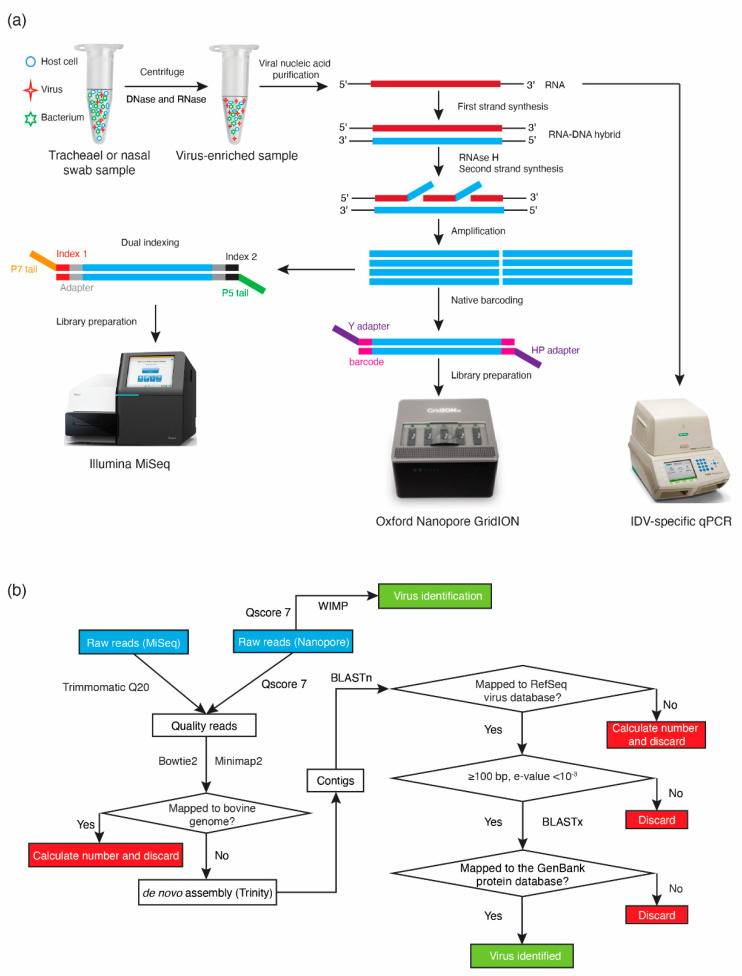
Figure 1.
Workflow for GridION Nanopore sequencing, MiSeq sequencing, and qPCR of bovine respiratory tract samples (BRD). (a) The workflow illustrates both the sample and library preparation. Extracted RNA was used directly for qPCR, while DNA randomly amplified from the same extracts was used for MiSeq sequencing and GridION Nanopore sequencing; (b) Bioinformatic workflow to identify viruses in BRD samples. WIMP (What’s In My Pot) analysis was used only for Nanopore data. The remaining analysis was the same for data from both MiSeq and GridION Nanopore sequencing except Minimap2 was used instead of Bowtie2 for host sequence subtraction.