Abstract
N6-Methyladenosine (m6A) is the most common RNA modification in eukaryotic mRNAs and growing evidence suggests the crucial roles of m6A and its regulators in human tumorigenesis. Recent studies have shown that the m6A regulators promote tumorigenesis of various types of cancer. However, the underlying molecular mechanisms of m6A regulators in breast cancer remain largely unknown. We therefore assessed the genetic alterations, expression and prognostic role of m6A regulators in breast cancer using openly available data from The Cancer Genome Atlas (TCGA). Analysis of TCGA data revealed that m6A regulators including KIAA1429, YTHDF1, and YTHDF3 were upregulated in breast cancer tissues, and the expression level significantly correlated with intrinsic subclasses and nodal metastasis. Importantly, we found for the first time that YTHDF1 and YTHDF3 were frequently amplified which contribute to the overexpression of YTHDF1 and YTHDF3 transcripts, thereby promoting breast cancer progression. Moreover, overexpression of YTHDF1 and YTHDF3 were associated with poor prognosis of breast cancer patients. Therefore, YTHDF1 and YTHDF3 serve a crucial role in the pathogenesis of breast cancer, which are potentially useful for prognosis stratification and therapeutic target for breast cancer.
Keywords: Breast cancer, m6A readers aberrations, prognostic signature, survival analysis
Introduction
Breast cancer is the second most common form of cancer afflicting women world-wide. According to the prediction by the American Cancer Society (ACS), breast cancer was responsible for about 41,760 cancer-related deaths in the United States in 2019 [1]. Despite remarkable advancements in the therapeutic modalities during recent years, the prognosis of breast cancer still remains poor. Therefore, further studies are required to determine the potential mechanism of oncogenesis and progression of breast cancer.
N6-methyladenosine (m6A) modification is one of the most common modification in eukaryotic mRNA that influences mRNA splicing, export, localization, translation, decay, and stability [2-5]. m6A RNA modification is reversible and catalyzed by a recently discovered multicomponent protein complex, including methyltransferases (METTL3, METTL14, and KIAA1429), demethylases (FTO and ALKBH5), and m6A binding proteins (YTHDF1-3, YTHDC1 and YTHDC2) [2-5]. A growing body of evidence suggested that m6A modification is involved in diverse biological processes including tissue development, embryonic stem cell self-renewal, and fate determination [2,6]. Recent studies have shown that dysregulation of m6A modification is associated with cancer progression [2,6].
YTH N6-methyladenosine RNA binding protein 1 (YTHDF1) is a member of the YTH domain-containing proteins including YTHDF1-3, YTHDC1 and YTHDC2. Many studies recently, demonstrated that YTHDF1 function as “reader” of m6A-modified mRNAs and promotes translation in the cytosol [2,5]. However, the relationship between YTHDF1 and different cancers are largely unknown, the more recent studies reported that YTHDF1 is closely related with initiation and progression of ovarian [2], HCC [7], and colon [8] cancers.
Therefore, in the present study, we first compared the transcriptional levels of m6A regulators in breast cancer and adjacent breast tissues using UALCAN database. In addition, the cBioPortal database was used to analyze the genetic alterations of YTHDF1 and YTHDF3, and the correlation with transcriptional levels. Furthermore, the Kaplan-Meier plotter database was used to assess the prognostic effects of YTHDF1 and YTHDF3 mRNAs expression in patients with breast cancer. Overall, our results demonstrated that m6A regulators are the crucial participants in the malignant progression of breast cancer, which are potentially useful for prognosis stratification and therapeutic target for breast cancer.
Materials and methods
Gene expression and survival analysis
In the current study, UALCAN (http://ualcan.path.uab.edu/) [9] was applied to analyze the transcriptional levels of YTHDF1 in primary breast cancer tissues and their association with intrinsic subclasses as well as to analyze the patient survival. In addition, we used the CCLE dataset (https://www.broadinstitute.org/ccle) [10] to analyze the YTHDF1 transcript level in cancer cell lines. The prognostic value of YTHDF1 in breast cancer patients were analyzed using Kaplan-Meier plotter (http://kmplot.com/analysis/).
Analysis of YTHDF1 alterations in breast cancer samples
The cBioPortal (www.cbioportal.org/) TCGA dataset is a user-friendly, interactive website resource and provides visualization, analysis, and download of large-scale cancer genomics datasets [11,12]. In the present study, the c-Bio-Portal was utilized to analyze YTHDF1 alterations in breast cancer samples.
Correlation analysis
Pearson correlation analysis was employed using the GEPIA dataset to reveal the association among different m6A RNA methylation regulators and their target genes.
In silico functional analysis
To evaluate the potential functional impact of missense mutations in YTHDF1 gene, we utilised a variety of pathogenicity prediction programs, including SIFT (http://sift.jcvi.org/), and PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2//).
Results
YTHDF1 is up-regulated in the breast cancer tissue and cells
UALCAN database analysis revealed that YTHDF1 mRNA expression of breast cancer significantly increased compared to the normal tissues (P=1.62e-12) (Figure 1A). In additions, YTHDF1 mRNA level also was increased in breast cancer cells (Figure 1B). The mRNA of YTHDF1 expression was significantly correlated with intrinsic subclasses of breast cancer (Figure 1C). The high expression of YTHDF1 was also crucial for breast cancer metastasis (Figure 1D).
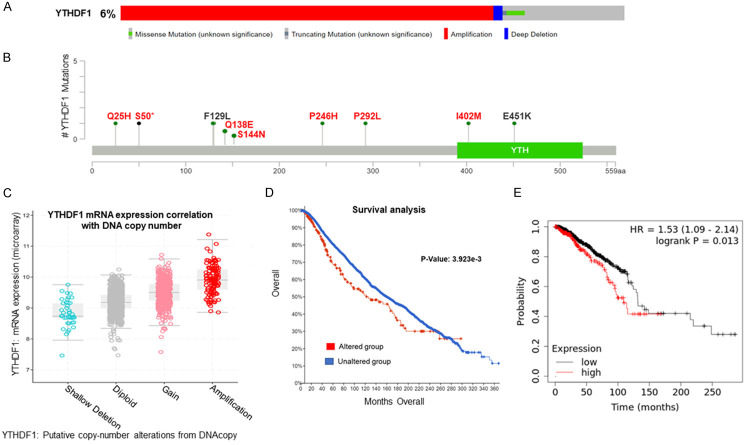
Figure 1.
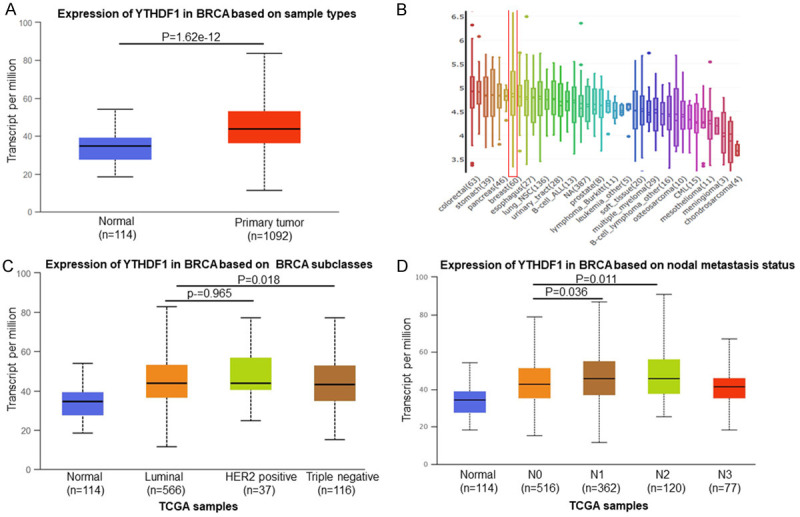
YTHDF1 was overexpressed in breast cancer. Expression of YTHDF1 significantly increased in breast cancer tissues compared with adjacent normal tissues (P=1.62e-12) (A) and breast cancer cells (B). The increased expression of YTHDF1 correlated with intrinsic subclasses (C) and nodal metastasis (D).
Genetic alterations of YTHDF1 is associated with poor prognosis in breast cancer patients
We analysed the genetic alterations and expression levels of YTHDF1 by using the cBioPortal TCGA dataset. Importantly, this gene displayed different mutations and expression patterns, among which YTHDF1 gene was most frequently amplified (Figure 2A). Interestingly, we also observed that many YTHDF1 mutations were novel (Figure 2B) may have strong functional consequences as predicted by in silico functional analysis (Table 1). Further, the copy number status of YTHDF1 positively correlated with its mRNA expression in breast cancer patients (Figure 2C). Moreover, breast cancer patients with YTHDF1 alterations have worse overall survival than breast cancer patients without YTHDF1 alterations (P=3.923e-3), (Figure 2D). In addition, we found that the overexpression of YTHDF1 correlated with the poor prognosis of breast cancer patients (P=0.013) (Figure 2E).
Figure 2.
Correlation between the genetic alterations of YTHDF1 and mRNA level in breast cancer tissues. Oncoprint in cBioPortal database exhibited the proportion and distribution of specimens with genetic alterations in YTHDF1 (A). Schematic diagram of the YTHDF1 protein and the positions of mutations (red letters are novel mutations, that were not presented in gnomAD) (B). Copy gain (gain and amplification) of YTHDF1 was associated with notably increased YTHDF1 mRNA levels compared with the copy-neutral (diploid) and copy-loss (shallow deletion) cases (C). Survival analysis of breast cancer patients with and without YTHDF1 gene alteration (D). The prognostic value of mRNA level of YTHDF1 in breast cancer patients, analyzed by Kaplan-Meier plotter (P=0.013) (E).
Table 1.
Summary of YTHDF1 mutations
| Mutation | SIFT | Polyphen-2 | gnomAD |
|---|---|---|---|
| Q25H | Deleterious | Probably Damaging | No |
| S50* | NA | NA | No |
| F129L | Deleterious | Probably Damaging | 1.732E-05 |
| Q138E | Deleterious | Probably Damaging | No |
| S144N | Tolerated | Probably Damaging | No |
| P246H | Deleterious | Probably Damaging | No |
| P292L | Deleterious | Benign | No |
| I402M | Deleterious | Probably Damaging | No |
| E451K | Tolerated | Probably Damaging | 7.074E-06 |
Abbreviations are follows: gnomAD, The Genome Aggregation database; NA, Not Applicable.
Truncating mutation.
Genetic alterations of YTHDF3 is associated with poor prognosis in breast cancer patients
We also analysed the genetic alterations and expression levels of YTHDF3 by using the cBioPortal TCGA dataset. Surprisingly, YTHDF3 gene was most frequently amplified (14%) (Figure 3A). Further amplification of YTHDF3 positively correlated with its mRNA expression in breast cancer patients (Figure 3B). Moreover, breast cancer patients with YTHDF3 alterations have worse overall survival than breast cancer patients without YTHDF3 alterations (P=0.0165) (Figure 3C). In addition, we found that the overexpression of YTHDF3 correlated with the poor prognosis of breast cancer patients (P=1.5e-11) (Figure 3D).
Figure 3.
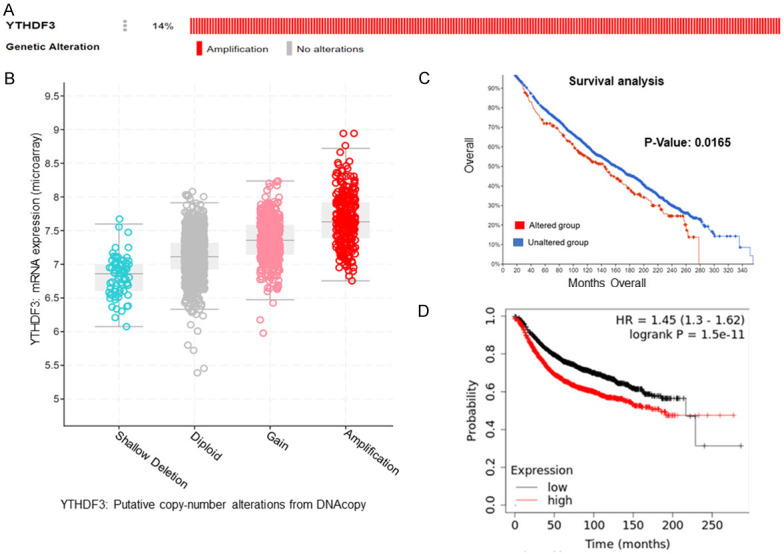
Correlation between the genetic alterations of YTHDF3 and mRNA level in breast cancer tissues. Oncoprint in cBioPortal database exhibited YTHDF3 amplification in breast cancer (A). Correlation between YTHDF3 amplification and its mRNA level (B). Survival analysis of breast cancer patients with and without YTHDF3 gene alteration (C). The prognostic value of mRNA level of YTHDF3 in breast cancer patients, analyzed by Kaplan-Meier plotter (P=1.5e-11) (D).
The m6A regulators target genes expression in breast cancer
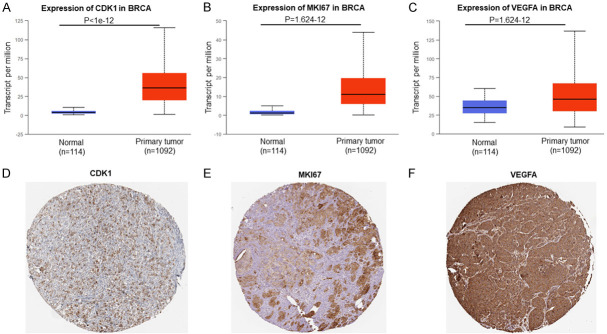
UALCAN database and the Human Protein Atlas (https://www.proteinatlas.org/) analysis revealed that m6A regulators target mRNAs (CDK1, P<1e-12; MKI67, P=1.624e-12; VEGFA, P=1.624e-12) (Figure 4A-C) and their proteins (Figure 4D-F) were highly expressed in breast cancer tissues.
Figure 4.
The m6A regulators target genes expression in breast cancer. CDK1 (A), MKI67 (B) and VEGFA (C) mRNAs expression were analysed using UALCAN database. CDK1 (D), MKI67 (E) and VEGFA (F) proteins expression were analysed in breast cancer tissue using the Human Protein Atlas (https://www.proteinatlas.org/).
Correlation of m6A regulators in breast cancer
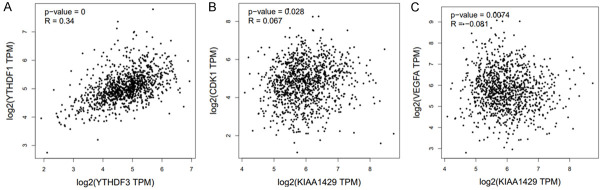
We analyzed the association among m6A regulators and their target genes using the GEPIA dataset. We found that the YTHDF1 positively corrected with YTHDF3 (R=0.34, P<0.05) (Figure 5A). Surprisingly, highly expressed KIAA1429 was positively corrected with CDK1 (R=0.067, P<0.05) (Figure 5B), and VEGFA (R=0.081, P<0.05) (Figure 5C).
Figure 5.
Correlation between the expression of YTHDF1 and YTHDF3 (A). Correlation between the expression of KIAA1429 and cancer associated genes CDK1 (B) and VEGFA (C).
Discussion
Emerging evidence support that dysregulation of m6A methylation is tightly associated with different cancers development. Recently, the results of multiple studies have shown that m6A regulators such as methyltransferases (METTL3, METTL14 and KIAA1429) and demethylases (FTO and ALKBH5) play important roles in various cancers including lung cancer, breast cancer, acute myeloid leukemia, endometrial cancer, and glioma [13-17]. For example, overexpression of METTL3 and ALKBH5 were associated with poor survival time of breast cancer and glioma [18,19].
Previous studies have reported that YTHDF1 was up-regulated in ovarian cancer [2], hepatocellular carcinoma [7], colorectal cancer [8], and lung cancer [13]. The up-regulated YTHDF1 correlated with the poor prognosis of ovarian cancer patients [2] and hepatocellular carcinoma [7]. In the present study, we found that YTHDF1 mRNA was up-regulated in breast cancer compared to the normal tissues and up-regulated YTHDF1 correlated with the poor prognosis of breast cancer patients. Liu et al. recently reported that YTHDF1 was crucial for proliferation and metastasis of ovarian cancer using in vitro and in vivo studies [2]. Our results also showed that overexpression of YTHDF1 significantly correlated with intrinsic subclasses and nodal metastasis of human breast cancer. Therefore, our findings suggest that high expressions of YTHDF1 mRNA remarkably correlated with the poor prognosis of breast cancer patients, which might be identified as promising biomarkers to predict the survival of breast cancer patients.
Gene copy number alterations are an important cause that drives aberrant up-regulation of oncogenes in cancer [20,21]. A recent study has shown that YTHDF1 copy number gain was a major mechanism that contributes to high expression of YTHDF1 in human colorectal carcinoma [6]. In addition, Liu et al. more recently reported that YTHDF1 gene was frequently amplified and increased in high-grade serous ovarian cancer [2], suggesting that YTHDF1 might be an important oncogene, which is selected during cancer evolution. Our finding is consistent with these reports that YTHDF1 gene is frequently amplified and contributes to high expression of YTHDF1 in human breast cancer. Surprisingly, YTHDF3 is also frequently amplified (14%) and contributes to high expression of YTHDF3 in human breast cancer. Importantly, we also found that breast cancer patients with YTHDF1 and YTHDF3 alterations have worse overall survival than breast cancer patients without YTHDF1 and YTHDF3 alterations. Recent studies have demonstrated that YTHDF1 and YTHDF3 enhance the translation efficiency of m6A-modified mRNAs [22,23]. Our findings demonstrated that YTHDF1 and YTHDF3 mRNAs were up-regulated in breast cancer, further YTHDF1 was positively correlated with YTHDF3. Therefore, YTHDF1 and YTHDF3 amplifications, and their transcripts upregulation may promote the translation of oncogene targets in m6A-dependent manner that results in breast cancer development.
Multiple lines of evidence suggested that METTL3 and KIAA1429 were increased m6A level of cancer associated genes including CDK1, MKI67 and VEGFA [24-26]. Qian et al. recently reported that KIAA1429 played oncogenic role in breast cancer by regulating cyclin-dependent kinase 1 (CDK1) in m6A-independent manner [24]. Moreover, KIAA1429 also played a critical role in liver cancer development by regulating GATA3 in m6A-dependent manner [27]. In the present study, we found that KIAA1429, CDK1, MKI67, and VEGFA were highly expressed in breast cancer, further KIAA1429 was positively corrected with CDK1, and VEGFA. Therefore, we speculated that KIAA1429 plays an oncogenic role in breast cancer by regulating CDK1, MKI67, and VEGFA in m6A-dependent or independent manner.
The studies showed that YTHDF1 and YTHDF3 promotes protein synthesis by interacting with the translation machinery. Moreover, YTHDF1 regulated oncogenes translation by recruiting the initiation factor EIF3 [28] and elongation factor eEF-2 [29] via an m6A-dependent manner. Liu et al. recently reported that YTHDF1 facilitates the translation of EIF3C in an m6A-dependent manner and concomitantly enhanced the overall translational output, thereby promoting tumorigenesis and metastasis of ovarian cancer [2]. In addition, EIF3C was associated with malignant behavior of human breast cancer [30], we also observed that mRNA (UALCAN; P=9.765e-08) and protein (The Human Protein Atlas; https://www.proteinatlas.org/) of EIF3C was up-regulated in breast cancer. Therefore, our findings suggest that up-regulated YTHDF1 might enhance the translation of EIF3C in an m6A-dependent manner and promotes translation of oncogenic transcripts, thus facilitating oncogenesis and metastasis of breast cancer.
Most human cancers, including breast cancer contain many different types of mutations and majority of these are missense mutations whi-ch can dramatically increase the risk of developing certain cancers. BRCA1 and BRCA2 genes are the most frequently mutated genes that are associated with high breast cancer risk [31]. A recent study found that point mutations spread over the coding sequence of the BRCA1 and BRCA2 were attributed to an increased risk of breast cancer [31]. Interestingly, we found several novel point mutations in YTHDF1 gene. The following lines of evidence support the pathogenic effects of these YTHDF1 gene mutation including novel mutations: 1) conservation data suggests these mutations were highly conserved amino acid residues of YTHDF1 protein; 2) all novel mutations were absent in gnomAD; 3) in silico functional analyses predict the mutations to be pathogenic with high probability scores. Therefore, our findings suggest that point mutations in YTHDF1 gene can vastly increase the risk of developing breast cancer.
In conclusion, the recent studies reported that YTHDF1 copy number gain was most common genetic alterations and correlated with high expression of YTHDF1 which promoted development of cancers by regulating cancer-related gene expression. Importantly, we reported for the first time that YTHDF1 and YTHDF3 copy number gains were strikingly higher than other cancer which contributes to the overexpression of YTHDF1 and YTHDF3, thereby promoting breast cancer progression. Therefore, YTHDF1 and YTHDF3 could be a promising prognostic biomarker and potential therapeutic target for breast cancer.
Acknowledgements
This work was supported by the Science and Engineering Research Board (SERB), Government of India (CRG/2019/003756 and EEQ/2019/000411).
Disclosure of conflict of interest
None.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Liu T, Wei Q, Jin J, Luo Q, Liu Y, Yang Y, Cheng C, Li L, Pi J, Si Y, Xiao H, Li L, Rao S, Wang F, Yu J, Yu J, Zou D, Yi P. The m6A reader YTHDF1 promotes ovarian cancer progression via augmenting EIF3C translation. Nucleic Acids Res. 2020;48:3816–3831. doi: 10.1093/nar/gkaa048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paramasivam A, Priyadharsini JV, Raghunandhakumar S. Implications of m6A modification in autoimmune disorders. Cell Mol Immunol. 2019;17:550–551. doi: 10.1038/s41423-019-0307-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paramasivam A, Vijayashree Priyadharsini J. Novel insights into m6A modification in circular RNA and implications for immunity. Cell Mol Immunol. 2020;17:668–669. doi: 10.1038/s41423-020-0387-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paramasivam A, Vijayashree Priyadharsini J, Raghunandhakumar S. N6-adenosine methylation (m6A): a promising new molecular target in hypertension and cardiovascular diseases. Hypertens Res. 2019;43:153–154. doi: 10.1038/s41440-019-0338-z. [DOI] [PubMed] [Google Scholar]
- 6.Bai Y, Yang C, Wu R, Huang L, Song S, Li W, Yan P, Lin C, Li D, Zhang Y. YTHDF1 regulates tumorigenicity and cancer stem cell-like activity in human colorectal carcinoma. Front Oncol. 2019;9:332. doi: 10.3389/fonc.2019.00332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao X, Chen Y, Mao Q, Jiang X, Jiang W, Chen J, Xu W, Zhong L, Sun X. Overexpression of YTHDF1 is associated with poor prognosis in patients with hepatocellular carcinoma. Cancer Biomark. 2018;21:859–868. doi: 10.3233/CBM-170791. [DOI] [PubMed] [Google Scholar]
- 8.Nishizawa Y, Konno M, Asai A, Koseki J, Kawamoto K, Miyoshi N, Takahashi H, Nishida N, Haraguchi N, Sakai D, Kudo T, Hata T, Matsuda C, Mizushima T, Satoh T, Doki Y, Mori M, Ishii H. Oncogene c-Myc promotes epitranscriptome m(6)A reader YTHDF1 expression in colorectal cancer. Oncotarget. 2018;9:7476–7486. doi: 10.18632/oncotarget.23554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK, Varambally S. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19:649–58. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehár J, Kryukov GV, Sonkin D, Reddy A, Liu M, Murray L, Berger MF, Monahan JE, Morais P, Meltzer J, Korejwa A, Jané-Valbuena J, Mapa FA, Thibault J, Bric-Furlong E, Raman P, Shipway A, Engels IH, Cheng J, Yu GK, Yu J, Aspesi P Jr, de Silva M, Jagtap K, Jones MD, Wang L, Hatton C, Palescandolo E, Gupta S, Mahan S, Sougnez C, Onofrio RC, Liefeld T, MacConaill L, Winckler W, Reich M, Li N, Mesirov JP, Gabriel SB, Getz G, Ardlie K, Chan V, Myer VE, Weber BL, Porter J, Warmuth M, Finan P, Harris JL, Meyerson M, Golub TR, Morrissey MP, Sellers WR, Schlegel R, Garraway LA. The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:l1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi Y, Fan S, Wu M, Zuo Z, Li X, Jiang L, Shen Q, Xu P, Zeng L, Zhou Y, Huang Y, Yang Z, Zhou J, Gao J, Zhou H, Xu S, Ji H, Shi P, Wu DD, Yang C, Chen Y. YTHDF1 links hypoxia adaptation and non-small cell lung cancer progression. Nat Commun. 2019;10:4892. doi: 10.1038/s41467-019-12801-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yi D, Wang R, Shi X, Xu L, Yilihamu Y, Sang J. METTL14 promotes the migration and invasion of breast cancer cells by modulating N6-methyladenosine and hsa-miR-146a-5p expression. Oncol Rep. 2020;43:1375–1386. doi: 10.3892/or.2020.7515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu L, Wu D, Ning J, Liu W, Zhang D. Changes of N6-methyladenosine modulators promote breast cancer progression. BMC Cancer. 2019;19:326. doi: 10.1186/s12885-019-5538-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Niu Y, Lin Z, Wan A, Chen H, Liang H, Sun L, Wang Y, Li X, Xiong XF, Wei B, Wu X, Wan G. RNA N6-methyladenosine demethylase FTO promotes breast tumor progression through inhibiting BNIP3. Mol Cancer. 2019;18:46. doi: 10.1186/s12943-019-1004-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu L, Liu X, Dong Z, Li J, Yu Y, Chen X, Ren F, Cui G, Sun R. N6-methyladenosine-related genomic targets are altered in breast cancer tissue and associated with poor survival. J Cancer. 2019;10:5447–5459. doi: 10.7150/jca.35053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang H, Xu B, Shi J. N6-methyladenosine METTL3 promotes the breast cancer progression via targeting Bcl-2. Gene. 2020;722:144076. doi: 10.1016/j.gene.2019.144076. [DOI] [PubMed] [Google Scholar]
- 19.Zhang S, Zhao BS, Zhou A, Lin K, Zheng S, Lu Z, Chen Y, Sulman EP, Xie K, Bögler O, Majumder S, He C, Huang S. m(6)A demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell. 2017;31:591–606. e596. doi: 10.1016/j.ccell.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carvalho B, Postma C, Mongera S, Hopmans E, Diskin S, van de Wiel MA, van Criekinge W, Thas O, Matthäi A, Cuesta MA, Terhaar Sive Droste JS, Craanen M, Schröck E, Ylstra B, Meijer GA. Multiple putative oncogenes at the chromosome 20q amplicon contribute to colorectal adenoma to carcinoma progression. Gut. 2009;58:79–89. doi: 10.1136/gut.2007.143065. [DOI] [PubMed] [Google Scholar]
- 21.Camps J, Nguyen QT, Padilla-Nash HM, Knutsen T, McNeil NE, Wangsa D, Hummon AB, Grade M, Ried T, Difilippantonio MJ. Integrative genomics reveals mechanisms of copy number alterations responsible for transcriptional deregulation in colorectal cancer. Genes Chromosomes Cancer. 2009;48:1002–17. doi: 10.1002/gcc.20699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li A, Chen YS, Ping XL, Yang X, Xiao W, Yang Y, Sun HY, Zhu Q, Baidya P, Wang X, Bhattarai DP, Zhao YL, Sun BF, Yang YG. Cytoplasmic m6A reader YTHDF3 promotes mRNA translation. Cell Res. 2017;27:444–447. doi: 10.1038/cr.2017.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jin H, Ying X, Que B, Wang X, Chao Y, Zhang H, Yuan Z, Qi D, Lin S, Min W, Yang M, Ji W. N6-methyladenosine modification of ITGA6 mRNA promotes the development and progression of bladder cancer. EBioMedicine. 2019;47:195–207. doi: 10.1016/j.ebiom.2019.07.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qian JY, Gao J, Sun X, Cao MD, Shi L, Xia TS, Zhou WB, Wang S, Ding Q, Wei JF. KIAA1429 acts as an oncogenic factor in breast cancer by regulating CDK1 in an N6-methyladenosine-independent manner. Oncogene. 2019;38:6123–6141. doi: 10.1038/s41388-019-0861-z. [DOI] [PubMed] [Google Scholar]
- 25.Zhang S, Zhao BS, Zhou A, Lin K, Zheng S, Lu Z, Chen Y, Sulman EP, Xie K, Bögler O, Majumder S, He C, Huang S. m 6 A demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell. 2017;31:591–606. e6. doi: 10.1016/j.ccell.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tian C, Huang Y, Li Q, Feng Z, Xu Q. Mettl3 regulates osteogenic differentiation and alternative splicing of vegfa in bone marrow mesenchymal stem cells. Int J Mol Sci. 2019;20:551. doi: 10.3390/ijms20030551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lan T, Li H, Zhang D, Xu L, Liu H, Hao X, Yan X, Liao H, Chen X, Xie K, Li J, Liao M, Huang J, Yuan K, Zeng Y, Wu H. KIAA1429 contributes to liver cancer progression through N6-methyladenosine-dependent post-transcriptional modification of GATA3. Mol Cancer. 2019;18:186. doi: 10.1186/s12943-019-1106-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, Weng X, Chen K, Shi H, He C. N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell. 2015;161:1388–1399. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin X, Chai G, Wu Y, Li J, Chen F, Liu J, Luo G, Tauler J, Du J, Lin S, He C, Wang H. RNA m(6)A methylation regulates the epithelial mesenchymal transition of cancer cells and translation of Snail. Nat Commun. 2019;10:2065. doi: 10.1038/s41467-019-09865-9. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.Zhao W, Li X, Wang J, Wang C, Jia Y, Yuan S, Huang Y, Shi Y, Tong Z. Decreasing eukaryotic initiation factor 3C (EIF3C) suppresses proliferation and stimulates apoptosis in breast cancer cell lines through mammalian target of rapamycin (mTOR) pathway. Med Sci Monit. 2017;23:4182–4191. doi: 10.12659/MSM.906389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lima ZS, Ghadamzadeh M, Arashloo FT, Amjad G, Ebadi MR, Younesi L. Recent advances of therapeutic targets based on the molecular signature in breast cancer: genetic mutations and implications for current treatment paradigms. J Hematol Oncol. 2019;12:38. doi: 10.1186/s13045-019-0725-6. [DOI] [PMC free article] [PubMed] [Google Scholar]