Graphical abstract
Keywords: COVID-19, Enteric virus, Disinfection, Aerosols, Sewage, Enveloped virus
Abbreviations: ACE2, Angiotensin-converting enzyme 2; AH, Absolute Humidity; AOPs, Advanced Oxidation Processes; ASP, Activate Sludge Process; A-WWTS, Algal-WWTS; BCoV, Bovine Enteric Coronavirus); BoRv, Bovine Rotavirus Group A; BSL, Biosafety Level; BVDV1, Bovine Viral Diarrhea Virus Type 1; BVDV2, Bovine Viral Diarrhea Virus Type 2; CCA, Carbon Covered Alumina; Cl−, Chlorine; ClO2, Chlorine dioxide; CNT, Carbon Nanotubes; COVID-19, Coronavirus Disease 2019; CRFK, Crandell Reese feline kidney cell line (CRFK); CVE, Coxsackievirus B5; Cys, Cysteine; DBP, Disinfection by-products; DBT, L2 and Delayed Brain Tumor Cell Cultures; DMEM, Dulbecco’s Modified Eagle Medium; DNA, deoxyribose nucleic acid; dPCR, Digital PCR; ds, Double Stranded; dsDNA, Double Stranded DNA; E gene, Envelope protein gene; EV, Echovirus 11; FC, Free Chlorine; FFP3, Filtering Face Piece; FIPV, Feline infectious peritonitis virus; GI, Gastrointestinal tract; H2O2, Hydrogen Peroxide; H3N2, InfluenzaA; H6N2, Avian influenza virus; HAdV, Human Adenovirus; HAV, Hepatitis A virus (HAV); HCoV, Human CoV; HEV, Hepatitis E virus; HKU1, Human CoV1; ICC-PCR, Integrated Cell Culture with PCR; JCV, JCV polyomavirus; log10, logarithm with base 10; MALDI-TOF MS, Mass Spectrometry; MBR, Membrane Bioreactor (MBR); MERS-CoV, Middle East Respiratory Syndrome Coronavirus; Met, Methionine; MHV, Murine hepatitis virus; MNV-1, Murine Norovirus; MWCNTs, Multiwalled Carbon Nanotubes; N gene, Nucleocapsid protein gene; STP, Sewage Treatment Plant; NCoV, Novel coronavirus; NGS, Next generation sequencing; NTP, Non-Thermal Plasma; O2, Singlet Oxygen; O3, Ozone; ORF, Open Reading Frame; PAA, Para Acetic Acid; PCR, Polymerase Chain Reaction; PEC, Photoelectrocatalytical; PEG, Polyethylene Glycol; PFU, Plaque Forming Unit; PMMoV, Pepper Mild Mottle Virus; PMR, Photocatalytic Membrane Reactors; PPE, Personal Protective Equipment; PTAF, Photocatalytic Titanium Apatite Filter; PV-1, Polivirus-1; PV-3, Poliovirus 3; PVDF, Polyvinylidene Fluoride; qRT-PCR, quantitative RT-PCR; Qβ, bacteriophages; RH, Relative Humidity; RNA, Ribose nucleic acid; RONS, Reactive Oxygen and/or Nitrogen Species; RT-PCR, Real Time Polymerase Chain Reaction; RVA, Rotaviruses A; SARS-CoV-1, Severe Acute Respiratory Syndrome Coronavirus 1; SARS-CoV-2, Severe Acute Respiratory Syndrome Coronavirus 2; SBR, Sequential Batch Reactor; SODIS, Solar water disinfection; ss, Single Stranded; ssDNA, Single Stranded DNA; ssRNA, Single Stranded RNA; T90, First order reaction time required for completion of 90%; T99.9, First order reaction time required for completion of 99.9%; TGEV, Porcine Coronavirus Transmissible Gastroenteritis Virus; TGEV, Transmissible Gastroenteritis; Trp, Tryptophan; Tyr, Tyrosine; US-EPA, United States Environmental Protection Agency; UV, Ultraviolet; WBE, Wastewater-Based Epidemiology; WWT, Wastewater Treatment; WWTPs, Wastewater Treatment Plants; αCoV, Alphacoronavirus; βCoV, Betacoronavirus; (h+), Photoholes; +ssRNA, Positive Sense Single-Stranded RNA
Abstract
The unprecedented global spread of the severe acute respiratory syndrome (SARS) caused by SARS-CoV-2 is depicting the distressing pandemic consequence on human health, economy as well as ecosystem services. So far novel coronavirus (CoV) outbreaks were associated with SARS-CoV-2 (2019), middle east respiratory syndrome coronavirus (MERS-CoV, 2012), and SARS-CoV-1 (2003) events. CoV relates to the enveloped family of Betacoronavirus (βCoV) with positive-sense single-stranded RNA (+ssRNA). Knowing well the persistence, transmission, and spread of SARS-CoV-2 through proximity, the faecal-oral route is now emerging as a major environmental concern to community transmission. The replication and persistence of CoV in the gastrointestinal (GI) tract and shedding through stools is indicating a potential transmission route to the environment settings. Despite of the evidence, based on fewer reports on SARS-CoV-2 occurrence and persistence in wastewater/sewage/water, the transmission of the infective virus to the community is yet to be established. In this realm, this communication attempted to review the possible influx route of the enteric enveloped viral transmission in the environmental settings with reference to its occurrence, persistence, detection, and inactivation based on the published literature so far. The possibilities of airborne transmission through enteric virus-laden aerosols, environmental factors that may influence the viral transmission, and disinfection methods (conventional and emerging) as well as the inactivation mechanism with reference to the enveloped virus were reviewed. The need for wastewater epidemiology (WBE) studies for surveillance as well as for early warning signal was elaborated. This communication will provide a basis to understand the SARS-CoV-2 as well as other viruses in the context of the environmental engineering perspective to design effective strategies to counter the enteric virus transmission and also serves as a working paper for researchers, policy makers and regulators.
1. Genesis – virions and pandemic
The ongoing global spread of the pandemic due to novel coronavirus (CoV) called COVID-19 or SARS-CoV-2 causing severe acute respiratory syndrome (SARS) is posing unprecedented repercussion on human health and economy. A virus is an infectious agent (non-cellular parasite) with genetic material (either deoxyribose nucleic acid (DNA) or ribose nucleic acid (RNA); single (ss) or double-stranded (ds) or non-enveloped/enveloped (lipoproteins)) encompassed by a protein capsid. It can replicate (or make copies) only with a host cell (living) either of animal, plant, or bacteria. Viruses depend on the biochemical machinery of the host cell to inject genetic information and express through transcription (DNA to RNA) and translation (RNA to protein) [1]. Capsid made of similar sub-units called capsomere are arranged tightly together in a pattern and serves as an impenetrable shell protecting the nucleic acid giving it a defined structure (helical and icosahedral) [2].
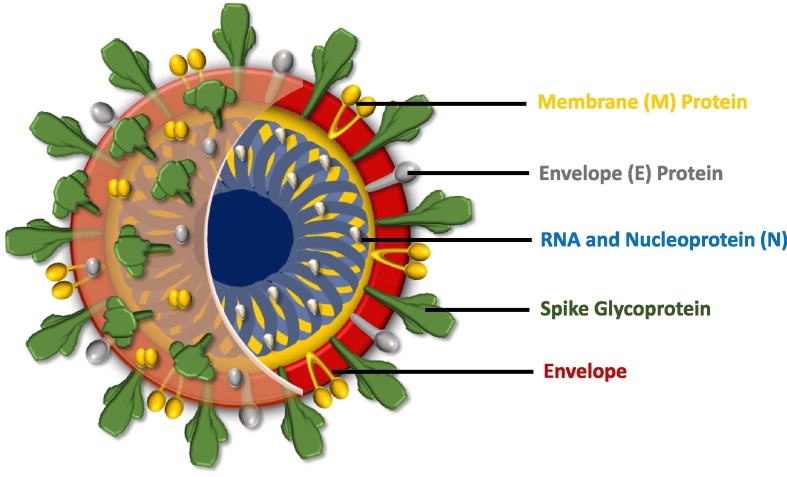
CoV (60–220 nm size; first identified in 1960) belongs to a large family of the enveloped virus with a + ssRNA and crown-like spikes on their spherical surfaces. CoV virion is classified as highly pathogenic virus and associated with SARS-CoV-1 (2003), MERS-CoV (2012) and SARS-CoV-2 so far [3], [4], [5], [6]. SARS-CoV and MERS-CoV belong to βCoV and are reported to have a high mortality rate [7]. The genome sequence of SARS-CoV-2 (25–32 kb) showed 82% of similarity with SARS-CoV-1 [8], [9], [10] and has two open reading frames (ORF1a and ORF1b, located at the 5′ end) coded for polyproteins and one-third of the genome encoding (terminal) for the proteins (spike, envelope, and capsid) [5]. The capsid (outer protein shell) confers specificity to the virus (Fig. 1 ) and the inner core confers to the infectivity and enveloped proteins associated with a virus life cycle (assembly, envelope formation and pathogenesis) [5]. Enveloped viruses are diverse with reference to genome, structure, replication, pathogenicities and persistence [11], [12], [13], [14]. The lipid bilayer with glycoproteins (protein coded and carbohydrates are added by cellular glycosyl transferase) helps the virus to identify the host cell and fuses with its membrane. Peplomers (spike-like projections; glycoproteins) helps in virus attachment to the host [15]. Angiotensin-converting enzyme 2 (ACE2) is the host receptor that interacts with the spike protein to facilitate entry of SARS-CoV-2 into the host cell and replicate mostly in the lungs apart from or besides heart, intestines, blood vessels and muscles [16], [17], [18], [19], [20]. Envelope allows the virus to exit using host cellular machinery and increases the virus particle packaging capacity with the additional viral proteins, hides the capsid antigen from freely circulating antibodies and has more structural flexibility and persistence [21]. Most of the zoonotic infecting human relate to the enveloped virus and their host choice vary depending on their surface proteins [22].
Fig. 1.
Structure of Corona Virus (SARS-CoV-2).
Outbreaks of SARS-CoV-2 evidenced the global vulnerability to emerging and infectious diseases [23]. The anthropogenic activities and unprecedented resource consumption manifested by the population explosion is causing ecological imbalance creating stress on the ecosystem and is being considered as one of the causing factors for this kind of epidemics/pandemics. According to an estimate, 1.67 million viral species exist on Earth and among them nearly 40% can infect humans due to the increased frequency of human interactions with the pristine nature [23], [24]. The rate of zoonotic viral transmission among humans is accelerating due to increment in the global footprint leading to a non-linear rise in pandemic risk [25], [26], [27].
SARS-CoV-2 transmits via contact through human-to-human (infected patients or incubation/asymptomatic individuals) or contact to infected surfaces (fomites or skin-to-skin) or mediated through the mouth, nose, eyes or through inhalation of the exhaled virus in the respiratory droplets (coughs or sneezes from an infected person) [7], [28], which eventually recommends the need for ‘social distancing’ to reduce the virus spread. The replication and persistence of SARS-CoV-2 in gastrointestinal (GI) tract and shedding through stools is now evidenced as a new potential transmission route to the environmental matrix, which is a major concern. Virions that persist in water, wastewater or air contacts host for further onward indirect transmission [12]. In this context, this communication attempts to review the virus influx in the environmental settings regarding its occurrence, persistence, detection, and inactivation based on the published literature so far.
2. Enteric virus – an emerging concern to the environment
Shedding of CoV in faeces was reported before the outbreak of SARS-CoV-2 pandemic [29], [30], [31], [32]. The viral genetic material detected in the stool does not necessarily indicate the viable infectious virions [18], [33]. The replication of SARS-CoV-1 was detected in the GI tract of the patients along with stools [29]. Cultivable SARS-CoV-1 virus was also detected in urine along with stool samples for a longer period (29–36 days) indicating its persistence in the patient excreta [30]. SARS-CoV-1 shares 82% genetic homology with SARS-CoV-2 in a subset of patients [18], [30], [34]. Patients infected with SARS-CoV-2 reported GI symptoms such as abdominal discomfort, GI bleeding, nausea/vomiting, and diarrhea apart from the respiratory infection [8], [18], [35]. Gastric, duodenal and rectal epithelia showed positive for ACE2 receptor and nucleocapsid protein providing evidence for GI infection of SARS-CoV-2 [36]. The expression of ACE2 was observed high in the upper oesophagus and stratified epithelial cells and absorptive enterocytes of ileum and colon, which enumerates the digestive system as a potential transmission route for SARS-CoV-2 infection [37]. Biopsy (colonoscopy/autopsy) studies on small and large intestine samples showed the presence of the virus and further their persistence in the faeces samples for more than 70 days after the symptom onset [38]. SARS-CoV-2 RNA was detected in faeces, respiratory specimens or blood from 6 patients, while 7 patients excreted virus in respiratory tract specimens and faeces or blood [35]. Viable SARS-CoV-2 occurrence was observed in the stool samples of 2 patients who did not suffer from diarrhea. High frequency of virus (83.3%) in faeces of mild patients was observed along with prolonged virus RNA shedding in faeces for one month [39]. Patients (18 no) samples with a mild respiratory tract infection diagnosed with SARS-CoV-2 resulted in the presence of virus in stools but not in urine [40]. SARS-CoV-2 genomic RNA was detected on the seventh day of illness in stool specimen [41]. The stool samples of SARS-CoV-2-infected patients (73 no; hospitalized) showed positive to viral RNA (53.42%) and remained in feces (23.29%) even after the viral RNA decreased to an undetectable level in the respiratory tract [36]. The virological analysis of stool samples of nine SARS-CoV-2 cases showed a combination of high virus RNA concentrations and occasional detection of single guide (sg) RNA containing cells for longer periods indicating active replication in the GI tract [42]. SARS-CoV-2 recovered patients (66 no) after treatment were detected positive for viral RNA in both stool (16.7%) and urine (6.9%) samples [43]. Viral RNA persisted in faeces for 33 days after the patient was tested negative for viral RNA in the respiratory tract [44]. Viral shedding through the digestive system can last longer than shedding from the respiratory tract [19]. The presence of fragments of viral RNA was also detected in the faeces of infected patients [5], [36], [41]. The presence of both viable virion and viral RNA of SARS-CoV-2 (Supplementary Table 1) was detected in stool samples [Supplementary References 1-11].
3. Persistence of virus
To transfer via the water cycle, the virus should be able to persist in waste and retain its infectivity before contact [45]. The fate of CoV persistence in wastewater (Table 1 ) was tested at varying temperatures using bacteriophages as indicators [46], [47], [48], [49]. Bacteriophages share morphological and biological properties with the enteric viruses and serve as surrogates [50]. Temperature is one of the critical factors that govern the virons survival in the environment. Various kinds of wastewaters were spiked with SARS-CoV-1, Human CoV (HCoV 229E) and surrogates of animal CoV as transmissible gastroenteritis (TGEV), Feline infectious peritonitis virus (FIPV) and Murine hepatitis virus A59 (MHV) to study their persistence at variable temperature conditions (4–25 °C) [46], [47], [48], [49], [51]. In-vitro studies on SARS-CoV-1 spiked in stool and urine sample, hospital and domestic wastewater and tap water showed virus persistence at relatively lower temperature (4 °C) [46]. SARS-CoV-1 persisted for 2 days at 20 °C, while for 14 days at 4 °C. On the contrary, urine samples showed longer persistence (17 days) at 20 °C. Salt content present in urine helped the virus to maintain osmotic pressure that is needed for their persistence [46]. Similar persistent period of HCoV and FIPV in filtered and unfiltered tap water, primary effluent and secondary effluent at 23 °C was observed contrary to 100 days at 4 °C [47]. The indicator organism (polivirus-1 (PV-1)) showed six times longer persistence than HCov and FIPV [47]. The persistence of the virus decreased with the increase in temperature. Surrogates of CoV was used to study the persistence [48], [49]. Spiking water (reagent grade), lake water, and human sewage (pasteurized) with surrogates of CoV (TGEV and MHV) showed the decline in virulence rapidly at 25 °C than 4 °C [48]. Wastewater and pasteurized wastewater spiked with MHV A59 was incubated at 25 °C and 10 °C (typical summer and winter temperatures) [49]. MHV A59 persisted for 19 ± 8 h at 25 °C in pasteurized wastewater which was less when compared to Pseudomonas phage ϕ6 (53 ± 8 h), MS2 and T3 enterobacteria phage (indicator organism) (121 ± 55 h). The decline in the infectious virus was more rapid and effective at higher temperatures [46], [47], [48], [49]. Enveloped viruses also showed rapid inactivation than the non-enveloped ones (>100 h) [49]. CoV is more sensitive than PV-1 due to the presence of envelope and low transmission rate observed with the CoV due to rapid inactivation at ambient temperatures [47].
Table 1.
Persistence and occurrence of coronavirus in wastewater tested at varied temperature.
| Virus | Cell line/Host/Media | Purification of virus particles | Environment matrix | Condition | Persisting Time | Findings | Reference | |
|---|---|---|---|---|---|---|---|---|
| SARS-CoV-1 | Detected using culture methods on Vero E6 cell. Cells were grown in Eagle’s growth medium with 8% fetal bovine serum and 0.015 M DMEM buffer and kanamycin+gentamycin 50 µg/mL each. | Centrifugation | Stool sample | Incubation at 20 °C and 4 °C | At 20 °C | At 4 °C |
|
[46] |
| 3 days | >17 days | |||||||
| Urine samples | 17 days | >17 days | ||||||
| Hospital wastewater | 2 days | 14 days | ||||||
| Domestic sewage | 2 days | 14 days | ||||||
| Tap water | 2 days | 14 days | ||||||
| E.coli and f2 phage | As an indicator microorganism for evaluating disinfection | |||||||
| Transmissible gastroenteritis (TGEV) | Grown in swine testicular cell cultures (ST) | Cells propagated by infecting with confluent layer of host cell cultures followed by harvesting and clarifying by centrifugation. The supernatant is used as viral stock | Reagent-grade water | Incubation at 23-25 °C and 4 °C | At 23–25 °C | At 4 °C |
|
[48] |
| 33 days | TGEV reduced after 49 days | |||||||
| Lake water | 13 days | 14 days | ||||||
| Pasteurized settled human sewage (70 °C - 3h) | 14 days | 73 days | ||||||
| Mouse hepatitis (MHV) | Grown in delayed brain tumor cell cultures (DBT) | Reagent-grade water | 26 days | MHV reduced after 49 days | ||||
| Lake water | 10 days | No decline | ||||||
| Pasteurized settled human sewage (70 °C - 3h) | 10 days | 105 days | ||||||
| Feline infectious peritonitis virus (FIPV) | Propagated and assayed in Crandell Reese feline kidney cell line (CRFK) | Centrifugation and addition of 9% PEG and 0.5 M NaCl followed by overnight stirring at 4 °C. The suspension was centrifuged and the resultant pellet was resuspended in 0.01 M PBS and stored. Whereas, poliovirus was purified by extraction with Vertrel XF. The resultant was emulsified, centrifuged and stored | Filtered tap water | Incubation at23 °C and 4 °C | At 23 °C | At 4 °C |
|
[47] |
| 10.1 days | >100 days | |||||||
| Unfiltered tap water | 12.5 days | |||||||
| Filtered Primary effluent | 2.40 days | |||||||
| Unfiltered Primary effluent | 2.56 days | |||||||
| Secondary effluent | 2.42 days | |||||||
| Human coronavirus 229E (HCoV) | Propagated and assayed in the fetal human lung fibroblast, MRC-5 cell line | Filtered tap water | 10.1 days | |||||
| Unfiltered tap water | 12.1 days | |||||||
| Filtered Primary effluent | 2.35 days | |||||||
| Unfiltered Primary effluent | 3.54 days | |||||||
| Secondary effluent | 2.77 days | |||||||
| Poliovirus-1 (PV-1) | Propagated and assayed in Buffalo green monkey kidney cell (BGM) | Filtered tap water | 64.9 days | |||||
| Unfiltered tap water | 71.3 days | |||||||
| Filtered Primary effluent | 35.5 days | |||||||
| Unfiltered Primary effluent | 10.9 days | |||||||
| Secondary effluent | 5.74 days | |||||||
| Murine hepatitis virus, strain A59 (MHV) (enveloped) | L2 and delayed brain tumor cell culture (DBT) grown in Dulbecco’s modified Eagle Medium (DMEM) + 10% new born calf serum + 1% L-glutamine + 1% penicillin/streptomycin incubated at 37 °C with 5% CO2 | Centrifugation followed by filtration using 0.22 µm poly-ether sulfone-(PES) membrane | Wastewater | Incubation at 25 °C and 10 °C | At 25 °C | At 10 °C |
|
[49] |
| 13 ± 1 h | 36 ± 5 h | |||||||
| Pasteurized wastewater | 19 ± 8 h | 149 ± 103 h | ||||||
| Pseudomonas phage ϕ6 (enveloped) | Luria-Bertani medium with NaCl at 26 °C | Wastewater | 7 ± 0.4 h | 28 ± 2 h | ||||
| Pasteurized wastewater | 53 ± 8 h | 146 ± 103 h | ||||||
| Enterobacteria phage MS2 and Enterobacteria phage T3 (non-enveloped) | MS2 and T3 propagated and assayed in E.coli hosts | Fast protein liquid chromatography (FPLC) using Sephacryl S-400 HR column followed by filtration using 0.22 µm poly-ether sulfone-(PES) membrane | Wastewater | 121 ± 36 h | 175 ± 33 h | |||
| Pasteurized wastewater | 121 ± 55 h | 212 ± 88 h | ||||||
4. Occurrence of virus in environmental matrix
4.1. Wastewater
Earlier studies [51], [52], [53], [54], [55], [56], [57], [58], [59], [60] showed the occurrence of CoV in wastewater samples (Table 2 ). Wastewater originating from the hospital during the SARS-CoV-1 outbreak was tested for its virulence/occurrence before and after disinfection using a symptomatic patient sample as a benchmark [51]. Infectious SARS-CoV-1 was found in the sewage of two hospitals before disinfection [51]. Influent and effluent sludge samples studied for the occurrence of HCoV 229E and HKU1 using virome as a benchmark detected 83% of CoV with HKU1 prevalence [52]. The relative abundance of CoV was higher in the influent sample than the effluent sample. Reports around the globe demonstrated the occurrence of SARS-CoV-2 in wastewater [56], [57], [58], [59], [60], [61], [62], [63], [64], [65], [66], [67], [68], [69] (Table 2). Sewage samples collected from seven sites detected nucleocapsid protein gene (N1-3) and E gene [60]. SARS-CoV-2 was detected before three weeks of the first confirmed case, which is very important as early warning signal. N1 and N3 fragments were detected in five sites and E fragment in 4 sites [60]. In another study, SARS-CoV-2 RNA fragments were detected in raw sewage sample [59] and showed positive for SARS-CoV-2 [58]. Bioinformatics with Monte Carlo simulation study detected SARS-CoV-2 from wastewater as inputs estimated the infected people in the selected catchment [38], [56]. Increasing the circulation of the virus in the population will increase the virus load into the sewer systems [60]. The presence of solvents and detergents in wastewater can compromise the viral envelope and therefore affects its persistence [47], [68], [70].
Table 2.
Occurrence of Coronavirus in wastewater.
| Virus | Bench-mark | Environmental Matrix | Concentration of virus particles | Detection Method | Primers/Probe used | Findings | Reference |
|---|---|---|---|---|---|---|---|
| SARS-CoV-1 | Stool from symptomatic patients from two hospitals | Sewage water from two hospitals receiving SARS patients | 3X nutrient broth (pH 7.2) was used to elute the adsorbed viruses, followed by reconcentration by PEG | RT-qPCR |
|
|
[51] |
| Human Coronavirus 229E | Virome | Class B biosolids from wastewater treatment | US EPA Method | PCR |
|
|
[53] |
| Human Coronavirus HKU1 | |||||||
| Human Coronavirus HKU1 | Virome | Influent and effluent sludge | 250 ml liquid sludge mixed with 0.25 M glycine (pH-9) and centrifuged followed by filtration using 5 µm and 0.45 µm sterile membrane filter | PCR |
|
|
[52] |
| Human Coronavirus 229E | |||||||
| Coronaviridae | Virome | Surface water | Bacteriophages precipitated by ultracentrifugation and isolated nucleic acid was concentrated using absolute ethanol | qPCR |
|
|
[54] |
| Alphacoronavirus | Hepatitis A virus | Surface water | Modified glass wool filtration method (increasing pH, contact time of beef extract buffer with glass wool, addition of detergent, recirulation of the buffer) and reconcentration of viruses using PEG | Semi-nested RT-PCR RT-qPCR |
|
|
[55] |
| Betacoronavirus | |||||||
| SARS-CoV-2 | - | Wastewater | RNA extraction from electronegative membranes followed by ultrafiltration | RT-qPCR |
|
|
[56] |
| - | Raw sewage from wastewater treatment plant | Filtration followed by PEG 8000 (8% w/v) precipitation and addition of 0.9 g NaCl to the filtrate followed by its centrifugation to get a pellet | RT-qPCR |
|
|
[57] | |
| - | Raw wastewater Sample | Samples were homogenised, centrifuged to get a pellet. The pellet was resuspended in 1X PBS buffer and concentrated | RT-qPCR |
|
|
[58] | |
| - | 12 raw sewage sample | Using two phase PEG-dextran method as detailed in WHO, 2003 | Broad range PCR targeting ORF1ab and semi-nested PCR |
|
|
[59] | |
| PCR followed by Nested PCR |
|
||||||
| PCR followed by Nested PCR | PCR
|
||||||
| Fragments of the nucleocapsid protein gene (N1-3). One fragment of the envelope protein gene (E) | Sewage sample collected from 7 sites | Ultracentrifugation | RT-PCR |
|
|
[60] | |
| - | WWTPs | PEG precipitation followed by centrifugation and ultrafiltration | RT-PCR |
|
|
[61] | |
| - | 18 Grab samples in 3 WWTPs (both influent and effluent) | Ultrafiltration | RT-PCR |
|
|
[63] | |
| - | Influent, Secondary and tertiary treated samples from WWTPs serving major municipalities | Concentration using Al(OH)3 absorption followed by precipitation, centrifugation, resuspension of pellet in PBS | RT-PCR |
|
|
[64] | |
| - | WWTPs | Filtration followed by PEG precipitation | RT-PCR |
|
|
[65] | |
| - | 7 WWTPs and 2 Manholes | Filtration and PEG precipitation | RT-PCR |
|
|
[66] | |
| - | 6 WWTPs | Filtration and PEG precipitation and UV treatment | RT-PCR |
|
|
[67] | |
| - | Wastewater Sewage Sludge | Wastewater-ultrafiltrationSewage sludge-PEG precipitation | RT-PCR |
|
|
[62] | |
| - | Five influent and five effluent grab samples from WWTPs and three river samples | Concentration using the electronegative membrane-vortex (EMV) followed by filtration, adsorption (direct RNA extraction) and filtration through a mixed cellulose-ester membrane | qPCR Nested PCR |
|
|
[68] | |
| - | Fourteen influent and Fourteen treated effluent samples from 7 STPs (80% of STP capacity of Hyderabad, India) and two sewage samples from gated residential community | Filtration through 0.22 um followed by centrifugation (10 kDa cut-off) | RT-PCR |
|
|
[69] |
4.2. Water
The presence of human enteric viruses were reported on surface and groundwater, public water supply, freshwater and sediments apart from sludge and wastewater samples [71]. Surface water (river, lake, and water reservoir) detected 37 families with a genetic material (dsDNA, ssDNA, and ssRNA) of which dsDNA samples were mostly related to bacteriophages and the majority of sequences related to families of Coronaviridae, Reoviridae and Herpesviridae [54]. Another study on the detection of CoV from surface water using hepatitis A virus (HAV) as a benchmark using semi-nested, real-time reverse transcriptase-polymerase chain reaction (RT-PCR) and quantitative RT-PCR (qRT-PCR) found positive for CoV to the lineage A of alpha-coronavirus (αCoV) related to the rodent clade [55]. Water treatment plants are also susceptible to have viral contamination and pose the possibility to transmit through water distribution if not fully inactivated [45]. Viruses in surface waters are exposed to inactivating stresses such as sunlight and predations [45]. The time required for virus infectivity in pure water and pasteurized settled sewage was several days at ambient temperature, which adds another concern for this potential transmission route [7], [48].
4.3. Engineered wastewater systems
Wastewater treatment plants (WWTPs), in general, have high density and diversity of viruses [72]. Human enteric viruses studied (rotavirus and enterovirus) in the final effluents of five WWTPs showed the persistence of Rotavirus (up to 105 genome copies (GC)/L in 41.7% of the samples) [73]. CoV persisted in primary wastewater for a longer period than secondary wastewater due to the presence of suspended solids [47]. The transmission of COV in the aqueous environment is less susceptible due to inactivation at ambient temperatures [47]. SARS-CoV-2 RNA was detected within six days from WWTPs indicating their persistence in water [56]. The indigenous microbial population generally affects virus survival in wastewater/water [74], [75]. Avian influenza virus (H6N2) survived in methanogenic landfill leachates and water showed rapid inactivation at elevated temperature and non-neutral pH while conductivity did not show any influence on the survival [76]. Viruses abundance in wastewater streams also influenced the bacterial community structure and catalyzed virus-mediated horizontal gene transfer [72]. Bacteriophages associated with the human GI tract are commonly reported in wastewater and phages infecting Bacteroides are used as an indicator for assessing the wastewater quality and contamination [77]. The inactivation time of virus in landfill leachates varied between 30 and 600 days indicating the viable viral infection during and after the waste disposal [76].
Viruses survive in WWTPs absorb/attach to solid surfaces (activated sludge) resulting in virus-laden sludge/biosolids [71]. The affinity of SARS-CoV-2 with biosolids of the sewage line acts as an indicator of incidence as they act as concentrators of SARS-CoV-2 genetic material [62]. The enveloped virus was found to adsorb on the solid fraction (26%) of wastewater, which indicates the virus removal by solid settling [49]. Biosolids as per United States Environemntal Protecton Agency (US-EPA) classified into two-namely Class A or pathogen-free biosolids and Class B biosolids, which may have some pathogens [78]. Class A biosolids can be used for gardening, while Class B biosolids can be applied on agricultural and forest lands which showed the presence of relatively large numbers of viable viruses [79]. Both enveloped and non-enveloped viruses (CoV, Herpesvirus, Torque Teno virus, and Parechovirus) were detected in Class B biosolids [52], [53]. Sewage sludge (influents and effluents) was detected about emerging viruses (CoV, Klassevirus, and cosavirus), wherein 83% of samples were detected for CoV followed by CoV-HKU1 [52]. WWTPs shed viruses through treated effluent discharge as well as biosolids [71]. HCoV inactivation was reported faster in filtered tap water than unfiltered tap water, suggesting that the suspended solids present in water infuse protection for viruses by adsorption [47]. The aerosolization potential of viruses in WWTPs evaluated with the partitioning of bacteriophages (MS2 and Phi6) with reference to sludge (synthetic and anaerobically digested) showed 94% of virions partition with the liquid phase [80]. The biofilm system (trickling filter) showed the abundances of viral sequences compared to the suspended growth system (activated sludge) [72]. Biofilm fosters multiplication of viruses due to the prevalence of higher density of microflora or due to the entrapment of viruses in the extracellular exopolysaccharide matrix that might contribute to viral abundance in the biofilm system. The sediment can function as enteric virus reservoir and viruses can survive for several months in soil and groundwater when temperatures are low and soils are moist, increasing risk because of water contamination [71]. The treatment capacity of WWTPs in the elimination of SARS-CoV-2 genome were reported by few studies (Table 2). SARS-CoV-2 RNA was detected in the sewage influent samples and secondary treated samples, whereas it was not detected in effluent samples of WWTPs stating that the treatment plants are capable of removing the genetic material of SARS-CoV-2 [61], [62], [63], [64], [65], [66], [67], [68], [69].
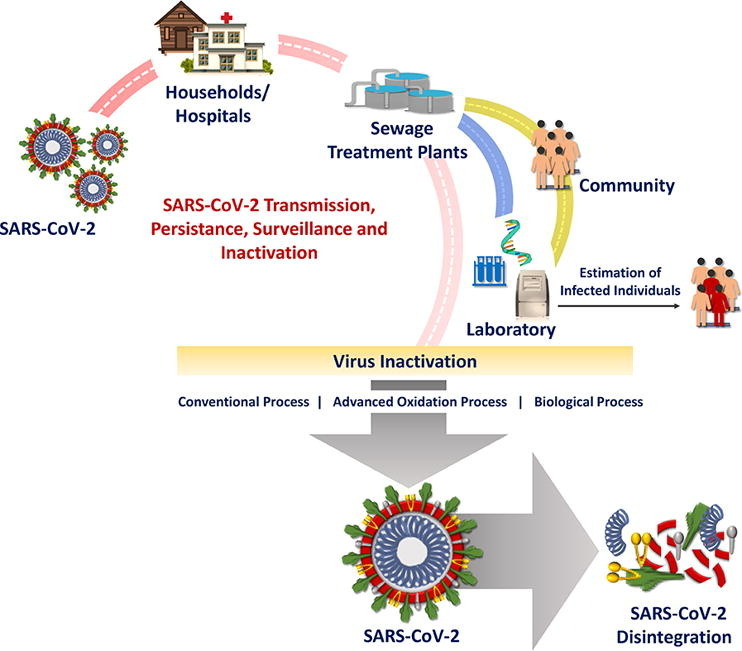
5. Environmental exposure routes – virus transmission
It is evident from studies reported so far that the GI tract is another site of SARS-CoV-2 replication and facilitates shedding through stools [18], [19] (Supplementary Table 1). The transmissibility of SARS-CoV-2 in the community is a major concern that cannot be ignored knowing the ability of other viruses spread to cause waterborne transmission through faecal-oral route [5], [12], [18] which resulted in many sporadic cases and outbreaks that are severe or sometimes fatal [81]. Enteric viruses infect GI tract, multiply and excrete through faecal-route contaminating water, food, and air through sewage discharges, septic tanks, water supplies, and runoff [82]. The range of severity depends on the virus survival and replication capability and most of the time an asymptotic individual unknowingly can infect and thus spreads [69], [83]. So far low SARS-CoV-2 loads in stool samples were reported [84]. The persistence of viral RNA in faeces for 33 days after the patient was tested negative [44] cannot be ignored. In general, enteric viruses facilitate transmission at a very low infectious dose (<20 particles) required to cause illness [153]. In some reports, high titers of virus in the faeces and occasionally, at lower concentrations in urines were detected in infected humans [5], [85].
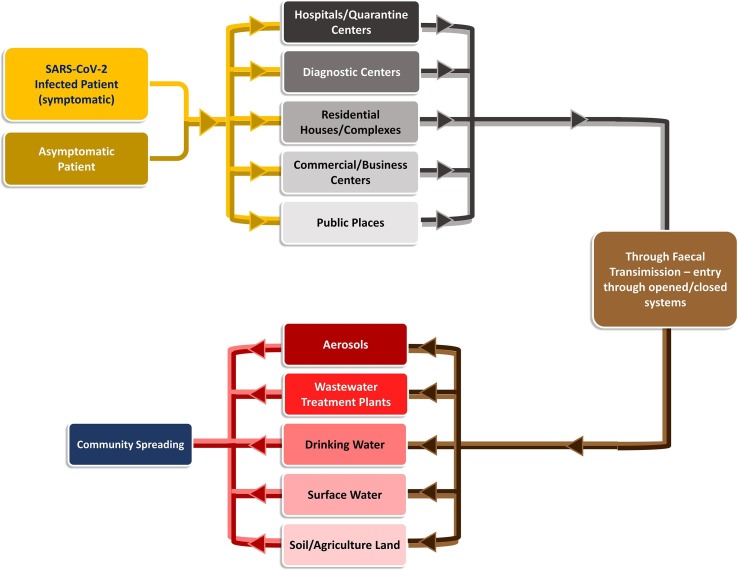
Enteric virus transmission to the community with wastewater streams is now a major point of focus and concern [12], [70] even though SARS-CoV-2 transmission via the faecal-oral route is still unclear with the limited information [19], [44], [86], [87]. The possibility of community transmissibility spread cannot be ignored based on the previous experience [5], [18], [12], [81], [88]. Viruses excreted through faecal-oral route usually link with the existing water infrastructure connected with drains, sewage, water, treatment plants, hospitals/clinics, diagnostic centres, etc. (Fig. 2 ) and manifest the transmission. Enteric virus occurrence was reported in surface water, sewage, WWTP discharges, septic tank outflow, sewer overflows, runoffs, and infiltration during rainfall which eventually leads to contaminate surface and groundwater [71]. The majority of the population will exhibit mild symptoms or carry viruses asymptomatically and are still capable of shedding the virus through faeces [69], [70], [89], [90], which eventually spread the virus through sewerage infrastructure [6], [91].
Fig. 2.
Possible Transmission Route of SARS-CoV-2.
5.1. Airborne transmission – virus laden aerosols
Given, the spread of SARS-CoV-2, the interconnectedness of the wastewater infrastructure, the concentrations of infected people pose a greater threat for airborne transmission due to aerosolization of the virus [92], [93]. Infectious viruses, including CoV, can survive for long periods outside of its host organism [94]. Contaminated water/wastewater will also function as a potential vehicle for human exposure if virus-laden aerosols are formed [48]. Infected stools further transmit through virus-laden aerosols during flushing operation [7]. The faulty sewage-drainage system was linked to the SARS-CoV-1 outbreak among the residents living in the surrounding buildings [95] wherein aerosolized particles play a major role in virus spreading [96]. The transportation of CoV in water potentially increases the virus spread through the aerosol formation [97] particularly during wastewater discharge and flow in the drainage system, pumping stations and WWTPs.
Airborne dust is another transmission route linked to infectious diseases [7], [96]. Although SARS-CoV-2 is not an airborne virus, adsorption of the virus on airborne dust and particulate matters (PM) contributes to long-range transport of the virus and their inhalation could pass the virus into deeper alveolar and tracheobronchial regions, which could increase the chance of infection [7], [98]. The high levels of PM concentration in air pollution might increase the susceptibility of the population to more serious symptoms of the disease [7]. High COVID-19 infection rate was linked to the air pollution in the Italian cities that exceeds the PM10 levels [99] and the cities with >100 days of air pollution had shown a high number of infection rate which was further associated with low wind speed. In other air quality studies with reference to two regions in Italy namely Lombardy and Emilia Romagna reported for their highest level of virus lethality in the world was among the most polluted areas in Europe [100]. Larger respiratory droplets (>5 μm) in general stay in the air for a short period of time [101], [102], while small aerosolized droplets (<5 μm) can persist for a longer time in the air and also could travel for long distances (>1 m) [103]. Transmission via the inhalation of small, exhaled respiratory aerosol droplets remain airborne for prolonged periods, mediating long-range human via air movement [7]. The aerosols with PM2.5 transmit to the respiratory tract more easily [104], [105], [106], which can be correlated with the SARS outbreak in Hong Kong and MERS outbreak in South Korea [107]. Air transmission dynamics of COVID-19 suggests possible ‘pollution-to-human transmission’ associated with the airborne viral infectivity [98].
Atmospheric loading of CoV as aerosols from wastewater provides a more direct route for human exposure [47] which is so far less understood. The virus that enters the sewer systems forms virus-laden aerosols [7] through mechanisms of cross-contamination [108], [109]. Unit operation in WWTP such as biological oxidation normally employed for treatment of sewage/wastewater influences the aerosol formation due to aeration operation. Particulate matter emissions from aeration basins demonstrated that wastewater material is getting aerosolized and transported beyond the facilities [97]. The potential for viruses to be aerosolized was studied and monitored by spiking Ebola virus surrogates (MS2 and Phi6) in wastewater systems (toilets, lab-scale aeration basin, and lab-scale model of converging sewer pipes) [110]. Emission rates of MS2 and Phi6 were 547 PFU/min and 3.8 PFU/min, respectively, for the aeration basin and 79 PFU/min and 0.3 PFU/min for the sewer pipes. Rotavirus and Norovirus were detected above the ASP from WWTP higher than the threshold values recommended [111]. Even though no concrete evidence is reported so far on the spread of SARS-CoV-2 through air, it is important to investigate and understand [93].
6. Environmental factors influence on the virus transmission
Environmental (meteorological) factors such as temperature, humidity, precipitation, and air-flow will have a significant influence on virus transmission/infection [102], [112].
6.1. Temperature
Temperatures are prone to inactivation of enteric viruses under specified conditions [47], [49], [74]. Two surrogate CoV namely TGEV and MHV survival were studied in water (reagent-grade), lake water, and settled human sewage at room temperature (23–25 °C) and 4 °C for 6 weeks [48]. The virus infection declined rapidly at 25 °C, indicating the regulatory role of temperature on the viral inactivation in the water matrix. At 25 °C, 99.9% reduction (T99.9) in reagent-grade water was observed for 33 days (TGEV) and 26 days (MHV), while in settled sewage (pasteurized) between 10 and 14 days. The fate of a HCoV-229E and animal FIPV CoV was evaluated in tap water (filtered and non-filtered) and wastewater (primary and activated sludge effluents) [47]. HCoV inactivated rapidly in wastewater (T99.9 of 2.77; 3.54 days] and tap water [T99.9 of 12.1; 12.5 days] while FIPV survived 2 to 3 times longer for a comparable reduction in wastewater (10.9 days) and in secondary effluents (5.7 days). Wastewater (unpasteurized and pasteurized) spiked with the viral stocks [enveloped viruses (MHV and Pseudomonas phage φ6) and non-enveloped viruses (bacteriophages MS2 and T3) incubated at 25 °C (summer) or 10 °C (winter) showed rapid inactivation of enveloped viruses in unpasteurized wastewaters at 25 °C [T90 of 13 h (MHV) and 7 h (phage φ6)], compared to non-enveloped phage MS2 [121 h] [49]. The inactivation kinetics of both MHV and φ6 at 10 °C was relatively slower [T90 of 28–36 h]. In pasteurized wastewater, both MHV and phage φ6 lost infectivity at a significantly slower rate compared to unpasteurized wastewater (T90 of 19 h (MHV) and 53 h (phage φ6)) at 25 °C [49].
6.2. Humidity and precipitation
The relationship between relative humidity and disease transmission depends on the persistence of viral viability before infecting susceptible individuals [110]. The absolute/relative humidity (AH/RH) role in mediating virus infectivity still needs investigation. Lower RH than 33% showed good viability than the corresponding intermediate RHs [110]. The aerosols (submicron) and droplets (1 μL) showed the transmission of infectious diseases [110]. Enveloped bacteriophage (Phi6) survived good at both high (>85%) and low (<60%) RHs with a significant decrease in infectivity at mid-range RHs (~60 to 85%) [113] and suggested that RH is the important factor in controlling virus infectivity in droplets. The non-linear nature of the relationship of influenza A virus with temperature and influenza B virus with AH, RH, and temperature explained the system complexity on virulence [112]. Temperature (air/water) and RH showed marked influence in the inactivate rates of the enveloped virus [48], [110], [114]. Viral transmissibility also significantly depends on precipitation [102]. Rainfall events manifest the mixing of untreated sewage and wastewater and get discharged into surface water pruning to increased exposure risk [115]. The risk of exposure via the faecal-oral route is also of particular concern where open defecation or non-sewered sanitation is being practised [70] and will further intensify when rainfall events occur.
6.3. Animals and plants
Reports indicate that the CoV infects pets, livestock, and wildlife [116], [117]. Due to prevailing environmental conditions, the transmission possibility through animals might happen but still needs to be investigated. Several viral families (highly stable pathogens) remain infective and transport long distances in water [118], which increased the potential impact when reclaimed wastewater was used for irrigation and animal husbandry. Studies on water-mediated plant virus transmission with reference to WWTPs, virus inactivation need to be investigated thoroughly along with enteric virus-plant association [118], [119].
7. Detection of virus in environmental matrices
Detection of enteric viruses in wastewater, ground/surface water, sludge samples, and drinking water is challenging due to the presence of low virus concentration (traces of viral particles or virions) in huge volumes of water which also depends on the severity of the infection. For accurate detection environmental samples need to be concentrated using microbiological and molecular methods prior to the detection [77], [120]. However, virus recovery by concentrating methods are always challenging [77].
7.1. Concentration
Concentrating viruses by ultracentrifugation, ultrafiltration, adsorption and elution (VIRADEL), coagulation, size-exclusion and flocculation are often used either in alone or in combination [120], [121]. Wastewater spiked with bovine enteric coronavirus (BCoV) was recovered using glass powder adsorption with 0.05 M glycine solution and a 3% beef extract solution varied with acidic (pH 3.3) and alkaline (pH 9) pH solution yielded in the recovery of 24% (pH 3.3) and 28% (pH 9) [122]. Glass columns equipped with electropositive filter particles (silica gel with Al(OH)3)) were used for concentrating SARS-CoV-1 and f2 phage and their mixture from sewage [46]. The spiked sewage samples concentrated using glass column followed by polyethylene glycol (PEG) precipitation showed recovery efficiencies of 21.4% (SARS-CoV-1)) and >100% (f2 phage) [46]. The concentration of a variety of waterborne viruses was reported using glass wool filtration from runoff of agricultural fields [123]. Bovine origin viruses like bovine viral diarrhea virus type 1 and 2 (BVDV1 and BVDV2), bovine rotavirus group A (BoRv) and poliovirus 3 were spiked to the water samples prior to glass filtration followed by elution (3% beef extract-glycine buffer; pH 9.5) and PEG flocculation before qPCR analysis. A higher recovery of 57.9% was observed with non-enveloped poliovirus 3 [123]. The recovery of enveloped (murine hepatitis virus ((MHV)–rodent coronavirus) and non-enveloped (phage MS2) viruses spiked in untreated sewage was evaluated employing PEG precipitation, ultracentrifugation and ultrafiltration [49]. Ultrafiltration resulted in the highest recoveries of both MHV (25.1%) and MS2 (55.6%). HAV and TGEV spiked water samples were concentrated using glass wool filtration followed by PEG precipitation [55]. The pH of elution buffer (pH 11) showed an increase in recovery from 2.6% to 28.8% when 5 L sample is used. Elution buffer with pH 11 with the addition of tween 80 resulted in the recovery of 23.9% (HAV) and 18% (TGEV). Secondary concentration by PEG precipitation showed a recovery of 51.3% (TGEV) and 47.2% (HAV) [55]. SARS-CoV-2 and MHV [124] were also concentrated to observe their existence in wastewater samples employing electronegative membrane filtration followed by ultrafiltration (100–200 mL concentrated to ~250 μL) [56], [124], PEG precipitation followed by centrifugation (11 mL concentrated to 400 µL) [57], [124], homogenization (40 mL) followed by centrifugation to achieve a pellet which was resuspended in 1X PBS buffer [57], [124], and also two phase PEG-dextran method [59], [124] using ultrafiltration and ultracentrifugation was also adopted [60], [69].
7.2. Detection
Various methods like isothermal amplification of target genes [51], biosensors [125], microarrays [126], [127], culture-based assays [128], metagenomics [129] and polymerase chain reaction (PCR) and its types [130], [131] are employed for the detection and quantification of the virus in the environmental samples (Table 2). E. coli and f2 phage [51], Poliovirus-1 (PV-1) Virome [47], [52], [53], [54], and HAV [55] were used as indicators for the evaluation of persistence and occurrence of CoV in environment wastewaters (Table 1, Table 2). PCR and its types like quantitative PCR (qPCR), real-time RT-PCR, RT-qPCR, nested PCR and digital PCR (dPCR) have been implemented for detecting enteric virus contamination in the water/wastewater [132]. qPCR can able to multiplex and detect viral targets [80], [133]. dPCR quantifies the viral genome and its population count in wastewaters without depending on known and calibrated standards [134], [135]. RT-PCR, RT-qPCR, and nested PCR use specific primers and probes for detection and quantification [136]. The detection rates depend on the primer and probes used to detect target virus which should be revised frequently to assure the novel strain detection [77]. An integrated method, where integrated cell culture (ICC) is combined with PCR called ICC-PCR was used to monitor the viruses in wastewater matrices [137]. Molecular techniques possess the potential to detect the low concentration of viruses in a large volume of wastewater either individually or with integrity [138].
A model enveloped virus Murine hepatitis strain A59 (MHV) grown in L2 and delayed brain tumor cell cultures (DBT) were used to detect the persistence of CoV in waste environmental matrices [49]. Cell lines of swine testicular cell cultures [48], Crandell Reese feline kidney cell line (CRFK), fetal human lung fibroblast, MRC-5 cell line, Buffalo green monkey kidney cell line [47] and Vero E6 [46] were also used to detect CoV. The occurrence of CoV (SARS-CoV-1 and 2) was detected majorly using PCR based techniques as RT-PCR [55], [60], [69], RT-qPCR [46], [55], [56], [57], [58], qPCR [54], semi-nested PCR [49], [58], nested PCR [59] (Table 1).
Next-generation sequencing (NGS) facilitates to study of virus diversity using amplicon sequencing [139]. Data achieved from NGS has the potential to increase knowledge of the viral community and its diversity in the environment [140]. Apart from whole virus detection, genome, and capsid integrity of the viruses can also be detected using molecular techniques and markers [141]. The wastewater disinfection using chemical or physical treatments causes virus inactivation modifying viral genome and proteins. The quantitative data for their integrity can be detected using RT-qPCR and protein mass spectrometry (MALDI-TOF MS) [142]. These methods help to identify the point changes that occurred rather than observing the complete modification caused [142]. Inconsistency was most often observed due to inhibitory substances present in a stool sample (bile salts, polysaccharides, lipids, and urate) and other samples (debris, humic acids, and polyphenols) as they co-precipitates or adsorbs with nucleic acid during the detection [143]. Cloning, metagenomic analysis (virus), and RT-qPCR with the use of process controls help to detect viruses accurately by knowing the virus recovery rate and the level of inhibition [121], [144], [145], [146], [147]. The selectivity and choice of process controls depend on the molecular and biochemical structure of the virus and their route of infection [148], [149].
Although PCR based techniques are more sensitive, specific, consumes less time (in hours), can also detect virus which cannot be culturable providing quantitative data of viral genome and is sometimes not conducive and effective [150], [121]. Rapid detection methods using dry samples [151] and paper analytical devices [150], were reported as a diagnostic tool to detect SARS-CoV-2 from a patient sample. These modifications and developments in methods and use of portable devices detects virus on-site, track virus carriers and serves as an early warning signal to the community to prevent the spread of the outbreak [150]. Paper-based devices are portable, easy to store, transport and incinerate [150]. A pilot air sampling study conducted to detect SARS-CoV-2 RNA (0.87 virus genomes/L air) and the approach illustrate the feasibility of tracking the progression of the outbreak using environmental aerosol samples instead of human specimens [152].
8. Fate of enveloped virus
Outbreaks of enveloped viruses, such as CoV (MERS, SARS-COV-1, and SARS-CoV-2), Ebola, Hantavirus, Measles, Zika, Avian influenzas, and Lassa virus, persisted in the water environment warranted effective management of the infectious waste and wastewater [153], [162], [163]. Enveloped viruses are structurally diverse to the non-enveloped viruses, and behave differently in water environment [154]. The survivability of the viruses also varies based on the presence and absence of envelope [155]. Reports on the enveloped virus with reference to their fate, transport, and inactivation are limited [11]. The persistence of enveloped viruses in aqueous medium was studied with the enveloped bacteriophage Phi6 as a surrogate (influenza viruses and coronaviruses) and observed that T90 (time for 90% inactivation) of Phi6 varied between 24 min and 117 days [49] which depends on the temperature, biological activity, and aqueous media composition [12]. The capsid protein of non-enveloped viruses is less susceptible to lipid solvents, temperature, and pH [156]. On the contrary, enveloped viruses are more sensitive to disinfectants, heat, and solvents [156], [157]. Non-enveloped viruses displayed higher resistance in water environments compared to the enveloped viruses [47]. CoV is much more sensitive to temperature than Poliovirus 1 LSc-2ab (PV-1) and that there is a considerable difference in survivability between PV-1 and the CoV in wastewater attributed due to the fact that enveloped virus is less stable in the environment than non-enveloped viruses [47].
9. Inactivation of virus
The survivability of viruses in nature as well as engineered systems impacts the public health [158] and therefore, enteric viruses must be removed or inactivated by 4 logs (99.99%) during water treatment [158]. For enteric viruses, WWTPs will function as an important and key barrier [71], [159], [160], [161]. The viral pathogens load discharging to wastewater treatment plants through domestic wastewater is typically in the range of 106–108 GC/L [159]. The occurrence of human adenovirus (HAdV), JC polyomavirus (JCV), and A rotaviruses (RVA) was reported in the range of 106–108 GC/L [162]. Human viruses (adenovirus, polyomavirus, and torque teno virus) were detected in the influent (105–106 GC/L) [163].
9.1. Wastewater treatment plants
WWTPs functions as the main barriers to terminate the virus transmission. Unit operation in WWTPs typically includes primary (screening, equalization (optional), coagulation, or settling (primary and secondary), main or biological ASP or sequential batch reactor (SBR) or membrane bioreactors operated with aerobic, anoxic and/or anaerobic microenvironments) and tertiary treatment (filtration (sand/membrane), adsorption (activated carbon or disinfection (UV irradiation, chlorination, ozonation, etc.), constructed wetlands or stabilization ponds in addition to the anaerobic digestion (for stabilization of sludge). Unit operations play a crucial role in virus removal [159]. The placement of unit operations in a defined sequence will mainly depend on the inflow wastewater quality, treated water quality required for discharge, and intended use of the treated water. Four WWTPs were evaluated for the presence of HAdV, JCV, and RVA for one year [162]. In the secondary effluent, HAdV was detected in all the analyzed samples followed by JCV (85.4%) and RVA (97.9%) [162]. HAdV was the more persistent virus detected in the tertiary effluent (62.2%), while membrane bioreactor (MBR) showed relatively better virus removal. The fate of three human viruses (adenovirus, polyomavirus, and torque teno virus) in three WWTPs was evaluated mainly operating with ASP [161]. All three human viruses were consistently persisted in the secondary treated effluent (102–103 GC/L). ASP in sub-tropical climate condition which could function as an effective treatment barrier with >3 log10 removal efficiency of enteric virus and tertiary treatment, in addition, is essentially required prior to reuse for non-potable purposes or discharge [161]. A comprehensive assessment of the fate of eight viruses during multiple steps of full-scale WWTPs was also reported [160]. Viral communities in WWT include large numbers of bacteriophage specific to bacterial hosts that influence the bacterial community structure and dynamics [72], [163].
Enveloped virus partition to solids is more compared to non-enveloped viruses [49], wherein the virus gets absorbed to suspended particles in wastewater as well as sludge/biosolids [71]. Solids separation and activated sludge flocs majorly contribute to virus removal. Sedimentation and sand filtration also contributed to virus removal before disinfection [164], [165]. Coagulation followed by sand filtration showed improved virus removal capability [166]. Physical process (sedimentation and filtration) and biological process (ASP-Sludge) separates/transfer the infective viruses from liquid to solid phase without inactivating and therefore exists a potential risk of transmission through other pathways [159]. Application of sludge stabilization processes, such as heat treatment (50–75 °C), mechanical dehydration/desiccation (virus capsid rupture), liming process, composting process (temperature and antagonistic organisms) and anaerobic digestion (mesophile) showed variable viral inactivation capability [159], [167], [168], [169]. During the treatment process, the virus gets inactivated due to the prevailing environmental conditions as well as microbial antagonism [170].
9.2. Disinfection process & technologies
The conventional WWTPs at present are not specifically designed to remove viruses. Disinfection is the unit operation designed in existing WWTPs for intended inactivation of pathogens including viruses.
9.2.1. Conventional process
Disinfectants agents/Chemical oxidants (chlorine (Cl−), chlorine dioxide (ClO2), ozone (O3), hydrogen peroxide (H2O2), sodium hypochlorite (NaOCl) and paracetic acid (PAA)) and UV-irradiation are most commonly used for inactivation of viruses (Fig. 3 ) (Table 3 ). Solar irradiation, filtration (slow sand, porous ceramic), membrane (micro/ultra/nano and reverse osmosis), etc. along with stabilization ponds and constructed wetlands also reported inactivating virus to some extent depending on the operational conditions and wastewater nature. Chlorine (ClO2, free chlorine (FC)) is well established and achieves inactivation by oxidizing the cellular material of the virus [71]. Contact time, dose, pH, temperature, and wastewater/water composition are critical factors that influence the virus inactivation efficiency. Chlorine (dose, 10/20 mg/L; contact time, 10/1 min) documented complete inactivation of SARS-CoV-1 in spite of in-effective inactivation of f2 phage [51]. Free chlorine was reported effective in inactivating SARS-CoV-1 and f2 phage than chlorine dioxide. The free residual chlorine (0.5 mg/L from Cl− or 2.19 mg/L from ClO2) ensured near complete inactivation of SARS-CoV-1 in wastewater [51]. However, the requirement of high doses of Cl− and the possibility of formation of disinfection by-products (DBPs) warrants its usage [171], [172]. PAA is considered as an alternative to the chlorination process, wherein the H2O2 and acetic acid (CH3COOH) generates reactive hydroxyl radicals [173], [174]. PAA application as an inactivation agent eventually increases the carbon concentration in final treated effluent due to the presence of acetic acid [172]. In a study conducted to estimate the SARS-CoV-2 infected individuals real field influent samples from STPs were collected and subjected to various concentration of NaOCl (0.1%, 0.5%, 1%, 2%, 3% and 4%) [69]. The concentration ≥2% NaOCl did not show any detection of SARS-CoV-2 genome indicating its complete inactivation [69]. 0.5 mg/L of free chlorine with 30 min of exposure time at 22 ± 3 °C would inactivates SARS CoV-2 viruses completely [44].
Fig. 3.
Conventional disinfections methods of virus inactivation.
Table 3.
Methods of Virus Inactivation.
| Type | Method | Process/Mechanism | Action on virus | Reference |
|---|---|---|---|---|
| Conventional methods |
|
Inactivates by oxidizing the structure of DNA/RNA and protein layer of enveloped and non-enveloped viruses. Excess dosages may lead to the formation of residual by-products. | Inactivates upto 99% of viruses | [51], [69], [71], [189], [171], [172], [173], [174], [175], [176], [177], [178], [179], [180], [181], [182], [184], [185], [186] |
| Filtration methods |
|
Allows separation through a usage of physical barrier from water. | Remove virus and bacteriophages upto 0.001 m size | [6], [71], [172], [191] |
| Advanced oxidation |
|
Rapid release of high reactive species (OH* radicals) that oxidizes or destroys enteric viruses and nucleic acids, cell membrane/ capsid of bacteriophages without the release of byproducts. | Inactivates enveloped viruses upto 99.99% | [6], [142], [159], [171], [193], [195], [214], [237], [238], [198], [199], [200], [201], [202], [203], [204], [205], [206], [217], [218], [219], [220] |
| Other Processes |
|
Synergic functions of sunlight | Bacteriophages also gets inactivated | [188], [190] |
Ozone (oxidation-potential, 2.07 V) leads to the formation of secondary oxidants (hydroxyl radicals) with higher reactivity which can inactivate in shorter reaction times [171], [175], [176], in spite of its reactive/corrosive nature, and the cost. The susceptibility of the selected viruses towards ozone application in the increasing order: bacteriophages (Qβ) > coxsackievirus B5 (CVEnv2) > echovirus 11 (EV) > bacteriophages (MS2) > bacteriophages (Φ174 & T4) > human adenovirus (HAdV) > coxsackievirus B5 (CVF, CVEnv1) [176]. The UV irradiation (220–320 nm) is also one of the widely used methods to inactivate pathogens including viruses. Inactivation of enteric viruses (noroviruses, rotavirus, reovirus, sapovirus, astrovirus, enteroviruses, adenoviruses, and JC virus) was evaluated with UV during WWTPs operation over two years period [177]. Both pre-UV and post-UV samples showed a relatively higher load of selected viruses. Log10 reduction of total infectious viruses (1.46 and 1.67 log) by UV irradiation was comparable to reovirus reduction (1.23 and 1.75 log). UV254 irradiation primary damaged the nucleic acid while inactivating human Norovirus surrogate (HuNoV) [178]. MS2 (with ssRNA) showed resistant to UV253.7 inactivation (7 log; 1800 J/m2) compared to other ssRNA viruses [179]. Heterogeneous sensitivities were observed in the genome regions after UV254 treatment [180]. The efficiency of UV disinfection depends on the time of exposure, irradiation intensity along with suspended particles/colour/turbidity of water/wastewater. UV functions with few seconds of contact time without chemical addition (no residual or chemical formation). Viral aggregation enhances genome recombination (damaged viruses inside their host cells) and therefore, reduces viral inactivation during the UV treatment [181]. The inactivation of bacteriophages (MS2, fr, and GA) by UV254, singlet oxygen (O2), free chlorine (FC), and chlorine dioxide (ClO2) was also studied [182].
Solar water disinfection (SODIS) on the inactivation of HAV and HNoV, murine Norovirus (MNV-1) indicated that the sunlight radiation associated with temperature play a major role in the inactivation [183], [184]. Wastewater temperature manifest higher inactivation rates specifically at higher values [47], [49]. Six hours of exposure to sun inactivated enteric viruses [184], [185]. Sunlight inactivates viruses via endogenous inactivation (absorption of solar light in the UVB range) and exogenous processes (adsorption of sunlight by external chromophores/sensitizers, which generate reactive species) [186]. Irradiation of water with solar simulator showed inactivation of human enteric viruses (MS2 coliphage) [187]. Viruses that are not susceptible to exogenous inactivation use UVB wavelengths (280–320 nm) for inactivation [188]. Endogenous sunlight inhibits viral RNA synthesis [189]. Extended solar exposure time ensured both bacterial and viral inactivation and discharge of the solar treated wastewater could allow the viral replication when the host is present [190]. Sunlight irradiation in natural/engineered treatment systems (drains, stabilization/algal ponds, constructed wetlands, water bodies, etc.) also functions to inactivate pathogens to a certain extent [188].
The membrane-based process allows separation of the pathogen by a physical barrier. Microfiltration (0.1–10 m) enables removal of bacteria/protozoan cysts and ultrafiltration (0.01–0.1 m) and nanofiltration (0.001–0.01 m) allows removal of viruses [172]. Reverse osmosis (<0.001 m) reject viruses from water along with virus surrogates such as bacteriophage MS2 [191]. MBR is a hybrid system with the integration of ASP with membrane filtration for biomass separation in a submerged or side-stream configuration [6], [71]. Full-scale MBR (pore size 0.04 μm) showed good log removal of four pathogenic viruses [adenovirus (3.9–5.5), norovirus GII (4.6–5.7), F+ coliphage (5.4–7.1)] [192].
9.2.2. Emerging processes
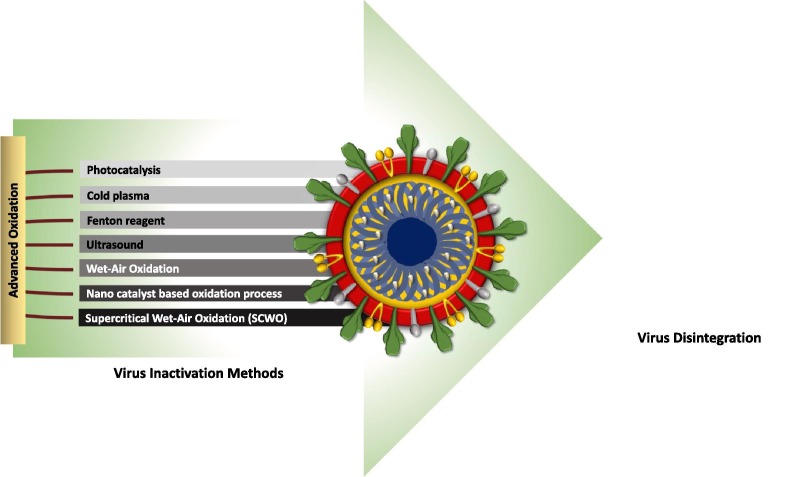
Although virus removal was achieved by conventional treatment processes to some extent, there is still emerging scope and need for effective virus inactivation technologies [6], [159]. The multibarrier wastewater and drinking water treatment systems are likely effective in protecting against SARS-CoV-2 [12]. The advanced and integrated process specifically targetting functional genome damage will aid in developing better methods for diverse viral pathogens [142]. UV-based advanced oxidation processes (AOPs) (Fig. 4 ) integrating with H2O2, Cl2, O3 and functional material facilitate enhanced reactive radical generation by photolysis [159]. AOPs employing UVC, UVC-H2O2, and UV-Fenton for inactivation of enteric surrogates (MS2 bacteriophage) in ultrapure water and synthetic matrices (wastewater and urine) was studied [193]. The occurrence of MS2 or E. coli notably decreased the antagonist microorganism inactivation kinetics, whereas, the addition of H2O2 improves the overall inactivation performance. The presence of H2O2 and Fe improved the inactivation rate significantly. Pulsed UV irradiation and low-pressure UV irradiation were applied for the inactivation of NoV and FRNA bacteriophage (GA) in secondary treated wastewaters [194]. At a high dose of UV (6.9 J/cm2) GA showed significant inactivation. Suspended solids concentration showed a direct impact on the process efficiency and settlement processes played an important role. The integration of UV irradiation with PAA yielded effective inactivation [195]. Ozone integrated with H2O2 or persulfate/monopersulfate promoted the production of hydroxyl radicals [196]. Ozone and photo-assisted integrated disinfection facilitated higher quality of water despite of cost [171].
Fig. 4.
Advanced oxidation methods used for virus inactivation
Photocatalytic disinfection due to its potential oxidative capability and ability to utilize solar energy is attracting significant attention in the disinfection domain [197]. Photocatalysis induces catalyst (semiconductor) function in the presence of irradiation source, sunlight or UV lamp [168], [198], [199]. Magnetic metals (iron (Fe), cobalt (Co) and nickel (Ni)), semiconductor nanoparticles (NP) such as metal oxides and doped structures (TiO2, ZnO, CuO, MgO, SnO2, WO3, SiO2, Fe2O3, Nb2O3, TiO2/CuO, Ag/TiO2, TiO2/Pt, Fe2O3/TiO2, Au/TiO2 and TiO2 doped with N-, C-, S-), noble metals (gold (Au), silver (Ag) and platinum (Pt)) based inorganic NPs and engineered carbon NPs (carbon nanotubes (CNTs), C60, and C70 fullerenes) [200]. TiO2 is a potential photocatalyst semiconductor material due to its strong oxidizing power [201], prolonged stability high reactivity and faster disinfection kinetics. Illumination of UV light at a wavelength (<385 nm), TiO2 generates an electron-hole on the surface pair by photon energy and generated hydroxyl radicals (*OH), and the electrons present in the conduction band produced superoxide ions (O2 −).*OH radicals are highly reactive species upon contacting the virus surface and enable inactivation rapidly [171], [198], [202], [203], [204], [205], [206], [207]. The generated oxidative species penetrate into the lipid layer of the virus which leads to inhibition or lysis [208]. Photocatalytic viral inactivation processes follow mainly three steps i) shape distortion, ii) protein oxidation, and iii) gene damage [209]. Photocatalytic membrane reactors (PMRs), using nano-TiO2–membrane coupled system inactivation of phage F2 model for a human enteric virus-like poliovirus, hepatitis A viruses, coxsackie virus, Norwalk, Rotavirus, Adriano virus, Herpes virus, and influenza virus was studied in flat membranes showed 1.8–5.9 log units of disinfection efficiency in wastewater [197], [210]. PMR (Nano-TiO2 P25 (10–25 mg/L); Flat-sheet polyvinylidene fluoride (PVDF) membrane (pore size of 0.15 µm), UV intensity, 0.16 mW/cm2) showed bacteriophage f2 (25 ± 1 nm) inactivation (5 log) primarily by photocatalysis where membrane functioned as a barrier [197].
Composite SiO2-TiO2 and silver-doped TiO2 NPs showed 2.7 times higher photocatalytic inactivation rates with bacteriophage MS2 than the unmodified TiO2 [211], [212]. TiO2 and ZnO showed efficient inactivation in the following order: viruses > prions > Gram-negative bacteria > Gram-positive bacteria > yeasts > molds [204]. TiO2 membrane with UV irradiation of airborne pathogens was noticed between photocatalyst sterilization and UV treatment process and the combination/impregnation of these TiO2 with various active metals such as Pt, Ag, and Cu. The TiO2/Ag nano-anti microbial materials used to study growth inhibition rates and found 97.9% inhibition [213]. Photocatalytic oxidation using undoped TiO2 and platinized sulfated TiO2 (Pt/TiO2) yielded 90% inactivation within 30 min using UV-irradiation on TiO2 and 90% to 99.8% with Pt/TiO2 over the viral and bacterial aerosols of influenza A (H3N2) virus, vaccinia virus, Mycobacterium smegmatis and Bacillus thuringiensis [214]. Photocatalytic titanium apatite filter (PTAF) absorbs and inactivates SARS-CoV-1 up to 99.99% within 6 h interaction under non-UV irradiation [203]. Photoelectrocatalytical (PEC) reactors perform the photocatalytic reactions enhancing the viral inactivation significantly (>90%) by applying a negative potential lead to damage of the virus due to the manifested electrostatic attraction between negatively charged viral capsid and catalyst surface [208], [215]. The virucidal activity in the presence of low concentrations of halides with reference to adenovirus inactivation by PEC systems showed positive influence by halide oxidation with a prolonged lifetime of photoholes (h+) [216].
Filters based on PVDF membranes (5 μm) coated with multiwalled carbon nanotubes (MWCNTs) layer showed effective removal of viral bacteriophages [217]. The main limitation of using MWCNTs for virus filtrations is the retention of the virus after the filtration [218]. MWCNT fictionalization with metals and metal oxides with anti-microbial silver, copper, and copper oxides would help an additional virus inactivation [219]. MWCNTs functionalized with silver-based filters showed complete removal of poliovirus, norovirus, and coxsackie virus [32]. Silver NPs deposition by electrochemical means over carbon covered alumina (CCA) showed effective inactivation of pathogens in water [220]. MWCNTs coated with copper(I) oxide (Cu2O) yielded good virus (MS2 bacteriophages) efficiency for specific water applications at relatively low cost [219]. Coagulation followed by ultrafiltration in pilot-scale wastewater reclamation plants (using secondary treated effluent as feed) using F-specific RNA bacteriophage MS2 showed stable at pH 5.5 with high virus removal rate [221].
Cold plasma (CP) is one strategy that mediated virus inactivation (Table 3). Plasma (partially or fully ionized gas) facilitates the emission of UV radiation along with reactive oxygen and/or nitrogen species (RONS) which eventually damage the nucleic acids and oxidize nucleic acids, proteins and lipids respectively [237]. The formation of ROS and/or RNS is the main limiting factor that contributes to virus inactivation. Viral aerosols inactivation in an air-stream with non-thermal plasma (NTP) exposure showed exponentially inactivation of aerosolized MS2 phage by increasing the applied voltage using a packed-bed dielectric barrier discharge reactor [239].
The removal of Enterovirus and Norovirus in algal-WWTS (A-WWTS) demonstrated feasibility in producing pathogenically-safe effluent with minimal post-disinfectant [240]. The virus community in the chlorinated-WWTS effluent showed ~250 diverse species while only 14 discrete non-pathogenic virus species were found in the non-chlorinated effluent of A-WWTS [240]. The fate of human enteric viruses (i.e. norovirus, adenovirus, Aichi virus 1, polyomaviruses, and enterovirus) and plant virus (pepper mild mottle virus (PMMoV)) were studied with reference to two surface flow wetlands [238]. Adenovirus and Aichi virus 1 was found in abundance in treated wastewater (an inlet of the wetlands) at concentrations of 102–105 GC/L [238]. Viral removal efficiencies by wetlands ranged between 1 and 3 log10. Polyomaviruses were removed to below detection limits while PMMoV was detected in a greater concentration in the inlet of both (104–107 GC/L), but exhibited little or no removal. Photosynthetic induced biological activity and temperature played a key role in virus removal in the wetlands [238].
9.3. Inactivation mechanism of virus
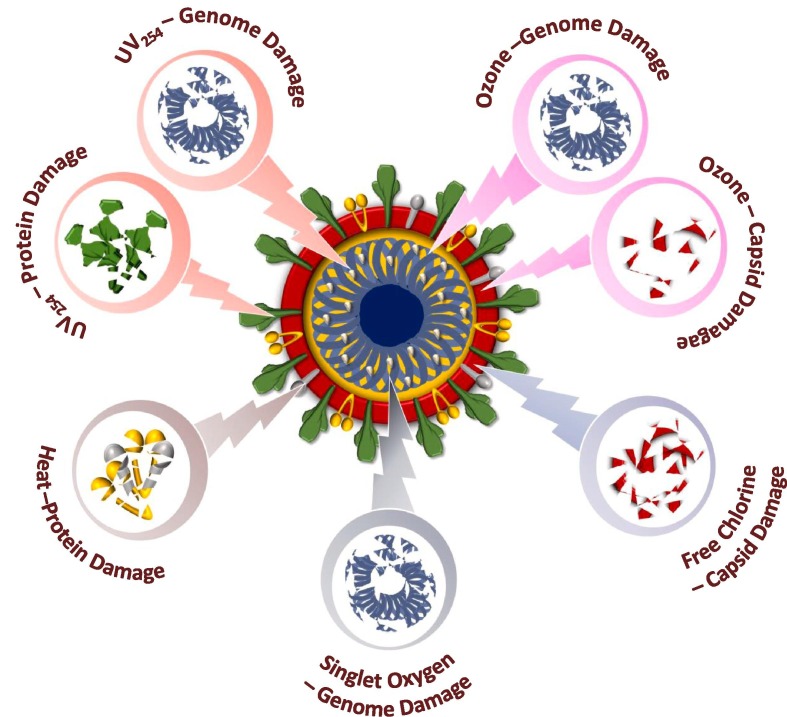
The extent and type of damage a virus can sustain before losing its ability to infect is important to understand in terms of the applied inactivation strategy. The ssRNA of SARS-CoV-2 likely renders it more susceptible to inactivation than enteric ssRNA viruses [12]. A virus to be infective, it must bind to the host, inject its genome inside the host, and replicate its genome within the host [142]. Oxidants and radiation cause damage to the viral proteins and nucleic acids, which is non-specific, however, the loss of specific virus functions (host recognition/binding, genome injection/replication) is important to understand [142]. UV254, singlet oxygen, and hypochlorous acid inhibit genome reliability, whereas, ClO2 and heat applications inhibit host-cell recognition/binding [142]. ClO2 inactivation induce damage to the proteins without inhibiting the genome function. On the protein level, ClO2 is a selective oxidant, which reacts mainly with Cys, Trp, and Tyr [222]. UV irradiation and chlorine treatment cause site-specific capsid protein backbone cleavage with the inhibition of both genome replication and injection [142]. UV254 application showed similar susceptibility for human norovirus and enteric (+) ssRNA viruses (Echovirus 12 and Feline calicivirus) with loss in genome functionality accounting for 60% of virus inactivation due to the protein damage [223]. Compared to ozone, inactivation by FC caused little or no loss of genome functionality, indicating its specific role in protein damage [223]. Ozone application to the E11 virus caused genome functionality loss proportionally to the infectivity [223]. Singlet oxygen impaired genome replication and host binding with a minor impairment associated with significant genome decay [142]. OH* radical oxidative action altered permeability and damage the capsid proteins [184].
The inactivation of three related bacteriophages (MS2, fr and GA) employing UV254, singlet oxygen, FC and ClO2 showed some interesting observations [182]. ClO2 did not induce genome damage in spite of variable inactivation kinetics, while all other inactivation processes showed more or less similar inactivation kinetics with marked genome damage [180]. On the protein level, UV254 damaged MS2 and capsid proteins, whereas, GA’s capsid remained intact. FC and ClO2 rapidly impaired the capsid proteins of all three viruses. Heat treatment to the E11 virus resulted in capsid protein denaturation rather than inhibiting genome functionality in spite of a high degree of infectivity loss [223]. MS2 inactivation by heat (72 °C) showed a loss inability to bind to the host cell and binding function decay rate correlated with inactivation rate, while injection and replication function rates were almost negligible [142]. Lack of replication function loss is in agreement with the lack of RNA degradation without peptide modifications. Heat induces structural changes due to attack on protease might cause inactivation by disrupting the specific structures needed to recognize and bind the host cells.
9.3.1. Enveloped virus inactivation
Enveloped viruses are less resistant to environmental conditions and inactivation compared to non-enveloped viruses [161], [207] and get inactivated at faster rates [47], [49], [224], [225], [226]. The inactivation/disinfection practices currently used are effective against non-enveloped viruses will also be effective for enveloped viruses also [99]. Enveloped viruses are more susceptible to oxidants than non-enveloped viruses [12], [158], [227]. Chemical-based disinfectants damage viral capsid, where UV irradiation affects nucleic acid [159]. Direct oxidation of reactive radicals damages the viral envelope manifesting the functional loss of the receptors [154]. The envelope does not impact the susceptibility of the virus to UV as the irradiation functionally impairs genomes [49]. To understand the enveloped virus inactivation mechanism of FC and UV irradiation was studied using model enveloped virus (Pseudomonas virus Phi6) [158]. FC reacts with proteins in the nucleocapsid and polymerase complex and Phi6 peptides which are more reactive when compared with the most reactive peptides with reference to non-enveloped coliphage MS2 [158]. Phi6 peptides contain a relatively large number of solvent-accessible Met and Cys residues which are responsible for rapid inactivation with FC [158]. UV254 photolysis kinetics of four model viral genomes (ssRNA, dsRNA, ssDNA, and dsDNA) showed high resistance of dsRNA [227]. UV and free chlorine applications cause protein backbone cleavage in MS2 in part inactivate the virus by inhibiting the genome injection function [142]. Inactivation of enveloped viruses in sewage by spiking bacteriophage Φ6 depicted that the enveloped viruses can undergo 6–7 log inactivation in sewage in 3–7 days, depending on the temperature [225]. Heat treatment induces denaturing of capsid protein rather than the genome functionality inhibition in spite of a high degree of infectivity loss [223].
10. Epidemiology Studies-Surveillance for early warning and spread
The detection of the virus in wastewater/sewage when the SARS-CoV-2 prevalence was low indicates the functional role of sewage for surveillance to monitor the circulation of the virus in the population [45], [60], [69] via WBE [56]. The wastewater will function both as a surveillance system and an early warning tool as shown previously for poliovirus [228], and Aichi virus [229]. The surveillance/monitoring of SARS-CoV-2 in wastewater can quantify the scale of infection prevailing within the community with a benefit of detecting virus for individuals who have not been tested, or are asymptomatic, potentially symptomatic, pre-symptomatic, or only have mild symptoms [73], [69], [188], [232]. It could provide an unbiased method of evaluating the spread of infection in different areas, even where resources for clinical diagnosis are limited and when reporting systems are unavailable [38], [230]. WBE approach will help to minimize the outbreak spread and also serve for future epidemics surveillance [150], [231]. WBE ability to detect low levels of viruses especially at early stages of an outbreak or when infection levels are decreasing following intervention is also critical [38]. Virome analysis of wastewater will enable to detect novel viruses before their clinical recognition in a community, allowing for early preventative measures and allocation of resources to potentially affected areas [52], [233], [234], [235]. The major limitations to WBE in establishing quantitative predictions from viral RNA results in over or underestimation of infected cases due to the complexity of wastewater, the dilute nature of biomarker in wastewater and inability to pinpoint specific locations [124], [150], [236]. Effective wastewater sampling techniques, effective virus concentration methods, robust and sensitive RT-qPCR assays asociated with source appropriation and transmission analysis through modelling approach are very much essential for successful WBE studies [150], [236].
11. Summary, challenges and recommendations
Since ancient times, viral-induced infective diseases are persistently posing a serious threat to human health as well on the economics at epidemic and pandemic scales. The CoV induced outbreaks were observed in three widespread cases viz., SARS-CoV-2 (2019), MERS-CoV (2012), and SARS-CoV-1 (2003) in the last two decades. CoV belongs to the enveloped family of βCoV with + ssRNA having crown-like spikes on its surfaces. SARS-CoV-2 genomic sequence showed more than 80% similarity with the SARS-CoV-1. Spike protein bind to the host cell by means of ACE2 receptor and replicates in the lungs, heart, intestines, blood vessels, and muscles. Data so far reported evidenced the replication and persistence of CoV (including SARS-CoV-2) in the human GI tract and shedding through stools showing the possibility of transmission through the faecal-oral route to the environment matrix. Along with virus-infected patients, pre-symptomatic and asymptomatic individuals also shed the virus. The faeces showed the presence of viral RNA and infectious virions in a few cases. The enteric virus in faeces persisted for a longer period of time (for many days) retaining its infectivity which is a major concern. The occurrence of SARS-CoV-2 in the water/wastewater/sewage was reported based and their persistence in the aqueous phase was also reported for days. However, the transmission of SARS-CoV-2 through faecal-oral route to the community is yet to be established in spite of the available evidence on the SARS-CoV-1 community spread through aerosol. Virus in general retains the infective property and infuses genetic information in the proximity of host leading to their replication. The presence of SARS-CoV-2 at an infectious dose and the duration of persistence in stools/wastewater are significant aspects that need to be studied and elucidated with a larger number of samples from the public health point of view. Routine and regular monitoring of viruses in the inflow to WWTPs if practiced along with other quality parameters will help in early detection of the outbreak with a warning. In this realm, water scientists are advocating the need for WBE for surveillance which could possibly enumerate the virus circulating flux in the community. However, rapid, effective and simple detection methods need to be developed specific to the environmental samples and implemented through a regulatory framework.
Detection, quantification, and determination of virulence in the environmental samples are of the major challenge for the scientist in this domain. Detection of the enteric virus in the environmental matrices is one of the important prerequisites to assess the fate and virulence of the pathogen as well as to design appropriate inactivation/treatment strategy. Virus detection protocol in environmental matrices will involve sample collection, transport, storage, concentration and detection, and inactivation which should ensure all the safety measures at each step of activity with Biosafety level (BSL) 2 (or above) procedure. Detection of enteric viruses in the environmental samples is extremely challenging because of their relatively low load (with reference to the quantity of wastewaters/water), interference caused due to the associated material in wastewater/water and sensitivity of the methods for precise quantification. The concentration of enveloped viruses from large volumes of environmental samples is an essential pre-requisite prior to detection. Electropositive filter particles, PEG precipitation and flocculation, ultrafiltration/ultracentrifugation are now practiced to concentrate to smaller volumes by simultaneously eliminating the associated interfaces. Culture (ICC-PCR) and molecular-based techniques (RT-PCR, qPCR, dPCR, Nested PCR, NGS and MALDI-TOF) were employed for the detection of virus from the concentrate solution which is considered to be sensitive and specific to provide quantitative information about the viral genome. Internal/external process controls are generally used to ensure accuracy in the detection. Rapid and cost-effective detection methods for virus monitoring with reference to environmental samples need to be developed.
Considering the fact that viruses pose a potential risk on human health, their inactivation in water and wastewater is an essential priority. Specific reports on the inactivation of SARS-CoV-2 in wastewater were not reported so far. Understanding the nature and fate of CoV are important to design an effective strategy for inactivation. An enveloped virus such as CoV is more unstable to oxidants and prevailing environmental conditions than the non-enveloped viruses. The primary and secondary unit operation in WWTPs only removes viruses to a marginal extent. Conventional disinfection methods such as chlorination, ozonation, H2O2, NaOCl and PAA along with solar and UV irradiation are the most frequently used unit operation in WWTPs to inactivate virus along with other pathogens. In general, reactive radicals produced during theses process damage either the envelope or/and capsid or/and viral RNA that ensures inactivation of virion in wastewater/water. However, the dosage of disinfectant agent/irradiation, contact time, pH and carbon of aqueous phase, presence of predators, etc. will have a regulatory role in the inactivation efficiency. The virus attachment to biofilm/sludge, interaction with a native microorganism (bacterial phases), plants, and animals is also a point of concern. The persistence of enveloped as well as non-enveloped viruses are detected after application of disinfection in treated water/wastewater, which warrants the necessity of advanced strategies such as AOP or integrated process to ensure virus free discharge. Optimized integration strategies of AOP and conventional disinfection process, design and development of large-scale disinfection unit operation, understanding the mechanism of viral inactivation, surveillance of discharges for the virus, the role of bacteriophages, etc. are some of the critical aspects to be investigated specifically on SARS-CoV-2 in diverse environmental settings. WWTPs need to be re-engineered keeping in the current and future pandemic situation by considering a multi-barrier approach. Integrating two or more disinfection strategies to strategically target to damage genome functional activity and envelope will able to provide an effective inactivation of the viral pathogens.
Enteric virus association with the water/wastewater infrastructure assuming the possible risk of transmission to the operators should take all the necessary protective measures against viral infection–wearing personal protective equipment (PPE) (protective clothing, gloves, boots, safety glasses, a face mask and/or respirator mask (FFP3)), face shield (to cover eyes, nose, and mouth), regular washing of common utility areas as well surfaces, frequent hand washing, virtual meetings, etc. According to the occupational health and safety protocols with safe working practices should be employed and should avoid direct contact with wastewater. Awareness needs to be created by the local governance by means of camps, leaflets, and social media by suggesting appropriate safety measures against the growing pandemics.
12. Conclusion
Past epidemiological experience showed frequent viral outbreaks and, in the future, there will also have epidemic/pandemic outbreaks due to the ever-changing anthropogenic activities affecting the ecological balance. In the environmental context, the possibility of transmission through water infrastructure is a major point of concern and its detection and inactivation will play a major role to curb their spread to the community. There is an imperative need to consider the virus as a regular parameter for routine monitoring along with other quality parameter with environmental samples to understand the early warning of the outbreaks and to effectively inactivate before discharge. Accordingly, a policy in the framework of regulation by appending to the environmental systems will help to safeguard the global community with future outbreak and transmissions.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgement
The authors wish to thank Director, CSIR-IICT (Manuscript No. IICT/Pubs./2020/146) for support.
Disclaimer
The authors declare no known competing financial interests or personal relationships that could have appeared to influence the review. The information discussed is purely based on the reported scientific literature.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cej.2020.126893.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Gale M., Tan S.L., Katze M.G. Translational control of viral gene expression in eukaryotes. Microbiol. Mol. Biol. Rev. 2000;64(2):239–280. doi: 10.1128/mmbr.64.2.239-280.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Angelescu D.G., Linse P. Viruses as supramolecular self-assemblies: modelling of capsid formation and genome packaging. Soft Matter. 2008;4(10):1981–1990. [Google Scholar]
- 3.La Rosa G., Fratini M., della Libera S., Iaconelli M., Muscillo M. Emerging and potentially emerging viruses in water environments. Ann. dell'Istituto Superiore Sanità. 2012;48:397–406. doi: 10.4415/ANN_12_04_07. [DOI] [PubMed] [Google Scholar]
- 4.Robinson C., Loeffelholz M.J., Pinsky B.A. Respiratory viruses. Clin. Virol. Man. 2016;19:255–276. [Google Scholar]
- 5.La Rosa G., Bonadonna L., Lucentini L., Kenmoe S., Suffredini E. Coronavirus in water environments: occurrence, persistence and concentration methods-a scoping review. Water Res. 2020 doi: 10.1016/j.watres.2020.115899. 115899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Naddeo V., Liu H. Editorial Perspectives: 2019 novel coronavirus (SARS-CoV-2): what is its fate in urban water cycle and how can the water research community respond? Environ. Sci. Water Res. Technol. 2020;6(5):1213–1216. [Google Scholar]
- 7.Qu G., Li X., Hu L., Jiang G. An imperative need for research on the role of environmental factors in transmission of novel coronavirus (COVID-19) Environ. Sci. Technol. 2020:3730–3732. doi: 10.1021/acs.est.0c01102. [DOI] [PubMed] [Google Scholar]
- 8.C. Yeo, S. Kaushal, D. Yeo, 2020. Enteric involvement of coronaviruses: is faecal–oral transmission of SARS-CoV-2 possible? Lancet Gastroentero. Hepatol. 5(4) (2020) 335–337. [DOI] [PMC free article] [PubMed]
- 9.Chan J.F.W., Kok K.H., Zhu Z., Chu H., To K.K.W., Yuan S., Yuen K.Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020;9(1):221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J. Virol. 2020;94(7) doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aquino de Carvalho N., Stachler E.N., Cimabue N., Bibby K. Evaluation of Phi6 persistence and suitability as an enveloped virus surrogate. Environ. Sci. Technol. 2017;51(15):8692–8700. doi: 10.1021/acs.est.7b01296. [DOI] [PubMed] [Google Scholar]
- 12.Wigginton K.R., Boehm A.B. Environmental engineers and scientists have important roles to play in stemming outbreaks and pandemics caused by enveloped viruses. Environ. Sci. Technol. 2020;54(7):3736–3739. doi: 10.1021/acs.est.0c01476. [DOI] [PubMed] [Google Scholar]
- 13.Salonen A.N.N.E., Ahola T.E.R.O., Kaariainen L.E.E.V.I. Membrane Trafficking in Viral Replication. Springer; Berlin, Heidelberg: 2004. Viral RNA replication in association with cellular membranes; pp. 139–173. [Google Scholar]
- 14.Tsai B. Penetration of nonenveloped viruses into the cytoplasm. Annu. Rev. Cell Dev. Biol. 2007;23:23–43. doi: 10.1146/annurev.cellbio.23.090506.123454. [DOI] [PubMed] [Google Scholar]
- 15.Cheng R.H., Kuhn R.J., Olson N.H., Rossmann M.G., Choi H.K., Smith T.J., Baker T.S. Nucleocapsid and glycoprotein organization in an enveloped virus. Cell. 1995;80(4):621–630. doi: 10.1016/0092-8674(95)90516-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.https://theconversation.com/ace2-the-molecule-that-helps-coronavirus-invade-your-cells-138369.
- 17.Ge X.Y., Li J.L., Yang X.L., Chmura A.A., Zhu G., Epstein J.H., Mazet J.K., Hu B., Zhang W., Peng C., Zhang Y.J. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503(7477):535–538. doi: 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amirian E.S. Potential fecal transmission of SARS-CoV-2: current evidence and implications for public health. Int. J. Infect. Dis. 2020;95:363–370. doi: 10.1016/j.ijid.2020.04.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu Y., Li X., Zhu B., Liang H., Fang C., Gong Y., Guo Q., Sun X., Zhao D., Shen J., Zhang H. Characteristics of pediatric SARS-CoV-2 infection and potential evidence for persistent fecal viral shedding. Nat. Med. 2020;26(4):502–505. doi: 10.1038/s41591-020-0817-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gu J., Han B., Wang J. COVID-19: gastrointestinal manifestations and potential fecal-oral transmission. Gastroenterology. 2020;158(6):1518–1519. doi: 10.1053/j.gastro.2020.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crawford D. The invisible enemy: a natural history of viruses. OUP Oxford. 2002 [Google Scholar]
- 22.Wisskirchen K., Lucifora J., Michler T., Protzer U. New pharmacological strategies to fight enveloped viruses. Trends Pharmacol. Sci. 2014;35(9):470–478. doi: 10.1016/j.tips.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carroll D., Daszak P., Wolfe N.D., Gao G.F., Morel C.M., Morzaria S., Pablos-Mendez A., Tomori O., Mazet J.A.K. The global virome project. Science. 2018;359(6378):872–874. doi: 10.1126/science.aap7463. [DOI] [PubMed] [Google Scholar]
- 24.https://sustainabilitycommunity.springernature.com/users/98825-seeram-ramakrishna/posts/looking-through-covid-19-lens-for-a-sustainable-new-modern-society.
- 25.Jones K.E., Patel N.G., Levy M.A., Storeygard A., Balk D., Gittleman J.L., Daszak P. Global trends in emerging infectious diseases. Nature. 2008;451(7181):990–993. doi: 10.1038/nature06536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Geoghegan J.L., Senior A.M., Di Giallonardo F., Holmes E.C. Virological factors that increase the transmissibility of emerging human viruses. Proc. Natl. Acad. Sci. 2016;113(15):4170–4175. doi: 10.1073/pnas.1521582113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.http://nautil.us/issue/83/intelligence/the-man-who-saw-the-pandemic-coming.
- 28.Hellewell J., Abbott S., Gimma A., Bosse N.I., Jarvis C.I., Russell T.W., Munday J.D., Kucharski A.J., Edmunds W.J., Sun F., Flasche S. Feasibility of controlling COVID-19 outbreaks by isolation of cases and contacts. Lancet Glob. Health. 2020 doi: 10.1016/S2214-109X(20)30074-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leung W.K., To K.F., Chan P.K., Chan H.L., Wu A.K., Lee N., Yuen K.Y., Sung J.J. Enteric involvement of severe acute respiratory syndrome-associated coronavirus infection. Gastroenterology. 2003;125(4):1011–1017. doi: 10.1016/j.gastro.2003.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu D., Zhang Z., Jin L., Chu F., Mao Y., Wang H., Liu M., Wang M., Zhang L., Gao G.F., Wang F.S. Persistent shedding of viable SARS-CoV in urine and stool of SARS patients during the convalescent phase. Eur. J. Clin. Microbiol. Infect. Dis. 2005;24(3):165–171. doi: 10.1007/s10096-005-1299-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Corman V.M., Kallies R., Philipps H., Gopner G., Muller M.A., Eckerle I., Brunink S., Drosten C., Drexler J.F. Characterization of a novel beta-corona virus related to middle East respiratory syndrome coronavirus in European hedgehogs. J. Virol. 2014;88(1):717–724. doi: 10.1128/JVI.01600-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim H.K., Yoon S.W., Kim D.J., Koo B.S., Noh J.Y., Kim J.H., Choi Y.G., Na W., Chang K.T., Song D., Jeong D.G. Detection of severe acute respiratory syndrome-like, middle east respiratory syndrome-like bat coronaviruses and group h rotavirus in faeces of korean bats. Transbound. Emerg. Dis. 2016;63(4):365–372. doi: 10.1111/tbed.12515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De Graaf M., Beck R., Caccio S.M., Duim B., Fraaij P.L., Le Guyader F.S., Lecuit M., Le Pendu J., De Wit E., Schultsz C. Sustained fecal-oral human-to-human transmission following a zoonotic event. Curr. Opin. Virol. 2017;22:1–6. doi: 10.1016/j.coviro.2016.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheng P.K., Wong D.A., Tong L.K., Ip S.M., Lo A.C., Lau C.S., Yeung E.Y., Lim W.W. Viral shedding patterns of coronavirus in patients with probable severe acute respiratory syndrome. Lancet. 2004;363(9422):1699–1700. doi: 10.1016/S0140-6736(04)16255-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang W., Xu Y., Gao R., Lu R., Han K., Wu G., Tan W. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 2020;323(18):1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xiao F., Tang M., Zheng X., Liu Y., Li X., Shan H. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology. 2020;158(6):1831–1833. doi: 10.1053/j.gastro.2020.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang H., Kang Z., Gong H., Xu D., Wang J., Li Z., Cui X., Xiao J., Meng T., Zhou W., Liu J. The digestive system is a potential route of 2019-nCov infection: a bioinformatics analysis based on single-cell transcriptomes. BioRxiv. 2020 [Google Scholar]
- 38.Kitajima M., Ahmed W., Bibby K., Carducci A., Gerba C.P., Hamilton K.A., Haramoto E., Rose J.B. SARS-CoV-2 in wastewater: state of the knowledge and research needs. Sci. Total Environ. 2020:139076. doi: 10.1016/j.scitotenv.2020.139076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cai J., Xu J., Lin D., Xu L., Qu Z., Zhang Y., Zhang H., Jia R., Wang X., Ge Y., Xia A. A Case Series of children with 2019 novel coronavirus infection: clinical and epidemiological features. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Young B.E., Ong S.W.X., Kalimuddin S., Low J.G., Tan S.Y., Loh J., Ng O.T., Marimuthu K., Ang L.W., Mak T.M., Lau S.K. Epidemiologic features and clinical course of patients infected with SARS-CoV-2 in Singapore. JAMA. 2020;323(15):1488–1494. doi: 10.1001/jama.2020.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holshue M.L., DeBolt C., Lindquist S., Lofy K.H., Wiesman J., Bruce H., Spitters C., Ericson K., Wilkerson S., Tural A., Diaz G. First case of 2019 novel coronavirus in the United States. N. Engl. J. Med. 2020;382:929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Woelfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Mueller M.A., Niemeyer D., Vollmar P., Rothe C., Hoelscher M., Bleicker T. Clinical presentation and virological assessment of hospitalized cases of coronavirus disease 2019 in a travel-associated transmission cluster. MedRxiv. 2020 [Google Scholar]
- 43.Ling Y., Xu S.B., Lin Y.X., Tian D., Zhu Z.Q., Dai F.H., Wu F., Song Z.G., Huang W., Chen J., Hu B.J. Persistence and clearance of viral RNA in 2019 novel coronavirus disease rehabilitation patients. Med. J. Chin. 2020 doi: 10.1097/CM9.0000000000000774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chan J.F.W., Yuan S., Kok K.H., To K.K.W., Chu H., Yang J., Xing F., Liu J., Yip C.C.Y., Poon R.W.S., Tsoi H.W. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.N. McLellan, D. Pernitsky, A. Umble, Coronavirus and The Water Cycle — Here Is What Treatment Professionals Need to Know, Water Online (https://www.wateronline.com/doc/coronavirus-and-the-water-cycle-here-is-what-treatment-professionals-need-to-know-0001), (2020).
- 46.Wang X.W., Li J.S., Jin M., Zhen B., Kong Q.X., Song N., Xiao W.J., Yin J., Wei W., Wang G.J., Si B.Y. Study on the resistance of severe acute respiratory syndrome-associated coronavirus. J. Virol. Methods. 2005;126(1–2):171–177. doi: 10.1016/j.jviromet.2005.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gundy P.M., Gerba C.P., Pepper I.L. Survival of coronaviruses in water and wastewater. Food Environ. Virol. 2009;1(1):10. [Google Scholar]
- 48.Casanova L., Rutala W.A., Weber D.J., Sobsey M.D. Survival of surrogate corona viruses in water. Water Res. 2009;43(7):1893–1898. doi: 10.1016/j.watres.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ye Y., Ellenberg R.M., Graham K.E., Wigginton K.R. Survivability, partitioning, and recovery of enveloped viruses in untreated municipal wastewater. Environ. Sci. Technol. 2016;50(10):5077–5085. doi: 10.1021/acs.est.6b00876. [DOI] [PubMed] [Google Scholar]
- 50.McMinn B.R., Ashbolt N.J., Korajkic A. Bacteriophages as indicators of faecal pollution and enteric virus removal. Lett Appl. Microbial. 2017;65(1):11–26. doi: 10.1111/lam.12736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang X.W., Li J.S., Guo T.K. Concentration and detection of SARS coronoavirus in sewage from Xiao tang Shan hospital and the 309th hospital. J. Virol. Methods. 2005;128(91–20):156–161. doi: 10.1016/j.jviromet.2005.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bibby K., Peccia J. Identification of viral pathogen diversity in sewage sludge by metagenome analysis. Environ. Sci. Technol. 2013;47(4):1945–1951. doi: 10.1021/es305181x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bibby K., Viau E., Peccia J. Viral metagenome analysis to guide human pathogen monitoring in environmental samples. Lett. Appl. Microbiol. 2011;52(4):386–392. doi: 10.1111/j.1472-765X.2011.03014.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Alexyuk M.S., Turmagambetova A.S., Alexyuk P.G. Comparative study of viromes from freshwater samples of the Ile-Balkhash region of Kazakhstan captured through metagenomic analysis. Virus Dis. 2017;28:18–25. doi: 10.1007/s13337-016-0353-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Blanco A., Abid I., Al-Otaibi N., Perez-Rodriguez F.J., Fuentes C., Guix S., Pinto R.M., Bosch A. Glass wool concentration optimization for the detection of enveloped and non-enveloped waterborne viruses. Food Environ. Virol. 2019;11(2):184–192. doi: 10.1007/s12560-019-09378-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O'Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J., Tscharke B. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;138764 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu Y., Guo C., Tang L., Hong Z., Zhou J., Dong X., Yin H., Xiao Q., Tang Y., Qu X., Kuang L. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol. Hepatol. 2020;5(5):434–435. doi: 10.1016/S2468-1253(20)30083-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wurtzer S., Marechal V., Mouchel J.M., Maday Y., Teyssou R., Richard E., Almayrac J.L., Moulin L. Evaluation of lockdown impact on SARS-CoV-2 dynamics through viral genome quantification in Paris wastewaters. medRxiv. 2020 doi: 10.2807/1560-7917.ES.2020.25.50.2000776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.La Rosa G., Iaconelli M., Mancini P., Ferraro G.B., Veneri C., Bonadonna L., Lucentini L., Suffredini E. First detection of SARS-CoV-2 in untreated wastewaters in Italy. Sci. Total Environ. 2020;139652 doi: 10.1016/j.scitotenv.2020.139652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-Coronavirus-2 in sewage. MedRxiv. 2020 doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- 61.I.B. Or, K. Yaniv, M. Shagan, E. Ozer, O. Erster, E. Mendelson, B. Mannasse, R. Shi-razi, E. Kramarsky-Winter, O. Nir, H. Abu-Ali, Z. Ronen, E. Rinott, Y.E. Lewis, E.F. Friedler, Y. Paitan, E. Bitkover, Y. Berchenko, A. Kushmaro, Regressing SARS-CoV-2 sewage measurements onto COVID-19 burden in thepopulation: a proof-of-concept for quantitative environmental surveillance. (2020) MedRxiv.
- 62.S. Balboa, M. Mauricio-Iglesias, S. Rodríguez, L. Martínez-Lamas, F.J. Vasallo, B. Regueiro, J.M. Lema, The fate of SARS-CoV-2 in wastewater treatmentplants points out the sludge line as a suitable spot for incidence monitoring (2020), MedRxiv. [DOI] [PMC free article] [PubMed]
- 63.S.G. Rimoldi, F. Stefani, A. Gigantiello, S. Polesello, F. Comandatore, D. Mileto, M. Maresca, C. Longobardi, A. Mancon, F. Romeri, C. Pagani, L. Moja, M.R. Gismondo, F. Salerno. Presence and vitality of SARS-CoV-2virus in wastewaters and rivers (2020), MedRxiv. [DOI] [PMC free article] [PubMed]
- 64.W. Randazzo, E. Cuevas-Ferrando, R. Sanjuan, P. Domingo-Calap, G. Sanchez, Metropolitan wastewater analysis for COVID-19 epidemiological surveil-lance (2020), MedRxiv. [DOI] [PMC free article] [PubMed]
- 65.M. Kumar, A.K. Patel, A.V. Shah, J. Raval, N. Rajpara, M. Joshi, C.G. Joshi. Thefirst proof of the capability of wastewater surveillance for COVID-19 in India through the detection of the genetic material of SARS-CoV-2 (2020), MedRxiv. [DOI] [PMC free article] [PubMed]
- 66.B.A. Kocamemi, H. Kurt, S. Hacioglu, C. Yarali, A.M. Saatci, B. Pakdemirli. First data-set on SARS-CoV-2 detection for Istanbul wastewaters in Turkey, (2020), MedRxiv.
- 67.S. Arora, A. Nag, J. Sethi, J. Rajvanshi, S. Saxena, S.K. Shrivastava, A.B. Gupta. Sewage surveillance for the presence of SARS-CoV-2 genome as a useful wastew-ater based epidemiology (WBE) tracking tool in India (2020), MedRxiv. [DOI] [PubMed]
- 68.E. Haramoto, B. Malla, O. Thakali, M. Kitajima. First environmental surveil-lance for the presence of SARS-CoV-2 RNA in wastewater and river water inJapan (2020), MedRxiv. [DOI] [PMC free article] [PubMed]
- 69.M. Hemalatha, U. Kiran, S.K. Kuncha, H. Kopperi, C.G. Gokulan, S. Venkata Mohan, R.K. Mishra. Comprehensive Surveillance of SARS-CoV-2 Spread Using Wastewater-based Epidemiology Studies (2020), medRxiv. [DOI] [PMC free article] [PubMed]
- 70.Quilliam R.S., Weidmann M., Moresco V., Purshouse H., O'Hara Z., Oliver D.M. COVID-19: The environmental implications of shedding SARS-CoV-2 in human faeces. Environ. Int. 2020 doi: 10.1016/j.envint.2020.105790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xagoraraki I., Yin Z., Svambayev Z. Fate of viruses in water systems. J. Environ. Eng. 2014;140(7):04014020. [Google Scholar]
- 72.Petrovich M.L., Ben Maamar S., Hartmann E.M., Murphy B.T., Poretsky R.S., Wells G.F. Viral composition and context in metagenomes from biofilm and suspended growth municipal wastewater treatment plants. Microb. Biotechnol. 2019;12(6):1324–1336. doi: 10.1111/1751-7915.13464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Osuolale O., Okoh A. Human enteric bacteria and viruses in five wastewater treatment plants in the Eastern Cape, South Africa. J. Infect. Public Heal. 2017;10(5):541–547. doi: 10.1016/j.jiph.2016.11.012. [DOI] [PubMed] [Google Scholar]
- 74.Pinon A., Vialette M. Survival of viruses in water. Intervirology. 2018;61(5):214–222. doi: 10.1159/000484899. [DOI] [PubMed] [Google Scholar]
- 75.Rzeżutka A., Cook N. Survival of human enteric viruses in the environment and food. FEMS Microbiol. Rev. 2004;28(4):441–453. doi: 10.1016/j.femsre.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 76.Graiver D.A., Topliff C.L., Kelling C.L., Bartelt-Hunt S.L. Survival of the avian influenza virus (H6N2) after land disposal. Environ. Sci. Technol. 2009;43(11):4063–4067. doi: 10.1021/es900370x. [DOI] [PubMed] [Google Scholar]
- 77.Farkas K., Walker D.L., Adriaenssens E.M., McDonald J.E., Hillary L.S., Malham S.K., Jones D.L. Viral indicators for tracking domestic wastewater contamination in the aquatic environment. Water Res. 2020;11:5926. doi: 10.1016/j.watres.2020.115926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.USEPA (US Environmental Protection Agency), Pesticide Fact Sheet, Boscalid, http://www.epa.gov/opprd001/factsheets/boscalid.pdf (2003).
- 79.Wong K., Onan B.M., Xagoraraki I. Quantification of enteric viruses, pathogen indicators, and Salmonella bacteria in class B anaerobically digested biosolids by culture and molecular methods. Appl. Environ. Microbiol. 2010;76(19):6441–6448. doi: 10.1128/AEM.02685-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lee D.Y., Leung K.T., Lee H., Habash M.B. Simultaneous detection of selected enteric viruses in water samples by multiplex quantitative PCR. Water Air Soil Pollut. 2016;227:107. [Google Scholar]
- 81.Ooi M.H., Wong S.C., Lewthwaite P., Cardosa M.J., Solomon T. Clinical features, diagnosis, and management of enterovirus 71. Lancet Neurol. 2010;9(11):1097–1105. doi: 10.1016/S1474-4422(10)70209-X. [DOI] [PubMed] [Google Scholar]
- 82.Gerba C.P. Virus occurrence and survival in the environmental waters. Perspect. Med. Virol. 2007;17:91–108. [Google Scholar]
- 83.Gerba C.P. Environmental Microbiology. Academic Press; 2009. Environmentally transmitted pathogens; pp. 445–484. [Google Scholar]
- 84.Lee P.I., Hsueh P.R. Emerging threats from zoonotic corona viruses-from SARS and MERS to 2019-nCoV. J. Microbiol. Immunol. Infect. 2020 doi: 10.1016/j.jmii.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.M. Rusinol, R. Girones, Summary of excreted and waterborne viruses. Global Water Pathogens Project Part 3 Viruses. (2017).
- 86.Foladori P., Cutrupi F., Segata N., Manara S., Pinto F., Malpei F., Bruni L., La Rosa G. SARS-CoV-2 from faeces to wastewater treatment: what do we know? a review. Sci. Total Environ. 2020 doi: 10.1016/j.scitotenv.2020.140444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Thompson J.R., Nancharaiah Y.V., Gu X., Lee W.L., Rajal V.B., Haines M.B., Girones R., Ng L.C., Alm E.J., Wuertz S. Making waves: wastewater surveillance of SARS-CoV-2 for population-based health management. Water Res. 2020 doi: 10.1016/j.watres.2020.116181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.J. Liu, X. Liao, S. Qian, J. Yuan, F. Wang, Y. Liu, Z. Wang, F.S. Wang, L. Liu, Z. Zhang, Community transmission of severe acute respiratory syndrome coronavirus 2, Shenzhen, China, 2020. Emerg. Infect. Dis. 26(6) (2020). [DOI] [PMC free article] [PubMed]
- 89.Kam K.Q., Yung C.F., Cui L. A well infant with coronavirus disease 2019 (COVID-19) with high viral load. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tang A., Tong Z.D., Wang H.L., Dai Y.X., Li K.F., Liu J.N., Wu W.J., Yuan C., Yu M.L., Li P., Yan J.B. Detection of novel coronavirus by RT-PCR in stool specimen from asymptomatic child, China. Emerg. Infect. Dis. 2020;26(6):1337–1339. doi: 10.3201/eid2606.200301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lodder W., de Roda Husman A.M. SARS-CoV-2 in wastewater: potential health risk, but also data source. Lancet Gastroenterol. Hepatol. 2020;5(6):533–534. doi: 10.1016/S2468-1253(20)30087-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.A.D. Workman, D.B. Welling, B.S. Carter, W.T. Curry, E.H. Holbrook, S.T. Gray, G.A. Scangas, and B.S. Bleier, 2020, Endonasal instrumentation and aerosolization risk in the era of COVID‐19: simulation, literature review, and proposed mitigation strategies, In International Forum of Allergy & Rhinology. (2020). [DOI] [PubMed]
- 93.Gormley M., Aspray T.J., Kelly D.A. COVID-19: mitigating transmission via wastewater plumbing systems. Lancet Glob. Health. 2020;8(5):643. doi: 10.1016/S2214-109X(20)30112-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Weber D.J., Rutala W.A., Fischer W.A., Kanamori H., Sickbert-Bennett E.E. Emerging infectious diseases: focus on infection control issues for novel coronaviruses (Severe Acute Respiratory Syndrome-CoV and Middle East Respiratory Syndrome-CoV), hemorrhagic fever viruses (Lassa and Ebola), and highly pathogenic avian influenza viruses, A (H5N1) and A (H7N9) Am. J. Infect. Control. 2016;44(5):91–100. doi: 10.1016/j.ajic.2015.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Peiris J.S.M., Chu C.M., Cheng V.C.C., Chan K.S., Hung I.F.N., Poon L.L., Law K.I., Tang B.S.F., Hon T.Y.W., Chan C.S., Chan K.H. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet. 2003;361(9371):1767–1772. doi: 10.1016/S0140-6736(03)13412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yu I.T., Li Y., Wong T.W., Tam W., Chan A.T., Lee J.H., Leung D.Y., Ho T. Evidence of airborne transmission of the severe acute respiratory syndrome virus. N. Engl. J. Med. 2004;350(17):1731–1739. doi: 10.1056/NEJMoa032867. [DOI] [PubMed] [Google Scholar]
- 97.Upadhyay N., Sun Q., Allen J.O., Westerhoff P., Herckes P. Characterization of aerosol emissions from wastewater aeration basins. J. Air Waste Manag. Assoc. 2013;63(1):20–26. doi: 10.1080/10962247.2012.726693. [DOI] [PubMed] [Google Scholar]
- 98.Arslan M., Xu B., El-Din M.G. Transmission of SARS-CoV-2 via fecal-oral and aerosols–borne routes: environmental dynamics and implications for wastewater management in underprivileged societies. Sci. Total Environ. 2020 doi: 10.1016/j.scitotenv.2020.140709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.M. Coccia, Factors determining the diffusion of COVID-19 and suggested strategy to prevent future accelerated viral infectivity similar to COVID, (2020). Sci. Total Environ. 729, 138474. [DOI] [PMC free article] [PubMed]
- 100.Conticini E., Frediani B., Caro D. Can atmospheric pollution be considered a cofactor in extremely high level of SARS-CoV-2 lethality in northern Italy? Environ. Pollut. 2020;261 doi: 10.1016/j.envpol.2020.114465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kutter J.S., Spronken M.I., Fraaij P.L., Fouchier R.A.M., Herfst S. Transmission routes of respiratory viruses among humans. Curr. Opin. Virol. 2018;28:142–151. doi: 10.1016/j.coviro.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pica N., Bouvier N.M. Environmental factors affecting the transmission of respiratory viruses. Curr. Opin. Virol. 2012;2(1):90–95. doi: 10.1016/j.coviro.2011.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Fernstrom A., Goldblatt M. Aerobiology and its role in the transmission of infectious diseases. J. Pathogens. 2013 doi: 10.1155/2013/493960. 493960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rychlik K.A., Secrest J.R., Lau C., Pulczinski J., Zamora M.L., Leal J., Langley R., Myatt L.G., Raju M., Chang R.C.A., Li Y. In utero ultrafine particulate matter exposure causes offspring pulmonary immunosuppression. Proc. Natl. Acad. Sci. 2019;116(9):3443–3448. doi: 10.1073/pnas.1816103116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wu G., Brown J., Zamora M.L., Miller A., Satterfield M.C., Meininger C.J., Steinhauser C.B., Johnson G.A., Burghardt R.C., Bazer F.W., Li Y. Adverse organogenesis and predisposed long-term metabolic syndrome from prenatal exposure to fine particulate matter. Proc. Natl. Acad. Sci. 2019;116(24):11590–11595. doi: 10.1073/pnas.1902925116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhang D., Ling H., Huang X., Li J., Li W., Yi C., Zhang T., Jiang Y., He Y., Deng S., Zhang X. Potential spreading risks and disinfection challenges of medical wastewater by the presence of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) viral RNA in septic tanks of fangcang hospital. Sci. Total Environ. 2020;741 doi: 10.1016/j.scitotenv.2020.140445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kim S.H., Chang S.Y., Sung M., Park J.H., Bin Kim H., Lee H., Choi J.P., Choi W.S., Min J.Y. Extensive viable Middle East Respiratory Syndrome (MERS) coronavirus contamination in air and surrounding environment in MERS isolation wards. Clin. Infect. Dis. 2016;63(3):363–369. doi: 10.1093/cid/ciw239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gormley M., Swaffield J., Sleigh P., Noakes C. An assessment of, and response to, potential cross-contamination routes due to defective appliance water trap seals in building drainage systems. Build. Serv. Eng. Res. Technol. 2012;33(2):203–222. [Google Scholar]
- 109.Gormley M., Templeton K., Kelly D., Hardie A. Environmental conditions and the prevalence of norovirus in hospital building drainage system wastewater and airflows. Build. Serv. Eng. Res. Technol. 2014;35(3):244–253. [Google Scholar]
- 110.Lin K., Marr L.C. Aerosolization of Ebola virus surrogates in wastewater systems. Environ. Sci. Technol. 2017;51(5):2669–2675. doi: 10.1021/acs.est.6b04846. [DOI] [PubMed] [Google Scholar]
- 111.Pasalari H., Ataei-Pirkooh A., Aminikhah M., Jafari A.J., Farzadkia M. Assessment of airborne enteric viruses emitted from wastewater treatment plant: Atmospheric dispersion model, quantitative microbial risk assessment, disease burden. Environ. Pollut. 2019;253:464–473. doi: 10.1016/j.envpol.2019.07.010. [DOI] [PubMed] [Google Scholar]
- 112.Peci A., Winter A.L., Li Y., Gnaneshan S., Liu J., Mubareka S., Gubbay J.B. Effects of absolute humidity, relative humidity, temperature, and wind speed on influenza activity in Toronto, Ontario, Canada. Appl. Environ. Microbiol. 2019;85(6):02426–2518. doi: 10.1128/AEM.02426-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Prussin A.J., Schwake D.O., Lin K., Gallagher D.L., Buttling L., Marr L.C. Survival of the enveloped virus Phi6 in droplets as a function of relative humidity, absolute humidity, and temperature. Appl. Environ. Microbiol. 2018;84(12):00551–618. doi: 10.1128/AEM.00551-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kim Y., Krishna V.D., Torremorell M., Goyal S.M., Cheeran M.C.J. Stability of porcine epidemic diarrhea virus on fomite materials at different temperatures. Vet. Sci. 2018;5(1):21. doi: 10.3390/vetsci5010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Donovan E., Unice K., Roberts J.D., Harris M., Finley B. Risk of gastrointestinal disease associated with exposure to pathogens in the water of the Lower Passaic River. Appl. Environ. Microbiol. 2008;74(4):994–1003. doi: 10.1128/AEM.00601-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wong A.C.P., Li X., Lau S.K.P., Woo P.C.Y. Global epidemiology of bat coronaviruses. Viruses. 2019;11:174. doi: 10.3390/v11020174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wang Q., Vlasova A.N., Kenney S.P., Saif L.J. Emerging and reemerging coronaviruses in pigs. Curr. Opin. virol. 2019;34:39–49. doi: 10.1016/j.coviro.2018.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bacnik K., Kutnjak D., Pecman A., Mehle N., Znidaric M.T., Aguirre I.G., Ravnikar M. Viromics and infectivity analysis reveal the release of infective plant viruses from wastewater into the environment. Water Res. 2020;115628 doi: 10.1016/j.watres.2020.115628. [DOI] [PubMed] [Google Scholar]
- 119.Thebo A.L., Drechsel P., Lambin E.F., Nelson K.L. A global, spatially-explicit assessment of irrigated croplands influenced by urban wastewater flows. Environ. Res. Lett. 2017;12(7) [Google Scholar]
- 120.Cashdollar J.L., Wymer L. Methods for primary concentration of viruses from water samples: a review and meta-analysis of recent studies. J. Appl. Microbiol. 2013;115:1–11. doi: 10.1111/jam.12143. [DOI] [PubMed] [Google Scholar]
- 121.Haramoto E., Kitajima M., Hata A., Torrey J.R., Masago Y., Sano D., Katayama H. A review on recent progress in the detection methods and prevalence of human enteric viruses in water. Water Res. 2018;135:168–186. doi: 10.1016/j.watres.2018.02.004. [DOI] [PubMed] [Google Scholar]
- 122.Collomb J., Laporte J., Vautherot J.F., Schwartzbrod L. Research on coronaviruses in water, adsorption and elution of the coronavirus on glass powder. Virologie. 1986;37(2):95–105. [PubMed] [Google Scholar]
- 123.Abd-Elmaksoud S., Spencer S.K., Gerba C.P., Tamimi A.H., Jokela, Borchardt M.A. Simultaneous concentration of bovine viruses and agricultural zoonotic bacteria from water using sodocalcic glass wool filters. Food Environ. Virol. 2014;6(4):253–259. doi: 10.1007/s12560-014-9159-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ahmed W., Bertsch P., Bivins A., Bibby K., Farkas K., Gathercole A., Haramoto E., Gyawali P., Korajkic A., McMinn B.R., Mueller J. Comparison of virus concentration methods for the RT-qPCR-based recovery of murine hepatitis virus, a surrogate for SARS-CoV-2 from untreated wastewater. Sci. Total Environ. 2020 doi: 10.1016/j.scitotenv.2020.139960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Liu W.T., Zhu L. Environmental microbiology on chip and its future impacts. Trends Biotechnol. 2005;23(4):174–178. doi: 10.1016/j.tibtech.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 126.Alhamlan F.S., Ederer M.M., Green T.L., Brinkman C.K., Coats E.R., Crawford R.L. A Novel screening tool using microarray and PCR to detect pathogens in agricultural impacted waters. Int. J. Environ. Protection. 2013;3(3):17–32. [Google Scholar]
- 127.M.A. Martinez, L. Soto-Del Rio Mde, R.M. Gutierrez, C.Y. Chiu, A.L. Greninger, J.F. Contreras, S. Lopez, C.F. Arias, P. Isa, DNA microarray for detection of gastrointestinal viruses, J. Clin. Microbiol. 53 (2015) 136–145. [DOI] [PMC free article] [PubMed]
- 128.Guo W., Wang S., Zhao C., Li J., Zhong J. An integrated cell absorption process and quantitative PCR assay for the detection of the infectious virus in water. Sci. Total Environ. 2018;635:964–971. doi: 10.1016/j.scitotenv.2018.04.223. [DOI] [PubMed] [Google Scholar]
- 129.Hjelmso M.H., Hellmer M., Fernandez-Cassi X., Timoneda N., Lukjancenko O., Seidel M., Elsasser D., Aarestrup F.M., Lofstrom C., Bofill-Mas S., Abril J.F., Girones R., Schultz A.C. Evaluation of methods for the concentration and extraction of viruses from sewage in the context of metagenomic sequencing. PLoS ONE. 2017;12 doi: 10.1371/journal.pone.0170199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lee S.H., Lee C., Lee K.W., Cho H.B., Kim S.J. The simultaneous detection of both enteroviruses and adenoviruses in environmental water samples including tap water with an integrated cell culture multiplex nested PCR procedure. J. Appl. Microbiol. 2005;98(5):1020–1029. doi: 10.1111/j.1365-2672.2004.02496.x. [DOI] [PubMed] [Google Scholar]
- 131.Hryniszyn A., Skonieczna M. Methods for detection of viruses in water and wastewater. Adv. Microbiol. 2013;3(05):442. [Google Scholar]
- 132.Bonadonna L., Briancesco R., La Rosa G. Innovative analytical methods for monitoring microbiological and virological water quality. Micro. Chem. J. 2019;150 [Google Scholar]
- 133.Farkas K., Malham S.K., Peters D.E., De Rougemont A., McDonald J.E., De Rougemont A., Malham S.K., Jones D.L. Evaluation of two triplex one step qRT-PCR assays for the quantification of human enteric viruses in environmental samples. Food Environ. Virol. 2017;9:343–349. doi: 10.1007/s12560-017-9293-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Sedji M.I., Varbanov M., Meo M., Colin M., Mathieu L., Bertrand I. Quantification of human adenovirus and norovirus in river water in the north-east of France. Environ. Sci. Pollut. Res. 2018:30497–30507. doi: 10.1007/s11356-018-3045-4. [DOI] [PubMed] [Google Scholar]
- 135.Monteiro S., Santos R. Enzymatic and viability RT-qPCR assays for evaluation of enterovirus, hepatitis A virus and norovirus inactivation: implications for public health risk assessment. J. Appl. Microbiol. 2018;124:965–976. doi: 10.1111/jam.13568. [DOI] [PubMed] [Google Scholar]
- 136.Barrios M.E., Blanco Fern andez M.D., Cammarata R.V., Torres C., Mbayed V.A. Viral tools for detection of fecal contamination and microbial source tracking in wastewater from food industries and domestic sewage. J. Virol. Methods. 2018;262:79–88. doi: 10.1016/j.jviromet.2018.10.002. [DOI] [PubMed] [Google Scholar]
- 137.Ming H.X., Zhu L., Zhang Y. Rapid quantification of infectious enterovirus from surface water in Bohai Bay, China using an integrated cell culture-qPCR assay. Mar. Pollut. Bull. 2011;62:2047–2054. doi: 10.1016/j.marpolbul.2011.07.024. [DOI] [PubMed] [Google Scholar]
- 138.Dhar B.C., Lee N.Y. Lab-on-a-Chip Technology for environmental monitoring of microorganisms. Biochip J. 2018;12:173–183. [Google Scholar]
- 139.Ogorzaly L., Walczak C., Galloux M., Etienne S., Gassilloud B., Cauchie H.M. Human adenovirus diversity in water samples using a next-generation amplicon sequencing approach. Food Environ. Virol. 2015;7:112. doi: 10.1007/s12560-015-9194-4. [DOI] [PubMed] [Google Scholar]
- 140.Hamza I.A., Bibby K. Critical issues in application of molecular methods to environmental virology. J. Virol. Methods. 2019;266:11–24. doi: 10.1016/j.jviromet.2019.01.008. [DOI] [PubMed] [Google Scholar]
- 141.Farkas K., Mannion F., Hillary L.S., Malham S.K., Walker D.I. Emerging technologies for the rapid detection of enteric viruses in the aquatic environment. Curr. Opin. Environ. Sci. Heal. 2020;16:1–6. [Google Scholar]
- 142.Wigginton K.R., Pecson B.M., Sigstam T., Bosshard F., Kohn T. Virus inactivation mechanisms: impact of disinfectants on virus function and structural integrity. Environ. Sci. Technol. 2012;46(21):12069–12078. doi: 10.1021/es3029473. [DOI] [PubMed] [Google Scholar]
- 143.Schrader C., Schielke A., Ellerbroek L., Johne R. PCR inhibitors e occurrence, properties and removal. J. Appl. Microbiol. 2012;113:1014–1026. doi: 10.1111/j.1365-2672.2012.05384.x. [DOI] [PubMed] [Google Scholar]
- 144.Teixeira P., Costa S., Brown B., Silva S., Rodrigues R., Valerio E. Quantitative PCR detection of enteric viruses in wastewater and environmental water sources by the Lisbon municipality: a case study. Water. 2020;12(2):544. [Google Scholar]
- 145.Simmons F.J., Xagoraraki I. Release of infectious human enteric viruses by full-scale wastewater utilities. Water Res. 2011;45(12):3590–3598. doi: 10.1016/j.watres.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 146.Hewitt J., Leonard M., Greening G.E., Lewis G.D. Influence of wastewater treatment process and the population size on human virus profiles in wastewater. Water Res. 2011;45(18):6267–6276. doi: 10.1016/j.watres.2011.09.029. [DOI] [PubMed] [Google Scholar]
- 147.Surbeck C.Q., Jiang S.C., Ahn J.H., Grant S.B. Flow fingerprinting fecal pollution and suspended solids in stormwater runoff from an urban coastal watershed. Environ. Sci. Technol. 2006;40(14):4435–4441. doi: 10.1021/es060701h. [DOI] [PubMed] [Google Scholar]
- 148.Diez-Valcarce M., Cook N., Hernandez M., Rodriguez-Lazaro D. Analytical application of a sample process control in detection of food borne viruses. Food Anal. Methods. 2011;4(4):614–618. [Google Scholar]
- 149.Wobus C.E., Thackray L.B., Virgin H.W. Murine norovirus: a model system to study norovirus biology and pathogenesis. J. Virol. 2006;80(11):5104–5112. doi: 10.1128/JVI.02346-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.K. Mao, H. Zhang, Z. Yang, Can a paper-based device trace COVID-19 sources with wastewater-based epidemiology? (2020). [DOI] [PubMed]
- 151.U. Kiran, C.G. Gokulan, S,K. Kuncha, D. Vedagiri, B.T. Chander, A.V. Sekhar, S. Dontamala, A.L. Reddy, K.B. Tallapaka, R.K. Mishra, K.H. Harshan. Easing diagnosis and pushing the detection limits of SARS-CoV-2. Biology Methods and Protocols (2020). [DOI] [PMC free article] [PubMed]
- 152.Lednicky J.A., Shankar S.N., Elbadry M.A., Gibson J.C., Alam M.M., Stephenson C.J., Eiguren-Fernandez A., Morris J.G., Mavian C.N., Salemi M., Clugston J.R. Collection of SARS-CoV-2 virus from the air of a clinic within a university student health care center and analyses of the viral genomic sequence. Aerosol Air Qual. Res. 2020;20 doi: 10.4209/aaqr.2020.02.0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.K. Bibby, N. Aquino de Carvalho, K. Wigginton, Research needs for wastewater handling in virus outbreak response. (2017) 2534–2535. [DOI] [PubMed]
- 154.Wigginton K.R., Ye Y., Ellenberg R.M. Emerging investigators series: the source and fate of pandemic viruses in the urban water cycle. Environ. Sci. Water Res. Technol. 2015;1(6):735–746. [Google Scholar]
- 155.Flint S.J., Racaniello V.R., Rall G.F., Skalka A.M. John Wiley & Sons; 2015. Principles of Virology. [Google Scholar]
- 156.D. Preda, Prevention, Treatment and Immunisation against COVID-19 and Other Encapsulated Viruses. (2020).
- 157.G. Dvorak, C. Petersen, Sanitation and disinfection. Infectious disease management in animal shelters, (2009) 49–60.
- 158.Ye Y., Chang P.H., Hartert J., Wigginton K.R. Reactivity of enveloped virus genome, proteins, and lipids with free chlorine and UV254. Environ. Sci. Technol. 2018;52:7698–7708. doi: 10.1021/acs.est.8b00824. [DOI] [PubMed] [Google Scholar]
- 159.Zhang M., Xu L.M., Xu P.C., Wang X.C. Elimination of viruses from domestic wastewater: requirements and technologies. World J. Microbiol. Biotechnol. 2016;32(4):69–78. doi: 10.1007/s11274-016-2018-3. [DOI] [PubMed] [Google Scholar]
- 160.Qiu Y., Lee B.E., Neumann N., Ashbolt N., Craik S., Maal-Bared R., Pang X.L. Assessment of human virus removal during municipal wastewater treatment in Edmonton, Canada. J Appl Microbiol. 2015;119(6):1729–1739. doi: 10.1111/jam.12971. [DOI] [PubMed] [Google Scholar]
- 161.Sidhu J.P.S., Sena K., Hodgers L., Palmer A., Toze S. Comparative enteric viruses and coliphage removal during wastewater treatment processes in a sub-tropical environment. Sci. Total Environ. 2018;616:669–677. doi: 10.1016/j.scitotenv.2017.10.265. [DOI] [PubMed] [Google Scholar]
- 162.Prado T., Bruni A.C., Barbosa M.R.F., Garcia S.C., Melo A.M.J., Sato M.I.Z. Performance of wastewater reclamation systems in enteric virus removal. Sci. Total Environ. 2019;678:33–42. doi: 10.1016/j.scitotenv.2019.04.435. [DOI] [PubMed] [Google Scholar]
- 163.Weinbauer M.G., Rassoulzadegan F. Are viruses driving microbial diversification and diversity? Environ. Microbiol. 2004;6(1):1–11. doi: 10.1046/j.1462-2920.2003.00539.x. [DOI] [PubMed] [Google Scholar]
- 164.Kitajima M., Iker B.C., Pepper I.L., Gerba C.P. Relative abundance and treatment reduction of viruses during wastewater treatment processes—identification of potential viral indicators. Sci. Total Environ. 2014;488:290–296. doi: 10.1016/j.scitotenv.2014.04.087. [DOI] [PubMed] [Google Scholar]
- 165.Zhou J., Wang X.C., Ji Z., Xu L., Yu Z. Source identification of bacterial and viral pathogens and their survival/fading in the process of wastewater treatment, reclamation, and environmental reuse. World J. Microb Biot. 2015;31(1):109–120. doi: 10.1007/s11274-014-1770-5. [DOI] [PubMed] [Google Scholar]
- 166.Shirasaki N., Matsushita T., Matsui Y., Oshiba A., Ohno K. Estimation of norovirus removal performance in a coagulation– rapid sand filtration process by using recombinant norovirus. VLPs Water Res. 2010;44:1307–1316. doi: 10.1016/j.watres.2009.10.038. [DOI] [PubMed] [Google Scholar]
- 167.Jofre G.J., Montemayor M., Lucena F. Occurrence and levels of indicators and selected pathogens in different sludges and biosolids. J. Appl. Microbiol. 2007;103:2420–2429. doi: 10.1111/j.1365-2672.2007.03487.x. [DOI] [PubMed] [Google Scholar]
- 168.Dumontet S., Dinel H., Baloda S.B. Pathogen reduction in sewage sludge by composting and other biological treatments: a review. Biol. Agric. Hortic. 1999;16:409–430. [Google Scholar]
- 169.Monpoeho S., Maul A., Bonnin C., Patria L., Ranarijaona S., Billaudel S., Ferre V. Clearance of human-pathogenic viruses from sludge: study of four stabilization processes by real-time reverse transcription-PCR and cell culture. Appl. Environ. Microbiol. 2004;70:5434–5440. doi: 10.1128/AEM.70.9.5434-5440.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Hao X.D., Wang Q.L., Zhu J.Y., Van Loosdrecht M.C. Microbiological endogenous processes in biological wastewater treatment systems. Crit. Rev. Environ. Sci. Technol. 2010;40(3):239–265. [Google Scholar]
- 171.Gomes J., Matos A., Gmurek M., Quinta-Ferreira R.M., Martins R.C. Ozone and photocatalytic processes for pathogens removal from water: a review. Catalysts. 2019;9(1):46. [Google Scholar]
- 172.Collivignarelli M.C., Abbà A., Benigna I., Sorlini S., Torretta V. Overview of the main disinfection processes for wastewater and drinking water treatment plants. Sustainability. 2018;10(1):86. [Google Scholar]
- 173.USEPA (US Environmental Protection Agency), Pesticide Product Information System http://www.epa.gov/pesticides/PPISdata/ (2012).
- 174.Liberti L., Lopez A., Notarnicola M. Disinfection with peracetic acid for domestic sewage re-use in agriculture. J. Water Environ. Manag. 1999;13:262–269. [Google Scholar]
- 175.Xu P., Janex M.L., Savoye P., Cockx A., Lazarova V. Wastewater disinfection by ozone: main parameters for process design. Water Res. 2002;36(4):1043–1055. doi: 10.1016/s0043-1354(01)00298-6. [DOI] [PubMed] [Google Scholar]
- 176.Wolf U., Von Gunten, Kohn T. Kinetics of inactivation of waterborne enteric viruses by ozone. Environ. Sci. Technol. 2018;52(4):2170–2177. doi: 10.1021/acs.est.7b05111. [DOI] [PubMed] [Google Scholar]
- 177.Qiu Y., Li Q., Lee B.E., Ruecker N.J., Neumann N.F., Ashbolt N.J., Pang X. UV inactivation of human infectious viruses at two full-scale wastewater treatment plants in Canada. Water Res. 2018;147:73–81. doi: 10.1016/j.watres.2018.09.057. [DOI] [PubMed] [Google Scholar]
- 178.Rockey N., Young S., Kohn T., Pecson B., Wobus C.E., Raskin L., Wigginton K.R. UV Disinfection of human norovirus: evaluating infectivity using a genome-wide PCR-based approach. Environ. Sci. Technol. 2020;54(5):2851–2858. doi: 10.1021/acs.est.9b05747. [DOI] [PubMed] [Google Scholar]
- 179.Calgua A., Carratala L., Guerrero-Latorre A., De Abreu Corrêa T., Kohn R., Sommer R., Girones R. UVC inactivation of dsDNA and ssRNA viruses in water: UV fluences and a qPCR-based approach to evaluate decay on viral infectivity. Food Environ. Viro. 2014;6(4):260–268. doi: 10.1007/s12560-014-9157-1. [DOI] [PubMed] [Google Scholar]
- 180.Pecson M., Ackermann M., Kohn T. Framework for using quantitative PCR as a nonculture based method to estimate virus infectivity. Environ. Sci. Technol. 2011;45:2257–2263. doi: 10.1021/es103488e. [DOI] [PubMed] [Google Scholar]
- 181.Mattle M.J., Kohn T. Inactivation and tailing during UV254 disinfection of viruses: contributions of viral aggregation, light shielding within viral aggregates, and recombination. Environ. Sci. Technol. 2012;46(18):10022–10030. doi: 10.1021/es302058v. [DOI] [PubMed] [Google Scholar]
- 182.Sigstam T., Gannon G., Cascella M., Pecson B.M., Wigginton K.R., Kohn T. Subtle differences in virus composition affect disinfection kinetics and mechanisms. Appl. Environ. Microbiol. 2013;79(11):3455–3467. doi: 10.1128/AEM.00663-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.M. Agulló-Barceló, M. I. Polo-López, F. Lucena, J. Jofre, P. Fernández-Ibáñez P, Solar advanced oxidation processes as disinfection tertiary treatments for real wastewater: implications for water reclamation. Appl. Catal. B: Environ. 136–137: (2013) 341–350.
- 184.Polo I., García-Fernández P., Fernández-Ibáñez, Romalde J.L. Solar water disinfection (SODIS): Impact on hepatitis A virus and on a human Norovirus surrogate under natural solar conditions. Int. Microbiol. 2015;18(1):41–49. doi: 10.2436/20.1501.01.233. [DOI] [PubMed] [Google Scholar]
- 185.Fisher M.B., Iriarte M., Nelson K.L. Solar water disinfection (SODIS) of Escherichia coli, Enterococcus spp., and MS2 coliphage: effects of additives and alternative container materials. Water Res. 2012;46:1745–1754. doi: 10.1016/j.watres.2011.12.048. [DOI] [PubMed] [Google Scholar]
- 186.Kohn T., Mattle M.J., Minella M., Vione D. A modeling approach to estimate the solar disinfection of viral indicator organisms in waste stabilization ponds and surface waters. Water Res. 2016;88:912–922. doi: 10.1016/j.watres.2015.11.022. [DOI] [PubMed] [Google Scholar]
- 187.Kohn T., Nelson K.L. Sunlight-mediated inactivation of MS2 coliphage via exogenous singlet oxygen produced by sensitizers in natural waters. Environ. Sci. Technol. 2007;41(1):192–197. doi: 10.1021/es061716i. [DOI] [PubMed] [Google Scholar]
- 188.Nelson K.L., Boehm A.B., Davies-Colley R.J., Dodd M.C., Kohn T., Linden K.G., Liu Y., Maraccini P.A., McNeill K., Mitch W.A., Nguyen T.H. Sunlight-mediated inactivation of health-relevant microorganisms in water: a review of mechanisms and modeling approaches. Environ Sci-Proc Imp. 2018;20(8):1089–1122. doi: 10.1039/c8em00047f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Romero-Maraccini O.C., Shisler J.L., Nguyen T.H. Solar and temperature treatments affect the ability of human rotavirus wa to bind to host cells and synthesize viral RNA. Appl. Environ. Microbiol. 2015;81:4090–4097. doi: 10.1128/AEM.00027-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Voumard M., Giannakis S., Carratala A., Pulgarin C. E. coli–MS2 bacteriophage interactions during solar disinfection of wastewater and the subsequent post-irradiation period. Chem. Eng. J. 2019;359:1224–1233. [Google Scholar]
- 191.Hornstra L.M., Da Silva T.R., Blankert B., Heijnen L., Beerendonk E., Cornelissen E.R., Medema G. Monitoring the integrity of reverse osmosis membranes using novel indigenous freshwater viruses and bacteriophages. Environ. Sci. 2019;5(9):1535–1544. [Google Scholar]
- 192.Chaudhry R.M., Nelson K.L., Drewes J.E. Mechanisms of pathogenic virus removal in a full-scale membrane bioreactor. Environ. Sci. Technol. 2015;49(5):2815–2822. doi: 10.1021/es505332n. [DOI] [PubMed] [Google Scholar]
- 193.Giannakis S., Androulaki B., Comninellis C., Pulgarin C. Wastewater and urine treatment by UVC-based advanced oxidation processes: implications from the interactions of bacteria, viruses, and chemical contaminants. Chem. Eng. J. 2018;343:270–282. [Google Scholar]
- 194.Barrett M., Fitzhenry K., O'Flaherty V., Dore W., Keaveney S., Cormican M., Rowan N., Clifford E. Detection, fate and inactivation of pathogenic norovirus employing settlement and UV treatment in wastewater treatment facilities. Sci. Total Environ. 2016;568:1026–1036. doi: 10.1016/j.scitotenv.2016.06.067. [DOI] [PubMed] [Google Scholar]
- 195.Lubello C., Caretti R., Gori Comparison between PAA/UV and H2O2/UV disinfection for wastewater reuse. Water Sci Tech – W Sup. 2002;2(1):205–212. [Google Scholar]
- 196.Zhang J., Tian B., Wang L., Xing M., Lei J. Springer; Singapore: 2018. Photo-Fenton Reaction. Photocatalysis; pp. 259–274. [Google Scholar]
- 197.Zheng X., Wang Q., Chen L., Wang J., Cheng R. Photocatalytic membrane reactor (PMR) for virus removal in water: performance and mechanisms. Chem. Eng. J. 2015;277:124–129. [Google Scholar]
- 198.Tondera K., Klaer K., Gebhardt J., Wingender J., Koch C., Horstkott M., Strathmann M., Jurzik L., Hamza I.A., Pinnekamp J. Reducing pathogens in combined sewer overflows using ozonation or UV irradiation. Int. J. Hyg. Environ. Health. 2015;218(8):731–741. doi: 10.1016/j.ijheh.2015.09.002. [DOI] [PubMed] [Google Scholar]
- 199.Nam Y., Lim J.H., Ko K.C., Lee J.Y. Photocatalytic activity of TiO2 nanoparticles: a theoretical aspect. J. Mater. Chem. A. 2019;7(23):13833–13859. [Google Scholar]
- 200.Yemmireddy V.K., Hung Y.C. Using photocatalyst metal oxides as antimicrobial surface coatings to ensure food safety—opportunities and challenges. Compr. Rev. Food Sci. 2017;16(4):617–631. doi: 10.1111/1541-4337.12267. [DOI] [PubMed] [Google Scholar]
- 201.Rueda-Marquez J.J., Levchuk I., Ibañez P.F., Sillanpää M. A critical review on application of photocatalysis for toxicity reduction of real wastewaters. J. Clean. Prod. 2020 120694. [Google Scholar]
- 202.Watts R.J., Kong S., Orr M.P., Miller G.C., Henry B.E. Photocatalytic inactivation of coliform bacteria and viruses in secondary wastewater effluent. Water Res. 1995;29(1):95–100. [Google Scholar]
- 203.W. Han, B. Zhang, W. Cao, D. Yang, I. TAIRA, Y. Okamoto, J. I. Arai, X. Yan, The inactivation effect of photocatalytic titanium apatite filter on SARS virus. Sheng wu hua xue yu sheng wu wu li jin zhan, 31(11) (2004) 982–985.
- 204.Bogdan J., Zarzyńska J., Pławińska-Czarnak J. Comparison of infectious agents susceptibility to photocatalytic effects of nanosized titanium and zinc oxides: a practical approach. Nanoscale Res. Lett. 2015;10(1):1–15. doi: 10.1186/s11671-015-1023-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Divyapriya G., Nambi I.M., Senthilnathan J. Nanocatalysts in Fenton based advanced oxidation process for water and wastewater treatment. J. Bionanosci. 2016;10(5):356–368. [Google Scholar]
- 206.Jain, Vaya D. Photocatalytic activity of TiO2 nanomaterial. J. Chil. Chem. Soc. 2017;62(4):3683–3690. [Google Scholar]
- 207.M.F. Brugnera, B.C. de Araújo Souza, and M.V.B. Zanoni, Advanced Oxidation Process Applied to Actinobacterium Disinfection. Actinobacteria: Basics and Biotechnological Applications, (2016) 353.
- 208.Zhang Y., Li D., Shuai Y., Shen D. Wang, Progress and challenges in photocatalytic disinfection of waterborne viruses: a review to fill current knowledge gaps. Chem. Eng. J. 2019;355:399–415. [Google Scholar]
- 209.Li Y., Zhang C., Shuai D., Naraginti S., Wang D., Zhang W. Visible-light-driven photocatalytic inactivation of MS2 by metal-free g-C3N4: virucidal performance and mechanism. Water Res. 2016;106:249–258. doi: 10.1016/j.watres.2016.10.009. [DOI] [PubMed] [Google Scholar]
- 210.Zheng X., Chen D., Lei Y., Cheng R. Nano-TiO2 membrane adsorption reactor (MAR) for virus removal in drinking water. Chem. Eng. J. 2013;230:180–187. [Google Scholar]
- 211.Liga M.V., Bryant E.L., Colvin V.L., Li Q. Virus inactivation by silver doped titanium dioxide nanoparticles for drinking water treatment. Water Res. 2011;45(2):535–544. doi: 10.1016/j.watres.2010.09.012. [DOI] [PubMed] [Google Scholar]
- 212.Reddy P.V.L., Kavitha B., Reddy P.A.K., Kim K.H. TiO2-based photocatalytic disinfection of microbes in aqueous media: a review. Environ. Res. 2017;154:296–303. doi: 10.1016/j.envres.2017.01.018. [DOI] [PubMed] [Google Scholar]
- 213.Su W., Wei S.S., Hu S.Q., Tang J.X. Preparation of TiO2/Ag colloids with ultraviolet resistance and antibacterial property using short chain polyethylene glycol. J. Hazard. Mater. 2009;172(2–3):716–720. doi: 10.1016/j.jhazmat.2009.07.056. [DOI] [PubMed] [Google Scholar]
- 214.Kozlova A., Safatov A.S., Kiselev S.A., Marchenko V.Y., Sergeev A.A., Skarnovich M.O., Emelyanova E.K., Smetannikova M.A., Buryak G.A., Vorontsov A.V. Inactivation and mineralization of aerosol deposited model pathogenic microorganisms over TiO2 and Pt/TiO2. Environ. Sci. Technol. 2010;44(13):5121–5126. doi: 10.1021/es100156p. [DOI] [PubMed] [Google Scholar]
- 215.Cho M., Cates E.L., Kim J.H. Inactivation and surface interactions of MS-2 bacteriophage in a TiO2 photoelectrocatalytic reactor. Water Res. 2011;45(5):2104–2110. doi: 10.1016/j.watres.2010.12.017. [DOI] [PubMed] [Google Scholar]
- 216.Li G., Liu X., Zhang H., Wong P., An T., Zhou W., Li B., Zhao H. Adenovirus inactivation by in situ photocatalytically and photoelectrocatalytically generated halogen viricides. Chem. Eng. J. 2014;253:538–543. doi: 10.1016/j.cej.2014.05.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Brady-Estévez A.S., Schnoor M.H., Vecitis C.D., Saleh N.B., Elimelech M. Multiwalled carbon nanotube filter: improving viral removal at low pressure. Langmuir. 2010;26:14975–14982. doi: 10.1021/la102783v. [DOI] [PubMed] [Google Scholar]
- 218.Rahaman M.S., Vecitis C.D., Elimelech M. Electrochemical carbon-nanotube filter performance toward virus removal and inactivation in the presence of natural organic matter. Environ. Sci. Technol. 2012;46(3):1556–1564. doi: 10.1021/es203607d. [DOI] [PubMed] [Google Scholar]
- 219.Domagała K., Jacquin C., Borlaf M., Sinnet B., Julian T., Kata D., Graule T. Efficiency and stability evaluation of Cu2O/MWCNTs filters for virus removal from water. Water Res. 2020;115879 doi: 10.1016/j.watres.2020.115879. [DOI] [PubMed] [Google Scholar]
- 220.V. Shashikala, V. S. Kumar, A. H. Padmasri, B. D. Raju, S. Venkata Mohan, P. N. Sarma, K. R. Rao, 2007. Advantages of nano-silver-carbon covered alumina catalyst prepared by electro-chemical method for drinking water purification. J. Mol. Catal. A. Chem, 268(1-2) (2007). 95–100.
- 221.Lee S., Ihara M., Yamashita N., Tanaka H. Improvement of virus removal by pilot-scale coagulation-ultrafiltration process for wastewater reclamation: Effect of optimization of pH in secondary effluent. Water Res. 2017;114:23–30. doi: 10.1016/j.watres.2017.02.017. [DOI] [PubMed] [Google Scholar]
- 222.Ison, Odeh I.N., Margerum D.W. Kinetics and mechanisms of chlorine dioxide and chlorite oxidations of cysteine and glutathione. Inorg. Chem. 2006;45(21):8768–8775. doi: 10.1021/ic0609554. [DOI] [PubMed] [Google Scholar]
- 223.Torrey J., Gunten U., Kohn T. Differences in viral disinfection mechanisms as revealed by quantitative transfection of echovirus 11 genomes. Appl. Environ. Microb. 2019;85(14):00961–1019. doi: 10.1128/AEM.00961-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Bibby K., Fischer R.J., Casson L.W., Stachler E., Haas C.N., Munster V.J. Persistence of ebola virus in sterilized wastewater. Environ. Sci. Technol. Lett. 2015;2:245–249. doi: 10.1021/acs.estlett.5b00193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Brainard J., Pond K., Hunter P.R. Censored regression modeling to predict virus inactivation in wastewaters. Environ. Sci. Technol. 2017;51:1795–1801. doi: 10.1021/acs.est.6b05190. [DOI] [PubMed] [Google Scholar]
- 226.Casanova L.M., Weaver S.R. Inactivation of an enveloped surrogate virus in human sewage. Environ. Sci. Technol. Lett. 2015;2:76–78. [Google Scholar]
- 227.Rice W., Adcock N.J., Sivaganesan M., Brown J.D., Stallknecht D.E., Swayne D.E. Chlorine inactivation of highly pathogenic avian influenza virus (H5N1) Emerging Infect. Dis. 2007;13:1568–1570. doi: 10.3201/eid1310.070323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Lodder W.J., Buisman A.M., Rutjes S.A., Heijne J.C., Teunis P.F., de Roda Husman A.M. Feasibility of quantitative environmental surveillance in poliovirus eradication strategies. Appl. Environ. Microbiol. 2012;78(11):3800–3805. doi: 10.1128/AEM.07972-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Lodder W.J., Rutjes S.A., Takumi K., de Roda Husman A.M. Aichi virus in sewage and surface water, The Netherlands. Emerg. Infect. Dis. 2013;19(8):1222. doi: 10.3201/eid1908.130312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Mallapaty S. How sewage could reveal true scale of coronavirus outbreak. Nature. 2020;580(7802):176–177. doi: 10.1038/d41586-020-00973-x. [DOI] [PubMed] [Google Scholar]
- 231.Daughton C.G. Monitoring wastewater for assessing community health: sewage chemical-information mining (SCIM) Sci. Total Environ. 2018;619–620:748–764. doi: 10.1016/j.scitotenv.2017.11.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Lodder W., Husman A.M.D.R. SARS-CoV-2 inwastewater: potential health risk, but also data source. Lancet Gastroenterol. Hepatol. 2020;5(6):533–534. doi: 10.1016/S2468-1253(20)30087-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Bibby K., Crank K., Greaves J., Li X., Wu Z., Hamza I.A., Stachler E. Metagenomics and the development of viral water quality tools. npj Clean Water. 2019;2(1):1–13. [Google Scholar]
- 234.Fernandez-Cassi X., Timoneda N., Martínez-Puchol S., Rusiñol M., Rodriguez-Manzano J., Figuerola N., Bofill-Mas S., Abril J.F., Girones R. Metagenomics for the study of viruses in urban sewage as a tool for public health surveillance. Sci. Total Environ. 2018;618:870–880. doi: 10.1016/j.scitotenv.2017.08.249. [DOI] [PubMed] [Google Scholar]
- 235.Ng T.F.F., Marine R., Wang C., Simmonds P., Kapusinszky B., Bodhidatta L., Oderinde B.S., Wommack K.E., Delwart E. High variety of known and new RNA and DNA viruses of diverse origins in untreated sewage. J. Virol. 2012;86:12161–12175. doi: 10.1128/JVI.00869-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Hart O.E., Halden R.U. Computational analysis of SARS-CoV-2/COVID-19 surveillance by wastewater-based epidemiology locally and globally: feasibility, economy, opportunities and challenges. Sci. Total Environ. 2020 doi: 10.1016/j.scitotenv.2020.138875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Filipic I., Gutierrez-Aguirre G., Primc M., Mozetič D., Dobnik Cold plasma, a new hope in the field of virus inactivation. Trends Biotechnol. 2020 doi: 10.1016/j.tibtech.2020.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Rachmadi A.T., Kitajima M., Pepper I.L., Gerba C.P. Enteric and indicator virus removal by surface flow wetlands. Sci. Total Environ. 2016;542:976–982. doi: 10.1016/j.scitotenv.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 239.Xia, T., Kleinheksel, A., Lee, E.M., Qiao, Z., Wigginton, K.R., Clack, H.L. Inactivation ofairborne viruses using a packed bed non-thermal plasma reactor. J. Phys. D: Appl. Phys. 2019;52:255201–2552013. doi: 10.1088/1361-6463/ab1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Delanka-Pedige, H.M., Munasinghe-Arachchige, S.P., Zhang, Y., Nirmala khandan, N. Bacteria and virus reduction in secondary treatment: Potential for minimizing postdisinfectant demand. Water Res. 2020:115802. doi: 10.1016/j.watres.2020.115802. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.