Abstract
Patients with advanced-stage colon cancer often exhibit resistance against treatment and distant metastasis, both key contributors to poor prognosis. Emerging evidence indicates that cancer stem cells (CSCs), characterized by the enhanced ability to self-renew, resist therapeutics, and promote metastasis, represents a clinical challenge to target. Alternative therapeutic approaches are urgently required. Here, we explored the feasibility of disrupting the intracellular communications between CSCs and the tumor microenvironment by way of exosomes. First, we demonstrated that exosomes secreted by colon tumorspheres (Exosp) promoted 5-FU resistance, migration, and tumorsphere formation. Exosp also increased the generation of cancer-associated fibroblasts and M2 polarized macrophages in vitro. Oncogenic molecules, including IL-6, p-STAT3, TGF-β1, and β-catenin, were identified as the cargoes of Exosp. Furthermore, the public database revealed the high abundance of miR-1246 in serum exosomes from colon cancer patients, and we verified in the Exosp from HCT116 and HT29 cells. Therapeutically, we demonstrated the ovatodiolide treatment reduced exosomal cargoes from tumorspheres (Exosp_OV). Exosp_OV were significantly less capable of promoting 5-FU resistance, migration, and tumorsphere formation when co-cultured with HCT116 and HT29 cells. Notably, Exosp_OV was less CAF- and M2 TAM-transformative. Computational docking analysis revealed that OV could bind and significantly reduced β-catenin activity. Finally, mouse xenograft data indicated that ovatodiolide suppressed tumor growth via down-regulating IL-6, STAT3, β-catenin expression, and serum exosomal miR-1246. In conclusion, our findings provided preclinical supports for ovatodiolide as a colon CSC inhibitor by reducing β-catenin/STAT3/miR-1246 signaling conveyed by CSC derived exosomes.
Keywords: Colon cancer stem cells, tumor microenvironment, 5-FU resistance, tumorsphere derived exosomes, β-catenin/STAT3/miR-1246 signaling, and ovatodiolide
Introduction
Despite the advance in the treatment modalities of colorectal cancer (CRC), the clinical outcome and the disease-free survival for metastatic CRC patients remains poor; two major contributing factors are the development of drug resistance and distant metastasis [1]. The existence of colon cancer stem cells (CSC) has been shown to drive the progression of colon cancer. CSCs are generally accepted as a subpopulation of cancer cells, residing within the tumor microenvironment (TME), with the enhanced ability to self-renew and differentiate into progenies with different functionalities, which directly participate in distant metastases and resistance to therapy [2].
Emerging evidence strongly indicates the complex intracellular communications within the TME is likely to be orchestrated by the CSCs and instrumental for tumorigenesis [3]. Based on these premises, a model has been proposed that the intercellular exchange of free or packaged signaling molecules may determine the oncogenic potential of the TME and cancer cells. Thus, to gain more insights into the molecular properties of the TME would lead to the identification of specific biomarkers and/or therapeutic targets.
Recently extracellular vesicles, termed exosomes (Exo), have been shown to be one of the major routes for intracellular communications not only within the TME but also at the distant metastatic sites [4]. Exosomes are characterized as membranous vesicles with the average size of 20-1000 nm encapsulating signaling molecules, including nucleic acids, lipids, and proteins, that are transferred among donor and respective recipient cells [5]. Exosomes of the tumor and stromal cell origins are trafficked within the TME and contributing significantly to tumor development and progression [6]. A growing body of literature suggests tumor-derived exosomal signaling facilitates the acquisition of drug-resistance and metastatic potential in different cancers [7]. However, the role of exosomes derived from CSC remains underappreciated.
Ovatodiolide, a small-molecule phytochemical isolated and purified from Anisomeles indica, has been traditionally used to treat inflammation-associated diseases and implicated for anticancer functions by our previous reports [8,9]. Various inflammatory and tumorigenic markers such as TNF-α, NF-κB, β-catenin, MMPs were shown to be suppressed by ovatodiolide treatment [9,10]. To further explore the therapeutic potentials of ovatodiolide as an inhibitor of colon cancer stem cells, we examined its effects on the CSC-derived exosomal cargoes and the TME.
Exosomes derived from tumor cells have recently gained much attention. Tumor exosomes are vital participants for intracellular communications in different stages of tumorigenesis. Essentially, exosomes contain signaling molecules, including phospholipids, nucleic acids, and proteins, that are transferred between donor and recipient cells [11,12]. Non-coding RNAs such as microRNAs (miRs) have been shown to be one of the most abundantly signaling molecules packaged within tumor-derived exosomes [13]. Accumulating evidence suggests that the exosomal miRs may function as prognostic and therapeutic biomarkers for many cancer types, including colon [14,15]. In this study, we provided evidence that CSC-derived exosomes facilitated the malignant transformation of the TME in part by generating cancer-associated fibroblasts (CAF) and M2 polarized tumor-associated macrophages (M2 TAM). We found that colon tumorspheres secreted exosomes containing oncomiR-1246 and signaling molecules, including IL-6, STAT3, β-catenin, and TGF-β1. Ovatodiolide treatment reduced colon cancer tumorigenesis by reducing exosomal cargoes released from the colon tumorspheres. Our findings provide essential preclinical evidence supporting the further development of ovatodiolide as an inhibitor of colon cancer stem cells.
Materials and methods
Cell culture and reagents
Human colon cancer cell lines, HCT116, HT29, monocytic progenitor cell line, THP1, and normal fibroblasts (NF) were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA). All cells were cultured and expanded according to the protocols recommended by the ATCC. The generation of colon tumorspheres (Sp) from HCT116 and HT29 was performed according to our previously published protocol [9]. In brief, HT29 and HCT116 cells were seeded (2000 cells) in six-well ultra-low attachment plates (Corning, Corning, NY, USA) in serum-free medium composing of Dulbecco’s modified Eagle medium (DMEM)/Ham’s F12 (1:1), human epidermal growth factor (hEGF, 20 ng/ml), basic fibroblast growth factor (bFGF; 10 ng/ml, PeproTech, Rocky Hill, NJ, USA), 2 μg/ml 0.2% heparin (Sigma, St. Louis, MO, USA), and 1% penicillin/streptomycin (P/S, 100 U/ml, Hyclone, Logan, UT, USA). Colon cancer cells were allowed to grow for 5-7 days. Spheroid cells with a diameter >100 µm (suspended) were considered a tumorsphere and counted. Ovatodiolide crystals were generous gifts from Professor Yew-Min Tzeng (Department of Life Science, National Taitung University, Taitung, Taiwan). The crystals were first dissolved in DMSO to make a stock solution and kept at -20°C until further use.
Isolation of tumor sphere-derived exosomes
Tumorsphere derived exosomes (Exosp) were isolated from the culture media collected from HCT116 and HT29 tumorspheres. The culture media were mixed with the Total Exosome Isolation Reagent (Thermo Fisher Scientific, Taipei, Taiwan) in a 10:1 ratio and processed according to the protocol provided by the vendor. Cluster of differentiation 9 (CD9) and CD63 antibodies were used to identify the exosomes isolated. Under our experimental conditions, 15 mL of culture medium (from the tumorsphere culture), approximately 0.9 to 1 mg of total protein (isolated Exosp) were obtained, as determined by Bradford protein assay.
In vitro cell migration assay
Cell migration assay was carried out using a Transwell apparatus (ThermoFIsher, Taipei, Taiwan). In brief, HCT116 and HT29 colon cancer cells (2 × 104 cells/well) incubated with or without (30 µg Exosp, 12 h) were seeded into the upper chambers which contained 200 μL serum-free DMEM, and 500 μL DMEM with 10% FBS in the lower chambers to generate a serum concentration gradient. The cells were then incubated for another 12 h, and then cells were fixed with formaldehyde (10%) followed by crystal violet staining. Cells on the upper side of the membrane were discarded, and colon cancer cells on the opposite of the membranes were counted.
Exosome-induced transformation of stromal cells
The transformation of normal fibroblasts (NF) to cancer-associated fibroblasts (CAF) was performed by co-culturing NF (2 × 104 cells/well) with Exosp (30 ug) in a six-well plate (Costar®, Corning, Taipei, Taiwan) for 48 h. The resultant fibroblasts were subjected to CAF characterization. NFs and Exosp transformed CAFs were first seeded in six-well chamber slides (Nunc™, Thermo Fisher Scientific, Rochester, NY, USA) for 24 h. Subsequently, immunofluorescence imaging was performed. The primary antibody, α-smooth muscle actin (α-SMA, 1:100, cat no. 48938) was added to the slides and incubated at room temperature for 1 h followed by another incubation with matched secondary antibody anti-mouse immunoglobulin G (IgG) (H+L), F(ab)2 fragment (1:800, AlexaFluor 488 conjugate, cat no. 4408). Stained cells were mounted using Vectashield mounting medium with 4’,6-diamidino-2-phenylindole (DAPI) for nuclear staining. Immunofluorescence images were captured with a Zeiss Axiophot (Carl Zeiss, Germany) fluorescence microscope and processed using AxioVision Zeiss software (Carl Zeiss, Germany).
A similar protocol was used for macrophage polarization. In short, uncommitted macrophages (MΦ) were obtained by treating naïve THP1 cells (1 × 105/well) with 320 nM PMA for 6 h in a six-well plate. MΦ (were then washed with PBS (3 ×) followed by a 48 h incubation with Exosp (30 µg). The resultant macrophages were then subjected to qPCR analysis based on their expression of M1 and M2 TAM markers, as well as an ELISA test of M2 cytokine, TGF-β1.
Cell viability assay
The cell viability was tested using an established sulforhodamine B (SRB) assay protocol [16]. Briefly, colon cancer cells, HT29, and HCT116 (8000 cells/well) were seeded in 96-well plates and received different concentrations of 5-FU (in the case of parental cells) or co-cultured with Exosp and Exosp_OV (30 µg) for the period of 48 h. After the 48 h culture time, cells were fixed with 10% cold trichloroacetic acid (TCA) for 1 h followed by several washes with ddH2O and stained with 0.4% SRB (Sigma, Taipei, Taiwan) in 1% acetic acid for 30 min at room temperature. Excess SRB was removed by with 1% acetic acid washes. Plates were air-dried at room temperature and washed in 20 mM Tris-base solution for 15 min with shaking. The absorbance was finally measured using a microplate reader (at 515 nm). The cell viability was then represented in ratio normalized with the untreated control (set at 100%).
Colony formation
The ability to generate colonies of both HCT116 and HT29 colon cancer cells under different treatments were examined using an established colony formation assay. Briefly, 500 HCT116 and HT29 cells were plated in six-well plates (Corning, Taipei, Taiwan) under different treatments (Exosp or Exosp_OV, 30 µg). Subsequently, the cells were allowed to grow for one week and fixed, stained, and counted.
In-silico molecular modeling
We utilized the AutoDock Vina [17] software to simulate the molecular docking and examine the potential molecular interactions between ovatodiolide and β-catenin. Molecular docking studies were performed using crystal structures of β-catenin. (PDB ID: 2Z6H). Protein Structures were further prepared using an open-source program, AutoDock Vina. First, the water molecule was removed. Second, the hydrogens (polar only) and missing side chains were added. The chemical structure of ovatodiolide was downloaded from PubChem (CID: 6451060) and was converted into a 3D structure using Pymol software. Finally, the geometry of each ligand was optimized using Autodock Vina. The ligand-binding pockets of β-catenin were specified by the β-catenin crystal and ovatodiolide, and a 3D box was formed around each crystal ligand to enclose the orthosteric ligand-binding pocket. The 3D box (40Å × 40Å × 40Å) centered at (95.033, 33.729, 47.576) Å for ovatodiolide. Each ligand was first dock-ed using AutoDock Vina scoring function with default procedures and parameters.
TOPFlash/FOPFlash assay
The canonical Wnt/β-catenin signaling was determined using the TOP/FOP Flash system (TCF Reporter Plasmid Kit, Cat#17-285, Merck, Taipei, Taiwan). In brief, HCT116 cells were seeded in 6-well plates (6 × 104 cells/well) and allowed to grow up to 60-65% confluence, followed by TCF-reporter plasmid (1 µg), TOPflash, FOPflash. A 50 ng pRL-CMV served as a control for transfection efficiency. 24 h after the transfection, cells were treated with control medium, Wnt-3a-conditioned media (50%, diluted in 5% FBS/DMEM), or 5 μM ovatodiolide for another 24 h. Cells were lysed and assayed for Firefly luciferase (from TOPflash or FOPflash) or Renilla luciferase activity (from pRL-CMV). The readouts were expressed as a ratio of Firefly luciferase over Renilla luciferase activity. Each data point represents the mean ± standard deviation.
Real-time polymerase chain reaction (qPCR)
Total RNA was extracted and purified using a TRIzol-based method (Life Technologies, Taipei, Taiwan). Total RNA (500 ng) was reverse-transcribed by a QIAGEN OneStep RT-PCR Kit (QIAGEN, Taipei, Taiwan), and the PCR reactions were carried out using a Rotor-Gene SYBR Green PCR kit. Primer sequences used in this study are all listed in Supplementary Table 1. GAPDH was used as a reference for normalization.
microRNA transfections
For determination of miR-1246 level in colon cancer cells and/or exosomes, MystiCq® microRNA qPCR Assay Primer (Cat# MIRAP00759-250RXN, Merck, Taipei, Taiwan) was purchased. Up and downregulation of miR-1246 in colon cancer cells was performed using MISSION® microRNA Mimic has-miR-1246 (Cat# HMI0094), MISSION® Synthetic microRNA Inhibitor (Cat# HSTUD0094) and negative control (Cat# NCSTUD001) respectively. The transfection experiments were carried out according to the vendor’s instructions.
Enzyme-linked immunosorbent assays (ELISA)
The culture media from the control (uncommitted macrophages, MΦ), M2 TAM (transformed by co-culturing with Exosp and Exosp_OV), or NF (control) and CAF (transformed from co-culturing with Exosp and Exosp_OV) were collected for analysis. ELISA kits for TGF-β1 (Cat# ab100647, Abcam, Taipei, Taiwan) and IL-6 (Cat# ab46027, Abcam, Taipei, Taiwan) and were used for quantitative measurements of secreted TGF-β1 and IL-6 respectively.
SDS-PAGE and immunoblotting
Cancer cells, tumorspheres, and exosomes harvested before and after different experimental conditions were collected and lysed. Total proteins were separated by SDS-PAGE with a Mini-Protean III system (Bio-Rad, Taiwan) and transferred onto PVDF membranes using the Trans-Blot Turbo Transfer System (Bio-Rad, Taiwan). Transferred membranes were then first incubated with primary antibodies overnight at 4°C followed by 3 washes with PBST, and incubation with respective secondary antibodies (room temperature, 1 h). The immunosignals were detected using an ECL substrate kit (Cat# ab133406, Abcam, Taipei, Taiwan). Blot images were captured and analyzed using the UVP BioDoc-It system (Upland, CA, USA). Please refer to Supplementary Table 2 for the list of primary antibodies and experimental conditions.
In vivo efficacy evaluation of ovatodiolide
Female NOD/SCID mice (6 weeks old) were purchased from BioLASCO Taiwan Co., Ltd. HCT116 tumorspheres (1 × 105 cells/injection) were injected subcutaneously in the right flank of the mice. Mice were allowed to recover for one week. The mice were randomly distributed to 4 different groups after the tumor had become palpable, and the treatments commenced. Four groups were defined as follows. Control (sham injection with PBS), ovatodiolide (10 mg/kg, 5 ×/week, i.p), 5-FU (30 mg/kg, 2 ×/week, i.p), and the combination (ovatodiolide, 10 mg/kg, 5 ×/week plus 5-FU, 30 mg/kg, 2 ×/week, respectively). Tumor volume and body weight were monitored weekly. Tumor volume was measured using a standard caliper. Tumor volume was calculated using the established formula. Tumor volume = 1/2 (length × width2) in mm3. Upon the completion of the experiments, all animals were humanely sacrificed, and tumor samples were collected for further analyses.
Immunohistochemistry
Tumor samples were collected from mice after in vivo experiments were completed. The staining was carried out according to the vendor’s instructions (UltravVision Quanto Detection System HRP DAB manual, Thermo Scientific, CA, USA). Briefly, the immunostaining was performed on 5-μm-thick tumor sections. The sections were first dewaxed and deparaffinized in xylene and rehydrated in graded alcohol solutions. The antigen-retrieval protocol was applied to the sections using a microwave (set at high power, 1 min) in Tris-EDTA buffer. Slides were subsequently stained with primary antibodies, STAT3 (Cat# ab119352, 1:400), CD44 (Cat# ab157107, 1:800), IL-6 (Cat# ab6672, 1:500) and β-catenin (Cat# ab6302, 1:400) (All purchased from Abcam, Taiwan) and their respective secondary antibodies. The sections were then counterstained with hematoxylin, dehydrated and mounted. The micrographs were captured and recorded using Tissue FAXS viewer software (TissueGnotics, GmBH, Vienna, Austria).
For immunofluorescence imaging, NF (2 × 104 cells/well) were incubated with Exosp (30 µg, 48 h) and Exosp_OV (30 µg, 48 h) and harvested for imaging. Anti-α-smooth muscle actin (α-SMA, Cat# ab5694, 1:100) and matched secondary antibody, goat Anti-Rabbit IgG H&L (Alexa Fluor® 488, Cat# ab150077) were used. The immunofluorescence experiment was performed using the established protocol published by the vendor (https://www.abcam.com/protocols/immunocytochemistry-immunofluorescence-protocol). Stained cells were mounted using Vectashield mounting medium with 4’,6-diamidino-2-phenylindole (DAPI) as the nuclear counterstain. Stained cells were finally imaged using a Zeiss Axiophot fluorescence microscope (Carl Zeiss, Germany). Microphotographs were captured by AxioCam MRc digital video camera and analyzed using AxioVision Zeiss software.
Statistical analysis
All experiments were independently repeated three times. Reported values shown in the graphs were the means and the error bars indicating the standard deviation (s.d). The Student’s t-test was performed using GraphPad Prism software. The experimental data were considered statistically significant when *P<0.05, **P<0.001, and ***P<0.001, respectively.
Results
Exosomes derived from colon tumorspheres enhanced tumorigenic properties of colon cancer cells in vitro
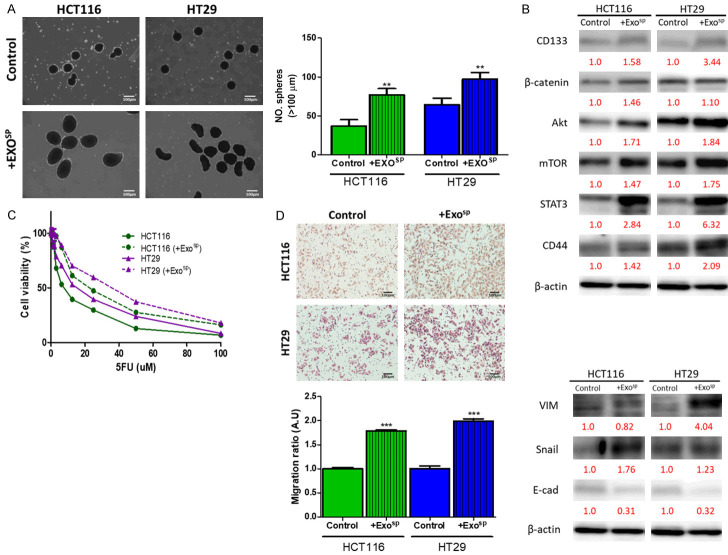
Cancer stem cells (CSCs) play an essential role in virtually all aspects of tumorigenesis; tumorspheres cultured in vitro is an established surrogate for CSC characterization [11]. We aimed to explore an emerging molecular route, namely exosomal communication, by which CSCs utilize to promote tumorigenesis with the tumor microenvironment (TME). First, we isolated exosomes secreted by colon tumorspheres of HT29 and HCT116 cells (Exosp). Exosp were added into the culture medium of parental HT29 and HCT116 cells (30 ug Exosp, 48 h). Exosp educated, HT29, and HCT116 cells were harvested and assayed for their oncogenic properties. Both Exosp educated HT29, and HCT116 cells showed a significantly increased ability to generate tumorspheres as compared to their parental selves (Figure 1A); these cells demonstrated an increased expression of stemness markers such as β-catenin, CD133 and CD44 and oncogenic makers such as mTOR, Akt, and STAT3 (Figure 1B). Additionally, Exosp-educated HT29 and HCT116 cells exhibited an apparent increase in 5-FU resistance as compared to their parental counterparts (Figure 1C). Furthermore, Exosp-educated HT29 and HCT116 cells exhibited an enhanced metastatic potential, as indicated by increased migratory ability and mesenchymal markers, vimentin (VIM), Snail, while the reduced epithelial marker, E-cadherin (Figure 1D).
Figure 1.
Tumor sphere derived exosomes promoted tumorigenic properties in colon cancer cells. A. Colon cancer cell lines, HT29 and HCT116 co-cultured with Exosp showed a significantly increased self-renewal ability, reflected by an increased number of tumorspheres generated. B. Comparative Western blots of Exosp-educated and parental HT29 and HCT116 cells. Exosp-educated colon cancer cells showed a markedly increased expression of stemness markers, β-catenin, CD133, and CD44; increased level of oncogenic molecules, mTOR, Akt, and STAT3. C. SRB assay showed that Exosp-educated HT29 and HCT116 became more resistant against 5-FU treatment. D. Increased migratory ability post-Exosp co-culture in both HT29 and HCT116 cells and increased expression of Vimentin, Snail, while decreased E-cadherin. Numbers in red indicate the relative expression ratio. **P<0.01; ***P<0.001.
Colon tumorspheres derived exosomes (Exosp) promoted the generation of CAFs and M2 TAMs
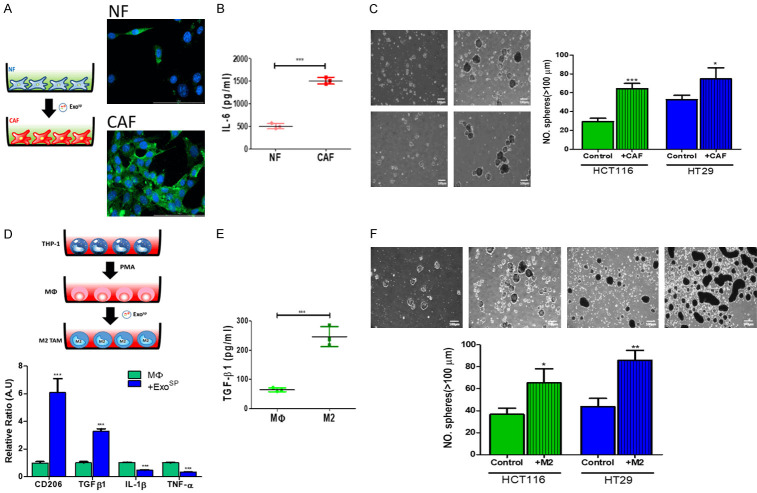
Next, we examined the CSC’s role in transforming the tumor microenvironment. First, isolated exosomes from tumorspheres of HT29 and HCT116. Exosp cultured normal fibroblasts (NF) showed an increased expression of α-SMA, a marker for cancer-associated fibroblasts (CAF) (Figure 2A); this was accompanied by significantly increased secretion of IL-6, one of the major cytokines secreted by tumor-promoting CAF (Figure 2B). Consistently, Exosp-transformed CAF promoted the ability to generate tumorspheres in both HT29 and HCT116 cells (Figure 2C). In a parallel experiment, we added Exosp into the culture medium of uncommitted macrophages (MΦ). Post-Exosp culture, these macrophages showed an elevated mRNA level of CD206 and TGF-β1, both of which are M2-polarized tumor-associated macrophages (M2 TAMs) markers whiled a significantly reduced M1 marker, TNF-α, and IL-1β (Figure 2D). In support, Exosp transformed macrophages showed an increased level of M2 cytokine, TGF-β1, as compared with their uncommitted macrophages (Figure 2E). Similarly, M2 TAM co-cultured HCT116 and HT29 cells resulted in a significantly increased ability to form tumorspheres, as compared to their naïve counterparts (Figure 2F).
Figure 2.
Tumor sphere derived exosomes promoted CAF transformation and M2 polarization in vitro. A. Immunofluorescence images of CAF-transformation. Normal fibroblasts (NF) incubated with Exosp showed an increase in α-SMA immunofluorescence (Green, α-SMA, Blue, DAPI). The insert depicts the experimental setup where NF (normal fibroblasts) were cultured with and without Exosp. B. IL-6 ELISA analysis. CAFs transformed Exosp secreted a significantly higher amount of IL-6 into the culture medium as compared to their normal fibroblasts (NF) counterparts. C. Tumor sphere formation assay. Both HCT116 and HT29 cells co-cultured with CAF (48 h), generated a significantly higher number of tumorspheres as compared to their parental counterparts. The insert shows the experimental setup where THP-1 monocytes were first Treated with PMA to generate uncommitted macrophages (MΦ), followed by the incubation of Exosp. M2 TAM, M2 polarized tumor-associated macrophages. D. Real-time PCR analysis of Exosp co-cultured uncommitted macrophages (MΦ). Post Exosp incubation, macrophages expressed a significantly higher M2 marker, CD206, and TGF-β1 while decreased M1 markers, IL-1β and TNF-α, as compared to the naïve counterparts. E. TGF-β1 ELISA test indicated that M2 TAMs generated by Exosp co-culture released a significantly higher amount of TGF-β1 than MΦ. F. A co-culture system of Exosp transformed M2 TAMs and parental HCT116 and HT29 colon cancer cells. Exosp transformed M2 TAM promoted tumor sphere-forming ability in parental HCT116 and HT29 colon cancer cells. *P<0.05; **P<0.01; ***P<0.001.
Exosomes derived from colon tumorspheres contained oncogenic signaling cargoes
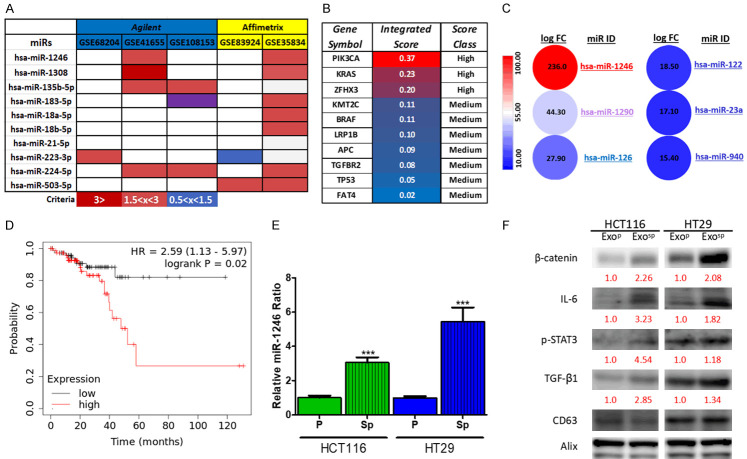
Noncoding RNAs such as microRNAs (miRs) have been shown to play key roles in tumorigenesis, including colon cancer, and emerged to become prognostic markers. Based on these premises, our bioinformatics research from several public miR databases, comparing the healthy and colon cancer samples, identified miR-1246 as one of the most abundantly expressed miRs across different databases and sequencing platforms (Figure 3A); these data were adapted from Falzone et al. [18]. This analysis showed the most upregulated miRs (colon cancer versus normal tissues), identified in at least 3 out of 10 total datasets. Also, ten different genes obtained from COSMIC and the mirDIP analysis indicated that miR-1246 was associated with colon cancer-associated oncogenes such as KRAS and PI3K with high levels of specificity (Figure 3B). Subsequently, we explored further using another database [19] where the level of miRs in serum exosomes was analyzed from 88 primary CRC patients and 11 healthy controls. Again, miR-1246 was one of the most abundant miRs (Figure 3C). More importantly, we found that a higher level of miR-1246 was associated with a significantly lower survival ratio in patients of colorectal cancer (Figure 3D) from the TCGA database (N = 395) [20]. In vitro, we compared the level of miR-1246 from the exosomes isolated from the parental and tumorspheres of HCT116 and HT29 colon cancer cell lines. Notably, exosomes from the tumorspheres contained a significantly higher level of miR-1246 (Figure 3E). Furthermore, exosomes from tumorspheres (Exosp) contained a relatively higher level of stromal transforming molecules, including β-catenin, IL-6, TGF-β1, and p-STAT3 as compared to exosomes isolated from the parental cancer cells (Exop) (Figure 3F).
Figure 3.
Increased exosomal miR-1246 in colon cancer patients and correlated with poor prognosis. A. MicroRNA profiling of clinical samples from colon cancer patients. MiR-1246 appeared to be the most abundantly expressed microRNA in tumors samples from colon cancer patients versus normal tissues in two databases (GSE41655 and GSE35834). The color bars represent the logFC (log fold-change) values. High (logFC ≥3), moderate (logFC 1.5<x<3), and slight (logFC 0.5<x<1.5). B. mirDIP gene target analysis. The association between miR-1246 and the top 10 mutated genes in colon cancer. The intensity of association is represented by color-scale ranging from red (high interaction) to pink (medium interaction). Data adapted and modified from Falzone [18]. C. Comparative profiling of serum exosomal microRNAs between colon cancer patients (n = 88) and healthy individuals (n = 11). MiR-1246 was the most abundant miR species in serum exosomes of colon cancer patients. D. TCGA survival analysis of colon cancer patients (N = 160). A higher miR-1246 predicts a significantly lower survival probability in patients of colon cancer. E. Comparative Real-time PCR analysis of miR-1246 of exosomal cargo between parental cells and tumorspheres of HCT116 and HT29. F. The exosomal cargo of colon tumorspheres were higher in IL-6, β-catenin, TGF-β1, and p-STAT3 than their parental counterparts. Numbers in red indicate the relative expression ratio. ***P<0.001.
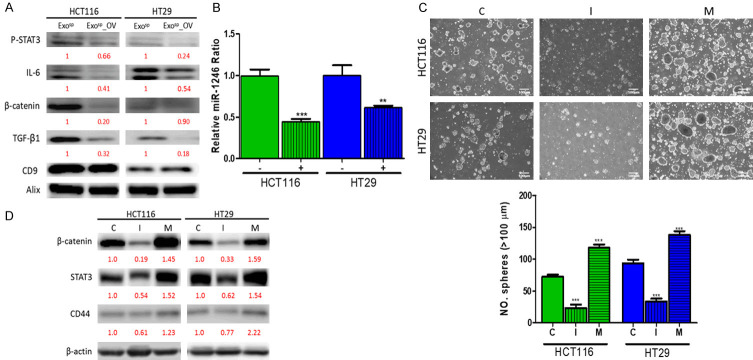
Ovatodiolide treatment resulted in reduced oncogenic properties in colon sphere-derived exosomes
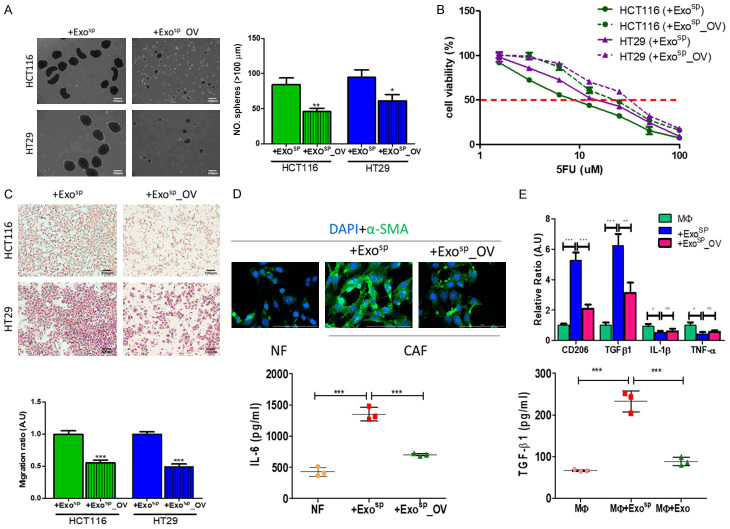
After establishing that tumorsphere derived exosomes (Exosp) contained oncogenic and stroma-transforming potential, we aimed to evaluate the therapeutic potential of ovatodiolide in a similar experimental setup. Exosomes were isolated from tumorspheres generated from ovatodiolide treated HT29 and HCT116 cells (Exosp_OV). Both Exosp_OV were less capable of promoting tumorspheres generation after co-cultured with parental HT29 and HCT116 cells (Figure 4A). Similarly, HT29 and HCT116 cells treated with Exosp_OV showed reduced 5-FU resistance as compared to their counterparts, which were co-cultured with Exosp (Figure 4B); Exosp_OV induced a comparatively lower migratory ability in both HT29 and HCT116 cells as to their counterparts cultured with Exosp (Figure 4C). Subsequently, we examined the stromal normalizing effects of ovatodiolide. We found that Exosp_OV lost a substantial ability to transform NF to CAF. For instance, NF co-cultured with Exosp_OV exhibited markedly lower immunofluorescence of α-SMA and secreted a significantly lower amount of IL-6, as compared to their counterparts co-cultured with Exosp (Figure 4D). Similarly, Exosp_OV exhibited a lower capacity to induce M2 polarization in macrophages, comparing to Exosp (Figure 4E); and the resultant M2 TAMs secreted a significantly lower level of TGF-β1 (Figure 4E).
Figure 4.
Ovatodiolide treatment resulted in less oncogenic exosomes produced by colon cancer tumorspheres. Exosomes, isolated from tumorspheres generated from ovatodiolide-treated (Exosp_OV) HCT116 and HT29, showed a significantly reduced ability to promote tumorsphere formation (A), 5-FU resistance (B), and migration (C) when co-cultured with parental HCT116 and HT29 cells. (D) Exosp_OV cultured normal fibroblasts (NF) showed a lower a-SMA expression (upper panel) and reduced amount of IL-6 released (lower panel) as compared to their Exosp-cultured counterparts. (E) Real-time PCR analysis of M1 M2 markers post macrophages co-cultured with Exosp and Exosp_OV (upper panel). Comparative ELISA of TGF-β1 released by macrophages after co-cultured with Exosp and Exosp_OV (lower panel). MΦ, uncommitted macrophages; M1 TAM markers, TNF-α, IL-1β; M2 TAM markers, CD206, TGF-β1. a, P<0.001; b, P<0.01; NS, no significant difference (all compared to MΦ). *P<0.05; **P<0.01; ***P<0.001.
Ovatodiolide treatment was associated with reduced exosomal cargoes
After observing the decreased oncogenic potential in Exosp_OV, we then analyzed and compared the exosomal cargoes before and after ovatodiolide treatment. According to the comparative Western blots, Exosp_OV demonstrated a significantly lower amount of β-catenin, IL-6, TGF-β1, and p-STAT3 (Figure 5A), including a lower level of miR-1246 as compared to Exosp (Figure 5B). Previous studies indicated that miR-1246 positively regulated β-catenin signaling [21,22]. Here, we explored its role in colon tumorigenesis by silencing miR-1246 level using inhibitor molecules in both HCT116 and HT29 cells. There was a significantly reduced number of tumorspheres generated in both miR-1246 silenced HCT116 and HT29 cells (Figure 5C), accompanied by the reduction of stemness markers such as β-catenin and CD44 as well as STAT3 (Figure 5D); this observation was reversed when the miR-1246 level was increased by miR-1246 mimic molecules (Figure 5C and 5D).
Figure 5.
Ovatodiolide treatment reduced the oncogenic potential of exosomes isolated from colon tumorspheres. A. Comparative western blots of exosomes isolated from tumorspheres generated from parental (Exosp) and ovatodiolide-treated (Exosp_OV) HCT116 and HT29 cells. A marked reduction in IL-6, β-catenin, TGF-β1, and p-STAT3 were observed in Exosp_OV. B. Real-time PCR analysis indicated that a significantly lower level of miR-1246 was found in Exosp_OV (lanes marked by +) as compared to Exosp (lanes marked by -). C. Tumor sphere formation assay in relationship with the miR-1246 level in colon cancer cells. C, scramble control; I, inhibitor; M, mimic. D. Comparative western blots of HCT116 and HT29 cells with miR-1246 level silenced or amplified. Numbers in red indicate the relative expression ratio. **p<0.01; ***p<0.001.
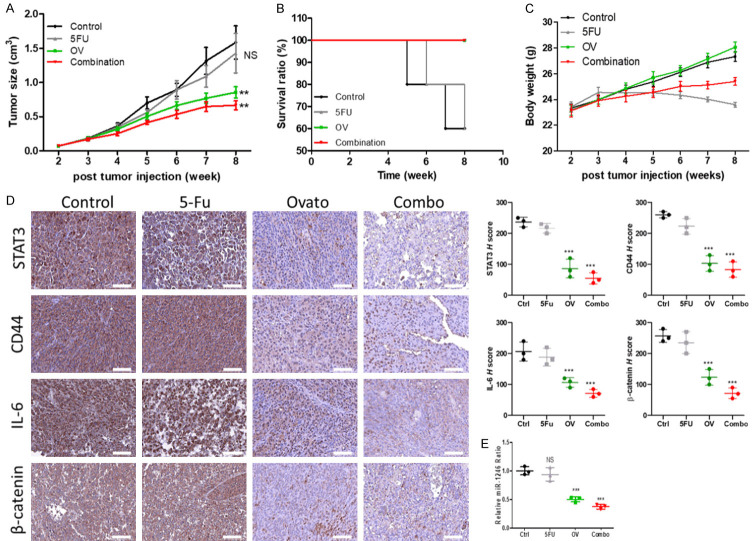
Ovatodiolide suppressed tumorsphere-initiated cancer growth and overcame 5-FU resistance
We examined the potential of ovatodiolide to serve as a colon CSC inhibitor by inoculating mice with HCT116 tumorspheres. Ovatodiolide treatment (10 mg/kg, five times/week) effectively suppressed the tumor growth. Notably, the combination of ovatodiolide and 5-FU (10 mg/kg, three times a week), the most significant inhibitory effect was observed (Figure 6A). The survival curve also showed that the mice which received the combination regimen and ovatodiolide only treatment had the highest survival ratio, followed by the control and 5-FU treatment groups (Figure 6B). The average weight of the mice was tracked over time and showed no significant difference among all groups (Figure 6C). Immunohistochemical sections revealed that tumors from the ovatodiolide only and the combination treatment, expressed a markedly lower level of β-catenin, CD44, IL-6, and STAT3 (Figure 6D). We also examined the level of miR-1246 from the pooled serum exosomes. The level of exosomal miR-1246 was significantly lower in both ovatodiolide and the combination treatment samples, as compared with the control and 5-FU samples (Figure 6E).
Figure 6.
Ovatodiolide treatment suppressed colon tumor growth and overcame 5-FU resistance in vivo. A. Change in tumor burden over time curve. Ovatodiolide alone and ovatodiolide + 5 FU (combination) regimen provided the most pronounced inhibitory effect on tumor growth. B. Kaplan-Meier survival curve. C. Average body weight over time curve. D. Immunohistochemical profiling of tumor samples harvested from all groups (left panels); the corresponding H-score of each marker examined is shown (right panels). E. Comparative qPCR profiling of serum exosomal miR-1246 abundance. **P<0.01; ***P<0.001; NS, no significant difference.
Ovatodiolide functioned as a potential β-catenin inhibitor
To provide mechanistic evidence to support ovatodiolide as a β-catenin inhibitor, we performed a computational docking analysis. The targeted compound (ovatodiolide) was docked into the active site of the β-catenin single subunit (Figure 7A), and the docking energy for each docking mode was calculated (Figure 7B). Amino acid residues within the active site of β-catenin (Asn516, Ser473, and Lys508) were shown to interact with ovatodiolide. The docking study indicated that ovatodiolide could fit and interact within the β-catenin active site where it approximately occupies the active site. The ester site of dixatricyclo ring appears to occupy the polar interaction with the amino acid active site (bond length 1.8 and 2.0 Å). This short length of the docking energy indicates a possible high drug interaction. The presence of hydrogen bond acceptor on ring A or B may increase the interaction between ovatodiolide and β-catenin through the hydrogen bond formation at the active site (Figure 7C). Also, the results from our TCF assay demonstrated that ovatodiolide treatment significantly reduced β-catenin activity in HCT116 cells (Figure 7D).
Figure 7.
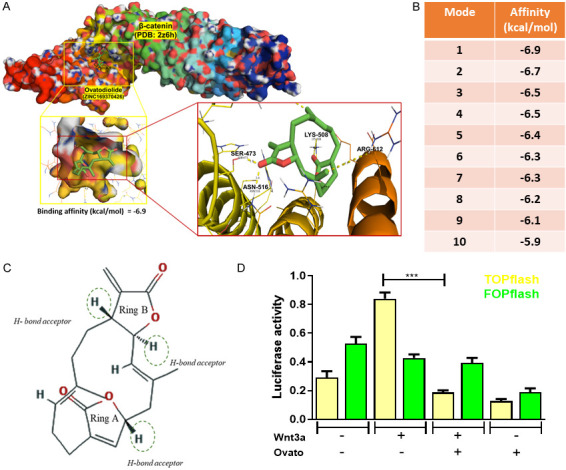
Assessment of ovatodiolide as an inhibitor of β-catenin. A. AutoDock Vina Computational docking model for ovatodiolide and β-catenin. The highest binding affinity scenario is depicted. The amino acid residues (Asn516, Ser473, and Lys508) participate in binding with ovatodiolide are shown. B. The top ten different binding modes are ranked by the free energy (binding affinity). C. The chemical structure of ovatodiolide depicts three potential H-bond acceptors for reinforcing its binding with β-catenin. D. TCF assay. Ovatodiolide treatment (5 µM) significantly reduced β-catenin activity, as determined by the reduced luciferase activity of the TOP/FOP flash system.
Discussion
Cancer stem cells (CSCs) are a subpopulation of cancer cells characterized by enhanced self-renewal ability, infinite dividing, and multi-lineage differentiation potential. CSCs contribute to virtually every aspect of tumorigenesis, including initiation, progression, drug resistance, distant metastasis, and relapse via complex networks of cellular communication within the tumor microenvironment (TME) [23]. Thus, targeting signaling networks involving the generation of CSCs and suppression of CSC-mediated TME transformation is a rational approach for the development of more effective therapeutics.
Here, we first demonstrated that exosomes derived from tumorspheres (Exosp) co-cultured with parental HCT116 and HT29 cells, significantly increased the malignant phenotypes, including increased 5-FU resistance, migratory ability, and self-renewal potential. These phenomena were accompanied by the markedly increased expression of colon CSC markers such as CD133, β-catenin, and CD44, as well as the oncogenic Akt/mTOR/STAT3 axis. In support, a previous study demonstrated that tumor-derived exosomes contributed to the development of 5-FU resistance in colon cancer via exosomal p-STAT3 transfer [24]. Besides, Wang et al., showed that tumor-derived exosomes enhanced both the proliferative and migratory abilities in colon cancer cells via activating WNT1/Akt signaling [25]. Furthermore, CD133 expressing exosomes obtained from both melanoma and colon cancer cells were shown to promote invasiveness in mesenchymal stem cells after a short-term co-culture [26]. Collectively, exosomes derived from colon cancer cells and tumorspheres (Exosp, from our observations) represents an essential route for disease progression.
We further examined the transformative effects of the Exosp on two major stromal cell populations, namely the cancer-associated fibroblasts and macrophages. According to our findings, the Exosp co-culture promoted CAF and M2 TAM transformation. Consequently, Exosp-transformed CAF and M2 TAMs significantly enhanced the ability to generate tumorspheres in parent HCT116 and HT29 cells. In corroboration, a previous study showed that serum exosomes isolated from patients of metastatic colorectal cancer promoted the differentiation of monocytes to M2 macrophages in vitro [27]; Webber et al., demonstrated that exosomes from bladder, prostate, breast, and colon cancer cells contained a high level of TGF-β1 and transformed NF to CAF [28]. Our observations added another dimension in which colon CSCs transform the TME to a pro-tumor niche.
To further characterize colon Exosp, we searched established evidence that provides insights into the exosomal cargoes derived from colon cancer patients and cell lines. Several datasets indicated that miR-1246 as one of the most abundant microRNA species expressed by patients of colorectal cancer [18], strongly associated with KRAS and PI3K signaling; clinically, a higher miR-1246 level predicted a significantly lower survival ratio. Another study corroborated that miR-1246 was highest among the seven most upregulated miRs in serum exosomes isolated from patients of colon cancer, and down-regulated after surgical intervention [19]. Furthermore, miR-1246 was shown to promote cancer stemness and chemoresistance in oral cancer [29]. Uniquely, we found Exosp contained a significantly higher level of miR-1246 than that in Exop, further supporting its role as a colon CSC marker. We also compared the protein cargoes from Exosp and Exop. As expected, Exosp contained a higher amount of β-catenin, TGF-β1, IL-6, and p-STAT3. The identification of p-STAT3 was of importance as the transfer of exosomal p-STAT3 was shown to contribute 5-FU resistance in colon cancer [24]. The enrichment of β-catenin, TGF-β1, IL-6, and p-STAT3 in Exosp reinforces our view that CSCs transform the TME via exosomal signaling. For instance, TGF-β1 and IL-6 enriched Exosp respectively promoted CAF transformation and M2 polarization of TAMs. Consistently, the downregulation of miR-1246 using inhibitor molecules resulted in the decreased expression of β-catenin, STAT3, and CD44 in both HCT116 and HT29 cells and the reverse was confirmed when mimic molecules amplified the miR-1246 level.
Of therapeutic importance, ovatodiolide treatment reduced the exosomal cargoes of Exosp, such as the signaling molecules mentioned above. This finding provided another therapeutic function of ovatodiolide, where it targets not only colon CSCs but also the TME via exosomal communication. A recent study reported that inhibiting β-catenin signaling prevented the CAF generation and suppression of melanoma growth [30]. Our docking simulation suggested that ovatodiolide bound to the Armadillo repeat domain of β-catenin and could inhibit its activity. This finding lent additional support to ovatodiolide as a colon CSC inhibitor. A previous report indicated that exosomal β-catenin could promote cytoskeletal rearrangement, cellular mobility in the recipient cells by activating wnt/β-catenin signaling [31]. These findings strongly suggested that ovatodiolide could suppress the generation of colon CSCs on two levels. First, ovatodiolide suppressed the intrinsic β-catenin expression and activity by reducing the cancer stemness; this subsequently leads to the reduced β-catenin cargo packaged into the CSC-derived exosomes. At present, the exact mechanism by which ovatodiolide reduced the exosomal abundance of oncogenic molecules remains unclear and is under our investigation.
Furthermore, miR-1246 expressing mesenchymal stem cells secrete high levels of IL-6 and induces Jak-Stat and NF-kB signaling pathways [32]; IL-6 induced STAT3 activation in CAFs was associated with colon cancer progression and poor prognosis [33]. More importantly, we demonstrated that ovatodiolide and, in combination with 5-FU, effectively suppressed HCT116 tumor sphere-initiated tumorigenesis. Consistent with the in vitro observations, tumor samples from ovatodiolide and combination treatments showed a markedly reduced expression of STAT3, CD44, IL-6, and β-catenin as compared to the vehicle control and 5-FU treatment groups. The serum exosomal miR-1246 level was significantly lower in both ovatodiolide and combination groups. Our observation and others [22] together suggested the prognostic potential of exosomal miR-1246 in colon cancer. In short, ovatodiolide mediated anti-colon cancer functions in part was associated with the reduced exosomal cargo produced by tumorspheres. The reduced oncogenic cargo examined in our study were all essential factors for the generation of CAF and colon cancer stemness. These findings were collectively summarized in Figure 8.
Figure 8.
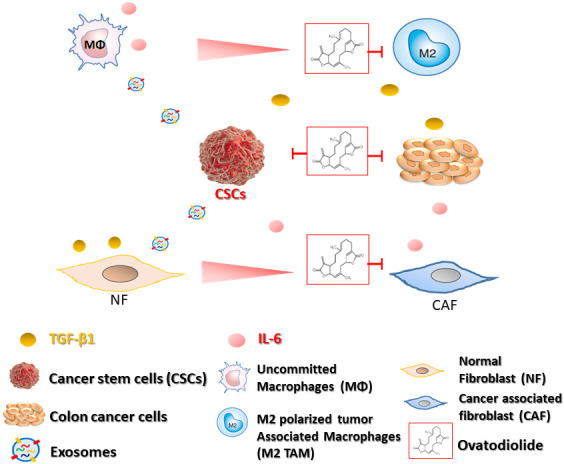
Colon cancer stem cells secrete exosomes containing signaling molecules, including miR-1246, TGF-β1, p-STAT3, β-catenin, and transform uncommitted macrophages and normal fibroblasts to M2 polarized tumor-associated macrophages (M2 TAM) and cancer-associated fibroblasts (CAF) respectively. M2 TAM and CAF promote cancer stemness and 5-FU resistance by secreting TGF-β1 and IL-6. TGF-β1 secreted by M2 TAM also facilitates CAF transformation; CAF secretes IL-6 to enhance M2 polarization and tumorigenesis. Ovatodiolide treatment prevents the generation of M2 TAM, CAF, and CSCs via reducing CSC derived exosomes and their cargoes.
In summary, we have provided preclinical evidence for ovatodiolide as a dual functional therapeutic agent which acts to suppress the generation of colon cancer stem cells, and the transformation of malignant stromal cells via reducing cancer stem cell-derived exosomal β-catenin/STAT3/miR-1246 cargoes.
Acknowledgements
Yan-Jiun Huang and Jing-Wen Shih were supported by the research fund provided by Taipei Medical University (Grant number: 108TMU-TMUH-26). Yan-Jiun Huang was additionally supported by the research grant from the Ministry of Science and Technology, Taiwan (MOST, 108-2314-B-038-013).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Zeuner A, Todaro M, Stassi G, De Maria R. Colorectal cancer stem cells: from the crypt to the clinic. Cell Stem Cell. 2014;15:692–705. doi: 10.1016/j.stem.2014.11.012. [DOI] [PubMed] [Google Scholar]
- 3.Bissell MJ, Hines WC. Why don’t we get more cancer? A proposed role of the microenvironment in restraining cancer progression. Nat Med. 2011;17:320–329. doi: 10.1038/nm.2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Becker A, Thakur BK, Weiss JM, Kim HS, Peinado H, Lyden D. Extracellular vesicles in cancer: cell-to-cell mediators of metastasis. Cancer Cell. 2016;30:836–848. doi: 10.1016/j.ccell.2016.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weidle UH, Birzele F, Kollmorgen G, Rüger R. The multiple roles of exosomes in metastasis. Cancer Genomics Proteomics. 2017;14:1–15. doi: 10.21873/cgp.20015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Milane L, Singh A, Mattheolabakis G, Suresh M, Amiji MM. Exosome mediated communication within the tumor microenvironment. J Control Release. 2015;219:278–294. doi: 10.1016/j.jconrel.2015.06.029. [DOI] [PubMed] [Google Scholar]
- 7.Mashouri L, Yousefi H, Aref AR, Ahadi AM, Molaei F, Alahari SK. Exosomes: composition, biogenesis, and mechanisms in cancer metastasis and drug resistance. Mol Cancer. 2019;18:75. doi: 10.1186/s12943-019-0991-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bamodu OA, Huang WC, Tzeng DT, Wu A, Wang LS, Yeh CT, Chao TY. Ovatodiolide sensitizes aggressive breast cancer cells to doxorubicin, eliminates their cancer stem cell-like phenotype, and reduces doxorubicin-associated toxicity. Cancer Lett. 2015;364:125–134. doi: 10.1016/j.canlet.2015.05.006. [DOI] [PubMed] [Google Scholar]
- 9.Huang YJ, Yang CK, Wei PL, Huynh TT, Whang-Peng J, Meng TC, Hsiao M, Tzeng YM, Wu AT, Yen Y. Ovatodiolide suppresses colon tumorigenesis and prevents polarization of M2 tumor-associated macrophages through YAP oncogenic pathways. J Hematol Oncol. 2017;10:60. doi: 10.1186/s13045-017-0421-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liao YF, Rao YK, Tzeng YM. Aqueous extract of Anisomeles indica and its purified compound exerts anti-metastatic activity through inhibition of NF-kappaB/AP-1-dependent MMP-9 activation in human breast cancer MCF-7 cells. Food Chem Toxicol. 2012;50:2930–2936. doi: 10.1016/j.fct.2012.05.033. [DOI] [PubMed] [Google Scholar]
- 11.Whiteside TL. Tumor-derived exosomes and their role in cancer progression. Adv Clin Chem. 2016;74:103–141. doi: 10.1016/bs.acc.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kalluri R. The biology and function of exosomes in cancer. J Clin Invest. 2016;126:1208–1215. doi: 10.1172/JCI81135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang J, Li S, Li L, Li M, Guo C, Yao J, Mi S. Exosome and exosomal microRNA: trafficking, sorting, and function. Genomics Proteomics Bioinformatics. 2015;13:17–24. doi: 10.1016/j.gpb.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoshii S, Hayashi Y, Iijima H, Inoue T, Kimura K, Sakatani A, Nagai K, Fujinaga T, Hiyama S, Kodama T, Shinzaki S, Tsujii Y, Watabe K, Takehara T. Exosomal microRNAs derived from colon cancer cells promote tumor progression by suppressing fibroblast TP53 expression. Cancer Sci. 2019;110:2396–2407. doi: 10.1111/cas.14084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ahmed FE. miRNA as markers for the diagnostic screening of colon cancer. Expert Rev Anticancer Ther. 2014;14:463–485. doi: 10.1586/14737140.2014.869479. [DOI] [PubMed] [Google Scholar]
- 16.Vichai V, Kirtikara K. Sulforhodamine B colorimetric assay for cytotoxicity screening. Nat Protoc. 2006;1:1112–1116. doi: 10.1038/nprot.2006.179. [DOI] [PubMed] [Google Scholar]
- 17.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Falzone L, Scola L, Zanghì A, Biondi A, Di Cataldo A, Libra M, Candido S. Integrated analysis of colorectal cancer microRNA datasets: identification of microRNAs associated with tumor development. Aging (Albany NY) 2018;10:1000–1014. doi: 10.18632/aging.101444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ogata-Kawata H, Izumiya M, Kurioka D, Honma Y, Yamada Y, Furuta K, Gunji T, Ohta H, Okamoto H, Sonoda H, Watanabe M, Nakagama H, Yokota J, Kohno T, Tsuchiya N. Circulating exosomal microRNAs as biomarkers of colon cancer. PLoS One. 2014;9:e92921. doi: 10.1371/journal.pone.0092921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y, Zhang Z. Potential microRNA-mediated oncogenic intercellular communication revealed by pan-cancer analysis. Sci Rep. 2014;4:7097. doi: 10.1038/srep07097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang JL, Fu YP, Gan W, Liu G, Zhou PY, Zhou C, Sun BY, Guan RY, Zhou J, Fan J, Yi Y, Qiu SJ. Hepatic stellate cells promote the progression of hepatocellular carcinoma through microRNA-1246-RORα-Wnt/β-Catenin axis. Cancer Lett. 2020;476:140–151. doi: 10.1016/j.canlet.2020.02.012. [DOI] [PubMed] [Google Scholar]
- 22.Desmond BJ, Dennett ER, Danielson KM. Circulating extracellular vesicle microRNA as diagnostic biomarkers in early colorectal cancer-a review. Cancers (Basel) 2019;12:52. doi: 10.3390/cancers12010052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsunedomi R, Yoshimura K, Suzuki N, Hazama S, Nagano H. Clinical implications of cancer stem cells in digestive cancers: acquisition of stemness and prognostic impact. Surg Today. 2020 doi: 10.1007/s00595-020-01968-x. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 24.Zhang Q, Liu RX, Chan KW, Hu J, Zhang J, Wei L, Tan H, Yang X, Liu H. Exosomal transfer of p-STAT3 promotes acquired 5-FU resistance in colorectal cancer cells. J Exp Clin Cancer Res. 2019;38:320. doi: 10.1186/s13046-019-1314-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang FW, Cao CH, Han K, Zhao YX, Cai MY, Xiang ZC, Zhang JX, Chen JW, Zhong LP, Huang Y, Zhou SF, Jin XH, Guan XY, Xu RH, Xie D. APC-activated long noncoding RNA inhibits colorectal carcinoma pathogenesis through reduction of exosome production. J Clin Invest. 2019;129:727–743. doi: 10.1172/JCI122478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rappa G, Mercapide J, Anzanello F, Pope RM, Lorico A. Biochemical and biological characterization of exosomes containing prominin-1/CD133. Mol Cancer. 2013;12:62. doi: 10.1186/1476-4598-12-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takano Y, Masuda T, Iinuma H, Yamaguchi R, Sato K, Tobo T, Hirata H, Kuroda Y, Nambara S, Hayashi N, Iguchi T, Ito S, Eguchi H, Ochiya T, Yanaga K, Miyano S, Mimori K. Circulating exosomal microRNA-203 is associated with metastasis possibly via inducing tumor-associated macrophages in colorectal cancer. Oncotarget. 2017;8:78598–78613. doi: 10.18632/oncotarget.20009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Webber J, Steadman R, Mason MD, Tabi Z, Clayton A. Cancer exosomes trigger fibroblast to myofibroblast differentiation. Cancer Res. 2010;70:9621–9630. doi: 10.1158/0008-5472.CAN-10-1722. [DOI] [PubMed] [Google Scholar]
- 29.Lin SS, Peng CY, Liao YW, Chou MY, Hsieh PL, Yu CC. miR-1246 targets CCNG2 to enhance cancer stemness and chemoresistance in oral carcinomas. Cancers (Basel) 2018;10:272. doi: 10.3390/cancers10080272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou L, Yang K, Wickett RR, Kadekaro AL, Zhang Y. Targeted deactivation of cancer-associated fibroblasts by β-catenin ablation suppresses melanoma growth. Tumour Biol. 2016;37:14235–14248. doi: 10.1007/s13277-016-5293-6. [DOI] [PubMed] [Google Scholar]
- 31.Dovrat S, Caspi M, Zilberberg A, Lahav L, Firsow A, Gur H, Rosin-Arbesfeld R. 14-3-3 and β-catenin are secreted on extracellular vesicles to activate the oncogenic Wnt pathway. Mol Oncol. 2014;8:894–911. doi: 10.1016/j.molonc.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bott A, Erdem N, Lerrer S, Hotz-Wagenblatt A, Breunig C, Abnaof K, Wörner A, Wilhelm H, Münstermann E, Ben-Baruch A, Wiemann S. miRNA-1246 induces pro-inflammatory responses in mesenchymal stem/stromal cells by regulating PKA and PP2A. Oncotarget. 2017;8:43897–43914. doi: 10.18632/oncotarget.14915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heichler C, Scheibe K, Schmied A, Geppert CI, Schmid B, Wirtz S, Thoma OM, Kramer V, Waldner MJ, Büttner C, Farin HF, Pešić M, Knieling F, Merkel S, Grüneboom A, Gunzer M, Grützmann R, Rose-John S, Koralov SB, Kollias G, Vieth M, Hartmann A, Greten FR, Neurath MF, Neufert C. STAT3 activation through IL-6/IL-11 in cancer-associated fibroblasts promotes colorectal tumour development and correlates with poor prognosis. Gut. 2020;69:1269–1282. doi: 10.1136/gutjnl-2019-319200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.