Abstract
Establishment of a physiologically relevant human hepatocyte‐like cell system for in vitro translational research has been hampered by the limited availability of cell models that accurately reflect human biology and the pathophysiology of human disease. Here we report a robust, reproducible, and scalable protocol for the generation of hepatic organoids from human induced pluripotent stem cells (hiPSCs) using short exposure to nonengineered matrices. These hepatic organoids follow defined stages of hepatic development and express higher levels of early (hepatocyte nuclear factor 4A [HNF4A], prospero‐related homeobox 1 [PROX1]) and mature hepatic and metabolic markers (albumin, asialoglycoprotein receptor 1 [ASGR1], CCAAT/enhancer binding protein α [C/EBPα]) than two‐dimensional (2D) hepatocyte‐like cells (HLCs) at day 20 of differentiation. We used this model to explore the biology of the pleiotropic TRIB1 (Tribbles‐1) gene associated with a number of metabolic traits, including nonalcoholic fatty liver disease and plasma lipids. We used genome editing to delete the TRIB1 gene in hiPSCs and compared TRIB1‐deleted iPSC‐HLCs to isogenic iPSC‐HLCs under both 2D culture and three‐dimensional (3D) organoid conditions. Under conventional 2D culture conditions, TRIB1‐deficient HLCs showed maturation defects, with decreased expression of late‐stage hepatic and lipogenesis markers. In contrast, when cultured as 3D hepatic organoids, the differentiation defects were rescued, and a clear lipid‐related phenotype was noted in the TRIB1‐deficient induced pluripotent stem cell HLCs. Conclusion: This work supports the potential of genome‐edited hiPSC‐derived hepatic 3D organoids in exploring human hepatocyte biology, including the functional interrogation of genes identified through human genetic investigation.
We have developed a robust growth factor–defined methodology for generating 3D hiPSC‐derived hepatic organoids in order to permit functional assessment of GWAS loci. The developed approach reflects human biology more faithfully than conventional cell models and is amiable for high‐throughput studies.

Abbreviations
- 2D
two‐dimensional
- 3D
three‐dimensional
- AFP
alpha‐fetoprotein
- ACACA
acetyl‐coenzyme A carboxylase
- ALB
albumin
- APOB
apolipoprotein B
- APOC3
apolipoprotein CIII
- ASGR1
asialoglycoprotein receptor 1
- BMP4
bone morphogenetic protein 4
- cDNA
complementary DNA
- C/EBPα
CCAAT/enhancer binding protein α
- DAPI
4′,6‐diamidino‐2‐phenylindole
- DE
definitive endoderm
- DGAT
diacylglycerol acyltransferase
- DLK1
delta‐like 1 homolog
- DPBS
Dulbecco’s phosphate buffered saline
- FACS
fluorescence‐activated cell sorting
- FASN
fatty acid synthase
- FGF2
fibroblast growth factor 2
- FOXA2
forkhead box A2
- GAPDH
glyceraldehyde 3‐phosphate dehydrogenase
- GFP
green fluorescent protein
- GWAS
genome‐wide association studies
- hC/EBPα
human CCAAT/enhancer binding protein α
- HGF
human growth factor
- hHEX
hematopoietically expressed homeobox protein
- hiPSC
human‐induced pluripotent stem cell
- HNF4A
hepatocyte nuclear factor 4 alpha
- iPSC
induced pluripotent stem cell
- KO
knockout
- LDL
low‐density lipoprotein
- LsKO
liver‐specific KO mice
- MDR1
multidrug resistance protein 1
- mRNA
messenger RNA
- PAS
periodic acid–Schiff
- PCR
polymerase chain reaction
- PFA
paraformaldehyde
- PROX1
prospero‐related homeobox 1
- RT
room temperature
- SCD
stearoyl‐coenzyme A desaturase
- ssODN
single‐stranded oligo nucleotide
- TG
triglyceride
- TRIB1
Tribbles‐1
- WT
wild‐type
- ZO‐1
tight junction protein 1
Large‐scale genome‐wide association studies (GWAS) have identified thousands of genomic loci associated with human traits and diseases, most of which lack a clear causal gene or mechanism.( 1 , 2 , 3 ) Model systems that reflect human biology are needed to systematically examine these findings and translate them into meaningful biology. Human induced pluripotent stem cells (hiPSCs) offer the opportunity to provide an inexhaustible supply of human cells that reflect naturally occurring genetic variation or can be genome‐edited for experimental purposes.( 4 ) Genetic variation in liver‐expressed genes and the effect on phenotype and disease has been of major interest, given the major impact of the many physiological processes performed by hepatocytes. Differentiation of hiPSCs into hepatocyte‐like cells has proved to be an important approach to understanding human biology, and there are several established protocols available to date.( 5 , 6 , 7 ) These hiPSC‐derived hepatocytes in a standard two‐dimensional (2D) monolayer culture system efficiently express hepatic markers and mimic some hepatocyte functions, enabling their use in drug toxicity screening( 8 ) and understanding biology.( 9 ) However, this approach suffers from specification defects. Most of the derived monolayer hepatocytes either lack expression of some key transcription factors or express them at suppressed levels, in turn making them metabolically immature.( 10 , 11 , 12 ) Another limitation is that many gene mutations in this system are studied in isolation, which does not reflect the true underlying biology.( 13 , 14 )
Recently, an induced pluripotent stem cell (iPSC)–derived hepatocyte three‐dimensional (3D) organoid culture system has been reported to establish heterotypic interactions and mimic the in vivo environment more closely in terms of distinct cellular behavior.( 14 , 15 , 16 , 17 , 18 , 19 , 20 ) However, most of the 3D approaches involve complex bioengineered matrices and synthetic scaffolds that need skilled handling expertise. Moreover, these different 3D models have not been rigorously evaluated in terms of their ability to functionally assess genes at GWAS loci in a way that reflects human biology more faithfully than conventional 2D iPSC hepatocytes.
To address these limitations, we developed a robust, growth factor–defined, scalable hepatic 3D organoid generation method from hiPSCs with short exposure to a laminin‐enriched, nonengineered matrix for the purpose of interrogating lipid GWAS loci. We then applied it to understanding the role of the pleiotropic lipid/metabolic GWAS gene TRIB1. Variants at the TRIB1 locus are associated with liver transaminases and nonalcoholic fatty liver disease (NAFLD), plasma triglycerides, total cholesterol, low‐density lipoprotein (LDL) cholesterol, high‐density lipoprotein cholesterol, adiponectin, and coronary artery disease.( 3 , 21 , 22 ) Studies in mice have indicated that the protein product of TRIB1, namely Tribbles‐1, posttranslationally regulates the degradation of CCAAT/enhancer binding protein α (C/EBPα) in the liver, which in turn contributes to the dysregulation of lipid‐related gene expression and thus lipid phenotypes.( 23 ) However, the role of TRIB1 in liver lipid biology remains uncertain, and an in vitro model is needed for more detailed functional investigation. We used CRISPR/Cas9 to delete TRIB1 in a human iPSC line, and show that iPSC‐derived 3D hepatic organoids lacking TRIB1 more faithfully expressed an expected lipid phenotype than 2D hepatocytes derived from the same iPSCs. This study emphasizes the potential advantages of hiPSC‐derived 3D hepatic organoids to interrogate the effect of genetic variation on human liver lipid metabolism.
Results
Generation of Self‐Organizing Functional Hepatic Organoids From hiPSCs
To generate hepatic organoids from hiPSCs, we developed a simple approach by making modest modifications to a conventional growth factor–induced protocol and providing them the 3D environment to overcome specification and maturation defects. Detailed methodology is described in the Methods section, but in brief, hiPSCs were passaged and aggregated in 96‐well U‐bottom low‐attachment plates. Later, laminin‐rich matrix was added only in the definitive endoderm (DE) stage for 4 days. To achieve homogeneous and robust DE induction, the DE Kit (STEMCELL Technologies, Vancouver, Canada) was used, which resulted in robust DE differentiation and greater than 90% forkhead box A2+ (FOXA2+) and GATA binding protein 4+ (GATA4+) cells (Supporting Fig. S1). On day 5, these organoids were moved to a medium containing bone morphogenetic protein 4 (BMP4), fibroblast growth factor 2 (FGF2), and keratinocyte growth factor (KGF) to promote foregut differentiation. On day 10, the developing foregut organoids were shifted to a medium containing human growth factor (HGF) and 0.5% DMSO to promote hepatoblast formation. For maturation on day 15, aggregates were moved to commercial hepatocyte culture medium along with Oncostatin M and Dexamethasone (R&D Systems, Minneapolis, MN) (Fig. 1A). To compare the functional capacity of the organoids with 2D monolayer cultures, a previously reported timeline was followed,( 5 , 24 , 25 , 26 ) and organoids were harvested at day 20 for comparison with 2D HLCs (Supporting Fig. S2). Immunostaining showed robust expression of early and mature hepatic markers such as hepatocyte nuclear factor 4 alpha (HNF4A), alpha‐fetoprotein (AFP), and albumin (ALB), in day 20 organoids (Fig. 1Bi‐iii). These organoids also possess characteristic hepatocyte features and express mature markers including arginase, a canalicular marker multidrug resistance protein 1 (MDR1), and tubular marker HNF1β (Fig. 1Biv‐vi). Functional assays were performed, which indicated efficient BODIPY‐LDL (Fisher Scientific, Waltham, MA) and CDFDA (Invitrogen, Carlsbad, CA) uptake (Fig. 1Ci‐ii). Glycogen and lipid droplet accumulation was also observed as analyzed by periodic acid–Schiff (PAS), Oil Red O (ORO) staining with corresponding electron microscopy images (Fig. 1Ciii‐vi), establishing the generation of functional hiPSC‐derived hepatic organoids.
FIG. 1.
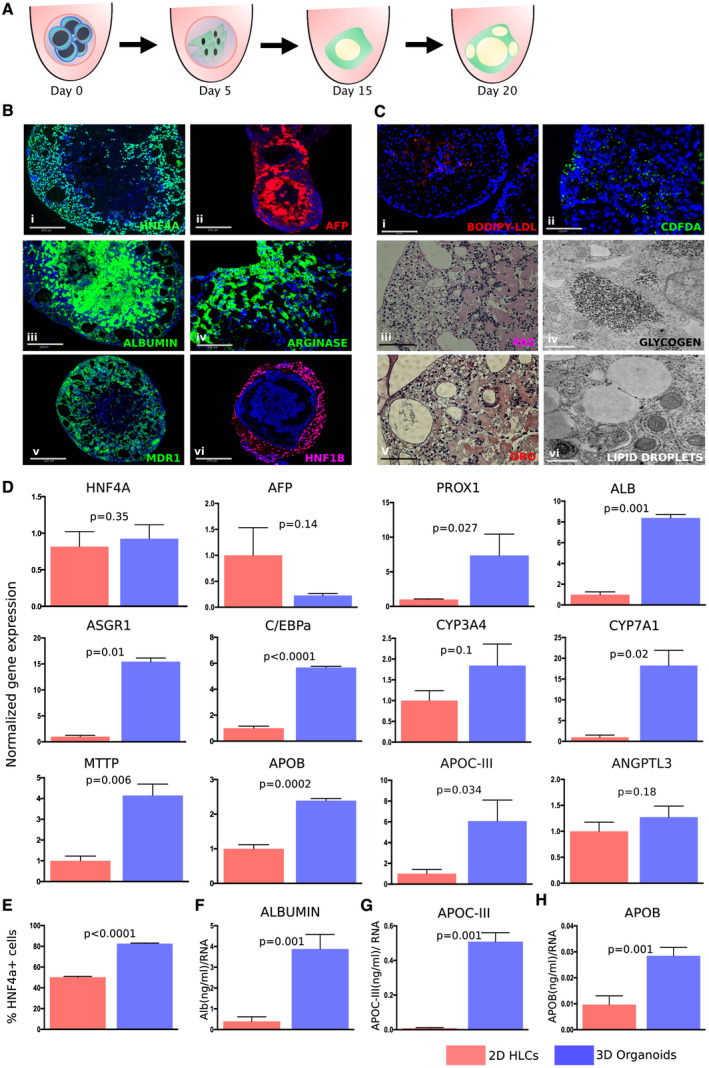
Hepatic organoids generated from hiPSCs exhibit liver‐like features more effectively than 2D HLCs. (A) Schematic showing directed differentiation of 3D hepatic organoids from hiPSCs. (B) Representative images of immunostaining showing expression of early (i‐ii; HNF4A, green; AFP, red) and mature (iii‐v; ALB, green; Arginase, green; MDR1, green) hepatic and tubular (vi; HNF1β, pink) markers at day 20 of organoid differentiation. (Ci‐ii) LDL‐uptake and bile‐uptake assays were performed with hepatic organoids at day 20, showing uptake of fluorescent BODIPY‐LDL (red) and fluorescent substrate CDFDA (green). Hepatic organoids exhibit functional liver‐like properties such as glycogen accumulation as shown by PAS staining (pink) (iii‐iv) and lipid droplet formation as shown by ORO (red) (v‐vi) with corresponding electron microscopy images showing glycogen and lipid droplets. (D) Hepatic organoids efficiently express early, late, and metabolic markers at day 20 organoids versus 2D HLCs calculated relative to GAPDH (reference gene) using the 2−ddCt method, normalized to a mean expression level in 2D HLCs. Data are representative of three independent experiments. (E) Increased HNF4A+ cells in day 20 organoids versus 2D HLCs when measured by FACS. (F‐H) Hepatic organoids secrete higher amounts of ALB, APOB, and APOCIII versus 2D HLCs when compared at day 20 of differentiation by enzyme‐linked immunosorbent assay. Values represent the mean ± SEM; P values were calculated with Welch’s t tests. DAPI (4′,6‐diamidino‐2‐phenylindole) is shown in blue in all of the immunofluorescence images. Scale bar: 125 µm (Biii,v; Ci‐iii,v) and 275 µm (Bi,ii,iv,vi); and electron microscopy: 500 nm (Civ,vi).
Hepatic Organoids Are More Effectively Differentiated Than Monolayer 2D HLC Cultures
To determine the differentiation efficiency of hiPSCs to hepatocytes, organoids were analyzed for gene expression of early hepatic markers such as HNF4A, AFP, and prospero‐related homeobox 1 (PROX1); maturation markers such as ALB, asialoglycoprotein receptor 1 (ASGR1), and C/EBPα; and metabolic markers including apolipoprotein B (APOB), apolipoprotein CIII (APOC3), angiopoietin‐like 3 (ANGPTL3), microsomal triglyceride transfer protein (MTTP), cytochrome P450 3A4 (CYP3A4), and cholesterol 7 alpha‐hydroxylase (CYP7A1) at day 20 of differentiation (Fig. 1D). Hepatic organoids express these markers at a higher level than 2D HLCs. Similar observations were made with a K3 hiPSC line derived from a different genetic background (Supporting Fig. S3). Interestingly, hepatic 3D organoids showed enhanced expression of canonical hepatocyte and metabolic genes than 2D HLCs and even primary human hepatocytes, but not when compared with human liver samples (Supporting Fig. S3A‐C). Fluorescence‐activated cell sorting (FACS) quantification on day 20 hepatic 3D organoids demonstrated an approximate 1.5‐fold higher HNF4A compared with day 20 2D HLCs (Fig. 1E). Following continuation to day 30, we noted an approximate 2‐5‐fold increase in expression of hepatic markers over day 20 3D organoids (Supporting Fig. S1B); in marked contrast, the 2D HLCs started to dedifferentiate after day 20 and began to detach around day 22‐25.( 27 ) Secretory proteins such as albumin, APOC3, and APOB were measured in the media, and were observed to be significantly higher in the media from hepatic organoids versus 2D HLCs (Fig. 1F‐H).
Hepatic Organoids for Modeling the Role of TRIB1
To determine whether hiPSC hepatocytes can recapitulate TRIB1 lipid biology, we generated TRIB1 knockout (KO) hiPSCs using genome engineering. We took our in‐house‐derived and fully characterized wild‐type (WT) hiPSC line, Sv20. Using CRISPR/Cas9, a stop codon was introduced in the first exon by frameshift mutation, eight amino acids after the start codon. Homology‐directed repair was used by designing a single‐stranded oligo nucleotide (ssODN), which introduced a stop codon and HindIII restriction site to screen the clones (Fig. 2A). Three clones each for WT and KO were selected for further characterization. KO clones showed restriction by HindIII, and later Sanger sequencing confirmed the insertion of ssODN (Fig. 2B,C). Due to the lack of availability of any reliable human TRIB1 antibody, we performed Sanger sequencing on the generated complementary DNA (cDNA) to confirm the genotype of KO clones. Fully characterized WT and KO clones were progressed for differentiation studies (Fig. 2D and Supporting Fig. S4).
FIG. 2.
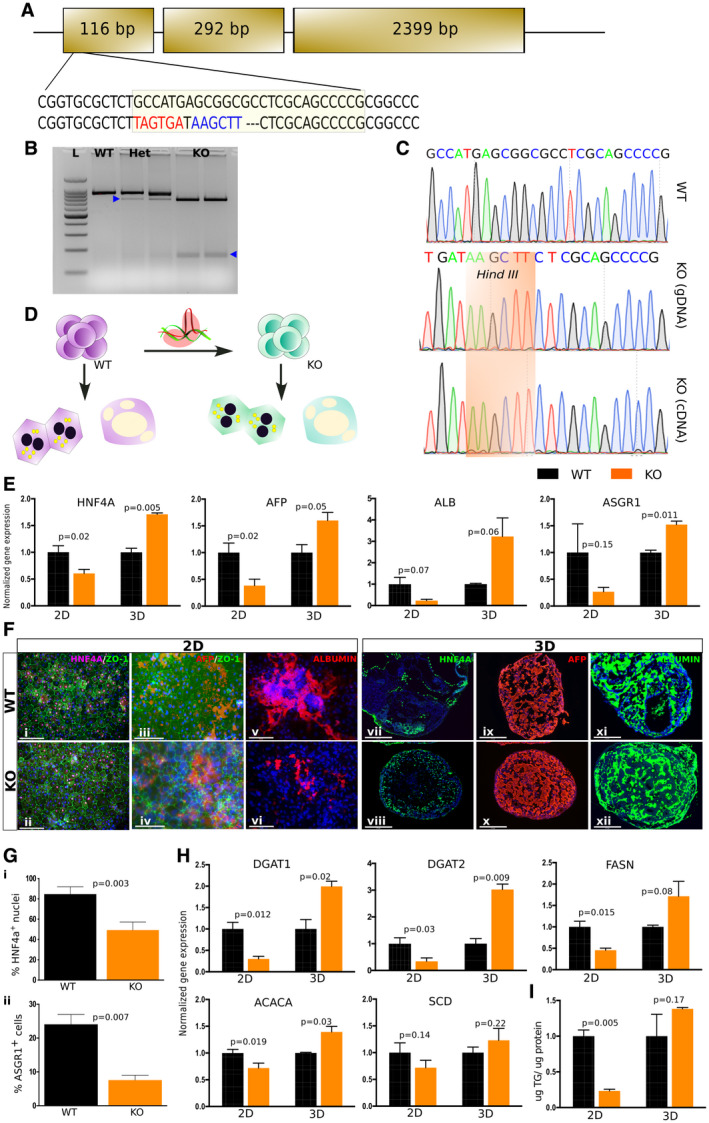
TRIB1 KO 2D HLCs failed to mimic TRIB1 lipid biology versus 3D hepatic organoids. (A) Strategy for generation of TRIB1 KO hiPSCs by CRISPR‐Cas9 using ssODN knock‐in approach. A stop codon was introduced after eight amino acids from the start codon in exon 1. Hind III site was introduced to screen for ssODN‐edited clones. (B) Representative image shows WT and HindIII positive TRIB1 heterozygous and KO clones in undifferentiated hiPSCs. (C) Sequence confirmation shows inclusion of knocked‐in ssODN in genomic DNA and cDNA versus WT clone. (D) Schematic shows parallel differentiation of WT and TRIB1 KO hiPSCs to 2D and 3D hepatocytes, which were used for characterization. (E,F) TRIB1 KO 2D HLCs exhibit lower levels of hepatic markers than WT, whereas 3D hepatic organoids exhibit no defect in hepatic differentiation in WT and KO as shown by quantitative PCR (E) and immunostaining (F). (G) TRIB1 KO 2D HLCs showed decreased HNF4A+ (i) and ASGR1+ (ii) cells as quantified by immunofluorescence image quantification and FACS sorting, respectively. (H) TRIB1 KO 2D HLCs express low DNL (DGAT1, DGAT2, FASN, ACACA, and SCD) markers versus WT, whereas TRIB1 KO 3D hepatic organoids express higher DNL markers and TG (I), mimicking TRIB1 lipid biology. Values represent the mean ± SEM shown for one representative experiment, calculated relative to GAPDH (reference gene) using the 2−ddCt method, normalized to the mean expression level in WT cells (n = 4 WT and four KO wells; two wells per clone with 24 organoids per well. For the 2D HLCs, n = 3 WT and seven KO wells; and n = 1 WT and three KO clones). P values were calculated with Welch’s t tests. DAPI is shown in blue in all of the immunofluorescence images. Scale bar: 200 µm (Fi,ii), 75 µm (Fiii,iv), 125 µm (Fv,vi), and 275 µm (Fvii‐xii). Abbreviation: Het, heterozygous.
Both WT and KO hiPSCs were differentiated to hepatocytes in parallel from the same culture dish, following 2D and 3D protocols. To assess the differentiation efficiency, at day 20 cells were harvested for gene expression and lipid phenotyping. Significant decreases in the levels of hepatic markers such as HNF4A, AFP, ALB, and ASGR1 were observed in TRIB1 KO 2D HLCs when compared with WT 2D HLCs, whereas TRIB1 KO 3D hepatic organoids exhibited no such defect in differentiation compared with WT 3D organoids (Fig. 2E). Similar observations were made using immunofluorescence for hepatic markers (Fig. 2Fi‐vi,vii‐xii), which were further quantified by image quantification and FACS sorting for HNF4A+ and ASGR1+ cells (Fig. 2Gi‐ii).
Trib1 liver‐specific KO mice have increased expression of genes involved in lipid synthesis and increased liver triglyceride (TG) mass.( 23 ) We characterized the expression of lipid synthesis genes such as diacylglycerol acyltransferase (DGAT)1, DGAT2, fatty acid synthase (FASN), acetyl‐coenzyme A carboxylase (ACACA), and stearoyl‐coenzyme A desaturase (SCD)1 in TRIB1 KO hiPSC‐derived hepatocytes. At day 20, expression of lipid synthesis genes was significantly reduced in TRIB1 KO 2D HLCs compared with WT cultures, consistent with the reduction in other canonical hepatocyte genes (Fig. 2H). Furthermore, the amount of TG in the TRIB1 KO 2D HLCs was markedly lower than in WT 2D cultures (Fig. 2I). In marked contrast, TRIB1 KO 3D organoids displayed significantly increased expression of lipid synthesis genes including DGAT1, DGAT2, and ACACA and a trend toward an increase in FASN and SCD compared with WT 3D organoids (Fig. 2H). The accumulated TGs were dramatically higher in the TRIB1 KO 3D organoids than in TRIB1 KO 2D HLCs and trended to being slightly higher compared with WT 3D organoids (Fig. 2I). Secreted TGs in the media of TRIB1 KO 3D organoids also trended higher than in the media of WT 3D organoids (Supporting Fig. S5).
TRIB1 KO 2D HLCs, but Not 3D Organoids, Exhibit Developmental Arrest Associated With Low Expression of C/EBPα
To determine how early TRIB1 affects hepatocyte differentiation in the 2D differentiation protocol, we measured gene expression at different stages of differentiation. At day 5, no change in the expression of DE stage marked by FOXA2, SRY (sex determining region Y)‐box (SOX17), and chemokine (C‐X‐C motif) receptor 4 (CXCR4) was observed (Fig. 3A). At day 12, a trend toward an increased expression of hepatoblast markers, such as hematopoietically expressed homeobox protein (hHEX) and delta‐like 1 homolog (DLK1), was observed in TRIB1 KO 2D HLCs over WT day 12 HLCs (Fig. 3B). However, at day 20, a significant reduction in hepatic markers like HNF4A to AFP, ALB, and ASGR1 was observed, and increased expression of hepatoblast markers (hHEX, DLK1, and PROX1) was observed in day 20 HLCs (Figs. 2E and 3B). No such increase in expression of hepatoblast markers was observed in 3D TRIB1 KO organoids at day 20 of differentiation (Fig. 3C).
FIG. 3.
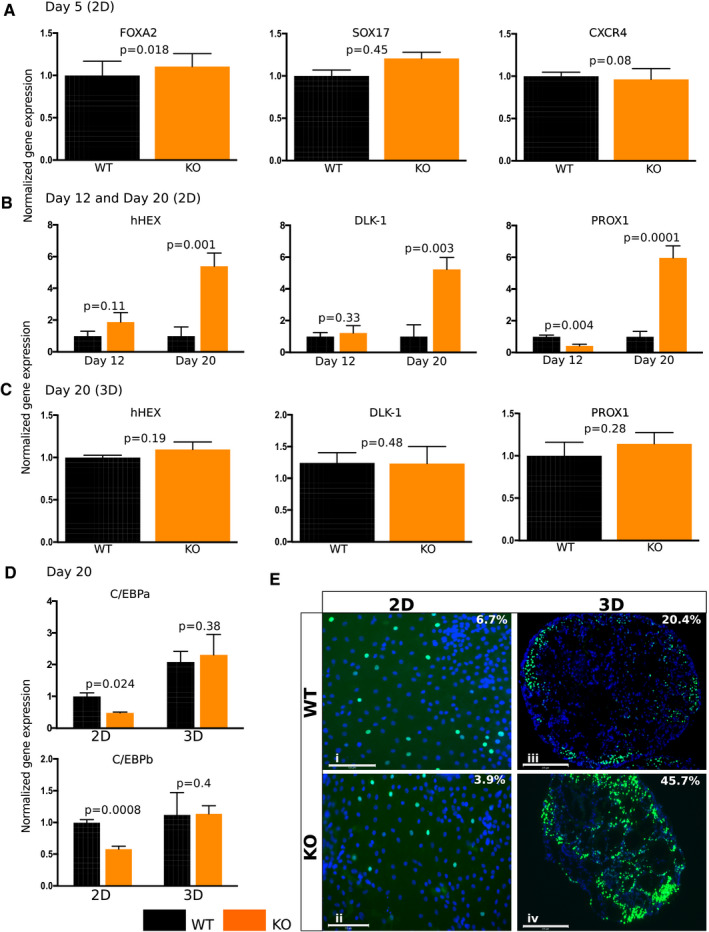
TRIB1 KO 2D HLCs exhibit developmental arrest due to the inherent low expression of C/EBPα. (A) TRIB1 KO 2D HLCs have equivalent DE markers (FOXA2, SOX17, and CXCR4) versus WT HLCs at day 5 of differentiation. (B) Increased expression of progenitor markers (hepatoblasts: hHEX, DLK1, and PROX1) were observed in TRIB1 KO HLCs at day 12 but predominantly at day 20 of differentiation versus WT HLCs. (C) Equivalent hepatoblast markers were observed in WT and TRIB1 KO 3D organoids at day 20 of differentiation. (D) Two‐dimensional HLCs have low expression of C/EBPα and C/EBPβ, which are further reduced in TRIB1 KO HLCs, whereas 3D organoids express adequate levels of C/EBPα and C/EBPβ, which increase in TRIB1 KO organoids as shown by quantitative PCR (D) and immunostaining for C/EBPα (E) (green) at day 20 of differentiation. Values represent the mean ± SEM. Data are representative of independent experiments, calculated relative to GAPDH (reference gene) using the 2−ddCt method, normalized to the mean expression level in WT cells (n = 4 WT and four KO wells; two wells per clone with 24 organoids per well. For the 2D HLCs, n = 8 WT and eight KO wells; and n = 2 WT and two KO clones). For C/EBPα and C/EBPβ quantitative PCR, normalized to the mean expression level in WT 2D HLCs. P values were calculated with Welch’s t tests. DAPI is shown in blue in all of the immunofluorescence images. Scale bar: 125 µm (Ei,ii) and 275 µm (Eiii,iv).
TRIB1 regulates levels of C/EBPα posttranslationally, and we reported increased levels of C/EBPα protein in Trib1 liver‐specific KO mice (LsKO).( 23 ) TRIB1 KO 2D HLCs exhibited decreased C/EBPα messenger RNA (mRNA) (Fig. 3D) and protein (3.9%) (Fig. 3E and Supporting Fig. S6) compared with WT 2D HLCs (6.7%). In contrast, TRIB1 KO 3D organoids displayed dramatically increased C/EBPα protein (45.7%) (Fig. 3E and Supporting Fig. S6) compared with WT 3D organoids (20.4%), despite C/EBPα mRNA. Similar observations were made for C/EBPß mRNA (Fig. 3D). Together, these data suggest the developmental arrest of hepatoblasts to mature hepatocytes in TRIB1 KO 2D HLCs, but that this does not exist in TRIB1 KO 3D organoids, which recapitulate the expected phenotype of increased C/EBPα protein without an increase in C/EBPß mRNA.
Overexpression of C/EBPα Rescues Hepatic and TG Phenotypes of TRIB1 KO hiPSC Hepatocytes
To determine whether increased C/EBPα protein was sufficient to rescue the maturation of hepatocytes and in turn drive increased lipid synthesis, we overexpressed human C/EBPα in both WT and TRIB1 KO hiPSC‐derived hepatocytes using an inducible lentivirus system (Fig. 4A and Supporting Fig. S7). The hiPSCs were differentiated to 2D HLCs and 3D hepatic organoids in parallel. hC/EBPα expression was induced starting at day 13 of differentiation and continued for 7 days. At day 20, cells were harvested for gene expression and lipid phenotyping (Fig. 4A). As expected, hC/EBPα‐induced WT and TRIB1 KO 2D HLCs had increased expression of C/EBPα (Fig. 4B) and increased C/EBPα protein (Fig. 4C); notably, while the TRIB1 KO 2D HLCs without overexpression of C/EBPα had reduced C/EBPα protein compared with WT 2D HLCs, those with overexpression had markedly more C/EBPα protein than the overexpressing WT 2D HLCs (Fig. 4C). C/EBPα overexpression “rescued” expression of hepatic markers HNF4A, AFP, ALB and ASGR1, and decreased the expression of hepatoblast markers at day 20 of differentiation (Fig. 4D). The lipid synthesis genes showed significantly increased expression in the C/EBPα‐overexpressing WT and TRIB1 KO 2D HLCs (Fig. 4E). Consistent with this, TG synthesis and TG mass were both significantly increased in the C/EBPα‐overexpressing WT and TRIB1 KO 2D HLCs compared with control cells (Fig. 4F). Interestingly, both TG synthesis and TG mass were significantly higher in the C/EBPα‐overexpressing TRIB1 KO 2D HLCs compared with the C/EBPα‐overexpressing WT 2D HLCs, despite being lower in the control cells (Fig. 4F). These data suggest that the low inherent expression of C/EBPα in 2D HLCs restricts the formation of mature hepatocytes and can be overcome by forced C/EBPα overexpression.
FIG. 4.
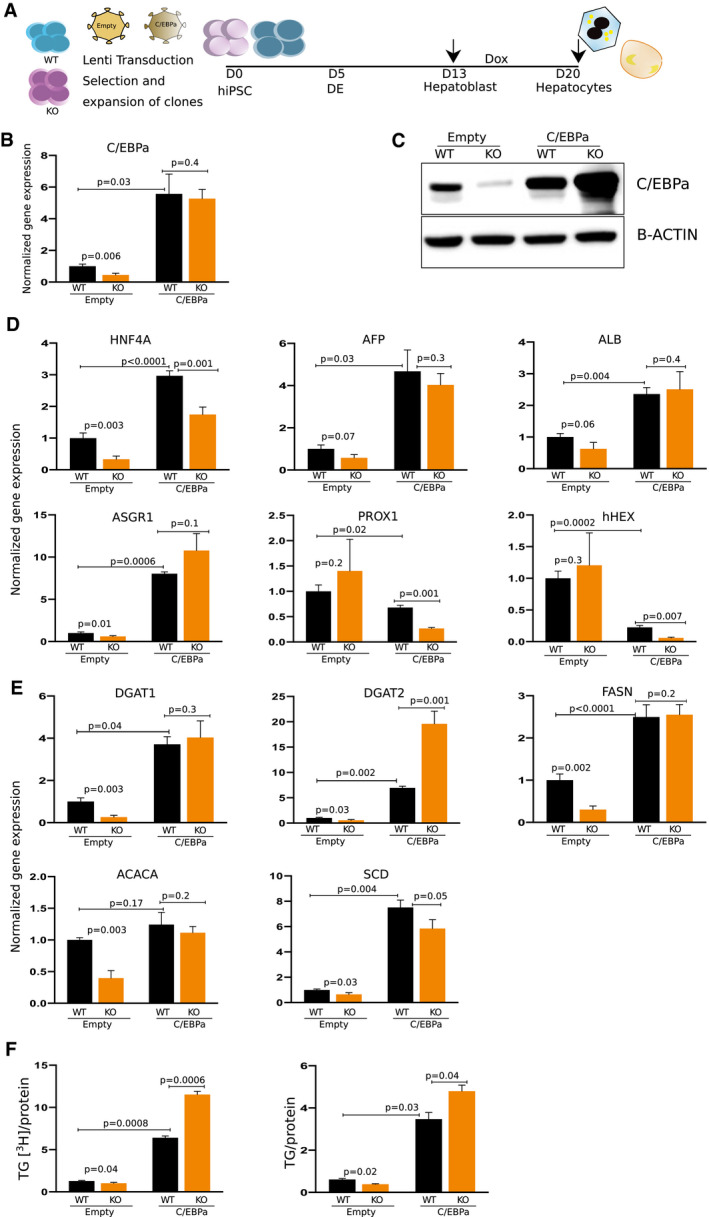
Overexpression of C/EBPα in WT and TRIB1 KO cells rescues the TRIB1 lipid phenotype in 2D HLCs. (A) Schematic showing experimental strategy in which WT and TRIB1 KO hiPSCs were exposed to empty vector and C/EBPα lentivirus. Differentiation was performed in parallel to 2D and 3D. At day 13, cells were induced with Dox for 7 days and at day 20, cells were harvested for downstream analysis. Shown here are the data for 2D HLCs; data for 3D are shown in Supporting Fig. S8. (B,C) C/EBPα was decreased in empty lentivirus‐transduced TRIB1 KO 2D HLCs versus WT HLCs, which were rescued both at the RNA and protein level on overexpression of C/EBPα. (D) Rescue of hepatic markers (HNF4A, AFP, ALB, and ASGR1) and decrease in progenitor markers (PROX1 and hHEX) were observed in day 20 WT and TRIB1 KO HLCs on overexpression of C/EBPα. Rescue of DNL gene expression (E) and TG phenotype (F) was observed in 2D TRIB1 KO HLCs on overexpressing C/EBPα. Values represent the mean ± SEM. Fold change calculated relative to GAPDH (reference gene) using the 2−ddCt method, normalized to the mean expression level in WT cells transduced with empty lenti vector as control (n = 6 WT + empty vector, four KO + empty vector; and n = 3 WT + C/EBPα and six KO + C/EBPα wells). P values were calculated with Welch’s t tests.
A similar observation was made with the 3D hepatic organoids, in which overexpression of hC/EBPα further increased the expression of hepatic (ALB) and lipid synthesis (DGAT1, DGAT2, FASN, ACACA, and SCD) genes as well as TG synthesis, and suppressed the expression of hepatoblast progenitor markers (hHEX and PROX1) at day 20 of differentiation (Supporting Fig. S8).
Discussion
Here we describe a 3D hiPSC‐derived hepatic organoid model that temporally recapitulates the stages of hepatocyte development more effectively than 2D hiPSC‐derived hepatocytes. We show that hiPSCs deleted in TRIB1 and differentiated to monolayer 2D HLCs are immature and have little of the expected lipid phenotype, but when differentiated to 3D, hiPSC‐derived hepatic organoids express more mature hepatocyte biology and recapitulate the expected biology of hepatocyte TRIB1 deficiency based on mouse studies, providing even further insights into the role of hepatocyte TRIB1 in lipid metabolism.
The early development and dynamic metabolic functions of a complex organ like the liver cannot be fully understood by analysis of a single type of cell in isolation, nor can the pathogenesis of genetic diseases affecting liver development and its function be fully recapitulated using individual cells.( 14 , 16 , 17 ) Recently, liver organoids have been generated from a myriad of cells including progenitor cells obtained from human liver tissue,( 28 ) hiPSCs,( 15 , 29 , 30 ) a pool of hiPSC‐derived progenitors( 31 ) with endothelial cells, mesenchymal stromal cells, human umbilical vein endothelial cells involving complex engineered matrices( 32 ) or perfused systems,( 33 ) or aggregation only in later stages of differentiation.( 30 , 31 ) We made the same observation that organoids have enhanced expression of hepatic markers and are more mature than 2D cultured cells, as reported previously.( 24 , 34 , 35 ) Here we describe an efficient protocol using hiPSCs for hepatic 3D organoid differentiation by using short exposure to a nonengineered matrix. In contrast to the engineered matrices such as polyethylene glycol, polyglycolic/poly‐l‐lactic acid, glycosaminoglycans, pullulan, synthetic scaffolds with collagen, fibrinogen and laminin commonly used for 3D organoid studies,( 36 , 37 ) we used a short exposure to nonengineered laminin‐enriched matrigel and CultureX (CultureX, Cambridge, MA) as a matrix for hepatic organoid differentiation. Studies are ongoing to further characterize these 3D organoids and improve the strategies to better obtain defined populations of nonparenchymal cells. One needs further detailed analysis about possible central cell death, its relationship to organoid size, and the potential use of high oxygen conditions for application of this 3D approach in understanding the liver injury disease modeling.
We used this approach to explore the pleiotropic cardiometabolic gene TRIB1. GWAS has identified a locus at chromosome 8q24, near the TRIB1 gene, at which variants are associated with NAFLD, plasma lipids, adiponectin, and coronary artery disease.( 23 ) Previously, we showed that targeted disruption of TRIB1 in mice results in an increased TG phenotype.( 23 ) However, considering the appreciable differences in mice versus human lipid biology and the absence, thus far, of any individual with a homozygous KO mutation in the Tribbles locus, led us to generate TRIB1 KO hiPSC using genome editing to understand its role in human hepatic and lipid biology. This approach is far better and offers several advantages over using teratogenically transformed hepatoma cell lines and primary cells, which dedifferentiate in a culture dish, or tissues from diseased biopsies, which do not reflect the true metabolic state.( 38 ) Moreover, of the other model systems discussed previously, only hiPSCs provide the opportunity to understand the role of a gene or rare mutation in early development.
To model TRIB1 lipid biology in a dish, WT and KO hiPSCs were differentiated to 2D HLCs, and surprisingly, TRIB1 KO HLCs showed reduced expression of hepatic markers and de novo lipogenesis (DNL) markers over WT HLCs in contrast to our earlier observations made with TRIB1 LsKO mice.( 23 ) Interestingly, no defect in the induction of DE from hiPSCs was observed. However, expression of hepatoblast markers was observed to be maintained at a higher level in TRIB1 KO HLCs, even in later stages of differentiation when compared with WT HLCs. We also observed a decrease in the expression of C/EBPα in TRIB1 KO 2D HLCs over WT HLCs in contrast to TRIB1 LsKO mice.
Organoid systems have previously been reported to offer several advantages regarding maturation and function of iPSC‐derived cell types for several organ systems including hepatocytes.( 17 , 30 ) Taking advantage of our 3D hepatic organoid method, we differentiated WT and TRIB1 KO hiPSCs in parallel and observed no decrease in the hepatic markers in contrast to the 2D HLCs. However, a trend toward an increase in gene expression of hepatic markers as early as HNF4A was observed in TRIB1 KO organoids over WT organoids. Following this increase in C/EBPα, its downstream target lipid synthesis gene expression (DGAT1, DGAT2, FASN, ACACA, and SCD) and TGs were observed in TRIB1 KO organoids, mimicking the phenotype observed in TRIB1 LsKO mice.( 23 ) Considering the shortcomings of conventional lipid measurement assays, stringent fluorescent‐based quantitative approaches are required to measure changes in miniscule lipid accumulation in small cultures like organoids. As reported previously in mice, an increase in the expression of genes responsible for fatty acid synthesis (FASN, ACACA, and SCD) and incorporation of di acyl to tri acyl glycerol (DGAT1 and DGAT2) were observed. This further reflects that TRIB1 is not responsible for hepatic specification, but is involved with hepatocyte maturation by affecting C/EBPα. This establishes a role of a metabolic gene, TRIB1, in hepatocyte development.
Monolayer 2D HLCs have been explored extensively for their robustness in understanding development and as an essential tool in drug discovery, but they lack the expression of critical late‐stage hepatic markers and are therefore considered to be metabolically immature in nature, either lacking or expressing suppressed levels of vital specification markers such as PROX1 and C/EBPα.( 10 ) Leveraging our previously generated 2D HLC database,( 9 ) consisting of approximately 90 HLCs of diverse background, we analyzed the gene expression of PROX1 and C/EBPα and found them to be expressed at significantly reduced levels when compared with other liver hepatic and metabolic genes such as ALB and APOC3 (Supporting Fig. S9). Interestingly, these HLCs also express low levels of peroxisome proliferator‐activated receptor (PPARα and PPAR delta) (Supporting Fig. S9), expressed robustly in metabolically mature hepatocytes. This highlights that although 2D cultures exhibit robust expression of hepatocyte markers and are a good model for some studies, they still lack the expression of key hepatocyte markers. Therefore, the need for 3D organoid cultures prevails to overcome this limitation and to produce metabolically mature hepatocyte‐like cells.
TRIB1 is reported to posttranscriptionally regulate levels of C/EBPα by ubiquitination.( 23 , 39 ) It has been observed that 2D HLCs express only low levels of C/EBPα, and in the absence of TRIB1, there is a further decrease in C/EBPα RNA. This affects the overall protein turnover of C/EBPα, which is reflected as reduced levels of the C/EBPα protein. This decrease in C/EBPα resulted in maturation defects of TRIB1 KO 2D HLCs, which fail to up‐regulate late‐stage hepatic markers and maintain the hepatoblast stage markers, as previously shown in primary hepatocytes( 40 ) and C/EBPα KO mice.( 41 ) Along similar lines, overexpression of C/EBPα rescued the maturation defects in TRIB1 KO 2D HLCs, further emphasizing that the TRIB1‐C/EBPα relationship holds true in hiPSC hepatocytes, which was responsible for influencing TG levels.( 23 ) However, overexpression of C/EBPα in WT HLCs also increased hepatic and DNL gene expression, indicating the attainment of a more mature state, emphasizing the definite need of C/EBPα for maturation of hepatocytes.( 42 , 43 , 44 )
Previous reports have established that C/EBPα regulates HNF4A, and both C/EBPα and HNF4A co‐operatively act to regulate downstream genes.( 45 , 46 ) Low levels of C/EBPα in 2D HLCs, in turn, resulted in decreased expression of HNF4A, which further explains the effect on hepatocyte differentiation. Recently, primary hepatocytes, in which TRIB1 was knocked down using small interfering RNA,( 47 ) showed a decrease in expression of HNF4A, which further mechanistically explains the influence on hepatocyte differentiation in 2D HLCs at the level of HNF4A in TRIB1 KO 2D HLCs.
Supporting our observation in hiPSC hepatic organoids, recent reports in HepG2 and rat primary hepatocytes have shown that hepatic genes such as PROX1, HNF4α, and C/EBPα are induced in 3D hepatocyte cultures.( 48 , 49 ) TRIB1 liver‐specific KO mice has increased C/EBPα protein (due to reduced protein degradation) and decreased C/EBPα transcript (due to negative feedback from the increased protein).( 23 ) We also know that TRIB1 is a transcriptional target of C/EBPα, leading to a negative feedback loop in which excess C/EBPα induces TRIB1, which then promotes the degradation of C/EBPα. In 2D cultures, we suggest that poor expression of C/EBPα fails to effectively induce TRIB1 expression, whereas consistent with the recent reports, we suggest that C/EBPα is more efficiently induced in 3D organoids, which more efficiently activates TRIB1 expression. This led us to speculate that in 3D organoids, enhanced expression of hepatic markers—especially C/EBPα—is maintained, which allowed the Trib1‐ C/EBPα feedback loop to establish and recapitulate the phenotype.
In summary, we report a simplified and defined 3D iPSC‐hepatocyte organoid system. We show, using TRIB1‐deleted iPSCs as proof of principle, that this 3D system more faithfully recapitulates the expected biology of TRIB1 deletion in hepatocytes than the 2D system. In this model, the effects of TRIB1 KO are more clearly seen in the 3D organoids than in the 2D cultures. This study establishes the importance of having a robust 3D model system to understand the role of genes, which are poorly reflected by conventional monolayer cultures. Furthermore, this organoid approach has the potential to permit expansion of our understanding of genetic diseases affecting the liver and has substantial implications for drug discovery.
Materials and Methods
hiPSC Culture
Established hiPSC lines were maintained in feeder‐free culture conditions in either mTeSR1 (STEMCELL Technologies) or iPS‐Brew (Miltenyi Biotec, Bergisch Gladbach, Germany) on Geltrex coated dishes (Thermo Fisher Scientific, Waltham, MA) as described previously.( 9 ) In‐house‐derived hiPSC line Sv20 derived from peripheral blood mononuclear cells and the K3 hiPSC line produced from foreskin fibroblasts were used for the studies, which were characterized previously in detail.( 5 , 9 )
Hepatocyte Differentiation
2D Monolayer
hiPSC lines were passaged with Accutase (Innovative Cell Technologies, San Diego, CA) for 5 minutes at room temperature (RT) and plated for differentiation on Geltrex coated dishes following Stephen Duncan’s protocol with modifications.( 5 ) For efficient DE differentiation, the DE kit (STEMCELL Technologies) was used for the first 4 days in ambient O2/5% CO2. Later, cells were cultured in RPMI‐B27 (with insulin) supplemented with BMP4 (20 ng/mL; PeproTech, Rocky Hill, NJ) and FGF2 (10 ng/mL; R&D Systems) for 5 days in 5% O2/5% CO2, followed by RPMI‐B‐27 (with insulin) supplemented with recombinant human HGF (20 ng/mL; PeproTech) for 5 days in 5% O2/5% CO2. The last stage required culturing cells in HCM Hepatocyte Culture Medium (Lonza Group AG, Basel, Switzerland) that was supplemented with recombinant human Oncostatin M (20 ng/mL; R&D Systems) for 5 days in ambient O2/5% CO2.( 9 )
3D Organoid
hiPSC lines were passaged with Accutase for 5 minutes at RT and plated in 96‐well U‐bottom low adherent plates using the DE kit with 10 µM Rock inhibitor and continued in ambient O2/5% CO2 throughout the differentiation. Approximately 3,000‐5,000 cells per 100 µL per 96‐well plate gave efficient aggregation to organoids with no to minimal evidence of central darkening suggestive of cell death. However, standardization from line to line is necessary to make sure the organoid size does not become limiting, in which case the potential for central cell death is higher. After 24 hours, half the medium was removed, and fresh medium with 4% matrigel and CultureX was added and cultured for 2 more days with daily medium addition. At day 5, aggregates were harvested and cultured in low‐adherent 6‐well plates in RPMI‐B27 (with insulin) supplemented with BMP4 (20 ng/mL), FGF2 (10 ng/mL), and KGF (20 ng/mL; PeproTech) for 5 days, followed by RPMI‐B‐27 (with insulin) supplemented with recombinant human HGF (20 ng/mL; PeproTech) and 0.5% DMSO for 5 days. Later, aggregates were cultured in HCM Hepatocyte Culture Medium and supplemented with recombinant human Oncostatin M (20 ng/mL) and Dexamethasone for 5 days.
CRISPR‐Mediated Genome Engineering of hiPSCs
Guide RNAs were designed and evaluated for potential off‐target activity using the online CRISPR Design tool at http://crispr.mit.edu. Genome editing was performed using SpCas9_GFP (Addgene plasmid #48138), which co‐expresses a human codon–optimized Cas9 nuclease and green fluorescent protein (GFP) using a viral 2A sequence. To generate the knockout using a knock‐in approach, a 200‐bp ssODN was designed in Exon 1 with a stop codon introduced after eight amino acids and a HindIII site, to confirm the targeted clones.
For nucleofection, cells in a 70%‐80% confluent 10‐cm plate were dissociated into single cells with Accutase, resuspended in phosphate‐buffered saline, and combined with 5 µg SpCas9‐GFP and 1 µg of ssODN C*G*C*A*GCGTCTTCCCGCGCGGATCCCGGGACTTAAAAAGCCGGGGCCACCCCGGCCCAGGACGGGATGCGGGTCGGTCCGGTGCGCTCTTAGTGATAAGCTTCTCGCAGCCCCGCGGCCCGGCCCTGCTCTTCCCAGCCACCCGAGGCGTCCCGGCCAAACGCCTGCTGGACGCCGACGACGCGGC*G*G*C*T. A single pulse was delivered at 250 V/500 mF (Lonza), and the cells were recovered and plated in mTeSR1 with 10 μM ROCK inhibitor (Y‐27632; Cayman Chemical Co., Ann Arbor, MI). Cells were dissociated with Accutase 48 hours after electroporation, and GFP‐positive cells were isolated by FACS (FACS AriaII; BD Biosciences, San Jose, CA) and replated onto 10‐cm Geltrex‐coated plates (~10,000 cells/plate) with conditioned medium and 10 µM ROCK inhibitor to facilitate recovery. Single cell–derived clones were picked after 2 weeks, expanded, and evaluated for presence of HindIII site by restriction digestion after polymerase chain reaction (PCR) amplification of a small region surrounding the targeting site, using primers F‐5′AAGACCAGTCTGCAAACTCCAT3′ and R‐ 5′ CTGAGGAAAGGAACAGATAGAGAAAG 3′. Clones were further confirmed with Sanger sequencing of the PCR products, before selecting them for differentiation.
Lentiviral Mediated Overexpression of hC/EBPα
To generate the lentivirus construct, hC/EBPα cDNA was purchased from the DNASU Plasmid Repository (Tempe, AZ) and cloned into pEn_TTmcs vector with Acc65I and XhoI, and gel purified the 3.8‐kb vector band. C/EBPα was ligated as insert and transformed into DH5α followed by digestion with XbaI and NcoI. To generate the empty pEn_TTmcs control vector, the XbaI‐digested vector was recircularized. Vectors were confirmed with Sanger sequencing and transferred to a pSLIK‐puro lenti backbone using Gateway LR clonase (Thermo Fisher Scientific).
Human embryonic kidney cells were seeded in 10‐cm lysine‐coated dishes and transfected using Lipofectamine 3000 with 12 µg lenti backbone, 4.5 µg PMD2, and 7.7 µg pSPAX2 in the presence of Polybrene (Sigma Aldrich, St. Louis, MO) according to the manufacturer’s protocol. Media were collected every 24 hours following transfection for 3 consecutive days, filtered with 0.45‐µ filters and concentrated 10 times, then added to hiPSCs. After 24 hours the regular iPS‐Brew medium was changed to hiPSCs and selected with Puromycin. Puromycin‐resistant cells were passaged with Accutase for 5 minutes to single cells and replated on 10‐cm Geltrex dishes for expansion. Single cell–derived clones were picked manually and cultured in the presence and absence of Doxycycline (Dox). hiPSCs were then analyzed for induction of C/EBPα by gene‐expression analysis.
Quantitative Real‐Time PCR
RNA isolation was performed using Qiagen miRNeasy and Zymo kits (Hilden, Germany) according to the manufacturer’s instructions, and cDNA was prepared using a High Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific) with an equimolar mixture of random hexamers and oligo‐dT. Gene expression was measured using the TaqMan Gene Expression Assays (Thermo Fisher Scientific) in 10 µL quantitative real‐time PCR reaction with 20 ng cDNA performed in technical duplicate. Reactions were carried out on a 7900HT QuantStudio Fast Real‐Time PCR System (Thermo Fisher Scientific). Relative expression levels were quantified by the 2–DDCt method using glyceraldehyde 3‐phosphate dehydrogenase (GAPDH) as reference gene.
Immunofluorescence Staining
Cells were fixed for 15 minutes at RT with 4% paraformaldehyde (PFA) and incubated overnight with antibodies anti‐FOXA2 (sc‐6554), anti‐GATA4 (sc‐1237), anti‐HNF4A (sc‐6556), anti‐AFP (A‐8452), anti‐albumin (CL‐2513A), anti‐C/EBPα (8178S), anti‐MDR1, anti‐arginase, anti‐HNF1ß (12533‐1‐AP), and anti‐ZO‐1 (40‐2200) as described previously.( 5 )
Flow Cytometry
Day 20 hepatocytes were dissociated using enzyme mixture (Accutase:collagenase:trypsin in 1: 1:1), washed with Permeabilization Wash Buffer (BioLegend, San Diego, CA) and stained for antibodies against asialoglycoprotein receptor 1 (563655) for 1 hour in the dark. Flow cytometry was performed using an Accuri Flow Cytometer (BD Biosciences) with unstained cells and only secondary stained as controls.
Bile Acid and LDL Uptake
The organoids were washed with Dulbecco’s phosphate buffered saline (DPBS), then incubated with 5 μg/mL CDFDA and 5 μg/mL Dil‐Ac‐LDL (Invitrogen) following the manufacturer’s instructions, and washed again before examination with fluorescence microscopy. Later, organoids were fixed with 4% PFA and mounted in optimal cutting temperature for sectioning.
PAS Staining
The organoids were fixed with 4% PFA in DPBS and the sections were stained with a PAS staining kit from Sigma‐Aldrich according to the manufacturer’s instructions.
ORO Staining
For detecting accumulated lipids, organoids were fixed with 4% PFA in DPBS and the sections were treated with 60% isopropanol for 5 minutes. Then the isopropanol was removed and ORO (Sigma‐Aldrich) was added following the manufacturer’s instructions and incubated for 15 minutes at RT. ORO was then removed and sections were rinsed with water until clear before imaging.
Enzyme‐Linked Immunosorbent Assay for Secretory Proteins
The human albumin, APOB, and APOC3 content in the cultured media was measured using enzyme‐linked immunosorbent assay kits from Bethyl Laboratories (Montgomery, TX), MABtech AB (Nacka Strand, Sweden), and Thermo Fisher Scientific, respectively, following the manufacturers’ instructions. The secretion was normalized to total cell RNA isolated from Zymo kits.
TG Measurement
Total TG was measured by colorimetric enzymatic assay. Briefly, lipids were extracted from day 20 HLCs and organoids using (3:2) hexane:isopropanol, and pellet was resuspended in water after 15% Triton‐chloroform treatment. Absorbance was measured at 500 nm after incubating 10 µL of extracted lipid or 10 µL of media with 190 µL TG Infinity enzymatic solution for 15 minutes at 37°C (Thermo Fisher Scientific). To measure newly synthesized TGs, day 20 cells were pre‐incubated with [3H]‐1,2,3‐Glycerol (10 µCi/mL) in the presence of Oleic Acid (0.4 mM) for 4 hours. To measure radiolabeled TG, lipids were extracted from cells, fractionated by thin‐layer chromatography, and quantified by beta counter (Beckman Coulter, Brea, CA). TG counts in cells was normalized to the total cellular protein for 2D HLCs, as measured by BCA. For organoids, equal number and sized organoids per treatment were used for the assay.( 50 )
Statistics
All of the quantitative data represent mean + SEM. The average gene‐expression levels or changes in lipid levels were compared among groups using the Welch’s t test, due to the groups having unequal sizes. Graphs were plotted using GraphPad Prism 8 for Mac OS X (GraphPad Software, San Diego, CA).
Supporting information
Fig S1
Fig S2
Fig S3
Fig S4
Fig S5
Fig S6
Fig S7
Fig S8
Fig S9
Acknowledgment
The authors thank Dawn Marchadier and Jamie Ifkovits for editing and proofreading the manuscript. We want to thank members of the Rader laboratory for the valuable discussions. We thank Cardiology Histology Core (UPenn) and MPIC (UPenn) for their help with sectioning and staining of organoids, as well as Wenli Yang, Kate Slovik, and Rachel Truitt from iPSC Core (UPenn) for their help and discussions.
Supported by the Foundation for the National Institutes of Health (R01HL134853 and U01HG006398).
Potential conflict of interest: Nothing to report.
Contributor Information
Deepti Abbey, Email: deeptiab@pennmedicine.upenn.edu.
Daniel J. Rader, Email: rader@pennmedicine.upenn.edu.
References
Author names in bold designate shared co‐first authorship.
- 1. Klarin D, Damrauer SM, Cho K, Sun YV, Teslovich TM, Honerlaw J, et al. Genetics of blood lipids among ~300,000 multi‐ethnic participants of the Million Veteran Program. Nat Genet 2018;50:1514‐1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. de Vries PS, Brown MR, Bentley AR, Sung YJ, Winkler TW, Ntalla I, et al. Multiancestry genome‐wide association study of lipid levels incorporating gene‐alcohol interactions. Am J Epidemiol 2019;188:1033‐1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nikpay M, Goel A, Won HH, Hall LM, Willenborg C, Kanoni S, et al. A comprehensive 1,000 Genomes‐based genome‐wide association meta‐analysis of coronary artery disease. Nat Genet 2015;47:1121‐1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Corbett JL, Duncan SA. iPSC‐derived hepatocytes as a platform for disease modeling and drug discovery. Front Med (Lausanne) 2019;6:265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Si‐Tayeb K, Noto FK, Nagaoka M, Li J, Battle MA, Duris C, et al. Highly efficient generation of human hepatocyte‐like cells from induced pluripotent stem cells. Hepatology 2010;51:297‐305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hay DC, Zhao D, Fletcher J, Hewitt ZA, McLean D, Urruticoechea‐Uriguen A, et al. Efficient differentiation of hepatocytes from human embryonic stem cells exhibiting markers recapitulating liver development in vivo. Stem Cells 2008;26:894‐902. [DOI] [PubMed] [Google Scholar]
- 7. Cai J, Zhao Y, Liu Y, Ye F, Song Z, Qin H, et al. Directed differentiation of human embryonic stem cells into functional hepatic cells. Hepatology 2007;45:1229‐1239. [DOI] [PubMed] [Google Scholar]
- 8. Cayo MA, Mallanna SK, Di Furio F, Jing R, Tolliver LB, Bures M, et al. A drug screen using human iPSC‐derived hepatocyte‐like cells reveals cardiac glycosides as a potential treatment for hypercholesterolemia. Cell Stem Cell 2017;20:478‐489.e475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pashos EE, Park Y, Wang X, Raghavan A, Yang W, Abbey D, et al. Large, diverse population cohorts of hiPSCs and derived hepatocyte‐like cells reveal functional genetic variation at blood lipid‐associated loci. Cell Stem Cell 2017;20:558‐570.e510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhao D, Chen S, Duo S, Xiang C, Jia J, Guo M, et al. Promotion of the efficient metabolic maturation of human pluripotent stem cell‐derived hepatocytes by correcting specification defects. Cell Res 2013;23:157‐161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Baxter M, Withey S, Harrison S, Segeritz CP, Zhang F, Atkinson‐Dell R, et al. Phenotypic and functional analyses show stem cell‐derived hepatocyte‐like cells better mimic fetal rather than adult hepatocytes. J Hepatol 2015;62:581‐589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Berger DR, Ware BR, Davidson MD, Allsup SR, Khetani SR. Enhancing the functional maturity of induced pluripotent stem cell‐derived human hepatocytes by controlled presentation of cell‐cell interactions in vitro. Hepatology 2015;61:1370‐1381. [DOI] [PubMed] [Google Scholar]
- 13. Asai A, Aihara E, Watson C, Mourya R, Mizuochi T, Shivakumar P, et al. Paracrine signals regulate human liver organoid maturation from induced pluripotent stem cells. Development 2017;144:1056‐1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vyas D, Baptista PM, Brovold M, Moran E, Brovold M, Gaston B, et al. Self‐assembled liver organoids recapitulate hepato‐biliary organogenesis in vitro. Hepatology 2017;67:750‐761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sgodda M, Dai Z, Zweigerdt R, Sharma AD, Ott M, Cantz T. A scalable approach for the generation of human pluripotent stem cell‐derived hepatic organoids with sensitive hepatotoxicity features. Stem Cells Dev 2017;26:1490‐1504. [DOI] [PubMed] [Google Scholar]
- 16. Camp JG, Sekine K, Gerber T, Loeffler‐Wirth H, Binder H, Gac M, et al. Multilineage communication regulates human liver bud development from pluripotency. Nature 2017;546:533‐538. [DOI] [PubMed] [Google Scholar]
- 17. Fatehullah A, Tan SH, Barker N. Organoids as an in vitro model of human development and disease. Nat Cell Biol 2016;18:246‐254. [DOI] [PubMed] [Google Scholar]
- 18. Takayama K, Kawabata K, Nagamoto Y, Kishimoto K, Tashiro K, Sakurai F, et al. 3D spheroid culture of hESC/hiPSC‐derived hepatocyte‐like cells for drug toxicity testing. Biomaterials 2013;34:1781‐1789. [DOI] [PubMed] [Google Scholar]
- 19. Wang B, Jakus AE, Baptista PM, Soker S, Soto‐Gutierrez A, Abecassis MM, et al. Functional maturation of induced pluripotent stem cell hepatocytes in extracellular matrix—a comparative analysis of bioartificial liver microenvironments. Stem Cells Transl Med 2016;5:1257‐1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gieseck Iii RL, Hannan NRF, Bort R, Hanley NA, Drake RAL, Cameron GWW, et al. Maturation of induced pluripotent stem cell derived hepatocytes by 3D‐culture. PLoS One 2014;9:e86372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kathiresan S, Melander O, Guiducci C, Surti A, Burtt NP, Rieder MJ, et al. Six new loci associated with blood low‐density lipoprotein cholesterol, high‐density lipoprotein cholesterol or triglycerides in humans. Nat Genet 2008;40:189‐197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chambers JC, Zhang W, Sehmi J, Li X, Wass MN, Van der Harst P, et al. Genome‐wide association study identifies loci influencing concentrations of liver enzymes in plasma. Nat Genet 2011;43:1131‐1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bauer RC, Sasaki M, Cohen DM, Cui J, Smith MA, Yenilmez BO, et al. Tribbles‐1 regulates hepatic lipogenesis through posttranscriptional regulation of C/EBPalpha. J Clin Invest 2015;125:3809‐3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Leclerc E, Kimura K, Shinohara M, Danoy M, Le Gall M, Kido T, et al. Comparison of the transcriptomic profile of hepatic human induced pluripotent stem like cells cultured in plates and in a 3D microscale dynamic environment. Genomics 2017;109:16‐26. [DOI] [PubMed] [Google Scholar]
- 25. Sullivan GJ, Hay DC, Park IH, Fletcher J, Hannoun Z, Payne CM, et al. Generation of functional human hepatic endoderm from human induced pluripotent stem cells. Hepatology 2010;51:329‐335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Song Z, Cai J, Liu Y, Zhao D, Yong J, Duo S, et al. Efficient generation of hepatocyte‐like cells from human induced pluripotent stem cells. Cell Res 2009;19:1233‐1242. [DOI] [PubMed] [Google Scholar]
- 27. Hay DC, Pernagallo S, Diaz‐Mochon JJ, Medine CN, Greenhough S, Hannoun Z, et al. Unbiased screening of polymer libraries to define novel substrates for functional hepatocytes with inducible drug metabolism. Stem Cell Res 2011;6:92‐102. [DOI] [PubMed] [Google Scholar]
- 28. Huch M, Gehart H, van Boxtel R, Hamer K, Blokzijl F, Verstegen MM, et al. Long‐term culture of genome‐stable bipotent stem cells from adult human liver. Cell 2015;160:299‐312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Guan Y, Xu D, Garfin PM, Ehmer U, Hurwitz M, Enns G, et al. Human hepatic organoids for the analysis of human genetic diseases. JCI Insight 2017;2:94954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ouchi R, Togo S, Kimura M, Shinozawa T, Koido M, Koike H, et al. Modeling steatohepatitis in humans with pluripotent stem cell‐derived organoids. Cell Metab 2019;30:374‐384.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Takebe T, Sekine K, Enomura M, Koike H, Kimura M, Ogaeri T, et al. Vascularized and functional human liver from an iPSC‐derived organ bud transplant. Nature 2013;499:481‐484. [DOI] [PubMed] [Google Scholar]
- 32. Willemse J, Lieshout R, van der Laan LJW, Verstegen MMA. From organoids to organs: bioengineering liver grafts from hepatic stem cells and matrix. Best Pract Res Clin Gastroenterol 2017;31:151‐159. [DOI] [PubMed] [Google Scholar]
- 33. Schepers A, Li C, Chhabra A, Seney BT, Bhatia S. Engineering a perfusable 3D human liver platform from iPS cells. Lab Chip 2016;16:2644‐2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Starokozhko V, Hemmingsen M, Larsen L, Mohanty S, Merema M, Pimentel RC, et al. Differentiation of human‐induced pluripotent stem cell under flow conditions to mature hepatocytes for liver tissue engineering. J Tissue Eng Regener Med 2018;12:1273‐1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nora F, Fanny K, Nadja S, Leila A, Petra S, Sebastian B, et al. Hepatic differentiation of human induced pluripotent stem cells in a perfused three‐dimensional multicompartment bioreactor. BioRes Open Access 2016;5:235‐248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yin X, Mead BE, Safaee H, Langer R, Karp JM, Levy O. Engineering stem cell organoids. Cell Stem Cell 2016;18:25‐38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kim G‐A, Spence JR, Takayama S. Bioengineering for intestinal organoid cultures. Curr Opin Biotechnol 2017;47:51‐58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Musunuru K. Stem cell modeling of lipid genetics. Curr Opin Lipidol 2018;29:151‐155. [DOI] [PubMed] [Google Scholar]
- 39. Soubeyrand S, Martinuk A, Lau P, McPherson R. TRIB1 is regulated post‐transcriptionally by proteasomal and non‐proteasomal pathways. PLoS One 2016;11:e0152346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gerlach JC, Over P, Foka HG, Turner ME, Thompson RL, Gridelli B, et al. Role of transcription factor CCAAT/enhancer‐binding protein alpha in human fetal liver cell types in vitro. Hepatol Res 2015;45:919‐932. [DOI] [PubMed] [Google Scholar]
- 41. Tomizawa M, Garfield S, Factor V, Xanthopoulos KG. Hepatocytes deficient in CCAAT/enhancer binding protein alpha (C/EBP alpha) exhibit both hepatocyte and biliary epithelial cell character. Biochem Biophys Res Commun 1998;249:1‐5. [DOI] [PubMed] [Google Scholar]
- 42. Akai Y, Oitate T, Koike T, Shiojiri N. Impaired hepatocyte maturation, abnormal expression of biliary transcription factors and liver fibrosis in C/EBPalpha(Cebpa)‐knockout mice. Histol Histopathol 2014;29:107‐125. [DOI] [PubMed] [Google Scholar]
- 43. Du Y, Wang J, Jia J, Song N, Xiang C, Xu J, et al. Human hepatocytes with drug metabolic function induced from fibroblasts by lineage reprogramming. Cell Stem Cell 2014;14:394‐403. [DOI] [PubMed] [Google Scholar]
- 44. Tomizawa M, Shinozaki F, Motoyoshi Y, Sugiyama T, Yamamoto S, Ishige N. Transcription factors and medium suitable for initiating the differentiation of human‐induced pluripotent stem cells to the hepatocyte lineage. J Cell Biochem 2016;117:2001‐2009. [DOI] [PubMed] [Google Scholar]
- 45. Stefflova K, Thybert D, Wilson MD, Streeter I, Aleksic J, Karagianni P, et al. Cooperativity and rapid evolution of cobound transcription factors in closely related mammals. Cell 2013;154:530‐540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jakobsen JS, Waage J, Rapin N, Bisgaard HC, Larsen FS, Porse BT. Temporal mapping of CEBPA and CEBPB binding during liver regeneration reveals dynamic occupancy and specific regulatory codes for homeostatic and cell cycle gene batteries. Genome Res 2013;23:592‐603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Soubeyrand S, Martinuk A, McPherson R. TRIB1 is a positive regulator of hepatocyte nuclear factor 4‐alpha. Sci Rep 2017;7:5574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hiemstra S, Ramaiahgari SC, Wink S, Callegaro G, Coonen M, Meerman J, et al. High‐throughput confocal imaging of differentiated 3D liver‐like spheroid cellular stress response reporters for identification of drug‐induced liver injury liability. Arch Toxicol 2019;93:2895‐2911. [DOI] [PubMed] [Google Scholar]
- 49. Borlak J, Singh PK, Rittelmeyer I. Regulation of liver enriched transcription factors in rat hepatocytes cultures on collagen and EHS sarcoma matrices. PLoS One 2015;10:e0124867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol 1959;37:911‐917. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1
Fig S2
Fig S3
Fig S4
Fig S5
Fig S6
Fig S7
Fig S8
Fig S9


