Abstract
NFYA (nuclear transcription factor Y, subunit A) is a CCAAT-binding transcription factor. Accumulating evidence suggests that NFYA plays an important role in breast, ovarian, lung and gastric cancer. However, the role of NFYA in clear cell renal cell carcinoma (ccRCC) remains unclear. In this study, it was discovered that the expression of NFYA is elevated in tissues of ccRCC patient and high NFYA expression is linked to poor overall survival in ccRCC patient. Inhibition of G1/S cell cycle transition and decreased cell proliferation were observed upon NFYA knockdown in ccRCC cells. Moreover, further investigation revealed that NFYA binds directly to the promoter region of both CDK4 and cyclin D1 (CCND1) thus transactivating their expression, resulting in RB phosphorylation and the activation of subsequent E2F pathway activation. Taken together, these findings imply the oncogenic role of NFYA in ccRCC progression and its potential as a target for ccRCC therapy.
Keywords: NFYA, G1/S transition, cyclin D1, CDK4, ccRCC
Introduction
Renal cell carcinoma (RCC) is one of the most common types of malignancy. Its incidence and mortality rates have ascended steadily in the past decade [1,2]. RCC can be classified into three major subtypes based on its histologically characteristic: chromophobe RCC (chRCC), papillary RCC (pRCC) and clear cell RCC (ccRCC) [3]. Among them, ccRCC accounts for more than 80% of all RCCs and represents the most malignant subtype [4]. Advanced RCC shows poor prognosis with 5-year survival of only 11.7% [5], and it usually exhibits resistance to traditional radiotherapy and chemotherapy [6,7]. Immunotherapy and tyrosine kinase inhibitor have been developed for RCC therapy in recent years, however, the prognosis for advanced RCC remains suboptimal [8,9]. Thus, identification of genes involved in the pathological procession of RCC might helpful for uncovering novel target.
Transcription factors are protein molecules that bind with specific DNA sequence to control the transcription of genetic information [10]. Emerging evidences have shown that transcription factors play important role in physiological and pathological processes, including neurodegenerative pathologies, diabetes and cancers [11]. Transcription factors are dysregulated in various types of cancers and play key roles in regulating self-renewal, differentiation, chemotherapy resistance, immune evasion, EMT (epithelial-mesenchymal transition), and proliferation of cancer cells [12]. Several transcription factors have been identified to involve in the progression of ccRCC [13]. The most famous one is HIF1α, which is induced in oxygen-deficient tumor microenvironment and promotes ccRCC aggressiveness by activating a number of oncogenes, such as VEGFA, HK2 and PGK1 [14]. Hence, the HIF1α inhibitor CRLX101 is designed for ccRCC therapy and shows beneficial in clinical trials [15]. Thus, transcription factor is potential ccRCC therapy target [12] and identifying novel ccRCC-associtated transcription factors will be helpful for ccRCC treatment.
As a subunit of nuclear transcription factor Y (NF-Y), NFYA usually forms a complex with NFYB and NFYC and specifically binding to the CCAAT cis-elements [16]. NFYA binds directly to DNA elements through its DNA-binding domain and is differentially expressed, severing as the regulatory subunit, while NFYB and NFYC form heterodimers to interact with NFYA and are constitutively expressed [17]. More and more evidences have emerged to suggest that NFYA plays an important role in various cancers, with its expression being elevated in lung cancer, ovarian cancer, breast cancer and gastric cancer [18-22]. Besides, it also regulates the metastasis, proliferation and progression of cancer cells through transcriptional mechanism [23-26]. For instance, NFYA binds to the CCAAT sites of EZH2 promoter, which would then upregulate the expression of EZH2 and H3K27me3, resulting in accelerated proliferation of human epithelial ovarian cancer (EOC) cells [27]. Despite serving as an oncoprotein in several cancers, the role of NFYA in ccRCC is still poorly characterized.
To identify ccRCC-associated transcription factors, we analyzed the expression patterns of 1665 human transcription factors in two ccRCC microarray cohorts, GSE6344 (10 ccRCC and paired-matched adjacent normal tissues) and GSE66272 (27 ccRCC and paired-matched adjacent normal tissues). The upregulation of NFYA was discovered in tissues of ccRCC patients. Furthermore, it was observed that NFYA promotes G1/S cell cycle transition and cell proliferation in ccRCC cell lines. Interestingly, NFYA binds to the promoter region of both CDK4 and cyclin D1, which then promotes the transcription of CDK4/cyclin D1 and subsequently leading to activation of RB-E2F pathways. These findings strongly suggest the involvement of the NFYA-CDK4/cyclin D1-RB-E2F axis in ccRCC progression.
Material and methods
Cell culture and siRNA transfection
769-P, ACHN, Caki-1, 786-O and 293T cells were obtained from the Cell Bank of the Chinese Academy of Science; while, A498 cells were derived from ATCC. Cells were cultivated in DMEM supplemented with 10% FBS and 1% penicillin/streptomycin under standard cell culture environment (37°C with 5% CO2). Small-interfering RNA (Si-RNA) transfection was performed using Lipofectamine RNAiMAX Reagent (Invitrogen) according to its manual. siRNAs targeting NFYA (NFYA-1: CTACGTGAATGCCAAACAA; NFYA-2: GGAGGCCAGCTAATCACAT), and negative control si-RNAs were purchased from RiboBio.
Cell proliferation assay
siRNA transfected ccRCC cells were seeded in 96-well plate at 1000 cells/well and cultured in DMEM (10% FBS) for specific period of time as indicated. 10 μL of MTT (5 mg/mL) solution was added into each well and was incubated for 3-4 hours in a humidified chamber. The reaction was stopped by adding 100 μL of 20% SDS and was mixed thoroughly until formazan dissolved completely. The absorbance was then measured at 570 nm.
For colony formation assay, ccRCC cells were seeded in 96-well plate at 100 cells/well and cultured in DMEM (10% FBS) for approximately 10 days. After that, cells were fixed with 4% paraformaldehyde for 20 minutes and stained with crystal violet. The clones then dissolved with 200 μL glacial acetic acid. The absorbance was measured at 570 nm.
Cell cycle analysis
ccRCC cells were digested and harvested by trypsin, washed with PBS and fixed in 70% ice-cold ethanol overnight. Fixed cells were washed with PBS for two times, followed by RNase A treatment and PI staining (50 mg/mL). PI labeled samples were analyzed with a CytoFLEX (Beckman) flow cytometer. The ModFit LT 5.0 software was used to calculate cell cycle distribution.
Cell synchronization and release
Control or NFYA si-RNAs were transfected into ACHN and 769-P cells. 24 hours later, the ACHN and 769-P cells were synchronized in FBS-free DMEM for 48 hours. After that, cells were digested by trypsin and seeded into 6-cm dishes for culturing in 10% FBS medium. Cells were harvested at the specific time (0, 15, 20 and 25 hours) and fixed in 70% ice-cold ethanol overnight. Cell cycle distribution was subsequently analyzed as previously described.
RNA-sequencing
To explore the transcriptome changes after NFYA knockdown, ACHN, 769P and A498 cells were transfected with si-NFYA and then cultured in humidified chamber for 60 hours. After that, total RNA was extracted from cells using Trizol reagent. mRNA library construction, single end sequencing and high throughput data analysis were executed by BGI. Differentially expressed genes (DEGs) were set as fold change ≥ 1.5 and FDR ≤ 0.001.
Bioinformatics and data analysis
NFYA expression profiles in GSE53757, GSE14994, GSE66272, GSE781 and GSE6344 [28-34] datasets were downloaded in GEO. NFYA mRNA expression and survival information of GSE4125, GSE3538 and GSE2109 [35,36] were downloaded in the Oncomine platform. The NFYA median expression was used as the cutoff value in determining NFYA expression level as being high or low. Expression profiles of NFYA in TCGA-KIRC (kidney renal cell carcinoma) and its expression correlation with cyclin D1/CDK4 was obtained from GEPIA2 (http://gepia2.cancer-pku.cn/#index) [37]. The protein expression levels of NFYA and SSRP1 in ccRCC and normal tissues was derived from UALCAN (http://ualcan.path.uab.edu/index.html) [38]. H3K27ac (ENCFF388WMD), H3K4me3 (ENCFF045NNJ) and NFYA (ENCFF000XIR) ChIP-seq bigwig files were downloaded in ENCODE (https://www.encodeproject.org/) [39,40] and visualized using IGV (Integrative Genomics Viewer) software.
RNA isolation and real time qPCR
Total RNA was purified using RNA Quick Purification kit (ESscience) according to the standard protocol. cDNA was synthesized with PrimerScript RT-PCR kit (Takara). The cDNA templates were amplified using CFX96 real time instrument (Biorad). The expression of each gene was calculated using the 2-ΔΔCt method. GAPDH was used as the internal control. The qPCR primers are listed in Table 1.
Table 1.
The primers used in real time qPCR are listed as follows
| Primer Name | Sequence 5’-3’ |
|---|---|
| CCND1 Forward | GCTGCGAAGTGGAAACCATC |
| CCND1 Reverse | CCTCCTTCTGCACACATTTGAA |
| CDK4 Forward | GGACATGTGGAGTGTTGGCT |
| CDK4 Reverse | CAACTGGTCGGCTTCAGAGT |
| NFYA Forward | CAGTGGAGGCCAGCTAATCAC |
| NFYA Reverse | CCAGGTGGGACCAACTGTATT |
| c-Myc Forward | GTCAAGAGGCGAACACACAAC |
| c-Myc Reverse | TTGGACGGACAGGATGTATGC |
| GAPDH Forward | CGACCACTTTGTCAAGCTCA |
| GAPDH Reverse | TTACTCCTTGGAGGCCATGT |
Abbreviations: CCND1: cyclin D1.
Western blot
Western blot was performed as previously described [41,42]. Primary antibodies specific to CDK4 (CST, mouse), CCND1 (CST, mouse), RB (CST, mouse), p-RB S795 (CST, rabbit), p-RB S807/811 (CST, rabbit), C-Myc (Transgene, mouse), NFYA (Santa Cruze, mouse) and GAPDH (Transgene, mouse) were used. The blots were incubated with goat anti-rabbit or anti-mouse secondary antibody (Transgene, 1:10000), and subsequently visualized using chemiluminescence (Beyotime).
ChIP-qPCR
About 1×107 ccRCC cells were fixed with 1% formaldehyde (Thermofisher) for 10 minutes at room temperature and the reaction was then terminated by glycine. Fixed cells were harvested and lysed with lysis buffer (NaCl 4.38 g, EDTA 0.731 g, Tris-base 3.03 g, NP40 2.5 mL, pH 7.5, H2O up to 500 mL), followed by centrifugation at 13000 rpm for 5 minutes to aspirate the supernatant. Cell nuclear pellet was suspended with 300 μL shearing buffer (20% SDS 25 mL, EDTA 1.461 g, Tris-base 3.03 g, pH 8.0, H2O up to 500 mL) and subjected to Bioruptor ultrasonicator in order to obtain the fragmented chromatin. For immunoprecipitation, precleared solubilized chromatin was incubated with 5 μg anti-NFYA (Santa Cruze, mouse) or IgG antibody overnight at 4°C, followed by the addition of 30 μL Dynabeads Protein G (Invitrogen). Immunoprecipitates were washed with lysis buffer for 6 times and DNA was eluted with elution buffer (1% SDS, 0.1 M NaHCO3). After reverse-crosslink, RNase A and Proteinase K treatment, immunoprecipitated DNA was purified with PCR purification kit (Thermofisher), followed by qPCR analysis. The ChIP-qPCR primers are listed in Table 2.
Table 2.
The primers used in ChIP-qPCR are listed as follows
| Primer Name | Sequence 5’-3’ |
|---|---|
| CCND1-P1 Forward | ACTGGTCAAGGTAGGAAGGC |
| CCND1-P1 Reverse | CAACCCCTGTGCAAGTTTCA |
| CCND1-P2 Forward | ATGACCCTCAAAAGCCCAGA |
| CCND1-P2 Reverse | GGTTAGCGAGCGTAAAGAGC |
| CCND1-P3 Forward | CCGGTCCGCCTAGTAACAG |
| CCND1-P3 Reverse | GCTATCACCCGGCCTCTC |
| CCND1-NC Forward | CTGCCCCGTGTTATCCTTTG |
| CCND1-NC Reverse | CGAAGACAGCAACATCCCAG |
| CDK4-P1 Forward | CGCTTGACATTGCTCTGAGG |
| CDK4-P1 Reverse | TTCAGCCTTCAGACCGGTAG |
| CDK4-P2 Forward | CATGTGACCAGCTGCCAAAG |
| CDK4-P2 Reverse | TAGCAACAGATCACGTGGCT |
| CDK4-P3 Forward | GCACTGGTTCTCATTCCTGG |
| CDK4-P3 Reverse | CTTTGGCAGCTGGTCACATG |
| CDK4-NC Forward | AATCTGGGAGGTGGAGGTTG |
| CDK4-NC Reverse | CAGTGGCCCAATTACAGCTC |
Abbreviations: CCND1: cyclin D1.
Dual-luciferase reporter assay
CCND1 promoter (-2570 to +421) and CDK4 promoter (-175 to +265) regions were cloned into firefly luciferase reporter vector, pGL3-basic (Promega). pGL3 and renilla luciferase control plasmid (pRL-TK) were co-transfected into 786-O and ACHN cells by using ViaFect reagent (Promega). Cells were harvested at 36 hours after transfection and luciferase activity was measured using Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s manual.
Gene ontology and GSEA
Gene ontology analysis of the differentially expressed genes (DEGs) upon NFYA knockdown was conducted in consensusPath DB (http://cpdb.molgen.mpg.de/) [43]. Expression matrixes of control and ACHN, 769-P and A498 with NFYA knockdown were build, followed by gene set enrichment analysis (GSEA) using GSEA desktop standalone program [44,45].
Results
Upregulation of NFYA in ccRCC patients
To investigate ccRCC-associated transcription factors, we analyzed the expression patterns of 1665 human transcription factors [46] in two ccRCC microarray cohorts, GSE6344 (10 ccRCC and paired-matched adjacent normal tissues) [29,31] and GSE66272 (27 ccRCC and paired-matched adjacent normal tissues) [33,34]. We discovered that 142 and 267 transcription factors were unregulated in GSE6344 and GSE66272 cohorts with > 1.5 fold changes (Figure 1A). In order to identify the key transcription factor that modulated ccRCC progression, we analyzed the Behan’s study [47], which performed genome-scale screen for fitness gene (defined as gene involved in cell growth or viability) in 339 pan-cancer cell lines (including 3 renal cancer cell lines), and discovered 1492 fitness genes (of which 49 are transcription factors) in RCC cell lines. Based on these results, we identified three fitness transcription factors (NFE2L3, NFYA and SSRP1) were unregulated in paired ccRCC tissues (Figure 1B). Interestingly, a recent study also identified NFE2L3 is overexpressed and correlates with poor prognosis in clear cell renal cell carcinoma [48]. We focused on NFYA for the further study since increased protein expression level of NFYA in ccRCC was more significant than SSRP1 (Figure 1C).
Figure 1.
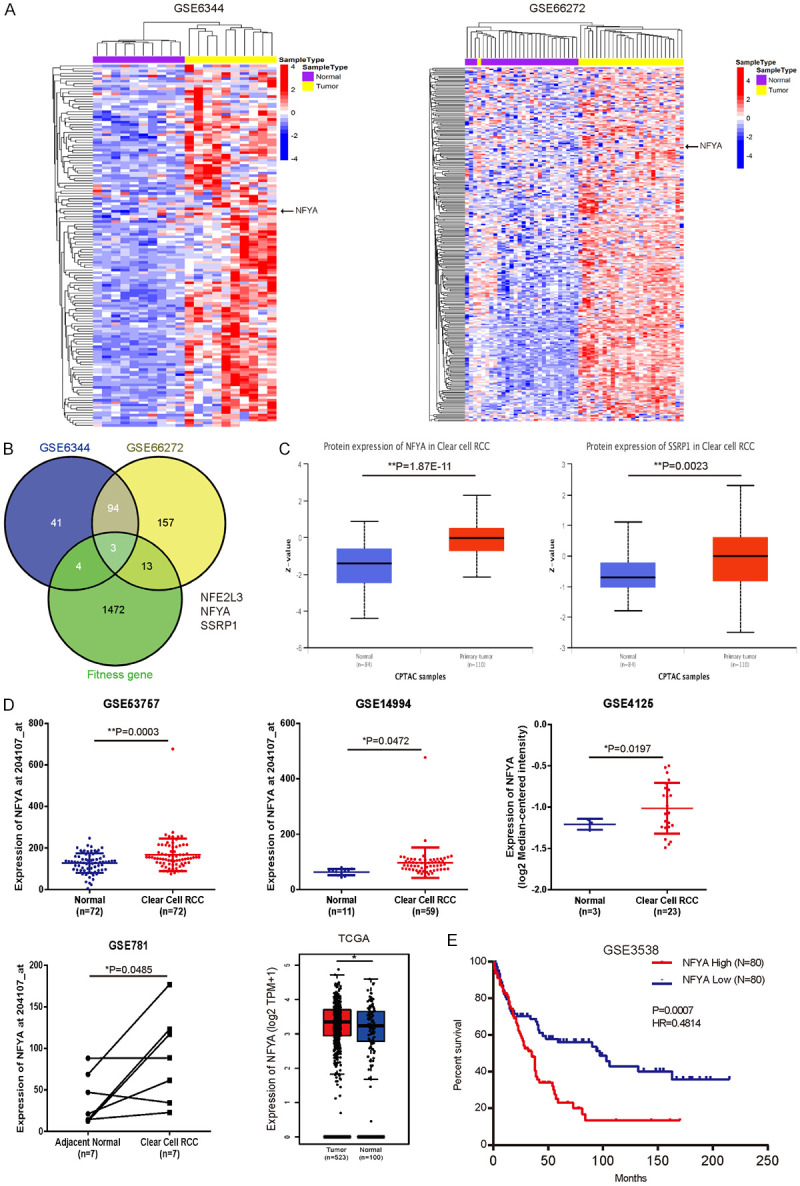
Upregulation of NFYA in ccRCC patients. A. Heatmap of upregulated transcription factors (fold change > 1.5) in ccRCC versus paired-matched adjacent normal tissues (GSE6344 and GSE66272). B. Veen plot of the overlapping genes based on GSE6344 (142 upregulated transcription factors), GSE66272 (267 upregulated transcription factors) and RCC fitness genes. RCC fitness gene was defined as appeared at least 2 out of 3 RCC cell lines in Behan’s study [47]. C. The protein level of NFYA and SSRP1 in primary ccRCC and normal tissues was analyzed from UALCAN [38]. D. mRNA expression of NFYA between ccRCC and normal kidney tissues was analyzed from GSE53757, GSE14994 and GSE4125 cohort. Results were presented as the mean ± standard deviation (SD). TCGA analysis of NFYA expression between kidney renal clear cell carcinoma (KIRC) and normal kidney tissues (including TCGA and GTEx data) was derived from GEPIA [37]. E. Overall survival rates between NFYA-low and NFYA-high groups of the 160 ccRCC patients from GSE3538 were compared. Median NFYA expression was used as the cutoff in survival analysis (log-rank test). *P < 0.05, **P < 0.01.
In an attempt to further evaluate the NFYA expression level in ccRCC tissues, we analyzed the expression pattern of several ccRCC cohorts (GSE53757, GSE14994, GSE4125 and GSE781). The mRNA level of NFYA was significantly upregulated in these ccRCC studies (Figure 1D). Similarly, TCGA cohort (TCGA kidney renal clear cell carcinoma) analysis showed that NFYA expression was also increased in ccRCC (Figure 1D). These findings clearly verify that NFYA expression is upregulated in ccRCC tissues. Moreover, Kaplan-Meier survival analysis of the GSE3538 dataset revealed that the upregulation of NFYA is significantly correlated with poor overall survival in ccRCC patients (Figure 1E). Collectively, these data suggest that NFYA might be an important regulator in the progression of ccRCC.
Knockdown of NFYA inhibits ccRCC cell proliferation
The expression level of NFYA in ccRCC cell lines was analyzed to further examine its role in ccRCC progression. Two NFYA isoforms were identified in ccRCC cell lines through western blot, which is consistent with previous reports [49,50]. The expression level of NFYA was noticed to be relatively high in A498, ACHN, 786-O and 769-P cells (Figure 2A). Hence, we knocked down NFYA expression in these four cells using two siRNAs. NFYA expression was decreased notably in A498, ACHN, 786-O and 769-P cells as a consequence to the gene knockdown (Figure 2B). This depletion in NFYA expression subsequently reduced cell proliferation in all four ccRCC cell lines (Figure 2C). In addition, NFYA knockdown cells formed fewer colonies compared with the control group (Figure 2D), an outcome that is consistent with the MTT assay data. Taken together, these data indicate that NFYA depletion inhibits ccRCC cell growth in vitro.
Figure 2.
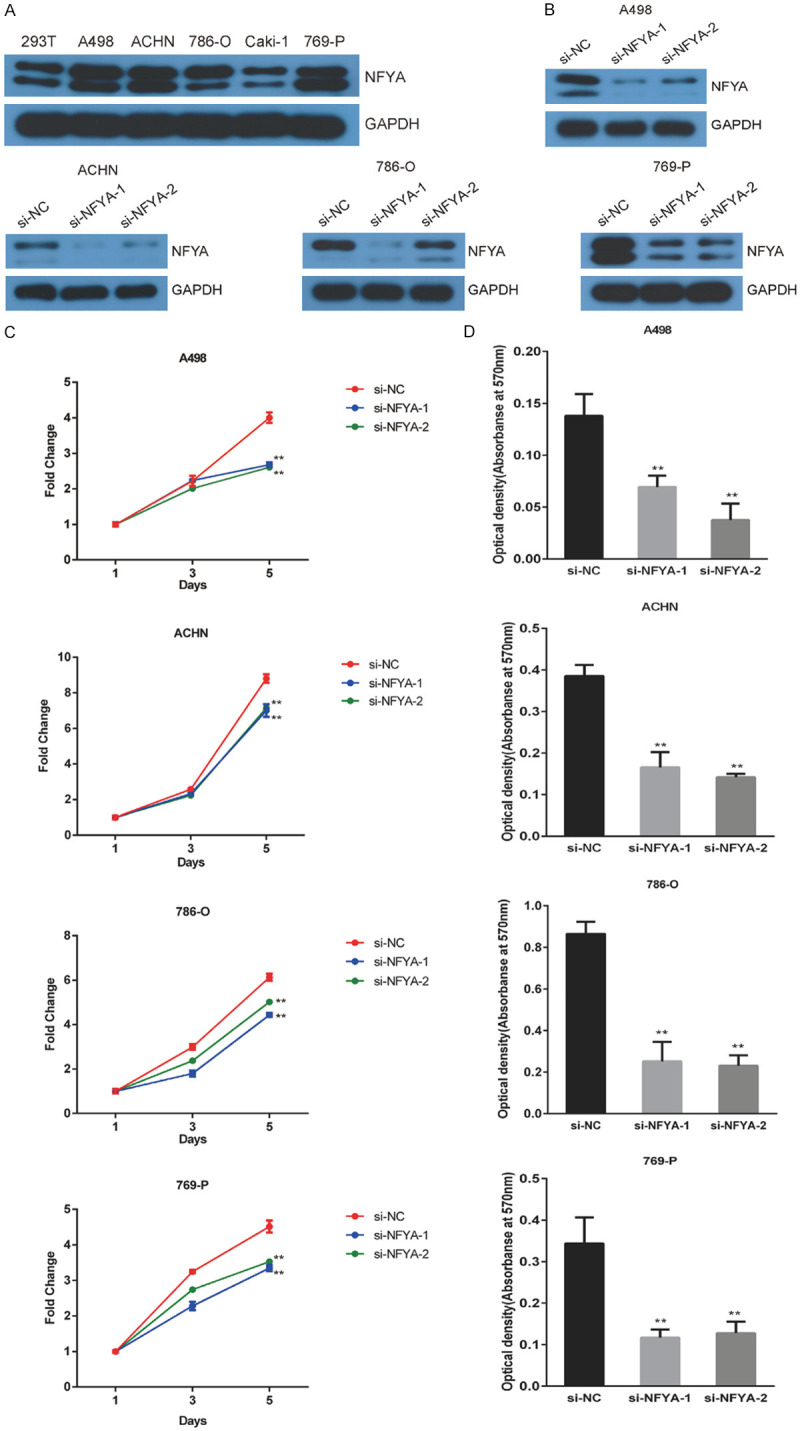
Knockdown of NFYA inhibits ccRCC cell proliferation. A. The expression of NFYA was measured in 293T, A498, ACHN, 786-O, Caki-1 and 769-P cells using western blot. B. The efficiency of NFYA knockdown in A498, ACHN, 786-O and 769-P cells by two si-RNAs was verified by western blot. C. Control or NFYA si-RNAs were transfected into ccRCC cells and cell viability was evaluated with MTT assays. Results were presented as the mean ± standard deviation (SD). D. The effect of NFYA knockdown on colony formation was measured in ccRCC cells. The clones were dissolved with glacial acetic acid and the absorbance was measured at 570 nm. The histogram shows the mean ± standard deviation (SD). **P < 0.01.
Transcriptome analysis of NFYA-regulated genes and processes
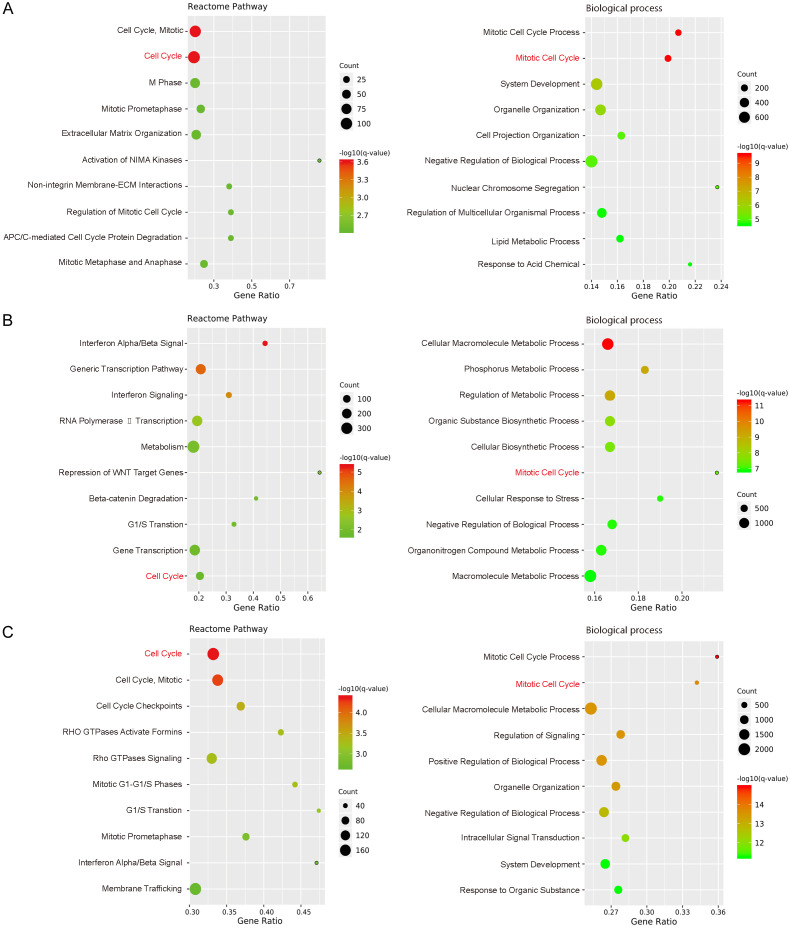
To further explore the role of NFYA in regulating cell proliferation, whole transcriptome sequencing (RNA-seq) was performed to identify the target genes of NFYA in ACHN, 769-P and A498 cells. Differentially expressed genes (DEGs) were set as fold change ≥ 1.5 and FDR ≤ 0.001 upon NFYA knock down. Pathway enrichment analysis revealed that NFYA-regulated genes were highly enriched in the cell cycle of all three cells (Figure 3). Meanwhile, gene ontology analysis of the differentially expressed genes indicated that NFYA depletion affected process enrichment mainly in mitotic cell cycle and metabolism (Figure 3). These analyses imply the potentially crucial role of NFYA in mitotic cell cycle process.
Figure 3.
Transcriptome analysis of NFYA-regulated genes and processes. A-C. Differentially expressed genes (fold change ≥ 1.5 and FDR ≤ 0.001) in ACHN (A), 769-P (B) and A498 (C) cells upon NFYA knockdown were analyzed in ConsensusPathDB. The top 10 Reactome pathways and biological processes were displayed.
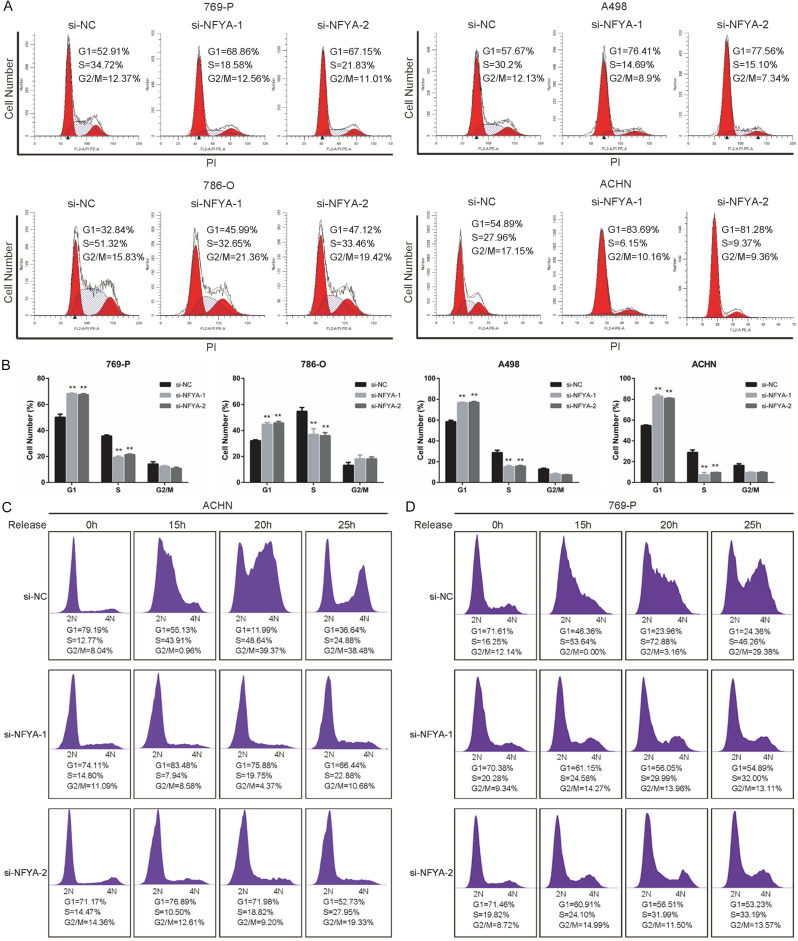
NFYA is required for G1/S cell cycle transition
In order to validate the result of the RNA-seq analysis, flow cytometer assay was performed to evaluate the involvement of NFYA in cell cycle control. It can be observed from the results that, NFYA depletion decreased A498, ACHN, 786-O and 769-P cell population in S phase whereas the proportion of G1 phase markedly increased (Figure 4A and 4B), suggesting the requirement of NFYA for G1/S cell cycle transition. To further assess the effect of NFYA on G1/S transition, control or NFYA si-RNA transfected ACHN and 769-P cells were synchronized in serum-free medium for 48 hours and release into normal medium (10% FBS) for culturing. As is shown in Figure 4C and 4D, most cells (more than 70%) were synchronized during G1 phase. Notably, the percentage of ACHN and 769-P control cells in S phase showed a dramatic increase at 15 and 20 hours, whereas increase in the NFYA knockdown group was minimal (Figure 4C and 4D). These results imply that the knockdown of NFYA inhibits ccRCC cell proliferation by inducing G1/S cell cycle arrest.
Figure 4.
NFYA is required for G1/S cell cycle transition. A, B. 769-P, A498, 786-O and ACHN cells were transfected with NFYA siRNAs or control siRNA for 72 hours, and the cell cycle distribution was analyzed with flow cytometry. The histogram showed the mean ± standard deviation (SD). **P < 0.01. C, D. siRNA transfected ACHN and 769-P cells were synchronized in FBS free DMEM for 48 hours and released into 10% FBS medium for culturing. The cell cycle was analyzed by flow cytometry at the indicated time.
NFYA activates the RB-E2F pathway by upregulating CDK4/CCND1
To investigate the molecular mechanism of NFYA in regulating cell cycle and cell proliferation, we performed gene set enrichment analysis of the RNA-seq data. NFYA-regulated genes were significantly enriched in E2F target gene set in all three ccRCC cell lines (Figure 5A). It has been widely reported that transcription factors E2F play an essential role in G1/S transition by activating downstream genes. There are mainly five members of the E2F family (E2F1, E2F2, E2F3, E2F4 and E2F5) that are involved in G1/S transition. During G0 and early G1 phase, E2F4 or E2F5 forms complex with RB to repress the transcription of target genes. When cells enter late G1 phase, RB is phosphorylated by cyclin D1/CDK4/6 and cyclin E/CDK2 complex, resulting in the disassociation of E2F4/E2F5/RB complex at the promoter region of target genes and the subsequent displacement by E2F1/E2F2/E2F3, followed with transactivation of E2F target genes at the G1/S boundary while entering S phase [51-53]. Therefore, the expression level of RB, E2F1, E2F2, E2F3, E2F4, E2F5, CDK2, CDK4, CDK6, cyclin E and cyclin D1 in the RNA-seq data were analyzed. Cyclin D1 was found to significantly decrease upon NFYA knockdown in all three cell lines alongside its binding partner, CDK4, which had showed a slight decline as well (Figure 5B). Furthermore, we validated the findings by real-time qPCR. As is shown in Figure 5C, NFYA knockdown decreased the mRNA level of cyclin D1 (CCND1) and CDK4, as well as C-Myc, which coherent with previous studies [54,55] (Figure 5C).
Figure 5.
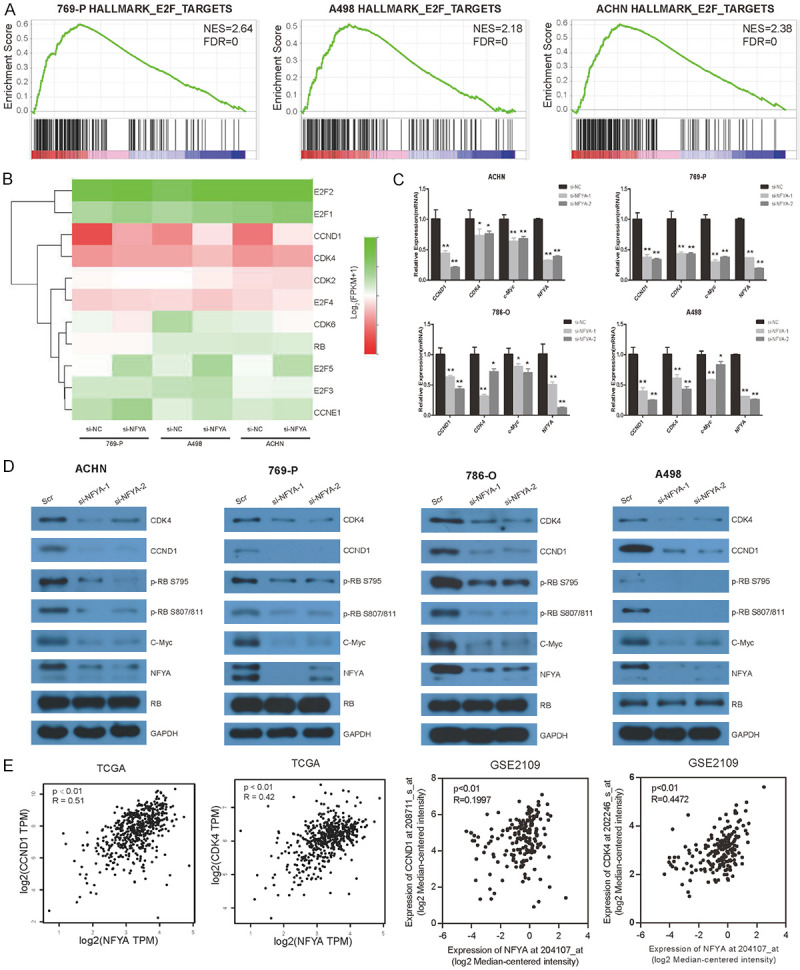
NFYA activates the RB-E2F pathway by upregulating CDK4/CCND1. A. GSEA of NFYA-regulated genes in 769-P, A498 and ACHN cells. B. The RNA-seq data of cyclin D1, cyclin E, CDK4, CDK2, CDK6, RB, E2F1, E2F2, E2F3, E2F4 and E2F5 in 769-P, A498 and ACHN cells upon NFYA knockdown. C. mRNA expression of CCND1, CDK4 and c-Myc upon NFYA knockdown were analyzed by real-time qPCR. Results were presented as the mean ± standard deviation (SD). D. Western blotting analysis of NFYA target gene and pathway in response to NFYA knockdown in ACHN, 769-P, 786-O and A498 cells. E. Co-expression analysis of NFYA and cyclin D1/CDK4 based on TCGA and GSE2109 dataset. R: spearman score. *P < 0.05, **P < 0.01.
The protein expression level of cyclin D1 and CDK4 were also analyzed upon NFYA depletion. Cyclin D1 and CDK4 were seen to be remarkably downregulated in NFYA knocked down ACHN, 769-P, 786-O and A498 cells as compared to the control group, along with a decrease in CDK4/cyclin D1 mediated p-RB (Figure 5D). It can be inferred from these data that NFYA promotes G1/S cell cycle transition by activating the expression of CDK4/cyclin D1 and subsequent E2F pathway. The expression profile of NFYA, cyclin D1 and CDK4 in ccRCC were analyzed to further validate the regulative relation between NFYA and CDK4/cyclin D1. Upon interrogating the TCGA-KIRC and GEO (GSE 2109) datasets, a positive correlation is shown between the mRNA levels of NFYA and cyclin D1/CDK4 in ccRCC patient samples (Figure 5E).
NFYA regulates the promoter activity of CDK4/CCND1
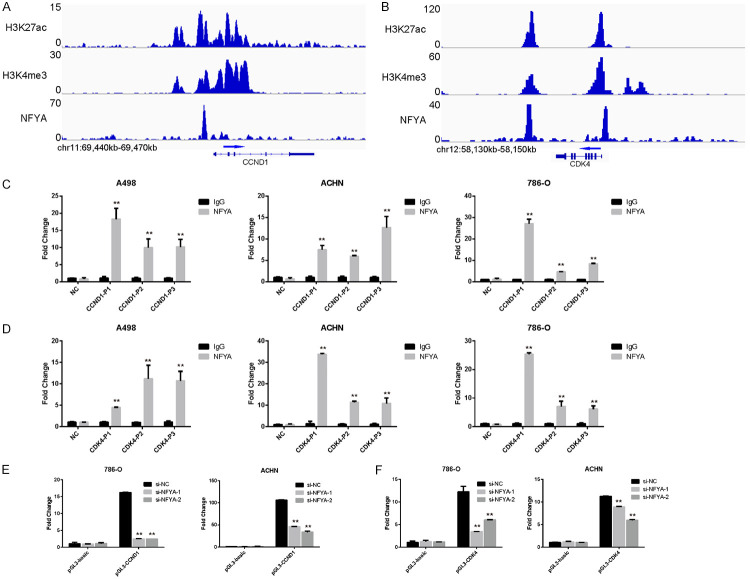
As a transcription factor, NFYA usually binds to the promoter or enhancer region of its target gene [56]. The NFYA ChIP-seq file is thus analyzed in the ENCODE platform to verify the association of NFYA to the cis-element of cyclin D1/CDK4. Based on the results in Figure 6A and 6B, strong NFYA binding peaks were found upstream of the CCND1/CDK4 transcription start site (TSS) in HeLa cell (Figure 6A and 6B). It is noteworthy that these two peaks are co-occupied by both the H3K27ac and H3K4me3 histone modifications, which are considered as active promoter (H3k27ac+/H3K4me3+) [57,58] (Figure 6A and 6B). These findings were further validated by ChIP-qPCR assay in ccRCC cell lines. Results showed that DNA immunoprecipitated by anti-NFYA antibody can be amplified effectively using primers specific to either the cyclin D1 or CDK4 promoter region, while poor efficiency was exhibited by the anti-IgG group. These data suggest that NFYA binds to the promoter region of CDK4/cyclin D1 (Figure 6C and 6D).
Figure 6.
NFYA regulates the promoter activity of CDK4/CCND1. A, B. The promoter region of cyclin D1 and CDK4 were co-occupied by NFYA, H3K27ac and H3K4me3 in HeLa cells. Data were derived from ENCODE. C, D. Confirmation of NFYA binding ability to cyclin D1 (C) and CDK4 (D) promoter through ChIP-qPCR. Results were presented as the mean ± standard deviation (SD). E, F. The promoter activity of cyclin D1 (E) and CDK4 (F) in both the control and NFYA knockdown ccRCC cells were analyzed using dual luciferase reporter assay. Results were presented as the mean ± standard deviation (SD). **P < 0.01.
Despite being associated with the promoter regions of CDK4/cyclin D1, the role of NFYA in regulating the promoter activity of CDK4/cyclin D1 is still unknown. To this end, the promoter elements were cloned into the pGL3-basic luciferase reporter vector (pGL3-CDK4 and pGL3-CCND1) and their activities were measured. Luciferase activity of both pGL3-CDK4 and pGL3-CCND1 showed significantly increased compared with the pGL3-basic empty vector (Figure 6E), implying the efficiency of these two DNA elements in driving gene expression. Moreover, NFYA knockdown remarkably decreased luciferase activity of CDK4/CCND1 promoter in 786-O and ACHN cells (Figure 6E and 6F). Taken together, these data suggest that NFYA promotes the expression of CDK4/CCND1 by regulating their promoter activity.
Discussion
This study first demonstrated the upregulation of transcription factor NFYA in clear cell renal cell carcinoma (ccRCC) tissue, and its correlation with poor prognosis. Furthermore, a novel network model was also proposed wherein NFYA promotes G1/S cell cycle transition by transactivating CDK4 and cyclin D1, which consequently facilitate the proliferation of ccRCC. These findings imply the oncogenic role of NFYA in ccRCC progression and its potential as a target for ccRCC therapy.
Cell cycle can be divided into four phases: G1, S, G2 and M. For most somatic cells, they are maintained in a quiescent condition, called G0. These cells re-enter the cell cycle upon mitogenic induction. Cancer cells by contrast, are independent of external stimulation, resulting in unscheduled division and proliferation. Cyclin D1 has been reported as a sensor that links mitogenic stimulation and cell cycle machinery [59]. Cyclin D1 is maintained at low levels in cells at quiescent state. The addition of growth factor stimulation results in the accumulation of cyclin D1, which consequently facilitates the phosphorylation of RB and elicits S phase initiation [60]. Cyclin D1 overexpression has been widely reported in a large fraction of human cancers, as such, allowing cancer cells to enter the cell cycle continuously regardless of external stimulate [61]. It has been proven that these factors may lead to cyclin D1 dysregulation: cyclin D1 genomic rearrangement, gene mutation, alternative splicing, and microRNA regulation [62]. For example, t(11;14)(q13;q32) rearrangement occurs in more than 90% of mantle cell lymphoma (MCL), leading to cyclin D1 overexpression [63]. In this study, cyclin D1 was found to be transactivated by transcription factor NFYA. More importantly, NFYA showed remarkable expression correlation with cyclin D1 in ccRCC patient samples. By taking the overexpression of NFYA in ccRCC into consideration, the NFYA-CCND1 axis may be presented as a novel cause for cyclin D1 dysregulation in cancers.
Based on literature review, the role of NFYA in hematopoietic disorder diseases and cancers was well documented. It has been reported that NFYA is involved in hematopoietic disorder by recruiting transcription activator or repressor to modulate the expression of HBG1, HBG2, HOXB4 and MHC, which are considered as hemogenesis related genes [64]. Additionally, NFYA also acts as an oncogene in various cancers by regulating cell proliferation, migration, transformation, apoptosis and cell cycle [23-27,65]. Several studies showed that the deletion of NFYA leads to decreased expression of cyclin B1 and cyclin B2 [23,65], which are important for the G2-M cell cycle transition. In the RNA-seq data, it was discovered that the mRNA level of cyclin B1 and cyclin B2 was also reduced in ACHN, 769-P and 786-O cells upon NFYA knockdown, proving the reliability of this experiment. However, among all four ccRCC cell lines, only in 786-O cells showed G2/M arrest upon NFYA depletion. It was demonstrated in this investigation that NFYA is required for G1/S cell cycle transition in ccRCC, although further exploration is needed to determine its role in G2/M transition.
Recent studies reveal that transcription factor NFYA usually cooperates with cofactors to activate its target gene. For instance, Zhu et al. demonstrated that NFYA interacts with USF1/2 on the promoter of HOXB4 to co-activate HOXB4 expression [66]. Henceforth, it is still unknown whether NFYA requires cofactors to transcriptionally activate cyclin D1 and CDK4. Early studies pointed out the interaction between NFYA and histone acetyl-transferases p300/CBP [67,68]. It was later learned that NFYA acts as a transcriptional activator by recruiting p300, which promotes target gene expression by facilitating histone H3 acetylation at the promoter region [69,70]. Furthermore, the acetyl-transferase activity of p300/CBP was also considered to be crucial for G1/S cell cycle transition [71]. In this study, the promoter region of cyclin D1 and CDK4 were co-occupied by both H3K27ac and NFYA, evidently raising the possibility of cooperation between NFYA and p300 to promote the H3K27ac modification at cyclin D1/CDK4 promoter, subsequently leading to transcriptional activation of these two genes.
The regulation of NFYA has been reported to work in multiple ways. Silvia et al. showed that NFYA is transcriptionally repressed by the other two NF-Y subunits (NFYB and NFYC) [72], while another study demonstrated that the expression of NFYA can be reduced by signaling activation of the PKC-p38-ERK and AKT-mTOR pathways [73]. Since the activation of ERK/AKT pathway and NFYB/NFYC were also linked to oncogenic roles in several cancers [25], it might not be the reason for NFYA dysregulation in cancers. The cause for NFYA overexpression in cancers remains unclear and requires further investigation.
Acknowledgements
This research is supported by the National Natural Science Foundation of China (81872140, 81572484 to DY, 81903069 to YL, 81772821 to KH) and the China Postdoctoral Science Foundation (2019M663292 to YL).
Disclosure of conflict of interest
None.
References
- 1.Chow WH, Devesa SS, Warren JL, Fraumeni JF Jr. Rising incidence of renal cell cancer in the United States. JAMA. 1999;281:1628–1631. doi: 10.1001/jama.281.17.1628. [DOI] [PubMed] [Google Scholar]
- 2.Moore LE, Wilson RT, Campleman SL. Lifestyle factors, exposures, genetic susceptibility, and renal cell cancer risk: a review. Cancer Invest. 2005;23:240–255. doi: 10.1081/cnv-200055962. [DOI] [PubMed] [Google Scholar]
- 3.Hsieh JJ, Purdue MP, Signoretti S, Swanton C, Albiges L, Schmidinger M, Heng DY, Larkin J, Ficarra V. Renal cell carcinoma. Nat Rev Dis Primers. 2017;3:17009. doi: 10.1038/nrdp.2017.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Linehan WM, Srinivasan R, Schmidt LS. The genetic basis of kidney cancer: a metabolic disease. Nat Rev Urol. 2010;7:277–285. doi: 10.1038/nrurol.2010.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 6.Rini BI, Campbell SC, Escudier B. Renal cell carcinoma. Lancet. 2009;373:1119–1132. doi: 10.1016/S0140-6736(09)60229-4. [DOI] [PubMed] [Google Scholar]
- 7.Goyal R, Gersbach E, Yang XJ, Rohan SM. Differential diagnosis of renal tumors with clear cytoplasm: clinical relevance of renal tumor subclassification in the era of targeted therapies and personalized medicine. Arch Pathol Lab Med. 2013;137:467–480. doi: 10.5858/arpa.2012-0085-RA. [DOI] [PubMed] [Google Scholar]
- 8.Makhov P, Joshi S, Ghatalia P, Kutikov A, Uzzo RG, Kolenko VM. Resistance to systemic therapies in clear cell renal cell carcinoma: mechanisms and management strategies. Mol Cancer Ther. 2018;17:1355–1364. doi: 10.1158/1535-7163.MCT-17-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parikh M, Lara PN Jr. Modern systemic therapy for metastatic renal cell carcinoma of the clear cell type. Annu Rev Med. 2018;69:209–221. doi: 10.1146/annurev-med-041916-124132. [DOI] [PubMed] [Google Scholar]
- 10.Latchman DS. Transcription factors: an overview. Int J Biochem Cell Biol. 1997;29:1305–1312. doi: 10.1016/s1357-2725(97)00085-x. [DOI] [PubMed] [Google Scholar]
- 11.Lambert M, Jambon S, Depauw S, David-Cordonnier MH. Targeting transcription factors for cancer treatment. Molecules. 2018;23:1479. doi: 10.3390/molecules23061479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bushweller JH. Targeting transcription factors in cancer - from undruggable to reality. Nat Rev Cancer. 2019;19:611–624. doi: 10.1038/s41568-019-0196-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schodel J, Grampp S, Maher ER, Moch H, Ratcliffe PJ, Russo P, Mole DR. Hypoxia, hypoxia-inducible transcription factors, and renal cancer. Eur Urol. 2016;69:646–657. doi: 10.1016/j.eururo.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu CJ, Wang LY, Chodosh LA, Keith B, Simon MC. Differential roles of hypoxia-inducible factor 1alpha (HIF-1alpha) and HIF-2alpha in hypoxic gene regulation. Mol Cell Biol. 2003;23:9361–9374. doi: 10.1128/MCB.23.24.9361-9374.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keefe SM, Heitjan D, Hennessey M, Robinson J, Mykulowicz K, Marshall A, Gunnarsson O, Mamtani R, Vaughn DJ, Hoffman-Censits JH, Nathanson KL, Lal P, Pryma DA, Eliasof S, Garmey EG, Cohen RB, Haas NB. Interim results of a phase 1b/2a study evaluating the nano pharmaceutical CRLX101 with bevacizumab (bev) in the treatment of patients (pts) with refractory metastatic renal cell carcinoma (mRCC) J. Clin. Oncol. 2014;32:412–412. [Google Scholar]
- 16.Mantovani R. The molecular biology of the CCAAT-binding factor NF-Y. Gene. 1999;239:15–27. doi: 10.1016/s0378-1119(99)00368-6. [DOI] [PubMed] [Google Scholar]
- 17.Marziali G, Perrotti E, Ilari R, Coccia EM, Mantovani R, Testa U, Battistini A. The activity of the CCAAT-box binding factor NF-Y is modulated through the regulated expression of its a subunit during monocyte to macrophage differentiation: regulation of tissue-specific genes through a ubiquitous transcription factor. Blood. 1999;93:519–526. [PubMed] [Google Scholar]
- 18.Mamat S, Ikeda J, Tian T, Wang Y, Luo W, Aozasa K, Morii E. Transcriptional regulation of aldehyde dehydrogenase 1A1 gene by alternative spliced forms of nuclear factor Y in tumorigenic population of endometrial adenocarcinoma. Genes Cancer. 2011;2:979–984. doi: 10.1177/1947601911436009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cicchillitti L, Corrado G, Carosi M, Dabrowska ME, Loria R, Falcioni R, Cutillo G, Piaggio G, Vizza E. Prognostic role of NF-YA splicing isoforms and Lamin A status in low grade endometrial cancer. Oncotarget. 2017;8:7935–7945. doi: 10.18632/oncotarget.13854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bezzecchi E, Ronzio M, Dolfini D, Mantovani R. NF-YA overexpression in lung cancer: LUSC. Genes (Basel) 2019;10:937. doi: 10.3390/genes10110937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bie LY, Li D, Mu Y, Wang S, Chen BB, Lyu HF, Han LL, Nie CY, Yang CC, Wang L, Ren CC, Zhang WJ, Guo P, Shi F, Fan QX, Wang LX, Chen XB, Luo SX. Analysis of cyclin E co-expression genes reveals nuclear transcription factor Y subunit alpha is an oncogene in gastric cancer. Chronic Dis Transl Med. 2019;5:44–52. doi: 10.1016/j.cdtm.2018.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dolfini D, Andrioletti V, Mantovani R. Overexpression and alternative splicing of NF-YA in breast cancer. Sci Rep. 2019;9:12955. doi: 10.1038/s41598-019-49297-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bungartz G, Land H, Scadden DT, Emerson SG. NF-Y is necessary for hematopoietic stem cell proliferation and survival. Blood. 2012;119:1380–1389. doi: 10.1182/blood-2011-06-359406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo J, Kong LM, Peng AF, Long XH, Zhou Y, Shu Y. Transcription factor NFYA promotes a malignant phenotype by upregulating fatty acid synthase expression. Mol Med Rep. 2016;14:5007–5014. doi: 10.3892/mmr.2016.5897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gurtner A, Manni I, Piaggio G. NF-Y in cancer: impact on cell transformation of a gene essential for proliferation. Biochim Biophys Acta Gene Regul Mech. 2017;1860:604–616. doi: 10.1016/j.bbagrm.2016.12.005. [DOI] [PubMed] [Google Scholar]
- 26.Hu Q, Lu JF, Luo R, Sen S, Maity SN. Inhibition of CBF/NF-Y mediated transcription activation arrests cells at G2/M phase and suppresses expression of genes activated at G2/M phase of the cell cycle. Nucleic Acids Res. 2006;34:6272–6285. doi: 10.1093/nar/gkl801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garipov A, Li H, Bitler BG, Thapa RJ, Balachandran S, Zhang R. NF-YA underlies EZH2 upregulation and is essential for proliferation of human epithelial ovarian cancer cells. Mol Cancer Res. 2013;11:360–369. doi: 10.1158/1541-7786.MCR-12-0661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lenburg ME, Liou LS, Gerry NP, Frampton GM, Cohen HT, Christman MF. Previously unidentified changes in renal cell carcinoma gene expression identified by parametric analysis of microarray data. BMC Cancer. 2003;3:31. doi: 10.1186/1471-2407-3-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gumz ML, Zou H, Kreinest PA, Childs AC, Belmonte LS, LeGrand SN, Wu KJ, Luxon BA, Sinha M, Parker AS, Sun LZ, Ahlquist DA, Wood CG, Copland JA. Secreted frizzled-related protein 1 loss contributes to tumor phenotype of clear cell renal cell carcinoma. Clin Cancer Res. 2007;13:4740–4749. doi: 10.1158/1078-0432.CCR-07-0143. [DOI] [PubMed] [Google Scholar]
- 30.Beroukhim R, Brunet JP, Di Napoli A, Mertz KD, Seeley A, Pires MM, Linhart D, Worrell RA, Moch H, Rubin MA, Sellers WR, Meyerson M, Linehan WM, Kaelin WG Jr, Signoretti S. Patterns of gene expression and copy-number alterations in von-hippel lindau disease-associated and sporadic clear cell carcinoma of the kidney. Cancer Res. 2009;69:4674–4681. doi: 10.1158/0008-5472.CAN-09-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tun HW, Marlow LA, von Roemeling CA, Cooper SJ, Kreinest P, Wu K, Luxon BA, Sinha M, Anastasiadis PZ, Copland JA. Pathway signature and cellular differentiation in clear cell renal cell carcinoma. PLoS One. 2010;5:e10696. doi: 10.1371/journal.pone.0010696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.von Roemeling CA, Radisky DC, Marlow LA, Cooper SJ, Grebe SK, Anastasiadis PZ, Tun HW, Copland JA. Neuronal pentraxin 2 supports clear cell renal cell carcinoma by activating the AMPA-selective glutamate receptor-4. Cancer Res. 2014;74:4796–4810. doi: 10.1158/0008-5472.CAN-14-0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liep J, Kilic E, Meyer HA, Busch J, Jung K, Rabien A. Cooperative effect of miR-141-3p and miR-145-5p in the regulation of targets in clear cell renal cell carcinoma. PLoS One. 2016;11:e0157801. doi: 10.1371/journal.pone.0157801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wotschofsky Z, Gummlich L, Liep J, Stephan C, Kilic E, Jung K, Billaud JN, Meyer HA. Integrated microRNA and mRNA signature associated with the transition from the locally confined to the metastasized clear cell renal cell carcinoma exemplified by miR-146-5p. PLoS One. 2016;11:e0148746. doi: 10.1371/journal.pone.0148746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Higgins JP, Shinghal R, Gill H, Reese JH, Terris M, Cohen RJ, Fero M, Pollack JR, van de Rijn M, Brooks JD. Gene expression patterns in renal cell carcinoma assessed by complementary DNA microarray. Am J Pathol. 2003;162:925–932. doi: 10.1016/S0002-9440(10)63887-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao H, Ljungberg B, Grankvist K, Rasmuson T, Tibshirani R, Brooks JD. Gene expression profiling predicts survival in conventional renal cell carcinoma. PLoS Med. 2006;3:e13. doi: 10.1371/journal.pmed.0030013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen F, Chandrashekar DS, Varambally S, Creighton CJ. Pan-cancer molecular subtypes revealed by mass-spectrometry-based proteomic characterization of more than 500 human cancers. Nat Commun. 2019;10:5679. doi: 10.1038/s41467-019-13528-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davis CA, Hitz BC, Sloan CA, Chan ET, Davidson JM, Gabdank I, Hilton JA, Jain K, Baymuradov UK, Narayanan AK, Onate KC, Graham K, Miyasato SR, Dreszer TR, Strattan JS, Jolanki O, Tanaka FY, Cherry JM. The Encyclopedia of DNA elements (ENCODE): data portal update. Nucleic Acids Res. 2018;46:D794–D801. doi: 10.1093/nar/gkx1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hu K, Li Y, Wu W, Chen H, Chen Z, Zhang Y, Guo Y, Dong Y. High-performance gene expression and knockout tools using sleeping beauty transposon system. Mobile DNA. 2018;9:33. doi: 10.1186/s13100-018-0139-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu K, Wu W, Li Y, Lin L, Chen D, Yan H, Xiao X, Chen H, Chen Z, Zhang Y, Xu S, Guo Y, Koeffler HP, Song E, Yin D. Poly(ADP-ribosyl)ation of BRD7 by PARP1 confers resistance to DNA-damaging chemotherapeutic agents. EMBO Rep. 2019;20:e46166. doi: 10.15252/embr.201846166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kamburov A, Pentchev K, Galicka H, Wierling C, Lehrach H, Herwig R. ConsensusPathDB: toward a more complete picture of cell biology. Nucleic Acids Res. 2011;39:D712–D717. doi: 10.1093/nar/gkq1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstråle M, Laurila E, Houstis N, Daly MJ, Patterson N, Mesirov JP, Golub TR, Tamayo P, Spiegelman B, Lander ES, Hirschhorn JN, Altshuler D, Groop LC. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 45.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hu H, Miao YR, Jia LH, Yu QY, Zhang Q, Guo AY. AnimalTFDB 3.0: a comprehensive resource for annotation and prediction of animal transcription factors. Nucleic Acids Res. 2019;47:D33–D38. doi: 10.1093/nar/gky822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Behan FM, Iorio F, Picco G, Goncalves E, Beaver CM, Migliardi G, Santos R, Rao Y, Sassi F, Pinnelli M, Ansari R, Harper S, Jackson DA, McRae R, Pooley R, Wilkinson P, van der Meer D, Dow D, Buser-Doepner C, Bertotti A, Trusolino L, Stronach EA, Saez-Rodriguez J, Yusa K, Garnett MJ. Prioritization of cancer therapeutic targets using CRISPR-Cas9 screens. Nature. 2019;568:511–516. doi: 10.1038/s41586-019-1103-9. [DOI] [PubMed] [Google Scholar]
- 48.Wang J, Zhao H, Dong H, Zhu L, Wang S, Wang P, Ren Q, Zhu H, Chen J, Lin Z, Cheng Y, Qian B, Zhang Y, Jia R, Wu W, Lu J, Tan J. LAT, HOXD3 and NFE2L3 identified as novel DNA methylation-driven genes and prognostic markers in human clear cell renal cell carcinoma by integrative bioinformatics approaches. J Cancer. 2019;10:6726–6737. doi: 10.7150/jca.35641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jin S, Fan F, Fan W, Zhao H, Tong T, Blanck P, Alomo I, Rajasekaran B, Zhan Q. Transcription factors Oct-1 and NF-YA regulate the p53-independent induction of the GADD45 following DNA damage. Oncogene. 2001;20:2683–2690. doi: 10.1038/sj.onc.1204390. [DOI] [PubMed] [Google Scholar]
- 50.Yang WT, Zhao ZX, Li B, Zheng PS. NF-YA transcriptionally activates the expression of SOX2 in cervical cancer stem cells. PLoS One. 2019;14:e0215494. doi: 10.1371/journal.pone.0215494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bracken AP, Ciro M, Cocito A, Helin K. E2F target genes: unraveling the biology. Trends Biochem Sci. 2004;29:409–417. doi: 10.1016/j.tibs.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 52.Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: a changing paradigm. Nat Rev Cancer. 2009;9:153–166. doi: 10.1038/nrc2602. [DOI] [PubMed] [Google Scholar]
- 53.Duronio RJ, Xiong Y. Signaling pathways that control cell proliferation. Cold Spring Harb Perspect Biol. 2013;5:a008904. doi: 10.1101/cshperspect.a008904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Benatti P, Basile V, Dolfini D, Belluti S, Tomei M, Imbriano C. NF-Y loss triggers p53 stabilization and apoptosis in HPV18-positive cells by affecting E6 transcription. Oncotarget. 2016;7:45901–45915. doi: 10.18632/oncotarget.9974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Benatti P, Belluti S, Miotto B, Neusiedler J, Dolfini D, Drac M, Basile V, Schwob E, Mantovani R, Blow JJ, Imbriano C. Direct non transcriptional role of NF-Y in DNA replication. Biochim Biophys Acta. 2016;1863:673–685. doi: 10.1016/j.bbamcr.2015.12.019. [DOI] [PubMed] [Google Scholar]
- 56.Fleming JD, Pavesi G, Benatti P, Imbriano C, Mantovani R, Struhl K. NF-Y coassociates with FOS at promoters, enhancers, repetitive elements, and inactive chromatin regions, and is stereo-positioned with growth-controlling transcription factors. Genome Res. 2013;23:1195–1209. doi: 10.1101/gr.148080.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, Wang W, Weng Z, Green RD, Crawford GE, Ren B. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 58.Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, Ching KA, Antosiewicz-Bourget JE, Liu H, Zhang X, Green RD, Lobanenkov VV, Stewart R, Thomson JA, Crawford GE, Kellis M, Ren B. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sherr CJ. Cancer cell cycles. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 60.Shipley J, Santarius T, Stratton MR, Brewer D, Cooper CS. A census of amplified and overexpressed human cancer genes. Nat Rev Cancer. 2010;10:59–64. doi: 10.1038/nrc2771. [DOI] [PubMed] [Google Scholar]
- 61.Cheng M, Sexl V, Sherr CJ, Roussel MF. Assembly of cyclin D-dependent kinase and titration of p27Kip1 regulated by mitogen-activated protein kinase kinase (MEK1) Proc Natl Acad Sci U S A. 1998;95:1091–1096. doi: 10.1073/pnas.95.3.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Qie S, Diehl JA. Cyclin D1, cancer progression, and opportunities in cancer treatment. J Mol Med (Berl) 2016;94:1313–1326. doi: 10.1007/s00109-016-1475-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bertoni F, Rinaldi A, Zucca E, Cavalli F. Update on the molecular biology of mantle cell lymphoma. Hematol Oncol. 2006;24:22–27. doi: 10.1002/hon.767. [DOI] [PubMed] [Google Scholar]
- 64.Ly LL, Yoshida H, Yamaguchi M. Nuclear transcription factor Y and its roles in cellular processes related to human disease. Am J Cancer Res. 2013;3:339–346. [PMC free article] [PubMed] [Google Scholar]
- 65.Park SH, Yu GR, Kim WH, Moon WS, Kim JH, Kim DG. NF-Y-dependent cyclin B2 expression in colorectal adenocarcinoma. Clin Cancer Res. 2007;13:858–867. doi: 10.1158/1078-0432.CCR-06-1461. [DOI] [PubMed] [Google Scholar]
- 66.Zhu J, Giannola DM, Zhang Y, Rivera AJ, Emerson SG. NF-Y cooperates with USF1/2 to induce the hematopoietic expression of HOXB4. Blood. 2003;102:2420–2427. doi: 10.1182/blood-2003-01-0251. [DOI] [PubMed] [Google Scholar]
- 67.Currie RA. NF-Y is associated with the histone acetyltransferases GCN5 and P/CAF. J Biol Chem. 1998;273:1430–1434. doi: 10.1074/jbc.273.3.1430. [DOI] [PubMed] [Google Scholar]
- 68.Li Q, Herrler M, Landsberger N, Kaludov N, Ogryzko VV, Nakatani Y, Wolffe AP. Xenopus NF-Y pre-sets chromatin to potentiate p300 and acetylation-responsive transcription from the Xenopus hsp70 promoter in vivo. EMBO J. 1998;17:6300–6315. doi: 10.1093/emboj/17.21.6300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jin S, Scotto KW. Transcriptional regulation of the MDR1 gene by histone acetyltransferase and deacetylase is mediated by NF-Y. Mol Cell Biol. 1998;18:4377–4384. doi: 10.1128/mcb.18.7.4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gurtner A, Fuschi P, Magi F, Colussi C, Gaetano C, Dobbelstein M, Sacchi A, Piaggio G. NF-Y dependent epigenetic modifications discriminate between proliferating and postmitotic tissue. PLoS One. 2008;3:e2047. doi: 10.1371/journal.pone.0002047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ait-Si-Ali S, Polesskaya A, Filleur S, Ferreira R, Duquet A, Robin P, Vervish A, Trouche D, Cabon F, Harel-Bellan A. CBP/p300 histone acetyl-transferase activity is important for the G1/S transition. Oncogene. 2000;19:2430–2437. doi: 10.1038/sj.onc.1203562. [DOI] [PubMed] [Google Scholar]
- 72.Belluti S, Semeghini V, Basile V, Rigillo G, Salsi V, Genovese F, Dolfini D, Imbriano C. An autoregulatory loop controls the expression of the transcription factor NF-Y. Biochim Biophys Acta Gene Regul Mech. 2018;1861:509–518. doi: 10.1016/j.bbagrm.2018.02.008. [DOI] [PubMed] [Google Scholar]
- 73.Chatterjee B, Ghosh K, Suresh L, Kanade SR. Curcumin ameliorates PRMT5-MEP50 arginine methyltransferase expression by decreasing the Sp1 and NF-YA transcription factors in the A549 and MCF-7 cells. Mol Cell Biochem. 2019;455:73–90. doi: 10.1007/s11010-018-3471-0. [DOI] [PubMed] [Google Scholar]