Abstract
Recently, the nuclear receptor farnesoid X receptor (FXR) has been considered to be a liver tumor suppressor. However, the role of FXR in liver cancer invasion and metastasis remains unclear. The results of the current study demonstrated that FXR suppressed the migratory and invasive capacities of SK-Hep-1 cells in vitro and that FXR overexpression inhibited local invasion and lung metastasis of SK-Hep-1 ×enografts in vivo. Bioinformatics analysis of the gene expression profile of SK-Hep-1 cells with different FXR levels indicated that FXR may regulate the Wnt/β-catenin pathway. Compared with controls, FXR-overexpressing SK-Hep-1 cells exhibited decreased expression of β-catenin target genes and reduced nuclear translocation of β-catenin proteins in vitro and in vivo. In conclusion, these results indicated that FXR may suppress SK-Hep-1 cell invasion and metastasis by suppressing the Wnt/β-catenin signaling pathway. The current study provided novel insight into the diagnosis and treatment of liver cancer.
Keywords: FXR, SK-Hep-1 cell, invasion and metastasis, Wnt/β-catenin
Introduction
Globally, liver cancer was the sixth most commonly diagnosed cancer and the fourth leading cause of cancer-associated mortality in 2018 (1). Early tumor recurrence and metastasis are the main causes of high mortality; however, the signaling pathways that regulate these processes remain unclear (2).
FXR was initially regarded as a multifunctional metabolic regulator involved in maintaining the hepatic homeostasis of bile acid, glucose and lipids (3,4). It has been well documented that FXR also suppresses liver carcinogenesis and progression (5–8). FXR may serve an indispensable role in inhibiting the activity of signaling pathways that are dysregulated in liver cancer cells (9).
The Wnt/β-catenin pathway serves a key role in liver biology and liver cancer pathogenesis (10). Aberrant activation of Wnt/β-catenin signaling has been observed in the majority of hepatocellular carcinomas (HCCs) and in hepatoblastomas, and is closely associated with HCC metastasis and prognosis (11,12). The canonical Wnt/β-catenin pathway interacts with the epithelial-mesenchymal transition (EMT) signaling network. The EMT is considered one of the most important initial steps of cancer invasion (13,14). Wolfe et al (15) demonstrated that the Wnt/β-catenin pathway is activated in the liver of FXR-knockout mice during spontaneous HCC development and that FXR deletion may result in the loss of E-cadherin expression.
It has been previously reported that FXR downregulation was correlated with the presence of multiple malignant clinicopathological features, including tumor size, advanced BCLC stage, poor differentiation and absence of encapsulation, in patients with HCC (7). FXR inhibited liver cancer cell proliferation in vivo and in vitro via inactivation of the mammalian target of rapamycin (mTOR)/ribosomal S6 kinase (S6K) pathway (9). The current study aimed to investigate whether FXR influenced the regulation liver cancer metastasis and the possible underlying mechanism.
Materials and methods
Cell culture
SK-Hep-1 cells were obtained from the cell bank of the Chinese Academy of Sciences. Mycoplasma detection and cell line authentication indicating no mycoplasma or other human cell line contamination were performed using quantitative PCR detection for mycoplasma and 10 short tandem repeat identification via capillary electrophoresis for cell line authentication. SK-Hep-1 cells were cultured in DMEM supplemented with 10% FBS (both Gibco; Thermo Fisher Scientific, Inc.) and 1% penicillin-streptomycin (HyClone; Cytiva) in a humidified atmosphere at 37°C with 5% CO2. The FXR agonist GW4064 was obtained from Sigma-Aldrich (Merck KGaA).
Construction of FXR recombinant lentiviral vectors and transduction of the lentivirus into SK-Hep-1 cells
The construction of the human FXR overexpression lentiviral vectors and negative controls, and transducing lentivirus into SK-Hep-1 cells were performed as previously described (7). Briefly, the human FXR gene NR1H4 PCR-amplified from plasmid pCR4-FXR-TOPO (Open BioSystems, Inc.) was cloned into the pGC-FU-GFP lentiviral vector (Shanghai GeneChem Co., Ltd.). A total of 10 µg of the resulting pGC-FU-FXR-GFP vector or the control empty vector pGC-FU-GFP was co-transfected with the packaging vectors using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) into 293T cells to produce the lentiviruses that were used to infect SK-Hep-1 cells. The cells were used for subsequent experimentation 48 h post infection. SK-Hep-1 cells transfected with recombinant vectors that overexpressed FXR or the corresponding controls were designated as SK-Hep-1-FXR or SK-Hep-1-NC, respectively.
RNA isolation and reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from SK-Hep-1-FXR and SK-Hep-1-NC cells using TriPure Isolation reagent (Roche Applied Science) and converted to cDNA using a PrimeScript RT reagent kit (Takara Bio, Inc.), according to the manufacturer's protocol. RT-qPCR was performed using SYBR Premix Ex Taq (Takara Bio, Inc.) and the Applied Biosystems 7500 Fast Real-Time PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol, with the following thermocycling conditions: Denaturation at 95°C for 3 min, followed by 40 cycles of denaturation at 95°C for 10 sec, annealing at 60°C for 30 sec and elongation at 72°C for 30 sec, and a final extension step at 72°C for 30 sec. β-actin was used as the internal reference gene. Relative mRNA levels to β-actin were analyzed using the 2−ΔΔCq method (16). The paired forward and reverse primers for the amplification of the specific genes were as follows: FXR forward, 5′-ATGCCTGTAACAAAGAAGCCCC-3′ and reverse, 5′-CACACAGTTGCCCCCGTTTTTAC-3′; low-density lipoprotein receptor-related protein 5 (LRP5) forward, 5′-GTACCCGCCGATCCTGAAC-3′ and reverse, 5′-TGTAGTCGCTGTCACACACG-3′; secreted Fzd-related protein (SFRP1) forward, 5′-CTCAACAAGAACTGCCACGC-3′ and reverse, 5′-CTCGTTGTCACAGGGAGGAC-3′; matrix metalloproteinase 9 (MMP9) forward, 5′-TCTATGGTCCTCGCCCTGAA-3′ and reverse, 5′-CATCGTCCACCGGACTCAAA-3′; cyclin D1 (CCND1) forward, 5′-CGGTGTCCTACTTCAAATGTGTGC-3′ and reverse 5′-AGAGGCCACGAACATGCAAGTGG-3′; β-catenin (CTNNB1) forward, 5′-CTGAGGACAAGCCACAAGATTA-3′ and reverse, 5′-ATCCACCAGAGTGAAAAGAACG-3′; c-MYC forward, 5′-AGCTTGTACCTGCAGGATCTGAGC-3′ and reverse, 5′-AGCCTGCCTCTTTTCCACAGAAAC-3′; c-JUN forward, 5′-GTTGAGCTCGGGCTGGATAA-3′ and reverse, 5′-GACTATACTGCCGACCTGGC-3′; Dishevelled 1 (DVL1) forward, 5′-TGAACCTCAACAGTGGCTCC-3′ and reverse, 5′-GTGTGATCCGATTCACTGCC-3′; and Frizzled 5 (FZD5) forward, 5′-TGTGCTTCATCTCCACGTCC-3′ and reverse, 5′-CCGTGGTCTCGTAGTGGATG-3′.
Microarray and bioinformatics analysis
SK-Hep-1-FXR cells and SK-Hep-1-NC were pretreated with 2 µM GW4064 for 24 h at 37°C, and total RNA from the cells was isolated according to the aforementioned method and sent to Kangchen BioTech Co., Ltd., for mRNA microarray analysis. The samples were labeled and hybridized using a Whole Human Genome Microarray kit (cat. no. 4×44K one-color v2; Agilent Technologies, Inc.), according to the manufacturer's protocol. Raw data was extracted using Agilent Feature Extraction software (v9.5.3; Agilent Technologies, Inc.). Gene ontology (http://geneontology.org/) and Kyoto Encyclopedia of Genes and Genomes databases (https://www.kegg.jp/) were used for bioinformatics analysis. The microarray data were deposited to the Gene Expression Omnibus (GEO) repository with the access no. GSE149111 (ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE149111).
Transwell migration and invasion assays
The migratory and invasive capabilities of SK-Hep-1-FXR and SK-Hep-1-NC cells were assessed using Transwell BioCoat™ Matrigel™ Invasion Chambers (BD Biosciences), according to the manufacturer's protocol. After pretreatment with the FXR agonist GW4064 for 24 h at 37°C, 2×104 cells in 200 µl of serum-free DMEM were loaded into the upper chamber of the Transwell system and 750 µl DMEM containing 10% FBS were added to the lower chamber. Following incubation for 24 h at 37°C, the cells that did not migrate through the pores of the Transwell inserts in the upper chamber were removed with a sterile cotton swab. Cells in the lower chamber were fixed in 4% paraformaldehyde for 10 min at room temperature and stained with hematoxylin for 3 min and eosin for 15 sec at room temperature. Cells were then counted and imaged in 10 randomly selected microscopic fields under an Olympus BX41 light microscope (Olympus Corporation) at a magnification of ×200. Experiments were performed in replicate inserts and mean values were calculated from 3 independent experiments. Inserts coated with Matrigel at room temperature for 10 min were used in the invasion assay to determine the invasive abilities of the cells.
Wound healing and scratch assays
After pretreatment with the FXR agonist GW4064 for 24 h at 37°C, a total of 2.5×105 SK-Hep-1-FXR and SK-Hep-1-NC cells were seeded onto 6-well plates and scratched with a sterile pipette tip to the confluent monolayer when cells reached 100% confluence. Following washing with PBS, serum-free DMEM was added and cell migration was observed at 0 and 24 h. Images of cells migrating across the scratched field were captured using an Olympus fluorescence inverted microscope (Olympus Corporation) at ×100 magnification, and the number of cells that migrated into the scratched area was counted (17) and statistical analysis was performed using SPSS software (v22.0; IBM Corp.) for 10 randomly selected fields within the scratched area from 3 independent experiments.
Preparation of protein extracts and western blotting
To prepare the total cellular proteins, SK-Hep-1-FXR and SK-Hep-1-NC cell pellets were lysed in Cell Lysis Buffer (Beyotime Institute of Biotechnology) with 1% phenylmethanesulfonyl fluoride (PMSF; Beyotime Institute of Biotechnology). Nuclear proteins were isolated using a Nuclear and Cytoplasmic Protein Extraction kit (Beyotime Institute of Biotechnology), according to the manufacturer's protocol. Briefly, cells were harvested and the pellets were dissolved with cytoplasmic protein extraction agent A (Beyotime Institute of Biotechnology) with PMSF. Cells were vortexed at room temperature for 5 sec and incubated for 10–15 min at 4°C on ice. Cytoplasmic protein extraction agent B (Beyotime Institute of Biotechnology) was then added and the lysates were centrifuged at 14,000 × g for 10 min at 4°C. The insoluble pellet fraction was resuspended in nuclear protein extraction agent supplemented with PMSF and the supernatants of the nuclear extract were collected immediately. Total and nuclear proteins were quantified using a bicinchoninic acid (BCA) protein assay (Beyotime Institute of Biotechnology), aliquoted and stored at −80°C until use. For western blotting, 30–70 µg of the proteins were separated via 12% SDS-PAGE, transferred to the PDVF membranes (Beyotime Institute of Biotechnology), blocked at room temperature with 5% non-fat milk for 1 h. The members were incubated overnight at 4°C with diluted mouse anti-human FXR monoclonal antibodies (1:500; R&D Systems, Inc.; cat. no. PP-A9033A-00), mouse anti-human monoclonal β-actin antibodies (1:2,500; Beyotime Institute of Biotechnology; cat. no. AF0003), rabbit anti-human polyclonal β-catenin (1:2,000; Abcam; cat. no. ab6302), rabbit anti-human cyclin D1 monoclonal antibodies (1:1,000; Cell Signaling Technology, Inc.; cat. no. 55506) and mouse anti-human Histone H3 monoclonal antibodies (1:1,000; Beyotime Institute of Biotechnology; cat. no. AF0009). The membranes were washed with TBS-Tween (0.1% Tween 20; Beyotime Institute of Biotechnology) three times for 10 min each and incubated with HRP-labeled secondary antibodies (goat anti-mouse, cat. no. 14709; or goat anti-rabbit, cat. no. 14708; both 1:1,000 dilution; Cell Signaling Technology, Inc.) at room temperature for 1 h. The bands were detected using a BeyoECL Plus kit (Beyotime Institute of Biotechnology) according to the manufacturer's protocol. Image Lab software (version 5.0; Bio-Rad Laboratories, Inc.) was used for the quantitative analysis. β-actin and Histone H3 were used as loading controls for whole cell extracts and nuclear extracts, respectively.
Animal studies
A total of 40 male BALB/c nu/nu mice (age, 4–6 weeks; weight, 16–18 g) were purchased from the Shanghai Laboratory Animals Center and housed under controlled illumination (12 h light and 12 h dark cycle), at a temperature of 22±2°C and humidity of 40–60% for 7 days with food and water available ad libitum. To observe local invasion, 6 mice per group were injected subcutaneously with 5×106 SK-Hep-1-FXR or SK-Hep-1-NC cells resuspended in serum-free DMEM into the flank. Tumors were palpable on day 7 post-transplantation. The mice were sacrificed on day 21 post-inoculation by cervical dislocation after anesthesia via intraperitoneal injection of a mixture of ketamine (100 mg/kg) and xylazine (10 mg/kg). Tumors (1,210±120 mm3 for SK-Hep-1-NC and 678±91 mm3 for SK-Hep-1-NC) and the surrounding tissues deep into the muscle wall (~0.5 cm from the tumor) were harvested and fixed in 10% neutral buffered formalin at room temperature for 24 h. Sections of 3-µm thickness were then stained at room temperature with hematoxylin for 5 min and eosin for 1 min. Images were captured using an Olympus BX41 light microscope (Olympus Corporation) at ×40 magnification.
For the in vivo lung metastasis study, 5×106 SK-Hep-1-FXR or SK-Hep-1-NC cells were injected into mice through the caudal vein (n=4/group). Lungs were harvested 30 days later. The lung tissues were fixed in 10% neutral buffered formalin at room temperature for 24 h, embedded into paraffin and stained with hematoxylin and eosin as aforementioned. Immunohistochemistry staining was performed in the lung tissues containing metastatic foci. The histopathologically defined metastatic foci were analyzed under a light microscope (Olympus BX41; Olympus Corporation; magnification, ×100) after the lung tissues were fixed and stained with hematoxylin and eosin, as aforementioned. The number of the metastases with a maximum diameter >0.2 mm was counted.
All experimental procedures involving the animals were performed in accordance with the National Institutes of Health Guide for the Care and Use of Animals (18) and were approved by the Institutional Animal Care and Use Committee of Fujian Medical University (Fuzhou, China).
Immunohistochemistry (IHC)
IHC staining was performed using an Envision Plus System (Fuzhou Maixin Biotech Co., Ltd.) according to the manufacturer's protocol. Briefly, the paraffin-embedded lung tissues were cut into 3-µm-thick sections that were heated at 67°C for 2 h. Sections were then deparaffinized twice with xylene for 5 min, rehydrated using a gradient ethanol series (100% for 5 min twice, 95% for 5 min twice, 90% for 5 min, 85% for 5 min and 80% for 5 min) and washed with double-distilled water for 1 h (all at room temperature). Subsequently, sections were pretreated with 3% H2O2 for 15 min at 4°C to block endogenous peroxidase activity. Antigens were retrieved by pressure cooker treatment at 120°C for 100 sec in 0.01 mmol/l citrate buffer (pH 6). To block non-specific antibody binding, sections were treated with 10% goat serum (cat. no. ZLI-9021; OriGene Technologies, Inc.) for 30 min at room temperature. The primary antibodies against β-catenin (1:2,000; Abcam; cat. no. ab6302) and FXR (1:500; R&D Systems, Inc.; PP-A9033A-00) were incubated with tissue sections overnight at 4°C. The next day, tissue sections were incubated with horseradish peroxidase-conjugated secondary antibodies (1:50; Beyotime Institute of Technology; cat. no. AF0216) for 2 h at room temperature. Images were captured using a fluorescence inverted microscope (Olympus Corporation) at ×100 magnification. For β-catenin, nuclear staining with or without cytoplasmic staining were considered as positive. For FXR, cytoplasmic plus nuclear staining was considered as positive. A 5-tiered scale was used to assess the degree of FXR and β-catenin staining based on the average percentage of positively stained cells (0, ≤5%; 1, 6–25%; 2, 26–50%; 3, 51–75%; 4, 76–100%) multiplied by the staining intensity judged by two experienced pathologists [0, no staining; 1, weak staining (light yellow); 2, moderate staining (yellow-brown); and 3, strong staining (brown)] to calculate a final score ranging 0–12.
Statistical analysis
Statistical analysis was performed using SPSS software (version 19; IBM Corp.). Values were expressed as the mean ± standard error of the mean of ≥3 independent experiments. Unpaired Student's t-test or χ2 test was used to determine the difference between groups. P<0.05 was considered to indicate a statistically significant difference.
Results
FXR suppresses liver cancer cell migration and invasion in vitro
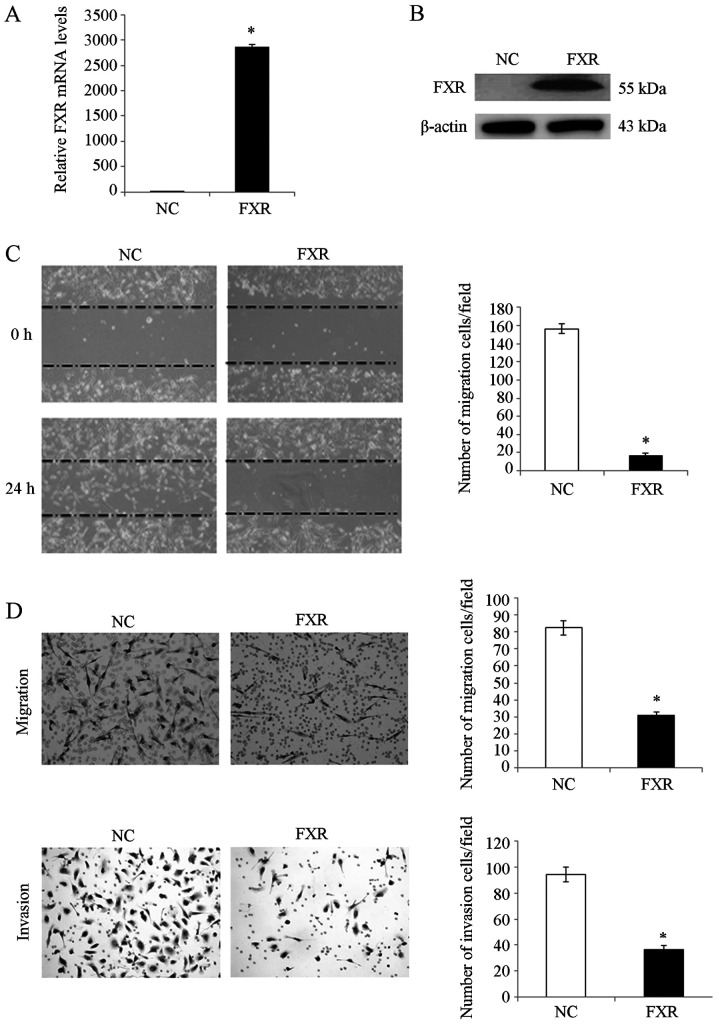
To investigate the effect of FXR on liver cancer cell migration and invasion in vitro, SK-HEP-1 cell line was used as the cell model since SK-HEP-1 expresses FXR at very low levels (7,9) and has a high metastatic capacity (19). Therefore, the metastatic suppressive effect of FXR could be determined when it is overexpressed. SK-HEP-1 is an immortal human cell line derived from the ascitic fluid of a patient with liver adenocarcinoma (20). Although these cells are of endothelial origin and do not exhibit hepatocyte properties, they have been generally considered as a human hepatoma cell line with mesenchymal characteristics (19,20). The isogenic paired FXR-overexpressing SK-HEP-1-FXR cell line was established from SK-HEP-1 using a lentivirus expression system as previously described (7). FXR overexpression in SK-HEP-1-FXR cells was confirmed using RT-qPCR (Fig. 1A) and western blotting analysis (Fig. 1B). After pretreating SK-Hep-1-FXR and control cells with the FXR agonist GW4064 for 24 h at 37°C, wound healing and Transwell assays were performed to determine cell migratory and invasive capacities, respectively. SK-Hep-1-FXR cells demonstrated significantly impaired wound healing ability (Fig. 1C; P<0.05) and significantly inhibited migration and invasion abilities (Fig. 1D; P<0.05), compared with controls. These data indicated that FXR negatively regulated liver cancer cell migration and invasion in vitro.
Figure 1.
FXR represses the migration and invasion capacities of SK-Hep-1 cells in vitro. FXR expression in SK-Hep-1-FXR and SK-Hep-1-NC cells were detected by (A) reverse transcription-quantitative PCR and (B) western blotting. SK-Hep-1-FXR and SK-Hep-1-NC cells were pretreated with 2 µM of the FXR agonist GW4064 for 24 h at 37°C, and representative images were captured of the (C) wound healing assay and the (D) Transwell assay for measurement of cell migration and invasion, respectively. The histograms indicate the quantification of migrated or invaded cell numbers determined from three independent experiments. *P<0.05. The ‘FXR’ group represents FXR-overexpressing SK-Hep-1-FXR cells and the ‘NC’ group represents the control SK-Hep-1-NC cells. FXR, farnesoid X receptor; NC, negative control.
FXR inhibits local invasion and metastasis of SK-Hep-1 ×enografts in vivo
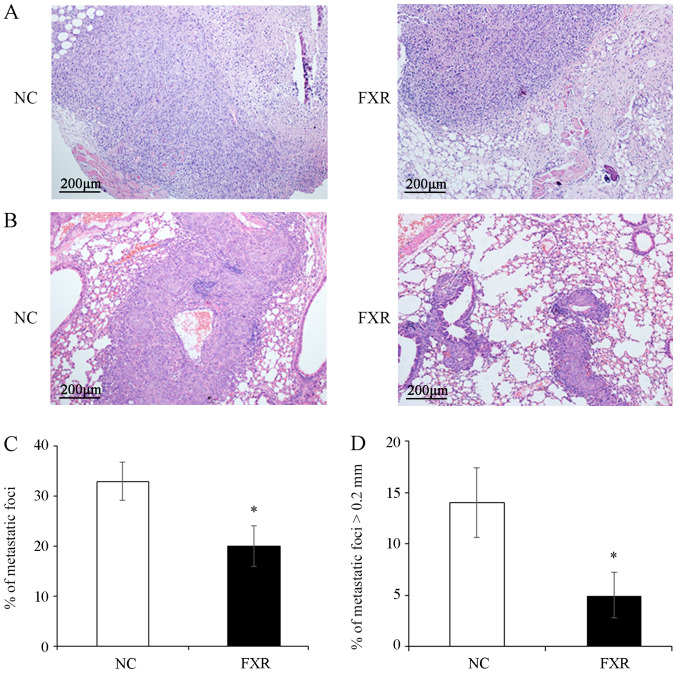
The effect of FXR overexpression on the metastatic potential in vivo was examined by subcutaneous injection of the SK-Hep-1-FXR or SK-Hep-1-NC cells into nude mice to observe the depth of local invasion or by tail vein injection to detect lung metastasis. The results demonstrated that the invasion depth of SK-Hep-1-FXR xenograft was limited to the subcutaneous loose connective tissue layer (Fig. 2A, right panel). By contrast, the tumors xenografted from controls invaded deeply into the muscle layer (Fig. 2A, left panel). Furthermore, significantly fewer and smaller pulmonary metastatic nodules, as shown in the representative images of hematoxylin and eosin staining on the lung tissue sections (Fig. 2B) and evidenced by the total number of metastatic foci (Fig. 2C) and the number of metastases with the maximum diameter >0.2 mm (Fig. 2D), were observed in the lungs of mice injected with SK-Hep-1-FXR cells compared with controls (both P<0.05). However, since most of lung metastatic foci are tiny and not located on the surface of the lung (21), it is a technical challenge to measure the volume or weight of such micro-metastases. These results demonstrated that FXR inhibited local invasion and lung metastasis of SK-Hep-1 ×enografts in vivo.
Figure 2.
FXR inhibits the local invasion and metastasis of the SK-Hep-1 ×enograft in vivo. After pretreating with the FXR agonist GW4064 2 µM for 24 h at 37°C, SK-Hep-1-FXR cells and controls were subcutaneously injected into nude mice (n=6 in each group) and tumors and surrounding tissues were harvested three weeks after transplantation. (A) Representative images of hematoxylin and eosin staining. Lung metastasis was induced by tail vein injections of SK-Hep-1-FXR cells and controls pretreated with GW4064 for 24 h (n=4 in each group) into nude mice. (B) Representative images of lung tissue sections. Scale bar, 200 µm (magnification, ×100). (C) Total number of metastatic foci. (D) Number of the metastases with the maximum diameter >0.2 mm. Data are presented as the mean ± standard error of the mean from two experiments with 6 or 4 mice per group for each experiment. *P<0.05. The ‘FXR’ group represents FXR-overexpressing SK-Hep-1-FXR cells and the ‘NC’ group represents the control SK-Hep-1-NC cells. FXR, farnesoid X receptor; NC, negative control.
FXR may inhibit Wnt/β-catenin pathway activation
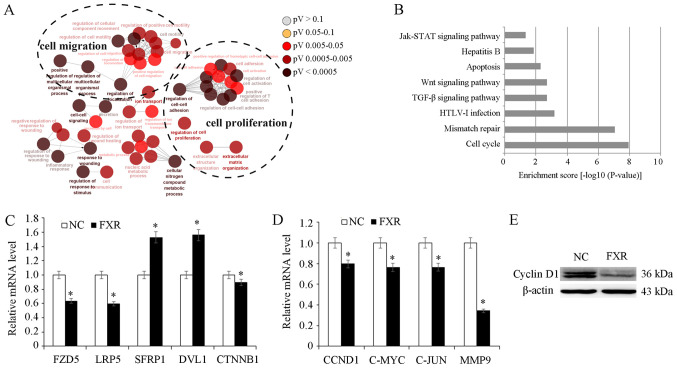
To explore possible molecular mechanisms underlying the metastatic suppressive effect of FXR, SK-Hep-1-FXR and SK-Hep-1-NC cells were pretreated with the FXR agonist GW4064 and microarray analysis was conducted to identify differentially expressed genes. Gene ontology analysis demonstrated that differentially expressed genes resulting from FXR overexpression in SK-Hep-1-FXR cells were involved in diverse biological processes including ‘cell migration’ and ‘cell proliferation’ (Fig. 3A). Pathway analysis demonstrated that FXR upregulation in SK-Hep-1 cells altered the activity of different cellular signaling pathways, including ‘cell cycle’, ‘mismatch repair’, ‘transforming growth factor β’ and ‘Wnt’ and ‘Janus kinase/signal transducers and activators of transcription’ (JAK-STAT; Fig. 3B). The Wnt pathway was selected for further study due to its well-established role in liver cancer cell proliferation and migration (22). The microarray data screened out several differentially expressed genes of Wnt pathway including FZD5, LRP5, sFRP1, DVL1 and CTNNB1. RT-qPCR was used to confirm microarray data. The results demonstrated that FXR overexpression downregulated Wnt/β-catenin upstream genes, including FZD5 and LRP5 (P<0.05; Fig. 3C). However, the mRNA levels of sFRP and DVL were upregulated in SK-Hep-1-FXR cells, while CTNNB1 expression was downregulated (P<0.05; Fig. 3C). sFRP is an antagonist of the Wnt pathway which binds directly to Wnt ligands, preventing Wnt from binding its receptors (23). DVL was considered as a dual function adaptor and has been reported to be a negative regulator of cell surface FZD level (24). Furthermore, β-catenin target genes CCND1, c-MYC, c-JUN and MMP9 were downregulated in response to FXR overexpression, as assessed via RT-qPCR (P<0.05; Fig. 3D) and western blotting (Fig. 3E). These results indicated that FXR may function as a suppressor of the Wnt/β-catenin pathway.
Figure 3.
Role of FXR in Wnt/β-catenin pathway. SK-Hep-1-FXR and SK-Hep-1-NC cells were pretreated with GW4064 2 µM for 24 h at 37°C, and harvested for microarray analysis. (A) Gene ontology analysis demonstrated that the differentially expressed genes resulting from FXR upregulation in SK-Hep-1-FXR cells were involved in diverse biological processes. (B) Kyoto Encyclopedia of Genes and Genomes pathway analysis revealed the alteration of diverse cellular signaling pathways due to the FXR overexpression. (C) mRNA expression levels of CTNNB1 and its upstream genes in SK-Hep-1-FXR and SK-Hep-1-NC was detected via RT-qPCR. (D) CTNNB1 target genes were detected using RT-qPCR. (E) Cyclin D1 protein expression was determined via western blotting. Results are presented as mean ± standard error of the mean from three independent experiments. *P<0.05 vs. NC. The ‘FXR’ group represents FXR-overexpressing SK-Hep-1-FXR cells and the ‘NC’ group represents the control SK-Hep-1-NC cells. FXR, farnesoid X receptor; NC, negative control; RT-qPCR, reverse transcription-quantitative PCR; Jak/STAT, Janus kinase/signal transducers and activators of transcription; TGF-β, transforming growth factor β; HTLV-1, human T-cell leukemia virus type 1; FZD5, Frizzled 5; LRP5, low-density lipoprotein receptor-related protein 5; SFRP1, secreted Fzd-related protein; DVL1, Dishevelled 1; CCND1, cyclin D1; CTNNB1, β-catenin.
FXR blocks the nuclear translocation of β-catenin in SK-Hep-1 cells in vitro and in vivo
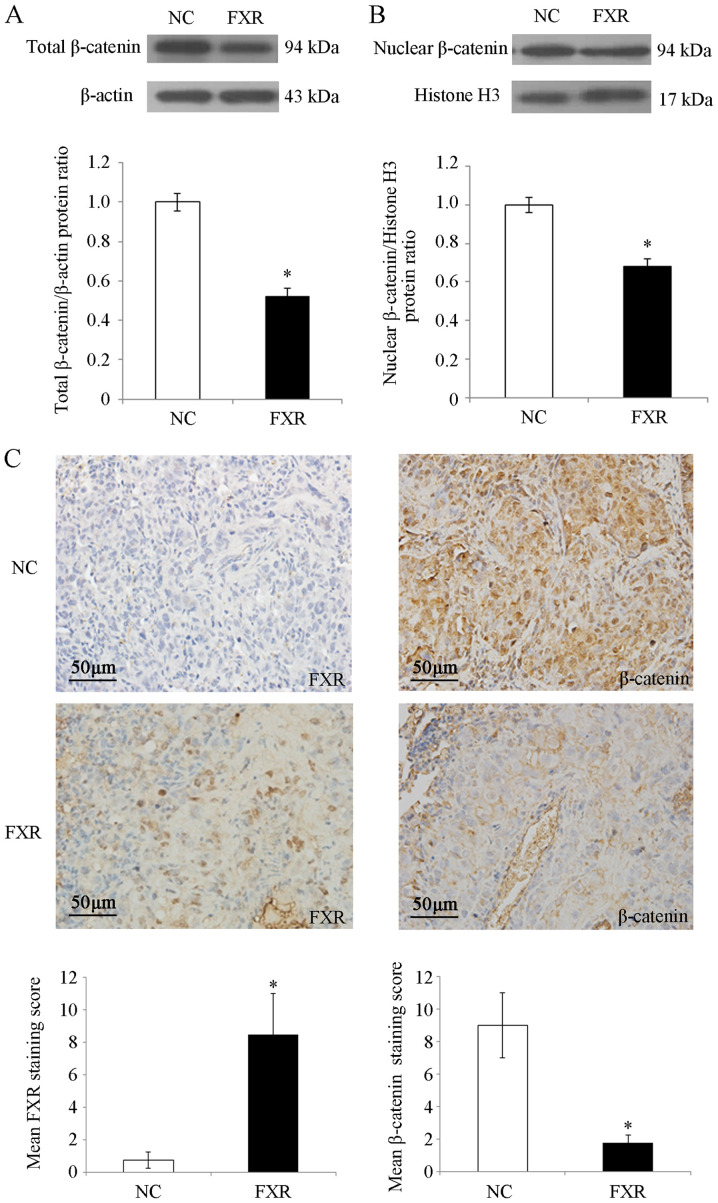
β-catenin is the nuclear effector of the Wnt/β-catenin pathway (25). To investigate the possible mechanisms by which FXR inhibited Wnt/β-catenin signaling, whether FXR blocked β-catenin nuclear translocation in SK-Hep-1 cells was investigated. The results demonstrated that total β-catenin protein levels were significantly decreased by FXR overexpression (P<0.05; Fig. 4A), which was consistent with the reduced CTNNB1 (the gene encoding β-catenin) mRNA levels in SK-Hep-1-FXR cells (P<0.05; Fig. 3C). Furthermore, nuclear β-catenin levels were significantly decreased in SK-Hep-1-FXR cells compared with controls (P<0.05; Fig. 4B). As shown in Fig. 4C, IHC staining demonstrated strong nuclear and moderate cytoplasmic staining for β-catenin protein in the lung metastases of nude mice injected with SK-Hep-1-NC cells, whereas β-catenin exhibited moderate membrane staining and weak nuclear and cytoplasmic staining in the lung metastases of the mice injected with SK-Hep-1-FXR cells (staining score, 9±2 for SK-Hep-1-NC vs. 1.75±0.5 for SK-Hep-1-FXR; P<0.05). These results indicated that when FXR was upregulated and activated, total and nuclear β-catenin protein levels were concurrently reduced in vitro and in vivo. FXR may inactivate the Wnt/β-catenin pathway by blocking the nuclear translocation of β-catenin, which may have been caused by the decreasing expression or increasing degradation of β-catenin.
Figure 4.
β-catenin expression in SK-Hep-1 cells and lung metastases. After SK-Hep-1-NC and SK-Hep-1-FXR cells were pretreated with GW4064 2 µM for 24 h at 37°C, total and nuclear proteins were isolated. (A) Expression of total β-catenin proteins and (B) nuclear β-catenin proteins were assessed via western blot analysis. The histograms indicate the levels of the protein determined from three independent experiments expressed as the mean ratio relative to that in the SK-Hep-1-NC cells following normalization to β-actin. Data are presented as mean ± standard error of the mean. (C) Representative images of FXR and β-catenin staining in lung metastases of nude mice via immunohistochemistry. Scale bar, 50 µm. Magnification, ×100. The immunoreactivity of tumor cells was quantified using scores based on the staining intensity and the proportion of positively stained tumor cells. Values are presented as mean ± standard error of the mean. *P<0.05. The ‘FXR’ group represents FXR-overexpressing SK-Hep-1-FXR cells and the ‘NC’ group represents the control SK-Hep-1-NC cells. FXR, farnesoid X receptor; NC, negative control.
Discussion
Nuclear receptor FXR is a ligand-mediated transcription factor (26). FXR was first identified as an endogenous bile acid sensor in the liver (27) and its role in protecting the liver via metabolic regulation and tumor inhibition is well established (5). However, the molecular mechanisms by which FXR protects against liver cancer are yet to be elucidated.
A previous study reported that FXR-knockout mice spontaneously developed liver tumors as they aged (28). Moreover, FXR re-expression or treatment with FXR agonists resulted in the repression of liver cancer cell proliferation and xenograft growth in nude mice (7,9). The results of the current study indicated that, in addition to suppressing liver cancer cell proliferation, FXR also inhibited migration and invasion in vitro. Furthermore, the results demonstrated that FXR inhibited local subcutaneous invasion and lung metastasis in the SK-Hep-1 ×enograft models. Impaired cell migration and reduced cell growth rate induced by FXR overexpression may contribute to the less aggressive response in these cells. These results are consistent with previous findings in human HCC specimens (7). Notably, whether other organs, in addition to the lungs, had metastases in the in vivo mouse model remains unclear. The site of injection largely defines where metastases will develop. For instance, tail vein injections of tumor cells results primarily in pulmonary metastases, while intrasplenic or portal vein injections are the most common sites employed to develop metastasis in the liver (29). Therefore, in the current study, the lungs were the major site of metastasis development following tail vein injections of SK-Hep-1-NC or SK-Hep-1-FXR cells; however, it would be worthwhile to investigate the other organs in future studies.
To further explore the possible molecular mechanism by which FXR exerts its metastatic suppressor function in liver cancer cells, gene expression profiles were analyzed in SK-Hep-1-FXR and SK-Hep-1-NC cells. Pathway analysis on differentially expressed genes revealed that FXR-upregulation in SK-Hep-1 cells significantly enriched various cellular signaling pathways, including ‘cell cycle’, ‘mismatch repair’, ‘TGF-β’, ‘JAK-STAT’ and ‘Wnt’. The role of FXR on the regulation of liver cancer cell cycle has been previously studied and the results demonstrated that FXR prevented the growth of liver cancer cells by inhibiting the mTOR-s6K pathway (9). The current study selected the Wnt/β-catenin pathway for further investigation, as the dysregulation of this pathway is a key molecular event in liver carcinogenesis and liver cancer progression (30).
The canonical Wnt-β/catenin network controls various biological processes in the liver. For example, when Wnt ligands bind the FZD and LRP 5/6 coreceptor complex, β-catenin phosphorylation and subsequent degradation is inhibited, resulting in β-catenin accumulation in the cytoplasm and translocation into the nucleus (21). In the nucleus, β-catenin binds lymphoid enhancing factor/T cell factor to form a transcriptional complex that promotes the transcription of its target genes (31). In the current study, mRNA microarray data revealed that FXR overexpression resulted in significant changes of mRNA levels of certain core components of this pathway, including downregulation of FZD5 and LRP5, upregulation of sFRP and DVL, and significantly decreased expression of β-catenin target genes (including CCND1, c-MYC, c-JUN and MMP9). RT-qPCR and western blotting confirmed the results of the microarray. These data support the hypothesis that FXR may represent an inhibitor of the Wnt/β-catenin pathway. Furthermore, the results demonstrated that FXR overexpression resulted in the concurrent reduction of total and nuclear β-catenin proteins in vitro and in vivo. Additionally, β-catenin mRNA levels were downregulated in SK-Hep-1-FXR cells. It should be noted that a pharmacological intervention on FXR expression to confirm these results was not performed since there is not a specific and potent FXR antagonist that is commercially available. Regardless, the impaired nuclear translocation of β-catenin by FXR upregulation may be caused by decreased expression or increased degradation of β-catenin; however, this requires further research. In summary, the results of the current study indicated that FXR inhibited the Wnt/β-catenin pathway, which may lead to a reduction of the migratory and invasive capabilities of Sk-Hep-1 cells.
Aberrant activation of the Wnt-β/catenin pathway has been reported in a variety of liver diseases, such as liver malignancies (hepatoblastoma, hepatocellular carcinoma and cholangiocarcinoma) and non-tumor liver diseases (liver fibrosis, liver steatosis, cholestasis and cystogenesis) (22). Increased nuclear β-catenin and c-MYC expression has been revealed to be associated with vascular invasion, which promotes HCC recurrence (11). Wnt/β-catenin signaling drives EMT, which is considered the key initial event in metastasis (32). Therefore, further investigation into the association between FXR, Wnt/β-catenin signaling and EMT is required. In addition to the Wnt/β-catenin pathway, the JAK-STAT pathway is a well-known oncogenic signaling pathway that promoted the invasive ability of HCC cells by modulating the expression of MMPs, including MMP-9 (10). Additionally, MMP-9 is a target gene of β-catenin (33). The results of the current study demonstrated that MMP9 mRNA was downregulated in response to FXR overexpression in SK-Hep-1 cells, which indicated a potential crosstalk between FXR, Wnt/β-catenin and JAK-STAT. Furthermore, the TGF-β pathway maintains liver homeostasis and participates in various liver diseases, including liver fibrosis and liver cancer (34). TGF-β serves a dual role in liver carcinogenesis, serving as a suppressor in early stages and a contributor to later progression by enhancing the migratory and invasive capacities of tumors (35). Whether other pathways such as TGF-β, JAK-STAT and mismatch repair also participate in FXR-regulated liver cancer cell invasion and metastasis will be researched in future studies.
Tumor cell dissemination via invasion or metastasis is the primary cause for the early recurrence of liver cancer in patients following curative resection, and is also the main obstacle to improving the prognosis of this disease (11). Due to the critical roles of FXR and the Wnt/β-catenin pathway in liver cancer tumorigenesis and progression, a more detailed understanding of the crosstalk between these factors is required for the development of effective liver cancer therapies.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- FXR
farnesoid X receptor
- EMT
epithelial- mesenchymal transition
- HCC
hepatocellular carcinoma
- LRP
low-density lipoprotein receptor-related protein
- FZD
frizzled
- sFRP
secreted FZD-related protein
Funding
The current work was supported by the Natural Science Foundation of Fujian Province (grant no. 2018J01827).
Availability of data and materials
Raw mRNA microarray data on the differentially expressed genes resulting from FXR overexpression was deposited into the Gene Expression Omnibus repository and available with the access no. ‘GSE149111’ via the following link: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE149111.
Authors' contributions
XH designed the study, interpreted data and wrote the manuscript. QL, NL, JL, HS and MG performed the experiments, analyzed the data and helped with the critical revision for important intellectual content. YZ and XW analyzed the data and prepared the figures. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The current study was approved by the Institutional Animal Care and Use Committee of Fujian Medical University (approval no. FJMUIRB-2011007).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Shang N, Wang H, Bank T, Perera A, Joyce C, Kuffel G, Zilliox MJ, Cotler SJ, Ding X, Dhanarajan A, et al. Focal adhesion kinase and β-catenin cooperate to induce hepatocellular carcinoma. Hepatology. 2019;70:1631–1645. doi: 10.1002/hep.30707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Massafra V, Pellicciari R, Gioiello A, van Mil SW. Progress and challenges of selective farnesoid X receptor modulation. Pharmacol Ther. 2018;191:162–177. doi: 10.1016/j.pharmthera.2018.06.009. [DOI] [PubMed] [Google Scholar]
- 4.Tran M, Liu Y, Huang W, Wang L. Nuclear receptors and liver disease: Summary of the 2017 basic research symposium. Hepatol Commun. 2018;2:765–777. doi: 10.1002/hep4.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang XF, Zhao WY, Huang WD. FXR and liver carcinogenesis. Acta Pharmacol Sin. 2015;36:37–43. doi: 10.1038/aps.2014.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang Y, Iakova P, Jin J, Sullivan E, Sharin V, Hong IH, Anakk S, Mayor A, Darlington G, Finegold M, et al. Farnesoid X receptor inhibits gankyrin in mouse livers and prevents development of liver cancer. Hepatology. 2013;57:1098–1106. doi: 10.1002/hep.26146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Su H, Ma C, Liu J, Li N, Gao M, Huang A, Wang X, Huang W, Huang X. Downregulation of nuclear receptor FXR is associated with multiple malignant clinicopathological characteristics in human hepatocellular carcinoma. Am J Physiol Gastrointest Liver Physiol. 2012;303:G1245–1253. doi: 10.1152/ajpgi.00439.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu N, Meng Z, Lou G, Zhou W, Wang X, Zhang Y, Zhang L, Liu X, Yen Y, Lai L, et al. Hepatocarcinogenesis in FXR−/− mice mimics human HCC progression that operates through HNF1alpha regulation of FXR expression. Mol Endocrinol. 2012;26:775–785. doi: 10.1210/me.2011-1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang X, Zeng Y, Wang X, Ma X, Li Q, Li N, Su H, Huang W. FXR blocks the growth of liver cancer cells through inhibiting mTOR-s6K pathway. Biochem Biophys Res Commun. 2016;474:351–356. doi: 10.1016/j.bbrc.2016.04.106. [DOI] [PubMed] [Google Scholar]
- 10.Aravalli RN, Steer CJ, Cressman EN. Molecular mechanisms of hepatocellular carcinoma. Hepatology. 2008;48:2047–2063. doi: 10.1002/hep.22580. [DOI] [PubMed] [Google Scholar]
- 11.Chen J, Rajasekaran M, Xia H, Zhang X, Kong SN, Sekar K, Seshachalam VP, Deivasigamani A, Goh BK, Ooi LL, et al. The microtubule-associated protein PRC1 promotes early recurrence of hepatocellular carcinoma in association with the Wnt/β-catenin signalling pathway. Gut. 2016;65:1522–1534. doi: 10.1136/gutjnl-2015-310625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dong LW, Yang GZ, Pan YF, Chen Y, Tan YX, Dai RY, Ren YB, Fu J, Wang HY. The oncoprotein p28GANK establishes a positive feedback loop in β-catenin signaling. Cell Res. 2011;21:1248–1261. doi: 10.1038/cr.2011.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fu Y, Zheng S, An N, Athanasopoulos T, Popplewell L, Liang A, Li K, Hu C, Zhu Y. β-catenin as a potential key target for tumor suppression. Int J Cancer. 2011;129:1541–1551. doi: 10.1002/ijc.26102. [DOI] [PubMed] [Google Scholar]
- 14.Schmalhofer O, Brabletz S, Brabletz T. E-cadherin, beta-catenin, and ZEB1 in malignant progression of cancer. Cancer Metastasis Rev. 2009;28:151–166. doi: 10.1007/s10555-008-9179-y. [DOI] [PubMed] [Google Scholar]
- 15.Wolfe A, Thomas A, Edwards G, Jaseja R, Guo GL, Apte U. Increased activation of the Wnt/β-catenin pathway in spontaneous hepatocellular carcinoma observed in farnesoid X receptor knockout mice. J Pharmacol Exp Ther. 2011;338:12–21. doi: 10.1124/jpet.111.179390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 17.Meng Z, Fu X, Chen X, Zeng S, Tian Y, Jove R, Xu R, Huang W. miR-194 is a marker of hepatic epithelial cells and suppresses metastasis of liver cancer cells in mice. Hepatology. 2010;52:2148–2157. doi: 10.1002/hep.23915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.National Research Council (US) Committee for the Update of the Guide for the Care and Use of Laboratory Animals, corp-author. 8th. National Academies Press; Washington, DC: 2011. The National Academies Collection: Reports Funded by National Institutes of Health: Guide for the Care and Use of Laboratory Animals. [Google Scholar]
- 19.Eun JR, Jung YJ, Zhang Y, Zhang Y, Tschudy-Seney B, Ramsamooj R, Wan YJ, Theise ND, Zern MA, Duan Y. Hepatoma SK Hep-1 cells exhibit characteristics of oncogenic mesenchymal stem cells with highly metastatic capacity. PLoS One. 2014;9:e110744. doi: 10.1371/journal.pone.0110744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heffelfinger SC, Hawkins HH, Barrish J, Taylor L, Darlington GJ. SK HEP-1: A human cell line of endothelial origin. In Vitro Cell Dev Biol. 1992;28A:136–142. doi: 10.1007/BF02631017. [DOI] [PubMed] [Google Scholar]
- 21.Shang X, Lin X, Alvarez E, Manorek G, Howell SB. Tight junction proteins claudin-3 and claudin-4 control tumor growth and metastases. Neoplasia. 2012;14:974–985. doi: 10.1593/neo.12942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perugorria MJ, Olaizola P, Labiano I, Esparza-Baquer A, Marzioni M, Marin JJG, Bujanda L, Banales JM. Wnt-β-catenin signalling in liver development, health and disease. Nat Rev Gastroenterol Hepatol. 2019;16:121–136. doi: 10.1038/s41575-018-0075-9. [DOI] [PubMed] [Google Scholar]
- 23.Kawano Y, Kypta R. Secreted antagonists of the Wnt signalling pathway. J Cell Sci. 2003;116:2627–2634. doi: 10.1242/jcs.00623. [DOI] [PubMed] [Google Scholar]
- 24.Jiang X, Charlat O, Zamponi R, Yang Y, Cong F. Dishevelled promotes Wnt receptor degradation through recruitment of ZNRF3/RNF43 E3 ubiquitin ligases. Mol Cell. 2015;58:522–533. doi: 10.1016/j.molcel.2015.03.015. [DOI] [PubMed] [Google Scholar]
- 25.Clevers H, Nusse R. Wnt/β-catenin signaling and disease. Cell. 2012;149:1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 26.Koutsounas I, Giaginis C, Theocharis S. Farnesoid X receptor (FXR) from normal to malignant state. Histol Histopathol. 2012;27:835–853. doi: 10.14670/HH-27.835. [DOI] [PubMed] [Google Scholar]
- 27.Makishima M, Okamoto AY, Repa JJ, Tu H, Learned RM, Luk A, Hull MV, Lustig KD, Mangelsdorf DJ, Shan B. Identification of a nuclear receptor for bile acids. Science. 1999;284:1362–1365. doi: 10.1126/science.284.5418.1362. [DOI] [PubMed] [Google Scholar]
- 28.Yang F, Huang X, Yi T, Yen Y, Moore DD, Huang W. Spontaneous development of liver tumors in the absence of the bile acid receptor farnesoid X receptor. Cancer Res. 2007;67:863–867. doi: 10.1158/0008-5472.CAN-06-1078. [DOI] [PubMed] [Google Scholar]
- 29.Khanna C, Hunter K. Modeling metastasis in vivo. Carcinogenesis. 2005;26:513–523. doi: 10.1093/carcin/bgh261. [DOI] [PubMed] [Google Scholar]
- 30.Liang Y, Feng Y, Zong M, Wei XF, Lee J, Feng Y, Li H, Yang GS, Wu ZJ, Fu XD, Feng GS. β-catenin deficiency in hepatocytes aggravates hepatocarcinogenesis driven by oncogenic β-catenin and MET. Hepatology. 2018;67:1807–1822. doi: 10.1002/hep.29661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Macdonald BT, Semenov MV, He X. SnapShot: Wnt/beta-catenin signaling. Cell. 2007;131:1204. doi: 10.1016/j.cell.2007.11.036. [DOI] [PubMed] [Google Scholar]
- 32.Gonzalez DM, Medici D. Signaling mechanisms of the epithelial-mesenchymal transition. Sci Signal. 2014;7:re8. doi: 10.1126/scisignal.2005189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu W, Li L, Zheng F, Yang W, Zhao S, Tian C, Yin W, Chen Y, Guo W, Zou L, Deng W. β-catenin cooperates with CREB binding protein to promote the growth of tumor cells. Cell Physiol Biochem. 2017;44:467–478. doi: 10.1159/000485013. [DOI] [PubMed] [Google Scholar]
- 34.Fabregat I, Moreno-Càceres J, Sánchez A, Dooley S, Dewidar B, Giannelli G, Ten Dijke P. TGF-β signalling and liver disease. FEBS J. 2016;283:2219–2232. doi: 10.1111/febs.13665. [DOI] [PubMed] [Google Scholar]
- 35.Fabregat I, Caballero-Díaz D. Transforming growth factor-β-induced cell plasticity in liver fibrosis and hepatocarcinogenesis. Front Oncol. 2018;8:357. doi: 10.3389/fonc.2018.00357. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Raw mRNA microarray data on the differentially expressed genes resulting from FXR overexpression was deposited into the Gene Expression Omnibus repository and available with the access no. ‘GSE149111’ via the following link: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE149111.