downregulated miRNAs. (A) Baseline analysis and (B) delta analysis. Category 1, we divided BC patients into two groups with clinical integrity, clinical concordance versus discordance. Clinical concordance was defined as a group of patients who were pCR/not relapsed and non-pCR/relapsed, according to NAC. On the other hand, clinical discordance was defined as a group of patients who were pCR/relapsed and non-pCR/not relapsed, according to NAC. Category 2, we divided NAC treated BC patients according to whether they relapsed in the group having pCR. Two microarray data sets GSE32603 and GSE87455 were downloaded from the GEO database. The data were processed by unpaired t-test (P<0.05, |log2FC| >0.17). Abbreviations: GO, Gene Ontology. BC, breast cancer; pCR, pathological complete response; NAC, neoadjuvant chemotherapy; GEO, Gene Expression Omnibus.
In Delta analysis, the GO terms of the target genes of the top 10 mRNAs with more changes are shown in Figure 3B. In Category 1, several pathways (CXCR chemokine receptor binding, Heparin binding, Glycosaminoglycan binding, Sulfur compound binding, Chemokine activity, and Cytokine receptor binding in the GSE32603 and Menocarboxylic acid binding alone in the GSE87455) were significant, but no pathway was common in Category 2. In Category 2 GSE32603 demonstrated Protein tag, ATP-dependent microtubule motor activity, minus-end-directed, Dynein intermediate chain binding, Dynein light intermediate chain binding, Metallocarboxypeptidase activity, WW domain binding, ATP-dependent microtubule motor activity, and Carboxypeptidase activity. The GSE87455 showed Copper ion binding, Receptor ligand activity, and Growth factor activity. There was no significant pathway in KEGG pathway analysis in both categories of both cohorts. Thus, in both Baseline and Delta analyses, there was no common pathway between Category 1 and 2.
Some genes involved in recurrence after pCR to NAC were also useful as prognostic factors in primary BC
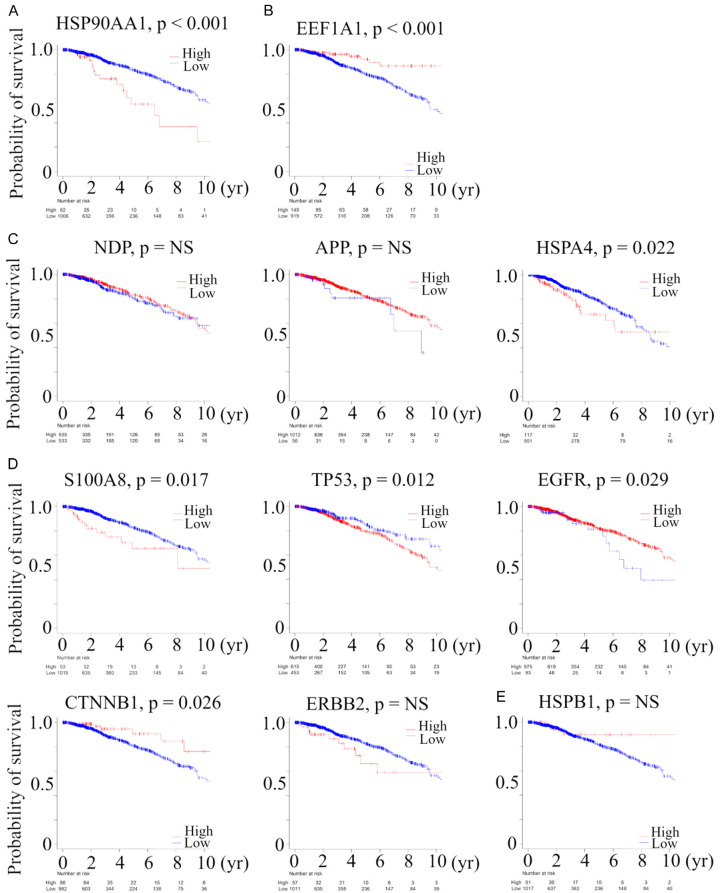
Next, we explored the prognostic relevance of genes extracted by the above analysis (Gene common to Category 1 and 2 in Baseline and Delta analysis: HSP90AA1, genes common to Category 1 and 2 in Baseline or Delta analysis: EEF1A1, NDP, APP, and HSPA4, genes identified only in Category 2 in Baseline or Delta analysis: S100A8, TP53, EGFR, CTNNB1, ERBB2, and HSPB1) utilizing a large BC cohort, TCGA (Figure 4). A total of 150 (14%) of 1068 BC patients in TCGA died, which were regarded as events when analyzing OS. Patients with a high expression of HSP90AA1 (P<0.001), HSPA4 (P = 0.022), S100A8 (P = 0.0017), and TP53 (P = 0.012) mRNA and patients with low expression of EEF1A1 (P<0.001), EGFR (P = 0.029), and CTNNB1 (P = 0.026) mRNA were significantly associated with worse OS, as evaluated by the Kaplan-Meier method and verified by the log-rank test. There was no statistically significant correlation between NDP, APP, ERBB2, and HSPB1 and OS in TCGA. These data suggest that some genes involved in recurrence after pCR due to NAC were also useful as prognostic factors in primary BC.
Figure 4.
Kaplan-Meier plots of the association of the presence of (A) gene common to Category 1 and 2 in baseline and delta analysis: HSP90AA1, (B) gene common to Category 1 and 2 in baseline analysis: EEF1A1, and (C) in delta analysis: NDP, APP, and HSPA4, (D) genes identified only in Category 2 in baseline: S100A8, TP53, EGFR, CTNNB1, and ERBB2 and (E) in delta analysis: HSPB1, with OS in TCGA BC cohort. The cutoff of each mRNA expression was defined as optimal cutoff to show the difference of OS. Category 1, we divided BC patients into two groups with clinical integrity, clinical concordance versus discordance. Clinical concordance was defined as a group of patients who were pCR/not relapsed and non-pCR/relapsed, according to NAC. On the other hand, clinical discordance was defined as a group of patients who were pCR/relapsed and non-pCR/not relapsed, according to NAC. Category 2, we divided NAC treated BC patients according to whether they relapsed in the group having pCR. Abbreviations: OS, overall survival; TCGA, The Cancer Genome Atlas; BC, breast cancer; NS, not significant.
Discussion
NAC is widely used to treat early-stage BC and provides an improvement in the breast conservation rate by tumor volume reduction and the early identification of sensitivity to treatment. While the achievement of pCR became the goal of NAC with the expectation of a concomitant improvement in patient survival [2], the predictive value of pCR for patient outcomes remains controversial due to variances according to the different biological subtypes [8-10] and because even after pCR after NAC cancer cells killed were not completely eradicated but instead merely lay dormant until BC recurrence [11]. We identified the genes and the pathways involved in the relationship between pCR and survival in BC cohorts by identifying differentially expressed mRNAs, by construction and analysis of the mRNA-hub gene network, and by functional and pathway enrichment analysis.
This study generated three interesting results with clinical implications. First, in Baseline analysis and Delta analysis, we identified genes involved in the relationship between pCR and survival. In mRNA expression profiling, NDP was a gene involved in recurrence after pCR, and it was related to clinical discordance and recurrence after pCR in Delta analysis (Table 2). NDP gene encodes a protein called Norrin that plays a role in Wnt signaling, one of the key signaling pathways for cell proliferation, adhesion, migration, and many other cellular activities including cancer stem cell biology [21]. We also found that S100A8 may behave differently in each NAC regimen since S100A8 related to no recurrence after pCR in GSE32603, but related to recurrence after pCR in GSE87455 in Baseline analysis (Table 1). S100A8 gene encodes S100 calcium-binding protein A8, which is involved in the regulation of several cellular processes such as cell cycle progression and differentiation. Yang et al reported that S100A8 was associated with BC drug resistance by proteomics/bioinformatics approach [22]. In the analysis of the mRNA hub gene network, HSP90AA1 was involved in recurrence after pCR due to EDB in baseline and delta analyses (Tables 3 and 4; Figures 1 and 2). HSP90AA1 (Heat Shock Protein 90 Alpha Family Class A Member 1) is a Protein Coding gene. HSP90 is required for the stabilization of many proteins in pathways that play key roles in BC growth and survival, such as estrogen receptor (ER), progesterone receptor (PR), essential components of HER2 signaling (HER2, AKT, c-SRC, RAF and HIF-1α), and EGFR [23]. Only in Baseline analysis, EEF1A1 was involved in recurrence after pCR to AC followed by OTC (Table 3 and Figure 1). EEF1A1 gene encodes an isoform of the alpha subunit of the eukaryotic elongation factor 1 (EEF1) complex, which is responsible for the enzymatic delivery of aminoacyl tRNAs to the ribosome [24]. Only in Delta analysis, APP and HSPA4 were involved in recurrence after pCR to EDB (Table 4 and Figure 2). High expression of APP (Amyloid-β precursor protein) mRNA is causally linked to tumorigenicity as well as the invasion of aggressive BC [25]. Cao and colleagues reported that HSPA4 indirectly promoted lymph node metastasis by targeting pathogenic IgG produced by B cells [26]. To our knowledge this is the first report that NDP, S100A8, HSP90AA1, EEF1A1, APP, and HSPA4 are involved in recurrence after pCR due to NAC. In addition, TP53, EGFR, ERBB2, CTNNB1, and HSPB1 may play a significant role in the survival of patients after pCR. Only in Category 2, TP53, EGFR, ERBB2, and CTNNB1 were recognized only in Baseline analysis (Table 3 and Figure 1) and HSPB1 was recognized only in Delta analysis (Table 4 and Figure 2). TP53, EGFR, and ERBB2 are genes that are deeply involved in BC development, biology, and BC treatment. A high level of expression of CTNNB1 (the gene that codes β-catenin) mRNA is a strong predictor for a favorable prognosis in gastric carcinoma, without any reported clinical role for CTNNB1 expression in BC [27]. HSPB1 (Heat Shock Protein Family B (Small) Member 1) upregulation is associated with tumor growth and resistance to chemo- and radio-therapeutic treatments. Interestingly, Gibert and colleagues demonstrated that HSPB1 silencing led to reduced cell migration and invasion in vitro and that in vivo it correlated with a decreased ability of BC cells to metastasize and grow in the skeleton [28].
Second, some genes involved in recurrence after pCR due to NAC were also useful as prognostic factors in primary BC. Patients with a high level of expression of HSP90AA1 (P<0.001), HSPA4 (P = 0.022), S100A8 (P = 0.0017), and TP53 (P = 0.012) mRNA and patients with low level of expression of EEF1A1 (P<0.001), EGFR (P = 0.029), and CTNNB1 (P = 0.026) mRNA were significantly associated with worse OS (Figure 4). High-level expression of HSP90AA1, one of two cytoplasmic HSP90 isoforms, was driven by chromosome coding region amplifications and was ab independent factors that led to death from BC among patients with TN and HER2-/ER+ subtypes, respectively [29]. High serum anti-HSPA4 IgG was correlated with high tumor HSPA4 expression and a poor prognosis for BC subjects [26]. Thus, HSP90AA1 and HSPA4 were compatible with previous reports. On the other hand, high EEF1A2 expression, one of two EEF1A isoforms, was a marker for good outcome in BC [30]. However, it has never been examined whether EEF1A1 expression has any prognostic value in BC.
Third, in functional and pathway enrichment analysis, we found multiple pathways involved in survival after pCR to NAC. In both Baseline and Delta analyses, there was no common pathway between Category 1 and 2, and some significant pathways in Category 2 were associated with survival after pCR to NAC. In Baseline analysis, in the GSE87455, RAGE receptor binding, Haptoglobin binding, Organic acid binding, Oxygen carrier activity, Antioxidant activity, Oxygen binding, Molecular carrier activity, Peroxidase activity, Oxidoreductase activity, acting on peroxide as receptor, and Toll-like receptor binding may be the pathways involved in clinical outcome after pCR (Figure 3A). Of note, Peroxidase activity and Oxidoreductase activity have been demonstrated to be associated with chemotherapy resistance in the course of cancer treatment [31,32]. In Delta analysis, in the GSE32603, Protein tag, ATP-dependent microtubule motor activity, minus-end-directed, Dynein intermediate chain binding, Dynein light intermediate chain binding, Metallocarboxypeptidase activity, WW domain binding, ATP-dependent microtubule motor activity, and Carboxypeptidase activity and in the GSE87455, Copper ion binding, Receptor ligand activity, and Growth factor activity may be the pathways involved in survival after pCR (Figure 3B). All these findings suggested that the functional pathways extracted by delta analysis have never been demonstrated to be associated with chemotherapy resistance in the course of cancer treatment and they are worth studying as new therapeutic targets.
Although the study demonstrates promising results, it has limitations. First, this is a retrospective study utilizing publicly available datasets (GSE32603, GSE87455, and TCGA), thus it is prone to selection bias. Second, this study does not include any in vitro or in vivo experiments that proves the mechanism of our results to further understand the correlations reported. Third, due to the small number of patients in NAC cohorts, we were unable to evaluate the data by subtype. It is known that achievement of pCR strongly predicted improved survival in TNBC and HER2-enriched BC subtypes, while data remain controversial for the luminal subtypes. Fourth, our dataset allowed us to evaluate only a single point of gene expression during NAC. Liquid biopsy, which is a non-invasively conducted genetic test using genes extracted from body fluids such as blood or urine, has been developed as a way of providing equivalent or better information obtained from genes in tumor tissue as previously demonstrated [33-37]. If transcriptomes can be monitored by liquid biopsy, it is expected to deepen the understanding of the relationship between drug efficacy and clinical outcomes in BC patients.
In conclusion, we demonstrated the genes and functional pathways involved in survival after pCR to NAC utilizing collected data from public BC cohorts with a bioinformatics approach. We found the genes involved in the relationship between pCR and survival utilizing Baseline and Delta analysis, some of which genes were also useful as prognostic factors in primary BC. Based on these reported results, we anticipate that further research can be conducted to establish a greater understanding of the relationship between the effect of NAC and survival.
Acknowledgements
This work was supported by NIH grant R01CA160688 to K.T, and NCI grant P30CA016056 involving the use of Roswell Park Cancer Institute’s Bioinformatics and Biostatistics Shared Resources.
Disclosure of conflict of interest
None.
Abbreviations
- BC
breast cancer
- NAC
neoadjuvant chemotherapy
- DFS
disease free survival
- OS
overall survival
- HR
hormone receptor
- pCR
pathological complete response
- TN
triple negative
- HER2
human epidermal growth factor receptor 2
- GEO
Gene Expression Omnibus
- AC
anthracycline based chemotherapy
- OTC
optimal taxane based chemotherapy
- EDB
epirubicin + docetaxel + bevacizumab
- FC
fold change
- GO
Gene Ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- TCGA
the cancer genome atlas
- NCI
National Cancer Institute
- ER
estrogen receptor
- PR
progesterone receptor
- EEF1
eukaryotic elongation factor 1
- APP
Amyloid-β precursor protein
- HSPB1
Heat Shock Protein Family B (Small) Member 1
Supporting Information
References
- 1.American Cancer Society. Cancer Facts & Figures 2019. Atlanta: American Cancer Society; 2019. [Google Scholar]
- 2.Wang-Lopez Q, Chalabi N, Abrial C, Radosevic-Robin N, Durando X, Mouret-Reynier MA, Benmammar KE, Kullab S, Bahadoor M, Chollet P, Penault-Llorca F, Nabholtz JM. Can pathologic complete response (pCR) be used as a surrogate marker of survival after neoadjuvant therapy for breast cancer? Crit Rev Oncol Hematol. 2015;95:88–104. doi: 10.1016/j.critrevonc.2015.02.011. [DOI] [PubMed] [Google Scholar]
- 3.von Minckwitz G, Blohmer JU, Costa SD, Denkert C, Eidtmann H, Eiermann W, Gerber B, Hanusch C, Hilfrich J, Huober J, Jackisch C, Kaufmann M, Kummel S, Paepke S, Schneeweiss A, Untch M, Zahm DM, Mehta K, Loibl S. Response-guided neoadjuvant chemotherapy for breast cancer. J. Clin. Oncol. 2013;31:3623–3630. doi: 10.1200/JCO.2012.45.0940. [DOI] [PubMed] [Google Scholar]
- 4.Prowell TM, Pazdur R. Pathological complete response and accelerated drug approval in early breast cancer. N Engl J Med. 2012;366:2438–2441. doi: 10.1056/NEJMp1205737. [DOI] [PubMed] [Google Scholar]
- 5.Huober J, von Minckwitz G, Denkert C, Tesch H, Weiss E, Zahm DM, Belau A, Khandan F, Hauschild M, Thomssen C, Hogel B, Darb-Esfahani S, Mehta K, Loibl S. Effect of neoadjuvant anthracycline-taxane-based chemotherapy in different biological breast cancer phenotypes: overall results from the GeparTrio study. Breast Cancer Res Treat. 2010;124:133–140. doi: 10.1007/s10549-010-1103-9. [DOI] [PubMed] [Google Scholar]
- 6.von Minckwitz G, Kümmel S, Vogel P, Hanusch C, Eidtmann H, Hilfrich J, Gerber B, Huober J, Costa SD, Jackisch C, Loibl S, Mehta K, Kaufmann M German Breast Group. Intensified neoadjuvant chemotherapy in early-responding breast cancer: phase III randomized GeparTrio study. J Natl Cancer Inst. 2008;100:552–562. doi: 10.1093/jnci/djn089. [DOI] [PubMed] [Google Scholar]
- 7.Untch M, Konecny GE, Paepke S, von Minckwitz G. Current and future role of neoadjuvant therapy for breast cancer. Breast. 2014;23:526–537. doi: 10.1016/j.breast.2014.06.004. [DOI] [PubMed] [Google Scholar]
- 8.von Minckwitz G, Untch M, Blohmer JU, Costa SD, Eidtmann H, Fasching PA, Gerber B, Eiermann W, Hilfrich J, Huober J, Jackisch C, Kaufmann M, Konecny GE, Denkert C, Nekljudova V, Mehta K, Loibl S. Definition and impact of pathologic complete response on prognosis after neoadjuvant chemotherapy in various intrinsic breast cancer subtypes. J. Clin. Oncol. 2012;30:1796–1804. doi: 10.1200/JCO.2011.38.8595. [DOI] [PubMed] [Google Scholar]
- 9.Houssami N, Macaskill P, von Minckwitz G, Marinovich ML, Mamounas E. Meta-analysis of the association of breast cancer subtype and pathologic complete response to neoadjuvant chemotherapy. Eur J Cancer. 2012;48:3342–3354. doi: 10.1016/j.ejca.2012.05.023. [DOI] [PubMed] [Google Scholar]
- 10.Cortazar P, Zhang L, Untch M, Mehta K, Costantino JP, Wolmark N, Bonnefoi H, Cameron D, Gianni L, Valagussa P, Swain SM, Prowell T, Loibl S, Wickerham DL, Bogaerts J, Baselga J, Perou C, Blumenthal G, Blohmer J, Mamounas EP, Bergh J, Semiglazov V, Justice R, Eidtmann H, Paik S, Piccart M, Sridhara R, Fasching PA, Slaets L, Tang S, Gerber B, Geyer CE Jr, Pazdur R, Ditsch N, Rastogi P, Eiermann W, von Minckwitz G. Pathological complete response and long-term clinical benefit in breast cancer: the CTNeoBC pooled analysis. Lancet. 2014;384:164–172. doi: 10.1016/S0140-6736(13)62422-8. [DOI] [PubMed] [Google Scholar]
- 11.Zhang XH, Giuliano M, Trivedi MV, Schiff R, Osborne CK. Metastasis dormancy in estrogen receptor-positive breast cancer. Clin Cancer Res. 2013;19:6389–6397. doi: 10.1158/1078-0432.CCR-13-0838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Magbanua MJ, Wolf DM, Yau C, Davis SE, Crothers J, Au A, Haqq CM, Livasy C, Rugo HS I-SPY 1 TRIAL Investigators. Esserman L, Park JW, van’t Veer LJ. Serial expression analysis of breast tumors during neoadjuvant chemotherapy reveals changes in cell cycle and immune pathways associated with recurrence and response. Breast Cancer Res. 2015;17:73. doi: 10.1186/s13058-015-0582-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Palazon A, Tyrakis PA, Macias D, Velica P, Rundqvist H, Fitzpatrick S, Vojnovic N, Phan AT, Loman N, Hedenfalk I, Hatschek T, Lovrot J, Foukakis T, Goldrath AW, Bergh J, Johnson RS. An HIF-1alpha/VEGF-A axis in cytotoxic T cells regulates tumor progression. Cancer Cell. 2017;32:669–683. e665. doi: 10.1016/j.ccell.2017.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lou W, Liu J, Ding B, Xu L, Fan W. Identification of chemoresistance-associated miRNAs in breast cancer. Cancer Manag Res. 2018;10:4747–4757. doi: 10.2147/CMAR.S172722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takeshita T, Yan L, Asaoka M, Rashid O, Takabe K. Late recurrence of breast cancer is associated with pro-cancerous immune microenvironment in the primary tumor. Sci Rep. 2019;9:16942. doi: 10.1038/s41598-019-53482-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takeshita T, Asaoka M, Katsuta E, Photiadis SJ, Narayanan S, Yan L, Takabe K. High expression of polo-like kinase 1 is associated with TP53 inactivation, DNA repair deficiency, and worse prognosis in ER positive Her2 negative breast cancer. Am J Transl Res. 2019;11:6507–6521. [PMC free article] [PubMed] [Google Scholar]
- 20.Pham VV, Zhang J, Liu L, Truong B, Xu T, Nguyen TT, Li J, Le TD. Identifying miRNA-mRNA regulatory relationships in breast cancer with invariant causal prediction. BMC Bioinformatics. 2019;20:143. doi: 10.1186/s12859-019-2668-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mohammed MK, Shao C, Wang J, Wei Q, Wang X, Collier Z, Tang S, Liu H, Zhang F, Huang J, Guo D, Lu M, Liu F, Liu J, Ma C, Shi LL, Athiviraham A, He TC, Lee MJ. Wnt/beta-catenin signaling plays an ever-expanding role in stem cell self-renewal, tumorigenesis and cancer chemoresistance. Genes Dis. 2016;3:11–40. doi: 10.1016/j.gendis.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang WS, Moon HG, Kim HS, Choi EJ, Yu MH, Noh DY, Lee C. Proteomic approach reveals FKBP4 and S100A9 as potential prediction markers of therapeutic response to neoadjuvant chemotherapy in patients with breast cancer. J Proteome Res. 2012;11:1078–1088. doi: 10.1021/pr2008187. [DOI] [PubMed] [Google Scholar]
- 23.Zhang H, Burrows F. Targeting multiple signal transduction pathways through inhibition of Hsp90. J Mol Med (Berl) 2004;82:488–499. doi: 10.1007/s00109-004-0549-9. [DOI] [PubMed] [Google Scholar]
- 24.Thornton S, Anand N, Purcell D, Lee J. Not just for housekeeping: protein initiation and elongation factors in cell growth and tumorigenesis. J Mol Med (Berl) 2003;81:536–548. doi: 10.1007/s00109-003-0461-8. [DOI] [PubMed] [Google Scholar]
- 25.Lim S, Yoo BK, Kim HS, Gilmore HL, Lee Y, Lee HP, Kim SJ, Letterio J, Lee HG. Amyloid-beta precursor protein promotes cell proliferation and motility of advanced breast cancer. BMC Cancer. 2014;14:928. doi: 10.1186/1471-2407-14-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gu Y, Liu Y, Fu L, Zhai L, Zhu J, Han Y, Jiang Y, Zhang Y, Zhang P, Jiang Z, Zhang X, Cao X. Tumor-educated B cells selectively promote breast cancer lymph node metastasis by HSPA4-targeting IgG. Nat Med. 2019;25:312–322. doi: 10.1038/s41591-018-0309-y. [DOI] [PubMed] [Google Scholar]
- 27.Rosa M, Han HS, Ismail-Khan R, Allam-Nandyala P, Bui MM. Beta-catenin expression patterns in matched pre- and post-neoadjuvant chemotherapy-resistant breast cancer. Ann Clin Lab Sci. 2015;45:10–16. [PubMed] [Google Scholar]
- 28.Gibert B, Eckel B, Gonin V, Goldschneider D, Fombonne J, Deux B, Mehlen P, Arrigo AP, Clezardin P, Diaz-Latoud C. Targeting heat shock protein 27 (HspB1) interferes with bone metastasis and tumour formation in vivo. Br J Cancer. 2012;107:63–70. doi: 10.1038/bjc.2012.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng Q, Chang JT, Geradts J, Neckers LM, Haystead T, Spector NL, Lyerly HK. Amplification and high-level expression of heat shock protein 90 marks aggressive phenotypes of human epidermal growth factor receptor 2 negative breast cancer. Breast Cancer Res. 2012;14:R62. doi: 10.1186/bcr3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kulkarni G, Turbin DA, Amiri A, Jeganathan S, Andrade-Navarro MA, Wu TD, Huntsman DG, Lee JM. Expression of protein elongation factor eEF1A2 predicts favorable outcome in breast cancer. Breast Cancer Res Treat. 2007;102:31–41. doi: 10.1007/s10549-006-9315-8. [DOI] [PubMed] [Google Scholar]
- 31.Jardim BV, Moschetta MG, Leonel C, Gelaleti GB, Regiani VR, Ferreira LC, Lopes JR, Zuccari DA. Glutathione and glutathione peroxidase expression in breast cancer: an immunohistochemical and molecular study. Oncol Rep. 2013;30:1119–1128. doi: 10.3892/or.2013.2540. [DOI] [PubMed] [Google Scholar]
- 32.Korde LA, Lusa L, McShane L, Lebowitz PF, Lukes L, Camphausen K, Parker JS, Swain SM, Hunter K, Zujewski JA. Gene expression pathway analysis to predict response to neoadjuvant docetaxel and capecitabine for breast cancer. Breast Cancer Res Treat. 2010;119:685–699. doi: 10.1007/s10549-009-0651-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takeshita T, Yamamoto Y, Yamamoto-Ibusuki M, Inao T, Sueta A, Fujiwara S, Omoto Y, Iwase H. Prognostic role of PIK3CA mutations of cell-free DNA in early-stage triple negative breast cancer. Cancer Sci. 2015;106:1582–9. doi: 10.1111/cas.12813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takeshita T, Yamamoto Y, Yamamoto-Ibusuki M, Inao T, Sueta A, Fujiwara S, Omoto Y, Iwase H. Clinical significance of monitoring ESR1 mutations in circulating cell-free DNA in estrogen receptor positive breast cancer patients. Oncotarget. 2016;7:32504–32518. doi: 10.18632/oncotarget.8839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takeshita T, Yamamoto Y, Yamamoto-Ibusuki M, Tomiguchi M, Sueta A, Murakami K, Omoto Y, Iwase H. Analysis of ESR1 and PIK3CA mutations in plasma cell-free DNA from ER-positive breast cancer patients. Oncotarget. 2017;8:52142–52155. doi: 10.18632/oncotarget.18479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takeshita T, Yamamoto Y, Yamamoto-Ibusuki M, Tomiguchi M, Sueta A, Murakami K, Iwase H. Clinical significance of plasma cell-free DNA mutations in PIK3CA, AKT1, and ESR1 gene according to treatment lines in ER-positive breast cancer. Mol Cancer. 2018;17:67. doi: 10.1186/s12943-018-0808-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takeshita T, Iwase H. dPCR mutational analyses in cell-free DNA: a comparison with tissues. Methods Mol Biol. 2019;1909:105–118. doi: 10.1007/978-1-4939-8973-7_8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.