Abstract
Dengue fever is an acute infectious disease caused by dengue virus (DENV) and transmitted by Aedes mosquitoes. There is no effective vaccine or antiviral drug available to date to prevent or treat dengue disease. Recently, RNA-dependent RNA polymerase (RdRp), a class of polymerases involved in the synthesis of complementary RNA strands using single-stranded RNA, has been proposed as a promising drug target. Hence, we screened new molecules against DENV RdRp using our previously constructed virtual screening method. Mol-5, [1,2,4]triazolo[1,5-a]pyrimidine derivative, was screened out from an antiviral compound library (~8000 molecules). Using biophysical methods, we confirmed the direct interactions between mol-5 and purified DENV RdRp protein. In luciferase assay, mol-5 inhibited NS5-RdRp activity with an IC50 value of 1.28 ± 0.2 μM. In the cell-based cytopathic effect (CPE) assay, mol-5 inhibited DENV2 infectivity with an EC50 value of 4.5 ± 0.08 μM. Mol-5 also potently inhibited DENV2 RNA replication as observed in immunofluorescence assay and qRT-PCR. Both the viral structural (E) and non-structural (NS1) proteins of DENV2 were dose-dependently decreased by treatment with mol-5 (2.5–10 μM). Mol-5 treatment suppressed DENV2-induced inflammation in host cells, but had no direct effect on host defense (JAK/STAT-signaling pathway). These results demonstrate that mol-5 could be a novel RdRp inhibitor amenable for further research and development.
Keywords: Dengue virus; antiviral agent; RdRp inhibitor; [1,2,4]triazolo[1,5-a]pyrimidine derivative; inflammation; JAK/STAT signaling pathway
Introduction
Dengue is a rapidly spreading vector-borne viral disease that is endemic in more than 100 countries [1–3]. Approximately 54 million dengue fever cases were reported in Latin America and the Caribbean [4]. Mosquito-borne members of the Flavivirus family include four serotypes of dengue virus (DENV1–4), West Nile virus (WNV) and the recently emerging Zika virus (ZIKV). These viruses are classified as re-emerging pathogens due to the frequency and severity of recent epidemics. The disease burden due to dengue has recently been revised, and accordingly, ~390 million infections occur annually in tropical and subtropical regions of the world. Infection by any one of the serotypes could be either asymptomatic or cause flu-like symptoms, which are known as dengue fever. Although primary infection by a single serotype results in life-long immunity, there is no cross protection against infections by other serotypes, which could lead to severe dengue disease due to an antibody-dependent enhancement mechanism [5, 6]. Recently, a vaccine against DENV named Dengvaxia was associated with increased death and was thus prohibited in the Philippines at the end of last year [7]. Therefore, currently, there is no effective vaccine or antiviral drug available to control or treat dengue disease.
Compared with the receptor of DENV and other critical molecular determinants [8, 9], the NS5 flavivirus RNA-dependent RNA polymerase (RdRp) is an important drug target [10, 11]. A number of allosteric inhibitors were described previously that could inhibit DENV RdRp function, and many of these show much promise for further development to target DENV RdRp [12] and virus fusion [13]. However, these inhibitors are still under development for clinical usage. Using fragment-based screening via X-ray crystallography, scientists identified a promising RdRp (DENV)-binding site in the thumb/palm interface, which is close to the active site (termed the “N pocket”) [14]. Structure-guided optimization yielded nanomolar inhibitors of RdRp de novo initiation activity with low micromolar EC50 values, as determined by DENV cell-based assays. However, the clinical effects of these compounds are still unknown. Thus, the design of new lead compounds is urgently needed for DENV2 treatment.
From our previously published virtual screening model of DENV NS5 [15, 16], we developed an interaction model for predicting the optimal binding characteristics at the DENV RdRp site [17]. From the in silico screening of the antiviral library of the ChemDiv Company (~8000 molecules, http://www.chemdiv.com/antiviral-library), mol-5 was identified as a promising molecule. Mol-5 was found to directly interact with purified DENV2 NS5-RdRp in vitro. In addition, the levels of DENV2 viral proteins and mRNA were reduced by mol-5. These results clearly demonstrate the potential of mol-5 as a novel RdRp inhibitor that is amenable to further development.
Materials and methods
Experimental materials
Baby hamster Syrian kidney (BHK-21) and C6/36 mosquito cells were cultured in RPMI-1640 (Thermo Fisher Scientific, MA, USA) with 10% calf serum (Thermo Fisher Scientific, MA, USA) at 37 °C. DENV2 (Dengue virus 2 New Guinea C derivative strain) was provided by Professor Xiaoguang Chen [18]. DENV2 was amplified in C6/36 cells and stored at −80 °C until use. DENV1, 3, and 4 were kindly gifts from by Professor Wei Zhao. Dr. Min Zou contributed the ZIKV used for our study.
Virtual screening assay
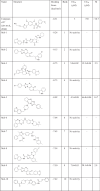
In this study, molecular docking was performed using AutoDock 4.2. [19]. The crystal structure of DENV 2 RdRp complexed with inhibitor compound 27 (PDB: 5K5M) [20] was selected and downloaded from the RCSB protein data bank (http://www.rcsb.org/pdb). To prepare the receptor, polar hydrogens and Gasteiger charges were added to the protein, which was obtained by deleting all the crystallographic waters and the inhibitor. Then, a 3D search grid was created by the use of the AutoGrid algorithm, which utilized the center of compound 27 as the grid center, and the size of the docking grid was set to 40 Å × 40 Å × 40 Å [20]. The antiviral compound library (~8000 compounds, ChemDiv) was prepared by openbabel software version 2.3.0 [21] and prepare_ligand4.py in AutoDockTools. The docking parameters were the same as those used in our previous research [15]: ga_pop_size, 1500; ga_num_evals, 25,000,000; ga_num_generations, 27000; ga_run, 30. The other parameters were kept at their default values. After docking, the top 10 compounds with the best scores were selected for experimental validation (Table 1). Both the chemical structures and docking scores are listed in Table 1. Schrödinger software was used to account for the protein–ligand interactions and the hydrophobic interactions [22].
Table 1.
The structure, docking score of the top 10 compounds
 |
Absorption, distribution, metabolism, and excretion (ADME) properties prediction
The ADME properties of the 10 selected compounds were predicted using the QikProp module version 3.4 in Schrödinger software. QikProp provides the rapid prediction of the ADME properties of drug candidate molecules. For each successfully processed molecule, QikProp predicts the pharmaceutically relevant descriptors. It evaluated the acceptability of the filtered compounds based on the Lipinski rule of 5, and the results are listed in Table 2.
Table 2.
ADME properties of top 10 compounds
| Name | MWa | QP log Po/wb | QP log Sc | hERGd | QPPCacoe | #metaf | % of human oral absorptiong | PSAh |
|---|---|---|---|---|---|---|---|---|
| Mol-1 | 495.5 | 3.101 | −5.937 | −4.501 | 445.183 | 1 | 92.508 | 129.074 |
| Mol-2 | 440.8 | 4.729 | −7.098 | −7.386 | 672.57 | 2 | 100 | 83.906 |
| Mol-3 | 469.5 | 5.595 | −7.359 | −5.051 | 2027.07 | 4 | 100 | 73.209 |
| Mol-4 | 440.8 | 4.718 | −7.11 | −7.402 | 669.552 | 2 | 100 | 83.9 |
| Mol-5 | 468.5 | 4.492 | −5.53 | −7.054 | 852.599 | 5 | 100 | 66.617 |
| Mol-6 | 430.5 | 3.65 | −5.578 | −7.343 | 218.116 | 4 | 90.175 | 98.849 |
| Mol-7 | 478.5 | 4.987 | −9.002 | −7.986 | 220.322 | 3 | 100 | 115.009 |
| Mol-8 | 485.4 | 3.724 | −6.294 | −6.838 | 599.355 | 3 | 100 | 111.596 |
| Mol-9 | 431.8 | 3.214 | −4.394 | −6.302 | 1487.57 | 2 | 100 | 87.507 |
| Mol-10 | 483.5 | 4.217 | −5.901 | −8.664 | 218.599 | 4 | 93.515 | 93.087 |
aMolecular weight (<500)
bPredicted octanol/water partition coefficient (from −2.0 to 6.5)
cPredicted aqueous solubility; S in mol/L (acceptable range: −6.5 to 0.5)
dPredicted blockage of hERG K+ channel (reasonable value < −6)
ePredicating Caco-2 cell permeability in nm·s−1 (<25 poor, >500 great)
fNumber of likely metabolic reactions (1–8)
g% of human oral absorption (<25% is weak and >80% is strong)
hPolar surface area (140 needed for compound to permeate cell membranes)
Purification of His-tagged recombinant DENV NS5-RdRp
DENV2 NS5-RdRp (amino acid 274–900) was cloned into the pET15b plasmid with an N-terminal His-tagged label and was expressed in E. coli BL21 cells [23]. After the extraction of the total protein, the RdRp protein was purified with a nickel-immobilized metal ion affinity chromatography column, gel filtration chromatography (SuperdexTM 75), and an ÄKTA purifier (GE Company, CT, USA). Finally, the purity of RdRp protein was tested using SDS–PAGE and Coomassie blue staining, and a purity > 90% was observed.
Surface plasmon resonance imaging (SPRi) analysis
SPRi, a highly sensitive analytical technique, was used to monitor the interaction between mol-5 and NS5-RdRp. In an experiment to monitor the interaction of DENV2 NS5-RdRp with mol-5 using a PlexArray HT A100 SPRi imager apparatus (Plexera, WA, USA), DMSO was used as the negative control, and the interaction between rapamycin and FKBP12 protein (100 nM) was used as the positive control. First, 0.5 μL of each sample was applied to the chip at 25 °C. When the indicated concentration of RdRp flowed through the chip, the reflectivity change at each point was measured, and the calibration coefficient was calculated to standardize the point response of the dynamic measurement. The average of the reflectance measurements that were repeated at several points, after subtracting the initial reactivity from the original data, was used to provide the value of the reflectance offset obtained when the molecules bound to the chip. Then, the Kd value was calculated by using a nonlinear regression fit model.
Determination of the 50% tissue culture infective dose (TCID50)
DENV virus was first propagated in C6/36 cells, and the supernatant was collected, aliquoted, and frozen at −80 °C until use. The TCID50 was determined using BHK-21 cells. The DENV stocks were diluted serially (10-fold), and 100 μL of each dilution was added to duplicate wells in a 96-well plate containing a confluent monolayer of BHK-21 cells. The cells were incubated for 4 days to allow the development of the cytopathic effects (CPEs). The dilution inducing CPE in half of the cultures was considered to indicate the TCID50 value per unit volume of viral suspension. In this study, virus at a concentration of 101 TCID50/mL was used to infect BHK-21 cells.
Measurement of the virus-induced CPE and viral infection
BHK-21 cells were cultured in a 96-well plate (1 × 105 cells/well) overnight. Before being infected, the cells were washed with PBS once to remove the serum-containing medium. Then, the virus was diluted 1:1000 with RPMI-1640 medium without fetal calf serum to obtain a concentration of 101 TCID50. The culture medium was removed after 1 h of infection with DENV2 viral inoculum (100 μL/well) at 37 °C, and maintenance medium (2% calf serum) was added. The cells were incubated for 96 h until lesions were clearly observed under a microscope (Nikon, Tokyo, Japan). At that time, the lactate dehydrogenase (LDH) levels could be measured in the supernatants (Beyotime Biotechnology, Shanghai, China) at OD450.
Plaque assay
C6/36 cells were cultured in a 12-well plate (5 × 105 cells/well) overnight. One hour after adding DENV2 (200 μL/well) to the cells (101 TCID50), the C6/36 cells were further incubated with maintenance medium containing 2% serum and 1% methyl cellulose. The cells were cultured for 96 h until lesions were clearly visible under microscope (Nikon, Tokyo, Japan). Next, the cells were immobilized with phosphate-buffered saline (PBS) containing 3.7% formaldehyde and were stained by methylene blue according to a procedure reported earlier [24].
Time-dependent administration assay
BHK-21 cells were cultured in a 12-well plate (105 cells/well) overnight. Before being infected, the cells were washed with PBS once to remove the serum-containing medium. Then, the DENV2 (101 TCID50) was diluted 1:1000 with RPMI-1640 (Thermo Fisher Scientific, MA, USA) containing fetal calf serum (Gibco, USA). The culture medium was removed after 1 h of infection with DENV2 viral inoculum (100 μL/well). At 0, 4, 8, and 12 h postinfection (hpi), maintenance medium (2% serum) containing mol-5 (10 μM, solubilized in 100% DMSO) was added to the infected cells. Cells treated with DMSO only were used as a control group. Forty-eight hpi, the cell culture supernatant was collected to measure the LDH levels.
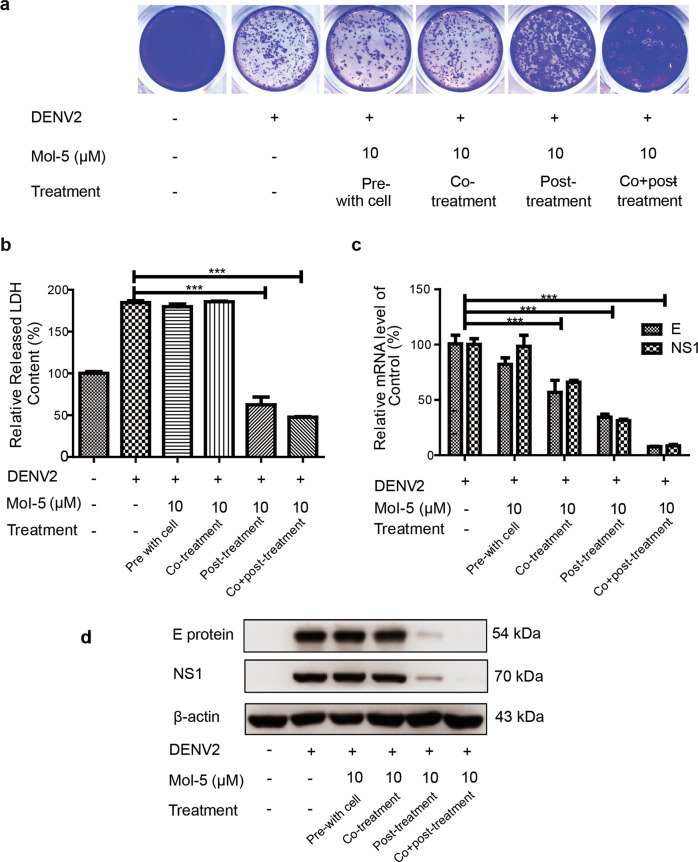
BHK-21 cells were cultured in a 12-well plate (105 cells/well) overnight. DENV2 (101 TCID50) was diluted 1:1000 with RPMI-1640 (Thermo Fisher Scientific, MA, USA) containing fetal calf serum (Gibco, USA). Pretreatment means that the cells were treated with mol-5 (10 μM, solubilized in 100% DMSO) dissolved in serum-free medium 1 h earlier. After removing the drug, the medium was washed away with PBS, and then DENV2 (in serum-free medium) was added for infection for 1 h. After removing the virus, the cells were cultured for 48 h with 2% FBS in culture medium. During cotreatment, DENV2 (serum-free medium) was mixed with mol-5 (10 μM) and added to the cells for 1 h, and then DENV2 and mol-5 were removed. Medium with 2% FBS was added, and the cells were cultured for 48 h. During posttreatment, DENV2 was added to the cells for 1 h, and then DENV2 was removed and medium with 2% FBS containing mol-5 (10 μM) was added. “Cotreatment” indicates that the cells were infected by DENV2 with mol-5 (10 μM), and “posttreatment” indicates that the cells were cultured with mol-5 (10 μM) in medium (2% FBS). DMSO was used as a solvent control.
Western blotting
During Western blotting, NS3 antibody (Abcam Company, Cambridge, UK) and E and NS5 antibodies (Neobioscience Company, Beijing, China) were used to specifically detect and quantify the target proteins NS3, E and NS5 in protein extracts. The β-actin antibody (CST, MA, USA) was used to determine the total protein content.
Immunofluorescence assay
BHK-21 cells were infected with DENV2 (101 TCID50) for 1 h and treated with mol-5 (5 and 10 μM) for 48 h or were not treated, and then the cells were fixed with 4% paraformaldehyde for 15 min, permeabilized with 0.1% Triton X-100 for 10 min and blocked with PBGB (PBS containing 10% normal goat serum and 1% BSA) at room temperature for 1 h after discarding the supernatant and washing twice with PBS. Subsequently, the cells were mixed with NS1, E, or dsRNA (J2) antibodies (English & Scientific Consulting Kft, Budapest, Hungary) and incubated at 4 °C overnight. After washing with PBS three times, the cells were incubated with fluorescein isothiocyanate or tetramethylrhodamine isothiocyanate-conjugated secondary antibody (Thermo Fisher Scientific, MA, USA) at room temperature for 1 h, and images were obtained with a confocal microscope (Zeiss, Jena, Germany).
Quantitative real-time PCR (qRT-PCR)
BHK-21 cells were first infected with DENV2 (101 TCID50) for 1 h and then treated with mol-5 (2.5, 5 and 10 μM) for 48 h. RNA was extracted and purified using a viral RNA extraction kit (Qiagen, Dusseldorf, Germany), and then cDNA was synthesized and monitored with a SuperScript-III kit (Takara, DaLian, China). The housekeeping gene GAPDH was used as an internal reference. The following primers were used in our experiments: NS1 forward, 5′-TGCAGGCAGGAAAACGATCT-3′ and reverse, 5′-CATGGACGGCTCTGTTGTCT-3′; E protein Forward, 5′-TCGCTCCCCTCATTGTTGTC-3′ and Reverse, 5′-AGGGGAGCGAAGAGAATGG-3′; ISG56 Forward, 5′-ACACCTGAAAGGCCAGAATGAGGA-3′ and Reverse, 5′-TGCCAGTCTGCCCATGTGGTAATA-3′; ISG54 Forward, 5′-GACACGGTTAAAGTGTGGAG-3′ and Reverse, 5′-GGTACTGGTTGTCAGGATTC-3′; IFITM1 Forward, 5′-GTTACTGGTATTCGGCTCTG-3′ and Reverse, 5′- GGTGTGTGGGTATAAACTGC-3′; IFITM3 Forward, 5′-ATGTCTCTGCCGTC-3′ and Reverse, 5′-GTCATGAGGATGCCCAGAAT-3′; IL-1β Forward, 5′-CAACAGGCTGCTCTGGGATT-3′ and Reverse, 5′-GGGCCATCAGCTTCAAAGAAC-3′; TNF-α Forward, 5′-GCCCATGTTGTAGCAAACCC-3′ and Reverse, 5′-GGACCTGGGAGTAGATGAGGT-3′; IL-8 Forward, 5′- GAGAGTGATTGAGGGACCAC-3′ and Reverse, 5′- CAACCCTGCACCCAGTTT -3′; GAPDH Forward, 5′-TGACCTCAACTACATGGTCTACA-3′ and Reverse, 5′-CTTCCCATTCTCGGCCTTG-3′.
Cell cytotoxicity assay
BHK-21 cells were cultured in a 96-well plate (1 × 105 cells/well) overnight. The cells were then treated with the screened compounds (Mol-1–Mol-10) at the indicated concentrations (0.3125, 0.625, 1.25, 2.5, 5, 10, or 20 μM) for 48 h and incubated with 0.5 mg/mL of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) for 4 h to test cell viability. The formazan crystals that formed were dissolved in DMSO and analyzed using a Tecan microplate reader at 570 nm (Tecan, Mannedorf, Switzerland).
Evaluation of the anti-DENV NS5 polymerase activity of mol-5 in BHK-21 cells
The method was performed as previously reported [25]. BHK-21 cells were seeded in a 24-well plate at a density of 5 × 104 cells per well and cotransfected with the pcDNA3.1-NS5-RdRp (0.5 μg) and p(+)Rluc-(−)DV-UTRΔC-Fluc (0.5 μg) plasmids. The pcDNA3.1-NS5-RdRp plasmid was synthesized by Synbio Tech Company, and the p(+)Rluc-(−)DV-UTRΔC-Fluc (0.5 μg) plasmid was synthesized as previously reported. The sequences of NS5 and UTR were obtained from GenBank (GenBank: AF038402.1). The plasmid-transfected cells were then treated with different concentrations (0.625, 1.3, 2.5, 5, 10, and 20 μM) of mol-5 for 72 h. The reporter gene expression was measured with a luciferase assay kit (Promega, WI, USA) according to the manufacturer’s instructions. Rluc is regulated by the CMV promoter, and its fluorescent signal indicates the number of cells; Fluc is regulated by the NS5-RdRp promoter, and its fluorescent signal indicates RdRp activity. The ratio of the two represents the expression level of NS5-RdRp.
Statistical analysis
All the data are expressed as the mean ± standard deviation (SD). Multiple comparisons were performed by one-way ANOVA followed by a Bonferroni post hoc test (to compare all pairs of columns) when the variances were equal between groups. All statistical analyses were performed using Prism 5 GraphPad software. In all cases, if the P value determined by a statistical test was <0.05, the difference was considered statistically significant. Values are presented as follows: ns P > 0.05, *P < 0.05, **P < 0.01, and ***P < 0.001. All experiments were performed in triplicate.
Results
Mol-5 binding to DENV2 RdRp in silico and biophysical assays
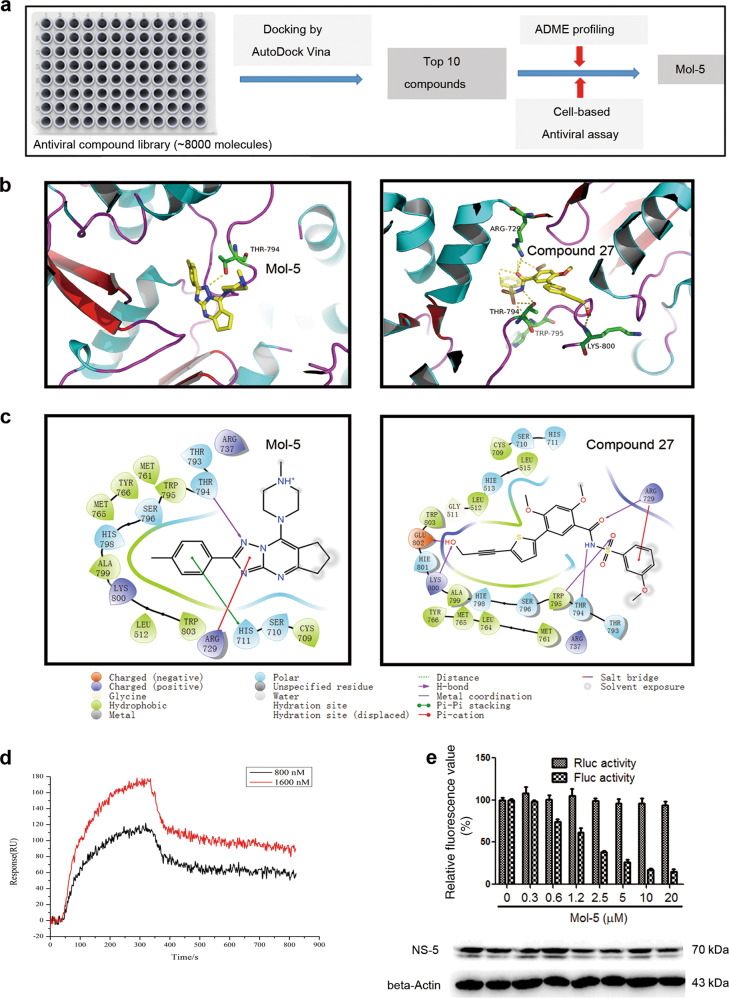
Unlike the previous study using the DENV-3 NS5-RdRp for drug screening, this study used virtual screening with the DENV2 NS5-RdRp crystal structure, which was resolved recently [20]. The workflow used for virtual screening is given in Fig. 1a. Water molecules, ions, and inhibitors were completely removed before performing the virtual screening procedure with the DENV2 RdRp protein. The inhibitor of RdRp (compound 27) was first docked to the protein to validate our calculation model (Fig. 1b); the important hydrogen bond interactions were found in the resulting crystal structure (Fig. 1c), such as those involving residues R729, T794, and E802, suggesting our docking model was reliable [20].
Fig. 1. Virtual screening of mol-5.
a The scheme used for the virtual screening strategy. b The binding modes of mol-5 and Compound 27 in the pocket of RdRP were determined with Docking Vina and PyMOL software. c The ligand interactions were analyzed by Schrödinger software. d The interaction effects between mol-5 and RdRP were detected by SPRi assays. e The reduction of DENV NS5 polymerase activity by mol-5 was shown in a cell-based reporter assay. BHK-21 cells were cotransfected with the p(+)Rluc-(−)DV-UTRΔC-Fluc (0.5 μg) and pcDNA3.1-NS5-RdRp plasmids (0.5 μg). The transfected cells were incubated with mol-5 at the indicated concentration for 72 h. The luciferase activity and NS5 protein levels in the cell lysates were analyzed as depicted in the figure. Each data point represents the mean ± S.D. of three separate experiments.
Through the virtual screening of the antiviral library (~8000 compounds), the top 10 compounds having the best scores were identified and ranked according to the docking scores (Table 1). These initial hits were evaluated for their antiviral effects by CPE assays (Fig. S1), and their cytotoxic effects were evaluated by MTT assays (Table 1). Initially, mol-3, mol-5, and mol-9 showed promising antiviral properties. In addition, the structures of these compounds were found to be similar and have good ADME characteristics (Table 2). However, mol-3 was found to produce high cytotoxicity. Based on all the tested antiviral properties, mol-5 was finally selected for further target validation (Table 1). Mol-5 is a [1,2,4]triazolo[1,5-a]pyrimidine derivative and its 1H-NMR, 13C-NMRA, HRMS, and IR spectra are shown in Fig. S2.
The docking scores for mol-5 were lower (−8.0 kcal/mol) than those for compound 27 (−8.31 kcal/mol), suggesting the decreased binding affinity of mol-5 for RdRp (Fig. 1b). However, these results also suggested that mol-5 does in fact form a hydrogen bond with residue T794 similar to that formed by compound 27 (Fig. 1c). These two molecules are shown overlapping in Fig. S3 for comparison. To confirm whether mol-5 could interact with RdRp, SPRi analysis, which is a label-free detection method suitable and reliable for the real time monitoring of biomolecular interactions [26], was performed. Consistent with the results of the in silico assay, mol-5 was found to bind RdRp with a Kd value of 1.0 ± 0.30 μM using a nonlinear regression program (Fig. 1d). We next performed a cell-based assay to examine the effect of mol-5 on DENV NS5 polymerase activity [25]. The pcDNA3.1-NS5-RdRp (274–900) and p(+)Rluc-(−)DV-UTRΔC-Fluc plasmids were cotransfected into BHK-21 cells and cultured in the presence of mol-5 for 72 h. The cell lysates were then harvested for analyzing the Rluc and Fluc activities. Rluc was regulated by the CMV promoter, and its fluorescent signal indicated the cell number. Fluc was regulated by NS5-RdRp promoter, and its fluorescent signal indicated the RdRp activity. We found that mol-5 reduced Fluc activity in a concentration-dependent manner but had no effect on Rluc activity (Fig. 1e), revealing that mol-5 inhibited DENV NS5 polymerase activity with an IC50 value of 1.28 ± 0.2 μM. These data suggested that mol-5 could directly interact with DENV2 RdRp and inhibit its activity.
Effects of mol-5 on host cells after DENV2 infection
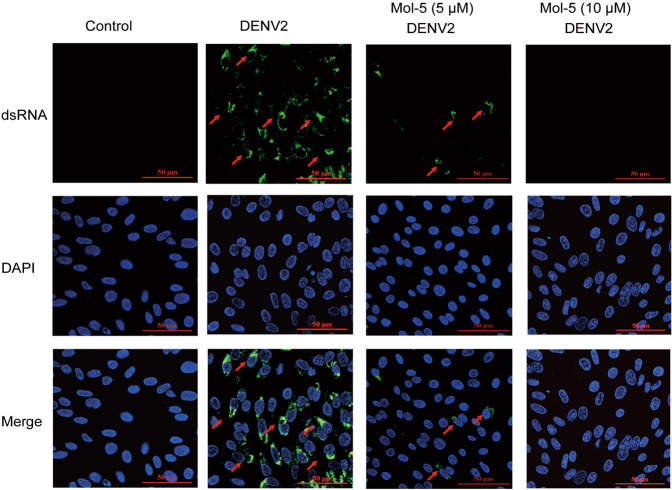
At the proliferation stage of DENV, RdRp synthesizes double-stranded RNA (dsRNA), which plays a vital role in the formation of progeny viral particles [27]. Hence, we analyzed whether mol-5 inhibited viral dsRNA synthesis. Immunofluorescence analysis was performed to detect dsRNA. As expected, mol-5 reduced dsRNA production, whereas dsRNA was clearly observed in the control group (Fig. 2). This result demonstrated that mol-5 inhibited DENV2 RNA replication after infection, indicating that it might be a potent inhibitor of DENV2 RdRp.
Fig. 2. Effects of mol-5 on DENV2 dsRNA.
BHK-21 cells infected with DENV2 (101 TCID50), washed with virus-free medium 1 hpi at 37 °C, and were then incubated with mol-5 (10 μM) for 24 h. The same amount of DMSO was added as a solvent control. The dsRNA intermediates were visualized by immunofluorescence with the primary antibody J2, which recognizes dsRNA (green). Nuclei were stained with DAPI (blue). dsRNA is indicated by arrows. Scale 50 μm.
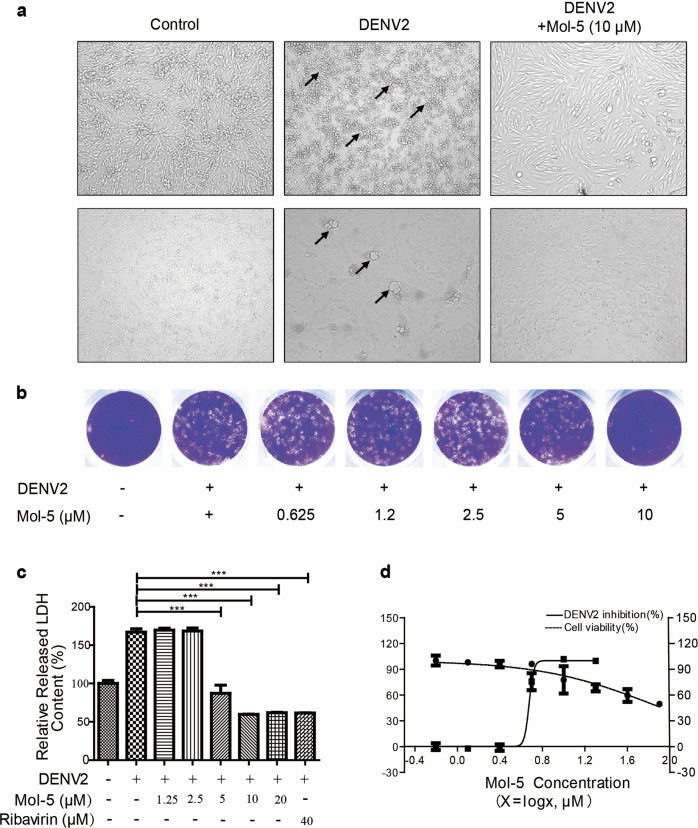
NS5, a nonstructural protein of the flavivirus, is a multifunctional protein containing two functional domains, a methyltransferase (MTase) and an RNA polymerase (RdRp), which play important roles in viral genome replication [28]. Mol-5 directly binds to RdRp, and the antiviral capacity of mol-5 was further evaluated at the cellular level. At first, the mosquito cell line C6/36 was chosen to confirm the antiviral effects of mol-5 because mosquitoes are natural hosts of DENV. C6/36 cells were incubated with DENV2 in the presence or absence of mol-5 for 24 h, and a significant CPE was observed in C6/36 cells after DENV2 infection. In contrast, the CPE of DENV was attenuated after mol-5 treatment (Fig. 3a). Furthermore, we investigated the effects of the anti-DENV activity of mol-5 in mammalian BHK-21 cells, which could be infected by DENV2 easily. BHK-21 cells were treated with mol-5 after DENV2 infection, and the result indicated that mol-5 could inhibit the DENV2-induced CPE in BHK-21 cells (Fig. 3a). In summary, the antiviral effect of mol-5 was consistently observed in both mosquito cells and mammalian cells. In addition, we have also found that the production of progeny viral particles was reduced by mol-5 treatment in a dose-dependent manner (Fig. 3b).
Fig. 3. Antiviral effect of mol-5 on BHK-21 cells.
BHK-21 or C6/36 cells were infected by DENV2 (101 TCID50), washed after infection at 37 °C for 1 h, and incubated with mol-5 (0.63, 1.25, 2.5, 5, or 10 μM) in the maintenance medium. Ribavirin (40 μM) was used as a positive control. At 96 hpi, a the CPE was observed under a microscope in both BHK-21 cells and C6/36 cells, as indicated; CPE is indicated by the arrow. b The extracellular virus yield was quantified by plaque assays. c The EC50 values of the extracellular culture media were determined by LDH assays. d BHK-21 cells were incubated with mol-5 (0.63, 1.25, 2.5, 5, or 10 μM) without DENV2, and the cell viability was measured using MTT assays. Each data point represents the mean ± S.D. of an average of three separate experiments. *** indicates P < 0.001.
DENV2 infection induced BHK-21 cell death 96 hpi, which increased LDH secretion into the culture supernatant [15]. To quantify the antiviral effect of mol-5, the LDH level in BHK-21 cell supernatant was measured after viral infection. Experiments have shown that mol-5 could inhibit DENV-induced LDH release in a dose-dependent manner with an EC50 of 4.5 ± 0.08 μM, as determined by nonlinear regression (Fig. 3c). In addition, the cytotoxic effect of mol-5 was measured by an MTT assay, and the data showed that mol-5 had a CC50 of 66.0 μM (Fig. 3d). As a result, the selective index (CC50/EC50) was 14.7, which showed that there is still potential for mol-5 modification to improve the selective index. Furthermore, the effects of mol-5 on other DENV serotypes, such as DENV1, DENV3, and DENV4, were tested, and EC50 values of 5.214 ± 0.15, 3.662 ± 0.13, and 9.95 ± 0.11 μM were determined, respectively (Fig. S3). However, mol-5 had a weak antiviral effect on ZIKV, with an EC50 of 34.30 ± 0.65 μM (Fig. S4). These data suggested mol-5 could be a pan-DENV inhibitor.
Effects of mol-5 on DENV2 RNA and protein levels
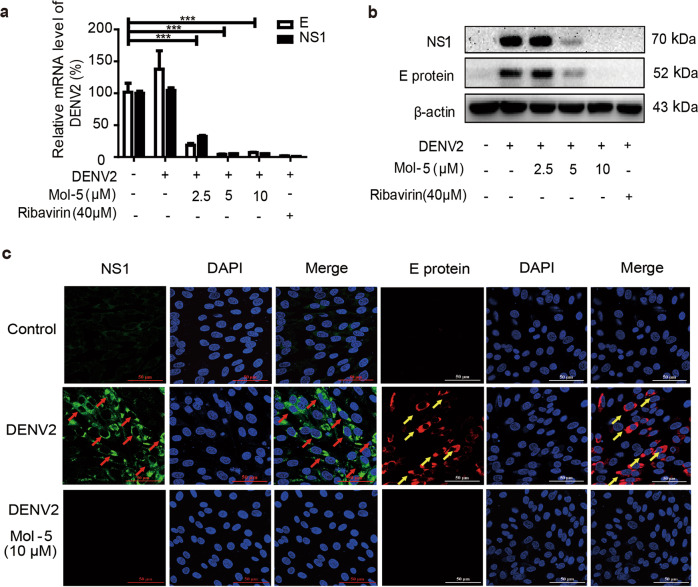
Viral replication involves viral gene transcription and protein expression [29]. To understand the antiviral effects of mol-5, we first analyzed viral-specific gene transcription by qRT-PCR. Both viral structure protein E (E) and non-structural protein 1 (NS1) were detected in BHK-21 cells after DENV2 infection. E and NS1 mRNA levels were reduced by ~90% by mol-5 treatment at 5 and 10 μM but not at 2.5 μM after 48 h of virus infection (Fig. 4a). Ribavirin (40 μM), which has strong antiviral effects, was chosen as the positive control. Next, the effect of mol-5 on viral proteins was evaluated by Western blotting experiments. Consistent with the above results, mol-5 (2.5, 5, and 10 μM) reduced E and NS1 protein levels in a dose-dependent manner (Fig. 4b). Immunofluorescence assays further confirmed the antiviral effects of mol-5. After BHK-21 cells were infected for 48 h, clear green (NS1) or red (E protein) fluorescence signals were observed compared to those in the control group, which comprised cells uninfected with DENV2 (Fig. 4c). However, the fluorescence signals were greatly reduced in the presence of mol-5 (Fig. 4c). These data strongly demonstrated the antiviral effect of mol-5.
Fig. 4. Effects of mol-5 on DENV2 mRNA and protein levels.
BHK-21 or C6/36 cells were infected by DENV2 (101 TCID50) at 37 °C for 1 h and then incubated with mol-5 (2.5, 5 or 10 μM) after washing with virus-free medium. Ribavirin (40 μM) was used as a positive control. a The mRNA levels of E and NS1 were detected by qRT-PCR at 48 hpi. b The protein levels of E and NS1 were detected by Western blotting. c The protein levels of the E (red, yellow arrow) and NS1 (green, red arrow) proteins were detected using immunofluorescence assays. DMSO used as a solvent control. Each data point represents the mean ± S.D. of an average of three separate trials. *** indicates P < 0.001.
Antiviral mode of mol-5
Given that RdRp plays an important role in early DENV RNA synthesis and proliferation after the virus has entered into host cells, a drug addition assay was performed to detect the antiviral stage of mol-5 activity. The three general treatment stages in relation to viral infection are the pretreatment, coincubation, and posttreatment phases [15, 30]. As shown in Fig. 5a, mol-5 had no effect at the pretreatment and coincubation stages, but it clearly reduced DENV2 effects at posttreatment stage, as determined by a plaque assay. These results indicated that mol-5 had neither virucidal activity nor effects on the viral attachment/entry stages. Mol-5 mainly inhibited DENV2 production in the postinfection stage, which was consistent with the RdRp inhibition effects of mol-5. Furthermore, DENV2-induced LDH secretion was decreased by mol-5 under posttreatment conditions (Fig. 5b), which was similar to the significant reductions in the viral mRNA (Fig. 5c) and protein levels (Fig. 5d). These results showed that mol-5 had antiviral effects in the postinfection stage of DENV2.
Fig. 5. Antiviral effects of mol-5 against DENV2.
BHK-21 cells with mol-5 (10 μM) pretreatment, cotreatment, or posttreatment were infected with DENV2 (101 TCID50) at 37 °C for 1 h after washing with virus-free medium, and the cells were then incubated with mol-5 (10 μM) in maintenance medium or without compound, followed by a plaque assays, b LDH assays, c qRT-PCR, or d Western blotting. DMSO was used as a control. Each data point represents the mean ± S.D. of an average of three separate trials. *** indicates P < 0.001.
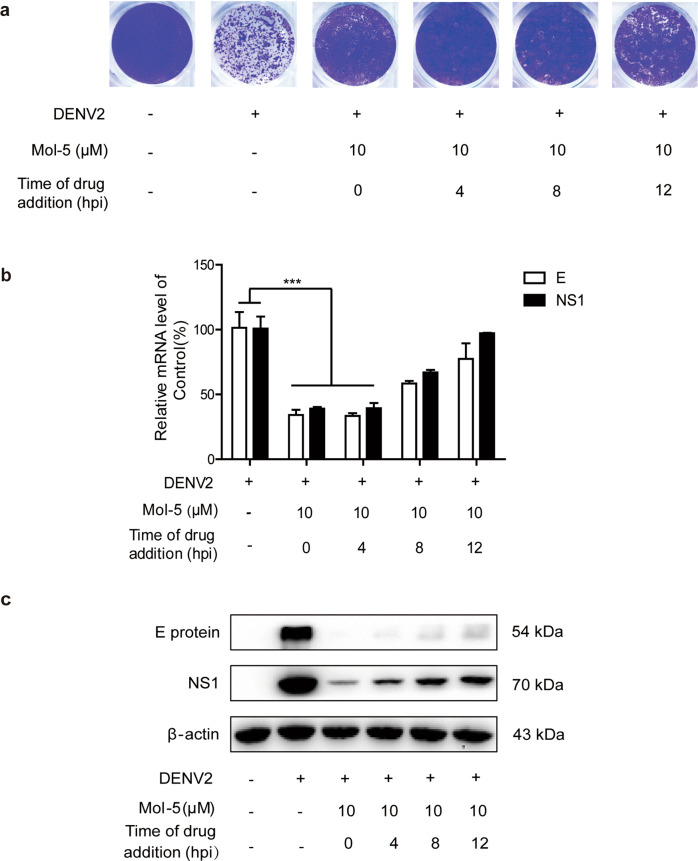
Since RdRp synthesizes viral RNA at an early stage, and progeny virions are assembled and released 12 hpi [30, 31], we next studied the exact time window of mol-5 action after DENV2 infection. Cells were incubated with mol-5 at the indicated time points (0, 4, 8, and 12 h) after DENV2 infection, and the number of progeny virions was assessed by plaque assay. Mol-5 treatment at 0, 4, and 8 hpi reduced the number of progeny virions (Fig. 6a), which was further confirmed using qRT-PCR (Fig. 6b) and Western blotting (Fig. 6c). The effective antiviral time window of mol-5 was between 0 and 4 hpi.
Fig. 6. Time-of-addition assay of mol-5 effects against DENV2.
BHK-21 cells were infected with DENV2 (101 TCID50) and washed after infection at 37 °C for 1 h. The cells were then washed, and mol-5 (10 μM) was added. At 0, 4, 8, and 12 hpi, a plaque assays, b qRT-PCR, and c Western blotting were performed. DMSO was used as a control. Each data point represents the mean ± S.D. of an average of three separate trials. *** indicates P < 0.001.
Taken together, these data clearly demonstrated that mol-5 blocked the early stages of the viral life cycle once the virus entered the host cells in the postinfection stages.
Effect of mol-5 on STAT1/ISGs activation
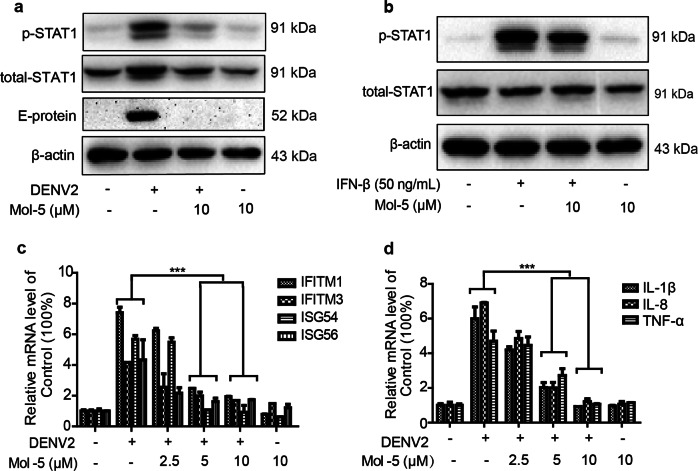
The recognition of viral ssRNA by cellular RIG-I leads to RIG-I activation and the subsequent secretion of type I interferon (IFN-α), which induces a potent antiviral state in the cell following the stimulation of two IFN receptor subunits (IFNAR1 and IFNAR2) to activate the Janus kinases (Jak1 and Tyk2) and signal transducers of transcription (STAT1 and STAT2) and leads to the upregulation of hundreds of IFN-stimulated genes (ISGs) [32]. In addition, the activation of STAT1/2 (especially STAT1) plays a major role in host defense against DENV infection [33]. Therefore, effects of mol-5 on STAT1 activation were investigated. As shown in Fig. 7a, after DENV2 infection, mol-5 treatment reduced the STAT1 phosphorylation level, which is increased by DENV2, when compared to that in the controls. Furthermore, the level of viral E protein was decreased by mol-5 treatment in THP-1 cells after DENV2 infection (Fig. 7a). On the other hand, mol-5 had no effect on IFN-β-induced STAT1 activation (Fig. 7b). These data excluded a direct effect of mol-5 on the JAK/STAT pathway.
Fig. 7. Effect of mol-5 on STAT1 activation and ISG expression.
THP-1 cells were infected with DENV2 (101 TCID50) and further incubated with mol-5 (2.5, 5, or 10 μM) in maintenance medium after washing with virus-free medium. a The protein levels of p-STAT1 (Tyr701), total STAT1, E protein and actin were determined at 48 hpi. b After THP-1 cells were incubated with IFN-β (50 ng/mL) or mol-5 (10 μM), the p-STAT1 (Tyr701), total STAT1 and actin levels were determined at 48 hpi. c The mRNA levels of ISG15, ISG54, IFITM1, and IFITM3 were detected by qRT-PCR as indicated. d The expression levels of IL-8, IL-1β, and TNF-α were quantified using qRT-PCR. DMSO was used as a solvent control. Each data point represents the mean ± S.D. of an average of three separate trials. *** indicates P < 0.001.
Many ISGs play antiviral roles downstream of the JAK/STAT pathway [34]. Hence, we used qRT-PCR to quantify the ISGs ISG54, ISG56, IFITM1, and IFITM3. DENV2 infection enhanced the mRNA levels of ISGs, whereas mol-5 treatment at 5 or 10 μM reduced the effect of DENV2 infection (Fig. 7c). Similarly, the pro-inflammatory mediators IL-8, IL-1β, and TNF-α play crucial roles in anti-infection, immune response regulation and anti-tumor immunity. In our study, IL-8 and IL-1β levels were enhanced by ~50% by infection, but TNFα did not appear to be affected (Fig. 7d). Mol-5 downregulated the expression of these inflammatory genes induced by DENV2 infection, suggesting the effective inhibition of DENV2 infection. Taken together, these data further highlighted the anti-DENV2 effect of mol-5.
Discussion
Because the cell-based screening of inhibitors against DENV was time consuming and costly [35], we used a virtual screening method to identify inhibitors of RdRp in DENV because the cost of in silico screening assays is much cheaper than that of cell-based assays. Using an in silico screening protocol and a commercially available antiviral compound library that had ~8000 small molecules, we identified 40 potential compounds. These compounds were ranked according to their docking scores, and the data obtained for the top 10 compounds are shown in Table 1. In addition, we also evaluated the ADME properties of these compounds. Based on these ADME characteristics and the cell-based antiviral assays, mol-5 was judged to have significant antiviral effects among the top 10 selected compounds. Furthermore, SPRi assays and cell-based NS5 polymerase activity assays confirmed the anti-NS5-RdRp effects of mol-5. Therefore, we identified a promising new pan-DENV inhibitor, mol-5, which could directly bind to NS5-RdRp both in silico and in vitro.
It is well documented that DENV NS5-RdRp is an attractive target for the discovery of new antiviral agents because it has no mammalian counterpart and its protein sequence is conserved across all four serotypes with more than 65% homology [36]. To date, RdRp structures from six Flaviviridae, DENV, WNV, JEV, bovine viral diarrhea virus, HCV, and ZIKV, have been determined. The catalytically active site of DENV RdRp is defined by a conserved GDD motif comprised of two aspartic acid residues (Asp663 and Asp664), which are located in the palm subdomain [37], and mol-5 could specifically bind to the palm subdomain of RdRp via Thr794. Several DENV RdRp inhibitors have been reported, such as pyridoxine-derived small molecule, allosteric NS5 polymerase inhibitor 27 and other derivatives, and these small molecules are still under development [38, 39]. Mol-5 contains a new chemical scaffold and is reported here for the first time. Although the IC50 of mol-5 is at the micromolar level at present, there is still potential for the improvement of its antiviral ability by using structure–activity relationship assays.
Our results demonstrate the reliability of mol-5 as a novel RdRp inhibitor having a novel structure. The antiviral effect of mol-5 was evaluated by a cell-based assay. Mol-5 inhibited DENV2-induced CPEs at a low EC50 value. However, mol-5 had cytotoxic effects on BHK-21 cells at or above 20 μM, which reduced the cell viability by ~20%. However, only a 5% reduction in cell viability was observed in cells treated with 10 μM mol-5, and interestingly, 100% of the antiviral effects of mol-5 were retained at this concentration. These data suggest that the antiviral effects of mol-5 were distinct from its cytotoxic effects. It is possible that future structure–activity relationship studies could very well improve the antiviral properties of mol-5, while maintaining the cytotoxic effects at very low levels. In this context, it is of interest to note that compounds having structures similar to mol-5 have already been reported as anti-tumor agents and as Mer-kinase or EGF inhibitors at 10 μM concentration. Moreover, many compounds have off-target effects, and at present, we cannot exclude the possibility of off-target effects for mol-5, which needs further study. Although the side effects of medications are always a concern, treatment for DENV infection typically continues for days or even weeks [40]. Thus, the risk/benefit assessment should be flexible when evaluating anti-DENV medications.
RdRp plays an important role in viral RNA synthesis [35], and we found that mol-5 reduced the dsRNA level of DENV based on immunofluorescence assays that were observed by a confocal microscope, which was consistent with the effects of RdRp inhibition. In cell culture, DENV was sensitive to type I IFN, and type I and II IFN receptor-deficient mice were highly susceptible to DENV infection [41, 42]. These observations suggest that the IFN system contributed to controlling DENV replication in vivo. Antiviral IFNs were increased upon viral infection, and they signal through the Janus-activated kinase/signal transducer and activator of transcription (JAK-STAT) pathway and induce the de novo transcription of hundreds of ISGs [43]. The products of these genes facilitate viral clearance and protect uninfected cells from further infection. Viral dsRNA-induced RIG-1 activation was the main cause of JAK-STAT activation and the changes in ISG expression [44]. Interestingly, mol-5 not only prevented dsRNA production but also decreased STAT activation and the expression of ISGs. As expected, the results in Fig. 7a indicated DENV2 infection enhanced the phosphorylation level of STAT1, and mol-5 could reverse the effect of DENV2. These results suggested that mol-5 exhibited anti-inflammatory ability in the presence of DENV2. It is worth noting that 2.5 μM mol-5 had no inhibitory effect on DENV-induced LDH release and viral replication. However, this mol-5 concentration suppressed the expression of ISG genes. This might reflect a delay between the transcriptional level and the translational level for the antiviral effects of mol-5. To exclude the effects of other anti-inflammatory mechanisms besides the inhibition of DENV RdRp, interferon-β (IFN-β) was used, which could induce STAT1 phosphorylation alone similar to DENV2 infection (Fig. 7b). However, mol-5 could not reverse the inflammatory induction effect of IFN-β. These results indicated that the anti-inflammatory effect of mol-5 was dependent on DENV inhibition rather than other anti-inflammatory effects.
In conclusion, we identified a small molecule, mol-5, with a new structure that is active against RdRp in DENV2 and that provided a new parent compound for further improvement. Cell-based experiments demonstrated the inhibitory effects of mol-5 on DENV infection not only in mosquito C6/36 cells but also in mammalian BHK-21 cells. Therefore, mol-5 is a promising inhibitor of RdRp and a new lead compound for further development to treat patients with DENV infections.
Supplementary information
Acknowledgements
The National Natural Science Foundation of China (81603118, 81700854), the Pearl River Nova Program of Guangzhou (201806010119), the Natural Science Foundation (2017A030313717), and the “New Drug Creation and Development” Major scientific and technological projects of Guangdong Province (2019B020202002) supported this work.
Author contributions
XGY, WYW, and SWL designed the research; YHW, WYW, SXG, and SJH performed the research; XGC, MZ, XYW, LL, and XDT contributed new reagents or analytical tools; XGY and SWL analyzed the data; XY and KSN wrote the paper.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Yi-hong Wan, Wen-yu Wu, Song-xin Guo
Contributor Information
Shu-wen Liu, Email: liusw@smu.edu.cn.
Xin-gang Yao, Email: yaoxingang@smu.edu.cn.
Supplementary information
The online version of this article (10.1038/s41401-019-0316-7) contains supplementary material, which is available to authorized users.
References
- 1.Rather IA, Parray HA, Lone JB, Paek WK, Lim J, Bajpai VK, et al. Prevention and control strategies to counter dengue virus infection. Front Cell Infect Microbiol. 2017;7:336. doi: 10.3389/fcimb.2017.00336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hong WX, Zhao H, Deng YQ, Jiang T, Yu XD, Song KY, et al. Severe dengue due to secondary DENV-1 infection in Mainland China. J Clin Virol. 2013;57:184–6. doi: 10.1016/j.jcv.2013.02.012. [DOI] [PubMed] [Google Scholar]
- 3.Teng HJ, Chen TJ, Tsai SF, Lin CP, Chiou HY, Lin MC, et al. Emergency vector control in a DENV-2 outbreak in 2002 in Pingtung City, Pingtung County, Taiwan. Jpn J Infect Dis. 2007;60:271–9. [PubMed] [Google Scholar]
- 4.Shankar MB, Rodriguez-Acosta RL, Sharp TM, Tomashek KM, Margolis HS, Meltzer MI. Estimating dengue under-reporting in Puerto Rico using a multiplier model. PLoS Negl Trop Dis. 2018;12:e0006650. doi: 10.1371/journal.pntd.0006650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boonnak K, Dambach KM, Donofrio GC, Tassaneetrithep B, Marovich MA. Cell type specificity and host genetic polymorphisms influence antibody-dependent enhancement of dengue virus infection. J Virol. 2011;85:1671–83. doi: 10.1128/JVI.00220-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen J, Wen K, Li XQ, Yi HS, Ding XX, Huang YF, et al. Functional properties of DENV EDIIIreactive antibodies in human DENV1infected sera and rabbit antiserum to EDIII. Mol Med Rep. 2016;14:1799–808. doi: 10.3892/mmr.2016.5454. [DOI] [PubMed] [Google Scholar]
- 7.Fatima K, Syed NI. Dengvaxia controversy: impact on vaccine hesitancy. J Glob Health. 2018;8:020312. doi: 10.7189/jogh.08-020312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fang S, Wu Y, Wu N, Zhang J, An J. Recent advances in DENV receptors. Sci World J. 2013;2013:684690. doi: 10.1155/2013/684690. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9.Yang J, Zou L, Hu Z, Chen W, Zhang J, Zhu J, et al. Identification and characterization of a 43 kDa actin protein involved in the DENV-2 binding and infection of ECV304 cells. Microbes Infect. 2013;15:310–8. doi: 10.1016/j.micinf.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 10.Iglesias NG, Filomatori CV, Gamarnik AV. The F1 motif of dengue virus polymerase NS5 is involved in promoter-dependent RNA synthesis. J Virol. 2011;85:5745–56. doi: 10.1128/JVI.02343-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang T, Wang ML, Zhang GR, Liu W, Xiao XQ, Yang YS, et al. Recombinant DENV 2 NS5: an effective antigen for diagnosis of DENV infection. J Virol Methods. 2019;265:35–41. doi: 10.1016/j.jviromet.2018.12.005. [DOI] [PubMed] [Google Scholar]
- 12.Lim SP, Noble CG, Nilar S, Shi PY, Yokokawa F. Discovery of potent non-nucleoside inhibitors of dengue viral RNA-dependent RNA polymerase from fragment screening and structure-guided design. Adv Exp Med Biol. 2018;1062:187–98. doi: 10.1007/978-981-10-8727-1_14. [DOI] [PubMed] [Google Scholar]
- 13.Kanyaboon P, Saelee T, Suroengrit A, Hengphasatporn K, Rungrotmongkol T, Chavasiri W, et al. Cardol triene inhibits dengue infectivity by targeting kl loops and preventing envelope fusion. Sci Rep. 2018;8:16643. doi: 10.1038/s41598-018-35035-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yokokawa F, Nilar S, Noble CG, Lim SP, Rao R, Tania S, et al. Discovery of potent non-nucleoside inhibitors of dengue viral RNA-dependent RNA polymerase from a fragment hit using structure based drug design. J Med Chem. 2016;59:3935–52. doi: 10.1021/acs.jmedchem.6b00143. [DOI] [PubMed] [Google Scholar]
- 15.Yao XG, Guo SX, Wu WY, Wang JN, Wu SG, He SJ, et al. Q63, a novel DENV2 RdRp non-nucleoside inhibitor, inhibited DENV2 replication and infection. J Pharm Sci. 2018;138:247–56. doi: 10.1016/j.jphs.2018.06.012. [DOI] [PubMed] [Google Scholar]
- 16.Yao XG, Ling Y, Guo SX, He SJ, Wang JN, Zhang Q, et al. Inhibition of dengue viral infection by diasarone-I is associated with 2’O methyltransferase of NS5. Eur J Pharmacol. 2018;821:11–20. doi: 10.1016/j.ejphar.2017.12.029. [DOI] [PubMed] [Google Scholar]
- 17.HS-j GuoSong-xin, Cui-hong Huang, Xin-gang Yao, Shu-wen Liu. Z1, a novel DENV2 NS5 RdRp small molecular inhibitor, inhibits DENV2 replication and infection. Chin Pharmacol Bull. 2018;34:790–6. [Google Scholar]
- 18.Wang YH, Jin BB, Liu PW, Li J, Chen XG, Gu JB. piRNA profiling of dengue virus type 2-infected Asian tiger mosquito and midgut tissues. Viruses. 2018;10:pii: E213. 10.3390/v10040213. [DOI] [PMC free article] [PubMed]
- 19.Trott O, Olson AJ. Software News and Update AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–61. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lim SP, Noble CG, Seh CC, Soh TS, El Sahili A, Chan GKY, et al. Potent allosteric dengue virus NS5 polymerase inhibitors: mechanism of action and resistance profiling. PLoS Pathog. 2016;12:e1005737. doi: 10.1371/journal.ppat.1005737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O’Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR. Open Babel: an open chemical toolbox. J Cheminform. 2011;3:33. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bhachoo J, Beuming T. Investigating protein–peptide interactions using the Schrodinger computational suite. Methods Mol Biol. 2017;1561:235–54. doi: 10.1007/978-1-4939-6798-8_14. [DOI] [PubMed] [Google Scholar]
- 23.Lei ZQ, Su YX, Zheng XL. [Expression, purification and identification of the domain III of DENV II envelop protein in Escherichia coli] Nan Fang Yi Ke Da Xue Xue Bao. 2010;30:1496–500. [PubMed] [Google Scholar]
- 24.Yao X, Ling Y, Guo S, Wu W, He S, Zhang Q, et al. Tatanan A from the Acorus calamus L. root inhibited dengue virus proliferation and infections. Phytomedicine. 2018;42:258–67. doi: 10.1016/j.phymed.2018.03.018. [DOI] [PubMed] [Google Scholar]
- 25.Lee JC, Tseng CK, Wu YH, Kaushik-Basu N, Lin CK, Chen WC, et al. Characterization of the activity of 2’-C-methylcytidine against dengue virus replication. Antivir Res. 2015;116:1–9. doi: 10.1016/j.antiviral.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 26.Puiu M, Bala C. SPR and SPR imaging: recent trends in developing nanodevices for detection and real-time monitoring of biomolecular events. Sensors (Basel) 2016;16:pii: E870. 10.3390/s16060870. [DOI] [PMC free article] [PubMed]
- 27.Brecher M, Zhang J, Li H. The flavivirus protease as a target for drug discovery. Virol Sin. 2013;28:326–36. doi: 10.1007/s12250-013-3390-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lim SP, Koh JHK, Seh CC, Liew CW, Davidson AD, Chua LS, et al. A crystal structure of the dengue virus non-structural protein 5 (NS5) polymerase delineates interdomain amino acid residues that enhance its thermostability and de novo initiation activities. J Biol Chem. 2013;288:31105–14. doi: 10.1074/jbc.M113.508606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lim SP, Wang QY, Noble CG, Chen YL, Dong H, Zou B, et al. Ten years of dengue drug discovery: progress and prospects. Antivir Res. 2013;100:500–19. doi: 10.1016/j.antiviral.2013.09.013. [DOI] [PubMed] [Google Scholar]
- 30.Quintana VM, Piccini LE, Panozzo Zenere JD, Damonte EB, Ponce MA, Castilla V. Antiviral activity of natural and synthetic beta-carbolines against dengue virus. Antivir Res. 2016;134:26–33. doi: 10.1016/j.antiviral.2016.08.018. [DOI] [PubMed] [Google Scholar]
- 31.Kato F, Ishida Y, Oishi S, Fujii N, Watanabe S, Vasudevan SG, et al. Novel antiviral activity of bromocriptine against dengue virus replication. Antivir Res. 2016;131:141–7. doi: 10.1016/j.antiviral.2016.04.014. [DOI] [PubMed] [Google Scholar]
- 32.MacMicking JD. Interferon-inducible effector mechanisms in cell-autonomous immunity. Nat Rev Immunol. 2012;12:367–82. doi: 10.1038/nri3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goh KCM, Tang CK, Norton DC, Gan ES, Tan HC, Sun B, et al. Molecular determinants of plaque size as an indicator of dengue virus attenuation. Sci Rep. 2016;6:26100. doi: 10.1038/srep26100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dai JF, Pan W, Wang PH. ISG15 facilitates cellular antiviral response to dengue and west nile virus infection in vitro. Virol J. 2011;8:468. doi: 10.1186/1743-422X-8-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boldescu V, Behnam MAM, Vasilakis N, Klein CD. Broad-spectrum agents for flaviviral infections: dengue, Zika and beyond. Nat Rev Drug Discov. 2017;16:565–86. doi: 10.1038/nrd.2017.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lim SP, Noble CG, Shi PY. The dengue virus NS5 protein as a target for drug discovery. Antivir Res. 2015;119:57–67. doi: 10.1016/j.antiviral.2015.04.010. [DOI] [PubMed] [Google Scholar]
- 37.Noble CG, Lim SP, Arora R, Yokokawa F, Nilar S, Seh CC, et al. A conserved pocket in the dengue virus polymerase identified through fragment-based screening. J Biol Chem. 2016;291:8541–8. doi: 10.1074/jbc.M115.710731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu HT, Colby-Germinario SP, Hassounah S, Quashie PK, Han YS, Oliveira M, et al. Identification of a pyridoxine-derived small-molecule inhibitor targeting dengue virus RNA-dependent RNA polymerase. Antimicrob Agents Chemother. 2016;60:600–8. doi: 10.1128/AAC.02203-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tarantino D, Cannalire R, Mastrangelo E, Croci R, Querat G, Barreca ML, et al. Targeting flavivirus RNA dependent RNA polymerase through a pyridobenzothiazole inhibitor. Antivir Res. 2016;134:226–35. doi: 10.1016/j.antiviral.2016.09.007. [DOI] [PubMed] [Google Scholar]
- 40.Lai JH, Lin YL, Hsieh SL. Pharmacological intervention for dengue virus infection. Biochem Pharmacol. 2017;129:14–25. doi: 10.1016/j.bcp.2017.01.005. [DOI] [PubMed] [Google Scholar]
- 41.Laurent-Rolle M, Morrison J, Rajsbaum R, Macleod JML, Pisanelli G, Pham A, et al. The interferon signaling antagonist function of yellow fever virus NS5 protein is activated by Type I interferon. Cell Host Microbe. 2014;16:314–27. doi: 10.1016/j.chom.2014.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li G, Feng T, Pan W, Shi X, Dai J. DEAD-box RNA helicase DDX3X inhibits DENV replication via regulating type one interferon pathway. Biochem Biophys Res Commun. 2015;456:327–32. doi: 10.1016/j.bbrc.2014.11.080. [DOI] [PubMed] [Google Scholar]
- 43.Lang J, Cheng Y, Rolfe A, Hammack C, Vera D, Kyle K, et al. An hPSC-derived tissue-resident macrophage model reveals differential responses of macrophages to ZIKV and DENV infection. Stem Cell Rep. 2018;11:348–62. doi: 10.1016/j.stemcr.2018.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nasirudeen AMA, Wong HH, Thien PL, Xu SL, Lam KP, Liu DX. RIG-I, MDA5 and TLR3 synergistically play an important role in restriction of dengue virus infection. PLoS Negl Trop Dis. 2011;5:e926. doi: 10.1371/journal.pntd.0000926. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.