Abstract
Objective
Dedifferentiation of pancreatic β-cells may reduce islet function in type 2 diabetes (T2D). However, the prevalence, plasticity and functional consequences of this cellular state remain unknown.
Methods
We employed single-cell RNAseq to detail the maturation program of α- and β-cells during human ontogeny. We also compared islets from non-diabetic and T2D individuals.
Results
Both α- and β-cells mature in part by repressing non-endocrine genes; however, α-cells retain hallmarks of an immature state, while β-cells attain a full β-cell specific gene expression program. In islets from T2D donors, both α- and β-cells have a less mature expression profile, de-repressing the juvenile genetic program and exocrine genes and increasing expression of exocytosis, inflammation and stress response signalling pathways. These changes are consistent with the increased proportion of β-cells displaying suboptimal function observed in T2D islets.
Conclusions
These findings provide new insights into the molecular program underlying islet cell maturation during human ontogeny and the loss of transcriptomic maturity that occurs in islets of type 2 diabetics.
Keywords: Human islet, α-Cell, β-Cell, Single cell RNAseq, Type 2 diabetes, Ontogeny
Highlights
-
•
Non-endocrine and developmental genes are repressed during maturation of human α- and β-cells.
-
•
Adult α-cells retain a less mature gene expression program than β-cells.
-
•
In T2D, both α- and β-cells re-establish an immature transcriptome.
1. Introduction
Multiple causes have been suggested to explain the lower functional β-cell mass present in the pancreas of type 2 diabetics (T2D) [1], which include increased susceptibility to apoptosis of β-cells caused by exposure to chronic glucolipotoxicity [2], excessive endoplasmic reticulum (ER) stress [3,4], oxidative stress [[5], [6], [7]] and accumulation of DNA damage in the form of double-strand DNA breaks [8]. Recently, an alternative pathway has been invoked that involves β-cell de-differentiation or even complete loss of β-cell identity to explain reduced functional β-cell mass in T2D [[9], [10], [11]]. However, the evidence supporting the existence of the de-differentiated cellular state in human islet cells of diabetics is limited, and the precise nature, level of plasticity and functional outcome of this altered cellular state are unclear [[12], [13], [14]]. Therefore, we posit that a better definition of the postnatal maturation process of human β-cells is key to understanding the detrimental changes observed in islet cells from T2D. Thus far, only limited analyses of the human islet transcriptome during ontogeny have been reported [[14], [15], [16]].
To define the maturation process of human islet cells, we expanded on our initial single cell RNAseq study [14] by increasing the islet sample number substantially from 7 to 22 organ donors, now spanning human ontogeny from early neonate (18 days) to adults (up to 65 years, details below). By first determining the natural maturation process of α and β-cells, we were able to define ‘immaturity signatures’ which we then employed for the analysis of the transcriptomes of α- and β-cells from T2D organ donors. Importantly, our analyses show that a large fraction of T2D β-cells acquire an immature expression profile characterised by de-repression of juvenile gene sets and activation of genes typically expressed in the exocrine compartment, supporting a global de-maturation process. The relaxation of the differentiated state of β-cells, which takes several years of postnatal development to achieve, likely contributes to islet dysfunction. These findings support the notion that while postnatal maturation of β-cells in parallel to increasing metabolic demand normally results in improved β-cell function, the dramatic increase in metabolic load accompanying the diabetic state is associated with transcriptional and functional reversal to the sub-optimal glucose response and hormone output characteristic of juvenile β-cells.
2. Materials and methods
2.1. Human pancreatic islet procurement and processing
Islets were obtained from Integrated Islet Distribution Program (IIDP) and the Human Pancreas Analysis Program (HPAP) consortium (https://hpap.pmacs.upenn.edu/), which is part of the Human Islet Research Network (https://hirnetwork.org/). Prior to organ retrieval, informed consent was provided by each donor's legal representative. Donor information is provided in Table S1. Prior to cell capture, islets were cultured in Prodo Islet Media (PIMS® Standard) with 5% human albumin serum and a glucose concentration of 5.8 mM. Islets were dissociated into single cells as described previously [17].
2.2. Single-cell RNA sequencing
Two integrated fluidic chips of 5–10 μm and 10–17 μm in size (Fluidigm 1006040 and 1006041) were used for cell capture of each islet sample. The SMART-seq method was used for first-strand cDNA synthesis and polymerase chain reaction (PCR) amplification (Clontech 634833). For two control adult donors, ArrayControl™ spike-in (Thermo Fisher Scientific AM1780) was applied during cell capture. Resulting cDNAs were pooled into a 96-well plate, and the Nextera XT DNA library preparation kit (Illumina FC-131-1096) was used for RNA-seq library preparation according to the Fluidigm protocol (Fluidigm 100-7168).
Single-cell RNAseq libraries were sequenced on an Illumina HiSeq 2500 with 100-bp single-end reads. Median read-depth prior to alignment was 4.4 million reads per cell. Read alignment and gene expression quantification was performed using RUM [18] with an alignment rate of approximately 85%. Considering just the RefSeq transcripts, the useful read count had a median depth of 1.7 million ± 1.3 million reads per cell. Cells with fewer than 500,000 uniquely aligned reads were excluded from downstream analysis. A total of 82% of sequenced cells passed initial technical quality control. All sequencing data are available in the Gene Expression Omnibus (GEO) repository (accession number GSE154126). Expression was initially quantified using raw read count. Normalised values reported later were computed in a trivial fashion using total aligned reads and/or the length of the transcript.
2.3. Cell type classification
Cell type classification was conducted as previously described with some modifications [14]. For this work, we leveraged the manual curation from Wang et al. [14] to calculate an average expression profile for each cell type, with the primary goal of eliminating double or multiple capture cells. To do this we calculated a set of in silico profiles for each potential combination of cell types where the level of mixing of the two cell types ranged from 0 to 1. Next, we calculated the relative distance of the expression profile of each cell to all of the mixture profiles. The cell was assigned the type corresponding to the closest mixture.
2.4. Differential expression analysis and derivation of cell type gene lists
We followed the method of Segerstolpe et al. [19] except as noted. We applied criteria 1 and 2 from Segerstolpe et al.; i.e., for each gene, the mean FPKM was ≥2 in at least one cell type, and the mean FPKM was ≥2 for two donors within some cell type. All three comparison types used the following common steps. Using the EdgeR package and starting with read count data, we removed any genes that did not reach 1 CPM in any cell. Then, we executed the following sequence of functions estimateGLMCommonDisp, estimateGLMTrendedDisp, estimateGLMTagwiseDisp, glmFit and glmLRT. Finally, genes with a false discovery rate (FDR) better than 5% were retained.
Methods for collecting the cells and preparing the design matrices varied depending on the comparison type. For the pairwise comparisons, we selected the cells corresponding to the two cell types in the comparison and employed a single-factor two-value design. This was iterated over all pairs of cell types. For cell-type vs compartment comparisons, we also selected the cells with the given cell type or from the given compartment (but with different cell types) and performed a single-factor two-value comparison. Finally, for the any-change comparison we down-weighted the extreme values for each gene. All weights were initially set to 1.0, and then the cells with the single highest and lowest values were weighted at 1E-6. Typically, there could be ties for undetected genes, so a single cell was down-weighted to avoid removing them all. In practice, ties for the maximum value do not occur, so the down-weighting had the effect of removing the single highest expression value. The design matrix was then generated using a constant plus a single-factor multi-value formula, i.e., a constant expression plus an effect for each cell type. An FDR cut-off of 5% was used in all comparisons.
Differential expression analysis between cells from adults and other conditions were performed using the same software and parameters as the pairwise comparisons.
Other gene sets were compiled from the MSigDB collections [20].
To analyse maturation and disease-related processes, all gene sets were used in a hypergeometric test for gene set enrichment analysis using the Genomica software (http://genomica.weizmann.ac.il/), considering gene sets with a P value <0.01 and an FDR <0.05 to be significant. Additionally, pathway analyses were performed with Gene Set Enrichment Analysis (GSEA; http://software.broadinstitute.org/gsea/index.jsp) and Ingenuity Pathway Analysis (IPA; http://www.ingenuity.com/).
2.5. Pseudotime analysis
We used all genes with expression levels above 2 CPM in at least 10 cells. We applied SCORPIUS using two dimensions, not the default three, prior to computing the trajectory. We corrected for multiple testing using the Benjamini–Hochberg method and required a q-value of 5%.
2.6. Immunofluorescence analysis
Paraffin sections of pancreata from ND adults, newborns and T2D donors were obtained from the Network for Pancreatic Organ donors with Diabetes (nPOD). Antigen retrieval was performed by pressure cooker heating (Prestige Medical, Northridge, CA) using citrate buffer (pH 6.0). Sections were blocked with CAS-Block (Invitrogen). Primary antibodies used in this study included guinea pig anti-insulin (1:500; DAKO), rat anti-CD44 (1:100; Invitrogen), rabbit anti-vimentin (1:100; Abcam), rabbit anti-FAP (1:100, Abcam), mouse anti-glucagon (1:400, Cell Signalling). For DNA counterstain, we used 4′,6-diamidino-2-phenylindole (DAPI; 1:100; Sigma–Aldrich). Secondary antibodies were all from Jackson Immunoresearch Laboratories (1:250; West Grove, PA, USA). For double staining, we used only affinity-purified secondary antibodies suitable for multiple labelling. All immunofluorescence images were captured on Olympus FV1000 confocal microscope.
2.7. ChIP-seq analysis
H3K4me3 and H3K27me3 ChIP-Seq data of sorted human β-cells was obtained previously [21].
3. Results
3.1. β-Cells demonstrate age-dependent maturation at the transcriptome level
Our initial single-cell RNAseq analysis of islet cells from seven donors of various ages and disease states suggested that both α- and β-cells from juvenile and T2D donors have an immature expression program [14]. To improve our understanding of the molecular program that dictates α- and β-cell maturation, we significantly expanded our previous single-cell RNAseq analysis of human islet cells from donors of different ages. The 22 donors studied here are divided into four groups of non-diabetics by age as follows: newborn (one donor), toddler (five donors), adolescent (two donors) and non-diabetic (ND) adult (four donors), in addition to an adult group with clinically diagnosed T2D (ten donors) (Table S1). Dissociated islet cells were sequenced using the Fluidigm C1 96-well platform as described previously [14] without pre-selection for particular cell types except for the newborn cells that were pre-sorted to enrich for β-cells [17]. Briefly, cDNAs were indexed, pooled, and further processed to construct RNA-seq libraries using the Nextera XT DNA library preparation kit according to the Fluidigm protocol. Each sample was sequenced to a median depth of 1.7 million ± 1.3 million reads aligned to RefSeq transcripts. Gene expression was quantified as fragments per kilobase of transcript per million mapped reads (FPKM) with an average of 4764 distinct transcripts detected per sample at an FPKM ≥1 (Figure S1A). For cell type annotation, we employed our previously published pipeline with some refinements (see Materials and Methods) [14]. We successfully assigned a specific cell-type to 619 cells (Table S2) and excluded 662 cells from further analysis due to conflicting profiles, which could have resulted from technical issues, such as doublets or from a real hybrid cellular condition related to developmental or disease state, as we discussed in our initial study [14]. For the purpose of this study, we opted to limit our analysis to cells which could clearly and definitively be classified into a single cell type.
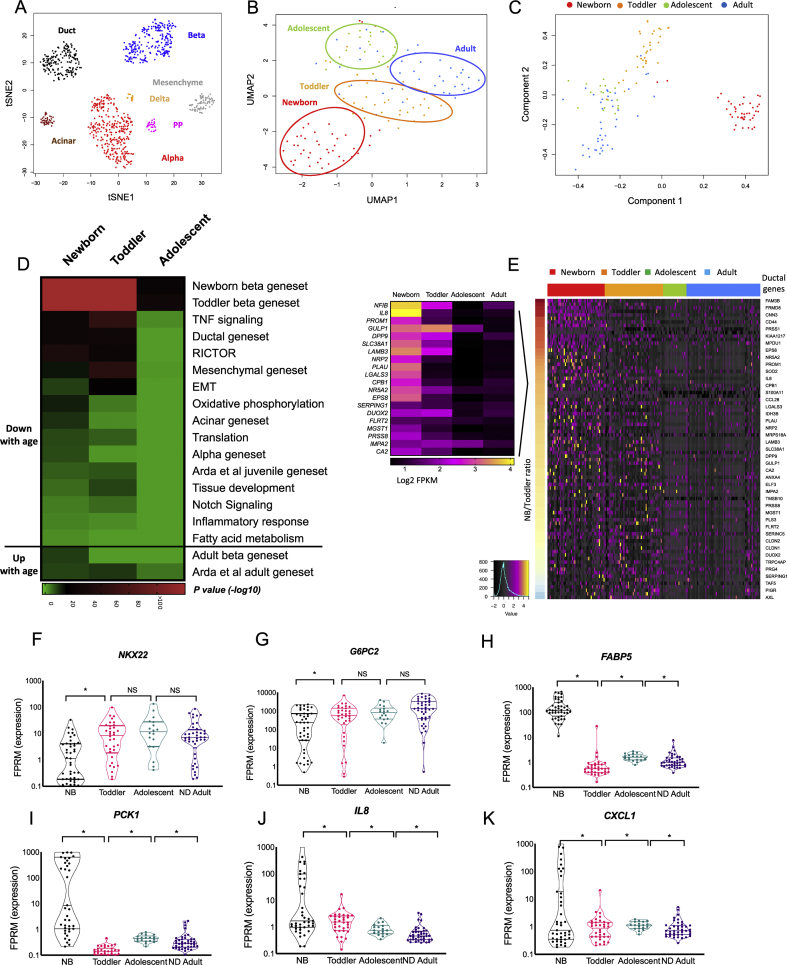
Unsupervised clustering of all annotated cells and visualisation using t-Distributed Stochastic Neighbour Embedding (tSNE) showed segregation of each cell type into distinct populations independent of donor age or health condition (Figure 1A), validating our filtering and annotation pipeline.
Figure 1.
Human β cells demonstrate age-dependent maturation. (A) Unsupervised clustering and visualisation of all annotated cells (n = 619 cells) using t-Distributed Stochastic Neighbour Embedding (tSNE) based on the expression values (log2 FPKM) of the top 20 genes with the largest range of expression values and including the major markers for each cell type. (B) Unsupervised clustering of β-cells from all non-diabetic donors and visualisation with UMAP using the top 22 principal components. (C) Pseudotime analysis using the SCORPIUS R package of all non-diabetic β-cells shows a temporal trajectory. (D) Heatmap displaying GSEA enrichment scores (p values) associated with β-cell maturation driven by the comparison between ND adults and each of the three younger age groups (newborn, toddler and adolescent), indicating biological pathways and gene sets typical of non-β-cell types which are being gradually silenced, while genes associated with mature β-cell function are being activated with age. p value <0.01 and an FDR <0.05 were considered as a significant enrichment. Green is low, black is intermediate and red is high enrichment. See Supplemental Table S5 for a complete list of genes. (E) Heatmap displaying relative expression levels of genes typical to the ductal gene set that show a gradual downregulation with age in single β-cells (SD of log2 FPKM). Genes (rows) are organised from high to low by the newborn/toddler relative expression ratio (colour bar on the left), cells (columns) are organised by age groups (newborn, toddler, adolescent and adult). Black is low, magenta is intermediate, and yellow is high expression. Only representative gene names are shown due to lack of space. For a complete list of genes, see Table S5. The relative expression levels of depicted genes by aggregation of all β-cells form each age group is displayed in the insert. (F–K) Violin plots of genes of interest with age-regulated gene expression in β-cells. Note the log10 scale. (∗) FDR <0.1 was considered significant for this analysis. NS, not significant. Lines represent median and quartiles.
Next, we identified differentially expressed genes between the different cell types (α, β, δ, pp, ductal, acinar and mesenchymal) in the four non-diabetic adult donors considering (1) pairwise cell-type comparisons, (2) cell-type to cell-type group comparisons (e.g., α to other endocrine cells) and (3) a multi-way analysis of variance (ANOVA)-like comparison (any change across all cell types) using a methodology similar to that employed by Segerstolpe et al. [19] with some modifications (Materials and Methods and Table S3). Briefly, we filtered genes for relevance by expression level and consistency across donors, then used the EdgeR algorithm [22] to identify genes that are differentially expressed in the three contexts listed above. To create stringent cell type-specific gene lists, genes were assigned based on differential expression between each given cell type and all other cell types detected from ND donors using an FDR (Benjamini–Hochberg) of ≤0.05 and a fold-change cutoff of >3 (Table S3).
Having obtained the expression profiles of pure α, β, δ, pp, ductal, acinar and mesenchymal cells present in the adult ND human pancreas, we proceeded to analyse changes to the β-cell transcriptome during human ontogeny. Unsupervised clustering of β-cells from all ages using Uniform Manifold Approximation and Projection techniques (UMAP) resulted in segregation into distinct clusters based on age, supporting a maturation process (Figure 1B). This notion was reinforced by ‘Pseudotime’ analysis (SCORPIUS R package, [23]) (Figure 1C), which revealed a trajectory corresponding to the donor ages. The pseudotime trajectory of β-cells displays five modules composed of the genes driving the age-dependent trajectory (Figure S1B and Table S4 for a complete list of genes). To exclude the possibility that the age-related clustering of β-cells (Figure 1B) was driven solely by the pre-sorting of the newborn sample rather than a real age-regulated process, and to match the analysis to that of α maturation which does not include the newborn state due to a low number of cells, we repeated the dimension reduction and pseudotime analyses omitting the newborn β-cells. This analysis verified that β-cells from young children (toddlers; ten months to four years old) still cluster separately from adolescent and adult cells, suggesting that even at four years of age, β-cells are not completely mature, being more similar to ten months old than to adult cells, and supporting the idea that the maturation process takes several years to complete (Figure S1C–E). Based on these results, we aggregated the expression data of single β-cells into four groups by age: newborn, toddler, adolescent and adult, and compared their expression profiles to create age-regulated gene sets based on an FDR of ≤0.05 and a fold change >2 (Table S4). Analysis of the age-related differentially expressed genes for enrichment of biological pathways and specific pancreatic cell signatures revealed a maturation process that proceeds from birth throughout childhood as clearly depicted in Figure 1D and Table S5, displaying GSEA enrichment scores (p values) associated with β-cell maturation driven by the comparison between non-diabetic adults and each of the three younger age groups (newborn, toddler and adolescent). Biological pathways associated with early developmental stages, such as NOTCH and tumour necrosis factor (TNF) signalling and tissue development, as well as genesets typical of non-β-cell types are gradually silenced, while genes associated with mature β-cell function are activated with age. For example, β-cells of newborn and young children express genes normally active in ductal cells such as PROM1 and NFIB, which are gradually silenced during maturation, as are S100A11 and S100A6, which are downregulated with age toward the optimal low adult level (Figure 1D and E, Figure S1F–I and Table S5). S100A11 and S100A6 belong to the EF-hand calcium binding protein gene family recently shown to be regulated by membrane depolarization and to be upregulated following increase in intracellular calcium levels in ABCC8 null mice, which exhibit a loss of β-cell identity [24]. Our findings of a gradual maturation process are further supported by the observation that our newborn and toddler β-cell gene sets are enriched for a juvenile geneset previously defined by Arda et al. by comparing bulk transcriptomes of α- and β-cells from children (0.8–6 years of age) and adults (p = 1.57E-06 and 2.93E-07 enrichment scores respectively), while our adult β-cell gene set is enriched for the adult gene set obtained from the same study (‘Arda at al adult gene set’) [16] (p = 7.2E-09, 4.68E-10 and 9.07E-05 enrichment scores comparing adult to newborn, toddler and adolescent β-cells, respectively, Figure 1D and Table S5).
Among the β-cell genes that are activated with age is the transcription factor NKX2-2 (Figure 1F), shown in mice to be necessary for maintaining the differentiated state of β-cells by directly activating key β-cell genes and repressing non-β-cell expression programs [25]. Our data support a similar role for NKX2-2 in human β-cells. In addition, G6PC2, encoding the catalytic subunit 2 of glucose-6-phosphatase, which is a negative regulator of basal glucose-stimulated insulin secretion that affects fasting blood glucose levels [26], was also significantly upregulated with age (Figure 1G), supporting a tighter control on insulin secretion in adulthood, especially at low glucose levels. Furthermore, expression of the transcription factors SIX2, SIX3 and ONECUT2 was induced in adulthood in human β-cells (Table S4), as was previously reported by Arda et al. [16]. These findings are in agreement with prior observations that β-cell function is enhanced with maturation [16,27]. Additional genes that are activated with age, but with currently unknown roles in β-cells, are neuronal pentraxin 2 (NPTX2) [28], previously suggested to act as a tumour suppressor in pancreatic cancer cells [29] (Figure S1J), and CDH2, encoding N-cadherin. The latter may play a role in islet cell connectivity and architecture and has been shown to be required for proper insulin secretion in mice [30] (Figure S1K).
Multiple genes that are downregulated during β-cell maturation are involved in metabolism of fatty acids, suggesting that newborn β-cells are in a different metabolic state compared to their adult counterparts. For example, FABP5, (fatty acid binding protein 5), which functions in fatty acid transport [31], is highly expressed in neonatal β-cells and quickly repressed during the first years of life (Figure 1D and H). A similar metabolic switch was previously reported to influence postnatal functional maturation and regeneration capacity of β-cells in mice [32]. Interestingly, FABP5 was previously shown to impact insulin secretion [33] and polymorphisms in FABP5 are associated with T2D [34]. Other fatty acid metabolism genes that are active in the newborn state encode enzymes important for β oxidation of fatty acids, such as ECH1, encoding enoyl-CoA hydratase 1 and ECI2, encoding enoyl-CoA delta isomerase 2 (see Table S5).
A key property of immature β-cells is the expression of metabolic genes that are strictly silenced in the mature β-cell state, and which have been previously termed ‘disallowed’ [[35], [36], [37]]. Among these is PCK1, encoding phosphoenolpyruvate carboxykinase 1 (PEPCK), a key enzyme in gluconeogenesis (Figure 1I). PEPCK catalyses the formation of phosphoenolpyruvate from oxaloacetate, the first step in gluconeogenesis. Given the reliance on glycolytic flux as a key component triggering glucose dependent insulin secretion, adult β-cells have to fully repress PCK1 in order to allow for precise regulation of insulin secretion [38]. At present, it is unknown why perinatal β-cells express PEPCK; however, it could contribute to the minimal glucose responsiveness of newborn β-cells recently reported by Helman et al. [39]. Other disallowed genes that are similarly silenced shortly after birth in our dataset are LDHA, encoding lactate dehydrogenase and SLC16A1, encoding the monocarboxylate carrier MCT1, a lactate and pyruvate transporter [40], the latter was detected in 25% of newborn β-cells and none of the β-cells from the other age groups (Figure S1L–M).
In addition, most of the young β cells express interleukin 8 (IL8, 74% of newborn β-cells and 64% of toddler β-cells, FPKM>1) and other inflammatory chemokines, such as CXCL1 (Figure 1J, K). Although chemokines are mainly known for their role in inflammation, they also provide directional guidance to migrating cells, such as neurons, during embryonic development [[41], [42], [43]]. The production of neuronal guidance cytokines by newborn and early childhood β-cells is intriguing, as islet innervation has been shown to increase dramatically postnatally in rodents [37]. Chemokines can also promote proliferation, cellular survival and angiogenesis [44] and could exert similar roles during postnatal islet assembly. To summarise, the maturation trajectory of human β cell transcriptomes support a process of functional maturation during ontogeny.
3.2. α-Cells retain immature characteristics into adulthood
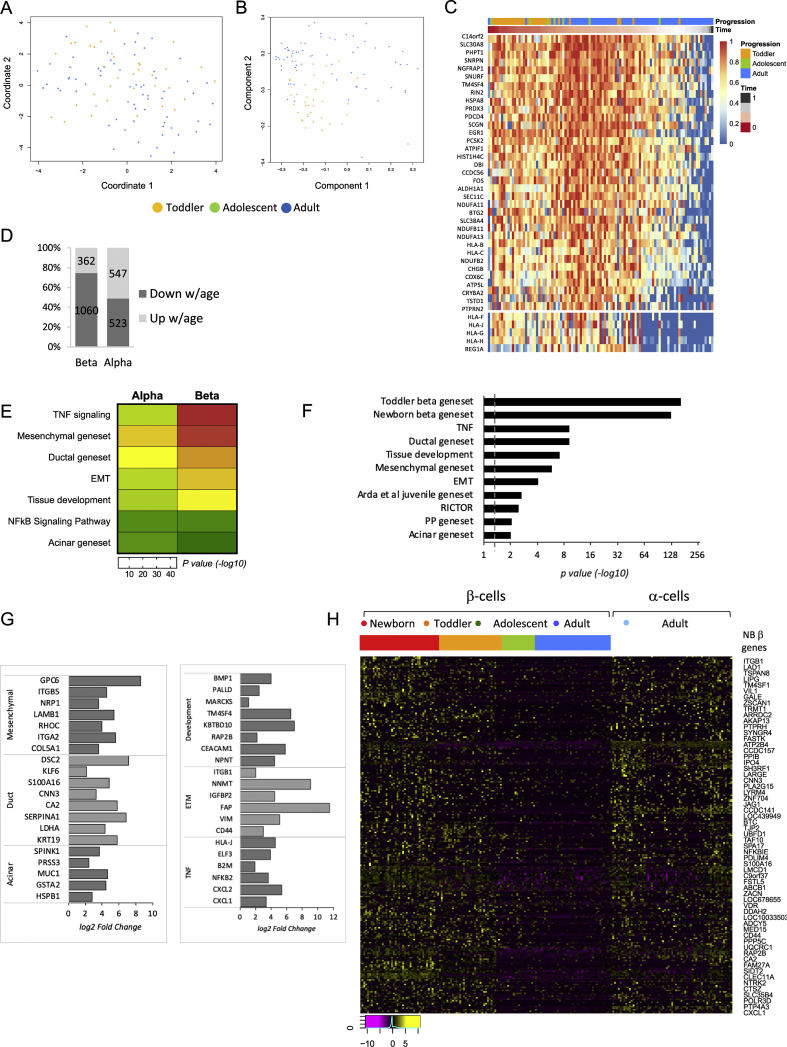
Clustering analysis of α and β-cells from our four age groups of ND donors by UMAP embedding revealed that while β-cells clearly cluster by age (Figures 1B and S1C), α-cells do not separate well into age-dependent subpopulations (Figure 2A). In concordance with this observation, the suggested pseudotime trajectory correlates only partially with the correct donor age (Figure 2B). This difference is further demonstrated by the lack of robust age-dependent silencing of the genes driving the α-cell pseudotime (Figure 2C, α-cell pseudotime modules as compared with Figure S1B and S1E for β-cell pseudotime modules) and by the lower number of differentially expressed genes between toddler and ND adult α-cells relative to β-cells of the same donors (Figure 2D and Table S6). In addition, although the α- and β-cell maturation program share many characteristics, such as silencing of the TNF signalling pathway and exocrine genes (Tables S5 and S6), the degree of change is less pronounced in α-cells as compared with β-cells (Figure 2E). For instance, 64 genes associated with the TNF signalling pathway are significantly downregulated with age in β-cells, as oppose to 14 genes in α-cells, and 51 genes typical to ductal cells are being silenced with age in β-cells compared to only 19 in α-cells. This observation implies that compared with β-cells, α-cells retain certain immature characteristics throughout life or alternatively attain their adult phenotype early in development. To differentiate between these possibilities, we compared the expression profile of α- and β-cells from ND adults. This comparison further supports the notion that adult α-cells are in a less differentiated state than adult β-cells, based on an expression program which is enriched for ductal, mesenchymal and developmental genes, as well as for the previously published juvenile gene set mentioned above [16] and referred to as ‘Arda et al. juvenile gene set’ (Figure 2F–G and Table S7). In fact, adult α-cells share many characteristics with newborn β-cells, determined by their high enrichment for the immature β-cell gene set (p = 1.97E-127, Figure 2F, H and Table S7).
Figure 2.
Human postnatal α-cell maturation follows a different path than that of β-cells. (A) Unsupervised clustering of all α-cells from toddler, adolescent and adult age groups and visualisation by UMAP embedding using 33 principal components reveals that unlike β-cells, α-cells do not separate into clear age-dependent subpopulations. (B) Pseudotime analysis of all non-diabetic α cells using the SCORPIUS R package confirms lack of clear temporal pattern. (C) Heatmap of the genes driving the trajectory seen in (B), organised into modules. (D) Number of genes up- and downregulated with age in α- and β-cells comparing toddler to ND adult cells (FDR < 10%). (E) Heatmap displaying the difference in magnitude of gene silencing with age in α versus β cells based on enrichment scores (p values) of each gene category. Green is low and red is high enrichment. p value <0.05 and an FDR <0.05 were considered as a significant enrichment. (F) Gene set enrichment analysis indicating biological pathways and gene sets typical to exocrine cell types that are enriched in α-cells relative to β-cells of ND adults. p value <0.01 and an FDR <0.05 were considered as a significant enrichment. Dashed line indicates p value = 0.05. See Supplemental Table S7 for a complete list of genes. (G) Representative genes from gene categories in (F) that are significantly upregulated in adult α-cells compared with adult β-cells. FC >1.5 and FDR <0.05 were considered as a significant change. (H) Heatmap displaying relative expression levels of genes highly expressed in newborn β-cells in single β cells (Log2 FPKM) organised by age group (newborn, toddler, adolescent and adult) and single ND adult α-cells. Magenta is low, black is intermediate and yellow is high expression.
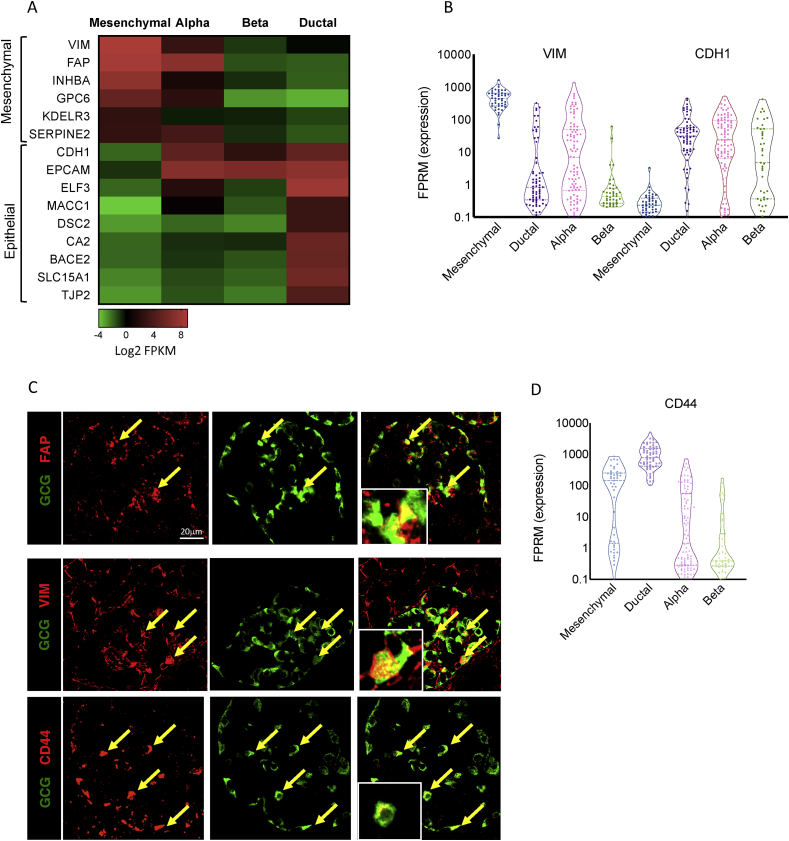
Interestingly, multiple genes active in mature α-cells are associated with the epithelial-to-mesenchymal transition (EMT) pathway (p = 7.7E-5, Figure 2F, G and Table S7), a process in which epithelial cells are transcriptionally reprogrammed to lose some of their characteristics and acquire a mesenchymal cell phenotype, which includes enhanced migratory capacity and invasiveness (for review on EMT see [45]). Specifically, adult α-cells present a mixed epithelial and mesenchymal expression program, resembling the hybrid epithelial/mesenchymal or partial EMT cellular state found during development and among breast cancer cells [46,47], which is characterised by increased stemness, regeneration and self-renewal properties [47,48]. Previously believed to be a transient phenomenon, recent reports have shown that this cellular state can be stably maintained [49]. An example of this mixed epithelial/mesenchymal-like expression program include the observation that mature α-cells express both VIM, encoding vimentin, a cytoskeletal component of mesenchymal cells that is widely used as a marker for mesenchyme-origin cells or epithelial cells undergoing EMT [50] and the epithelial marker CDH1, encoding E-cadherin, while β-cells express only CDH1 (Figure 3A–C). Vimentin has been previously suggested to indicate the plasticity level of islet cells [51]. Cheng et al. ablated β-cells by streptozotocin (STZ) and demonstrated the appearance of β-like cells, which were positive for both vimentin and MAFB, an α-cell specific transcription factor in mice [52].
Figure 3.
Adult α-cells present a mixed epithelial and mesenchymal expression prolife. (A) Heatmap displaying the expression levels (as median Log2 FPKM values) of mesenchymal and epithelial marker genes in aggregated mesenchymal, α-, β- and ductal cells. Green is low, black is intermediate and red is high expression levels. (B) Violin plot displaying the distribution of single cells expressing the mesenchymal marker VIM encoding vimentin and the epithelial marker CDH1 encoding E-cadherin in adult α- and β-single cells. Lines mark median and quartiles. (C) Immunostaining of human pancreatic sections obtained from non-diabetic donors for FAP, VIM, CD44 (red) and glucagon (green). Arrows point to α-cells exhibiting co-staining. Scale bars: 20 μm. (D) Violin plot displaying the expression of CD44 among single α- and β-cells from ND adults. Lines mark median and quintiles.
Another mesenchymal/EMT marker gene is FAP (fibroblast activation protein A, a cell surface serine protease, Figure 3A and C), previously reported to be highly expressed in human α-cells [19,53,54]. FAP plays an important role in modulating both the tumour microenvironment [55] as well as normal tissue biology during developmental and wound-healing processes through its gelatinase activity, which degrades extracellular matrix (for review see [56]). FAP belongs to the prolyl-oligopeptidase family, which includes dipeptidyl peptidase 4 (DPP4), also present on α- but not β-cells in humans [57,58], which is the enzyme that cleaves and inactivates the incretin, glucagon-like peptide 1 (GLP1). In contrast, FAP was shown to cleave and inactivate human FGF21, a liver-secreted hormone that reduces hepatic glucose output and stimulates glucose uptake [58]. Therefore, FAP in α-cells may be involved in a novel metabolic role yet to be determined. These findings provide an explanation for the increased α-cell plasticity seen in multiple mouse models [[59], [60], [61]] and are consistent with the high frequency of bivalently marked gene promoters in human α-cells [21]. The relatively immature state of adult α-cells suggested by our transcriptome profiling is also supported by the fact that human α-cells proliferate at approximately twice the rate of β-cells throughout life [62,63]. Finally, CD44, a transmembrane glycoprotein and receptor for hyaluronic acid, is likewise preferentially expressed in α-cells (8FC, Figure 3C and D). CD44 is involved in cell–cell interactions, cell adhesion, migration and tumour metastasis and serves as a marker for hematopoietic stem cells (for review see [64]). Altogether, these findings support the notion that α-cells possess a limited transcriptional resemblance with mesenchymal cells and retain a significant degree of cellular plasticity even in adulthood.
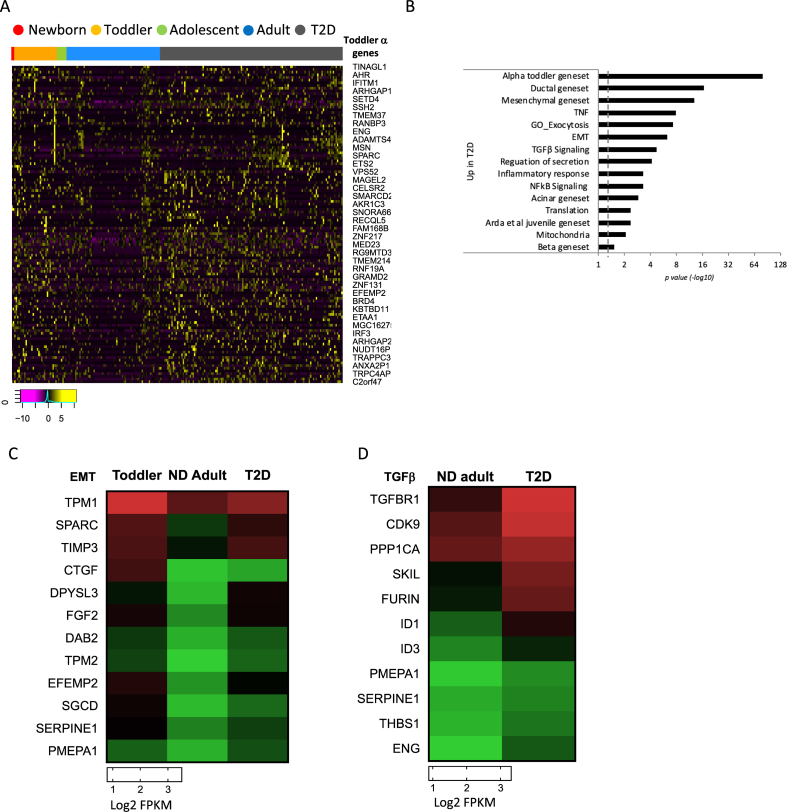
3.3. α- and β-cells from T2D relax their maturation state through de-repression of immature and non-endocrine expression programs
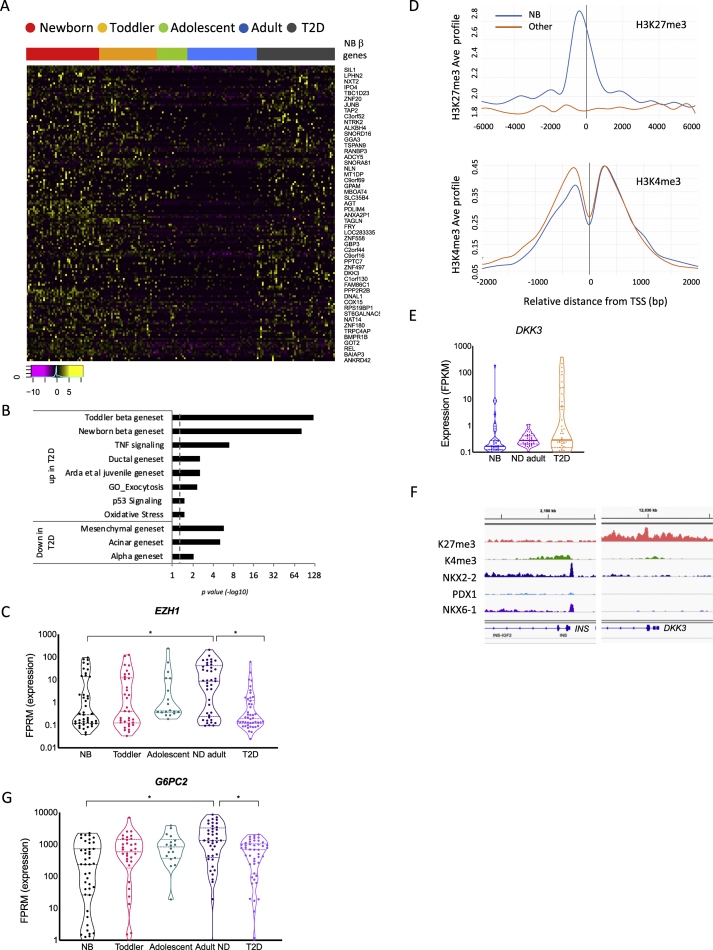
In our previous single cell-transcriptome analysis of human islet cells from normal and diabetic donors [14], we showed that the expression profiles of both α- and β-cells from donors with T2D are compromised and exhibit juvenile gene activation, supporting a partial de-maturation process, in line with mouse studies reporting partial de-differentiation of β-cells in diabetes [11,65]. To gain a better characterisation of the altered state of α- and β-cells from diabetic donors, we expanded our cohort and investigated the enrichment of our cell type-specific gene sets (ductal, acinar, mesenchymal, PP, delta, α and β) in addition to the immature signature obtained from juvenile α- and β-cells. According to our analysis, most β- and α-cells of diabetic donors undergo at least some degree of dematuration, evidenced by the upregulation of the newborn and toddler juvenile gene sets defined above as well as the previously published juvenile gene set ‘Arda et al. juvenile gene set’ [16] across T2D cells from all donors (Figure 4A–B and Table S8 for T2D β-cells and Figure 5A–B and Table S9 for T2D α-cells). Among the many juvenile genes that are downregulated with age and upregulated again in T2D β-cells are CAPS, encoding calcyphosin, a calmodulin superfamily calcium-binding protein which plays a role in cellular signalling events (4.3 and 7.6 fold higher on average in newborn and T2D than ND adult, respectively) [66], PRDX2, encoding the antioxidant protein peroxiredoxin II, shown to have a protective effect on oxidative stress-induced apoptosis in β-cells (6.7 and 2.2 fold higher on average in newborn and T2D than ND adult, respectively) [67] and the transcription factor JunB, which together with ATF3 activates a survival pathway as a mechanism against apoptosis in β-cells in response to cytokines [68] (4.2 and 5.5 fold higher on average in newborn and T2D than ND adult, respectively; Figure S2A). Additionally, NHP2 and RPS19BP1, both involved in ribosome biosynthesis, are upregulated in a large proportion of T2D β-cells as is the Alzheimer's disease risk gene PLD3, a lysosomal enzyme associated with amyloid formation [69], which could be relevant to the characteristic amyloid formation in diabetic islets [70]. Finally, SRA1, a steroid receptor transcription coactivator and long non-coding RNA, and FAT1, a member of the cadherin superfamily involved in adhesion and signalling [71] are re-activated to newborn state expression levels (5.8 and 4.9 fold higher in newborn and T2D than ND adult, respectively) (Figure S2A). The function of most of these age-regulated genes in β-cells and the contribution of their dysregulation to dysfunction in the diabetic state has yet to be determined.
Figure 4.
β-cells from T2D relax their mature transcriptional state through de-repression of immature and non-endocrine expression programs. (A) Heatmap of genes that are highly expressed in newborn β-cells, gradually being silenced toward adulthood, but are re-activated in T2D β-cells. Shown are relative expression levels (log2 FPKM) of single β-cells by age group and disease. Magenta is low, black is intermediate and yellow is high expression. Only representative gene names are shown due to lack of space. For a complete list of genes, see Table S8. (B) Gene set enrichment analysis demonstrating de-repression of juvenile and exocrine expression programs in β-cells of diabetic donors. p value <0.05 and an FDR <0.05 were considered as a significant enrichment. Dashed line indicates p value = 0.05. See Supplemental Table S8 for list of genes. (C) Violin plot demonstrating the downregulation of EZH1 in β cells of diabetics. (∗) FDR <0.1 and FC >1.5 was considered significant for this analysis. NS, not significant. Lines mark median and quartiles (D) Aggregation plots displaying the average ChIP signal of the active H3K4me3 and the repressive H3K27me3 histone modifications [21] at promoter regions of newborn genes that were found to be upregulated in T2D β-cells as compared with promoter regions of the remaining upregulated genes in T2D β-cells. (E) Violin plot demonstrating the activation of DKK3 in β-cells of diabetics. (F) Enrichment for the repressive histone mark H3K27me3 at the DDK3 promoter region in adult β-cells. (G) Violin plot demonstrating the downregulation of G6PC2 in β cells of diabetics. (∗) FDR <0.1 was considered significant for this analysis. NS, not significant.
Figure 5.
α-Cells from T2D de-repress their immature expression program and increase their mesenchymal characteristics. (A) Heatmap of genes that are high in toddler α-cells are gradually silenced toward adulthood but are re-expressed in T2D α-cells. Shown are relative expression levels of single α-cells by age group and disease (log2 FPKM). Magenta is low, black is intermediate and yellow is high expression. Only representative gene names are shown due to lack of space. For a complete list of genes, see Table S9. (B) Gene set enrichment analysis demonstrating de-repression of juvenile and exocrine and mesenchymal expression programs in α-cells of diabetic donors. p value <0.05 and an FDR <0.05 were considered as a significant enrichment. Dashed line indicates p value = 0.05. See Supplemental Table S9 for list of genes. (C) Heatmap displaying the expression levels in α-cells (as median log2 FPKM values of aggregated single α-cells by age and disease state) of genes related to the EMT pathway that are downregulated with age and de-repressed in diabetics. (D) Heatmap displaying the activation of TGFβ signalling pathway in diabetic α-cells (median log2 FPKM values). In both maps, green is low, black is intermediate and red is high expression.
One possible mechanism that could account for the relaxation of β-cell identity in diabetic islets is reduced gene silencing by the Polycomb-Repressive Complex PRC2. Indeed, EZH1, the polycomb group protein ‘enhancer of zeste homolog 1’, which functions as histone methyltransferase in PRC2 is downregulated in T2D β-cells (Figure 4C). Lu et al. have shown that the PRC2 complex helps to maintain β-cell identity by reinforcing gene silencing, and that elimination of PRC2 activity causes dedifferentiation and diabetes through loss-of-silencing at poised, or bivalent polycomb regulatory regions [72]. Therefore, we examined the chromatin state at promoters of genes upregulated in T2D β-cells using ChIP-seq maps of the repressive H3K27me3 and active H3K4me3 histone modifications that we previously obtained from sorted non-diabetic adult human β-cells [21]. Many of the newborn signature genes re-activated in diabetic β-cells are marked as bivalent, with both the repressive H3K27me3 and active H3K4me3 histone modifications enriched at their promoters in adult β-cells, while the remaining genes upregulated in T2D are marked by the active mark alone (Figure 4D). For example, DDK3, encoding Dickkopf-related protein 3, a developmental protein that is also important in maintaining the dedifferentiated state in the pancreatic cancer cell line PAM [73,74], is transcriptionally silenced, and carries bivalent histone marks in non-diabetic adult β-cells; however, this gene is de-repressed in β-cells from several diabetic donors (Figure 4E–F).
β-cells from T2D patients also upregulate genes typical to the pancreatic ductal compartment such as SERPINA1, encoding a serine protease inhibitor, and CTSD, encoding the lysosomal protease cathapsin D (Figure S2B) (both 5-fold upregulated). ST14 (matriptase), a type II transmembrane serine protease associated with increased invasiveness in pancreatic ductal adenocarcinoma (10-fold upregulated) [75] is another example of a typically ductal gene that is upregulated in T2D β-cells. Interestingly, we detected hyperexpression of the HLA class I antigens HLA-A and HLA-B as well as that of B2M (β2-microglobulin), the second component of MHC class I complex, among diabetic β-cells (Figure S2B), possibly as a response to pro-inflammatory conditions, supported by the upregulation of TNF-α signalling in T2D islet cells (Figure 4B) [76,77].
In addition to the activation of genes normally expressed in newborn and non-β-cells, our dataset shows increased expression of exocytosis genes important for insulin secretion in T2D β cells. These genes include TSPAN1, recently identified as a positive regulator of β-cell exocytosis [78] and RGS16, encoding a regulator of G-protein signalling, shown to stimulate insulin secretion and to be upregulated under hyperglycaemic conditions (both increased by 8 fold on average, Figure S2C) [79]. Interestingly, G6PC2, which limits insulin secretion at low glucose levels and is activated with age about 3-fold on average (Figure 1G), is downregulated in T2D back to the levels typical of young children (2-fold, Figure 4G), which could impact glucose-stimulated insulin secretion. The fact that other cell type-specific genes, such as the acinar REG1A and REG1B are dysregulated as well, albeit in the opposite direction, suggests an overall relaxing of the mature expression program of β-cells in T2D (Figure S2D). Notably, we did not detect activation of progenitor markers, such as NEUROG3, POU5F1 and NANOG in diabetic β-cells, in line with the findings of Guo et al. [80] and contrary to what was previously reported in diabetic mouse models [11]. Importantly, the dysregulation of many of the genes mentioned above, such as FAT1, HLA-A, HLA-B and NME7, as well as the exocytosis-related genes TSPAN1, ROBO2, DLK1 and SORL1 showed similar trends in other scRNAseq studies [19]. Differential expression of other genes identified here, which were not reported as such previously, is likely due to differences in sequencing depth, technologies used, and donor variation.
Similar to diabetic β-cells, α-cells from diabetic donors show partial loss of maturation characteristics based on their enrichment for juvenile and exocrine genes (p = 3.6E-80, 2.19E-17, 1.56E-13 and 1.2E-03, for toddler, ductal, mesenchymal and acinar gene sets respectively, Figure 5A, B and Table S9). As such, α-cells of both toddlers and T2D donors express on average 2-fold higher mRNA levels of PRSS1 (serine protease 1) compared to ND adult donors and over 50-fold more MGST1 (microsomal glutathione S-transferase 1), 5-fold more TSBH1, encoding Thrombospondin 1, and 4-fold more CTGF, encoding connective tissue growth factor, all typical exocrine genes (Figure S3). In addition, the expression profile of diabetic α-cells is enriched for the EMT gene set when compared with ND adult α-cells (p = 5.65E-07, Figure 5B; Table S9). The heatmap in Figure 5C demonstrates the relative expression level of genes associated with the EMT pathway, which are downregulated with age and re-activated in T2D α-cells. Many of these genes influence the capability of cells to migrate or to respond to mitogenic stimuli, are involved in extracellular matrix homeostasis [81,82] and can either promote or decrease EMT/mesenchymal characteristics depending on the cell type and disease state (cancerous as opposed to normal cells). For instance, TIMP3 and EFEMP2 suppress EMT in cancer cells and therefore tumour invasiveness through inhibition of metalloproteinase (MMP) genes (Figure 5C) [83,84], while elevation of TPM1/2 (encoding tropomyosins) in lens epithelial cells is associated with induction of EMT following injury through increased TGFβ (transforming growth factor) signalling [85], one of the main pathways to induce EMT [86]. Interestingly, in diabetic α-cells both EMT and TGFβ signalling marker genes are upregulated (Figure 5D). Overall, it is difficult to predict what would be the outcome of the dysregulation of EMT genes on α-cell function; however, one possibility is that these transcriptome changes could modulate the differentiated state of the cells. Thus, Efrat et al. showed that human β-cells expanded in vitro undergo dedifferentiation and require a block of TGF1β signalling for their re-differentiation [87]. Thus, increased TGF1β signalling could contribute to the dematuration of α-cells in T2D.
4. Discussion
In this study, we determined the postnatal maturation of human β- and α-cells at the transcriptome level and demonstrated how this process is perturbed in T2D. In our prior proof-of-principle study, we compared a limited number of single-cell transcriptomes across non-diabetic adults, T2D, and juvenile human islet samples and showed that β-cell gene signatures of adult T2D samples were less well defined compared to non-diabetic adults [14]. However, this dataset was inadequate to determine maturation-associated gene signatures or to identify specific pathways that contribute to islet dysfunction in T2D. We greatly expanded our data both by adding multiple datasets from additional individuals with and without diabetes and, importantly, by including a rare newborn organ donor. The main pancreatic cell types (α, β, δ, PP acinar and ductal cells) arise from common progenitor cells during embryonic development (for review see [88,89]). Thus, a comparative analysis of the gene expression program of adult pancreatic cells is useful to define their differentiated state in a developmental context and to investigate cellular processes, such as maturation in health and loss of identity in disease. Using this strategy, we were able to show that maturation of α and β-cells is associated with silencing of expression programs that characterize the other pancreatic cell types, expression of ‘disallowed’ genes, and activation of function genes.
Obtaining the extremely rare islet cells from a newborn donor was particularly revealing. β-cell function changes dramatically during the first years of life. In the foetus, the primary function of the β-cell is to produce insulin to stimulate foetal growth. Foetal β-cell do not play a role in regulating blood glucose levels, as this is controlled entirely by the mother. Post-natal β-cell must acquire regulatory mechanisms to control blood glucose levels and adjust their function after weaning to adapt to a high carbohydrates diet. Indeed, we show that the transition from infant to toddler is associated with change in the expression of at least three genes that control insulin secretion at low glucose levels. G6PC2 is activated, preventing any residual glycolytic flux at low glucose levels, thus contributing to the very tight control of insulin secretion in mature β-cells. Second, PCK1, a key regulator of gluconeogenesis, is quickly down-regulated in postnatal β-cells, preventing the generation of pyruvate that could trigger insulin secretion in the absence of glucose. Finally, FABP5, which facilitates intake and metabolism of fatty acids, is rapidly downregulated postnatally as well. Thus, while a basal level of insulin secretion in response to fatty acids might be beneficial in newborns where insulin serves to promote growth, fatty acid catabolism in the mature β-cell is disallowed to make insulin secretion solely dependent on glucose. Collectively, these changes in gene expression programs throughout postnatal development, contribute to the reduction of insulin secretion at low glucose levels or in response to pyruvate and fatty acids, thus preventing fasting hypoglycemia. Given that newborn organ donors are extremely rare, it is possible that some of the aging-related changes reported here will be modified in the future should more such donors become available for analysis. Our cell type-specific and age-related ‘immature’ gene signatures has revealed that numerous changes in the transcriptome of diabetic α and β-cells are associated with dematuration. Our analyses have shown that a substantial fraction of T2D β-cells revert to an immature expression profile characterised by de-repression of the newborn gene set. Interestingly, the promoters of these re-activated genes display a bivalent chromatin state in normal adult β-cells, which is defined by simultaneous enrichment for both the repressive and active histone modifications H3K27me3 and H3K4me3, respectively, and can therefore be rapidly reactivated by removal of the repressive H3K27me3 mark (Figure 4G), as was shown for the rapid induction of bivalent developmental genes in embryonic stem cells [90]. Stress-induced activation of bivalent, non-expressed gene is observed in Ewing's sarcoma [91], suggesting a model in which metabolic stress induces activation of bivalent genes in islet cells. Thus, while normal maturation is accompanied by repression of potentially harmful genes, this repression is tenuous, and these genes remain poised to re-initiate expression when placed under metabolic stress.
5. Conclusions
In conclusion, by significantly expanding our single-cell RNAseq dataset, and most importantly by adding data from a unique and rare neonatal pancreas donor, we have defined the postnatal maturation process of human pancreatic islet cells. We have generated discrete gene sets that are characteristic of immature α- and β-cells. Using these signatures, we show that while both α and β-cells mature with age, adult α-cells expression program suggest that these cells achieve a less differentiated state that might allow for cellular plasticity and self-renewal, consistent with the increased regeneration and transdifferentiation processes previously reported in numerous diabetes mouse models (reviewed in [92]). Importantly, we demonstrated that in T2D, both α- and β-cells undergo a process of dematuration. Further, we have identified several dysregulated genes that likely contribute to the α- and β-cell phenotype observed in T2D, and thus could qualify as potential targets for the development of novel therapeutics.
Contributions
DA, KHK and BG designed the experiments and wrote the manuscript. YJW, DA, and EF performed experiments. JS, DA and YJW developed the computational pipeline. JS and DA performed computational analyses. CL and AN provided rare human islet samples. KHK and BG directed the whole study.
Acknowledgements
We thank Olga Smirnova for excellent technical support and Ran Avrahami for his critical help with data analysis. We thank Yuval Dor and Agnes Klochendler for critical reading of the manuscript. This research was performed using resources and/or funding provided by the NIDDK-supported Human Islet Research Network (HIRN, RRID:SCR_014393; https://hirnetwork.org; UC4-DK104119 to KHK and BG). BG was supported by The BIRAX Regenerative Medicine Initiative (14BX14NHBG); Israel Science Foundation (ISF)—Juvenile Diabetes Research Foundation Joint Program in Type 1 Diabetes Research 1506/12; ISF grant 1782/18; EFSD award supported by EFSD/JDRF/Lilly Program on Type 1 Diabetes Research and the EU's Horizon 2020 research and innovation programme No 874710. This manuscript used data acquired from the Human Pancreas Analysis Program (HPAP-RRID:SCR_016202) Database (https://hpap.pmacs.upenn.edu), a Human Islet Research Network (RRID:SCR_014393) consortium (UC4-DK-112217, U01-DK-123594, UC4-DK-112232, and U01-DK-123716). We thank the University of Pennsylvania Diabetes Research Centre (DRC) for the use of the Functional Genomics, (P30-DK19525). We owe special gratitude to the pancreatic islet donors and their families. Without their altruistic contribution in times of great personal tragedy, human islet research would not be possible.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2020.101057.
Contributor Information
Benjamin Glaser, Email: ben.glaser@mail.huji.ac.il.
Klaus H. Kaestner, Email: kaestner@pennmedicine.upenn.edu.
Conflicts of interest
The authors declare that no conflicts of interest exist pertaining to the contents of this manuscript.
Appendix A. Supplementary data
The following are the supplementary data to this article:
Donor information.
Cell-type identification. This table contains the annotation of cells per condition.
This table contains the derivation of gene lists characteristic for each cell type in non-diabetic adult donors using multi-factor analysis of variance (ANOVA) and differential expression analysis. Genes appear in the file corresponding to the cell type in which they have the highest expression (ANOVA results sheets 3–9) and cell type specific gene sets composed of genes enriched for each cell type from non-diabetic (ND) adult donors derived from DE analysis of each cell type to all other cell types (sheet 10).
This table contains the lists of genes up- and downregulated in β- and α-cells with aging and the lists of genes driving the pseudotime modules displayed in Figure S1B and E.
Related to Figure 1D. Lists of gene sets that were found up- and down-regulated during β-cell maturation.
Related to Figure 2D. Lists of gene sets that were found up- and down-regulated during α-cell maturation and the lists of genes driving the pseudotime modules displayed in Figure 2C.
Related to Figure 2F. Lists of gene sets that were found enriched in α-cells as compared with β-cells from non-diabetic adults.
Related to Figure 4B. Lists of gene sets that were found up- and downregulated in T2D as compared with adult non-diabetic β-cells.
Related to Figure 5B. Lists of gene sets that were found up- and downregulated in T2D as compared with adult non-diabetic α-cells.
Figure S1. Related to Figure 1. Characteristics of β-cell maturation. (A) Boxplots showing the number of genes detected at FPKM≥1 threshold per cell by donor and condition. (B) Modules driving the trajectory plotted in Figure 1C (for a complete gene list, see Table S4). (C) Unsupervised clustering of β-cells from all non-diabetic group ages except for newborn donor visualised by UMAP using the top 13 principal components. (D) Pseudotime analysis (SCORPIUS R package) of all non-diabetic group ages except for the newborn donor β-cells shows a temporal trajectory. (E) Modules driving the trajectory seen in Figure S1D (gene list in Table S4). (F–K) Violin plots of age-regulated genes in β-cells. Note the log10 scale. (∗) FDR <0.1 was considered significant for this analysis. NS, not significant. Lines mark median and quartiles.
Figure S2. Related to Figure 4. Age-related dysregulation of genes supports non-endocrine and immature expression profile of T2D β-cells. Violin plots demonstrating upregulation of β-cell juvenile genes (genes active in newborn and toddlers) in diabetic donors (A), upregulation of ductal genes in T2D β-cells (B), upregulation of genes involved in exocytosis in β-cells of diabetic donors (C) and typical acinar genes that are dysregulated in T2D β-cells (D). All genes shown are significantly dysregulated in T2D compared with ND adult β-cells (FDR<0.1). Lines mark median and quartiles.
Figure S3. Related to Figure 5. Elevated expression of exocrine gene in T2D α-cells support partial loss of maturation characteristics. Violin plots demonstrating upregulation of acinar genes in α-cells from T2D donors. All genes shown are significantly upregulated in T2D compared with ND adult α-cells (FDR < 0.1). Lines mark median and quartiles.
References
- 1.Butler A.E., Janson J., Bonner-Weir S., Ritzel R., Rizza R.A., Butler P.C. Beta-cell deficit and increased beta-cell apoptosis in humans with type 2 diabetes. Diabetes. 2003;52(1):102–110. doi: 10.2337/diabetes.52.1.102. [DOI] [PubMed] [Google Scholar]
- 2.Donath M.Y., Ehses J.A., Maedler K., Schumann D.M., Ellingsgaard H., Eppler E. Mechanisms of beta-cell death in type 2 diabetes. Diabetes. 2005;54(Suppl 2):S108–S113. doi: 10.2337/diabetes.54.suppl_2.s108. [DOI] [PubMed] [Google Scholar]
- 3.Back S.H., Kaufman R.J. Endoplasmic reticulum stress and type 2 diabetes. Annual Review of Biochemistry. 2012;81:767–793. doi: 10.1146/annurev-biochem-072909-095555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fonseca S.G., Gromada J., Urano F. Endoplasmic reticulum stress and pancreatic beta-cell death. Trends in Endocrinology and Metabolism. 2011;22(7):266–274. doi: 10.1016/j.tem.2011.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaneto H., Kajimoto Y., Fujitani Y., Matsuoka T., Sakamoto K., Matsuhisa M. Oxidative stress induces p21 expression in pancreatic islet cells: possible implication in beta-cell dysfunction. Diabetologia. 1999;42(9):1093–1097. doi: 10.1007/s001250051276. [DOI] [PubMed] [Google Scholar]
- 6.Robertson R., Zhou H., Zhang T., Harmon J.S. Chronic oxidative stress as a mechanism for glucose toxicity of the beta cell in type 2 diabetes. Cell Biochemistry and Biophysics. 2007;48(2–3):139–146. doi: 10.1007/s12013-007-0026-5. [DOI] [PubMed] [Google Scholar]
- 7.Robertson R.P., Harmon J., Tran P.O., Tanaka Y., Takahashi H. Glucose toxicity in beta-cells: type 2 diabetes, good radicals gone bad, and the glutathione connection. Diabetes. 2003;52(3):581–587. doi: 10.2337/diabetes.52.3.581. [DOI] [PubMed] [Google Scholar]
- 8.Tornovsky-Babeay S., Dadon D., Ziv O., Tzipilevich E., Kadosh T., Schyr-Ben Haroush R. Type 2 diabetes and congenital hyperinsulinism cause DNA double-strand breaks and p53 activity in beta cells. Cell Metabolism. 2014;19(1):109–121. doi: 10.1016/j.cmet.2013.11.007. [DOI] [PubMed] [Google Scholar]
- 9.Hunter C.S., Stein R.W. Evidence for loss in identity, de-differentiation, and trans-differentiation of islet beta-cells in type 2 diabetes. Frontiers in Genetics. 2017;8:35. doi: 10.3389/fgene.2017.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salinno C., Cota P., Bastidas-Ponce A., Tarquis-Medina M., Lickert H., Bakhti M. β-cell maturation and identity in health and disease. International Journal of Molecular Sciences. 2019;20(21) doi: 10.3390/ijms20215417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Talchai C., Xuan S., Lin H.V., Sussel L., Accili D. Pancreatic beta cell dedifferentiation as a mechanism of diabetic beta cell failure. Cell. 2012;150(6):1223–1234. doi: 10.1016/j.cell.2012.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cinti F., Bouchi R., Kim-Muller J.Y., Ohmura Y., Sandoval P.R., Masini M. Evidence of beta-cell dedifferentiation in human type 2 diabetes. Journal of Clinical Endocrinology and Metabolism. 2016;101(3):1044–1054. doi: 10.1210/jc.2015-2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diedisheim M., Oshima M., Albagli O., Huldt C.W., Ahlstedt I., Clausen M. Modeling human pancreatic beta cell dedifferentiation. Molecular Metabolism. 2018;10:74–86. doi: 10.1016/j.molmet.2018.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Y.J., Schug J., Won K.J., Liu C., Naji A., Avrahami D. Single-cell transcriptomics of the human endocrine pancreas. Diabetes. 2016;65(10):3028–3038. doi: 10.2337/db16-0405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Enge M., Arda H.E., Mignardi M., Beausang J., Bottino R., Kim S.K. Single-cell analysis of human pancreas reveals transcriptional signatures of aging and somatic mutation patterns. Cell. 2017;171(2):321–330. doi: 10.1016/j.cell.2017.09.004. e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arda H.E., Li L., Tsai J., Torre E.A., Rosli Y., Peiris H. Age-dependent pancreatic gene regulation reveals mechanisms governing human beta cell function. Cell Metabolism. 2016;23(5):909–920. doi: 10.1016/j.cmet.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dorrell C., Abraham S.L., Lanxon-Cookson K.M., Canaday P.S., Streeter P.R., Grompe M. Isolation of major pancreatic cell types and long-term culture-initiating cells using novel human surface markers. Stem Cell Research. 2008;1(3):183–194. doi: 10.1016/j.scr.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 18.Grant G.R., Farkas M.H., Pizarro A.D., Lahens N.F., Schug J., Brunk B.P. Comparative analysis of RNA-Seq alignment algorithms and the RNA-Seq unified mapper (RUM) Bioinformatics. 2011;27(18):2518–2528. doi: 10.1093/bioinformatics/btr427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Segerstolpe A., Palasantza A., Eliasson P., Andersson E.M., Andreasson A.C., Sun X. Single-cell transcriptome profiling of human pancreatic islets in health and type 2 diabetes. Cell Metabolism. 2016;24(4):593–607. doi: 10.1016/j.cmet.2016.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bramswig N.C., Everett L.J., Schug J., Dorrell C., Liu C., Luo Y. Epigenomic plasticity enables human pancreatic alpha to beta cell reprogramming. Journal of Clinical Investigation. 2013;123(3):1275–1284. doi: 10.1172/JCI66514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cannoodt R., Saelens W., Saeys Y. Computational methods for trajectory inference from single-cell transcriptomics. European Journal of Immunology. 2016;46(11):2496–2506. doi: 10.1002/eji.201646347. [DOI] [PubMed] [Google Scholar]
- 24.Stancill J.S., Cartailler J.P., Clayton H.W., O'Connor J.T., Dickerson M.T., Dadi P.K. Chronic β-cell depolarization impairs β-cell identity by disrupting a network of Ca2+-regulated genes. Diabetes. 2017 doi: 10.2337/db16-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gutierrez G.D., Bender A.S., Cirulli V., Mastracci T.L., Kelly S.M., Tsirigos A. Pancreatic beta cell identity requires continual repression of non-beta cell programs. Journal of Clinical Investigation. 2017;127(1):244–259. doi: 10.1172/JCI88017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pound L.D., Oeser J.K., O'Brien T.P., Wang Y., Faulman C.J., Dadi P.K. G6PC2: a negative regulator of basal glucose-stimulated insulin secretion. Diabetes. 2013 doi: 10.2337/db12-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Avrahami D., Li C., Zhang J., Schug J., Avrahami R., Rao S. Aging-dependent demethylation of regulatory elements correlates with chromatin state and improved beta cell function. Cell Metabolism. 2015;22(4):619–632. doi: 10.1016/j.cmet.2015.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nica A.C., Ongen H., Irminger J.C., Bosco D., Berney T., Antonarakis S.E. Cell-type, allelic, and genetic signatures in the human pancreatic beta cell transcriptome. Genome Research. 2013;23(9):1554–1562. doi: 10.1101/gr.150706.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang L., Gao J., Li L., Li Z., Du Y., Gong Y. The neuronal pentraxin II gene (NPTX2) inhibit proliferation and invasion of pancreatic cancer cells in vitro. Molecular Biology Reports. 2011;38(8):4903–4911. doi: 10.1007/s11033-010-0632-y. [DOI] [PubMed] [Google Scholar]
- 30.Cirulli V. Cadherins in islet beta-cells: more than meets the eye. Diabetes. 2015;64(3):709–711. doi: 10.2337/db14-1662. [DOI] [PubMed] [Google Scholar]
- 31.Chmurzynska A. The multigene family of fatty acid-binding proteins (FABPs): function, structure and polymorphism. Journal of Applied Genetics. 2006;47(1):39–48. doi: 10.1007/BF03194597. [DOI] [PubMed] [Google Scholar]
- 32.Stolovich-Rain M., Enk J., Vikesa J., Nielsen F.C., Saada A., Glaser B. Weaning triggers a maturation step of pancreatic β cells. Developmental Cell. 2015;32(5):535–545. doi: 10.1016/j.devcel.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 33.Hormozdiari F., van de Bunt M., Segre A.V., Li X., Joo J.W.J., Bilow M. Identification of novel genes for glucose metabolism based upon expression pattern in human islets and effect on insulin secretion and glycemia. Diabetes. 2010;24(3):423–434. doi: 10.1172/JCI69519. [DOI] [PubMed] [Google Scholar]
- 34.Bu L., Salto L.M., De Leon K.J., De Leon M. Polymorphisms in fatty acid binding protein 5 show association with type 2 diabetes. Diabetes Research and Clinical Practice. 2011;92(1):82–91. doi: 10.1016/j.diabres.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rutter G.A., Pullen T.J., Hodson D.J., Martinez-Sanchez A. Pancreatic β-cell identity, glucose sensing and the control of insulin secretion. The Biochemical Journal. 2015 doi: 10.1042/BJ20141384. [DOI] [PubMed] [Google Scholar]
- 36.Quintens R., Hendrickx N., Lemaire K., Schuit F. Why expression of some genes is disallowed in beta-cells. Biochemical Society Transactions. 2008;36(Pt 3):300–305. doi: 10.1042/BST0360300. [DOI] [PubMed] [Google Scholar]
- 37.Lemaire K., Thorrez L., Schuit F. Disallowed and allowed gene expression: two faces of mature islet beta cells. Annual Review of Nutrition. 2016;36(1):45–71. doi: 10.1146/annurev-nutr-071715-050808. [DOI] [PubMed] [Google Scholar]
- 38.MacDonald M.J., McKenzie D.I., Walker T.M., Kaysen J.H. Lack of glyconeogenesis in pancreatic islets: expression of gluconeogenic enzyme genes in islets. Hormone and Metabolic Research. 1992;24(4):158–160. doi: 10.1055/s-2007-1003284. [DOI] [PubMed] [Google Scholar]
- 39.Helman A., Cangelosi A.L., Davis J.C., Pham Q., Rothman A., Faust A.L. A nutrient-sensing transition at birth triggers glucose-responsive insulin secretion. Cell Metabolism. 2020 doi: 10.1016/j.cmet.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pullen T.J., Rutter G.A. When less is more: the forbidden fruits of gene repression in the adult β-cell. Diabetes, Obesity and Metabolism. 2013 doi: 10.1111/dom.12029. [DOI] [PubMed] [Google Scholar]
- 41.Al-Alwan L.A., Chang Y., Mogas A., Halayko A.J., Baglole C.J., Martin J.G. Differential roles of CXCL2 and CXCL3 and their receptors in regulating normal and asthmatic airway smooth muscle cell migration. The Journal of Immunology. 2013;191(5):2731–2741. doi: 10.4049/jimmunol.1203421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lewellis S.W., Knaut H. Attractive guidance: how the chemokine SDF1/CXCL12 guides different cells to different locations. Seminars in Cell & Developmental Biology. 2012;23(3):333–340. doi: 10.1016/j.semcdb.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miyake M., Goodison S., Urquidi V., Gomes Giacoia E., Rosser C.J. Expression of CXCL1 in human endothelial cells induces angiogenesis through the CXCR2 receptor and the ERK1/2 and EGF pathways. Laboratory Investigation. 2013;93(7):768–778. doi: 10.1038/labinvest.2013.71. [DOI] [PubMed] [Google Scholar]
- 44.Ning Y., Manegold P.C., Hong Y.K., Zhang W., Pohl A., Lurje G. Interleukin-8 is associated with proliferation, migration, angiogenesis and chemosensitivity in vitro and in vivo in colon cancer cell line models. International Journal of Cancer. 2011;128(9):2038–2049. doi: 10.1002/ijc.25562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thiery J.P., Acloque H., Huang R.Y., Nieto M.A. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139(5):871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 46.Sha Y., Haensel D., Gutierrez G., Du H., Dai X., Nie Q. Intermediate cell states in epithelial-to-mesenchymal transition. Physical Biology. 2019 doi: 10.1088/1478-3975/aaf928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grosse-Wilde A., D’Hérouël A.F., McIntosh E., Ertaylan G., Skupin A., Kuestner R.E. Stemness of the hybrid epithelial/mesenchymal state in breast cancer and its association with poor survival. PLoS One. 2015;10(5) doi: 10.1371/journal.pone.0126522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chin V.L., Lim C.L. Epithelial-mesenchymal plasticity — engaging stemness in an interplay of phenotypes. Stem Cell Investigation. 2019 doi: 10.21037/sci.2019.08.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jolly M.K., Tripathi S.C., Jia D., Mooney S.M., Celiktas M., Hanash S.M. Stability of the hybrid epithelial/mesenchymal phenotype. Oncotarget. 2016;7(19):27067–27084. doi: 10.18632/oncotarget.8166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mani S.A., Guo W., Liao M.J., Eaton E.N., Ayyanan A., Zhou A.Y. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133(4):704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roefs M.M., Carlotti F., Jones K., Wills H., Hamilton A., Verschoor M. Increased vimentin in human α- and β-cells in type 2 diabetes. Journal of Endocrinology. 2017;233(3):217–227. doi: 10.1530/JOE-16-0588. [DOI] [PubMed] [Google Scholar]
- 52.Cheng Y., Kang H., Shen J., Hao H., Liu J., Guo Y. Beta-cell regeneration from vimentin+/MafB+ cells after STZ-induced extreme beta-cell ablation. Scientific Reports. 2015;5:11703. doi: 10.1038/srep11703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lawlor N., George J., Bolisetty M., Kursawe R., Sun L., Sivakamasundari V. Single-cell transcriptomes identify human islet cell signatures and reveal cell-type-specific expression changes in type 2 diabetes. Genome Research. 2017;27(2):208–222. doi: 10.1101/gr.212720.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xin Y., Kim J., Okamoto H., Ni M., Wei Y., Adler C. RNA sequencing of single human islet cells reveals type 2 diabetes genes. Cell Metabolism. 2016;24(4):608–615. doi: 10.1016/j.cmet.2016.08.018. [DOI] [PubMed] [Google Scholar]
- 55.Cheng J.D., Dunbrack R.L., Jr., Valianou M., Rogatko A., Alpaugh R.K., Weiner L.M. Promotion of tumor growth by murine fibroblast activation protein, a serine protease, in an animal model. Cancer Research. 2002;62(16):4767–4772. [PubMed] [Google Scholar]
- 56.Jacob M., Chang L., Pure E. Fibroblast activation protein in remodeling tissues. Current Molecular Medicine. 2012;12(10):1220–1243. doi: 10.2174/156652412803833607. [DOI] [PubMed] [Google Scholar]
- 57.Omar B.A., Liehua L., Yamada Y., Seino Y., Marchetti P., Ahren B. Dipeptidyl peptidase 4 (DPP-4) is expressed in mouse and human islets and its activity is decreased in human islets from individuals with type 2 diabetes. Diabetologia. 2014;57(9):1876–1883. doi: 10.1007/s00125-014-3299-4. [DOI] [PubMed] [Google Scholar]
- 58.Busek P., Hrabal P., Fric P., Sedo A. Co-expression of the homologous proteases fibroblast activation protein and dipeptidyl peptidase-IV in the adult human Langerhans islets. Histochemistry and Cell Biology. 2015;143(5):497–504. doi: 10.1007/s00418-014-1292-0. [DOI] [PubMed] [Google Scholar]
- 59.Chera S., Baronnier D., Ghila L., Cigliola V., Jensen J.N., Gu G. Diabetes recovery by age-dependent conversion of pancreatic δ-cells into insulin producers. Nature. 2014;514(7253):503–507. doi: 10.1038/nature13633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chung C.H., Hao E., Piran R., Keinan E., Levine F. Pancreatic beta-cell neogenesis by direct conversion from mature alpha-cells. Stem Cells. 2010;28(9):1630–1638. doi: 10.1002/stem.482. [DOI] [PubMed] [Google Scholar]
- 61.Thorel F., Nepote V., Avril I., Kohno K., Desgraz R., Chera S. Conversion of adult pancreatic alpha-cells to beta-cells after extreme beta-cell loss. Nature. 2010;464(7292):1149–1154. doi: 10.1038/nature08894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bensellam M., Jonas J.C., Laybutt D.R. Mechanisms of beta-cell dedifferentiation in diabetes: recent findings and future research directions. Journal of Endocrinology. 2018;236(2):R109–R143. doi: 10.1530/JOE-17-0516. [DOI] [PubMed] [Google Scholar]
- 63.Wang Y.J., Golson M.L., Schug J., Traum D., Liu C., Vivek K. Single-cell mass cytometry analysis of the human endocrine pancreas. Cell Metabolism. 2016;24(4):616–626. doi: 10.1016/j.cmet.2016.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Senbanjo L.T., Chellaiah M.A. CD44: a multifunctional cell surface adhesion receptor is a regulator of progression and metastasis of cancer cells. Frontiers in Cell and Developmental Biology. 2017;5:18. doi: 10.3389/fcell.2017.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang Z., York N.W., Nichols C.G., Remedi M.S. Pancreatic β cell dedifferentiation in diabetes and redifferentiation following insulin therapy. Cell Metabolism. 2014;19(5):872–882. doi: 10.1016/j.cmet.2014.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nemoto Y., Ikeda J., Katoh K., Koshimoto H., Yoshihara Y., Mori K. R2D5 antigen: a calcium-binding phosphoprotein predominantly expressed in olfactory receptor neurons. Journal of Cell Biology. 1993;123(4):963–976. doi: 10.1083/jcb.123.4.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhao F., Wang Q. The protective effect of peroxiredoxin II on oxidative stress induced apoptosis in pancreatic β-cells. Cell & Bioscience. 2012;2(1):22. doi: 10.1186/2045-3701-2-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gurzov E.N., Barthson J., Marhfour I., Ortis F., Naamane N., Igoillo-Esteve M. Pancreatic β-cells activate a JunB/ATF3-dependent survival pathway during inflammation. Oncogene. 2012;31(13):1723–1732. doi: 10.1038/onc.2011.353. [DOI] [PubMed] [Google Scholar]
- 69.Van Acker Z.P., Bretou M., Annaert W. Endo-lysosomal dysregulations and late-onset Alzheimer's disease: impact of genetic risk factors. Molecular Neurodegeneration. 2019 doi: 10.1186/s13024-019-0323-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cao P., Marek P., Noor H., Patsalo V., Tu L.H., Wang H. Islet amyloid: from fundamental biophysics to mechanisms of cytotoxicity. FEBS Letters. 2013:1106–1118. doi: 10.1016/j.febslet.2013.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hsu T.N., Huang C.M., Huang C.S., Huang M.S., Yeh C.T., Chao T.Y. Targeting FAT1 inhibits carcinogenesis, induces oxidative stress and enhances cisplatin sensitivity through deregulation of LRP5/WNT2/GSS signaling axis in oral squamous cell carcinoma. Cancers. 2019;11(12) doi: 10.3390/cancers11121883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lu T.T.H., Heyne S., Dror E., Casas E., Leonhardt L., Boenke T. The polycomb-dependent epigenome controls β cell dysfunction, dedifferentiation, and diabetes. Cell Metabolism. 2018;27(6):1294–1308. doi: 10.1016/j.cmet.2018.04.013. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Krupnik V.E., Sharp J.D., Jiang C., Robison K., Chickering T.W., Amaravadi L. Functional and structural diversity of the human Dickkopf gene family. Gene. 1999 doi: 10.1016/S0378-1119(99)00365-0. [DOI] [PubMed] [Google Scholar]
- 74.Zenzmaier C., Herman M., Hengster P., Berger P. Dickkopf-3 maintains the PANC-1 human pancreatic tumor cells in a dedifferentiated state. International Journal of Oncology. 2012;40(1):40–46. doi: 10.3892/ijo.2011.1180. [DOI] [PubMed] [Google Scholar]
- 75.Li C., Morvaridi S., Lam G., Chheda C., Kamata Y., Katsumata M. MSP-RON signaling is activated in the transition from pancreatic intraepithelial neoplasia (PanIN) to pancreatic ductal adenocarcinoma (PDAC) Frontiers in Physiology. 2019;10(FEB):147. doi: 10.3389/fphys.2019.00147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Richardson S.J., Rodriguez-Calvo T., Gerling I.C., Mathews C.E., Kaddis J.S., Russell M.A. Islet cell hyperexpression of HLA class I antigens: a defining feature in type 1 diabetes. Diabetologia. 2016;59(11):2448–2458. doi: 10.1007/s00125-016-4067-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Böni-Schnetzler M., Meier D.T. Islet inflammation in type 2 diabetes. Seminars in Immunopathology. 2019:501–513. doi: 10.1007/s00281-019-00745-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Soler J.C., Dai X., Hang Y., Kim S., Macdonald P. 1711-P: pancreas single-cell patch-seq links physiologic dysfunction in diabetes to transcriptomic phenotypes. Diabetes. 2019 doi: 10.2337/db19-1711-. [DOI] [Google Scholar]
- 79.Vivot K., Moullé V.S., Zarrouki B., Tremblay C., Mancini A.D., Maachi H. The regulator of G-protein signaling RGS16 promotes insulin secretion and β-cell proliferation in rodent and human islets. Molecular Metabolism. 2016;5(10):988–996. doi: 10.1016/j.molmet.2016.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Guo S., Dai C., Guo M., Taylor B., Harmon J.S., Sander M. Inactivation of specific β cell transcription factors in type 2 diabetes. Journal of Clinical Investigation. 2013;123(8):3305–3316. doi: 10.1172/JCI65390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Svystonyuk D.A., Ngu J.M., Mewhort H.E., Guzzardi D.G., Lipon B.D., Park D. Fibroblast growth factor (FGF-2) prevents human cardiac fibroblast-mediated extracellular matrix remodeling. Canadian Journal of Cardiology. 2014;30(10):S153. doi: 10.1016/j.cjca.2014.07.227. [DOI] [Google Scholar]
- 82.Ahn H.J., Lee W.J., Kwack K.B., Kwon Y. Do. FGF2 stimulates the proliferation of human mesenchymal stem cells through the transient activation of JNK signaling. FEBS Letters. 2009;583(17):2922–2926. doi: 10.1016/j.febslet.2009.07.056. [DOI] [PubMed] [Google Scholar]
- 83.Su C.W., Chang Y.C., Chien M.H., Hsieh Y.H., Chen M.K., Lin C.W. Loss of TIMP3 by promoter methylation of Sp1 binding site promotes oral cancer metastasis. Cell Death & Disease. 2019;10(11):1–17. doi: 10.1038/s41419-019-2016-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Song L., Li X., Liu X., Wang Z., Yu Y., Shi M. EFEMP2 suppresses the invasion of lung cancer cells by inhibiting epithelial-mesenchymal transition (EMT) and down-regulating MMPs. OncoTargets and Therapy. 2020;13:1375–1396. doi: 10.2147/OTT.S236111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kubo E., Shibata T., Singh D.P., Sasaki H. Roles of TGF β and FGF signals in the lens: tropomyosin regulation for posterior capsule opacity. International Journal of Molecular Sciences. 2018 doi: 10.3390/ijms19103093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gonzalez D.M., Medici D. Signaling mechanisms of the epithelial-mesenchymal transition. Science Signaling. 2014 doi: 10.1126/scisignal.2005189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Toren-Haritan G., Efrat S. TGFβ pathway inhibition redifferentiates human pancreatic islet β cells expanded in vitro. PLoS One. 2015;10(9) doi: 10.1371/journal.pone.0139168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pan F.C., Brissova M. Pancreas development in humans. Current Opinion in Endocrinology, Diabetes and Obesity. 2014:77–82. doi: 10.1097/MED.0000000000000047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Carolan P.J., Melton D.A. New findings in pancreatic and intestinal endocrine development to advance regenerative medicine. Current Opinion in Endocrinology, Diabetes and Obesity. 2013 doi: 10.1097/MED.0b013e32835bc380. [DOI] [PubMed] [Google Scholar]
- 90.Azuara V., Perry P., Sauer S., Spivakov M., Jørgensen H.F., John R.M. Chromatin signatures of pluripotent cell lines. Nature Cell Biology. 2006;8(5):532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- 91.Krook M.A., Hawkins A.G., Patel R.M., Lucas D.R., Van Noord R., Chugh R. A bivalent promoter contributes to stress-induced plasticity of CXCR4 in Ewing sarcoma. Oncotarget. 2016;7(38):61775–61788. doi: 10.18632/oncotarget.11240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Habener J.F., Stanojevic V. α-cell role in β-cell generation and regeneration. Islets. 2012:188–198. doi: 10.4161/isl.20500. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Donor information.
Cell-type identification. This table contains the annotation of cells per condition.
This table contains the derivation of gene lists characteristic for each cell type in non-diabetic adult donors using multi-factor analysis of variance (ANOVA) and differential expression analysis. Genes appear in the file corresponding to the cell type in which they have the highest expression (ANOVA results sheets 3–9) and cell type specific gene sets composed of genes enriched for each cell type from non-diabetic (ND) adult donors derived from DE analysis of each cell type to all other cell types (sheet 10).
This table contains the lists of genes up- and downregulated in β- and α-cells with aging and the lists of genes driving the pseudotime modules displayed in Figure S1B and E.
Related to Figure 1D. Lists of gene sets that were found up- and down-regulated during β-cell maturation.
Related to Figure 2D. Lists of gene sets that were found up- and down-regulated during α-cell maturation and the lists of genes driving the pseudotime modules displayed in Figure 2C.
Related to Figure 2F. Lists of gene sets that were found enriched in α-cells as compared with β-cells from non-diabetic adults.
Related to Figure 4B. Lists of gene sets that were found up- and downregulated in T2D as compared with adult non-diabetic β-cells.
Related to Figure 5B. Lists of gene sets that were found up- and downregulated in T2D as compared with adult non-diabetic α-cells.