Abstract
Recent advances in CRISPR/Cas9 based genome editing have considerably advanced genetic engineering of industrial yeast strains. In this study, we report the construction and characterization of a toolkit for CRISPR activation and interference (CRISPRa/i) for a polyploid industrial yeast strain. In the CRISPRa/i plasmids that are available in high and low copy variants, dCas9 is expressed alone, or as a fusion with an activation or repression domain; VP64, VPR or Mxi1. The sgRNA is introduced to the CRISPRa/i plasmids from a double stranded oligonucleotide by in vivo homology-directed repair, allowing rapid transcriptional modulation of new target genes without cloning. The CRISPRa/i toolkit was characterized by alteration of expression of fluorescent protein-encoding genes under two different promoters allowing expression alterations up to ~ 2.5-fold. Furthermore, we demonstrated the usability of the CRISPRa/i toolkit by improving the tolerance towards wheat straw hydrolysate of our industrial production strain. We anticipate that our CRISPRa/i toolkit can be widely used to assess novel targets for strain improvement and thus accelerate the design-build-test cycle for developing various industrial production strains.
Subject terms: Expression systems, Industrial microbiology, Metabolic engineering, Applied microbiology, Industrial microbiology, Biotechnology, Microbiology, Molecular biology
Introduction
The yeast Saccharomyces cerevisiae is one of the most commonly used microorganisms for industrial applications ranging from wine and beer fermentations to the production of biofuels and high-value metabolites1,2. However, some of the current production processes are compromised by low yields and productivities, thus further optimization is required3. In particular, the production of second-generation bioethanol and other biochemicals from lignocellulosic biomass, which provides an alternative to oil-based chemicals, suffers from sub-optimal productivity4. During the hydrolysis of the raw material, inhibitory compounds (e.g. organic acids and aromatic aldehydes) are formed or released, compromising the microbial performance5. While quite some work has been done on elucidating genes required for tolerance, much less work has been done on improving tolerance towards stress by altering expression of genes6. Some previous studies demonstrated deletion7–9 or overexpression10–14 of endogenous genes to improve tolerance of S. cerevisiae towards inhibitors commonly found in lignocellulosic hydrolysates. However, most of the published work focuses on improving the tolerance of laboratory yeast strains that generally have weaker tolerance to stress6, while translation of beneficial modifications to more robust, industrial strains often is very challenging. The choice of yeast strain to be engineered is crucial for the successful implementation of the engineered phenotype in an industrial production process15.
Yeast strains used in industrial processes tend to be genetically diverse, since they usually arise from hybridization between different species16. Hence, they typically present aneuploidy, polyploidy or other chromosomal rearrangements16–18. This makes the strain development schemes for industrial yeast much more time-demanding and laborious compared to the engineering of well-studied laboratory strains19. In order to select for superior strain variants, industrial production strains are often subjected to directed evolution techniques such as random mutagenesis, protoplast fusion or genome shuffling20. Although these techniques are less dependent on biological information and available genetic tools, they suffer from being non-directional, having the risk of improving one trait to the detriment of others20,21.
In recent years, CRISPR/Cas technologies have become an indispensable tool for genome editing (reviewed by Pickar-Oliver and Gersbach22). These technologies are based on DNA-binding proteins (Cas endonucleases) and the single guide RNA (sgRNA) duplexes that can be easily programmed to precisely target and cleave a specific locus on double-stranded DNA. Recent studies have shown the applicability of this technology in industrial yeast strains, with the simultaneous disruption of two alleles of a gene or even several genes simultaneously without the introduction of a selection marker23–26. Still, the alteration of gene expression levels is often a more desirable metabolic engineering strategy compared to complete gene inactivation, or promoter swapping for overexpression27. Dynamic gene alteration allows optimizing trade-offs between growth and production27.
Alteration of gene expression by a nuclease-deficient form of Cas9, dCas9, often fused to an activation or repression domain, has been widely shown in both prokaryotes and eukaryotes22. This so-called CRISPR activation/interference (CRISPRa/i) technology is a way to modulate the expression of genes targeted by an sgRNA, allowing also the expression alteration of essential genes and the manipulation of multiple traits without altering the target sequence28. In laboratory yeast strains, the CRISPRa/i technology has been successfully employed to e.g. generate metabolic flux sensitivity maps29, identify chemical-genetic interactions30 and optimize different production pathways31–33. CRISPR based repression (CRISPRi) has been shown using dCas9 or dCas9 fused to repressor domains like KRAB (Krüppel associated box) or Mxi1 (reviewed by Jensen34). Similarly, gene activation by fusion of transcriptional activators, such as VP16, VP64 or VPR to dCas9 has been reported34.
In this study, we report the construction and characterization of a set of CRISPRa/i plasmids for the alteration of gene expression in a polyploid industrial yeast strain. In the CRISPRa/i plasmids, to which the sgRNA was introduced to its expression cassette by in vivo homology-directed repair, dCas9 was expressed alone, or as a fusion with an activation or repression domain (i.e. VP64, VPR or Mxi1). The CRISPRa/i toolkit was used for the alteration of expression of fluorescent protein-encoding genes under two different promoters. Furthermore, we demonstrated the usability of the CRISPRa/i system by improving the tolerance towards growth in wheat straw hydrolysate, by targeting SSK2, a gene essential for tolerance to environmental stress35. We anticipate that our CRISPRa/i toolkit can be widely used to accelerate the design-build-test cycle for developing industrial production hosts.
Results
Modular design of the CRISPRa/i toolkit
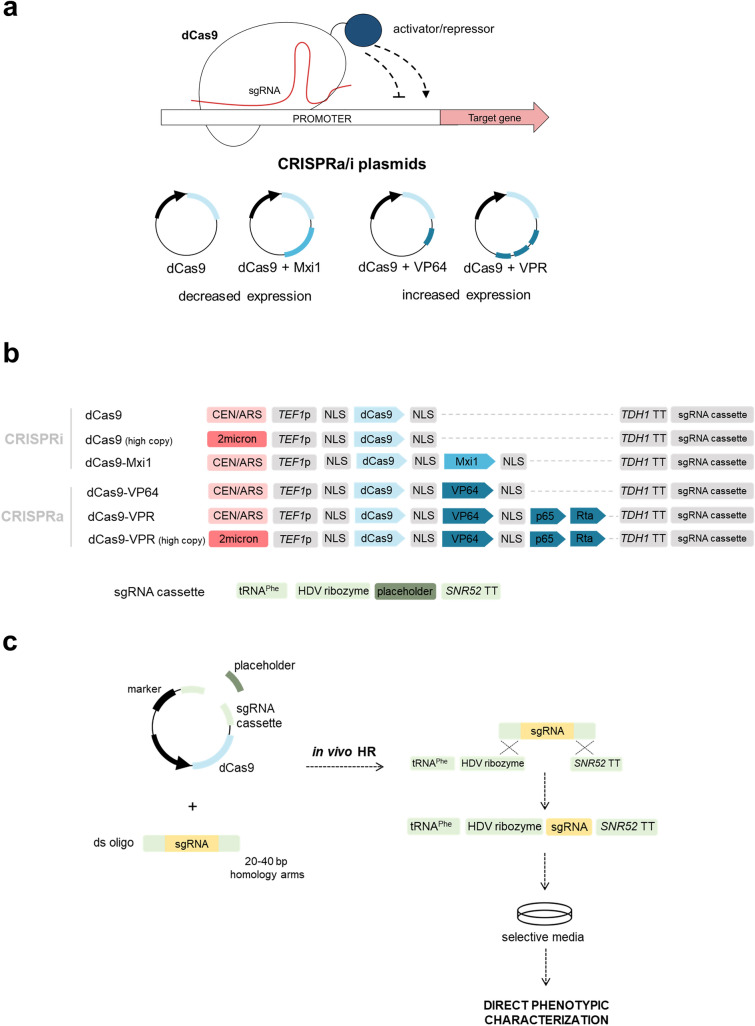
In order to obtain a CRISPRa/i toolkit for transcriptional modulation in industrial yeast, a set of plasmids was designed for the expression of dCas9 and the sgRNA (Fig. 1a). The dCas9 protein was expressed alone or fused to an activation or repression domain; the mammalian transcriptional repressor Mxi136, the VP64 activation domain containing 4 tandem copies of Herpes Simplex Viral Protein 1637 or VPR, a fusion of three different activation domains; VP64, p65, and Rta38.
Figure 1.
Design of the CRISPRa/i toolkit for transcriptional modulation. (a) The CRISPRa/i technology utilizes a catalytically inactive Cas9 (dCas9) to modulate the expression of genes targeted by an sgRNA. This can be further increased by fusing activation or repressor domains to dCas9. (b) Schematic representation of the CRISPRa/i components included in the CRISPRa/i plasmids and of the sgRNA expression cassette, present in all the plasmids. (c) The target-specific sgRNA from a double stranded oligonucleotide (ds oligo) was inserted into the CRISPRa/i plasmid through in vivo homologous recombination, allowing direct phenotypic characterization of strains with altered gene expression.
For all the CRISPRa/i plasmids, dCas9 was cloned under the strong TEF1 promoter and flanked by the SV40 nuclear localization sequence (Fig. 1b). The sgRNA cassette consisted of the phenylalanine tRNA promoter and an HDV ribozyme prior to a placeholder for the sgRNA, followed by the sgRNA scaffold and an SNR52 terminator39 (Fig. 1b). To insert the 20 bp specific sgRNA sequence targeting a gene of interest, the CRISPRa/i plasmids were digested at the placeholder prior to the yeast transformation. Therefore, the placeholder fragment was replaced by the sgRNA by in vivo homologous recombination (Fig. 1c).
In order to assess the impact of the CRISPRa/i toolkit on growth of the strains, strains expressing dCas9 or the dCas9-fusions (Fig. 1a) but no sgRNA were compared to a control strain carrying an empty plasmid lacking any dCas9 expression cassette (Fig. 2 and Supplementary Table S1). In the strains expressing dCas9-Mxi1 or dCas9-VPR, both the lag phase and the generation time were significantly (p < 0.05) increased compared to strains carrying an empty plasmid. Also the strain carrying an empty plasmid grew slightly slower than the parental strain (Fig. 2).
Figure 2.
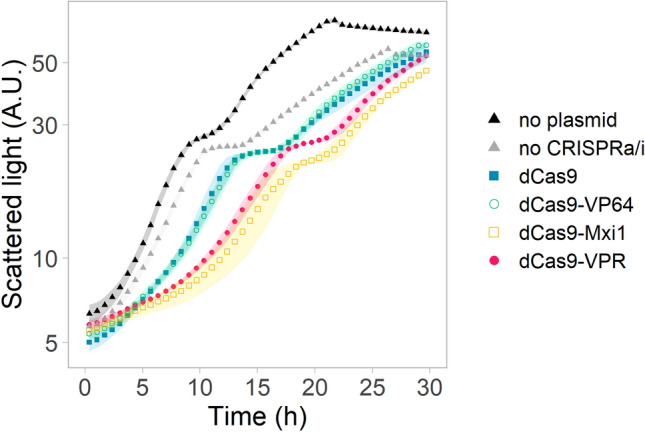
Growth of KE6-12-Ruby strains carrying different CRISPRa/i plasmids expressing dCas9 (blue solid squares), dCas9-VP64 (open turquoise circles), dCas9-Mxi1 (open yellow squares), dCas9-VPR (solid pink circles) or an empty plasmid lacking the dCas9 or sgRNA expression cassette (no CRISPRa/i strain, grey triangles). KE6-12-Ruby was added as a control to evaluate the plasmid effect (no plasmid strain, black triangles). The strains were grown in microbioreactors in YPD medium supplemented with geneticin for plasmid maintenance, and without antibiotic for the no plasmid strain. Data obtained from three biological replicates; shadowed regions show the standard deviation.
CRISPRa/i demonstration by changing expression of a fluorescence protein
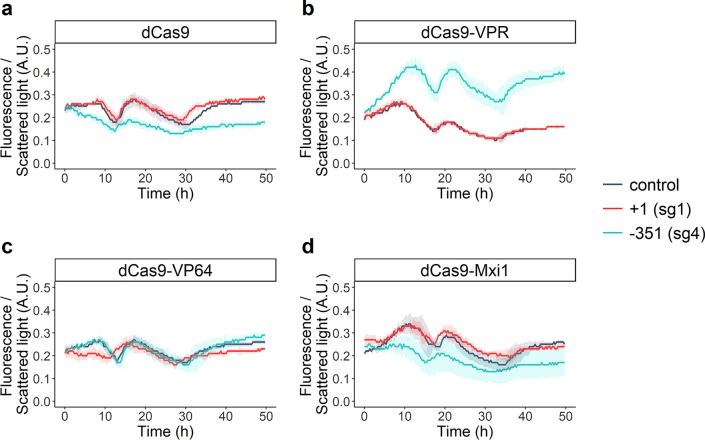
An expression cassette containing the red fluorescent protein-encoding gene mRuby2 under TDH3p was put into the PDR12 locus of the industrial strain KE6-12, a polyploid strain optimized for xylose consumption and ethanol production from lignocellulosic hydrolysates40,41, resulting in a strain named KE6-12-Ruby (Supplementary Fig. S1). Different CRISPRa/i plasmids and sgRNAs targeting the TDH3p promoter driving mRuby2, at positions + 1 or − 351 bp relative to the transcription starting site (TSS), were transformed into KE6-12-Ruby and the fluorescence of the strains was followed over time (Fig. 3 and Supplementary Fig. S2). Whereas no change in expression was observed when targeting position + 1, a significant (p = 6.5E−03) downregulation of 30% was obtained at the end of the culture when expressing dCas9 together with the sgRNA targeting a region at − 351 bp (Fig. 3a). In strains expressing this sgRNA and dCas9-VPR a significant (p = 7.6E−03) increase of 144% in fluorescence was measured at the end of the culture (Fig. 3b). In contrast, in strains expressing dCas9-VP64 or dCas9-Mxi1 and the same sgRNA, no significant changes in expression were observed (Fig. 3c, d).
Figure 3.
CRISPRa/i based change in expression of a fluorescent protein, measured over time in microbioreactors. Normalized fluorescence of strains expressing dCas9 (a), dCas9-VPR (b), dCas9-VP64 (c) or dCas9-Mxi1 (d) and sgRNAs targeting a region at + 1 (sg1; red line) or − 351 (sg4; turquoise line) bp relative to the TSS or the CRISPRa/i plasmid with a placeholder (control; grey line). Data obtained from three biological replicates; shadowed regions show the standard deviation.
Characterization of the CRISPRa/i toolkit
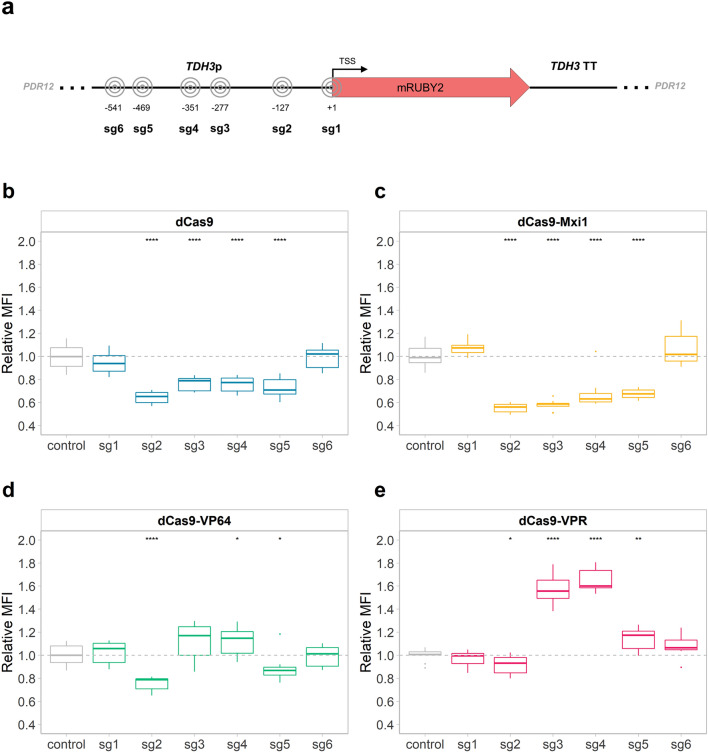
The impact of the sgRNA target region on the transcriptional modulation was evaluated using 6 different sgRNAs, targeting TDH3p at regions between − 541 and + 1 bp relative to the TSS (Fig. 4a). The mean fluorescence intensity (MFI) of the strains expressing mRuby2 was measured by flow cytometry from samples taken after 24 h of cultivation (Fig. 4b–e), during exponential growth (Supplementary Fig. S2).
Figure 4.
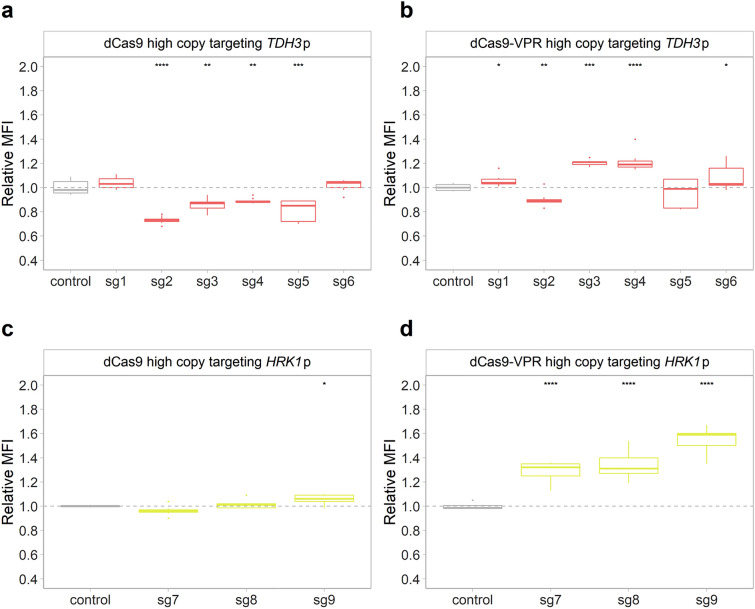
Modulation of mRuby2 fluorescence by different CRISPRa/i plasmids. (a) Six sgRNAs targeting different loci of TDH3p were tested. (b)–(e) Relative MFI of strains expressing dCas9 (b), dCas9-Mxi1 (c), dCas9-VP64 (d) and dCas9-VPR (e) together with either of the sgRNAs (sg1–6) or the placeholder (control). The box plots show the relative fluorescence, compared to KE6-12-Ruby expressing the corresponding CRISPRa/i plasmid with the placeholder (control, grey dashed line). Data obtained from three biological and three technical replicates. The line dividing the box represents the median of the data, the whiskers indicate the data outside the middle 50% and the outliers are shown as individual points. Statistical significance represented as “*” for p ≤ 0.05, “**” for p ≤ 0.01, “***” for p ≤ 0.001 and “****” for p ≤ 0.0001.
In strains expressing dCas9 alone, the fluorescence of mRuby2 was significantly reduced when targeting positions between − 127 and − 469 bp (sg2–5, p < 0.05) (Fig. 4b), reaching a maximal decrease of 35% with sg2. Likewise, expression of sg2, sg3, sg4, or sg5 together with dCas9-Mxi1 led to a reduction in fluorescence (Fig. 4c). When targeting positions at − 127 or − 277 bp, the repression was increased by 10 or 18% in strains expressing dCas9-Mxi1 compared to strains expressing dCas9 alone (Fig. 4b, c). Strains expressing dCas9-VP64 and sg4 showed a 10% increase in fluorescence compared to the no sgRNA control (Fig. 4d). In contrast, a decrease in mRuby2 expression was obtained with sg2 and sg5. Strains expressing dCas9-VPR and sg3, sg4 or sg5 displayed an increase in fluorescence, with sg3 and sg4 targeting positions at − 277 or − 351 bp leading to highest activation, displaying an increase in fluorescence of 57% or 65%, respectively (Fig. 4e). The strain expressing dCas9-VPR and sg2 displayed a significant (p = 0.02) decrease in expression of mRuby2 (Fig. 4e). Strains expressing any of the CRISPRa/i plasmids and sgRNAs sg1 or sg6 showed no change in expression compared to the control strain with a placeholder in the sgRNA cassette.
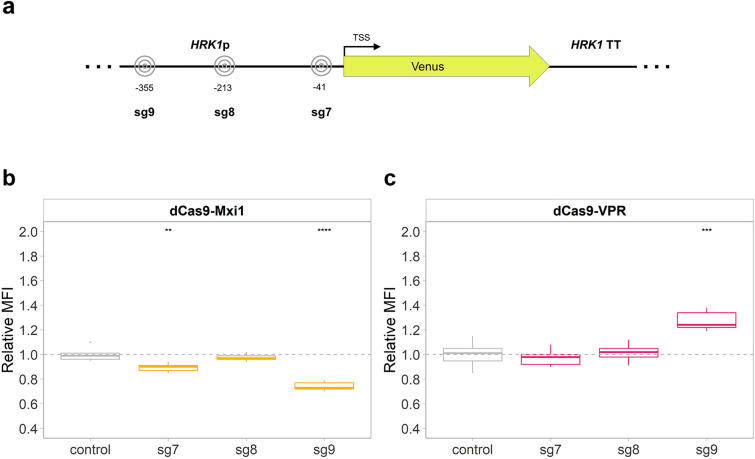
To further test the applicability of the CRISPRa/i toolkit, the most effective plasmids were tested with sgRNAs targeting a presumably weaker promoter42, HRK1p. In KE6-12-Ruby-Venus all native copies of HRK1 were replaced by a gene encoding the yellow fluorescent protein Venus (Fig. 5a and Supplementary Fig. S3). Three sgRNAs targeting positions at regions − 355, − 213 or − 40 bp relative to the TSS were tested, the most impactful being sg9, hybridizing at − 355 bp (Fig. 5b, c). The strain expressing dCas9-Mxi1 and sg9 showed a repression of fluorescence by 26% (Fig. 5b) whereas the strain expressing dCas9-VPR showed a 27% increase in expression of Venus.
Figure 5.
Modulation of fluorescence of Venus expressed under HRK1p by CRISPRa/i. (a) Three sgRNAs targeting different loci of HRK1p were tested. (b)–(c) Relative MFI of strains expressing dCas9-Mxi1 (b), and dCas9-VPR (c) together with either of the sgRNAs (sg7–9) or the placeholder (control). The box plots show the relative fluorescence, compared to KE6-12-Ruby-Venus expressing the corresponding CRISPRa/i plasmid with the placeholder (control, grey dashed line). Data obtained from three biological and three technical replicates. The line dividing the box represents the median of the data, the whiskers indicate the data outside the middle 50% and the outliers are shown as individual points. Statistical significance represented as “*” for p ≤ 0.05, “**” for p ≤ 0.01, “***” for p ≤ 0.001 and “****” for p ≤ 0.0001.
High copy variants of the CRISPRa/i plasmids
In an attempt to increase the impact of the CRISPRa/i system, high copy variants of the CRISPRa/i plasmids expressing dCas9 or dCas9-VPR, were used for targeting TDH3p in the KE6-12-Ruby strain or HRK1p in the KE6-12-Ruby-Venus strain. Nonetheless, the repression of mRuby2 expression was similar when using centromeric, low copy (Fig. 4b) or high copy plasmids (Fig. 6a) expressing dCas9 and sg2, sg3, sg4 or sg5 (Supplementary Table S2). Similarly, expressing dCas9 and sg7, sg8 or sg9 from the high copy plasmid led to strains with Venus expression similar to that of the corresponding centromeric plasmid carrying strains (Figs. 5a, 6c and Supplementary Table S3).
Figure 6.
Modulation of fluorescence using high copy CRISPRa/i plasmids. (a) Relative MFI of strains expressing dCas9 (a), or dCas9-VPR (b) together with either of the sgRNAs targeting TDH3p (sg1–6) or HRK1p (sg7–9) or the placeholder (control). The box plots show the relative fluorescence, compared to KE6-12-Ruby (a, b) or KE6-12-Ruby-Venus (c, d) expressing the corresponding high copy CRISPRa/i plasmid with the placeholder (control, grey dashed line). Data obtained from three biological and three technical replicates. The line dividing the box represents the median of the data, the whiskers indicate the data outside the middle 50% and the outliers are shown as individual points. Statistical significance represented as “*” for p ≤ 0.05, “**” for p ≤ 0.01, “***” for p ≤ 0.001 and “****” for p ≤ 0.0001.
CRISPRa strains expressing dCas9-VPR and sg3 or sg4 from high copy plasmids (Fig. 6b) showed a 20% increase in expression of mRuby2, which was a threefold lower activation compared to expressing dCas9-VPR and the same sgRNAs from the low copy plasmid (Fig. 4e). Conversely, strains carrying the high copy CRISPRa plasmid expressing dCas9-VPR and sg7, sg8 or sg9 showed an increase in expression of Venus of 29, 34 and 56%, respectively (Fig. 6d). Nonetheless, strains with the high copy CRISPRa plasmid expressing dCas9-VPR displayed a serious growth defect (Supplementary Table S1).
Application of the CRISPRa/i toolkit to improve tolerance towards lignocellulosic hydrolysate
Four genes reported to be involved in tolerance towards inhibitors found in lignocellulosic hydrolysate, HRK143, SSK235, ISC144 and BDH245, were selected as targets for applying the CRISPRa/I toolkit. Strains with 3 different sgRNAs were designed for locating dCas9 or dCas9-VPR to the promoter regions of these genes were tested in minimal medium with added inhibitors (Supplementary Fig. S4). Strains with sgRNAs targeting the promoters of ISC1 or BDH2 performed the same or worse than the strain with no sgRNA (Supplementary Fig. S4). Two sgRNAs targeting HRK1 led to higher biomass yield whereas one of the sgRNAs tested resulted in a slower growth in medium supplemented with acetate (Supplementary Fig. S4). However, all strains expressing dCas9 and sgRNAs targeting SSK2 grew better in medium supplemented with 20 mM furfural (Supplementary Fig. S4) and were therefore tested in lignocellulosic hydrolysate.
Downregulation of SSK2 has previously been suggested to improve tolerance towards furfural35. Therefore, targeting SSK2p with the CRISPRi system, using either dCas9 or dCas9-Mxi1 expressing plasmids were tested in lignocellulosic hydrolysate (Supplementary Fig. S5). The expression of dCas9-Mxi1 caused a high impairment on growth whereas strains expressing dCas9 alone grew comparably to the control strain or better (Fig. 7a–d).
Figure 7.
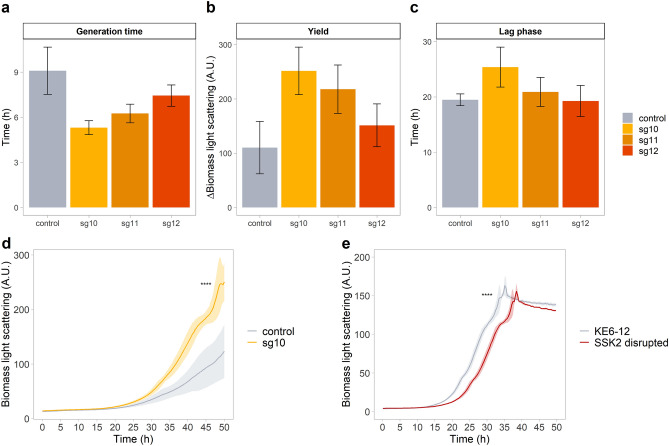
Application of CRISPRi for improving growth in wheat straw hydrolysate. (a) generation time, (b) biomass yield and (c) lag phase of KE6-12 expressing dCas9 and different sgRNAs targeting SSK2p. (d) Growth of KE6-12 strain expressing dCas9 and sg10 or the placeholder. (e) Growth of KE6-12 strain compared to KE6-12 with SSK2 gene disrupted. The strains were grown in microbioreactors in 64% wheat straw hydrolysate for 50 h. Data obtained from three biological replicates (a–d) or three technical replicates (e) are expressed as mean ± standard deviation (SD). Statistical significance represented as “*” for p ≤ 0.05, “**” for p ≤ 0.01, “***” for p ≤ 0.001 and “****” for p ≤ 0.0001.
The growth of strains expressing dCas9 from a low copy plasmid and sgRNAs targeting positions − 20, − 180 or − 380 relative to the TSS of SSK2 (sg10, sg11 and sg12, respectively), was compared to strains expressing dCas9 with the placeholder (Fig. 7a–d). The generation time of all strains expressing sgRNAs targeting SSK2p was reduced compared to the control strain (Fig. 7a). Moreover, the strain expressing sg10 showed a 2.3-fold increase in final biomass yield compared to the no sgRNA control (Fig. 7b, d). The lag phase of the strains was not affected by expression of any of the sgRNAs (Fig. 7c). When SSK2 was disrupted from KE6-12, this modification led to an approx. 5 h longer lag phase compared to KE6-12, when growth in hydrolysate was evaluated (Fig. 7e).
Discussion
In this study, we have successfully developed a CRISPRa/i toolkit for modulation of transcription in a polyploid, industrial yeast strain. The CRISPRa/i technology enables gradual modulation of gene expression allowing for screening of optimal expression levels. Regulation of transcription is a very sensitive balance and even small perturbations in gene expression can have great impact on the regulatory network and fitness of a cell subjected to stressful environments46.
We evaluated our CRISPRa/i toolkit by altering the expression of fluorescent proteins expressed under the strong TDH3 promoter or under the relatively weak, endogenous HRK1 promoter. An increased expression under TDH3p was observed for two out of six sgRNAs in strains expressing dCas9-VPR from a centromeric, low copy plasmid (Fig. 4e). In strains expressing dCas9 alone or the dCas9-Mxi fusion two additional sgRNAs could decrease the expression of the fluorescent protein expressed under TDH3p (Fig. 4b, c). The most efficient sgRNAs, sg3 and sg4, both targeted a region of TDH3p close to a known upstream activating sequence, UAS2, −255 –309 bp relative to the TSS47. The highest repression of TDH3p was found in strains expressing sg2 targeting position − 127 (Fig. 4c). Our results suggest that a critical regulation element could be located in this region, since a repression of the promoter was obtained with all the plasmids tested, including dCas9-VPR (Fig. 4b–e). Despite designed to function as a transcriptional activator, dCas9-VPR has also previously in combination with certain sgRNAs been shown to confer transcriptional repression32.
Our previous work has shown that robustness and tolerance can vary greatly between industrial strains and evolved mutants41,48, highlighting the need for a system that can be rapidly implemented in novel hosts. The benefit of plasmid-based expression of dCas9 and sgRNA in comparison with genomic integration is that the CRISPRa/i toolbox can rapidly be introduced to new strains and for performing novel genomic regulation events. Targeting a new gene and phenotyping the alteration introduced can be achieved in a week.
A recent study, where a plasmid-based CRISPRa/i-system was developed demonstrated a threefold difference in butanediol production when the same regulation perturbations were implemented in two different laboratory yeast49. With our CRISPRa/i toolkit, a 2.5-fold change in gene expression was observed, when comparing the best performing sgRNA for repression using dCas9-Mxi1 with the most potent sgRNA for activation using dCas9-VPR. This fold-change in expression level is similar to what was previously seen in laboratory yeast strains31,32,34. Some previous studies report even stronger modulation of expression by CRISPRa/i, but the strength of developed CRISPRa/i systems has been shown to be largely dependent on the sgRNAs used (reviewed by Jensen34). Our study confirmed that current state-of-the-art software tools fail to predict how specific sgRNAs and scRNAs quantitatively interfere with the activity of a native promoter. The systematic evaluation of the efficiency of different sgRNAs was beyond the scope of this study and as the genome sequence of KE6-12 is unknown, sequence variability may have a caused further decrease in targeting efficiency when targeting native promoters as was previously reported by Smith et al.30. Furthermore, as previous work30,32 has shown that up to two thirds of the sgRNAs designed fail to function for CRISPRa/i, it may be that a higher effect of our CRISPRa/i toolkit would be possible by testing a larger set of sgRNAs.
In order to improve transformation efficiency and genetic stability, we chose to express the genes encoding the different dCas9 variants and the sgRNA from the same plasmid. The protospacer sequence of the sgRNA was co-transformed as a double stranded oligonucleotide and the correct assembly of the CRISPRa/i plasmids in all strains was confirmed by colony PCR (Supplementary Fig. S7), demonstrating that the homologous recombination capacity of our strain was remarkable. The design where the protospacer sequence is added as a double stranded oligonucleotide allows rapid targeting of any loci and circumvents the need for cloning when altering the expression of a new gene is desired. In fact, the phenotypes of the new strains can be characterized directly after transformation. Similarly, during strain construction by CRISPR/Cas9 based genome editing, a highly efficient single-step gene disruption in the polyploid industrial strain was obtained by using marker-free donor DNA with 40 bp homology to the integration site. When Cas9 and the sgRNA were put on episomal plasmids, the plasmids could easily be cured from the yeast, effectively resulting in a marker-free, permanent modification.
High expression of Cas9 has previously been reported to be toxic for industrial yeast26 and high expression of dCas9 has been shown to be toxic for many bacteria (reviewed in50). Similarly, a few studies report that a high expression of the VP16 in yeast led to a slower growth51–53. Here, we did not observe any significant growth defect with strains expressing dCas9, but we observed a growth defect when strains were expressing dCas9-Mxi1 or dCas9-VPR, even without any sgRNA, (Fig. 2 and Supplementary Table S1). This effect was stronger when expressing the dCas9 fusions from high copy plasmids compared to when low copy plasmids were used. Still, in order to achieve an increase in expression under HRK1p, a higher expression of dCas9-VPR and/or the sgRNA appeared to be needed. In contrast, the high copy variant of the CRISPRa/i plasmids did not outperform the low copy plasmids when targeting TDH3p, further demonstrating that the effect of CRISPRa/i is highly target-dependent, which emphasizes the need for a versatile CRISPRa/i toolkit. The most suited for plasmid of the toolkit may depend on the strain and target gene chosen.
The applicability of the developed CRISPRa/i toolkit was demonstrated by successfully improving the resistance of our industrial yeast strain towards lignocellulosic hydrolysate (Fig. 7). A number of previous studies report engineering of resistance to phenolics, furfural and carboxylic acids, the most common inhibitors found in lignocellulosic hydrolysates. Still, the sensitivity towards inhibitors in hydrolysates is a major bottleneck in developing robust strains4, therefore even small improvements in tolerance may have big effects on the profitability of biorefineries. The MAP kinase Ssk2 has been shown to be involved in furfural tolerance35 and it was also shown to be essential for combatting environmental stress54. In our study, SSK2p was targeted with dCas9, resulting in an improvement in growth in wheat straw hydrolysate (Fig. 7d). When SSK2 was disrupted, growth was delayed (Fig. 7e), indicating that down-regulation of SSK2 is a better strategy compared to classical knockouts. This shows that the developed CRISPRa/i toolkit provides a rapid method for testing and validating gene targets for strain improvement. For industrial production strains it is likely beneficial to make the genetic targets identified using the CRISPRa/i toolkit stable, for instance through promoter substitution.
Collectively, this work demonstrates that CRISPRa/i can be used for transcriptional regulation in polyploid, industrial yeast strains. Moreover, this work has provided valuable information on transcriptional regulation using dCas9 and dCas9-fusions to transcriptional effectors. The CRISPRa/i plasmids have also been verified in laboratory CEN.PK strains (work in preparation), demonstrating that the toolkit can be expected to be widely applicable in different strain backgrounds. Even though there is still a need for further refining the toolkit, our results provide the framework for future work on CRISPRa/i based transcriptional modulation in polyploid yeast, accelerating the development of more robust strains to be used in biorefineries.
Conclusions
The CRISPRa/i toolkit was shown to lead to transcriptional activation and repression in a polyploid yeast strain. The repression of a gene essential for environmental stress, SSK2, by CRISPRi demonstrated the potential of our toolkit for balancing and optimizing gene expression without genome editing. We anticipate that our CRISPRa/i toolkit can be widely used to accelerate the design-build-test cycle for developing various industrial production strains.
Methods
Plasmid assembly
All primers and plasmids used in this study are listed in the Supplementary material online. Primers and oligonucleotides used were ordered from Eurofins. Phusion High-Fidelity and Phire Hot Start II DNA polymerases were used for PCR amplification. All PCR fragments used for plasmid assembly were column purified (using the GeneJET PCR Purification Kit) and sequenced (Eurofins) prior to the final assembly. Chemically competent Escherichia coli DH5α cells were used for cloning and transformed cells were selected on LB with the appropriate antibiotics. All chemicals and reagents used were purchased from ThermoScientific™ unless otherwise noted.
CRISPRa/i plasmids were assembled using the MoClo Yeast Toolkit55 (Addgene #1000000061), which allows for rapid creation of plasmids with different selection cassettes or auxotrophic markers for expanding the versatility of the toolkit. New genetic parts containing dCas9, Mxi1, VP64 or VPR where constructed by PCR amplification of the corresponding gene fragments from plasmids pTDH3-dCas9 (36, Addgene #46920) and pAG414GPD-dCas9-VPR (38 Addgene #63801). The parts were put into the entry plasmid pYTK001 using overhangs added to the amplification primers, resulting in plasmids EC0_1_dCas9, EC0_2_dCas9_VP64, EC0_3_dCas9_Mxi1 and EC0_4_dCas9_VPR. For the expression of the sgRNA, a placeholder sequence non-homologous to the yeast genome (primers hybridized by boiling followed by a gradual decrease in temperature) containing two BbsI sites was cloned into the plasmid #pYTK050 via BsmBI, resulting in the plasmid EC0_6_sgRNA, carrying a tRNA Phe promoter, the tSNR52 terminator and an HDV Ribozyme site. dCas9 was expressed under the ScTEF1 promoter and ScTDH1 terminator. The final multigene plasmids, EC2_1_dCas9_sgRNA, EC2_2_dCas9_VP64_sgRNA, EC2_3_dCas9_Mxi1_sgRNA and EC2_4_dCas9_VPR_sgRNA carried dCas9, activator/repressor domains, the sgRNA scaffold and the CEN6/ARS4 origin of replication. High copy plasmids EC2_7_dCas9_HC_sgRNA and EC2_10_dCas9_VPR_HC_sgRNA were assembled with the 2µ origin of replication.
Linear oligonucleotides with 20 nt protospacers and 40 bp overlaps on either side of the sgRNA scaffold were assembled into the plasmids through in vivo homologous recombination. Prior to transformation, plasmids were digested by BpiI to remove the placeholder region in the sgRNA cassette. Clones were confirmed with colony PCR.
For gene editing by CRISPR/Cas9, the plasmid YN2_1_Cas9 was constructed assembling several genetic elements from the MoClo Yeast Toolkit, resulting in the Cas9 gene from Streptococcus pyrogens under the ScPGK1 promoter and terminator, the tRNA Phe promoter, tSNR52 terminator, an HDV Ribozyme site for sgRNA expression and the sgRNA scaffold with a bacterial GFP expression cassette replacing the protospacer. The GFP expression cassette was amplified from pYTK050 using primers that added flanks compatible with the MoClo system. In plasmids YN2_1_Cas9_Ruby and YN2_1_Cas9_Venus for genome editing, the GFP cassette was removed by PstI digestion and replaced with the protospacer sequences.
A description of the parts assembled for each plasmid is provided in Supplementary Tables S4–S5.
Strain construction and maintenance
The polyploid industrial S. cerevisiae strain KE6-12 was used as a parental strain. KE6-12 is derived from TMB400 by evolutionary engineering, with XR and XDH from Pichia stipitis integrated into the genome (Albers et al., unpublished). Yeast strains were transformed according to the Gietz method56 and cells were selected in YPD media supplemented with G418.
KE6-12 expressing mRuby2 and Venus (KE6-12-Ruby and KE6-12-Ruby-Venus) were constructed by transformation of plasmids YN2_1_Cas9_Ruby or YN2_1_Cas9_Venus and a donor DNA containing the mRuby2 or Venus expression cassettes (Supplementary Table S6). The fluorescence protein-encoding genes were amplified from EC1_10 or pYTK033, adding 40 bp homologous to the PDR12 or HRK1 locus.
Correct transformants, with pTDH3-mRuby2-tTDH1 disrupting all the PDR12 ORFs or placing Venus under the control of the native HRK1 promoter at all chromosome copies, were selected. All the strains were verified by colony PCR (Supplementary Fig. S1,S3 and Table S7) and cured for plasmids before proceeding to transcriptional regulation using the CRISPRa/i toolkit or tolerance assays. Deletion of PDR12 or HRK1 was during non-stressful conditions previously shown not affect the growth of S. cerevisiae9,57. Strains were maintained in − 80 and the presence of plasmids expressing dCas9 was confirmed in all the strains at the end of the growth in hydrolysate (Supplementary Fig. S8).
KE6-12 strain with SSK2 disrupted was constructed by co-transformation of BpiI-digested EC2_5 plasmid (Supplementary Table S5) and a donor DNA containing a barcode sequence with flanking 40 bp homologous arms to SSK2 locus (Supplementary Table S6). Correct transformants with all the SSK2 ORFs disrupted were selected by colony PCR (Supplementary Fig. S6).
sgRNA design
The yeast CRISPRi webtool30 and CRISPR-ERA58 were used to design the sgRNAs, using as a criteria ATAC-seq values around 1 and nucleosome-free areas, to ensure accessibility to the target genes (Supplementary Table S7). Additionally, poly-N and presence of off-targets were avoided and GC content between 30 and 55 was preferred. For KE6-12-Ruby, positions at + 1, − 127, − 277, − 351, − 469 or − 541 bp relative to the TSS were chosen to cover most of the TDH3 promoter region. A spacing of around 200 base pairs was chosen to cover most of the promoter region. For KE6-12-Ruby-Venus, 3 sgRNAs were designed targeting HRK1p at − 41, − 213 or − 355 bp relative to the TSS, while for SSK2p positions targeting regions at − 20, − 180 or − 380 bp relative to the TSS were chosen. Positions for sgRNAs targeting BDH2p or ISC1p can be found in Supplementary Table S8. Verification of sgRNA integration into the expression plasmid was done by colony PCR for strains with sgRNAs targeting TDH3p and HRK1p (Supplementary Table S9). Oligonucleotides used as placeholder or sgRNA are provided in the Supplementary Table S8.
Media
The E. coli strain was grown in liquid or solid (supplemented with 15 g/L agar) Luria–Bertani (LB) medium (10 g/L bacto tryptone, 5 g/L yeast extract, 10 g/L NaCl) containing 25 mg/L chloramphenicol, 100 mg/L ampicillin or 50 mg/L neomycin. Yeast strains were grown in liquid or solid (supplemented with 15 g/L agar) yeast extract peptone dextrose (YPD) medium (10 g/L yeast extract, 20 g/L peptone, and 20 g/L glucose). Strains containing plasmids were grown on YPD medium supplemented with 200 mg/L geneticin. For the tolerance assays in minimal media, YNB media (1.9 g/L YNB, 0.79 g/L CSM and 5 g/L ammonium sulphate, 20 g/L glucose) containing geneticin (200 mg/L) for CRISPRa/i strains and supplemented with either acetic acid (100 mM) furfural (20 mM) or vanillin (12 mM) was used. The medium was adjusted at pH 4.5 and buffered with succinate (50 mM). For the tolerance assay in hydrolysates, wheat straw hydrolysate with a composition of 68.8 g/L glucose, 36.4 g/L xylose, 1.2 g/L formic acid, 4.7 g/L acetic acid, 0.6 g/L 5-(hydroxymethyl)furfural (HMF), and 3.0 g/L furfural45 was used. The hydrolysate was kindly provided by Dr M. Galbe at Lund University (Sweden).
Culture conditions
For growth characterization and flow cytometry analysis, transformants carrying CRISPRa/i plasmids were grown in liquid medium using the Growth Profiler 960 platform (Enzyscreen). Precultures were inoculated from a fresh colony and incubated overnight at 30 °C and 200 rpm in YPD, and then transferred into 96-well, transparent bottom, white-walled plates (CR1496d, EnzyScreen) with 250 µL of YPD at an initial OD600 of 0.05. Cultures were incubated at 30 °C and 250 rpm for 24 h. OD600 values were analyzed using the PRECOG software59 to extract the length of the lag phase, the generation time and the biomass (OD600) yield. The average and standard deviation of three biological replicates are reported.
For real-time monitoring of mRuby2 fluorescence and growth assessment of KE6-12 in wheat straw hydrolysate, strains were grown in microbioreactors, using the Biolector platform (m2p-Laboratories GmbH). Precultures were inoculated from a fresh colony and incubated overnight at 30 °C and 200 rpm in YPD medium in a 96-well microtiter plate. For KE6-12-Ruby strains, overnight cultures were transferred into a 48-well microtiter plate (48-B FlowerPlate, m2p-labs) with 1 mL of YPD at an initial OD600 of 0.05. For KE6-12, precultures were centrifuged and resuspended in sterile water to a final OD600 of 5 and 100 µL of cell suspension was transferred into the 48-well microtiter plate containing 900 µL of wheat straw hydrolysate (diluted to 70%). All cultures were incubated at 30 °C, 1200 rpm, at a humidity of 85%. On-line parameters were measured in 20 min intervals, using an excitation and emission filter at 600 nm for scattered light, with a filter gain of 10 and a filter gain of 20 for YPD and hydrolysate media, respectively. mRuby2 fluorescence was measured at 580/610 nm and a gain of 100.
Flow cytometry measurements
For fluorescence measurements, transformants carrying CRISPRa/i plasmids were grown for 24 h using the Growth Profiler, in 96-well microtiter plates containing 250 µL of YPD, incubated at 30 °C and 250 rpm. Fluorescence was analyzed by flow cytometry using the Guava easyCyte 8HT system (Merck Millipore). All samples were, prior to analysis by flow cytometry, diluted in sterile water to a final concentration of 200–500 cells/µL and a total amount of 5,000 cells were measured per analysis at a flow rate of 0.59 mL/s. An excitation wavelength of 488 nm and a detection wavelength of 583 nm and 525 nm were used for KE6-12-Ruby and KE6-12-Ruby-Venus, respectively. The average and standard deviation of the geometric mean for three biological and three technical replicates is reported.
Statistical analysis
Pairwise comparisons were carried out with R, using the unpaired Student’s T-test for normally distributed data. The normal distribution of the data was determined using the Shapiro–Wilk test. Statistical relevance due to the p-value (P) obtained by the statistical tests performed is shown with “ns” for p > 0.05, “*” for p ≤ 0.05, “**” for p ≤ 0.01, “***” for p ≤ 0.001 and “****” for p ≤ 0.0001.
Supplementary information
Acknowledgements
This work was supported by The Swedish Energy Agency, project number P43978-1 and Chalmers Area of Advance Energy. Taurus Energy AB is thanked for the yeast strain supplied. The authors wish to thank Vaskar Mukherjee for valuable help with developing high-throughput transformation schemes and for fruitful discussions during the project. Luca Torello Pianale is acknowledge for help with molecular biology work and Marlous van Dijk and Ignis Trollmann are acknowledged for help with setting up analysis of growth in hydrolysate using the microbioreactor platform.
Author contributions
E.C. and Y.N. designed the experiments and wrote the manuscript. E.C. and I.L. performed the experiments. All authors read and approved the final manuscript.
Funding
Open Access funding provided by Chalmers University of Technology.
Data availability
All data generated or analyzed during this study are included in this published article and its supplementary files. Plasmids generated will be publicly available through Addgene.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-71648-w.
References
- 1.Krivoruchko A, Nielsen J. Production of natural products through metabolic engineering of Saccharomyces cerevisiae. Curr. Opin. Biotechnol. 2015;35:7–15. doi: 10.1016/j.copbio.2014.12.004. [DOI] [PubMed] [Google Scholar]
- 2.Mattanovich D, Sauer M, Gasser B. Yeast biotechnology: Teaching the old dog new tricks. Microb. Cell. Fact. 2014;13:34. doi: 10.1186/1475-2859-13-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Du J, Shao Z, Zhao H. Engineering microbial factories for synthesis of value-added products. J. Ind. Microbiol. Biotechnol. 2011;38:873–890. doi: 10.1007/s10295-011-0970-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosales-Calderon O, Arantes V. A review on commercial-scale high-value products that can be produced alongside cellulosic ethanol. Biotechnol. Biofuels. 2019;12:240. doi: 10.1186/s13068-019-1529-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hemansi et al. Second generation bioethanol production: The state of art. In Sustainable Approaches for Biofuels Production Technologies: From Current Status to Practical Implementation (eds. Srivastava et al.) 121–46 (Springer, Berlin, 2019).
- 6.Deparis Q, Claes A, Foulquié-Moreno MR, Thevelein JM. Engineering tolerance to industrially relevant stress factors in yeast cell factories. FEMS Yeast Res. 2017;17:4. doi: 10.1093/femsyr/fox036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casatta N, Porro A, Orlandi I, Brambilla L, Vai M. Lack of Sir2 increases acetate consumption and decreases extracellular pro-aging factors. Biochim. Biophys. Acta. 2013;1833:593–601. doi: 10.1016/j.bbamcr.2012.11.008. [DOI] [PubMed] [Google Scholar]
- 8.Fujitomi K, Sanda T, Hasunuma T, Kondo A. Deletion of the PHO13 gene in Saccharomyces cerevisiae improves ethanol production from lignocellulosic hydrolysate in the presence of acetic and formic acids, and furfural. Bioresour. Technol. 2012;111:161–166. doi: 10.1016/j.biortech.2012.01.161. [DOI] [PubMed] [Google Scholar]
- 9.Nygård Y, et al. The diverse role of Pdr12 in resistance to weak organic acids. Yeast. 2014;31:219–232. doi: 10.1002/yea.3011. [DOI] [PubMed] [Google Scholar]
- 10.Adeboye PT, Bettiga M, Olsson L. The chemical nature of phenolic compounds determines their toxicity and induces distinct physiological responses in Saccharomyces cerevisiae in lignocellulose hydrolysates. AMB Express. 2014;4:1–10. doi: 10.1186/s13568-014-0046-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alriksson B, Sárvari I, Jönsson LJ. Overexpression of Saccharomyces cerevisiae transcription factor and multidrug resistance genes conveys enhanced resistance to lignocellulose-derived fermentation inhibitors. Process. Biochem. 2010;45:264–271. [Google Scholar]
- 12.Larsson S, Nilvebrant N, Jönson L. Effect of overexpression of Saccharomyces cerevisiae Pad1p on the resistance to phenylacrylic acids and lignocellulose hydrolysates under aerobic and oxygen-limited conditions. Appl. Microbiol. Biotechnol. 2001;57:167–174. doi: 10.1007/s002530100742. [DOI] [PubMed] [Google Scholar]
- 13.Wu G, Xu Z, Jönsson LJ. Profiling of Saccharomyces cerevisiae transcription factors for engineering the resistance of yeast to lignocellulose-derived inhibitors in biomass conversion. Microb. Cell. Fact. 2017;16:199. doi: 10.1186/s12934-017-0811-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim D, Hahn JS. Roles of the Yap1 transcription factor and antioxidants in Saccharomyces cerevisiae’s tolerance to furfural and 5-Hydroxymethylfurfural, which function as Thiol-Reactive electrophiles generating oxidative stress. Appl. Environ. Microbiol. 2013;79:5069–5077. doi: 10.1128/AEM.00643-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cagnin L, et al. Comparing laboratory and industrial yeast platforms for the direct conversion of cellobiose into ethanol under simulated industrial conditions. FEMS Yeast Res. 2019;19:2. doi: 10.1093/femsyr/foz018. [DOI] [PubMed] [Google Scholar]
- 16.Querol A, Bond U. The complex and dynamic genomes of industrial yeasts. FEMS Microbiol. Lett. 2009;293:1–10. doi: 10.1111/j.1574-6968.2008.01480.x. [DOI] [PubMed] [Google Scholar]
- 17.Borneman AR, et al. Whole-genome comparison reveals novel genetic elements that characterize the genome of industrial strains of Saccharomyces cerevisiae. PLoS Genet. 2011;7:e1001287. doi: 10.1371/journal.pgen.1001287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kong II, Turner TL, Kim H, Kim RS, Yin YS. Phenotypic evaluation and characterization of 21 industrial Saccharomyces cerevisiae yeast strains. FEMS Yeast Res. 2018;18:1–11. doi: 10.1093/femsyr/foy001. [DOI] [PubMed] [Google Scholar]
- 19.Le Borgne, S. Genetic engineering of industrial strains of Saccharomyces cerevisiae. In Recombinant Gene Expression (ed. Lorence, A.) 451–465 (Humana Press, London, 2011). [DOI] [PubMed]
- 20.Steensels J, et al. Improving industrial yeast strains: exploiting natural and artificial diversity. FEMS Microbiol. Rev. 2014;38:947–995. doi: 10.1111/1574-6976.12073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu Z, et al. Evolutionary engineering of industrial microorganisms-strategies and applications. Appl. Microbiol. Biotechnol. 2018;102:4615–4627. doi: 10.1007/s00253-018-8937-1. [DOI] [PubMed] [Google Scholar]
- 22.Pickar-Oliver A, Gersbach CA. The next generation of CRISPR–Cas technologies and applications. Nat. Rev. Mol. Cell Biol. 2019;20:490–507. doi: 10.1038/s41580-019-0131-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang GC, et al. Construction of a quadruple auxotrophic mutant of an industrial polyploid Saccharomyces cerevisiae strain by using RNA-guided Cas9 nuclease. Appl. Environ. Microbiol. 2014;80:7694–7701. doi: 10.1128/AEM.02310-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stovicek V, Borodina I, Forster J. CRISPR-Cas system enables fast and simple genome editing of industrial Saccharomyces cerevisiae strains. Metab. Eng. Commun. 2015;2:13–22. doi: 10.1016/j.meteno.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lian J, Bao Z, Hu S, Zhao H. Engineered CRISPR/Cas9 system for multiplex genome engineering of polyploid industrial yeast strains. Biotechnol. Bioeng. 2018;115:1630–1635. doi: 10.1002/bit.26569. [DOI] [PubMed] [Google Scholar]
- 26.Ryan OW, Cate JHD. Multiplex engineering of industrial yeast genomes using CRISPRm. Methods Enzymol. 2014;546:473–489. doi: 10.1016/B978-0-12-801185-0.00023-4. [DOI] [PubMed] [Google Scholar]
- 27.Brockman IM, Prather KLJ. Dynamic metabolic engineering: new strategies for developing responsive cell factories. Biotechnol. J. 2015;10:1360–1369. doi: 10.1002/biot.201400422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qi LS, et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deaner M, Alper HS. Systematic testing of enzyme perturbation sensitivities via graded dCas9 modulation in Saccharomyces cerevisiae. Metab. Eng. 2017;40:14–22. doi: 10.1016/j.ymben.2017.01.012. [DOI] [PubMed] [Google Scholar]
- 30.Smith JD, et al. Quantitative CRISPR interference screens in yeast identify chemical-genetic interactions and new rules for guide RNA design. Genome Biol. 2016;17:45. doi: 10.1186/s13059-016-0900-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vanegas KG, Lehka BJ, Mortensen UH. SWITCH: A dynamic CRISPR tool for genome engineering and metabolic pathway control for cell factory construction in Saccharomyces cerevisiae. Microb. Cell Fact. 2017;16:25. doi: 10.1186/s12934-017-0632-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jensen ED, et al. Transcriptional reprogramming in yeast using dCas9 and combinatorial gRNA strategies. Microb. Cell Fact. 2017;16:46. doi: 10.1186/s12934-017-0664-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kildegaard KR, et al. CRISPR/Cas9-RNA interference system for combinatorial metabolic engineering of Saccharomyces cerevisiae. Yeast. 2019;36:237–247. doi: 10.1002/yea.3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jensen, M.K. Design principles for nuclease-deficient CRISPR-based transcriptional regulators. FEMS Yeast Res.18, 4 (2018). [DOI] [PMC free article] [PubMed]
- 35.Kim H, Kim N, Kim W. Insertion of transposon in the vicinity of SSK2 confers enhanced tolerance to furfural in Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2012;95:531–540. doi: 10.1007/s00253-012-4022-3. [DOI] [PubMed] [Google Scholar]
- 36.Gilbert LA, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Farzadfard F, Perli SD, Lu TK. Tunable and multifunctional eukaryotic transcription factors based on CRISPR/Cas. ACS Synth. Biol. 2013;2:604–613. doi: 10.1021/sb400081r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chavez A, et al. Highly efficient Cas9-mediated transcriptional programming. Nat. Methods. 2015;12:326–328. doi: 10.1038/nmeth.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ryan OW, et al. Selection of chromosomal DNA libraries using a multiplex CRISPR system. eLife. 2014;3:e03703. doi: 10.7554/eLife.03703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Novy V, Wang R, Westman JO, Franzén CJ, Nidetzky B. Saccharomyces cerevisiae strain comparison in glucose-xylose fermentations on defined substrates and in high-gravity SSCF: Convergence in strain performance despite differences in genetic and evolutionary engineering history. Biotechnol. Biofuels. 2017;10:1–14. doi: 10.1186/s13068-017-0887-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tomás-Pejó E, Bonander N, Olsson L. Industrial yeasts strains for biorefinery solutions: Constructing and selecting efficient barcoded xylose fermenting strains for ethanol. Biofuels Bioprod. Biorefining. 2014;8:626–634. [Google Scholar]
- 42.Ho B, Baryshnikova A, Brown GW. Unification of protein abundance datasets yields a quantitative Saccharomyces cerevisiae proteome. Cell Syst. 2018;6:192–205.e3. doi: 10.1016/j.cels.2017.12.004. [DOI] [PubMed] [Google Scholar]
- 43.Mira NP, Becker JD, Sá-Correia I. Genomic expression program involving—The Haa1p-regulon in Saccharomyces cerevisiae response to acetic acid. OMICS. 2010;14:587–601. doi: 10.1089/omi.2010.0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rego A, et al. Modulation of mitochondrial outer membrane permeabilization and apoptosis by ceramide metabolism. PLoS ONE. 2012;7:e48571. doi: 10.1371/journal.pone.0048571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shen Y, et al. High vanillin tolerance of an evolved Saccharomyces cerevisiae strain owing to its enhanced vanillin reduction and antioxidative capacity. J. Ind. Microbiol. Biotechnol. 2020;41:1637–1645. doi: 10.1007/s10295-014-1515-3. [DOI] [PubMed] [Google Scholar]
- 46.Rodrigues-Pousada C, Menezes RA, Pimentel C. The Yap family and its role in stress response. Yeast. 2010;27:245–258. doi: 10.1002/yea.1752. [DOI] [PubMed] [Google Scholar]
- 47.Kuroda S, Otaka S, Fujisawa Y. Fermentable and nonfermentable carbon sources sustain constitutive levels of expression of yeast triosephosphate dehydrogenase 3 gene from distinct promoter elements. J. Biol. Chem. 1994;269:6153–6162. [PubMed] [Google Scholar]
- 48.van Dijk M, Erdei B, Galbe M, Nygård Y, Olsson L. Strain-dependent variance in short-term adaptation effects of two xylose-fermenting strains of Saccharomyces cerevisiae. Bioresour. Technol. 2019;292:121922. doi: 10.1016/j.biortech.2019.121922. [DOI] [PubMed] [Google Scholar]
- 49.Deaner M, Holzman A, Alper HS. Modular ligation extension of guide RNA operons (LEGO) for multiplexed dCas9 regulation of metabolic pathways in Saccharomyces cerevisiae. Biotechnol. J. 2018;13:1700582. doi: 10.1002/biot.201700582. [DOI] [PubMed] [Google Scholar]
- 50.Zhang S, Voigt CA. Engineered dCas9 with reduced toxicity in bacteria: Implications for genetic circuit design. Nucleic Acids Res. 2018;46:11115–11125. doi: 10.1093/nar/gky884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McIsaac RS, et al. Fast-acting and nearly gratuitous induction of gene expression and protein depletion in Saccharomyces cerevisiae. Mol. Biol. Cell. 2011;22:4447–4459. doi: 10.1091/mbc.E11-05-0466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Garí E, Piedrafita L, Aldea M, Herrero E. A set of vectors with a tetracycline-regulatable promoter system for modulated gene expression in Saccharomyces cerevisiae. Yeast. 1997;13:837–848. doi: 10.1002/(SICI)1097-0061(199707)13:9<837::AID-YEA145>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 53.Dossani ZY, et al. A combinatorial approach to synthetic transcription factor-promoter combinations for yeast strain engineering. Yeast. 2018;35:273–280. doi: 10.1002/yea.3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Posas F, Saito H. Activation of the yeast SSK2 MAP kinase kinase kinase by the SSK1 two-component response regulator. EMBO J. 1998;17:1385–1394. doi: 10.1093/emboj/17.5.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee ME, DeLoache WC, Cervantes B, Dueber JE. A highly characterized yeast toolkit for modular, multipart assembly. ACS Synth. Biol. 2015;4:975–986. doi: 10.1021/sb500366v. [DOI] [PubMed] [Google Scholar]
- 56.Gietz RD, Schiestl RH. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2007;2:31–34. doi: 10.1038/nprot.2007.13. [DOI] [PubMed] [Google Scholar]
- 57.Guerreiro JF, Mira NP, Santos AXS, Riezman H, Sá-Correia I. Membrane phosphoproteomics of yeast early response to acetic acid: Role of HRK1 kinase and lipid biosynthetic pathways, in particular sphingolipids. Front Microbiol. 2017;8:1302. doi: 10.3389/fmicb.2017.01302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu H, et al. CRISPR-ERA: A comprehensive design tool for CRISPR-mediated gene editing, repression and activation. Bioinformatics. 2015;31:3676–3678. doi: 10.1093/bioinformatics/btv423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fernandez-Ricaud L, Kourtchenko O, Zackrisson M, Warringer J, Blomberg A. PRECOG: A tool for automated extraction and visualization of fitness components in microbial growth phenomics. BMC Bioinform. 2016;17:1–15. doi: 10.1186/s12859-016-1134-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary files. Plasmids generated will be publicly available through Addgene.