Here, we report the genome-wide identification of transcription start sites (TSSs) from two Alphaproteobacteria grown under conditions that result in significant changes in gene expression. TSSs that were identified as present in one condition or both will be an important resource for future studies of these, and possibly other, Alphaproteobacteria.
ABSTRACT
Here, we report the genome-wide identification of transcription start sites (TSSs) from two Alphaproteobacteria grown under conditions that result in significant changes in gene expression. TSSs that were identified as present in one condition or both will be an important resource for future studies of these, and possibly other, Alphaproteobacteria.
ANNOUNCEMENT
Rhodobacter sphaeroides and Novosphingobium aromaticivorans are metabolically diverse and industrially relevant Alphaproteobacteria. R. sphaeroides is a facultative bacterium that can harvest solar energy, fix nitrogen, sequester CO2, and produce valuable chemicals (1–5), while N. aromaticivorans can convert aromatics found in contaminated environments, or derived from lignin, into bioproducts (6–9). Recently, genome-scale experiments have been performed to better understand the metabolic and regulatory networks of each organism, including an analysis of protein-DNA interactions (2, 10–13), global transcript abundance measurements (8, 10, 11, 13–16), and identification of conditionally essential genes using transposon-based sequencing of mutant libraries (9, 17). Here, we report on genome-wide transcription start site (TSS) identification using high-throughput sequencing (TSS-seq) during aerobic respiration and anaerobic photosynthetic growth of R. sphaeroides in Sistrom’s medium (18) at 30°C during mid-log phase and during aerobic growth of N. aromaticivorans in the presence and absence of the aromatic compound vanillic acid in modified Sistrom’s medium (8, 18) at 30°C during mid-log phase.
Three replicates of R. sphaeroides 2.4.1 or N. aromaticivorans DSM 12444 ΔsacB cultures were grown, and RNA was isolated as previously described (8, 18, 19). TSS-seq libraries were produced using RppH, which converts the 5′ triphosphates on unprocessed mRNA species to monophosphates, making them a substrate for ligation of the Illumina adapters (20). The resulting material was sequenced on an Illumina HiSeq 2500 instrument (1 × 50 bp; 117,189,686 total reads for R. sphaeroides and 63,260,190 total reads for N. aromaticivorans) (Table 1). The FASTQ files were split using the index barcode sequences to separate the sequences for the samples treated with or without RppH (RppH+ and RppH−, respectively) using fastx_barcode_splitter.pl version 0.0.13.2 (http://hannonlab.cshl.edu/fastx_toolkit/). The sequences were trimmed to remove any remaining adapter-derived bases using Trimmomatic version 0.3 (HEADCROP, 6; MINLEN, 25) (19) and were aligned to the R. sphaeroides genome (assembly ASM1290v2, GenBank accession number GCF_000012905.2) or the N. aromaticivorans genome (assembly ASM1332v1, GenBank accession number GCF_000013325.1) using Bowtie 2 version 2.3.5.1 (21), allowing for one mismatch (38,571,087 total aligned reads for R. sphaeroides and 29,552,504 total aligned reads for N. aromaticivorans) (Table 1). The aligned Bowtie 2 file was further processed with Picard tools version 2.10.0 (https://broadinstitute.github.io/picard/) and SAMtools (22). The genomeCov command from BEDtools version 2.27.0 (https://bedtools.readthedocs.io/en/latest/) was used to identify genomic locations of the first base in each aligned sequence read, which we defined as the TSS. A pseudocount of 1 was added to all TSS read values to prevent division by 0. The R package edgeR (version 3.10) (23) was used to map locations with a statistically significant increase in read abundance in the RppH+ samples compared to the RppH− samples. Locations with a significant increase in read count in the RppH+ samples compared to the RppH− samples (false discovery rate [FDR], ≤0.05) were retained, defined as TSSs, and associated with genes if the TSS was 350 bp upstream of the translation start site.
TABLE 1.
Summary of sequencing statistics for each sample
| Samplea by bacterial species | Total no. of sequence reads | No. of trimmed sequence reads | No. of aligned sequence reads |
|---|---|---|---|
| R. sphaeroides | |||
| Aerobic Rep A RppH− | 8,254,464 | 6,065,679 | 2,803,014 |
| Aerobic Rep A RppH+ | 8,383,862 | 6,080,322 | 2,795,258 |
| Aerobic Rep B RppH− | 6,174,801 | 4,622,659 | 2,456,095 |
| Aerobic Rep B RppH+ | 8,457,109 | 6,270,320 | 3,469,770 |
| Aerobic Rep C RppH− | 11,058,996 | 8,247,098 | 3,089,246 |
| Aerobic Rep C RppH+ | 13,653,189 | 10,112,149 | 4,366,100 |
| Photosynthetic Rep A RppH− | 10,038,077 | 7,340,387 | 1,565,952 |
| Photosynthetic Rep A RppH+ | 11,451,252 | 8,654,668 | 4,121,559 |
| Photosynthetic Rep B RppH− | 7,016,230 | 4,765,608 | 1,914,259 |
| Photosynthetic Rep B RppH+ | 8,489,377 | 6,181,210 | 3,726,327 |
| Photosynthetic Rep C RppH− | 11,377,067 | 8,247,308 | 2,631,811 |
| Photosynthetic Rep C RppH+ | 12,835,262 | 9,648,342 | 5,631,696 |
| N. aromaticivorans | |||
| Glucose Rep A RppH− | 6,147,801 | 4,690,677 | 2,542,350 |
| Glucose Rep A RppH+ | 5,912,261 | 4,531,213 | 2,262,352 |
| Glucose Rep B RppH− | 4,557,911 | 3,387,765 | 2,286,253 |
| Glucose Rep B RppH+ | 4,706,281 | 3,498,597 | 2,236,403 |
| Glucose Rep C RppH− | 5,300,718 | 4,054,452 | 2,728,261 |
| Glucose Rep C RppH+ | 5,099,093 | 3,891,366 | 2,492,722 |
| Vanillic Acid Rep A RppH− | 3,673,324 | 2,808,993 | 1,596,876 |
| Vanillic Acid Rep A RppH+ | 4,952,555 | 3,789,825 | 2,709,442 |
| Vanillic Acid Rep B RppH− | 3,773,367 | 2,873,551 | 1,725,337 |
| Vanillic Acid Rep B RppH+ | 5,212,702 | 3,992,016 | 3,044,911 |
| Vanillic Acid Rep C RppH− | 6,280,041 | 4,754,420 | 2,122,011 |
| Vanillic Acid Rep C RppH+ | 7,644,055 | 5,887,229 | 3,805,586 |
Each sample was split and treated either with (RppH+) or without (RppH−) RppH as described (20).
In total, 3,214 unique TSSs were identified from the two R. sphaeroides conditions, with 1,793 common TSSs, supporting a large core of promoters used under both conditions and a dramatic reprogramming of the transcriptional network under the two conditions (Fig. 1) (24–26). Of the 2,303 unique TSSs identified under the two N. aromaticivorans conditions, 1,784 were common to both growth conditions, suggesting that there is also a significant transcriptional reprogramming in the presence of an aromatic substrate (Fig. 1). These TSS data sets will serve as a valuable resource to the community, aiding in defining transcription units, identifying promoter elements, predicting binding sites for sigma and other transcription factors, and helping test predictions on the genome-scale metabolic and transcriptional changes associated with lifestyle changes in these and possibly other bacteria (9).
FIG 1.
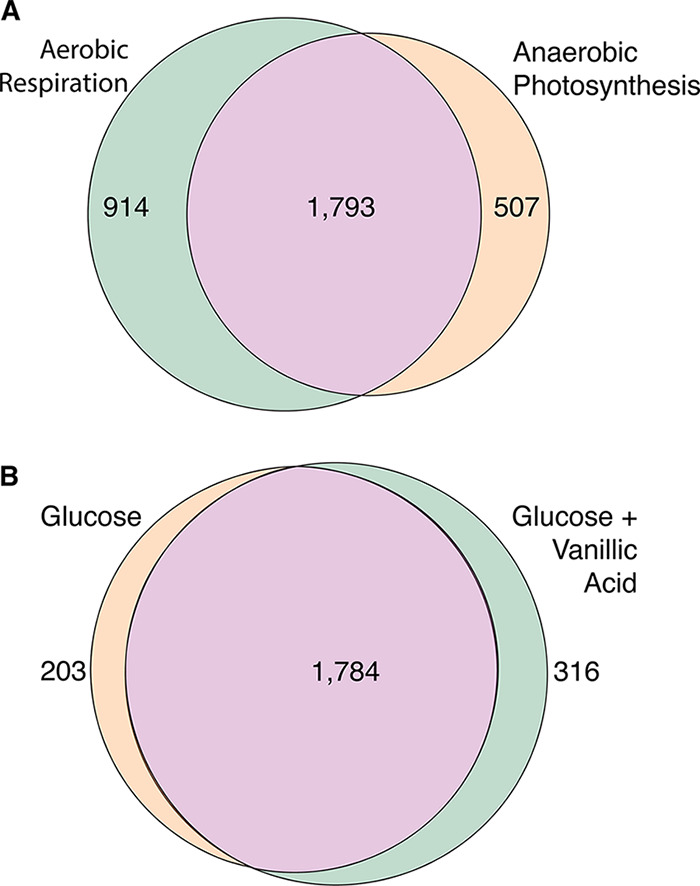
Condition-dependent transcription start site (TSS) identification. TSS populations from R. sphaeroides grown by aerobic respiration and anaerobic photosynthetic conditions (A) and N. aromaticivorans grown in glucose and glucose plus vanillic acid (B). The differences in TSSs in R. sphaeroides and N. aromaticivorans provide a new molecular view on previous reports of condition-dependent changes in gene expression in these Alphaproteobacteria (9, 24–26).
Data availability.
Data are publicly available at NCBI GEO (GSE150944) and SRA (SRP245572).
ACKNOWLEDGMENT
This material is based upon work supported by the Great Lakes Bioenergy Research Center, U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research under award number DE-SC0018409.
REFERENCES
- 1.Atsumi S, Higashide W, Liao JC. 2009. Direct photosynthetic recycling of carbon dioxide to isobutyraldehyde. Nat Biotechnol 27:1177–1180. doi: 10.1038/nbt.1586. [DOI] [PubMed] [Google Scholar]
- 2.Imam S, Noguera DR, Donohue TJ. 2013. Global insights into energetic and metabolic networks in Rhodobacter sphaeroides. BMC Syst Biol 7:89. doi: 10.1186/1752-0509-7-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kontur WS, Ziegelhoffer EC, Spero MA, Imam S, Noguera DR, Donohue TJ. 2011. Pathways involved in reductant distribution during photobiological H(2) production by Rhodobacter sphaeroides. Appl Environ Microbiol 77:7425–7429. doi: 10.1128/AEM.05273-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yilmaz LS, Kontur WS, Sanders AP, Sohmen U, Donohue TJ, Noguera DR. 2010. Electron partitioning during light- and nutrient-powered hydrogen production by Rhodobacter sphaeroides. Bioenerg Res 3:55–66. doi: 10.1007/s12155-009-9072-8. [DOI] [Google Scholar]
- 5.Khatipov E, Miyake M, Miyake J, Asada Y. 1998. Polyhydroxybutyrate accumulation and hydrogen evolution by Rhodobacter sphaeroides as a function of nitrogen availability, p 157–161. In Zaborsky OR, Benemann JR, Matsunaga T, Miyake J, San Pietry A (eds), BioHydrogen. Springer, Boston, MA. [Google Scholar]
- 6.Gall DL, Ralph J, Donohue TJ, Noguera DR. 2014. A group of sequence-related sphingomonad enzymes catalyzes cleavage of beta-aryl ether linkages in lignin beta-guaiacyl and beta-syringyl ether dimers. Environ Sci Technol 48:12454–12463. doi: 10.1021/es503886d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kontur WS, Olmsted CN, Yusko LM, Niles AV, Walters KA, Beebe ET, Vander Meulen KA, Karlen SD, Gall DL, Noguera DR, Donohue TJ. 2019. A heterodimeric glutathione S-transferase that stereospecifically breaks lignin's β(R)-aryl ether bond reveals the diversity of bacterial beta-etherases. J Biol Chem 294:1877–1890. doi: 10.1074/jbc.RA118.006548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kontur WS, Bingman CA, Olmsted CN, Wassarman DR, Ulbrich A, Gall DL, Smith RW, Yusko LM, Fox BG, Noguera DR, Coon JJ, Donohue TJ. 2018. Novosphingobium aromaticivorans uses a Nu-class glutathione S-transferase as a glutathione lyase in breaking the beta-aryl ether bond of lignin. J Biol Chem 293:4955–4968. doi: 10.1074/jbc.RA117.001268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cecil JH, Garcia DC, Giannone RJ, Michener JK. 2018. Rapid, parallel identification of catabolism pathways of lignin-derived aromatic compounds in Novosphingobium aromaticivorans. Appl Environ Microbiol 84:e01185-18. doi: 10.1128/AEM.01185-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dufour YS, Imam S, Koo BM, Green HA, Donohue TJ. 2012. Convergence of the transcriptional responses to heat shock and singlet oxygen stresses. PLoS Genet 8:e1002929. doi: 10.1371/journal.pgen.1002929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Imam S, Noguera DR, Donohue TJ. 2015. CceR and AkgR regulate central carbon and energy metabolism in Alphaproteobacteria. mBio 6:e02461-14. doi: 10.1128/mBio.02461-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Imam S, Yilmaz S, Sohmen U, Gorzalski AS, Reed JL, Noguera DR, Donohue TJ. 2011. iRsp1095: a genome-scale reconstruction of the Rhodobacter sphaeroides metabolic network. BMC Syst Biol 5:116. doi: 10.1186/1752-0509-5-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mackenzie C, Eraso JM, Choudhary M, Roh JH, Zeng X, Bruscella P, Puskas A, Kaplan S. 2007. Postgenomic adventures with Rhodobacter sphaeroides. Annu Rev Microbiol 61:283–307. doi: 10.1146/annurev.micro.61.080706.093402. [DOI] [PubMed] [Google Scholar]
- 14.Imam S, Noguera DR, Donohue TJ. 2014. Global analysis of photosynthesis transcriptional regulatory networks. PLoS Genet 10:e1004837. doi: 10.1371/journal.pgen.1004837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dufour YS, Kiley PJ, Donohue TJ. 2010. Reconstruction of the core and extended regulons of global transcription factors. PLoS Genet 6:e1001027. doi: 10.1371/journal.pgen.1001027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berghoff BA, Glaeser J, Sharma CM, Zobawa M, Lottspeich F, Vogel J, Klug G. 2011. Contribution of Hfq to photooxidative stress resistance and global regulation in Rhodobacter sphaeroides. Mol Microbiol 80:1479–1495. doi: 10.1111/j.1365-2958.2011.07658.x. [DOI] [PubMed] [Google Scholar]
- 17.Burger BT, Imam S, Scarborough MJ, Noguera DR, Donohue TJ. 2017. Combining genome-scale experimental and computational methods to identify essential genes in Rhodobacter sphaeroides. mSystems 2:e00015-17. doi: 10.1128/mSystems.00015-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sistrom WR. 1960. A requirement for sodium in the growth of Rhodopseudomonas spheroides. J Gen Microbiol 22:778–785. doi: 10.1099/00221287-22-3-778. [DOI] [PubMed] [Google Scholar]
- 19.Lemmer KC, Alberge F, Myers KS, Dohnalkova AC, Schaub RE, Lenz JD, Imam S, Dillard JP, Noguera DR, Donohue TJ. 2020. The NtrYX two-component system regulates the bacterial cell envelope. mBio 11:e00957-20. doi: 10.1128/mBio.00957-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vera JM, Ghosh IN, Zhang Y, Hebert AS, Coon JJ, Landick R. 2020. Genome-scale transcription-translation mapping reveals features of Zymomonas mobilis transcription units and promoters. mSystems 5:e00250-20. doi: 10.1128/mSystems.00250-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup . 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Callister SJ, Nicora CD, Zeng X, Roh JH, Dominguez MA, Tavano CL, Monroe ME, Kaplan S, Donohue TJ, Smith RD, Lipton MS. 2006. Comparison of aerobic and photosynthetic Rhodobacter sphaeroides 2.4.1 proteomes. J Microbiol Methods 67:424–436. doi: 10.1016/j.mimet.2006.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arai H, Roh JH, Kaplan S. 2008. Transcriptome dynamics during the transition from anaerobic photosynthesis to aerobic respiration in Rhodobacter sphaeroides 2.4.1. J Bacteriol 190:286–299. doi: 10.1128/JB.01375-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roh JH, Smith WE, Kaplan S. 2004. Effects of oxygen and light intensity on transcriptome expression in Rhodobacter sphaeroides 2.4.1. Redox active gene expression profile. J Biol Chem 279:9146–9155. doi: 10.1074/jbc.M311608200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data are publicly available at NCBI GEO (GSE150944) and SRA (SRP245572).


