Abstract
The high mortality and poor clinical prognosis of glioblastoma multiforme (GBM) are concerns for many GBM patients as well as clinicians and researchers. The lack of a preclinical model that can easily be established and accurately recapitulate tumour biology and the tumour microenvironment further complicates GBM research and its clinical translation. GBM organoids (GBOs) are promising high-fidelity models that can be applied to model the disease, develop drugs, establish a living biobank, mimic therapeutic responses and explore personalized therapy. However, GBO models face some challenges, including deficient immune responses, absent vascular system and controversial reliability. In recent years, considerable progress has been achieved in the improvement of brain tumour organoid models and research based on such models. In addition to the traditional cultivation method, these models can be cultivated via genetic engineering and co-culture of cerebral organoids and GBM. In this review, we summarize the applications of GBM organoids and related advances and provide our opinions on associated limitations and challenges.
Keywords: Glioblastoma, organoids, precision oncology, invasion, immunotherapy
Introduction
Glioblastoma multiforme (GBM) is the most common primary malignant brain tumour in adults [1]. Because of its aggressive growth and high heterogeneity, this kind of tumour is often fatal [2]. The main treatment methods for GBM include surgery combined with postoperative radiotherapy and chemotherapy, as well as targeted treatment, immunotherapy and tumour treating fields [3-5]. However, these treatments have not made satisfactory progress [6]. The molecular heterogeneity observed between and within tumours is the main reason for the poor effect of many clinical trials [7-9]. For basic trials and clinical applications, developing a model including tumour atypia in time to verify the effectiveness of individualized treatments for GBM is a major challenge. At present, several research models are helping researchers understand the biological mechanisms of GBM, but they have limitations. First, animal models cannot fully reflect the genetic characteristics of human cancer or capture the heterogeneity of tumours. Second, in vitro culture models, whether two-dimensional (2D) cell line cultures or tumour spheroid cultures, acquire additional mutations during the process of cloning and amplification, which is not conducive to maintaining the expression of key driving genes in multiple cell subtypes and parent tumours. Patient-derived xenotransplantation (PDX) models can retain the important features of the primary tumour. However, these PDX models have the disadvantages of a large workload, high cost, and variable tumour implantation efficiency, so they are difficult to use in high-throughput drug screening. Therefore, a better model system to study GBM is urgently needed. Organoids are three-dimensional structures that are derived from stem cells, have self-organization ability and can simulate the structure and function of human organs [10]. Compared with traditional 2D culture models, tumour organoids can more closely represent the cell composition and physiological behaviour of the human body and replicate the three-dimensional (3D) tumour microenvironment (TME) [11], especially with respect to hypoxia, which is the key element in cancer biology [12]. Many kinds of tumour organoids retain the heterogeneity of primary tumours and can be widely amplified in culture while maintaining the stability of the genome [13-15]. Therefore, organoid-based models of malignancy are emerging as an indispensable platform for cancer research [16].
Common research models for glioma
Cell line models
At present, brain glioma cell lines, including U87, U251 and GL261, are the most common two-dimensional cell research models due to their easy access and high cost-effectiveness [17,18]. However, due to the lack of tumour structure and heterogeneity and the absence of single-cell infiltration, tumour necrosis and microvascular proliferation, 2D cell models are quite different from tumour cells growing in vivo in terms of cell-cell/matrix interactions, proliferation, morphology and signal transduction; thus, these models cannot accurately simulate the clinical response [19]. Moreover, in 2016, Allen compared the original U87MG-GBM cell line with the same cell line from the American Type Culture Collection (ATCC) and Cell Line Service (CLS). The results showed that the original U87MG gene did not match the U87MG gene present in the ATCC and CLS lines, even though these U87MG cell lines came from the same institution [20]. Therefore, cell lines do not usually recapitulate cancer heterogeneity or reveal a credible mechanism for therapeutic resistance.
Genetically engineered mouse models
Huge amounts of animal models have been developed to study the occurrence and development of glioma and serve as a platform for preclinical potential therapies screening [21,22]. One of the most commonly used models are genetically engineered mouse (GEM) models, which are helpful for exploring the effect of specific mutations/oncogenes on the characteristics of glioma stem cells, as well as their roles in tumour formation, invasion and metastasis. For example, to evaluate the role of the p21ras pathway in GBM, a transgenic mouse model expressing the oncogene v-Ha-ras was developed [23]. This study found that the level of v-Ha-ras was in direct proportion to the development of astrocytoma. At the same time, inoculating these tumour cells into different hosts can induce tumour growth. However, even if these GEM models have combined with powerful new technologies, they cannot fully describe the genetic complexity of tumours. Additionally, due to the lack of non-neoplastic normal host cells, these models often fail to recapitulate many human-specific features of the TME [24], and there may be great obstacles when the treatment results are translated from animals to humans. Therefore, the therapy effect must be examined in human tumours rather than in mouse tumours when preparing human personal therapy regimens.
Patient-derived xenotransplantation models
PDX mouse models are increasingly used as personalized preclinical platforms because they are maintaining the characteristics of parental tumours. They can be used to analyse the genome map and clinical data of a specific tumour tissue as a whole, as well as allowing for the generation of direct correlative associations between the cancer genome and the outcome of drug treatment [25]. By predicting the clinical success rate of a new therapy, GBM-PDX models have been further proven to overcome the limitations of traditional cell lines. For example, bevacizumab has anticancer properties in a mouse model established by U87 cell line xenotransplantation [26]. However, Joo et al. [27] showed that bevacizumab did not change the overall survival (OS) rate using a GBM-PDX model. The survival rate of GBM patients was also not significantly prolonged by these drugs in phase 2 clinical trials [28]. The main challenges of GBM-PDX are its high cost, time-consuming approach, immunodeficient and varying transplant rates [29-31]. Although the use of “humanized” mice (expressing components of the human immune system), the coinjection of human stromal/immune cells and human cytokines and injection of human GBM into mouse embryos might address the problem of immunodeficient [32,33], some obstacles remain when using these models for high-throughput drug screening. What’s more, there has demonstrated that Patient-derived xenografts may follow mouse-specific evolutionary trajectories which potentially jeopardize cancer modeling researches.
3D-bioprinting models
3D-bioprinting of multiple cell types within optimised extracellular matrices has the potential to more closely mimic the 3D environments involved in human physiology and disease [34]. Using 3D-printing technology [35], Yi et al. [36] reported a bioprinted human glioblastoma-on-a-chip that could identify patient-specific responses to chemoradiotherapy. This ex vivo patient-derived tumour model can not only recapitulate the characteristics of the parental tumour but also has the potential advantage of biophysical manipulation [37,38]. More importantly, the model can replicate complex tumour structures [39]. As only 1-2 weeks is required to generate the human glioblastoma-on-a-chip, ex vivo GBM models may be good models for high-throughput drug screening. We summarize the characteristics of the four kinds of research models in Figure 1.
Figure 1.
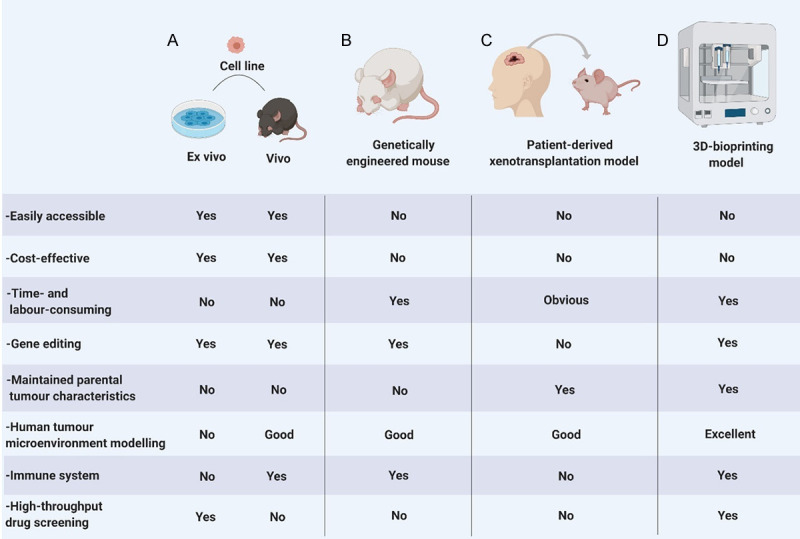
Comparison of common research models of glioma: A. Cell line models are divided into 2D ex vivo models and vivo models. Although they are easily accessible and cost-effective, 2D ex vivo models lack a 3D TME, and neither model type can recapitulate the characteristics of the parental tumour; B. Genetically engineered mouse models are conducive to studying specific gene mutations, but the characteristics of the parental tumour are not maintained; C. Patient-derived xenotransplantation models can recapitulate the characteristics of the parental tumour, but challenges include high cost, a time-consuming approach and varying transplant rates; D. 3D-bioprinting models can recapitulate the characteristics of the parental tumour and tumour structures and have the potential advantage of biophysical manipulation.
Generation of brain tumour organoids
Cerebral organoids, also called ‘mini-brains’, which are a more recent approach to modelling brain in vitro, can be obtained from two types of stem cells: (I) pluripotent stem cells (PSCs), such as embryonic stem cells (ESCs) or induced pluripotent stem cells (iPSCs), and (II) organ-specific adult stem cells (ASCs). Organoids can grow and differentiate after the addition of an intricate sequence of media and additives. Cerebral organoids are eventually cultured in rotating bioreactors, where they grow to contain functionally mature populations of various brain cell types and further mirror the regional differences observed in the human brain [40]. By replacing PSCs with patient-derived glioblastoma stem cells (GSCs), tumour organoids replicating the tumour cytoarchitecture rather than the brain architecture have been generated [41]. Several methods can be used to generate brain tumour organoids (Figure 2) (Table 1).
Figure 2.
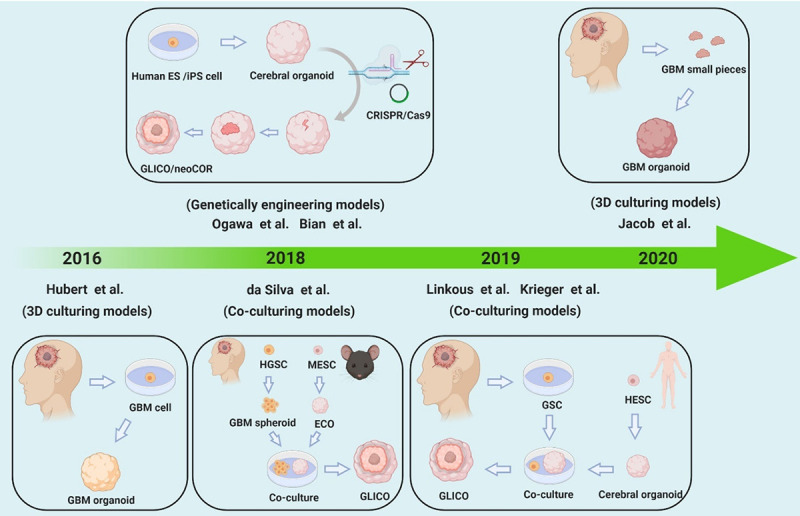
Classification of glioma organogenesis. We summarize three common methods used to generate brain tumour organoids, including traditional 3D culturing, genetic engineering and co-culturing.
Table 1.
A summary of the formation of brain tumour organoids
| Year/Author | Material | Method | Generation time | Tumour | Vessels and immune cells | Model |
|---|---|---|---|---|---|---|
| Traditional 3D culture | ||||||
| Hubert et al. 2016 [41] | patient-derived GBM CSCs | 3D culture | >2 m | GBM | No | |
| Jacob et al. 2020 [43] | GBM pieces | 3D culture; | 1-2 w | GBM | Yes | GBO |
| No extracellular matrix/EGF/bEGF | ||||||
| Gene engineered | ||||||
| Ogawa et al. 2018 [45] | hESCs | CRISPR/Cas9 | 3-4 m | GBM | No | |
| Bian et al. 2018 [44] | hESCs/iPSCs | Transposon- and CRISPR-Cas9-mediated mutagenesis | 1-2 m | GBM/PNET | No | neoCOR |
| Ballabio et al. 2020 [47] | hESCs/iPSCs | CRISPR/Cas9 | 35-60 d | G-3MB | No | |
| Co-culture | ||||||
| Da Silva et al. 2018 [48] | GBM cells/mESCs | Co-culture | 14 d | GBM | No | |
| Linkous et al. 2019 [46] | GSCs/hESCs/iPSCs | Co-culture | 1 m | GBM | No | GLICO |
| Krieger 2019 [49] | iPSCs/GBM cells | Co-culture | <4 w | GBM | No | |
Traditional 3D culture models
Traditional 3D culturing usually involves cutting human glioblastoma into small pieces and separating glioma stem cells for in vitro culture under the effects of matrix and exogenous factors. For example, in 2016, with the help of 3D culture methods, Hubert et al. [41] described a novel glioblastoma organoid (GBO) culture system using patient-derived glioblastoma CSCs that recapitulates the hypoxia gradients and stem cell heterogeneity found in tumours in vivo. Similar to Hubert et al.’s [41] research, most GBO studies have used Matrigel to obtain a 3D structure. However, this traditional material has some drawbacks, such as the difficulty of fine-tuning its properties to optimize cell growth. Gjorevski et al. [42] applied polyethylene glycol (PEG) matrix, a synthetic hydrogel system, to generate intestinal cancer organoids, which can serve as a substitute for Matrigel but does not affect biochemical signalling. Additionally, in a recent study, Jacob et al. [43] established a GBO model with the addition of Matrigel. This model directly cut the specimen into small pieces for culture without dissociating them into single cells or adding extracellular matrix or epidermal growth factor (EGF)/basic fibroblast growth factor (bEGF). This model well preserved the diversity, gene expression and mutations of the primary tumour cells. Moreover, only 1-2 weeks are needed to establish the model, which is beneficial for supporting the timeliness of preclinical studies of glioblastoma patients [43]. Continuous improvement of methods to construct organoids will lead to the development of a more realistic preclinical model for cancer research.
Genetically engineered models
Generating a genetically engineered model requires induction of glioma organogenesis by CRISPR/Cas9 gene editing technology or the addition of oncogenes [44]. In 2018, Ogawa et al. [45] combined technological advances in human organoid technology and CRISPR genome engineering to generate a genetically defined model of human glioblastoma, which was also called the glioma cerebral organoid (GLICO) model, in the study of Linkous et al. [46]. In this model, tumours can be induced in human cerebral organoids by introducing CRISPR/Cas9 and sgRNAs in combination with the activated oncogene HRasG12V and simultaneous disruption of the tumour suppressor TP53. The modified cells proliferate, show invasive phenotypes within organoids, and can be transplanted into immunodeficient mice. These tumours, when transplanted into mice, possess proliferative hallmarks of tumorigenesis, are invasive, and exhibit disease pathology. Additionally, Bian et al. [44] established a 3D in vitro model called a neoplastic cerebral organoid (neoCOR) in which brain tumourigenesis was replicated by introducing oncogenic mutations into cerebral organoids via transposon and CRISPR/Cas9 mediated mutagenesis. They defined mutation combinations that result in glioblastoma-like and central nervous system primitive neuroectodermal tumour (CNS-PNET)-like neoplasms. Therefore, neoCORs are suitable for use in investigations of aspects of tumour biology, such as invasiveness, and for evaluating drug effects in the context of specific DNA aberrations [44]. Moreover, using CRISPR/Cas9 technology, Ballabio et al. [47] recently validated these findings in human cerebellar organoids, where Otx2/c-MYC engendered MB-like organoids harbouring a DNA methylation signature that clusters with human Group 3 medulloblastoma.
Co-culturing models
Another approach is to form brain tumour organoids through cocultivation, such as coculture of glioblastoma and cerebral organoids to generate a GLICO model. For example, in 2018, da Silva et al. [48] established a glioma organoid invasion model by coculturing human GBM spheroids with mouse embryonic stem cell (mESC)-derived early-stage cerebral organoids (eCO). This approach may be used for the identification of anti-GBM invasion strategies. In 2019, Krieger et al. [49] presented an experimental model using human cerebral organoids as a scaffold for patient-derived glioblastoma cell invasion. This GLICO model has proven useful for studying GBM invasion and transcriptional heterogeneity in vitro, with applications for both pharmacological screens and patient-specific treatment selection at a time scale amenable to clinical practice. Additionally, in 2019, Linkous et al. [46] established a GLICO model to retro-engineer patient-specific GBMs using patient-derived glioma stem cells (GSCs) and human embryonic stem cell (hESC)-derived cerebral organoids. The model results showed that the tumours closely phenocopied GBMs from patients and were supported by tumour microtubes that promoted invasion into host tissue.
The various tumour organoid models have been widely implemented in glioma research, all of which have their unique advantages. For example, due to its fast production and inclusion of immune cells, the GBO model reported by Jacob et al. [43] is especially suitable for constructing a biobank and mimicking immunotherapy responses. Genetically engineered models are conducive to studying specific gene mutations and developing personalized therapy. Containing both tumour cells and normal brain tissue, GLICO models are suitable for exploring tumour biology and the TME and drug screening.
Applications of glioma organoid models
Glioma organoids have opened new avenues for the development of novel, more physiological human brain tumour models. Such preclinical models are essential for more efficient translation of basic cancer research into novel treatment regimens for patients with glioma (Figure 3) (Table 2). Wild-type GBOs can be grown from GSCs and display self-organizing capacity, phenocopying essential aspects of human-derived glioblastoma. Genetic modification of organoids allows disease modelling in a setting that approaches the physiological environment. Additionally, organoids can be grown with high efficiency from patient-derived normal and tumour tissues, potentially enabling patient-specific drug testing and the development of individualized treatment regimens [50].
Figure 3.
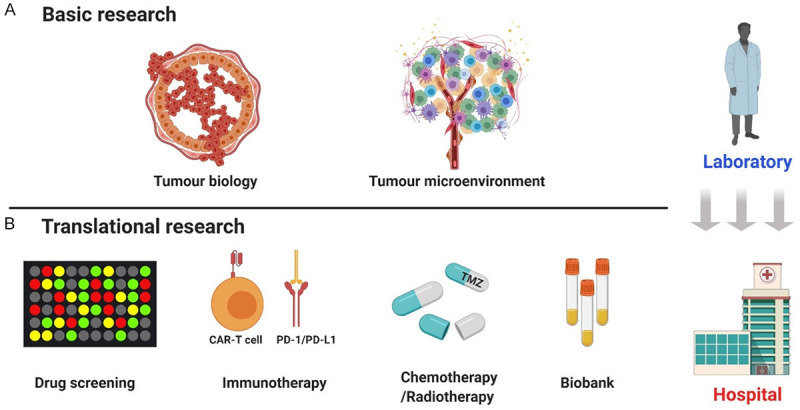
Applications of brain tumour organoids. Glioma organoid models have been widely implemented in basic and translational research: A. In basic research, we focus on tumour biology and the TME; B. In translational research, we summarize these models’ application in drug screening, chemotherapy, radiotherapy, and biobanks and focus more on immunotherapy, especially CAR-T cell therapy.
Table 2.
A summary of the application of brain tumour organoids
| Year/Author | Basic research | Drug screening | Immune-therapy | Chemotherapy/Radiotherapy | Biobank |
|---|---|---|---|---|---|
| Hubert et al. 2016 [41] | Exploration of microenvironmental influences and CSC biology | No reported | No | Yes | No reported |
| Ogawa et al. 2018 [45] | Model for tumour formation; Platform for tumour cell transplantation | No reported | No | No reported | No reported |
| Bian et al. 2018 [44] | Studying tumour biology | Afatinib, erlotinib, gefitinib, canertibib, pelitinib | No | No reported | No reported |
| Da Silva et al. 2018 [48] | Modelling the process of GBM invasion | No reported | No | No reported | No reported |
| Linkous et al. 2019 [46] | GBM biology, tumour microtubes | TMZ, BCNU | No | Yes | No reported |
| Krieger 2019 [49] | Studying GBM invasion and transcriptional heterogeneity | Yes | No | Yes | No reported |
| Jacob et al. 2020 [43] | Exploration of cellular heterogeneity, GBO engraftment and infiltration | TMZ, gefitinib, trametinib, everolimus | CAR-T | Yes | Yes |
| Ballabio et al. 2020 [47] | Studying medulloblastoma biology | Tazemetostat | No | No reported | No reported |
Organoids for basic research
Organoids to explore tumour biology
Organoid fusion is a new strategy to study the mechanism of GBM diffusion in the brain [34]. GSCs can fuse with cerebral organoids to evaluate the infiltration of tumour cells. Linkous et al. [46] observed the network of tumour microtubes in 923 and 827 GSC-formed GBMs growing in cerebral organoids, and these tumour microtubes promote the invasion of GLICO tumours into normal host tissue. These microtubules are also present in human glioblastomas, where they contribute to multicellular communication between cells and provide a pathway for invasion [51]. The detection of these microtubules in the GLICO model provides evidence that the behaviour and phenotype of cancer cells in this system are very similar to those of primary tumours. Thus, it is a good tool to study GBM biology in the human brain environment. Additionally, da Silva et al. [48] demonstrated that human GBM spheroids possess the ability to spontaneously infiltrate early-stage cerebral organoids (eCOs). The resulting formation of a hybrid organoid demonstrated the existence of an invasive tumour phenotype. These findings provide a basis for modelling and quantification of the GBM infiltration process using a stem-cell-based organoid approach and may be used for the identification of anti-GBM invasion strategies [48]. What’s more, Bian et al. [44] established a neoCOR model in which they recapitulated brain tumorigenesis by introducing oncogenic mutations into cerebral organoids via transposon- and CRISPR/Cas9 mediated mutagenesis. They demonstrated that neoCORs are suitable for use in investigations of aspects of tumour biology, such as invasiveness [44].
Modelling the tumour microenvironment
The TME is a self-sufficient ecosystem [52] involving interactions and networking between cells and between cells and non-cells. The main components of the TME include cancer cells, immune cells, stromal cells, and blood vessels as well as physical structures [53-55]. The focus of targeted therapy and immunotherapy for glioma has shifted from tumour cells to the TME [5]. Furthermore, tumour immune microenvironment normalization has the potential to transform a cold tumour into a hot tumour [56-58]. The TME is also critical for the maintenance of cellular states found in primary glioblastomas. By comparing single-cell RNA sequencing results for tumour cells from different GSC-derived model types, Allison R. Pine found that GSCs within the GLICO model were enriched for a neural progenitor-like cell (NPC) subpopulation and recapitulated the cellular states and plasticity found in the corresponding primary parental tumours [59]. They also found that the GLICO differentially expressed not only stemness marker SOX4 and glioma invasiveness marker BCAN but also prominent Notch pathway members (DLL3, HES1, and HES6) that are differentially expressed in other GLICO model cells. Thus, the GLICO model not only contained patient-derived GSCs but also closely resembled the TME physically, which is critical for studying cancer biology and drug screening.
Organoids for translational research
Drug development
Targeting tumour stem cells is the key to preventing tumour recurrence [60,61]. The therapeutic response to anticancer drugs, especially targeted drugs, strongly depends on the genetic and epigenetic background of the patient [62]. Patient-derived organoids can capture tumour-specific gene mutations, gene expression, and individual histopathological changes, thus representing an ideal preclinical model for individual drug screening [63,64]. For example, GLICO tumours can maintain the key genetic features, molecular signalling networks, and EGFR amplification found in the parental tumours, whereas EGFR amplification is lost in 2D cultures. Phosphorylation of EGFR, which is related to RTK, and ephrin type-B receptor 3 (EphB3) was observed in both parental tumours and the corresponding GLICO tumours from GSC lines [46]. This model can provide a basis for screening potential drug targets or therapies by measuring the effects of a drug or gene intervention on tumour cell proliferation, death and invasiveness. More importantly, organoids containing both normal and tumour tissues can be generated, enabling screening for drugs that specifically target tumour cells while leaving healthy cells unharmed. For example, Dijkstra et al. [65] established a modified patient-derived tumour organoid system that allows the expansion of tumour-specific T cells by coculture of peripheral blood lymphocytes (PBMCs) and tumour organoids derived from mismatch repair-deficient colorectal cancer and non-small-cell lung cancer. The induced T cell populations do not recognise healthy organoids or tissue and can be used to assess the efficiency of T cell-mediated tumour killing, thus establishing a foundation for exploration of individual antitumour drugs with functions similar to those of tumour-specific T cells.
Immunotherapy
Immune checkpoint inhibitors
Cancer immunotherapy, which has resulted in exciting and significant treatment outcomes for multiple cancer types, has attracted unprecedented research interest as a treatment for gliomas [66]. Retaining the genetic and phenotypic characteristics of the parent tumour, tumour organoids are a promising model to test immunotherapy effects ex vivo [67,68]. The newly developed coculture organoid and air-liquid interface (ALI) systems have provided new insights for studying epithelia-immune cell interactions based on their endogenous distributions. In 2018, Neal et al. [69] generated a tumour organoid model including the tumour immune microenvironment using ALI cultures. The model preserved CD8+ T cells, CD4+ T cells, B cells, natural killer cells, natural killer T cells, and macrophages. The conserved T cell receptor repertoire of the parental tumour was retained within the ALI PDOs. More importantly, PD-1/PD-L1 immune checkpoint blockade expanded and activated antigen-specific tumour-infiltrating lymphocytes (TILs) within ALI PDOs derived from melanoma, lung cancer, and renal cell carcinoma, and the results showed a clinical response rate similar to that observed in ALI PDOs [70]. ALI systems also enable oxygen to be transported in a more efficient manner, resulting in a more rational oxygen gradient [71], which is particularly significant for glioma research. Thus, ALI PDOs show potential as a tool to predict clinical responses to immune checkpoint therapies.
Chimeric antigen receptor-T cell therapy
Chimeric antigen receptor (CAR)-T cells represent a potent new approach to treat GBM. The novelty of CAR-T cells partly lies in the genetically engineered chimeric receptor [72,73]. In contrast to native T-cell receptors (TCRs), CAR-T cells can recognise any cell surface structure independent of MHC presentation [74]. An infusion of CAR-T cells targeting EGFRvIII, IL-13Rα2, or HER-2 has frequently been reported in glioblastomas [75-78]. However, experiments testing the efficacy of CAR-T cells have often relied on overexpression of target antigens in tumour cell lines, which may not recapitulate the endogenous condition [79]. Tumour organoids have recently been used as a model to test CAR-T cell killing due to their 3D architecture [80]. For example, CAR-T cell therapies comprise genetically engineered T cells that are redirected towards tumour cell-surface antigens [81]. The ability to label these lymphocytes will enable potential quantitative intra-GLICO tracking studies of CAR-T cells [82] as well as studies of interactions between CAR-T cells and tumour targets at the invasive front, which is the origin of recurrent disease. Additionally, by coculturing GBOs with 2173BBz CAR-T cells designed to react specifically with cells expressing EGFRvIII [78], Jacob et al. [43] demonstrated the utility of GBOs for rapid testing of antigen-specific CAR-T cell treatment responses with the endogenous target in culture. Although some limitations exist, such as immune components in organoid cultures that can be maintained only for a short period [83], with the development of several advanced technologies, such as microfluidics, organ-on-a-chip technology, 4D imaging technology, next-generation sequencing and transcriptomics [84-87], more potential advantages of brain tumour organoid use in immunotherapy research will be identified.
Radiotherapy and chemotherapy
In addition to applications in tumour immunotherapy, brain tumour organoids can also be used in the study of radiotherapy and chemotherapy [46,88]. For example, Linkous et al. [46] demonstrated that isogenic GSC lines are substantially more resistant to chemotherapeutic agents and radiation-induced genotoxic stress when grown within the microenvironment of a cerebral organoid (GLICO) than when grown under traditional 2D conditions. Hubert et al. [41] reported that CSCs in organoids are radioresistant, whereas adjacent non-stem tumour cells are radiosensitive. Furthermore, by using human brain organoids comprised of mature neural cell types as a 3D tissue substrate to model the invasive growth of glioma, Liu et al. [88] found that antisense oligonucleotides targeting lncRNA Glioma Radiation Sensitizer (lncGRS-1) selectively decreased tumour growth and sensitized glioma cells to radiation therapy. Therefore, glioma organoids can be used as a screening tool to identify tumour sensitivity to radiotherapy and chemotherapy.
Living biobank
In view of the urgent need for individualized cancer treatments, many studies are developing a “living organism bank” composed of patient-derived organoids [89]. Studies have shown that tumour organoid cultures can be established from prostate, colon, breast, pancreas, stomach, liver and endometrium cancer tissues [90-96]. Importantly, the genetic and phenotypic characteristics of tumour-derived organoids resemble those of the tumour epithelia from which they were derived. In a recent study, Jacob et al. [43] provided a resource of 70 biobank GBOs from different patients that captures major genomic alterations associated with glioblastoma pathogenesis. They provided detailed characterizations of many of these biobank GBOs, including the histology, RNA-seq, whole-exome sequencing, and responses to different drugs and CAR-T cell therapies. Therefore, these biobanks will be a useful resource for future biological studies and for testing therapeutic agents for glioblastomas.
Challenges and prospects for brain tumour organoids
Although glioma organoids have been widely used in glioma research, some limitations still exist. However, technological innovation and development may increase the potential of glioma organoids as preclinical models.
Deficiency in immune responses
As immunotherapy investigations require an intact immune microenvironment, the absence of blood vessels and immune cell types that act directly on antitumour immune responses will limit the utility of brain tumour organoid models for immunotherapy investigations. However, the improvement of the brain tumour organoid culture method also makes it possible to study immunotherapy ex vivo. For example, Jacob et al. [43] generated the GBO model, which provides a platform to test and optimize CAR-T cell therapies for solid tumours in vitro. Importantly, these GBOs recapitulate the endogenous expression of antigens, allowing for more accurate assessment of CAR-T cell target reactivity, response thresholds, and specificity. Furthermore, due to preservation of the primary tumour epithelium en bloc with native endogenous TILs, ALI PDOs can be used to predict the clinical response of the PD-1/PD-L1 immune checkpoint [69]. In addition, tumour organoids can be co-cultured with peripheral blood lymphocytes [65], and such models can be used to evaluate the sensitivity of T cells to tumour cell-mediated killing, which can also be extended to evaluate the effectiveness of immunotherapy at different time points.
Lack of a vascular system
The lack of endothelial cells and tumour vasculature is also a major problem that limits the utility of brain tumour organoids, as the microvascular system is a major component of the tumour environment [5], which can affect the growth of tumour cells and the antitumour immune effect [53,54,97,98].
Several methods to induce vascularization in cerebral organoids in vitro have been developed. First, coculture with endothelial cells may facilitate vascularization in cerebral organoids [99-101]; for example, human endothelial cells (hECs) can be embedded in Matrigel for coculture with early-stage organoids, and the hECs will assemble into capillaries around the organoids and generate a vascular network over time. Moreover, a recent study showed that treatment with VEGF from the beginning of hESC differentiation could successfully generate blood vessels to facilitate further maturation of the vasculature in cultured cerebral organoids [102]. Additionally, by engineering hESCs to ectopically express human ETS variant 2 (ETV2), B. Cakir found that ETV2-expressing cells in human cortical organoids (hCOs) contributed to forming a complex vascular-like network in hCOs. These vhCOs form vasculature-like structures that resemble the vasculature in the early prenatal brain and represent a robust model to study brain disease in vitro [103]. Moreover, these researchers reported that vhCOs acquired several blood-brain barrier (BBB) characteristics, including increases in the expression of tight junctions, nutrient transporters and trans-endothelial electrical resistance; these characteristics allow vhCOs to more closely and realistically replicate brain function.
Controversy in structural or genetic fidelity
In addition, some studies showed that the structural and genetic fidelity of brain organoids is still unclear [104-106]. Recently, Aparna and other researchers found that brain organoids do not produce unique cell subtypes or regional circuit structures unique to normal human brain circuits and that brain organoids express high levels of cell “stress” genes (including the glycolysis gene pgk132 and the endoplasmic reticulum stress genes arcn133 and gorasp2) [107]. Moreover, for some gene knock-in mouse strain-derived organoids, there is still a lack of robust knock-in methods for the precise integration of exogenous DNA sequences into human-derived organoids [108,109]. However, recently, the Hans Cleaver research group established gene editing technology independent of the homologous arm of DNA repair. Based on NHEJ, a novel and robust gene typing technology, CRISPR-hot, has been established, which provides a reliable gene editing method for adult stem cell-derived organoid visualization research [110]. With the improvement of culture methods and techniques, more realistic brain organoids and brain tumour organoids will be produced in the future.
Conclusion
The glioma organoid model is one of the most promising models for glioma research and can not only retain the genetic phenotype of the primary tumour but also provide a 3D TME similar to that of the parental tumour. Several methods can be used to generate glioma organoids, including traditional 3D culturing, genetic engineering and coculturing. These methods may be used to generate other brain tumour organoids, such as medulloblastoma and brain metastasis organoids. To date, brain glioma organoids have been widely used in basic research and clinical transformation research and have considerable potential particularly in the study of immunotherapy. By combining innovative technologies such as 3D bioprinting and 4D real-time imaging, a more realistic brain tumour organoid model will contribute to anti-cancer research in the near future.
Acknowledgements
We would like to thank everyone who take part in this study. Funding information, this work is supported by the Basic research foundation of Shanghai (19JC1415000), 12th undergraduate training programs for innovation of Shanghai Jiao Tong University School of Medicine (No. 1218201), Key research and development projects of Hainan Province (ZDYF2019173) and Hainan Provincial Natural Science Foundation of China (819QN351).
Disclosure of conflict of interest
None.
References
- 1.Ostrom QT, Cioffi G, Gittleman H, Patil N, Waite K, Kruchko C, Barnholtz-Sloan JS. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2012-2016. Neuro Oncol. 2019;21:v1–v100. doi: 10.1093/neuonc/noz150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paolillo M, Boselli C, Schinelli S. Glioblastoma under siege: an overview of current therapeutic strategies. Brain Sci. 2018;8:15. doi: 10.3390/brainsci8010015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gilbert MR, Dignam JJ, Armstrong TS, Wefel JS, Blumenthal DT, Vogelbaum MA, Colman H, Chakravarti A, Pugh S, Won M, Jeraj R, Brown PD, Jaeckle KA, Schiff D, Stieber VW, Brachman DG, Werner-Wasik M, Tremont-Lukats IW, Sulman EP, Aldape KD, Curran WJ, Mehta MP. A randomized trial of bevacizumab for newly diagnosed glioblastoma. N Engl J Med. 2014;370:699–708. doi: 10.1056/NEJMoa1308573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stupp R, Taillibert S, Kanner A, Read W, Steinberg D, Lhermitte B, Toms S, Idbaih A, Ahluwalia MS, Fink K, Di Meco F, Lieberman F, Zhu JJ, Stragliotto G, Tran D, Brem S, Hottinger A, Kirson ED, Lavy-Shahaf G, Weinberg U, Kim CY, Paek SH, Nicholas G, Bruna J, Hirte H, Weller M, Palti Y, Hegi ME, Ram Z. Effect of tumor-treating fields plus maintenance temozolomide vs maintenance temozolomide alone on survival in patients with glioblastoma: a randomized clinical trial. JAMA. 2017;318:2306–2316. doi: 10.1001/jama.2017.18718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mpekris F, Voutouri C, Baish JW, Duda DG, Munn LL, Stylianopoulos T, Jain RK. Combining microenvironment normalization strategies to improve cancer immunotherapy. Proc Natl Acad Sci U S A. 2020;117:3728–3737. doi: 10.1073/pnas.1919764117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perrin SL, Samuel MS, Koszyca B, Brown MP, Ebert LM, Oksdath M, Gomez GA. Glioblastoma heterogeneity and the tumour microenvironment: implications for preclinical research and development of new treatments. Biochem Soc Trans. 2019;47:625–638. doi: 10.1042/BST20180444. [DOI] [PubMed] [Google Scholar]
- 7.Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR, Zheng S, Chakravarty D, Sanborn JZ, Berman SH, Beroukhim R, Bernard B, Wu CJ, Genovese G, Shmulevich I, Barnholtz-Sloan J, Zou L, Vegesna R, Shukla SA, Ciriello G, Yung WK, Zhang W, Sougnez C, Mikkelsen T, Aldape K, Bigner DD, Van Meir EG, Prados M, Sloan A, Black KL, Eschbacher J, Finocchiaro G, Friedman W, Andrews DW, Guha A, Iacocca M, O’Neill BP, Foltz G, Myers J, Weisenberger DJ, Penny R, Kucherlapati R, Perou CM, Hayes DN, Gibbs R, Marra M, Mills GB, Lander E, Spellman P, Wilson R, Sander C, Weinstein J, Meyerson M, Gabriel S, Laird PW, Haussler D, Getz G, Chin L Tcga Research Network. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mandel JJ, Yust-Katz S, Patel AJ, Cachia D, Liu D, Park M, Yuan Y, Kent TA, de Groot JF. Inability of positive phase II clinical trials of investigational treatments to subsequently predict positive phase III clinical trials in glioblastoma. Neuro Oncol. 2018;20:113–122. doi: 10.1093/neuonc/nox144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Neftel C, Laffy J, Filbin MG, Hara T, Shore ME, Rahme GJ, Richman AR, Silverbush D, Shaw ML, Hebert CM, Dewitt J, Gritsch S, Perez EM, Gonzalez Castro LN, Lan X, Druck N, Rodman C, Dionne D, Kaplan A, Bertalan MS, Small J, Pelton K, Becker S, Bonal D, Nguyen QD, Servis RL, Fung JM, Mylvaganam R, Mayr L, Gojo J, Haberler C, Geyeregger R, Czech T, Slavc I, Nahed BV, Curry WT, Carter BS, Wakimoto H, Brastianos PK, Batchelor TT, Stemmer-Rachamimov A, Martinez-Lage M, Frosch MP, Stamenkovic I, Riggi N, Rheinbay E, Monje M, Rozenblatt-Rosen O, Cahill DP, Patel AP, Hunter T, Verma IM, Ligon KL, Louis DN, Regev A, Bernstein BE, Tirosh I, Suva ML. An integrative model of cellular states, plasticity, and genetics for glioblastoma. Cell. 2019;178:835–849. e21. doi: 10.1016/j.cell.2019.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li M, Izpisua Belmonte JC. Organoids - preclinical models of human disease. N Engl J Med. 2019;380:569–579. doi: 10.1056/NEJMra1806175. [DOI] [PubMed] [Google Scholar]
- 11.Jin MZ, Han RR, Qiu GZ, Ju XC, Lou G, Jin WL. Organoids: an intermediate modeling platform in precision oncology. Cancer Lett. 2018;414:174–180. doi: 10.1016/j.canlet.2017.11.021. [DOI] [PubMed] [Google Scholar]
- 12.Qiu GZ, Jin MZ, Dai JX, Sun W, Feng JH, Jin WL. Reprogramming of the tumor in the hypoxic niche: the emerging concept and associated therapeutic strategies. Trends Pharmacol Sci. 2017;38:669–686. doi: 10.1016/j.tips.2017.05.002. [DOI] [PubMed] [Google Scholar]
- 13.Behjati S, Huch M, van Boxtel R, Karthaus W, Wedge DC, Tamuri AU, Martincorena I, Petljak M, Alexandrov LB, Gundem G, Tarpey PS, Roerink S, Blokker J, Maddison M, Mudie L, Robinson B, Nik-Zainal S, Campbell P, Goldman N, van de Wetering M, Cuppen E, Clevers H, Stratton MR. Genome sequencing of normal cells reveals developmental lineages and mutational processes. Nature. 2014;513:422–425. doi: 10.1038/nature13448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huch M, Gehart H, van Boxtel R, Hamer K, Blokzijl F, Verstegen MM, Ellis E, van Wenum M, Fuchs SA, de Ligt J, van de Wetering M, Sasaki N, Boers SJ, Kemperman H, de Jonge J, Ijzermans JN, Nieuwenhuis EE, Hoekstra R, Strom S, Vries RR, van der Laan LJ, Cuppen E, Clevers H. Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell. 2015;160:299–312. doi: 10.1016/j.cell.2014.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duarte AA, Gogola E, Sachs N, Barazas M, Annunziato S, de Ruiter JR, Velds A, Blatter S, Houthuijzen JM, van de Ven M, Clevers H, Borst P, Jonkers J, Rottenberg S. BRCA-deficient mouse mammary tumor organoids to study cancer-drug resistance. Nat Methods. 2018;15:134–140. doi: 10.1038/nmeth.4535. [DOI] [PubMed] [Google Scholar]
- 16.Tuveson D, Clevers H. Cancer modeling meets human organoid technology. Science. 2019;364:952–955. doi: 10.1126/science.aaw6985. [DOI] [PubMed] [Google Scholar]
- 17.Tu Y, Gao X, Li G, Fu H, Cui D, Liu H, Jin W, Zhang Y. MicroRNA-218 inhibits glioma invasion, migration, proliferation, and cancer stem-like cell self-renewal by targeting the polycomb group gene Bmi1. Cancer Res. 2013;73:6046–6055. doi: 10.1158/0008-5472.CAN-13-0358. [DOI] [PubMed] [Google Scholar]
- 18.Qiu GZ, Mao XY, Ma Y, Gao XC, Wang Z, Jin MZ, Sun W, Zou YX, Lin J, Fu HL, Jin WL. Ubiquitin-specific protease 22 acts as an oncoprotein to maintain glioma malignancy through deubiquitinating B cell-specific Moloney murine leukemia virus integration site 1 for stabilization. Cancer Sci. 2018;109:2199–2210. doi: 10.1111/cas.13646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.da Hora CC, Schweiger MW, Wurdinger T, Tannous BA. Patient-derived glioma models: from patients to dish to animals. Cells. 2019;8:1177. doi: 10.3390/cells8101177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Allen M, Bjerke M, Edlund H, Nelander S, Westermark B. Origin of the U87MG glioma cell line: good news and bad news. Sci Transl Med. 2016;8 doi: 10.1126/scitranslmed.aaf6853. 354re3. [DOI] [PubMed] [Google Scholar]
- 21.Misuraca KL, Hu G, Barton KL, Chung A, Becher OJ. A novel mouse model of diffuse intrinsic pontine glioma initiated in Pax3-expressing cells. Neoplasia. 2016;18:60–70. doi: 10.1016/j.neo.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hennika T, Hu G, Olaciregui NG, Barton KL, Ehteda A, Chitranjan A, Chang C, Gifford AJ, Tsoli M, Ziegler DS, Carcaboso AM, Becher OJ. Pre-clinical study of panobinostat in xenograft and genetically engineered murine diffuse intrinsic pontine glioma models. PLoS One. 2017;12:e0169485. doi: 10.1371/journal.pone.0169485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ding H, Roncari L, Shannon P, Wu X, Lau N, Karaskova J, Gutmann DH, Squire JA, Nagy A, Guha A. Astrocyte-specific expression of activated p21-ras results in malignant astrocytoma formation in a transgenic mouse model of human gliomas. Cancer Res. 2001;61:3826–3836. [PubMed] [Google Scholar]
- 24.Chongsathidkiet P, Jackson C, Koyama S, Loebel F, Cui X, Farber SH, Woroniecka K, Elsamadicy AA, Dechant CA, Kemeny HR, Sanchez-Perez L, Cheema TA, Souders NC, Herndon JE, Coumans JV, Everitt JI, Nahed BV, Sampson JH, Gunn MD, Martuza RL, Dranoff G, Curry WT, Fecci PE. Sequestration of T cells in bone marrow in the setting of glioblastoma and other intracranial tumors. Nat Med. 2018;24:1459–1468. doi: 10.1038/s41591-018-0135-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pauli C, Hopkins BD, Prandi D, Shaw R, Fedrizzi T, Sboner A, Sailer V, Augello M, Puca L, Rosati R, McNary TJ, Churakova Y, Cheung C, Triscott J, Pisapia D, Rao R, Mosquera JM, Robinson B, Faltas BM, Emerling BE, Gadi VK, Bernard B, Elemento O, Beltran H, Demichelis F, Kemp CJ, Grandori C, Cantley LC, Rubin MA. Personalized in vitro and in vivo cancer models to guide precision medicine. Cancer Discov. 2017;7:462–477. doi: 10.1158/2159-8290.CD-16-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.von Baumgarten L, Brucker D, Tirniceru A, Kienast Y, Grau S, Burgold S, Herms J, Winkler F. Bevacizumab has differential and dose-dependent effects on glioma blood vessels and tumor cells. Clin Cancer Res. 2011;17:6192–6205. doi: 10.1158/1078-0432.CCR-10-1868. [DOI] [PubMed] [Google Scholar]
- 27.Joo KM, Kim J, Jin J, Kim M, Seol HJ, Muradov J, Yang H, Choi YL, Park WY, Kong DS, Lee JI, Ko YH, Woo HG, Lee J, Kim S, Nam DH. Patient-specific orthotopic glioblastoma xenograft models recapitulate the histopathology and biology of human glioblastomas in situ. Cell Rep. 2013;3:260–273. doi: 10.1016/j.celrep.2012.12.013. [DOI] [PubMed] [Google Scholar]
- 28.Lai A, Tran A, Nghiemphu PL, Pope WB, Solis OE, Selch M, Filka E, Yong WH, Mischel PS, Liau LM, Phuphanich S, Black K, Peak S, Green RM, Spier CE, Kolevska T, Polikoff J, Fehrenbacher L, Elashoff R, Cloughesy T. Phase II study of bevacizumab plus temozolomide during and after radiation therapy for patients with newly diagnosed glioblastoma multiforme. J. Clin. Oncol. 2011;29:142–148. doi: 10.1200/JCO.2010.30.2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ben-David U, Ha G, Tseng YY, Greenwald NF, Oh C, Shih J, McFarland JM, Wong B, Boehm JS, Beroukhim R, Golub TR. Patient-derived xenografts undergo mouse-specific tumor evolution. Nat Genet. 2017;49:1567–1575. doi: 10.1038/ng.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patrizii M, Bartucci M, Pine SR, Sabaawy HE. Utility of glioblastoma patient-derived orthotopic xenografts in drug discovery and personalized therapy. Front Oncol. 2018;8:23. doi: 10.3389/fonc.2018.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoshida GJ. Applications of patient-derived tumor xenograft models and tumor organoids. J Hematol Oncol. 2020;13:4. doi: 10.1186/s13045-019-0829-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bartucci M, Ferrari AC, Kim IY, Ploss A, Yarmush M, Sabaawy HE. Personalized medicine approaches in prostate cancer employing patient derived 3D organoids and humanized mice. Front Cell Dev Biol. 2016;4:64. doi: 10.3389/fcell.2016.00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoffmann N, Fernandez V, Pereira RC, Rancati S, Pelizzoli R, Tonelli D. A xenotransplant model of human brain tumors in wild-type mice. iScience. 2020;23:100813. doi: 10.1016/j.isci.2019.100813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hermida MA, Kumar JD, Schwarz D, Laverty KG, Di Bartolo A, Ardron M, Bogomolnijs M, Clavreul A, Brennan PM, Wiegand UK, Melchels FP, Shu W, Leslie NR. Three dimensional in vitro models of cancer: bioprinting multilineage glioblastoma models. Adv Biol Regul. 2020;75:100658. doi: 10.1016/j.jbior.2019.100658. [DOI] [PubMed] [Google Scholar]
- 35.Choi YJ, Yi HG, Kim SW, Cho DW. 3D cell printed tissue analogues: a new platform for theranostics. Theranostics. 2017;7:3118–3137. doi: 10.7150/thno.19396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yi HG, Jeong YH, Kim Y, Choi YJ, Moon HE, Park SH, Kang KS, Bae M, Jang J, Youn H, Paek SH, Cho DW. A bioprinted human-glioblastoma-on-a-chip for the identification of patient-specific responses to chemoradiotherapy. Nat Biomed Eng. 2019;3:509–519. doi: 10.1038/s41551-019-0363-x. [DOI] [PubMed] [Google Scholar]
- 37.Pedron S, Becka E, Harley BA. Spatially gradated hydrogel platform as a 3D engineered tumor microenvironment. Adv Mater. 2015;27:1567–1572. doi: 10.1002/adma.201404896. [DOI] [PubMed] [Google Scholar]
- 38.Lind JU, Busbee TA, Valentine AD, Pasqualini FS, Yuan H, Yadid M, Park SJ, Kotikian A, Nesmith AP, Campbell PH, Vlassak JJ, Lewis JA, Parker KK. Instrumented cardiac microphysiological devices via multimaterial three-dimensional printing. Nat Mater. 2017;16:303–308. doi: 10.1038/nmat4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee H, Cho DW. One-step fabrication of an organ-on-a-chip with spatial heterogeneity using a 3D bioprinting technology. Lab Chip. 2016;16:2618–2625. doi: 10.1039/c6lc00450d. [DOI] [PubMed] [Google Scholar]
- 40.Lancaster MA, Renner M, Martin CA, Wenzel D, Bicknell LS, Hurles ME, Homfray T, Penninger JM, Jackson AP, Knoblich JA. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hubert CG, Rivera M, Spangler LC, Wu Q, Mack SC, Prager BC, Couce M, McLendon RE, Sloan AE, Rich JN. A three-dimensional organoid culture system derived from human glioblastomas recapitulates the hypoxic gradients and cancer stem cell heterogeneity of tumors found in vivo. Cancer Res. 2016;76:2465–2477. doi: 10.1158/0008-5472.CAN-15-2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gjorevski N, Sachs N, Manfrin A, Giger S, Bragina ME, Ordonez-Moran P, Clevers H, Lutolf MP. Designer matrices for intestinal stem cell and organoid culture. Nature. 2016;539:560–564. doi: 10.1038/nature20168. [DOI] [PubMed] [Google Scholar]
- 43.Jacob F, Salinas RD, Zhang DY, Nguyen PTT, Schnoll JG, Wong SZH, Thokala R, Sheikh S, Saxena D, Prokop S, Liu DA, Qian X, Petrov D, Lucas T, Chen HI, Dorsey JF, Christian KM, Binder ZA, Nasrallah M, Brem S, O’Rourke DM, Ming GL, Song H. A patient-derived glioblastoma organoid model and biobank recapitulates inter- and intra-tumoral heterogeneity. Cell. 2020;180:188–204. e22. doi: 10.1016/j.cell.2019.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bian S, Repic M, Guo Z, Kavirayani A, Burkard T, Bagley JA, Krauditsch C, Knoblich JA. Genetically engineered cerebral organoids model brain tumor formation. Nat Methods. 2018;15:631–639. doi: 10.1038/s41592-018-0070-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ogawa J, Pao GM, Shokhirev MN, Verma IM. Glioblastoma model using human cerebral organoids. Cell Rep. 2018;23:1220–1229. doi: 10.1016/j.celrep.2018.03.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Linkous A, Balamatsias D, Snuderl M, Edwards L, Miyaguchi K, Milner T, Reich B, Cohen-Gould L, Storaska A, Nakayama Y, Schenkein E, Singhania R, Cirigliano S, Magdeldin T, Lin Y, Nanjangud G, Chadalavada K, Pisapia D, Liston C, Fine HA. Modeling patient-derived glioblastoma with cerebral organoids. Cell Rep. 2019;26:3203–3211. e5. doi: 10.1016/j.celrep.2019.02.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ballabio C, Anderle M, Gianesello M, Lago C, Miele E, Cardano M, Aiello G, Piazza S, Caron D, Gianno F, Ciolfi A, Pedace L, Mastronuzzi A, Tartaglia M, Locatelli F, Ferretti E, Giangaspero F, Tiberi L. Modeling medulloblastoma in vivo and with human cerebellar organoids. Nat Commun. 2020;11:583. doi: 10.1038/s41467-019-13989-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.da Silva B, Mathew RK, Polson ES, Williams J, Wurdak H. Spontaneous glioblastoma spheroid infiltration of early-stage cerebral organoids models brain tumor invasion. SLAS Discov. 2018;23:862–868. doi: 10.1177/2472555218764623. [DOI] [PubMed] [Google Scholar]
- 49.Krieger TG, Tirier SM, Park J, Jechow K, Eisemann T, Peterziel H, Angel P, Eils R, Conrad C. Modeling glioblastoma invasion using human brain organoids and single-cell transcriptomics. Neuro Oncol. 2020 doi: 10.1093/neuonc/noaa091. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Drost J, Clevers H. Organoids in cancer research. Nat Rev Cancer. 2018;18:407–418. doi: 10.1038/s41568-018-0007-6. [DOI] [PubMed] [Google Scholar]
- 51.Osswald M, Jung E, Sahm F, Solecki G, Venkataramani V, Blaes J, Weil S, Horstmann H, Wiestler B, Syed M, Huang L, Ratliff M, Karimian Jazi K, Kurz FT, Schmenger T, Lemke D, Gommel M, Pauli M, Liao Y, Haring P, Pusch S, Herl V, Steinhauser C, Krunic D, Jarahian M, Miletic H, Berghoff AS, Griesbeck O, Kalamakis G, Garaschuk O, Preusser M, Weiss S, Liu H, Heiland S, Platten M, Huber PE, Kuner T, von Deimling A, Wick W, Winkler F. Brain tumour cells interconnect to a functional and resistant network. Nature. 2015;528:93–98. doi: 10.1038/nature16071. [DOI] [PubMed] [Google Scholar]
- 52.Quail DF, Joyce JA. The microenvironmental landscape of brain tumors. Cancer Cell. 2017;31:326–341. doi: 10.1016/j.ccell.2017.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Peterson TE, Kirkpatrick ND, Huang Y, Farrar CT, Marijt KA, Kloepper J, Datta M, Amoozgar Z, Seano G, Jung K, Kamoun WS, Vardam T, Snuderl M, Goveia J, Chatterjee S, Batista A, Muzikansky A, Leow CC, Xu L, Batchelor TT, Duda DG, Fukumura D, Jain RK. Dual inhibition of Ang-2 and VEGF receptors normalizes tumor vasculature and prolongs survival in glioblastoma by altering macrophages. Proc Natl Acad Sci U S A. 2016;113:4470–4475. doi: 10.1073/pnas.1525349113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zheng X, Fang Z, Liu X, Deng S, Zhou P, Wang X, Zhang C, Yin R, Hu H, Chen X, Han Y, Zhao Y, Lin SH, Qin S, Wang X, Kim BY, Zhou P, Jiang W, Wu Q, Huang Y. Increased vessel perfusion predicts the efficacy of immune checkpoint blockade. J Clin Invest. 2018;128:2104–2115. doi: 10.1172/JCI96582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gutmann DH, Kettenmann H. Microglia/brain macrophages as central drivers of brain tumor pathobiology. Neuron. 2019;104:442–449. doi: 10.1016/j.neuron.2019.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stylianopoulos T, Munn LL, Jain RK. Reengineering the physical microenvironment of tumors to improve drug delivery and efficacy: from mathematical modeling to bench to bedside. Trends Cancer. 2018;4:292–319. doi: 10.1016/j.trecan.2018.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chauhan VP, Chen IX, Tong R, Ng MR, Martin JD, Naxerova K, Wu MW, Huang P, Boucher Y, Kohane DS, Langer R, Jain RK. Reprogramming the microenvironment with tumor-selective angiotensin blockers enhances cancer immunotherapy. Proc Natl Acad Sci U S A. 2019;116:10674–10680. doi: 10.1073/pnas.1819889116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Datta M, Coussens LM, Nishikawa H, Hodi FS, Jain RK. Reprogramming the tumor microenvironment to improve immunotherapy: emerging strategies and combination therapies. Am Soc Clin Oncol Educ Book. 2019;39:165–174. doi: 10.1200/EDBK_237987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pine AR, Cirigliano SM, Nicholson JG, Hu Y, Linkous A, Miyaguchi K, Edwards L, Singhania R, Schwartz TH, Ramakrishna R, Pisapia DJ, Snuderl M, Elemento O, Fine HA. Tumor microenvironment is critical for the maintenance of cellular states found in primary glioblastomas. Cancer Discov. 2020;10:964–979. doi: 10.1158/2159-8290.CD-20-0057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Qiu GZ, Sun W, Jin MZ, Lin J, Lu PG, Jin WL. The bad seed gardener: deubiquitinases in the cancer stem-cell signaling network and therapeutic resistance. Pharmacol Ther. 2017;172:127–138. doi: 10.1016/j.pharmthera.2016.12.003. [DOI] [PubMed] [Google Scholar]
- 61.Wang Z, Xu X, Liu N, Cheng Y, Jin W, Zhang P, Wang X, Yang H, Liu H, Tu Y. SOX9-PDK1 axis is essential for glioma stem cell self-renewal and temozolomide resistance. Oncotarget. 2018;9:192–204. doi: 10.18632/oncotarget.22773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wilding JL, Bodmer WF. Cancer cell lines for drug discovery and development. Cancer Res. 2014;74:2377–2384. doi: 10.1158/0008-5472.CAN-13-2971. [DOI] [PubMed] [Google Scholar]
- 63.Azzarelli R. Organoid models of glioblastoma to study brain tumor stem cells. Front Cell Dev Biol. 2020;8:220. doi: 10.3389/fcell.2020.00220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jin WL, Jin MZ, Tu YY. Organoids: a platform ready for glioblastoma precision medicine? Trends Cancer. 2020;6:265–267. doi: 10.1016/j.trecan.2020.01.016. [DOI] [PubMed] [Google Scholar]
- 65.Dijkstra KK, Cattaneo CM, Weeber F, Chalabi M, van de Haar J, Fanchi LF, Slagter M, van der Velden DL, Kaing S, Kelderman S, van Rooij N, van Leerdam ME, Depla A, Smit EF, Hartemink KJ, de Groot R, Wolkers MC, Sachs N, Snaebjornsson P, Monkhorst K, Haanen J, Clevers H, Schumacher TN, Voest EE. Generation of tumor-reactive T cells by co-culture of peripheral blood lymphocytes and tumor organoids. Cell. 2018;174:1586–1598. e12. doi: 10.1016/j.cell.2018.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang H, Xu T, Huang Q, Jin W, Chen J. Immunotherapy for malignant glioma: current status and future directions. Trends Pharmacol Sci. 2020;41:123–138. doi: 10.1016/j.tips.2019.12.003. [DOI] [PubMed] [Google Scholar]
- 67.Corte CMD, Barra G, Ciaramella V, Di Liello R, Vicidomini G, Zappavigna S, Luce A, Abate M, Fiorelli A, Caraglia M, Santini M, Martinelli E, Troiani T, Ciardiello F, Morgillo F. Antitumor activity of dual blockade of PD-L1 and MEK in NSCLC patients derived three-dimensional spheroid cultures. J Exp Clin Cancer Res. 2019;38:253. doi: 10.1186/s13046-019-1257-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ye W, Luo C, Li C, Huang J, Liu F. Organoids to study immune functions, immunological diseases and immunotherapy. Cancer Lett. 2020;477:31–40. doi: 10.1016/j.canlet.2020.02.027. [DOI] [PubMed] [Google Scholar]
- 69.Neal JT, Li X, Zhu J, Giangarra V, Grzeskowiak CL, Ju J, Liu IH, Chiou SH, Salahudeen AA, Smith AR, Deutsch BC, Liao L, Zemek AJ, Zhao F, Karlsson K, Schultz LM, Metzner TJ, Nadauld LD, Tseng YY, Alkhairy S, Oh C, Keskula P, Mendoza-Villanueva D, De La Vega FM, Kunz PL, Liao JC, Leppert JT, Sunwoo JB, Sabatti C, Boehm JS, Hahn WC, Zheng GXY, Davis MM, Kuo CJ. Organoid modeling of the tumor immune microenvironment. Cell. 2018;175:1972–1988. e16. doi: 10.1016/j.cell.2018.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sun L, Yang H, Mao Y. Programmed cell death protein 1/programmed death ligand-1 checkpoint blockade meets patient-derived organoids. Ann Transl Med. 2019;7:S287. doi: 10.21037/atm.2019.11.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ootani A, Li X, Sangiorgi E, Ho QT, Ueno H, Toda S, Sugihara H, Fujimoto K, Weissman IL, Capecchi MR, Kuo CJ. Sustained in vitro intestinal epithelial culture within a Wnt-dependent stem cell niche. Nat Med. 2009;15:701–706. doi: 10.1038/nm.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang C, Liu J, Zhong JF, Zhang X. Engineering CAR-T cells. Biomark Res. 2017;5:22. doi: 10.1186/s40364-017-0102-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.June CH, Sadelain M. Chimeric antigen receptor therapy. N Engl J Med. 2018;379:64–73. doi: 10.1056/NEJMra1706169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sanmamed MF, Chen L. A paradigm shift in cancer immunotherapy: from enhancement to normalization. Cell. 2019;176:677. doi: 10.1016/j.cell.2019.01.008. [DOI] [PubMed] [Google Scholar]
- 75.Brown CE, Badie B, Barish ME, Weng L, Ostberg JR, Chang WC, Naranjo A, Starr R, Wagner J, Wright C, Zhai Y, Bading JR, Ressler JA, Portnow J, D’Apuzzo M, Forman SJ, Jensen MC. Bioactivity and safety of IL13Ralpha2-redirected chimeric antigen receptor CD8+ T cells in patients with recurrent glioblastoma. Clin Cancer Res. 2015;21:4062–4072. doi: 10.1158/1078-0432.CCR-15-0428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Brown CE, Alizadeh D, Starr R, Weng L, Wagner JR, Naranjo A, Ostberg JR, Blanchard MS, Kilpatrick J, Simpson J, Kurien A, Priceman SJ, Wang X, Harshbarger TL, D’Apuzzo M, Ressler JA, Jensen MC, Barish ME, Chen M, Portnow J, Forman SJ, Badie B. Regression of glioblastoma after chimeric antigen receptor T-Cell therapy. N Engl J Med. 2016;375:2561–2569. doi: 10.1056/NEJMoa1610497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ahmed N, Brawley V, Hegde M, Bielamowicz K, Kalra M, Landi D, Robertson C, Gray TL, Diouf O, Wakefield A, Ghazi A, Gerken C, Yi Z, Ashoori A, Wu MF, Liu H, Rooney C, Dotti G, Gee A, Su J, Kew Y, Baskin D, Zhang YJ, New P, Grilley B, Stojakovic M, Hicks J, Powell SZ, Brenner MK, Heslop HE, Grossman R, Wels WS, Gottschalk S. HER2-specific chimeric antigen receptor-modified virus-specific T cells for progressive glioblastoma: a phase 1 dose-escalation trial. JAMA Oncol. 2017;3:1094–1101. doi: 10.1001/jamaoncol.2017.0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.O’Rourke DM, Nasrallah MP, Desai A, Melenhorst JJ, Mansfield K, Morrissette JJD, Martinez-Lage M, Brem S, Maloney E, Shen A, Isaacs R, Mohan S, Plesa G, Lacey SF, Navenot JM, Zheng Z, Levine BL, Okada H, June CH, Brogdon JL, Maus MV. A single dose of peripherally infused EGFRvIII-directed CAR T cells mediates antigen loss and induces adaptive resistance in patients with recurrent glioblastoma. Sci Transl Med. 2017;9:eaaa0984. doi: 10.1126/scitranslmed.aaa0984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Johnson LA, Scholler J, Ohkuri T, Kosaka A, Patel PR, McGettigan SE, Nace AK, Dentchev T, Thekkat P, Loew A, Boesteanu AC, Cogdill AP, Chen T, Fraietta JA, Kloss CC, Posey AD Jr, Engels B, Singh R, Ezell T, Idamakanti N, Ramones MH, Li N, Zhou L, Plesa G, Seykora JT, Okada H, June CH, Brogdon JL, Maus MV. Rational development and characterization of humanized anti-EGFR variant III chimeric antigen receptor T cells for glioblastoma. Sci Transl Med. 2015;7 doi: 10.1126/scitranslmed.aaa4963. 275ra22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schnalzger TE, de Groot MH, Zhang C, Mosa MH, Michels BE, Roder J, Darvishi T, Wels WS, Farin HF. 3D model for CAR-mediated cytotoxicity using patient-derived colorectal cancer organoids. EMBO J. 2019;38:e100928. doi: 10.15252/embj.2018100928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Brown MP, Ebert LM, Gargett T. Clinical chimeric antigen receptor-T cell therapy: a new and promising treatment modality for glioblastoma. Clin Transl Immunol. 2019;8:e1050. doi: 10.1002/cti2.1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pageon SV, Govendir MA, Kempe D, Biro M. Mechanoimmunology: molecular-scale forces govern immune cell functions. Mol Biol Cell. 2018;29:1919–1926. doi: 10.1091/mbc.E18-02-0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ashok A, Choudhury D, Fang Y, Hunziker W. Towards manufacturing of human organoids. Biotechnol Adv. 2020;39:107460. doi: 10.1016/j.biotechadv.2019.107460. [DOI] [PubMed] [Google Scholar]
- 84.Jin BJ, Battula S, Zachos N, Kovbasnjuk O, Fawlke-Abel J, In J, Donowitz M, Verkman AS. Microfluidics platform for measurement of volume changes in immobilized intestinal enteroids. Biomicrofluidics. 2014;8:024106. doi: 10.1063/1.4870400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160:48–61. doi: 10.1016/j.cell.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yin X, Mead BE, Safaee H, Langer R, Karp JM, Levy O. Engineering stem cell organoids. Cell Stem Cell. 2016;18:25–38. doi: 10.1016/j.stem.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Heitman N, Saxena N, Rendl M. Advancing insights into stem cell niche complexities with next-generation technologies. Curr Opin Cell Biol. 2018;55:87–95. doi: 10.1016/j.ceb.2018.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liu SJ, Malatesta M, Lien BV, Saha P, Thombare SS, Hong SJ, Pedraza L, Koontz M, Seo K, Horlbeck MA, He D, Birk HS, Jain M, Olsen HE, Akeson M, Weissman JS, Monje M, Gupta N, Raleigh DR, Ullian EM, Lim DA. CRISPRi-based radiation modifier screen identifies long non-coding RNA therapeutic targets in glioma. Genome Biol. 2020;21:83. doi: 10.1186/s13059-020-01995-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.van de Wetering M, Francies HE, Francis JM, Bounova G, Iorio F, Pronk A, van Houdt W, van Gorp J, Taylor-Weiner A, Kester L, McLaren-Douglas A, Blokker J, Jaksani S, Bartfeld S, Volckman R, van Sluis P, Li VS, Seepo S, Sekhar Pedamallu C, Cibulskis K, Carter SL, McKenna A, Lawrence MS, Lichtenstein L, Stewart C, Koster J, Versteeg R, van Oudenaarden A, Saez-Rodriguez J, Vries RG, Getz G, Wessels L, Stratton MR, McDermott U, Meyerson M, Garnett MJ, Clevers H. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell. 2015;161:933–945. doi: 10.1016/j.cell.2015.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bartfeld S, Bayram T, van de Wetering M, Huch M, Begthel H, Kujala P, Vries R, Peters PJ, Clevers H. In vitro expansion of human gastric epithelial stem cells and their responses to bacterial infection. Gastroenterology. 2015;148:126–136. e6. doi: 10.1053/j.gastro.2014.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Boj SF, Hwang CI, Baker LA, Chio II, Engle DD, Corbo V, Jager M, Ponz-Sarvise M, Tiriac H, Spector MS, Gracanin A, Oni T, Yu KH, van Boxtel R, Huch M, Rivera KD, Wilson JP, Feigin ME, Ohlund D, Handly-Santana A, Ardito-Abraham CM, Ludwig M, Elyada E, Alagesan B, Biffi G, Yordanov GN, Delcuze B, Creighton B, Wright K, Park Y, Morsink FH, Molenaar IQ, Borel Rinkes IH, Cuppen E, Hao Y, Jin Y, Nijman IJ, Iacobuzio-Donahue C, Leach SD, Pappin DJ, Hammell M, Klimstra DS, Basturk O, Hruban RH, Offerhaus GJ, Vries RG, Clevers H, Tuveson DA. Organoid models of human and mouse ductal pancreatic cancer. Cell. 2015;160:324–338. doi: 10.1016/j.cell.2014.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Drost J, Karthaus WR, Gao D, Driehuis E, Sawyers CL, Chen Y, Clevers H. Organoid culture systems for prostate epithelial and cancer tissue. Nat Protoc. 2016;11:347–358. doi: 10.1038/nprot.2016.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fujii M, Shimokawa M, Date S, Takano A, Matano M, Nanki K, Ohta Y, Toshimitsu K, Nakazato Y, Kawasaki K, Uraoka T, Watanabe T, Kanai T, Sato T. A colorectal tumor organoid library demonstrates progressive loss of niche factor requirements during tumorigenesis. Cell Stem Cell. 2016;18:827–838. doi: 10.1016/j.stem.2016.04.003. [DOI] [PubMed] [Google Scholar]
- 94.Broutier L, Mastrogiovanni G, Verstegen MM, Francies HE, Gavarro LM, Bradshaw CR, Allen GE, Arnes-Benito R, Sidorova O, Gaspersz MP, Georgakopoulos N, Koo BK, Dietmann S, Davies SE, Praseedom RK, Lieshout R, IJzermans JMJ, Wigmore SJ, Saeb-Parsy K, Garnett MJ, van der Laan LJ, Huch M. Human primary liver cancer-derived organoid cultures for disease modeling and drug screening. Nat Med. 2017;23:1424–1435. doi: 10.1038/nm.4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Turco MY, Gardner L, Hughes J, Cindrova-Davies T, Gomez MJ, Farrell L, Hollinshead M, Marsh SGE, Brosens JJ, Critchley HO, Simons BD, Hemberger M, Koo BK, Moffett A, Burton GJ. Long-term, hormone-responsive organoid cultures of human endometrium in a chemically defined medium. Nat Cell Biol. 2017;19:568–577. doi: 10.1038/ncb3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sachs N, de Ligt J, Kopper O, Gogola E, Bounova G, Weeber F, Balgobind AV, Wind K, Gracanin A, Begthel H, Korving J, van Boxtel R, Duarte AA, Lelieveld D, van Hoeck A, Ernst RF, Blokzijl F, Nijman IJ, Hoogstraat M, van de Ven M, Egan DA, Zinzalla V, Moll J, Boj SF, Voest EE, Wessels L, van Diest PJ, Rottenberg S, Vries RGJ, Cuppen E, Clevers H. A living biobank of breast cancer organoids captures disease heterogeneity. Cell. 2018;172:373–386. e10. doi: 10.1016/j.cell.2017.11.010. [DOI] [PubMed] [Google Scholar]
- 97.Tian L, Goldstein A, Wang H, Lo HC, Kim IS, Welte T, Sheng K, Dobrolecki LE, Zhang X, Putluri N, Phung TL, Mani SA, Stossi F, Sreekumar A, Mancini MA, Decker WK, Zong C, Lewis MT, Zhang XH. Mutual regulation of tumour vessel normalization and immunostimulatory reprogramming. Nature. 2017;544:250–254. doi: 10.1038/nature21724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Munn LL, Jain RK. Vascular regulation of antitumor immunity. Science. 2019;365:544–545. doi: 10.1126/science.aaw7875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ibrahim M, Richardson MK. Beyond organoids: in vitro vasculogenesis and angiogenesis using cells from mammals and zebrafish. Reprod Toxicol. 2017;73:292–311. doi: 10.1016/j.reprotox.2017.07.002. [DOI] [PubMed] [Google Scholar]
- 100.Lauschke K, Frederiksen L, Hall VJ. Paving the way toward complex blood-brain barrier models using pluripotent stem cells. Stem Cells Dev. 2017;26:857–874. doi: 10.1089/scd.2017.0003. [DOI] [PubMed] [Google Scholar]
- 101.Pham MT, Pollock KM, Rose MD, Cary WA, Stewart HR, Zhou P, Nolta JA, Waldau B. Generation of human vascularized brain organoids. Neuroreport. 2018;29:588–593. doi: 10.1097/WNR.0000000000001014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ham O, Jin YB, Kim J, Lee MO. Blood vessel formation in cerebral organoids formed from human embryonic stem cells. Biochem Biophys Res Commun. 2020;521:84–90. doi: 10.1016/j.bbrc.2019.10.079. [DOI] [PubMed] [Google Scholar]
- 103.Cakir B, Xiang Y, Tanaka Y, Kural MH, Parent M, Kang YJ, Chapeton K, Patterson B, Yuan Y, He CS, Raredon MSB, Dengelegi J, Kim KY, Sun P, Zhong M, Lee S, Patra P, Hyder F, Niklason LE, Lee SH, Yoon YS, Park IH. Engineering of human brain organoids with a functional vascular-like system. Nat Methods. 2019;16:1169–1175. doi: 10.1038/s41592-019-0586-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Camp JG, Badsha F, Florio M, Kanton S, Gerber T, Wilsch-Brauninger M, Lewitus E, Sykes A, Hevers W, Lancaster M, Knoblich JA, Lachmann R, Paabo S, Huttner WB, Treutlein B. Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. Proc Natl Acad Sci U S A. 2015;112:15672–15677. doi: 10.1073/pnas.1520760112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pollen AA, Bhaduri A, Andrews MG, Nowakowski TJ, Meyerson OS, Mostajo-Radji MA, Di Lullo E, Alvarado B, Bedolli M, Dougherty ML, Fiddes IT, Kronenberg ZN, Shuga J, Leyrat AA, West JA, Bershteyn M, Lowe CB, Pavlovic BJ, Salama SR, Haussler D, Eichler EE, Kriegstein AR. Establishing cerebral organoids as models of human-specific brain evolution. Cell. 2019;176:743–756. e17. doi: 10.1016/j.cell.2019.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Velasco S, Kedaigle AJ, Simmons SK, Nash A, Rocha M, Quadrato G, Paulsen B, Nguyen L, Adiconis X, Regev A, Levin JZ, Arlotta P. Individual brain organoids reproducibly form cell diversity of the human cerebral cortex. Nature. 2019;570:523–527. doi: 10.1038/s41586-019-1289-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bhaduri A, Andrews MG, Leon WM, Jung D, Shin D, Allen D, Jung D, Schmunk G, Haeussler M, Salma J, Pollen AA, Nowakowski TJ, Kriegstein AR. Cell stress in cortical organoids impairs molecular subtype specification. Nature. 2020;578:142–148. doi: 10.1038/s41586-020-1962-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mao XY, Dai JX, Zhou HH, Liu ZQ, Jin WL. Brain tumor modeling using the CRISPR/Cas9 system: state of the art and view to the future. Oncotarget. 2016;7:33461–33471. doi: 10.18632/oncotarget.8075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Haapaniemi E, Botla S, Persson J, Schmierer B, Taipale J. CRISPR-Cas9 genome editing induces a p53-mediated DNA damage response. Nat Med. 2018;24:927–930. doi: 10.1038/s41591-018-0049-z. [DOI] [PubMed] [Google Scholar]
- 110.Artegiani B, Hendriks D, Beumer J, Kok R, Zheng X, Joore I, Chuva de Sousa Lopes S, van Zon J, Tans S, Clevers H. Fast and efficient generation of knock-in human organoids using homology-independent CRISPR-Cas9 precision genome editing. Nat Cell Biol. 2020;22:321–331. doi: 10.1038/s41556-020-0472-5. [DOI] [PubMed] [Google Scholar]


