Abstract
The aim of the present study was to use The Cancer Genome Atlas (TCGA) database to identify tumor neoantigens, combined with a bioinformatics analysis to design and analyze antigen epitope peptides. Epitopes were screened using immunogenicity tests to identify the ideal epitope peptides to target tumor neoantigens, which can specifically activate the immune response of T cells. The high-frequency mutation loci (top 10) of colorectal, lung and liver cancer genes were screened using TCGA database. The antigenic epitope peptides with high affinity for major histocompatibility complex molecules were selected and synthesized using computer prediction algorithms, and were subsequently detected using flow cytometry. The cytotoxicity of specific cytotoxic T lymphocytes (CTLs) on peptide-loaded T2 cells was initially verified using interferon IFN-γ detection and a calcein-acetoxymethyl (Cal-AM) release assay. Tumor cell lines expressing point mutations in KRAS, TP53 and CTNNB1 genes were constructed respectively, and the cytotoxicity of peptide-induced specific CTLs on wild-type and mutant tumor cells was verified using a Cal-AM release assay and carboxyfluorescein succinimidyl ester-propidium iodide staining. The high-frequency gene mutation loci of KRAS proto-oncogene (KRAS) G12V, tumor protein p53 (TP53) R158L and catenin β1 (CTNNB1) K335I were identified in TCGA database. A total of 3 groups of wild-type and mutant peptides were screened using a peptide prediction algorithm. The CTNNB1 group had a strong affinity for the human leukocyte antigen-A2 molecule, as determined using flow cytometry. The IFN-γ secretion of specific CTLs in the CTNNB1 group was the highest, followed by the TP53 and the KRAS groups. The killing rate of mutant peptide-induced specific CTLs on peptide-loaded T2 cells in the CTNNB1 group was higher compared with that observed in the other groups. The killing rate of specific CTLs induced by mutant peptides present on tumor cells was higher compared with that induced by wild-type peptides. However, when compared with the TP53 and KRAS groups, specific CTLs induced by mutant peptides in the CTNNB1 group had more potent cytotoxicity towards mutant and wild-type tumor cells. In conclusion, point mutant tumor neoantigens screened in the three groups improved the cytotoxicity of specific T cells, and the mutant peptides in the CTNNB1 group were more prominent, indicating that they may activate the cellular immune response more readily.
Keywords: TCGA, neoantigen, mutation, epitope peptide, cytotoxicity
Introduction
Tumor immunotherapy has become an effective treatment option following surgery, chemotherapy and radiotherapy in some specific types of cancer, such as acute lymphoblastic leukemia (1). Cancer immunotherapy includes immune checkpoint inhibitors, adoptive cell transfer therapy (ACT), tumor-specific vaccines and small molecule inhibitors (1).
Tumor antigens can be divided into tumor-specific antigens (TSAs), tumor-associated antigens (TAAs) and cancer-testis antigens. TAAs are overexpressed in tumor cells; however, they are typically present in low amounts in healthy cells. During thymic development, T cells undergo positive and negative selection, in which T cells with high affinity to autoantigens are eliminated naturally, and only the T cells with low affinity to autoantigens develop and mature (2). At present, vaccines targeting TAAs are often affected by central or peripheral immune tolerance or cause serious side effects (2).
New tumor antigens (neoantigens), also known as TSAs, are novel antigens encoded by a mutation in tumor cells, which are not expressed in healthy host cells. Major somatic mutations, such as gene fusion, point and deletion mutations, can produce abnormal proteins (3). These proteins can be specifically recognized by T cells following presentation, resulting in an immune response (3). Therefore, immunotherapy targeting different tumor neoantigens has become a novel prospect in the treatment of solid tumors.
Individualized immunotherapy and precision medicine have emerged as the future of malignant tumor treatment. Individualized tumor immunotherapy is a novel type of treatment based on identifying specific tumor neoantigens expressed in each individual patient. This type of therapy uses sequencing and bioinformatic analyses on specific tumor tissues of patients (4), and includes the use of personalised vaccines or ACT to activate the immune response using a combination of experimental screening and synthesis of peptides (5).
In the present study, genes with a high frequency of point mutations in colorectal, lung and liver cancer were screened using The Cancer Genome Atlas (TCGA) database, and three groups of antigen epitope peptides with a strong affinity for major histocompatibility complex (MHC) molecules were selected using a computer prediction algorithm, which identified mutant peptides of KRAS proto-oncogene (KRAS) G12V, tumor protein p53 (TP53) R158L and catenin β1 (CTNNB1) K335I. The ideal epitope peptides that could specifically activate T-cell immune responses were screened using immunogenicity tests. The present study aimed to perform a preliminary screening of tumor neoantigens and immunogenic epitopes, to provide a foundation for individualized immunotherapy and precision medical treatment in the late stages of oncogenesis (6).
Materials and methods
Reagents
The human HCT116 colorectal cancer, T2 cell lines, human HepG2 liver cancer and human NCI-H292 lung cancer cell lines were obtained from and authenticated using short-tandem repeat analysis by The Cell Bank of Type Culture Collection of the Chinese Academy of Sciences. Human peripheral blood was collected from laboratory healthy volunteers [pre-determined human leukocyte antigen (HLA)-A2 positive] and written informed consent was provided by each volunteer.
The synthesis of wild-type and mutant peptides was performed by Shanghai Biology Co., Ltd. (http://www.chinapeptides.com/). The upstream and downstream primers reverse transcription-quantitative (RT-q)PCR were synthesized by Beijing Qingke Biotechnology Co., Ltd.. Lipofectamine® 3000 and TRIzol® were purchased from Invitrogen (Thermo Fisher Scientific, Inc.). The SYBR Green qPCR mix was purchased from Promega Corporation. BamH1 and Sal1 restriction endonucleases were purchased from New England BioLabs, Inc. CD3 (cat. no. 05131-20) and CD28 (cat. no. 10311-20) monoclonal antibodies were purchased from PeproTech, Inc. Carboxyfluorescein succinimidyl ester (CFSE)-fluorescein isothiocyanate (FITC) fluorescent dye, phycoerythrin (PE) anti-human CD3 (cat. no. 300307), FITC anti-human CD4 (cat. no. 357405), FITC anti-human CD8 (cat. no. 344703), PE-Cyanine 7 (Cy7) anti-human CD25 (cat. no. 302611), FITC anti-human HLA-A2 (cat. no. 343303) and granzyme B antibodies (cat. no. 515403) were purchased from BioLegend, Inc. X–VIVO culture medium was purchased from Lonza Group, Ltd.. The plasmid extraction kit was purchased from Omega Bio-Tek, Inc. The ELISA kit for IFN-γ (cat. no. 1110002) was purchased from Neobioscience Technology Co., Ltd.. Calcein-acetoxymethyl (Cal-AM) and propidium iodide (PI) fluorescent dyes were purchased from the Japan Research Institute, Ltd.. Recombinant human (rh)interleukin (IL)-2, rhIL-7, rhIL-15, rhIL-4, tumor necrosis factor (rhTNF)-α and granulocyte-macrophage colony-stimulating factor (rhGM-CSF) all were recombinant human cytokines, which were purchased from Sangon Biotech Co., Ltd.
TCGA database screening of gene mutation sites
The ‘mutant gene’ column of colon, lung and liver cancer was searched in TCGA database (https://portal.gdc.cancer.gov/), and the top 10 genes with the highest mutation frequency in these three types of cancer were identified. The mutation sites with a single base substitution were screened out and used for further evaluation.
The immune presentation ability of epitope peptides evaluated using the SYFPEITHI, BIMAS prediction system
The epitope peptide score of colon, lung and liver cancer containing a gene mutation site was predicted (9 aa, HLAA201). According to the location of mutation in wild-type and mutant peptides, Epitope peptides with higher scores (≥8 points) before (wild-type) and after gene locus mutation (mutant) were screened out for further evaluation. A total of 6 epitope peptides (Table I) were screened and divided into the following three groups: Wild-type and mutant peptides of the KRAS group, TP53 group, and the CTNNB1 group, respectively, which were synthesized by the aforementioned company. (SYFPEITHI prediction system: The immune presentation ability of epitope peptides could be evaluated by these prediction systems. The prediction score of epitope peptide indicated the ability of immune presentation (7,8).
Table I.
WT and mutant epitope peptides.
| Gene | Mutation locus | WT epitope peptide sequence (name) | Mutant epitope peptide sequence (name) |
|---|---|---|---|
| KRAS | G12V | YKLVVVGAG (KRAS WT) | YKLVVVGAV (KRAS mutant) |
| TP53 | R158L | VRAMAIYKQ, (TP53 WT) | VLAMAIYKQ (TP53 mutant) |
| CTNNB1 | K335I | IMRTYTYEK, (CTNNB1 WT) | IMRTYTYEI (CTNNB1 mutant) |
Underlined amino acid letter indicates the differences between the WT and mutant peptide. WT, wild-type; KRAS, KRAS proto-oncogene; TP53, tumor protein 53; CTNNB1, catenin β1.
Verifying the affinity of epitope peptides to HLA-A2 molecules
T2 cells are B cells with the HLA-A2 gene. These cells are deficient in the antigen polypeptide transporter, which is required for endogenous antigen presentation, as they are unable to present their own antigens on the cell surface. Therefore, MHC I molecules on the T2 cell surface are often used to present foreign antigens for immune cell recognition in immunological functional testing experiments (9,10).
T2 cells were inoculated in a 6-well plate with RPMI-1640 containing 10% FBS in 37°C and 5% CO2 incubators, at a density of 1×106 cells/ml. A total of 3 groups of the aforementioned wild-type and mutant epitope peptides (20 ug/ml) were added to the 6-well plate with a well for each peptide. Following a 4-h incubation at 37°C, the mean fluorescence intensity (MFI) was detected using a Gallios flow cytometer (Beckman Coulter, Inc.) using a labelled HLA-A2-FITC fluorescent antibody (ready to use, incubation in 4°C for 20 min). The affinity was measured using the flowing equation: Fluorescence index=(MFI of epitope peptide-background MFI)/background MFI. Fluorescence index >1.0 indicated the strong stability, fluorescence index <0.5 indicated the weakest stability.
Specific cytotoxic T lymphocytes (CTLs) induced by epitope peptides in vitro
Dendritic cells (DCs) cultured in vitro
Human peripheral blood mononuclear cells obtained from laboratory healthy volunteers (PBMCs; HLA-A2+, 300 × g centrifugation in 37°C for 30 min) were cultured in 37°C and 5% CO2 incubators for 4 h. The culture medium was removed and the adherent cells were isolated. rhGM-CSF [1,000 international units (IU)/ml] and rhIL-4 (500 IU/ml) were added to stimulate the growth of monocytes. On the 3rd and 6th day, 10 µg/ml rhTNF-α was added to promote the maturation of DCs and supplemented with cytokines [rhGM-CSF (1,000 IU/ml), rhIL-4 (500 IU/ml), rhTNF-α (10 µg/ml)]. From the 6th day, rhTNF-α (10 µg/ml) was added every day until the 9th day, and mature DCs were harvested by washing the cells with PBS.
Specific CTLs induced by wild-type and mutant peptides in each group
In the first round of stimulation, PBMCs (2×106 cells/well) were inoculated in a 6-well plate and the corresponding three groups of wild-type peptides (20 µg/ml), mutant peptides (20 µg/ml) and β2-microglobulin (PeproTech, 3 µg/ml) were added to each well. IL-2 (100 IU/ml) and TNF-α (800 IU/ml) were added to each well every 2 days for 7 days. In the second round of stimulation, mature DCs were added to the three groups of wild-type and mutant peptides (20 µg/ml) and subsequently co-cultured with PBMCs at a ratio of 1:10. The proliferation of T lymphocytes was stimulated by adding the cytokines, IL-2, IL-7 and IL-15 (10 ng/ml), every 3 days for 7 days. In the third round of stimulation, CD3 and CD28 monoclonal antibodies (2 µg/ml) were added to stimulate the proliferation of specific T lymphocytes. Specific effective T cells were collected on the 21st day (300 × g centrifugation in 37°C for 5 min) for functional detection. Therefore, six corresponding specific T lymphocyte subsets targeting three groups of wild-type and mutant peptides were obtained (Table II).
Table II.
Specific CTLs induced by WT and mutant peptides.
| Group | WT epitope peptide sequence to induce CTLs (name) | Mutant epitope peptide sequence to induce CTLs (name) |
|---|---|---|
| KRAS | YKLVVVGAG (CTLKRAS WT) | YKLVVVGAV (CTLKRAS mutant) |
| TP53 | VRAMAIYKQ (CTLTP53 WT) | VLAMAIYKQ (CTLTP53 mutant) |
| CTNNB1 | IMRTYTYEK (CTLCTNNB1 WT) | IMRTYTYEI (CTLCTNNB1 mutant) |
Underlined amino acid letter indicates the differences between the WT and mutant peptide. WT, wild-type; CTLs, cytotoxic T lymphocytes; KRAS, KRAS proto-oncogene; TP53, tumor protein 53; CTNNB1, catenin β1.
Detection of subsets of specific T lymphocytes
The CTLs from culture on day 21 were labelled with ready to use CD3-PE, CD8-FITC, CD4-FITC and CD25-PE-Cy7-flow fluorescent antibodies and incubated at 4°C for 20 min in the dark. Each tube was washed twice with PBS containing 2% FBS (Gibco; Thermo Fisher Scientific, Inc.). The supernatant was discarded following centrifugation at 400 × g in 37°C for 5 min. The cells were resuspended with PBS in flow tube and analysed using flow cytometry.
Detection of CTL cell proliferation
CTLs from culture on day 14 were placed into Eppendorf® tubes and incubated with CFSE fluorescent dye (final concentration, 1 µmol/l) for 10 min at 37°C. CTLs were washed twice with X–VIVO medium and cultured into a 6-well plate at a density of 1×106 cells/well in X–VIVO medium in an incubator at 37°C with 5% CO2. A total of 1×105 cells were detected using flow cytometry for 4 consecutive days from the 16th to the 19th day. The cell proliferation was analysed using the Modfit software (Verity Software House, v5.0.9). The software would automatically fit cell different generations, when the cells were in the state of proliferation and division.
ELISA for the detection of IFN-γ secretion in T lymphocytes stimulated by epitope peptides in vitro
T2 cells treated with 3 groups of epitope peptides and the peptide-induced specific T-lymphocytes were mixed and cultured at a density of 1×106 cells/well at 37°C for 24 h. The cell culture supernatant was extracted from each group (500 µl) following centrifugation at 400 × g at 37°C for 5 min. Detection of IFN-γ secretion using ELISA was performed according to the manufacturer's protocol. The experimental groups are shown in Table III.
Table III.
T2 cells loaded with WT or mutant peptides co-cultured with peptide-induced CTLs.
| Group | Peptide (WT) + CTL (WT) | Peptide (mutant) + CTL (mutant) |
|---|---|---|
| KRAS | T2peptide KRAS WT + CTLKRAS WT | T2peptide KRAS mutant + CTLKRAS mutant |
| TP53 | T2peptide TP53 WT + CTLTP53 WT | T2peptide TP53 mutant + CTLTP53 mutant |
| CTNNB1 | T2peptide CTNNB1 WT + CTLCTNNB1 WT | T2peptide CTNNB1 mutant + CTLCTNNB1 mutant |
WT, wild-type; CTL, cytotoxic T lymphocyte; KRAS, KRAS proto-oncogene; TP53, tumor protein 53; CTNNB1, catenin β1.
Preliminary detection of cytotoxic activity of specific CTLs using Cal-AM release assay
The greater release of Cal-AM from tumor cells, the stronger the cytotoxic activity. T2 cells were stained with 2 µg/ml Cal-AM at 37°C for 10 min in the dark. Following centrifugation in 300 × g at 37°C for 5 min, twice with sterile PBS, T2 cells were added to the three groups of epitope peptides (20 µg/ml) and incubated at 37°C for 4 h. Target cells were inoculated in 96-well plates (1×105 cells/ml; 100 µl/well). Effective CTLs were added according to the effective target ratio of 20:1 with a volume of 100 µl/well. The maximum release group (2% Triton X-100 treated T2 cells for 24 h) and the self-releasing group (T2 cells only) were prepared. The final volume of each well was 200 µl, and each group was analyzed in triplicate. Following co-culture at 37°C for 24 h (including the experimental group, the maximum release group, the self-releasing group), 80 µl/well supernatant was collected by centrifugation in 300 × g at 37°C for 5 min, and the optical density (OD) values of each group were measured using an automatic microplate reader (Thermo Fisher Scientific, Inc.) at an excitation wavelength of 485 nm and an emission wavelength of 536 nm. The killing rate was calculated as follows: Killing rate (%)=(ODexperimental group-ODself-release group)/(ODmaximum release group-ODself-release group) ×100. The experimental groups are shown in Table III.
For the antibody blocking experiments, homotypic control antibody (FITC Mouse lgG2b, Isotype Control, ready to use, cat. no. 555057, eBioscience; Thermo Fisher Scientific, Inc.) or HLA-A2 antibody were added to the target cells (T2 cells) and incubated at 37°C with 5% CO2 for 60 min. Target cells were added to 96-well plates (1×105 cells/ml; 100 µl/well) with the three groups of mutant peptide-induced CTLs (1×105 cells/ml; 100 µl/well), which were tested according to the aforementioned Cal-AM release assay.
Construction of recombinant eukaryotic expression plasmids
TRIzol® was used to extract RNA from HCT116 colon cancer cells, and the RNA was reverse transcribed into cDNA. The reverse transcription conditions were as follows: 25°C For 5 min, 42°C for 60 min, 70°C for 15 min, and 4°C for 10 min (GoScript™ Reverse Transcription Mix, Promega Corportation). Upstream and downstream primers for mutant genes were designed and synthesized (Table IV). Using the site-directed mutagenesis PCR method (11), the 12th amino acid translated from the KRAS gene was transformed from glycine (G) to valine (V) by changing the codon from GGT to GTT. The PCR amplification conditions were as follows: 98°C For 10 sec, 58°C for 5 sec and 72°C for 90 sec, for 35 cycles (PrimeSTAR Max DNA Polymerase, Takara Biotechnology Co., Ltd.). The high-fidelity enzyme amplification product was identified using 1% agarose gel electrophoresis with ethidium bromide. The mutant KRAS G12V gene fragment was ligated with the pIRES2-EGFP plasmid (Guangzhou Aiji Biotechnology Biological Co., Ltd.) using BamH1 and Sal1 restriction endonucleases (New England BioLabs, Inc.) at 16°C overnight. The recombinant plasmid, KRAS G12V-pIRES2-EGFP, was obtained from the correctly sequenced (sequenced by Guangzhou Qingke Biotechnology Co., Ltd.) genetically engineered bacteria (DH5α) using a removing endotoxin plasmid extraction kit (Omega Bio-Tek, Inc.).
Table IV.
Primers used in the present study.
| Primer name | Sequence, 5′→3′ | Product, bp |
|---|---|---|
| KRAS-G12V-F | ATGACTGAATATAAACTTGTGGTAGTTGGAGCTGTTGGCGTAGGCA | 46 |
| KRAS-G12V-R | TTACATTATAATGCATTTTTTAATTTTCACACAGC | 35 |
| TP53-R158L-F1 | ATGGAGGAGCCGCAGTCAGATCCTA | 25 |
| TP53-R158L-R1 | TAGATGGCCATGGCGAGGACGC | 22 |
| TP53-R158L-F2 | CGCGTCCTCGCCATGGCCATCT | 22 |
| TP53-R158L-R2 | TCAGTCTGAGTCAGGCCCTTCTGT | 24 |
| CTNNB1-K335I-F1 | ATGGCTACTCAAGCTGATTTGATGGAGTT | 29 |
| CTNNB1-K355I-R1 | TGGTCCACAGTAGTATTTCGTAAGTATAGGTCCT | 34 |
| CTNNB1-K335I-F2 | TATACTTACGAAATACTACTGTGGACCACAAGCAGAGT | 38 |
| CTNNB1-K355I-R2 | TTACAGGTCAGTATCAAACCAGGCCAGCT | 29 |
| KRAS-qPCR-F | ACTTGTGGTAGTTGGAGCTGGTGGCGTAGG | 30 |
| KRAS-qPCR-R | GCACTGTACTCCTCTTGACCTGCTGTGTCG | 30 |
| CTNNB1-qPCR-F | GGCTTGGAATGAGACTGCTGAT | 22 |
| CTNNB1-qPCR-R | GCTGATTGCTGTCACCTGGAG | 21 |
| TP53-qPCR-F | CCGTCTGGGCTTCTTGCATT | 20 |
| TP53-qPCR-R | CGCCTCACAACCTCCGTCAT | 20 |
| GAPDH-F | GGTGAAGGTCGGAGTCAACG | 20 |
| GAPDH-R | CAAAGTTGTCATGGATGHACC | 20 |
F, forward; R, reverse; KRAS, KRAS proto-oncogene; TP53, tumor protein 53; CTNNB1, catenin β1; G, glycine; V, valine; R, arginine; L, leucine K, lysine; I, isoleucine.
The RNA of NCI-H292 lung cancer cells was extracted and reverse transcribed into cDNA as aforementioned. Using the site-directed mutagenesis PCR method, TP53 mutation gene was needed to be amplified by two pairs PCR primers (TP53-F1,R1,F2,R2). The 158th amino acid translated from the TP53 gene was transformed from arginine (R) to leucine (L) by changing the codon from CGC to CTC. The steps of plasmid construction were the same as aforementioned, obtaining the recombinant plasmid TP53-R158L-pIRES2-EGFP.
The RNA of HepG2 liver cancer cells was extracted and reverse transcribed into cDNA as aforementioned. Using the site-directed mutagenesis PCR method, CTNNB1 mutation gene was needed to be amplified by two pairs PCR primers (CTNNB1-F1,R1,F2,R2). The 335th amino acid translated from the CTNNB1 gene was transformed from lysine (K) to isoleucine (I) by changing the codon from AAA to ATA. The steps of plasmid construction are the same as aforementioned, obtaining the recombinant plasmid CTNNB1-K335I-pIRES2-EGFP. The PCR primers used are shown in Table IV.
Sequencing comparison between the KRAS G12V, TP53 R158L, CTNNB1 K335I mutant gene clone and their respective wild-type gene of KRAS, TP53, CTNNB1, was analyzed through the tools of Sequence Alignment in the Vector builder analysis system (https://en.vectorbuilder.com/tool/sequence-alignment.html).
Transfection of tumor cells with recombinant plasmid
The extracted recombinant plasmids KRAS-G12V- pIRES2-EGFP, TP53-R158L-pIRES2-EGFP and CTNNB1-K335I-pIRES2-EGFP (800 ug/ul) were transfected into HCT116 colon cancer cells (HLA-A2+; 1×106 cells/well), NCI-H292 lung cancer cells (HLA-A2+; 1×106 cells/well) and HepG2 liver cancer cells (HLA-A2+; 1×106 cells/well), respectively, using Lipofectamine® 3000 according to the manufacturer's protocol. The expression of green fluorescent protein (GFP) was observed using an inverted fluorescence microscope at 200× magnification (Olympus IX51; Olympus Corporation) 24 h following transfection (maximum excitation wavelength, 490 nm), and the transfection efficiency was detected using flow cytometry. The positive cells expressing GFP were collected using a flow cell sorter (MoFlo XDP; Beckman Coulter, Inc.).
Expression of mutated genes in tumor cells using RT-qPCR
KRAS-G12V-pIRES2-EGFP, TP53-R158L-pIRES2-EGFP and CTNNB1-K335I-pIRES2-EGFP recombinant plasmids were transfected into HCT116, NCI-H292 and HepG2 cells, respectively. RNA was extracted from wild-type and mutant tumor cells expressing recombinant plasmids and reverse transcribed to cDNA, 24 h following transfection, as aforementioned. The expression of the mutant genes in transfected tumor cells was detected using RT-qPCR and the SYBR Green qPCR mix (primers are shown in Table IV). The following thermocycling conditions (Roche Diagnostics) were used: Initial denaturation at 95°C for 2 min, followed by 40 cycles of 95°C for 15 sec, 55°C for 30 sec and 72°C for 15 sec. GAPDH was used as the internal control. The mRNA expression levels of each gene were calculated using the 2−ΔΔCq method (12).
Cytotoxic effect of specific CTLs on tumor cells using the Cal-AM release assay
The 3 groups of mutant and wild-type tumor cells were collected into Eppendorf® tubes and incubated with 2 µg/ml Cal-AM for 10 min at 37°C in the dark. The target cells were seeded into a 96-well plate at a density of 1×105 cells/ml (100 µl/well). The corresponding peptide-induced CTLs were added for co-culture for 24 h at 37°C (1×105 cells/ml; 100 µl/well). The remaining steps of the Cal-AM release assay used was as aforementioned. The experimental groups are shown in Table V.
Table V.
Co-culture of CTLs induced by WT and mutant tumor cells and peptides.
| Group | Cancer cells (type) + CTLWT | Cancer cells (type) + CT mutant |
|---|---|---|
| KRAS | HCT116 (WT) + CTLKRAS WT | HCT116 (WT) + CTLKRAS mutant |
| HCT116 (mutant) + CTLKRAS WT | HCT116 (mutant) + CTLKRAS mutant | |
| TP53 | NCI-H292 (WT) + CTLTP53 WT | NCI-H292 (WT) + CTLTP53 mutant |
| NCI-H292 (mutant) + CTLTP53 WT | NCI-H292 (mutant) + CTLTP53 mutant | |
| CTNNB1 | HepG2 (WT) + CTLCTNNB1 WT | HepG2 (WT) + CTLCTNNB1 mutant |
| HepG2 (mutant) + CTLCTNNB1 WT | HepG2 (mutant) + CTLCTNNB1 mutant |
WT, wild-type; CTL, cytotoxic T lymphocyte; KRAS, KRAS proto-oncogene; TP53, tumor protein 53; CTNNB1, catenin β1.
Cytotoxic effect of specific CTLs on tumor cells using the CFSE-PI assay
The 3 groups of wild-type and mutant tumor cells were respectively collected into Eppendorf® tubes, stained with CFSE fluorescent dye (final concentration, 1 µmol/l), for 10 min at 37°C in the dark and subsequently washed twice with PBS. The tumor cells of each group labelled with CFSE dye were added to a 24-well plate at a density of 1×105 cells/ml in X–VIVO medium. The mature specific T lymphocytes (1×105 cells/ml) were co-cultured at 37°C and 5% CO2. The cells were collected and stained with PI dye (final concentration, 10 µg/ml, stained at 37°C for 10 min), following 24 h of culture. The ratio of double-positive cells was detected using flow cytometry. The experimental groups are shown in Table VI.
Table VI.
Co-culture of WT and mutant tumor cells with peptide-induced CTLs.
| Group | Peptide (WT) + CTL (WT) | Peptide (mutant) + CTL (mutant) |
|---|---|---|
| KRAS | HCT116 (WT) + CTLKRAS WT | HCT116 (mutant) + CTLKRAS mutant |
| TP53 | NCI-H292 (WT) + CTLTP53 WT | NCI-H292 (mutant) + CTLTP53 mutant |
| CTNNB1 | HepG2 (WT) + CTLCTNNB1 WT | HepG2 (mutant) + CTLCTNNB1 mutant |
WT, wild-type; CTL, cytotoxic T lymphocyte; KRAS, KRAS proto-oncogene; TP53, tumor protein 53; CTNNB1, catenin β1.
Statistical analysis
All data were expressed as the mean ± SD, which were repeated in triplicate. Comparisons among multiple groups were performed using two-way ANOVA and the Bonferroni's method (the post hoc test) as indicated using the GraphPad Prism software v5.0 (GraphPad Software, Inc.). Student's unpaired t-test was used to compare the means between two groups. P<0.05 was considered to indicate a statistically significant difference.
Results
Screening epitope peptides using TCGA database
The loci of single base substitutions in colon, lung and liver cancer were screened using TCGA database (Table VII). Based on SYFPEITHI and BIMAS prediction and evaluation system, three groups of epitope peptides containing wild-type and mutant gene loci, with increased immune-presentation capability, were selected and synthesized (9 aa, HLAA201). These included the epitope peptide, YKLVVVGAG, expressed by the KRAS wild-type gene; YKLVVVGAV, expressed by the KRAS mutant gene; the VRAMAIYKQ wild-type peptide, expressed by the TP53 gene; the VLAMAIYKQ mutant peptide, expressed by the TP53-mutant gene; the IMRTYTYEK wild-type peptide, expressed by the CTNNB1 gene; and the IMRTYTYEI mutant peptide, expressed by the CTNNB1-mutant gene (Table VIII).
Table VII.
Cancer gene mutation sites in The Cancer Genome Atlas database.
| A, Mutations in liver cancer | ||
|---|---|---|
| Gene | Mutant locus | Mutation frequency (the number of patient cases) |
| TP53 | R249S | 11/364 |
| CTNNB1 | S45P | 11/364 |
| CTNNB1 | D32G | 7/364 |
| CTNNB1 | K335I | 6/364 |
| CTNNB1 | S33C | 6/364 |
| B, Mutation in lung cancer | ||
| Gene | Mutant locus | Mutation frequency |
| KRAS | G12C | 62/1062 |
| KRAS | G12V | 39/1062 |
| EGFR | L858R | 23/1062 |
| PIK3CA | E545K | 21/1062 |
| TP53 | R158L | 20/1062 |
| KRAS | G12D | 20/1062 |
| PIK3CA | E542K | 18/1062 |
| C, Mutations in colon cancer | ||
| Gene | Mutant locus | Mutation frequency |
| KRAS | G12D | 60/537 |
| KRAS | G12V | 50/537 |
| BRAF | V600E | 50/537 |
| KRAS | G13D | 41/537 |
| TP53 | R175H | 39/537 |
| PIK3CA | E545K | 35/537 |
KRAS, KRAS proto-oncogene; TP53, tumor protein 53; CTNNB1, catenin β1; PIK3CA, phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit α; EGFR, epidermal growth factor receptor; BRAF, B-Raf proto-oncogene, serine/threonine kinase. R, arginine; S, serine; P, proline; D, aspartic acid; G, glycine; K, lysine; I, isoleucine; C, cysteine; V, valine; L, leucine; E, glutamic acid; H, histidine.
Table VIII.
Wild-type and mutant epitope peptides.
| Mutant locus | Wild-type epitope peptide | |||||
|---|---|---|---|---|---|---|
| Type of cancer | Gene | Mutant epitope peptide | Sequence | Score | Sequence | Score |
| Colon | KRAS | G12V | YKLVVVGAG | 9 | YKLVVVGAV | 19 |
| Lung | TP53 | R158L | VRAMAIYKQ | 10 | VLAMAIYKQ | 20 |
| Liver | CTNNB1 | K335I | IMRTYTYEK | 16 | IMRTYTYEI | 24 |
Underlined amino acid letter indicates the differences between the WT and mutant peptide. KRAS, KRAS proto-oncogene; TP53, tumor protein 53; CTNNB1, catenin β1; R, arginine; V, valine; G, glycine; K, lysine; I, isoleucine.
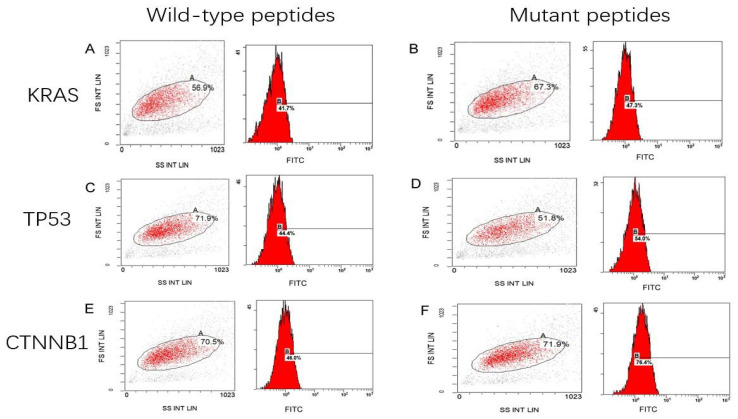
Affinity of epitope peptides with HLA-A2 molecule
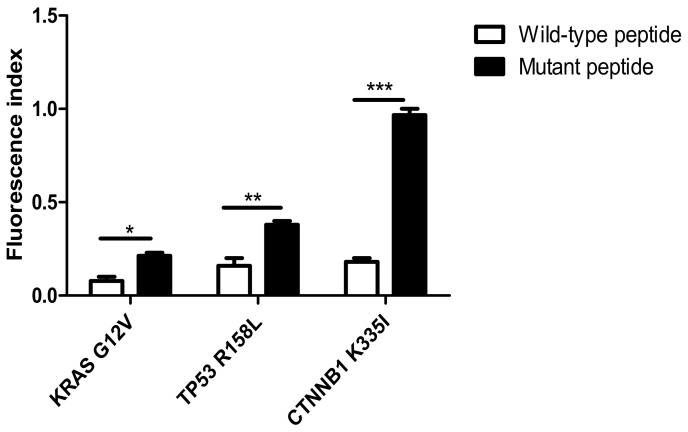
As shown in Fig. 1, the HLA-A2 concentration, as expressed by T2 cells treated with the KRAS mutant peptide (Fig. 1B), was increased by 5.6±4% compared with that in the cells with wild-type peptide (Fig. 1A). The expression of HLA-A2 was 9.6±3% higher in T2 cells with the TP53 mutant peptides (Fig. 1D) compared with that in the cells with the wild-type peptides (Fig. 1C). The expression of HLA-A2 was significantly increased by 30.4±2% in T2 cells loaded with CTNNB1 mutant peptides (Fig. 1F) compared with that with wild-type peptides (Fig. 1E). Notably, the fluorescence index of the CTNNB1 group increased the most (P<0.001; Fig. 2). The present results indicated that the mutant peptides promoted the higher expression of the HLA-A2 molecule in T2 cells, which formed a compound with high affinity and strong stability with MHC I molecules. The affinity observed in the TP53 mutant epitope peptides was weakest (fluorescence index <0.5), with the weakest occurring in the KRAS group (Fig. 2).
Figure 1.
Mean fluorescence intensity of peptide-treated T2 cells. T2 cells treated with (A) KRAS wild-type peptide (41.7%), (B) KRAS mutant peptide (47.3%), (C) TP53 wild-type peptide (44.4%), (D) TP53 mutant peptide (54.0%), (E) CTNNB1 wild peptide (46.0%) and (F) CTNNB1 mutant peptide (76.4%). KRAS, KRAS proto-oncogene; TP53, tumor protein 53; CTNNB1, catenin β1; FS, forward scatter; SS, side scatter; INT, intensity; LIN, linear; FITC, fluorescein isothiocyanate.
Figure 2.
FI in peptide-treated T2 cells. The FI in T2 cells treated with the KRAS mutant peptide increased by 5.6±4% compared with that in cells treated with the wild-type peptide. The FI in T2 cells loaded with TP53 mutant peptides was 9.6±3% higher compared with that in cells treated with the wild-type peptides. The FI in T2 cells treated with CTNNB1 mutant peptides was significantly increased by 30.4±2% compared with that in cells loaded with the wild-type peptides. *P<0.05; **P<0.01; ***P<0.001. FI, fluorescence index; KRAS, KRAS proto-oncogene; TP53, tumor protein 53; CTNNB1, catenin β1; G, glycine; V, valine; R, arginine; L, leucine K, lysine; I, isoleucine.
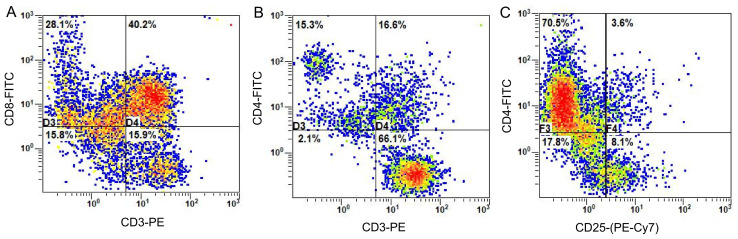
Flow cytometry for phenotypic detection of CTLs
CTLs were cultured for 21 days before being collected and the surface antigens were detected using flow cytometry. As shown in Fig. 3, the proportion of CD8+ T cells (CD3+ CD8+T) among the lymphocytes was 40.2% (Fig. 3A), whereas T helper cells (CD3+ CD4+T) accounted for 16.6% of lymphocytes (Fig. 3B), while 3.6% were comprised of regulatory T cells (CD4+CD25+T) (Fig. 3C).
Figure 3.
Detection of phenotypic CTLs (T-cell subsets) using flow cytometry. Detection of (A) cytotoxic T cells (CD3+CD8+) (B) T helper cells (CD3+CD4+) and (C) regulatory T cells (CD4+CD25+). FITC, fluorescein isothiocyanate; PE, phycoerythrin; Cy7, Cyanate 7.
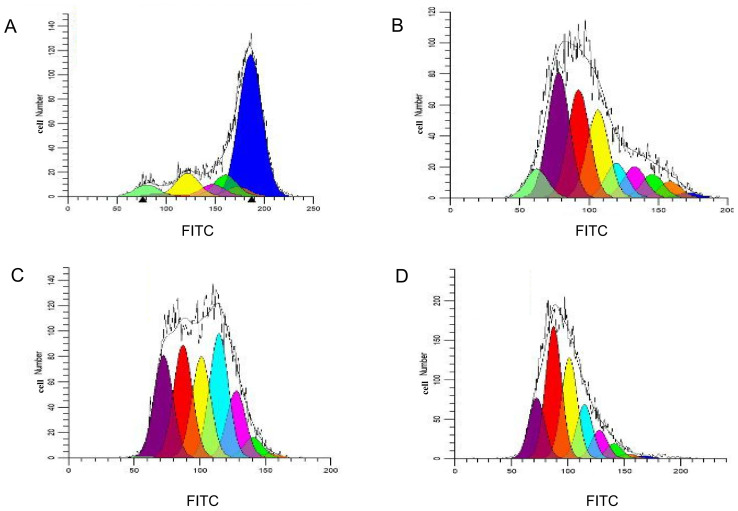
Proliferation of specific CTLs
Following CFSE staining and flow detection of CTLs cultured in vitro for 14 days, the Modfit software was used to analyze the results and determine the proliferation index of T lymphocytes. Between the 16 and the 19th day, the proliferation index of T cells increased gradually from 1.45 to 15.13, 18.55 and 18.86, respectively (Fig. 4). This indicated that T lymphocytes were in a state of proliferation and division. However, as the cells were in the third stage of differentiation (Fig. 4C and D for days 16–19), induced by antigenic peptides, the growth and proliferation rate of the cells tended to be flat (Fig. 4).
Figure 4.
Modfit analysis of cell proliferation and division. Cell proliferation and division on (A) the 1st, (B) the 2nd, (C) the 3rd and (D) the 4th day following carboxyfluorescein succinimidyl ester staining. Blue, the parental cell; orange, generation 2; green, generation 3; pink, generation 4; light blue, generation 5; yellow, generation 6; red, generation 7; purple, generation 8; and laurel-green, generation 9. FITC, fluorescein isothiocyanate.
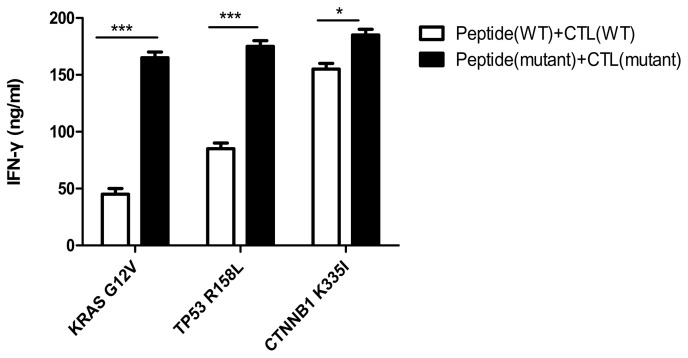
IFN-γ secretion of specific T cells stimulated by epitope peptides in vitro
As shown in Fig. 5, the secretion of IFN-γ from CTLs, induced by mutant peptides of the KRAS, TP53 and CTNNB1 groups, was 160±10, 174±5 and 180±6 ng/ml, respectively. The IFN-γ secretion of specific CTLs induced by mutant peptides in the three groups was higher compared with that in the CTLs induced by the wild-type peptides, and the difference was determined statistically significant using the Bonferroni's test (the post hoc test following an ANOVA, P<0.05). Notably, the results indirectly indicated that the mutant antigenic peptides of the three groups improved the cellular immune function of specific CTLs, especially in the CTNNB1 group.
Figure 5.
IFN-γ secretion of specific T cells stimulated by epitope peptides. IFN-γ secretion of CTLs induced by mutant peptides in the KRAS, TP53 and CTNNB1 groups, was 160±10, 174±5 and 180±6 ng/ml, respectively. Experiments were repeated three times. Date are presented as the mean ± SD. *P<0.05; ***P<0.001. IFN, interferon; CTL, cytotoxic T lymphocyte; KRAS, KRAS proto-oncogene; TP53, tumor protein 53; CTNNB1, catenin β1; WT, wild-type; G, glycine; V, valine; R, arginine; L, leucine K, lysine; I, isoleucine.
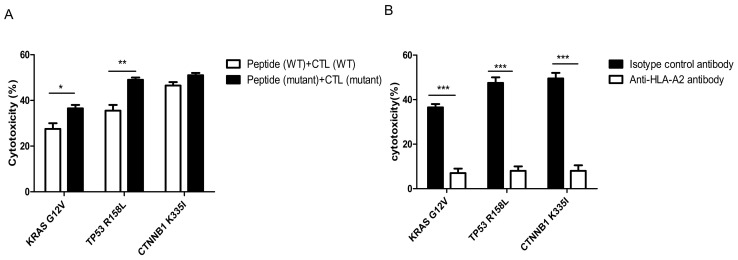
Preliminary detection of cytotoxic activity in specific CTLs against T2 cells with added peptide
As shown in Fig. 6A, the killing rates of CTLs induced by mutant peptides in the KRAS, TP53 and CTNNB1 groups were 35±3, 48±2 and 50±2%, respectively. The target cell killing rate of mutant peptide-specific CTLs in the CTNNB1 group was higher compared with that in the other two groups. However, compared with wild-type peptides, the increased killing rate of CTLs induced by the mutant peptides in vitro was the highest in the TP53 group, and the difference was determined to be statistically significant using Bonferroni's post hoc test following an ANOVA (P<0.01). It was initially determined that, compared with wild-type peptides, the induction of specific CTLs by the three groups of mutant peptides improved their affinity for target cells.
Figure 6.
Cytotoxicity of specific CTLs simulated by antigenic peptides. (A) Cytotoxicity of specific CTLs to T2 cells treated with peptides detected using the calcein-acetoxymethyl release assay. The killing rates of CTLs induced by mutated peptides in the KRAS, TP53 and CTNNB1 groups were 35±3, 48±2 and 50±2%, respectively. The amplified cytotoxic function of CTLs induced by mutant peptides in vitro was the largest in the TP53 group. (B) HLA-A2 antibody blocking assay. The difference between mutant peptide-loaded T2 cells treated with isotype control antibody and anti-HLA-A2 antibody in each group was statistically significant. Experiments were repeated three times. Date are presented as the mean ± SD. *P<0.05; **P<0.01; ***P<0.001. CTL, cytotoxic T lymphocyte; KRAS, KRAS proto-oncogene; TP53, tumor protein 53; CTNNB1, catenin β1; WT, wild-type; HLA, human leukocyte antigen; G, glycine; V, valine; R, arginine; L, leucine K, lysine; I, isoleucine.
The specificity of CTLs towards the mutant peptide-treated T2 cells in each group was significantly decreased in the cells treated with anti-HLA-A2 antibody compared with that in the isotype control antibody (P<0.001; Fig. 6B).
Construction of recombinant eukaryotic expression plasmids
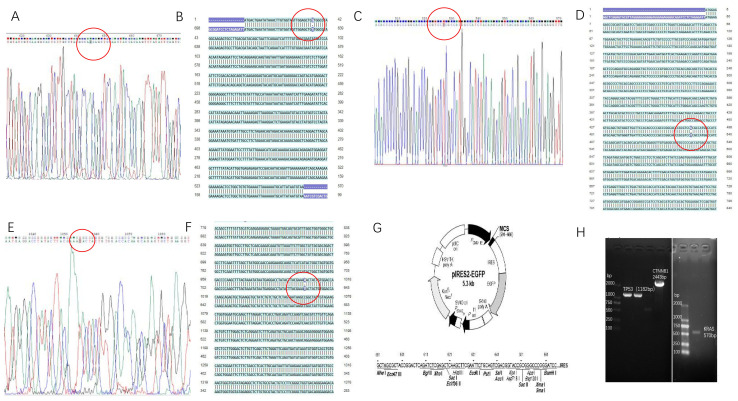
Using the site-directed mutagenesis PCR method, PCR products were determined to contain the KRAS G12V, TP53 R158L and CTNNB1 K335I mutated genes using agarose gel electrophoresis, yielding sizes of 570, 1,182 and 2,443 bp, respectively. The electrophoresis bands were consistent with the expected sizes of the target fragments (Fig. 7H).
Figure 7.
Construction of recombinant eukaryotic expression plasmids. (A) Sequencing chromatogram and (B) sequencing comparison between the KRAS G12V mutant gene clone and KRAS wild-type gene. The red circle indicates that the KRAS G12V mutant gene codon changed from GGT to GTT. (C) Sequencing chromatogram and (D) Sequencing comparison between the TP53 R158L mutant gene clone and TP53 wild-type gene. The red circle indicates that the TP53 R158L mutant gene coden changed from CGC to CTC. (E) Sequencing chromatogram and (F) sequencing comparison between the CTNNB1 K335I mutant gene clone and CTNNB1 wild-type gene. The red circle indicates that the CTNNB1 K335I mutant gene codon changed from AAA to ATA. (G) The pIRES2-EGFP plasmid map. (H) Agarose gel electrophoresis of the point mutations in the 3 genes (Red circle indicated the mutant base in mutant gene clone). KRAS, KRAS proto-oncogene; TP53, tumor protein 53; CTNNB1, catenin β1; GFP, green fluorescent protein.
Following sequencing of the mutant genetically engineered bacteria (about 5 clones were used for the sequencing of the mutant genes), the recombinant plasmid of the site-directed mutant KRAS G12V-pIRES2-EGFP was compared with that for the wild-type in the HCT116 cell line (Fig. 7A). The 12th amino acid encoding the KRAS protein was changed from G to V by substituting the codon from GGT to GTT, while none of the other bases were mutated in the KRAS gene (Fig. 7B). The TP53 R158L (Fig. 7C and D) and CTNNB1 K335I (Fig. 7E and F) mutant gene clones were also successfully constructed in the pIRES2-EGFP plasmid (Fig. 7G).
Transfection of tumor cells with recombinant plasmid and detection of transfer efficiency
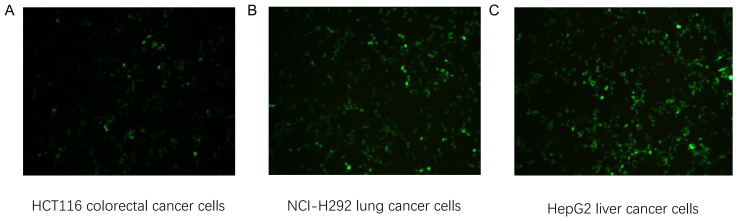
HCT116 colorectal (HLA-A2+), NCI-H292 lung (HLA-A2+) and HepG2 liver cancer cells (HLA-A2+) were transfected with KRAS G12V-EGFP-pIRES2, TP53 R158L-pIRES2-EGFP and CTNNB1 K335I-pIRES2-EGFP recombinant plasmids, respectively. After 24 h, GFP expression was confirmed in all three cell lines using an inverted fluorescence microscope, 24 h following transfection (Fig. 8).
Figure 8.
Inverted fluorescence microscope images of plasmid transfected tumor cells. (A) KRAS G12V-pIRES2-EGFP plasmid transfected into HCT116 cells. (B) TP53 R158L-pIRES2-EGFP plasmid transfected into NCI-H292 cells. (C) CTNNB1 K335I-pIRES2-EGFP plasmid transfected into HepG2 cells. Magnification, ×200.
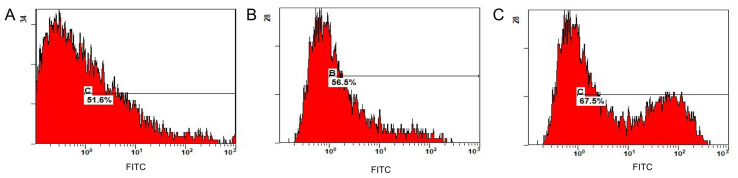
Flow cytometry revealed that the transfer efficiency of the KRAS G12V-pIRES2-EGFP, TP53 R158L-pIRES2-EGFP and CTNNB1 K335I-pIRES2-EGFP recombinant plasmids was 51.6, 56.5 and 67.5%, respectively (Fig. 9). The positive cells expressing GFP were separated and collected using a flow cell sorter for follow-up experiments.
Figure 9.
Transfer efficiency detected using flow cytometry. Transfer efficiency (%) in (A) HCT116 cells. Flow cytometry showed that the transfer efficiency (%) of the KRAS G12V-pIRES2-EGFP recombinant plasmid was 51.6%. (B) NCI-H292 cells. The transfer efficiency (%) of TP53 R158L-pIRES2-EGFP recombinant plasmid was 56.5% using flow detection. (C) HepG2 cells. The transfer efficiency (%) of CTNNB1 K335I-pIRES2-EGFP recombinant plasmid was 67.5% using flow detection. FITC, fluorescein isothiocyanate.
Expression of mutant genes in tumor cells
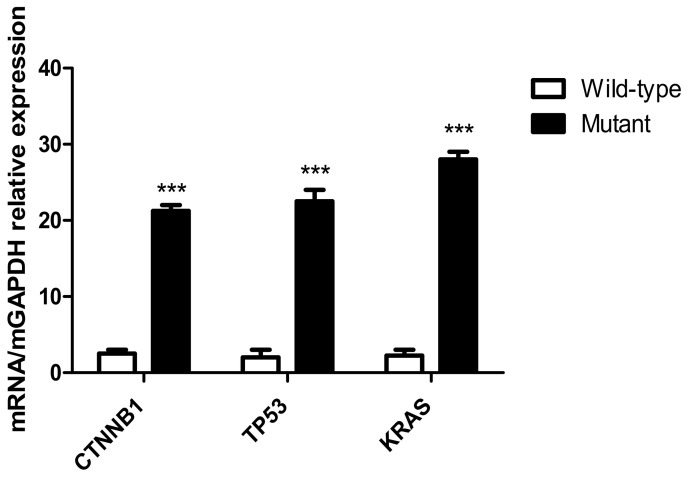
Following transfection of the recombinant plasmids, KRAS G12V-pIRES2-EGFP, TP53 R158L-pIRES2-EGFP and CTNNB1 K335I-pIRES2-EGFP into HCT116, NCI-H292 and HepG2 cells, respectively, RT-qPCR results indicated that the mutant gene was overexpressed in the transfected tumor cells compared with that in the wild-type cells (P<0.001; Fig. 10).
Figure 10.
Expression levels of mutant genes in tumor cells detected using RT-qPCR. Expression levels of mutant genes in the CTNNB1, TP53 and KRAS groups. The results of the RT-qPCR indicated that following transfection of KRAS G12V-pIRES2-EGFP, TP53 R158L-pIRES2-EGFP and CTNNB1 K335I-pIRES2-EGFP recombinant plasmids into HCT116, NCI-H292 and HepG2 tumor cells, respectively. The KRAS, TP53, CTNNB1 mutant gene was overexpressed in the transfected tumor cells compared with that in the wild-type cells. ***P<0.001. RT-qPCR, reverse transcription-quantitative PCR; KRAS, KRAS proto-oncogene; TP53, tumor protein 53; CTNNB1, catenin β1.
Cytotoxicity of peptide-induced specific CTLs on wild-type and mutant tumor cells
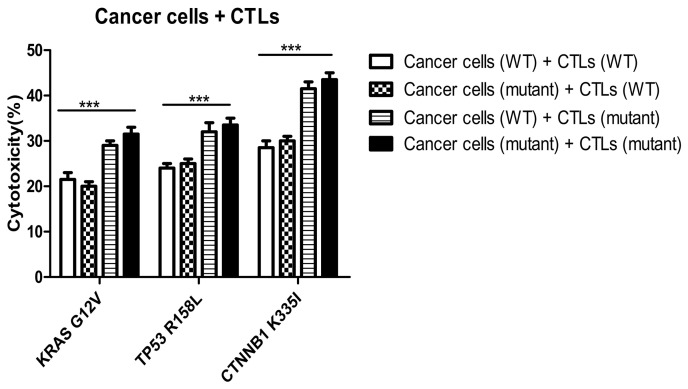
In the KRAS, TP53 and CTNNB1 groups, the killing rates of mutant peptide-induced specific CTLs towards mutant tumor cells [cancer cells (mutant) + CTLs (mutant)] were 30±3, 32±4 and 42±3%, respectively (Fig. 11). The killing rates of CTLs induced by mutant peptides in the three groups on wild-type and mutant tumor cells were similar. However, the cytotoxic specificity of wild-type peptide-induced CTLs to both wild-type and mutant tumor cells was low. In addition, the killing rates of specific CTLs induced by mutant peptides in the KRAS, TP53 and CTNNB1 groups were significantly higher compared with those induced by wild-type peptides. The difference was statistically significant between the cancer cell (WT)+CTL (WT) and cancer cell (mutant)+CTL (mutant) groups (P<0.0001; Fig. 11). The target cell killing rate of CTLs induced by CTNNB1 mutant peptides was higher compared with that in the KRAS and TP53 groups, and the results were consistent with the cytotoxicity of CTLs against T2 cells treated with peptides (Fig. 6A). Furthermore, the specificity and cytotoxicity of CTLs induced by mutant peptides in the CTNNB1 group were enhanced, when compared with those induced by wild-type peptides.
Figure 11.
Cytotoxicity of specific CTLs on wild-type and mutant tumor cells. The killing rates of mutant peptide-induced specific CTLs to mutant tumor cells were 30±3, 32±4 and 42±3% in the KRAS, TP53 and CTNNB1 groups, respectively. The killing rates of specific CTLs induced by mutant peptides in the KRAS, TP53, and CTNNB1 groups were higher compared with those induced by WT peptides. ***P<0.0001. KRAS, KRAS proto-oncogene; TP53, tumor protein 53; CTNNB1, catenin β1; CTL, cytotoxic T lymphocyte; WT, wild-type; G, glycine; V, valine; R, arginine; L, leucine K, lysine; I, isoleucine.
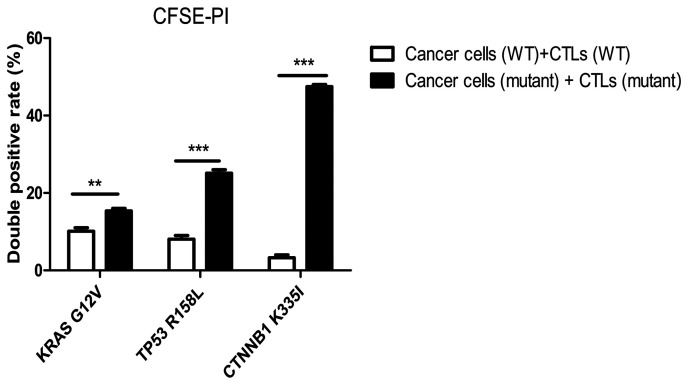
Detection of cytotoxicity of specific CTLs on tumor cells using CFSE-PI staining
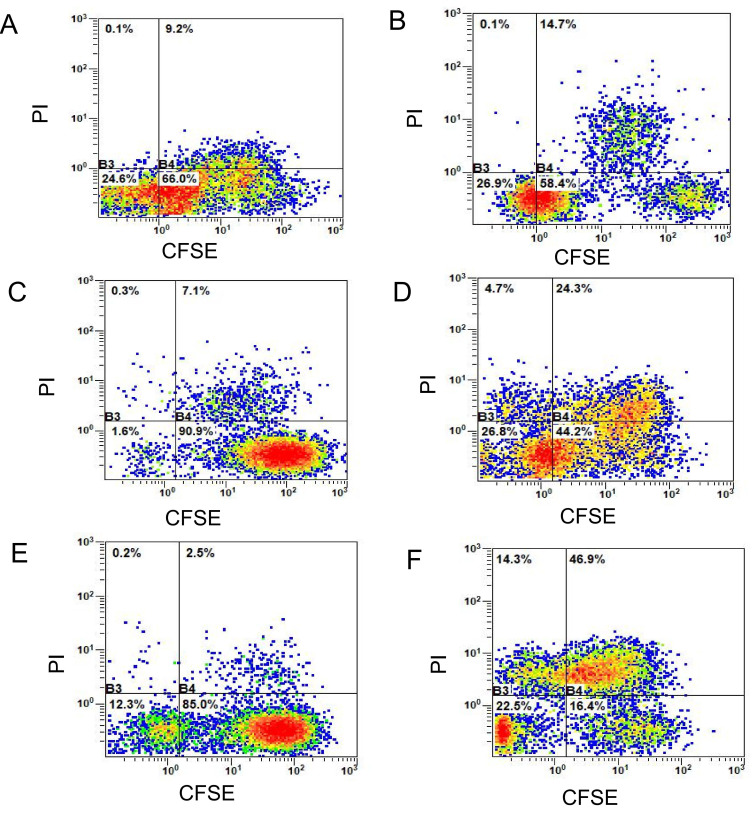
The double-positive rate (CFSE+PI+, the first quadrant) of mutant peptide-induced CTLs in the three groups (Fig. 12) was significantly higher compared with that in the wild-type peptide group, and this difference was statistically significant according to the Bonferroni post hoc test following an ANOVA. (P<0.0001; Fig. 13).
Figure 12.
Killing percentages of specific CTLs to wild-type and mutant tumor cells. (A) Wild-type HCT116 cells + KRAS wild-type peptide-induced CTLs. The double-positive percentage (CFSE+PI+) was 9.2%. (B) Mutant HCT116 cells + KRAS mutant peptide-induced CTLs. The double-positive percentage (CFSE+PI+) was 14.7%. (C) Wild-type NCI-H292 cells + TP53 wild-type peptide-induced CTLs. The double-positive percentage (CFSE+PI+) was 7.1%. (D) Mutant NCI-H292 cells + TP53 mutant peptide-induced CTLs. The double-positive percentage (CFSE+PI+) was 24.3%. (E) Wild-type HepG2 cells + CTNNB1 wild-type peptide-induced CTLs. The double-positive percentage (CFSE+PI+) was 2.5%. (F) Mutant HepG2 cells + CTLs induced by CTNNB1 mutant peptide. The double-positive percentage (CFSE+PI+) was 46.9%. CFSE, carboxyfluorescein succinimidyl ester; PI, propidium iodide; KRAS, KRAS proto-oncogene; TP53, tumor protein 53; CTNNB1, catenin β1; CTL, cytotoxic T lymphocyte.
Figure 13.
Detection of the killing rate of specific CTLs on wild-type and mutant tumor cells using CFSE-PI staining. The killing rate of the mutant peptide-induced CTLs on mutant target tumor cells in the KRAS, TP53 and the CTNNB1 groups increased by 5.5±3, 17.2±4 and 44.4±2.5%, respectively compared with that in their respective WT peptide-induced specific CTLs in the wild-type tumor cells. The differences were statistically significant according to the Bonferroni's post hoc test following an ANOVA. **P<0.01; ***P<0.001. CFSE, carboxyfluorescein succinimidyl ester; PI, propidium iodide; KRAS, KRAS proto-oncogene; TP53, tumor protein 53; CTNNB1, catenin β1; CTL, cytotoxic T lymphocyte; WT, wild-type; G, glycine; V, valine; R, arginine; L, leucine K, lysine; I, isoleucine.
When compared with the wild-type peptide-induced specific CTLs, the killing rate of mutant peptide-induced target cells in the KRAS group increased by 5.5±3.0% (Fig. 12A and B), whereas in the TP53 group it increased by 17.2±4.0% (Fig. 12C and D) and in the CTNNB1 group by 44.4±2.5% (Fig. 12E and F). The killing rates of target cells induced by mutant peptides in the CTNNB1 group were the highest, followed by those in the TP53 and KRAS groups (Figs. 12 and 13). The results were consistent with those of Cal-AM detection (Fig. 11), indicating that the CTLs induced by mutant peptides of the CTNNB1 group had a stronger cytotoxic effect on tumor cells compared with those induced by mutant peptides of the TP53 and KRAS groups.
Discussion
Tumorigenesis has been associated with gene mutations, such as gene fusion, point and deletion mutations (9). When genes encoding the normal proteins are mutated, it may lead to abnormal functions within the cell, such as exponential growth, proliferation and metastasis, as well as evading the immune system (10). TAAs are primarily derived from autoantigens, which are highly expressed by tumor cells, however these are expressed in low amounts in healthy somatic cells (13). Therefore, it is difficult to activate the low affinity, naive T cells involved in immune tolerance. During T-cell development in thymus and positive and negative selection, progenitor T cells will eliminate immature T cells with high affinity to autoantigens, and the remaining cells will become naive T cells with a low affinity to autoantigens to develop and mature (14). Therefore, it is difficult to activate naive T cells with low affinity involved in immune tolerance, and immunotherapy targeting TAAs cannot achieve the ideal curative effect, such as tumor regression. However, new tumor antigens, also known as neoantigens or TSAs, are epitope-specific antigens produced on the cell surface due to mutations within cancer cells (15). These antigenic peptides are tumor-specific and can therefore be recognized by T lymphocytes, directly inducing an immune response (15). Furthermore, TSAs are only expressed on the tumor cells and not on healthy cells. The binding affinity of the T-cell receptor (TCR) to T cells is much higher compared with that of TAA (16). T cell activation and cytotoxicity have been associated with the affinity of TCR-MHC antigenic peptide complexes, therefore TSAs have the potential to be effective targets for tumor immunotherapy (17).
During the cellular immune response, mutant peptides of tumor cells can be presented on the surface of cells by MHC I molecules, which are then recognized by the TCR of T cells (18). However, the majority of tumor cells evade recognition by the immune system, as they can downregulate or delete the expression of MHC I molecules (18). Therefore, at present, polypeptide synthesis has been used to produce mutated peptides of tumor cells in vitro, which are then introduced into the human body through individualized vaccines or ACT to stimulate the proliferation of specific T cells that recognize these neoantigens in vivo, thus specifically killing tumor cells (19).
TCGA database contains gene expression profiling, somatic mutations, copy number variation and DNA methylation data which has been produced by sequencing tumors and paracancerous tissues for >30 types of cancer (20). In the present study, point mutation gene loci of high-frequency mutations of colorectal, lung and liver cancer were identified from TCGA. A total of 3 short epitope peptides with high affinity to HLA-A201 molecules were screened using a peptide prediction algorithm and directed at these new tumor antigens. The tumor antigens of cancer cells were recognized by the immune system through the epitope peptides. The activation effect of the epitope peptide on T cells was analyzed using immunogenicity tests of specific CTLs in vitro, to identify the epitope peptides which could activate specific T cells to recognize tumor antigens, which may be used in future immunotherapy studies (21).
In the present study, KRAS G12V, TP53 R158L and CTNNB1 K335I gene point mutation loci were identified in colorectal, lung and liver cancer to design antigenic peptides, which were used to induce CTL proliferation. CD8+ T cells accounted for about 40% of all lymphocytes after 21 days. From the immunogenicity experiments of antigen epitope peptides containing point mutations, it was demonstrated that specific T cells induced by mutant antigen peptides in the CTNNB1 group had the strongest cytotoxic affinity towards mutant HepG2 liver cancer cells. The cytotoxic effects of the TP53 mutant antigen peptide-induced CTLs were weaker compared with those in the CTNNB1 group, and with the TP53 wild-type antigen peptide; however, the immunogenicity was markedly increased, and TP53 mutant peptideinduced specific T cells, which could target the tumor cells with this point mutation more effectively. The immunogenicity of the mutant antigenic peptide in the KRAS group was lower compared with that in the TP53 and CTNNB1 groups; however, the cytotoxiceffect of specific CTLs on HCT116 colorectal cancer cells was slightly improved. The present results suggest that point mutations in the antigenic peptides targeting tumor neoantigens may improve the stability of binding to the MHC molecules, enhancing immunogenicity and activating CD8+ T cells effectively. The cytotoxiceffect of CTLs induced by mutant peptides in the three groups was similar in both wild-type and mutant tumor cells, which may be associated with the cross-recognition effect of T cells. As there was only one amino acid difference between the mutant and wild-type peptides, there were two types of antigenic peptide-MHC complexes that were recognized by specific, effective CD8+ T cells (22). The present results indicate that specific CTLs, induced by mutant peptides, can kill both mutant and wild-type tumor cells. When the anti-HLA-A2 antibody was added, the cytotoxic effect of specific CTLs in the three groups was significantly reduced, indicating that the cytotoxic effect was performed in an HLA-A2-dependent manner. In summary, all three groups of the epitope peptides with higher affinity to MHC molecules may induce specific CTLs against the point mutant antigens and enhance the immune response. Among these peptides, the point mutant antigenic peptide in the CTNNB1 group demonstrated a potent ability to induce T-cell proliferation, activation, specific recognition and apoptosis of tumor cells, which may explain why the affinity of mutant peptides for MHC molecules in the CTNNB1 group was higher compared with that in the other two groups. In addition, the TCR gene family should be further analyzed in future studies. Currently, an ideal and effective TCR molecule is a key focus of research into TCR-T immunotherapy. Therefore, the present study provides preliminary screening data of effective TCR molecules, which may be beneficial to develop TCR-T immunotherapy in the future.
Among solid tumors, melanoma has the highest frequency of point mutations (such as BRAF V600E), followed by lung (KRAS G12C), colorectal (KRAS G12D), gastric (BRAF V600E) and liver cancer (TP53 R249S) (18). However, there are relatively few mutations in blood tumors, such as acute lymphocytic leukaemia and acute myelogenous leukaemia (23). Chimeric antigen receptor T-cell (CAR-T) therapy has been effective in the treatment of some blood tumors (such as acute lymphoblastic leukemia), but remains poorly effective in solid tumors (such as liver cancer) (24). CAR-T therapy recognizes membrane surface antigens (such as CD19 and B-cell maturation antigen); however, as solid tumors lack cell-surface specific targets, TCR T-cell (TCR-T) therapy may be more effective compared with CAR-T in the treatment of solid tumors, since it recognizes TSAs that are produced due to genetic mutations in cancer cells (24). Therefore, TSAs have become the key targets for screening TCR molecules in TCR-T immunotherapy. At present, the key to TCR-T immunotherapy is to identify an ideal TCR molecule (24). Therefore, the present study synthesized epitope peptides based on tumor neoantigens and identified specific T-cell clones with significant cytotoxic effects, which is conducive to screening effective TCR molecules.
Recent preclinical studies have demonstrated that individualized cancer vaccines based on new antigens have been used effectively in melanoma and glioblastoma. Ott et al (25) revealed that tumor regression without recurrence was observed in patients with melanoma injected with a long peptide (15 aa) vaccine targeting individualized new antigens. Keskin et al (26) observed that patients with glioblastoma inoculated with multi-epitope new antigen vaccines exhibited an increase in the number of new antigen-specific CD4 and CD8 tumor infiltrating lymphocytes (TILs). With regards to ACT and based on new antigens, the study by Cafri et al (27) used TILs to identify KRAS G12D mutants to treat patients with colorectal and breast cancer. The results revealed that patients with colorectal cancer treated high activity CD8+ TILs, could identify KRAS G12D, and a complete regression of the tumor was observed. Therefore, tumor neoantigen-specific T cells serve an important role in tumor immunotherapy (28). Previous studies also suggest that transfecting the TCR gene, which recognizes tumor neoantigens, into naive CD8+ T cells may provide tumor neoantigen-specific TILs therapy and TCR-T immunotherapy for patients which have the same antigen (29,30). At present, vaccines and ACT based on neoantigens have achieved some success (31), while immune checkpoint inhibitors and combination therapies towards solid tumors remain in the development stage (32,33).
With the development of next-generation sequencing technology and peptide prediction algorithms (34), it is now possible to sequence individual tumor tissues and identify non-synonymous mutations which occur in tumor cells but not in healthy somatic cells (35,36). To improve the accuracy of T-cell immunotherapy for cancer treatment, and to fully achieve the benefits of individual tumor immunotherapy, it is necessary to design antigen epitope peptides with high affinity for MHC molecules and high specificity to tumor neoantigens (37).
In conclusion, the point mutations in tumor neoantigens identified in the three groups may improve the cytotoxicity of specific T cells. The present study revealed that the mutant peptides in the CTNNB1 group were effective at activating the cellular immune response. Therefore, immunotherapies using this tumor neoantigen epitope peptide should be investigated in the future, to improve the accuracy of T-cell immunotherapy in cancer treatment and to fully achieve personalized immunotherapy and precision medical treatment.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- TCGA
The Cancer Genome Atlas
- CTL
cytotoxic T lymphocyte
- MHC
major histocompatibility complex
- HLA
human leukocyte antigen
- ACT
adoptive cell transfer therapy
- TAA
tumor-associated antigen
- TSA
tumor-specific antigen
- CFSE
carboxyfluorescein succinimidyl ester
- PI
propidium iodide
- MFI
mean fluorescence intensity
- DC
dendritic cell
- PBMC
peripheral blood mononuclear cell
- Cal-AM
calcein-acetoxymethyl
Funding
The present study was supported by the National Natural Science Foundation of China (grant nos. 81703053, 21771042 and 31300737). The present study was also supported by the Natural Science Foundation of Guangdong Province (grant nos. 2018A030313114, 2018A030313860 and 2016A030310298), and the Innovation and University Promotion Project of Guangdong Pharmaceutical University (grant nos. 2017KCXTD020 and 2017KZDXM049).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
WW prepared and wrote the manuscript. YC performed the experiments. LH and WL performed experimental data analysis. CT critically revised the manuscript. HS designed the experimental scheme, reviewed and edited the manuscript. WW, YC, LH, WL, CT and HS contributed to data analysis, drafting or revising of the manuscript, gave final approval of the version to be published and agree to be accountable for all aspects of the work.
Ethics approval and consent to participate
The present study follows international and national regulations in accordance with the Declaration of Helsinki. The present study was approved by the Ethics Committee of The First Affiliated Hospital of Guangdong Pharmaceutical University (Guangzhou, China; ref. no. 2017-003). Human peripheral blood was collected from laboratory healthy volunteers and written informed consent was provided by each volunteer.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Efremova M, Finotello F, Rieder D, Trajanoski Z. Neoantigens generated by individual mutations and their role in cancer immunity and immunotherapy. Front Immunol. 2017;8:1679. doi: 10.3389/fimmu.2017.01679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jiang T, Shi T, Zhang H, Hu J, Song Y, Wei J, Ren S, Zhou C. Tumor neoantigens: From basic research to clinical applications. J Hematol Oncol. 2019;12:93. doi: 10.1186/s13045-019-0787-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peng M, Mo Y, Wang Y, Wu P, Zhang Y, Xiong F, Guo C, Wu X, Li Y, Li X, et al. Neoantigen vaccine: An emerging tumor immunotherapy. Mol Cancer. 2019;18:128. doi: 10.1186/s12943-019-1055-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou C, Zhu C, Liu Q. Toward in silico identification of tumor neoantigens in immunotherapy. Trends Mol Med. 2019;25:980–992. doi: 10.1016/j.molmed.2019.08.001. [DOI] [PubMed] [Google Scholar]
- 5.Garcia-Garijo A, Fajardo CA, Gros A. Determinants for neoantigen identification. Front Immunol. 2019;10:1392. doi: 10.3389/fimmu.2019.01392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wirth TC, Kühnel F. Neoantigen targeting-dawn of a new era in cancer immunotherapy? Front Immunol. 2017;8:1848. doi: 10.3389/fimmu.2017.01848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rammensee H, Bachmann J, Emmerich NP, Bachor OA, Stevanović S. SYFPEITHI: Database for MHC ligands and peptide motifs. Immunogenetics. 1999;50:213–219. doi: 10.1007/s002510050595. [DOI] [PubMed] [Google Scholar]
- 8.Parker KC, Bednarek MA, Coligan JE. Scheme for ranking potential HLA-A2 binding peptides based on independent binding of individual peptide side-chains. J Immunol. 1994;152:163–175. [PubMed] [Google Scholar]
- 9.Richters MM, Xia H, Campbell KM, Gillanders WE, Griffith OL, Griffith M. Best practices for bioinformatic characterization of neoantigens for clinical utility. Genome Med. 2019;11:56. doi: 10.1186/s13073-019-0666-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tran E, Ahmadzadeh M, Lu YC, Gros A, Turcotte S, Robbins PF, Gartner JJ, Zheng Z, Li YF, Ray S, et al. Immunogenicity of somatic mutations in human gastrointestinal cancers. Science. 2015;350:1387–1390. doi: 10.1126/science.aad1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carey MF, Peterson CL, Smale ST. PCR-mediated site-directed mutagenesis. Cold Spring Harb Protoc. 2013;2013:738–742. doi: 10.1101/pdb.prot076505. [DOI] [PubMed] [Google Scholar]
- 12.Mazzei M, Vascellari M, Zanardello C, Melchiotti E, Vannini S, Forzan M, Marchetti V, Albanese F, Abramo F. Quantitative real time polymerase chain reaction (qRT-PCR) and RNAscope in situ hybridization (RNA-ISH) as effective tools to diagnose feline herpesvirus-1-associated dermatitis. Vet Dermatol. 2019;30:e491–e147. doi: 10.1111/vde.12787. [DOI] [PubMed] [Google Scholar]
- 13.Thaxton JE, Li Z. To affinity and beyond: Harnessing the T cell receptor for cancer immunotherapy. Hum Vaccin Immunother. 2014;10:3313–3321. doi: 10.4161/21645515.2014.973314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kisielow P. How does the immune system learn to distinguish between good and evil? The first definitive studies of T cell central tolerance and positive selection. Immunogenetics. 2019;71:513–518. doi: 10.1007/s00251-019-01127-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schumacher TN, Schreiber RD. Neoantigens in cancer immunotherapy. Science. 2015;348:69–74. doi: 10.1126/science.aaa4971. [DOI] [PubMed] [Google Scholar]
- 16.Kanaseki T. Proteogenomics HLA Ligandome analysis for cancer antigen research. Gan To Kagaku Ryoho. 2019;46:1377–1381. (In Japanese) [PubMed] [Google Scholar]
- 17.Matsushita H, Demachi-Okamura A, Takahashi Y. Neoantigens are critical targets in naturally and therapeutically induced immune responses to cancer. Gan To Kagaku Ryoho. 2019;46:1372–1376. (In Japanese) [PubMed] [Google Scholar]
- 18.Garrido F, Aptsiauri N. Cancer immune escape: MHC expression in primary tumors versus metastases. Immunology. 2019;158:255–266. doi: 10.1111/imm.13114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mandal R, Chan TA. Personalized oncology meets immunology: The path toward precision immunotherapy. Cancer Discov. 2016;6:703–713. doi: 10.1158/2159-8290.CD-16-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Z, Jensen MA, Zenklusen JC. A practical guide to the cancer genome atlas (TCGA) Methods Mol Biol. 2016;1418:111–141. doi: 10.1007/978-1-4939-3578-9_6. [DOI] [PubMed] [Google Scholar]
- 21.Lu YC, Robbins PF. Cancer immunotherapy targeting neoantigens. Semin Immunol. 2016;28:22–27. doi: 10.1016/j.smim.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ren L, Leisegang M, Deng B, Matsuda T, Kiyotani K, Kato T, Harada M, Park JH, Saloura V, Seiwert T, et al. Identification of neoantigen-specific T cells and their targets: Implications for immunotherapy of head and neck squamous cell carcinoma. Oncoimmunology. 2019;8:e1568813. doi: 10.1080/2162402X.2019.1568813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen YP, Zhang Y, Lv JW, Li YQ, Wang YQ, He QM, Yang XJ, Sun Y, Mao YP, Yun JP, et al. Genomic analysis of tumor microenvironment immune types across 14 solid cancer types: Immunotherapeutic implications. Theranostics. 2017;7:3585–3594. doi: 10.7150/thno.21471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vormehr M, Diken M, Türeci Ö, Sahin U, Kreiter S. Personalized neo-epitope vaccines for cancer treatment. Recent Results Cancer Res. 2020;214:153–167. doi: 10.1007/978-3-030-23765-3_5. [DOI] [PubMed] [Google Scholar]
- 25.Ott PA, Hu Z, Keskin DB, Shukla SA, Sun J, Bozym DJ, Zhang W, Luoma A, Giobbie-Hurder A, Peter L, et al. An immunogenic personal neoantigen vaccine for patients with melanoma. Nature. 2017;547:217–221. doi: 10.1038/nature22991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keskin DB, Anandappa AJ, Sun J, Tirosh I, Mathewson ND, Li S, Oliveira G, Giobbie-Hurder A, Felt K, Gjini E, et al. Neoantigen vaccine generates intratumoral T cell responses in phase Ib glioblastoma trial. Nature. 2019;565:234–239. doi: 10.1038/s41586-018-0792-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cafri G, Yossef R, Pasetto A, Deniger DC, Lu YC, Parkhurst M, Gartner JJ, Jia L, Ray S, Ngo LT, et al. Memory T cells targeting oncogenic mutations detected in peripheral blood of epithelial cancer patients. Nat Commun. 2019;10:449. doi: 10.1038/s41467-019-08304-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tran E, Turcotte S, Gros A, Robbins PF, Lu YC, Dudley ME, Wunderlich JR, Somerville RP, Hogan K, Hinrichs CS, et al. Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science. 2014;344:641–645. doi: 10.1126/science.1251102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tran E, Robbins PF, Lu YC, Prickett TD, Gartner JJ, Jia L, Pasetto A, Zheng Z, Ray S, Groh EM, et al. T-cell transfer therapy targeting mutant KRAS in cancer. N Engl J Med. 2016;375:2255–2262. doi: 10.1056/NEJMoa1609279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zacharakis N, Chinnasamy H, Black M, Xu H, Lu YC, Zheng Z, Pasetto A, Langhan M, Shelton T, Prickett T, et al. Immune recognition of somatic mutations leading to complete durable regression in metastatic breast cancer. Nat Med. 2018;24:724–730. doi: 10.1038/s41591-018-0040-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shindo Y, Hazama S, Nagano H. Cancer vaccine focused on neoantigens. Gan To Kagaku Ryoho. 2019;46:1367–1371. (In Japanese) [PubMed] [Google Scholar]
- 32.Chu Y, Liu Q, Wei J, Liu B. Personalized cancer neoantigen vaccines come of age. Theranostics. 2018;8:4238–4246. doi: 10.7150/thno.24387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou J, Zhao W, Wu J, Lu J, Ding Y, Wu S, Wang H, Ding D, Mo F, Zhou Z, et al. Neoantigens derived from recurrently mutated genes as potential immunotherapy targets for gastric cancer. Biomed Res Int. 2019;2019:8103142. doi: 10.1155/2019/8103142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vilimas T. Measuring tumor mutational burden using whole-exome sequencing. Methods Mol Biol. 2020;2055:63–91. doi: 10.1007/978-1-4939-9773-2_3. [DOI] [PubMed] [Google Scholar]
- 35.Smith CC, Chai S, Washington AR, Lee SJ, Landoni E, Field K, Garness J, Bixby LM, Selitsky SR, Parker JS, et al. Machine-learning prediction of tumor antigen immunogenicity in the selection of therapeutic epitopes. Cancer Immunol Res. 2019;7:1591–1604. doi: 10.1158/2326-6066.CIR-19-0155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Riley TP, Keller GLJ, Smith AR, Davancaze LM, Arbuiso AG, Devlin JR, Baker BM. Structure based prediction of neoantigen immunogenicity. Front Immunol. 2019;10:2047. doi: 10.3389/fimmu.2019.02047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brennick CA, George MM, Corwin WL, Srivastava PK, Ebrahimi-Nik H. Neoepitopes as cancer immunotherapy targets: Key challenges and opportunities. Immunotherapy. 2017;9:361–371. doi: 10.2217/imt-2016-0146. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.