Abstract
Aflatoxin B1 (AFB1) is a known toxic human carcinogen and can be detoxified by laccases, which are multicopper oxidases that convert several environmental pollutants and toxins. In this study, a new laccase that could catalyze AFB1 degradation was purified and identified from the white-rot fungus Cerrena unicolor 6884. The laccase was purified using (NH4)2SO4 precipitation and anion exchange chromatography, and then identified as Lac 2 through zymogram and UHPLC-MS/MS based on the Illumina transcriptome analysis of C. unicolor 6884. Six putative laccase protein sequences were obtained via functional annotation. The lac 2 cDNA encoding a full-length protein of 512 amino acids was cloned and sequenced to expand the fungus laccase gene library for AFB1 detoxification. AFB1 degradation by Lac 2 was conducted in vitro at pH 7.0 and 45 °C for 24 h. The half-life of AFB1 degradation catalyzed by Lac 2 was 5.16 h. Acetosyringone (AS), Syrinagaldehyde (SA) and [2,2′ -azino-bis-(3-ethylbenzothiazoline-6-sulfonic acid)] (ABTS) at 1 mM concentration seemed to be similar mediators for strongly enhancing AFB1 degradation by Lac 2. The product of AFB1 degradation catalyzed by Lac 2 was traced and identified to be Aflatoxin Q1 (AFQ1) based on mass spectrometry data. These findings are promising for a possible application of Lac 2 as a new aflatoxin oxidase in degrading AFB1 present in food and feeds.
Keywords: laccase, Cerrena unicolor, aflatoxin B1, biodegradation, detoxification, product, transcriptome
1. Introduction
Aflatoxins (AFs), which are commonly produced by Aspergillus fungi such as Aspergillus flavus and Aspergillus parasiticus, are toxoids leading to fluorescence, human carcinogenicity and mutagenicity. The toxicity of AFs is related to the C8-C9 double bond of the difuran ring and the lactone ring within the coumarin ring [1,2]. Based on these data, AFs can be structurally classified into two main groups: the cyclopentenone group (e.g., aflatoxin B1 (AFB1, Figure 1), aflatoxin M1 (AFM1), aflatoxin Q1 (AFQ1), and aflatoxicol), and the cyclo-lactone group (e.g., aflatoxin G1 (AFG1) and its derivatives); AFB1, among the various aflatoxins, is commonly considered to be the most carcinogenic, inhibiting the synthesis of DNA, RNA and proteins [3]. AFB1, a difuranocoumarin derivative, is not only a known human carcinogen, but it can also affect a wide range of commodities [4,5]. AFB1 and its metabolites are mainly distributed in corn, peanuts, rice, wheat, oil by-products, dairy products and condiments [6,7]. It is estimated that over 5 billion people worldwide are at risk of chronic exposure to AFs in food [4].
Figure 1.
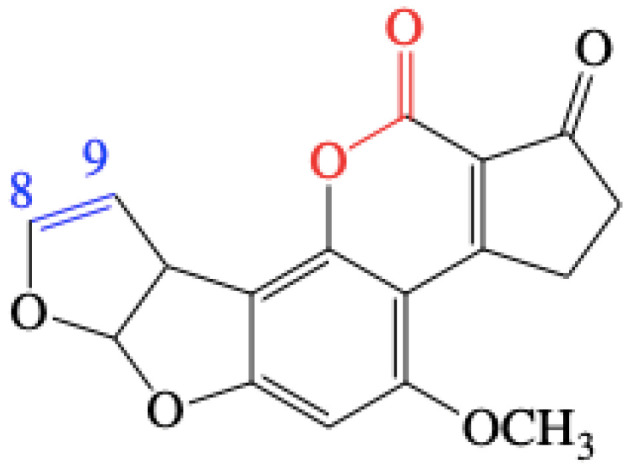
Chemical structure of aflatoxin B1 (AFB1). The C8-C9 double bond is highlighted in blue, while the lactone bond is indicated in red.
Traditional methods of degradation of aflatoxin B1, including physical [8], chemical [9], and microbial approaches [10], suffer from the drawbacks of being time consuming, the loss of nutrients, lowering of quality, increased cost, and formation of toxic residues or derivatives [11,12]. Enzymatic degradation of AFB1 is effective and environmentally friendly, especially in food and feed industries [13]. Enzymes reportedly employed for AFB1 degradation comprise mainly oxidoreductase, including aflatoxin oxidase enzyme (AFO) [14], peroxidase [15], F420H2-dependent reductases (FDR) [16], Mn peroxidase (MnP) [11,17], myxobacteria aflatoxin degrading enzyme (MADE) [18], bacillus aflatoxin-degrading enzyme (BADE) [19] and laccases [20,21,22]. The mechanisms of AFB1 degradation for these enzymes have been elucidated since they have different targets on AFB1 molecules and different active sites: AFO [23,24] and MnP [11] act on the bifuran ring; FDR catalyzes reduction of α, β-unsaturated ester bond [16]; MADE acts on aromatic lactone and the methoxy group of the coumarin ring [18]. However, the mechanism of laccase-catalyzed AFB1 degradation has not been fully unraveled.
Laccases (EC 1.10.3.2) are copper-containing polyphenol oxidases which catalyze the oxidation of an array of aromatic substrates concomitantly with the reduction of molecular oxygen to water [25]. Laccases are produced by fungi for degrading lignin, humic compounds (phenols) and pollutant. However, few reports have focused on their effectiveness on toxins, like AFB1. Alberts et al. first found that the pure and recombinant laccases from Trametes versicolor could directly oxidize AFB1 with significant loss of mutagenicity, and its degradation products could not be detected after treatment using LCMS and HPLC [21]. In their hypothesis, the C8-C9 double bond in the furofuran ring might be responsible for the loss of the fluorescence and mutagenic properties of AFB1 with treatment of fungal laccases. Furthermore, the prooxidant properties and mutagenicity of the detoxification products obtained via laccase from T. versicolor have been studied by Hamed et al. [26]. Moreover, the interaction between AFs and laccases (T. versicolor [27] and Saccharomyces cerevisiae [28]), were studied by using 3D molecular techniques to gain an insight into the detailed mechanism of catalysis and decipher the reasons underlying the formation of degraded products.
Apart from T. versicolor laccases, Lac 2 from Pleurotus pulmonarius and Ery 4 from Pleurotus eryngii were also demonstrated by Loi et al. to degrade AFB1 via the mediation of natural phenolic compounds [22,29]. In their research, a laccase-mediator system (LMS) proved to enhance the degradation properties of laccases towards multiple mycotoxins. However, no degradation product of AFB1 catalyzed via LMS was identified. They hypothesized that the deeper modification of the coumarin-like core of the AFB1 would be responsible for the fluorescence quenching [22], which was different from the findings of Alberts et al. [21]. In addition, Wang et al. investigated whether CotA laccase from Bacillus subtilis (BsCotA) was capable of detoxifying AFB1 and zearalenone (ZEN) using structurally defined chemicals and complex natural mediators, and the degraded products of AFB1 using the LMS were detoxified [20]. In more recent studies, Guo et al. found that recombinant CotA laccase from Bacillus licheniformis transformed AFB1 to AFQ1 and epi-AFQ1, which did not suppress cell viability or induce apoptosis [30]. As summarized above, both molecular docking and product analysis were employed to ascertain the mechanism of AFB1 degradation. Nevertheless, neither the mechanism of fungal laccase-mediated AFB1 degradation, nor the detoxification products, have been fully elucidated. Therefore, the tasks of screening new fungal laccases and possible degradation products still pose great challenges and will open up new perspectives at molecular and product levels to clarify the mechanism.
Cerrena unicolor, known as a white-rot fungus of the Polyporaceae family, was previously reported to be an excellent producer of fungal laccase with an ability to degrade environmental pollutants [31]. Many studies have been undertaken on laccases from C. unicolor, including purification, characterization, cloning, heterologous expression, transcriptomic analysis and application [32,33,34,35,36,37,38,39]. Since laccases from T. versicolor and C. unicolor always share similar enzymatic properties [21], the study of AFB1 degradation by C. unicolor laccase would be a practical project based on those mentioned above. Our hypothesis is that C. unicolor laccase would be a workable and efficient candidate for AFB1 detoxification. In this study, we purified and identified a new laccase from C. unicolor and investigated its capability for AFB1 degradation. Our results showed that laccase from C. unicolor was a valuable enzyme in AFB1 degradation. Moreover, efforts were undertaken to identity the laccase-AFB1 degraded products to hypothesize on the active sites.
2. Results
2.1. Species Identification
A fragment of the Internal Transcribed Spacer (ITS) gene of the target fungal strain 6884 was amplified by PCR and sequenced, and 6884 was classified based on its sequence. A phylogeny tree (Figure S1) was constructed, indicating that 6884 was closest to Cerrena unicolor (GenBank accession No. FN907915.1), Cerrena unicolor strain 042 (EU661887.1), and three more Cerrena unicolor strains (MK937761.1, MH855029.1, MH979248.1), sharing 100% sequence identity. The strain was named Cerrena unicolor 6884.
2.2. Preparation and Purification
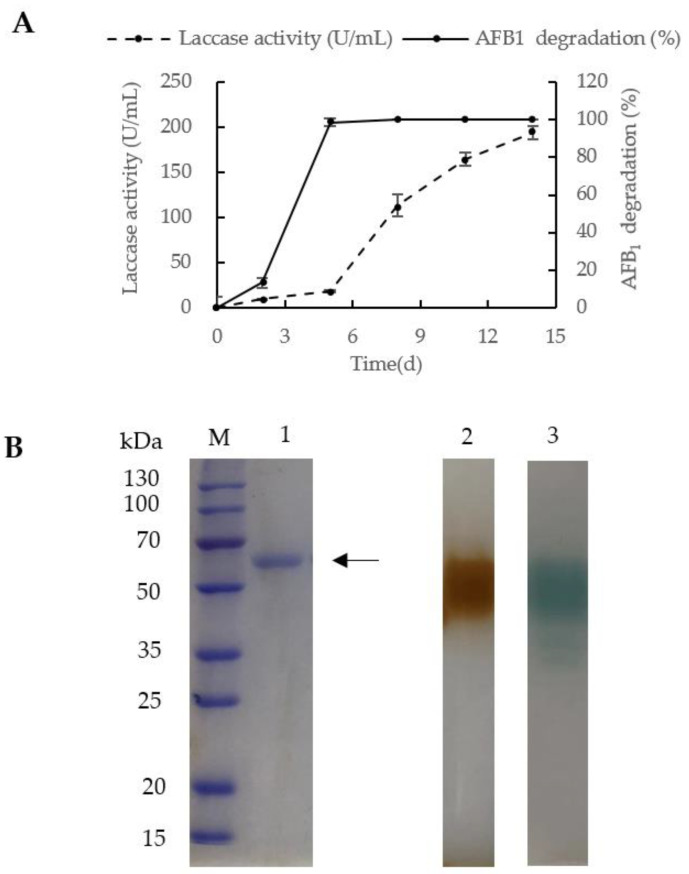
After fermentation of C. unicolor 6884 for 14 days, laccase activity attained a maximum yield of 194 U/mL in the shake flask at 30 °C. On the fifth day of fermentation, C. unicolor 6884 produced laccase with an activity of 18.03 U/mL, with which, AFB1 at the concentration of 5 µg/mL could almost be all degraded (98.56%, Figure 2A). Meanwhile, 13.25% of AFB1 was degraded with laccase of fermentation broth possessing an activity of 9.54 U/mL on the second day. Considering the existence of other oxidases (e.g., peroxidase and MnP, both of which have been shown to degrade AFB1 effectively [11,15,17]) besides laccase in the culture broth, the AFB1 degradation of the culture broth was significantly improved even though the laccase activity was still in its lag phase on the fifth day. Thus, it is difficult to prove a correlation between the laccase activity and AFB1 degradation with the culture broth. However, the purified laccase activity and AFB1 degradation were positively correlated as shown in Figure S2.
Figure 2.
Preparation and zymogram of purified laccase from C. unicolor 6884. (A) Time course of laccase activity and AFB1 degradation mediated by the laccase. (B) Zymographic analysis of the purified laccase. Lane M: protein molecular weight marker by Takara; lane 1: purified laccase in SDS-PAGE; lane 2: purified laccase stained with guaiacol after electrophoresis; lane 3: purified laccase stained with 2,2′ -azino-bis-(3-ethylbenzothiazoline-6-sulfonic acid) (ABTS) after electrophoresis. The samples applied to lanes 2 and 3 had not been heated before loading. The arrow indicates the purified laccase. In A, the values represent means ± standard errors (n = 3).
After 14 days of fermentation, laccase was purified from the fermentation supernatant by employing a protocol involving ammonium sulfate precipitation and ion exchange chromatography (DEAE) (Table 1). The specific activity of laccase was 68.07 U/mg in the fermentation medium. After ion exchange chromatography, the specific activity was increased to 1643.70 U/mg, and the purification fold was 24.15. A purified protein band with a molecular weight of approximately 60 kDa was detected in SDS-PAGE (Figure 2B, lane 1). Laccase activity was shown after native electrophoresis using guaiacol and 2,2′ -azino-bis-(3-ethylbenzothiazoline-6-sulfonic acid) (ABTS) as substrates in lanes 2 and 3, respectively (Figure 2B).
Table 1.
Summary of laccase purification from culture filtrate of Cerrena unicolor 6884.
| Purification Step | Total Activity (U) | Total Protein (mg) | Specific Activity (U/mg) | Purification Fold | Recovery (%) |
|---|---|---|---|---|---|
| Crude extract | 44,641.67 | 655.86 | 68.07 | 1.00 | 100 |
| NH4(SO4)2 | 33,653.33 | 163.49 | 205.84 | 3.02 | 75.39 |
| DEAE cellulose |
6592.00 | 4.01 | 1643.70 | 24.15 | 14.77 |
2.3. Laccase Identification
An mRNA-Seq approach was employed to gain insight into the laccase gene expression profile of C. unicolor 6884 during routine growth on PDA plates. A high-quality database containing a total of 39,419,310 clean reads were obtained with a Q 30 Bases Ratio 95.33%. The total number of reads was assembled to conduct the Transcript (numbers: 56591) and the Unigene (numbers: 13682, redundancy removal) of C. unicolor 6884. The whole sequences in Unigene were annotated against nine databases, including NR (NCBI non-redundant protein sequences, number of genes: 8732, percentage: 63.82%), NT (NCBI nucleotide sequences, 1410, 10.31%), PFAM (Protein family, 4103, 29.99), KOG (euKaryotic Ortholog Groups, 4267, 31.19%), Swiss-Prot (A manually annotated and reviewed protein sequence database, 5500, 40.2%), KEGG (Kyoto Encyclopedia of Genes and Genomes, 1849, 13.51%), CDD (Conserved Domain Database, 5060, 36.98%), TrEMBL (A supplement of Swiss-Prot, 8692, 63.53%) and GO (Gene Ontology, 6846, 50.05%), to find genes encoding putative laccases. There were 23 genes in the Unigene functionally annotated to be putative laccases, and only six different sequences were obtained after blasting and assembling, named lac 1-lac 6 (Table S1).
Based on the Illumina transcriptome analysis of C. unicolor 6884, six laccase cDNAs covering the four copper-binding motifs of fungal laccases (L 1–L 4) (Table S1) were annotated for the function of coding laccase. Information about the laccases is presented in Table S2. A database (DB) containing all six laccase protein sequences was customized for Proteome Discover software (version 2.2, 2018, Thermo Fisher Scientific, Germering, Germany).
Table 2 summarizes the identification of peptides that were digestion products from the single protein band in SDS-PAGE (Figure 2B, lane 1) with the highest score. As a result, 13 unique peptides were identified and assigned to Lac 2 from C. unicolor 6884 with 48% coverage. After purification and identification, laccase from C. unicolor 6884 was designated as Lac 2. The deduced Lac 2 protein with 512 amino acids demonstrated the greatest resemblance (79% identity) to laccase 2 precursor from Cerrena sp. HYB07 (Table S2). The 1536-bp cDNA of lac 2 was cloned and submitted to GenBank with the accession number MT232811.
Table 2.
Analysis of the excised band using UHPLC-MS/MS in combination with Protein Discovery.
| Assigned Protein | Coverage [%] | Peptides | Sequence | Confidence | Theo. MH+ [Da] |
|---|---|---|---|---|---|
| Lac 2 from Cerrena. unicolor 6884 |
48 | 13 | VVELVIPPLAVGGPHPFHLHGHNFWVVR | High | 3123.71556 |
| TVGGPAQSPLNEADLRPLVPAPVPGNAVPGGADINHR | 3653.91467 | ||||
| SQTGPADAELAVISVEHNKR | 2122.08872 | ||||
| SQTGPADAELAVISVEHNK | 1965.98761 | ||||
| SAGSDEYNFDDAILRDVVSIGAGTDEVTIR | 3185.52331 | ||||
| SAGSDEYNFDDAILR | 1672.74492 | ||||
| NASVEEPK | 873.43124 | ||||
| NAAILR | 657.40423 | ||||
| MLTPTSIHWHGFFQK | 1845.91049 | ||||
| YSFVLNANQPDDNYWIR | 2114.99303 | ||||
| MLTPTSIHWHGFFQK | 1829.91557 | ||||
| GAFVVYDPNDPHK | 1458.70120 | ||||
| DVVSIGAGTDEVTIR | 1531.79623 | ||||
| DLYDVDDESTVITLADWYHVLAQTVVGAATPDSTLINGLGR | 4404.18816 |
2.4. AFB1 Degradation by Lac 2 from Cerrena Unicolor 6884
Results of AFB1 degradation mediated by Lac 2 are shown in Figure 3. Fluorescence properties of the AFB1 molecule declined with the prolongation of the duration of incubation with Lac 2 as detected by a fluorescent detector (FLD) on HPLC (Agilent 1260 Infinity Ⅱ Series, Agilent Technologies, Waldbronn, Germany).
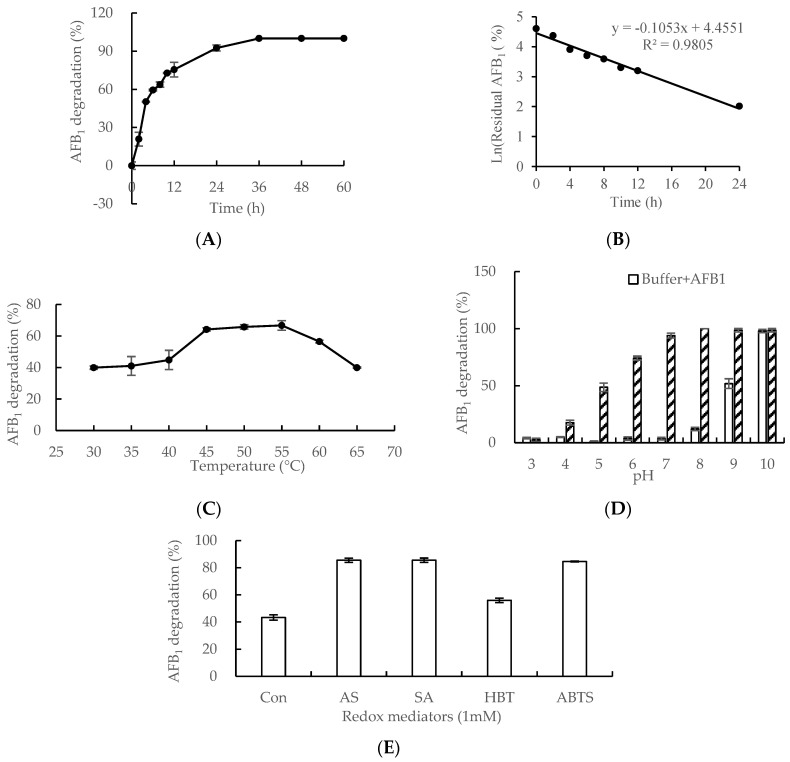
Figure 3.
Cerrena unicolor 6884 Lac 2-mediated aflatoxin B1 (AFB1) degradation. (A) and (B) Time-course analysis of AFB1 degradation mediated by Lac 2. AFB1 (5 µg/mL) was incubated with Lac 2 (50 U/mL) in MilliQ water at 45 °C over a period of 60 h. Samples were periodically taken for HPLC. Effects of temperature (C), pH (D) and redox mediators (E) on AFB1 degradation by Lac 2. AFB1 (5 µg/mL) incubation with Lac 2 (50 U/mL) for 0 h were included as a treatment control. The residual AFB1 of all samples above was determined by a fluorescent detector (FLD) on HPLC (Agilent 1260 Infinity Ⅱ Series, Agilent Technologies, Waldbronn, Germany). The values represent means ± standard errors (n = 4).
The time-course analysis of AFB1 degradation is shown in Figure 3A. The percentage of AFB1 degradation was 50% after an incubation of 4 h, increased steadily to 93% following an incubation of 24 h, and then gradually ascended to 100% at the end of an incubation period of 36 h. The half-life of AFB1 degradation catalyzed by Lac 2 was 5.16 h in this study (Figure 3B).
The optimal temperature for AFB1 degradation was investigated over a period of 8 h (Figure 3C). The percentage of AFB1 degradation was lower than 45% when the reaction temperature was below 45 °C. However, the AFB1 degradation rate increased rapidly from 45% to 65% when the temperature rose from 40 °C to 45 °C. The percentage of AFB1 degradation was maintained at about 65% in the temperature range of 45 °C to 55 °C. When the temperature was elevated to 60 °C and then to 65 °C, the percentage of AFB1 degradation fell to 55% and then to 41%. Therefore, the optimal temperature for AFB1 degradation mediated by laccase from C. unicolor 6884 lay between 45 °C and 55 °C, as shown in Figure 3C.
AFB1 degradation was analyzed over a range pH values (3.0–10.0) by incubation of AFB1 with Lac 2 for 24 h (Figure 3D). When the pH was lower than 7.0 (pH 3.0 to 7.0), AFB1 degradation could not proceed without Lac 2. The percentage of Lac 2-catalyzed AFB1 degradation was 94% when pH was 7.0. However, AFB1 was degradable in an alkaline buffer with a pH between 8.0 and 10.0. Thus, it is deduced that neutral pH was the optimal condition for AFB1 degradation by Lac 2 from C. unicolor 6884 (Figure 3D). The kinetic parameters, Km, Kcat and Vmax, of the Lac 2 towards AFB1 were 14.46 μM, 0.08 s−1and 1.30 μg min−1 mg−1, respectively.
Furthermore, different LMSs were studied for AFB1 degradation by Lac 2 over a period of 3 h (Figure 3E). AFB1 degradation by Lac 2 was almost doubled (85%, 85% and 83% vs. 44%, respectively) in the presence of 1 mM AS, SA and ABTS. Syringyl-type phenols (AS and SA) and ABTS enhanced AFB1 degradation mediated by Lac 2 to similar extents, while hydoxybenzotriazole (HBT) promoted AFB1 degradation mediated by Lac 2 less effectively.
2.5. Analysis of Products of AFB1 Degradation Catalyzed by Lac 2
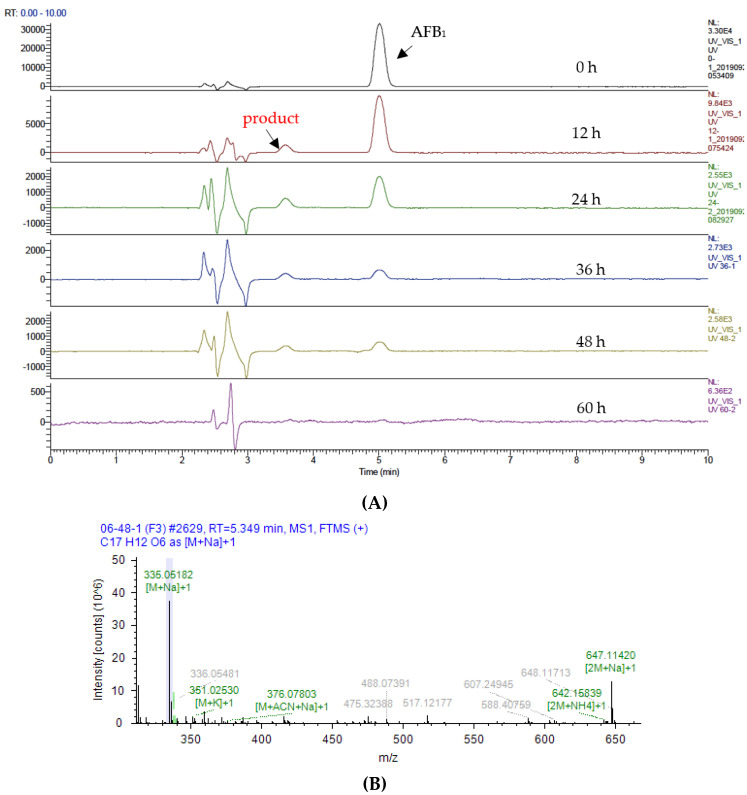
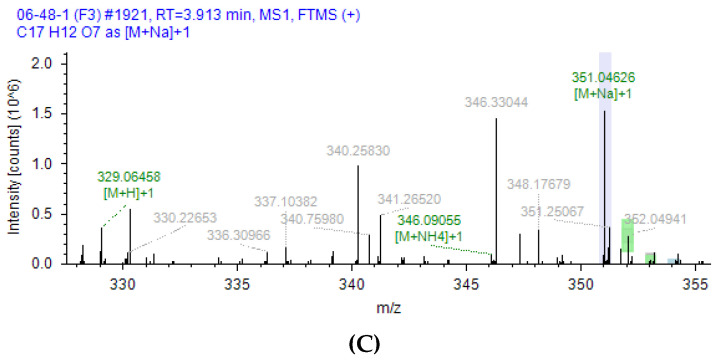
As shown in Figure 4A, the products of AFB1 formed as a result of catalysis by Lac 2 were analyzed by UHPLC (UltiMate 3000 series, Thermo Fisher Scientific, Germering, Germany) with a diode array detector (DAD) and mass detector (Q Exactive mass spectrometer, Thermo Fisher Scientific, Germering, Germany). With the prolongation of incubation time, the percentage of residual AFB1 gradually declined to zero. In the process of AFB1 degradation, a new fraction emerged in the chromatogram at an intermediate stage at about 3.57 min and subsequently vanished. The new fraction in the chromatogram exhibited the molecular formula C17H12O7 ((M + H), and demonstrated + ion peaks at m/z 329.06458, (M + NH4) + ion peaks at m/z 346.09055, and (M + Na) + ion peaks at m/z 351.04626) by UHPLC-MS/MS using Compound Discover software (version 3.0, 2018, Thermo Fisher Scientific, Germering, Germany). The mass spectrum of AFB1 and product are shown in Figure 4B and Figure 4C, respectively.
Figure 4.
Mass spectral analysis of Lac 2-mediated aflatoxin B1 degradation. NSI-MS experiments were performed using a Full MS in the positive ion mode. (A) Lac 2-mediated AFB1 degradation process over a period of 60 h. Samples were periodically taken for UHPLC-MS/MS. AFB1 and the product at the retention time of 5.349 min and 3.913 min, respectively. (B) Mass spectrum of AFB1 (C17H12O6). (C) Mass spectrum of the product from Lac 2-mediated AFB1 degradation (C17H12O7).
3. Discussion
There were six genes encoding a hypothetical laccase in the genome of C. unicolor 6884 based on the results of Illumina transcriptome analysis. After purification and identification, the secreted laccase from C. unicolor 6884 was designated as Lac 2. The open reading frame (ORF) of lac 2 gene encoded a full-length protein of 512 amino acids with an estimated molecular mass of about 60 kDa, which is in the range of molecular masses of most fungal laccases with regular three domains (50–70 kDa) [25]. Reports on laccases responsible for AFB1 degradation include the following: BsCotA from B. subtilis (Accession: AID81987.1) [20], laccase from T. versicolor (Accession: CAA77015.1 [21], PDB code: 1KYA [26,27]); Lac 2 from P. pulmonarius (Accession: AAX40733.1) [22]; and Ery4 from P. eryngii (Accession: CAO79915.1) [29]. Researchers have been trying to investigate the mechanism by employing both molecular techniques and direct analysis of degradation products. A homology model of AFB1 and laccase protein from T. versicolor has been analyzed by Dellafiora et al. [27]. The results showed that homology modeling was an effective analytical tool to assess the laccase–AFB1 interaction for explaining the mechanism of catalysis. It is necessary to acquire more laccase sequences to expand the library when studying the mechanism of laccase-catalyzed AFB1 degradation. In this study, we identified a new laccase from C. unicolor 6884, which demonstrated the ability to degrade AFB1.
After purification by (NH4)2SO4 precipitation and anion exchange chromatography, the specific activity of purified Lac 2 was 1643.7 U/mg with ABTS as substrate, which is similar to those reported from Cerrena sp. WR1 (1013.5 U/mg) [40], C. unicolor BBP6 (1215.9 U/mg) [39] and Cerrena sp. HYB07 (1952.4 U/mg) [31]. Results of SDS-PAGE and zymogram suggested that Lac 2 is a monomeric protein (Figure 2B) resembling most fungal laccases from C. unicolor GSM-019 (63.2 kDa) [41] and Lepista nuda (56 kDa) [42].
The plane-conjugated configuration of furofuran and cumarin rings is responsible for the AF fluorescence [43]. The fluorescence properties of the AFB1 molecule were determined with FLD on HPLC. Some laccases from white rot fungi are capable of degrading AFB1. Only 12.66% of AFB1 remained after catalysis by laccase from T. versicolor at pH 6.5 for 72 h [21]. C. unicolor, a strain of white rot fungi we studied, is a representative wood-degrading basidiomycete used to produce laccase for degradation of environmental pollutants due to its high enzymatic activity toward a broad range of substrates. In vitro, Lac 2 from C. unicolor 6884 catalyzed degradation of 92.5% AFB1 at 45 °C for 24 h, and then nearly 100% degradation after 36 h (Figure 3A).
AFB1 is thermostable for its conjugated structure formed by the C8-C9 double bond, and it would not be dilapidated even at a high temperature of 160 °C [44]. The enzyme-substrate reaction temperatures affect the enzyme activity and molecular motions. In our study, the optimal temperature range was between 45 °C and 55 °C (Figure 3C). The optimal temperature for laccase from T. versicolor was 35 °C [26]. In the case of CotA laccase from B. subtilis, the AFB1 degradation rate rose when temperature was raised from 20 to 50 °C, but the rate was not reduced when the temperature continued to increase to 80 °C [20]. The optimal temperature for laccase-mediated AFB1 degradation depends on the source and species of origin of the laccase.
Exposure of AFB1 to alkaline conditions caused the formation of nonfluorescent derivatives [45]. In an alkaline buffer, the lactone bond in AFB1 will be opened when attacked by nucleophiles, especially OHˉ, and then the detectability of AFB1 toxicity will be shielded, but the nonfluorescent compounds formed are strongly toxic based on results of the chick embryo’s test [12,46]. Our study showed similar results that buffers at pH 8.0, 9.0 and 10.0 were responsible for AFB1 degradation without laccase (Figure 3D). The extent of AFB1 degradation catalyzed by Lac 2 at pH 7.0 would be much greater than that in acidic buffers, which was similar to findings on CotA laccase from B. subtilis [20], but different from T. versicolor laccase with an optimal pH of 4.5. Furthermore, ABTS was studied as the substrate for the stability of Lac 2. After incubating Lac 2 at pH 6.0–9.0 and 25 °C for 24 h or 48 h, over 75% or 65% of the original enzyme activity was retained, respectively (Figure S3), indicating that Lac 2 was relatively more stable at pH of 6.0 or above. Therefore, the neutral condition was suitable for AFB1 degradation catalyzed by Lac 2. Considering the stability of Lac 2 and the better effect of Lac 2 on AFB1 degradation at higher pHs, the application of AFB1 degradation in the intestinal lumen of humans and non-ruminants presents an opportunity to investigate the potential applicability of feed enzymes in animal intestinal tracts. Therefore, in subsequent studies, Lac 2 will be immobilized to improve its resistance for further application.
LMS was efficient in improving degradation, as with purified laccases from the other white rot fungi. In this study, Lac 2 produced by C. unicolor 6884 degraded AFB1 (Figure 3E), which was similar to purified laccase from T. versicolor [21] and Lac 2 from P. pulmonarius [22]. As reported, Ery4-LMSs, 1 mM AS, SA and ABTS were able to double the degradation percentage compared to Lac 2 alone (85%, 85% and 83% vs. 44%, respectively), while HBT could improve degradation slightly. It took 13.86 h for the rate of AFB1 degradation catalyzed by Lac 2 to exceed 80% (Figure 3B), but only 3 h by addition of 1 mM AS, SA or ABTS in the reaction mixture (Figure 3E). Mediators efficiently improved AFB1 degradation. Considering the reaction modes, the AS, SA (natural 2,6-dimethoxy-substituted phenols) and HBT (artificial compounds) share the Hydrogen Atom Transfer (HAT) mechanism, while ABTS (artificial compounds) follows the Electron Transfer (ET) mechanism [47]. Therefore, the potential use of Lac 2 in treatment of raw materials could be advantageous due to its redox mediators and LMS high efficiency.
Few reports are available on the catalytic mechanism deployed by laccase in degradation of AFB1 and the degradation products. Several chemical reactions including epoxidation, hydroxylation, dehydrogenation and reduction, etc., [22], have been speculated as the potential mechanism of AFB1 degradation, but most are merely hypotheses and have not been corroborated. It is necessary to develop methods for AFB1 degradation debris tracking. A recent study on rCotA laccase from B. licheniformis revealed that AFB1 could be oxidized to AFQ1 and epi-AFQ1 [30]. However, studies on products derived from AFB1 formed by the catalytic action of a fungal laccase have not been described. We attempted to investigate the products of AFB1 degradation catalyzed by Lac 2 from C. unicolor 6884 in order to speculate a mechanism of action of fungal laccase.
In our study, a new conspicuous fraction eluted at about 3.57 min in the UHPLC chromatogram (Figure 4A) emerged at an intermediate stage but eventually became undetectable. The same results were observed when AFG1 was degraded with the catalytic assistance of Lac 2 (data not shown). Thus, it can be concluded that the cyclopentenone structure of AFB1 and the cyclo-lactone structure of AFG1 were not the target sites for Lac 2. The new fraction had the molecular formula C17H12O7, which showed the molecular ion peak at m/z 329.06458 ([M + H] +), 346.09055 ([M + NH4] +), and 351.04626 ([M + Na] +). Compared to AFB1 (C17H12O6), this formula shows addition of only one more oxygen, and it is considered to be an AFB1 metabolite. According to the results of UHPLC-MS/MS in Figure 4A and Figure S4, the concentration of AFB1 gradually decreased to zero after incubation for 60 h. In the whole process, the AFB1 metabolite increased in the first 4 h, and then declined as AFB1 appeared. Therefore, part of AFB1 would be converted to the metabolite, which would be slowly degraded, such as that of AFB1 via Lac 2 during the following incubation.
To date, AFB1, AFB2, AFM1, AFG1 and AFG2, can be detoxified with laccases [22,28]. As we know, AFB1 can undergo in vivo hydroxylation to its derivatives, AFM1, AFQ1, or aflatoxicol [1]. In our hypothesis, the molecular formula of C17H12O7 for the AFB1 metabolite is an AFB1-like structure, probably AFM1, AFQ1, epi-AFQ1, AFG1, AFG2 or 8,9-AFB1 epoxide, for these aflatoxins sharing the same m/z and molecular formula. According to Loi et al. [48], comparing the MS/MS spectrum of the unknown AFB1 metabolite to equivalent spectra of all known AFs can give a clue about the identity of this metabolite. AFQ1 is a known AFB1 metabolite; however, it is not commercially available. The MS/MS spectra of AFQ1 was cited as a reference by Loi et al. [48], in which Dr. De Boevre generously provided the MS/MS spectra of AFQ1.
A comparison of the MS/MS spectra of AFB1 metabolite obtained in this study and equivalent spectra of AFQ1 [48] revealed a matching of most diagnostic ions, e.g., m/z 174.99153, 176.98946, 206.05647, 283.05804, 311.05396 and 330.06827 (Figure S5). Based on these findings, the unknown AFB1 metabolite hydroxylated by Lac 2 was extremely likely to be AFQ1.
4. Conclusions
In this study, the next-generation sequencing technology (NGS, Illumina transcriptome analysis) was deployed, and a high-quality Unigene was conducted and annotated against nine databases to a customized database for direct identification of a new laccase and cloning of its cDNA sequence. Six laccase cDNAs covering the four copper-binding motifs of fungal laccases were annotated for the function of coding laccase. A new Lac 2 that catalyzed AFB1 degradation was purified and identified from the white-rot fungus Cerrena unicolor 6884. The cDNA of lac 2 was cloned and submitted to GenBank. The extent of AFB1 degradation achieved at pH 7.0 and 45 °C and an incubation duration of 24 h with laccase as the enzyme was 94%. The stability of Lac 2, and the more extensive degradation of AFB1 at higher pH values suggest the potential applicability of feed enzymes in the animal intestinal tract. The freely additional of redox mediators and LMS high efficiency enhanced the potential applicability of Lac 2 in the treatment of raw materials. Additional studies on the immobilization of Lac 2 and Lac 2-mediated AFB1 degradation in the animal intestinal tract and raw food material processing are required.
The Lac 2-mediated AFB1 degradation product is surmised to be an AFB1-like structure through UHPLC-MS/MS. AFB1 may be converted to AFB1 metabolite partially with the catalytic assistance of Lac 2, and all will then be transformed to degradation products with chemical properties that vastly differ from the AFs. Comparing the MS/MS spectra of AFB1 metabolite with equivalent spectra of AFQ1 led to the conclusion that the AFB1 metabolite was AFQ1. To our knowledge, this identification of the product represents the first characterization of the role of fungal laccase in AFB1 detoxification. The complete unraveling of the mechanism of AFB1 degradation induced by a laccase from white-rot fungus necessitates further investigations. Besides analysis of the degradation products, molecular technology will also be a tool for elucidating the reasons underlying the formation of degraded products.
5. Materials and Methods
5.1. Organism, Chemicals and Other Materials
The Cerrena unicolor 6884 strain was purchased from China Forestry Culture Collection Center (CFCC). pMD-18 T vector Kit was purchased from TAKARA (Dalian, Liaoning, China). E. coli Top10 and RevertAid First Strand cDNA Synthesis Kit was purchased from Thermo Scientific (Vilnius, Lithuania). Total RNA Extractor (Trizol) and BCA Protein Assay Kit were purchased from Sangon (Shanghai, China). Illumina transcriptome analysis and primer synthesis were performed by Sangon (Shanghai, China) and sequencing reactions were performed by Invitrogen (Guangzhou, Guangdong, China).
Chemicals for gel electrophoresis were supplied by Solarbio (Beijing, China) and TaKaRa (Dalian, Liaoning, China). Trypsin (proteomic grade), trizol, and SA (Syrinagaldehyde) were purchased from Thermo Fisher Scientific (Waltham, MA, USA). AFB1 standards (purify ≥ 98%) and ABTS (2-azino-di-[3-ethylbenzo-thiazolin-sulphonate]) were purchased from Sigma-Aldrich (St. Louis, MO, USA). AS (Acetosyringone) was purchased from Solarbio (Beijing, China). HBT (N Hydroxy benzorizole) was purchased from Aladdin (Shanghai, China). Acetonitrile (ACN), Methanol, formic acid (FA), and trifluoroacetic acid (TFA) used in mass spectrometry of HPLC grade were purchased from Merck (Darmstadt, Germany). Regenerable cellulose syringe filters, 0.22 μm (size 13 mm), were obtained from Jinteng (Tianjin, China). Other chemicals used were of analytical grade from Sinopharm (Shanghai, China).
5.2. Phylogenetic Analysis
Phylogeny of the strain was identified by using ITS sequencing. Genomic DNA was extracted with a DNA Quick Plant System (TIANGEN, Beijing, China), and universal primers ITS 1 and ITS 4 were used for amplification of ITS. The PCR product was sequenced. The ITS sequence has been submitted to GenBank with the accession number MT712199. The phylogenetic analysis (with 1000 bootstraps) was performed with MEGA version 7.0 by the neighbor-joining method. Other fungal ITS sequences used in this study were from GenBank.
5.3. Illumina Transcriptome Analysis of Cerrena Unicolor 6884
C. unicolor 6884 was routinely grown on PDA plates at 30 °C for 4 days. The extraction of total RNA and the Illumina transcriptome analysis were both performed by Sangon. An mRNA-Seq approach was employed to gain insight into the laccase gene expression profile of C. unicolor 6884 during routine growth on PDA plates. The total number of reads was assembled with Trinity (Version 2.4.0) to conduct the Transcript and Unigene, and the Unigene of C. unicolor 6884 was submitted to NCBI with the SRA accession PRJNA644218. The whole sequences in Unigene were blasted against nine databases, including NR, NT, PFAM, KOG, Swiss-Prot, KEGG, CDD, TrEMBL and GO, to find genes encoding putative laccases. These putative laccase genes have been submitted to GeneBank with accession numbers MT720691, MT720692, MT720693, MT720694, MT720695 and MT720696, respectively.
5.4. Production of Laccase
The production of laccase was carried out as previously described in [38]. Briefly, C. unicolor 6884 was grown in 50 mL medium (2% v/v glycerol, 1.5% w/v peptone, 6 g KH2PO4, 4.14 g MgSO4·7H2O, 0.3 g CaCl2, 0.18 g NaCl, 0.0625 g CuSO4·5H2O, 0.018 g ZnSO4·7H2O, 0.015 g VB1, 1000 mL H2O) in 250 mL Erlenmeyer flasks at 30 °C with shaking at 200 rpm. Samples were collected at regular time intervals (0, 2, 5, 8, 11 and 14 d, respectively) for enzyme activity assays and AFB1 degradation in vitro described below. The reaction mixture of AFB1 degradation contained cultured broth and AFB1 (5 µg/mL) incubated at 45 °C over a period of 60 h. The samples were removed for HPLC analysis (Agilent 1260 Infinity Ⅱ Series, Agilent Technologies, Waldbronn, Germany) as detailed underneath. The values represent means ± standard errors (n = 3).
5.5. Enzyme Assay
Laccase activity was measured photometrically at 420 nm (Ɛ = 36,000 M−1·cm−1) according to the procedure described in [38]. The 2 mL reaction system contained 100 mM pH 3.0 sodium acetate solution (975 μL), 0.5 mM ABTS (1000 μL) and an appropriate amount of enzyme solution (25 μL). One unit was defined as the amount of enzyme which oxidized 1 μmol of substrate per min. All measurements were carried out in triplicate.
5.6. Purification of Laccase
After cultivation for 14 d, the fermentation culture was harvested, collected, centrifuged and fractionated by addition of 50% to 90% (NH4)2SO4. The precipitate was resuspended and desalted by dialysis against 20 mM Tris-HCl buffer (pH 8.5) and applied to a HiTrap DEAE column (15 cm × 5 cm, 5mL, GE Healthcare Bio-Science Corp, Piscataway, USA) on an ÄKTATM Purifier (GE Healthcare Bio-Science AB, Uppsala, Sweden). Adsorbed proteins were eluted with 0.2 M NaCl in 20 mM Tris-HCl buffer (pH 8.5) and ultra-filtered with a membrane module (nominal MW cut-off 10-kDa) at 4000× g for 20 min. Electrophoretic analyses were performed according to Yang [31] with a little modification. Briefly, fractions with laccase activity were examined by SDS-PAGE on 12% gels stained with Coomassie brilliant blue R-250 (Sigma, St. Louis, MO, USA) and zymography (stained with 0.04 mM guaiacol and 0.25 mM ABTS, respectively). Protein concentration was quantified by BCA Protein Assay Kit.
5.7. Identification of Laccase by UHPLC-MS/MS
The identification of laccase from C. unicolor 6884 by UHPLC-MS/MS was performed by cutting the laccase bands obtained by SDS-PAGE analysis. Protein digestion was accomplished according to the manufacturer’s instructions with slight modifications, with trypsin incubation overnight at 37 °C [22]. Digestion products were subsequently analyzed by UHPLC-MS/MS for peptide sequences on UltiMate 3000 RSLCnano/Q Exactive mass spectrometer (Thermo Fisher Scientific, Germering, Germany). NSI-MS experiments were performed using a Full MS/ddMS2 mass spectrometer system in the positive ion mode. Full MS was in the range 100–5000 m/z. Peptide separation was performed on an Acclaim PepMapTM RSLC-C18 analytical column (75 μm × 15 cm, 2 μm, 100 Å, nanoViper, Thermo Fisher Scientific, USA). The injection volume was 5 μL. The following linear elution gradient was used for the analytical separation: solvent B remained at 2% for 3 min at the beginning of the elution, it was then varied from 2% to 6% in 3 min, from 6% to 28% in 45 min, from 28% to 34% in 4 min; then it was increased up to 95% in 2 min and this ratio was maintained constant for the following 13 min. The percentage of B suddenly decreased at 2% and kept stable for 10 min for column reconditioning. The two reserves used were: A = 2% ACN + 98% H2O + 0.04% FA and B = 80% ACN + 20% H2O + 0.08% FA; flow rate was set at 300 μL/min. The data were acquired and analyzed by Proteome Discover (version 2.2, 2017, Thermo Fisher Scientific, Germering, Germany).
5.8. Cloning of the Laccase cDNA from Cerrena Unicolor 6884
The strain C. unicolor 6884 was grown on PDA plates at 30 °C for 4 d. Total RNA was extracted with Total RNA Extractor (Trizol). The cDNA of C. unicolor 6884 was generated by RT-PCR from total RNA with RevertAid First Strand cDNA Synthesis Kit. Lac 2 was amplified by PCR with the primer pair (lac 2-F: 5′-ATGGGATTGAACTCGGCT-3′; lac 2-R: 5′-TTAAATAGCAGTTCCTTTCTTAGGC-3′) using total cDNA of C. unicolor 6884 as the template. PCR products were inserted into the pMD18-T vector and transformed into E. coli top10 competent cells which were then cultured on an LB plate with 100 mg/mL ampicillin. Three positive clones of each fragment were sequenced. The cDNA sequence of lac 2 studied in this paper has been submitted to GenBank with the accession number MT232811.
The sequences of C. unicolor 6884 laccases were analyzed online BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi). SignalP-5.0 was used for theoretical signal peptide determination (http://www.cbs.dtu.dk/services/SignalP/).
5.9. In Vitro Degradation of AFB1 with Lac 2
For AFB1 degradation by Lac 2, a time-course analysis was carried out in the dark. The 200 μL reaction mixture contained 30 μg/mL Lac 2 (50 U/mL) and AFB1 (5 µg/mL) in MilliQ water at 45 °C over a period of 60 h. The samples were periodically (at 2 h, 4 h, 6 h, 8 h, 10 h, 12 h, 24 h, 36 h, 48 h, and 60 h) taken for HPLC analysis.
To investigate the effect of temperature on Lac 2-mediated AFB1 degradation, AFB1 (5 µg/mL) was individually incubated with Lac 2 (50 U/mL) in Na2HPO4-citric acid buffer (pH 7.0) at various temperatures ranging from 30 to 65 °C (30, 35, 40, 45, 50, 55, 60, and 65 °C) over a period of 8 h.
To investigate the effect of pH on AFB1 degradation by Lac 2, AFB1 (5 µg/mL) was individually incubated with or without Lac 2 (50 U/mL) for 24 h at 45 °C in buffers at different pH values, including Na2HPO4–citric acid buffers (pH 3.0–8.0), and 50 mM glycine-NaOH buffers (pH 9.0–10.0), respectively.
To determine the kinetic parameters (Km, Kcat and Vmax) for AFB1 degradation, the initial reaction rate was investigated by monitoring the removal of AFB1 (initial concentration from 1, 2, 3, 4, 5, 10, 20, and 25 μg/mL) at 10 min intervals up to 50 min at 45 °C and pH 7.0 with 30 μg/mL of Lac 2 (50 U/mL). The kinetic parameters were determined by nonlinear regression of Michaelis- Menten plots using the software GraphPad Prism 8.2.1 (San Diego, CA, USA). One unit of AFB1-degratation activity was defined as the quantity of laccase that detoxified 1 μg of AFB1 per minute.
To investigate the effect of LMS on AFB1 degradation, ABTS, AS, SA or HBT was independently tested as redox mediators at 1 mM with Lac 2 (50 U/mL) in Na2HPO4–citric acid buffer (pH 7.0) at 45 °C for 3 h, while controls did not have any mediator.
Incubation of AFB1 (5 µg/mL) with Lac 2 (50 U/mL) for 0 h was included as a treatment control. The residual AFB1 in all aforementioned samples was determined by FLD on HPLC (Agilent 1260 Infinity Ⅱ Series, Agilent Technologies, Waldbronn, Germany). The values represent means ± standard errors (n = 4).
5.10. AFB1 Assay
AFB1 was extracted from the samples with an equal volume of dichloromethane three times and evaporated under nitrogen gas at 50 °C. The samples were dissolved in an equal volume of methanol and filtered (0.22 μm, Jinteng, China).
HPLC analyses of the samples were performed on an Agilent 1260 Infinity Ⅱ Series (Agilent Technologies, Waldbronn, Germany) using a Poroshell 120 EC-C18 column (2.1×50 mm, 1.9 μm, Agilent Technologies, Waldbronn, Germany). The mobile phase for elution was composed of methanol, aceto-nitrile and water (1:1:2, v/v/v) at a flow rate of 0.1 mL/min for 5 min at 40 °C. The sample injection volume was 2 μL. AFB1 was measured by a FLD Detector which was set at the wavelengths of 365 nm (excitation) and 450 nm (emission). The retention time of AFB1 was 2.5 min.
The percentage of AFB1 degradation was calculated according to the following formula:
| D (%) = (A0 − A)/A0 × 100, | (1) |
where D was the degradation efficiency (%), and A0 and A represented the peak area of AFB1 before and after degradation, respectively.
5.11. UPLC-MS/MS Analysis of AFB1 Degradation Products
The Lac 2-mediated AFB1 degradation products were detected by UHPLC-MS/MS. The degradation reaction was carried out in the dark at 45 °C for 0 h, 2 h, 4 h, 6 h, 8 h, 10 h, 12 h, 24 h, 48 h, and 60 h. The Lac 2-mediated AFB1 degradation products were analyzed by UHPLC (Thermo Scientific UltiMate 3000 System, Thermo Fisher Scientific, Germering, Germany) coupled with a Q Exactive mass spectrometer (Thermo Fisher Scientific, Germering, Germany). NSI-MS experiments were performed in the positive ionization mode. The mobile phase, which consisted of water, methanol and acetonitrile (1:1:2, v/v/v), was used for elution at a flow rate of 0.1 mL/min for 10 min at 40 °C. An Hypersil Gold C18 column (100 × 2.1 mm, 1.9 μm, Thermo Fisher Scientific, Germering, Germany) was used for separation at a flow rate of 0.1 mL/min for 10 min at 40 °C. The sample injection volume was 2 μL. AFB1 was measured by a DAD Detector at 365 nm. The retention time of AFB1 was 5.0 min. The AFB1 degradation products were identified and analyzed with the Compound Discover (version 3.0, 2018, Thermo Fisher Scientific, Germering, Germany).
Abbreviations
| ABTS | [2,2′ -azino-bis-(3-ethylbenzothiazoline-6-sulfonic acid)] |
| ACN | Acetonitrile |
| AFB1 | aflatoxin B1 |
| AFG1 | aflatoxin G1 |
| AFM1 | aflatoxin M1 |
| AFQ1 | aflatoxin Q1 |
| AS | Acetosyringone |
| DAD | Diode array detector |
| DEAE | diethylamino ethyl |
| FLD | Fluorescent detector |
| HBT | Hydroxy benzotriazole |
| Lac 2 | laccase 2 |
| LMS | laccase mediator system |
| SA | Syrinagaldehyde |
Supplementary Materials
The following are available online at https://www.mdpi.com/2072-6651/12/8/476/s1, Table S1. Amino acid sequence alignment of the four conserved copper-binding sites of laccases from Cerrena unicolor 6884, Table S2. Information about laccases from Cerrena unicolor 6884, Figure S1. Phylogenetic relationships of Cerrena unicolor 6884 and related species based on ITS sequences. The numbers following the strains are accession numbers of ITS sequences. Bootstrap values at nodes are percentages of 1000 replicates. Scale bar indicates base substitutions/100 bases, Figure S2. The correclation between Lac 2 from Cerrena unicolor 6884 and aflatoxin B1 (AFB1) degradation. AFB1 (5 µg/mL) was individually incubated with Lac 2 ranging from 0 U/mL to 50 U/mL in MilliQ water at 45 °C over a period of 60 h. Incubation for 0 h were included as treatment controls. The residual AFB1 of all samples above was determined by FLD on HPLC (Agilent 1260 Infinity Ⅱ Series, Agilent Technologies, Waldbronn, Germany). The values represent means ± standard errors (n = 4), Figure S3. Effects of pH on stability of Lac 2. pH stability was studied by incubating the purified enzyme at different pH values in 20 mM Na2HPO4–citric acid buffers (pH 2.0–7.0), or 20 mM Na2HPO4–NaOH buffers (pH 9.0–10.0) at 25 °C for 24 h or 48 h, and the residual laccase activity was quantified at the optimum pH and temperature using ABTS as the substrate. The activity of the untreated enzyme was taken as 100%. The values represent means ± standard errors (n = 3), Figure S4. Biological degradation of aflatoxin B1 (AFB1) by laccase Lac 2 (50U/mL) produced by Cerrena unicolor at pH 7.0 and at 45 °C over a period of 0, 2, 4, 6, 8, 10, 12, 24, 36, 48, and 60 h. The residual AFB1 and intermediate product were determined by UHPLC (Thermo Scientific UltiMate 3000 System, Thermo Fisher Scientific, Germering, Germany). The values represent means ± standard errors (n = 3), Figure S5. MS/MS spectra of unknown AFB1 metabolite.
Author Contributions
Data curation, Z.Z.; Formal analysis, Z.Z. and R.L.; Funding acquisition, J.Y. and X.Y.; Investigation, Z.Z. and Y.L.; Methodology, R.L. and J.Y.; Project administration, X.Y.; Resources, J.Y.; Supervision, R.L. and X.Y.; Writing–original draft, Z.Z.; Writing–review & editing, R.L., T.B.N. and X.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by National Natural Science Foundation of China, grant number 31671795.
Conflicts of Interest
The authors declare no conflict of interest.
Key Contribution
A product identified to be AFQ1 was found from Lac 2-mediated AFB1 degradation, indicating that AFB1 would be transformed during Lac 2-mediated AFB1 degradation.
References
- 1.Loi M., Fanelli F., Liuzzi V.C., Logrieco A., Mulè G. Mycotoxin Biotransformation by Native and Commercial Enzymes: Present and Future Perspectives. Toxins. 2017;9:111. doi: 10.3390/toxins9040111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schroeder T., Zweifel U., Sagelsdorff P., Friederich U., Luethy J., Schlatter C. Ammoniation of aflatoxin-containing corn: Distribution, in vivo covalent deoxyribonucleic acid binding, and mutagenicity of reaction products. J. Agric. Food Chem. 1985;33:311–316. doi: 10.1021/jf00062a038. [DOI] [Google Scholar]
- 3.Hussein H. Toxicity, metabolism, and impact of mycotoxins on humans and animals. Toxicology. 2001;167:101–134. doi: 10.1016/S0300-483X(01)00471-1. [DOI] [PubMed] [Google Scholar]
- 4.Strosnider H., Azziz-Baumgartner E., Banziger M., Bhat R.V., Breiman R., Brune M.-N., Decock K., Dilley A., Groopman J., Hell K., et al. Workgroup Report: Public Health Strategies for Reducing Aflatoxin Exposure in Developing Countries. Environ. Health Perspect. 2006;114:1898–1903. doi: 10.1289/ehp.9302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mao J., He B., Zhang L., Li P., Zhang Q., Ding X., Zhang W. A Structure Identification and Toxicity Assessment of the Degradation Products of Aflatoxin B1 in Peanut Oil under UV Irradiation. Toxins. 2016;8:332. doi: 10.3390/toxins8110332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abrar M., Anjum F.M., Butt M.S., Pasha I., Randhawa M.A., Saeed F., Waqas K. Aflatoxins: Biosynthesis, Occurrence, Toxicity, and Remedies. Crit. Rev. Food Sci. Nutr. 2013;53:862–874. doi: 10.1080/10408398.2011.563154. [DOI] [PubMed] [Google Scholar]
- 7.Williams J.H., Phillips T.D., Jolly P.E., Stiles J.K., Jolly C.M., Aggarwal D. Human aflatoxicosis in developing countries: A review of toxicology, exposure, potential health consequences, and interventions. Am. J. Clin. Nutr. 2004;80:1106–1122. doi: 10.1093/ajcn/80.5.1106. [DOI] [PubMed] [Google Scholar]
- 8.Luo X., Wang R., Wang L., Li Y., Bian Y., Chen Z. Effect of ozone treatment on aflatoxin B1 and safety evaluation of ozonized corn. Food Control. 2014;37:171–176. doi: 10.1016/j.foodcont.2013.09.043. [DOI] [Google Scholar]
- 9.Ji N., Diao E., Li X., Zhang Z., Dong H. Detoxification and safety evaluation of aflatoxin B1 in peanut oil using alkali refining. J. Sci. Food Agric. 2016;96:4009–4014. doi: 10.1002/jsfa.7592. [DOI] [PubMed] [Google Scholar]
- 10.Wang Y., Zhao C., Zhang D., Zhao M., Zheng D., Lyu Y., Cheng W., Guo P., Cui Z. Effective degradation of aflatoxin B1 using a novel thermophilic microbial consortium TADC7. Bioresour. Technol. 2017;224:166–173. doi: 10.1016/j.biortech.2016.11.033. [DOI] [PubMed] [Google Scholar]
- 11.Wang J., Ogata M., Hirai H., Kawagishi H. Detoxification of aflatoxin B1 by manganese peroxidase from the white-rot fungus Phanerochaete sordida YK-624. FEMS Microbiol. Lett. 2010;314:164–169. doi: 10.1111/j.1574-6968.2010.02158.x. [DOI] [PubMed] [Google Scholar]
- 12.Jard G., Liboz T., Mathieu F., Guyonvarc’H A., Lebrihi A. Review of mycotoxin reduction in food and feed: From prevention in the field to detoxification by adsorption or transformation. Food Addit. Contam. Part A. 2011;28:1590–1609. doi: 10.1080/19440049.2011.595377. [DOI] [PubMed] [Google Scholar]
- 13.Adebo O.A., Njobeh P.B., Gbashi S., Nwinyi O.C., Mavumengwana V. Review on microbial degradation of aflatoxins. Crit. Rev. Food Sci. Nutr. 2015;57:3208–3217. doi: 10.1080/10408398.2015.1106440. [DOI] [PubMed] [Google Scholar]
- 14.Liu D., Yao D., Liang R., Ma L., Cheng W., Gu L. Detoxfication of aflatoxin B1 by Enzymes Isolated from Armillariella tabescens. Food Chem. Toxicol. 1998;36:563–574. doi: 10.1016/S0278-6915(98)00017-9. [DOI] [PubMed] [Google Scholar]
- 15.Das C., Mishra H. In vitro Degradation of Aflatoxin B1 in Groundnut (Arachis hypogea) Meal by Horse Radish Peroxidase. LWT Food Sci. Technol. 2000;33:308–312. doi: 10.1006/fstl.2000.0655. [DOI] [Google Scholar]
- 16.Taylor M.C., Jackson C.J., Tattersall D.B., French N., Peat T.S., Newman J., Briggs L.J., Lapalikar G.V., Campbell P.M., Scott C., et al. Identification and characterization of two families of F420H2-dependent reductases from Mycobacteria that catalyse aflatoxin degradation. Mol. Microbiol. 2010;78:561–575. doi: 10.1111/j.1365-2958.2010.07356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yehia R.S. Aflatoxin detoxification by manganese peroxidase purified from Pleurotus ostreatus. Braz. J. Microbiol. 2014;45:127–133. doi: 10.1590/S1517-83822014005000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao L.H., Guan S., Gao X., Ma Q.G., Lei Y.P., Bai X.M., Ji C. Preparation, purification and characteristics of an aflatoxin degradation enzyme from Myxococcus fulvus ANSM068. J. Appl. Microbiol. 2010;110:147–155. doi: 10.1111/j.1365-2672.2010.04867.x. [DOI] [PubMed] [Google Scholar]
- 19.Xu L., Eisa Ahmed M.F., Sangare L., Liu Y., Selvaraj J.N., Xing F., Wang Y., Yang H., Liu Y. Novel Aflatoxin-Degrading Enzyme from Bacillus shackletonii L7. Toxins. 2017;9:36. doi: 10.3390/toxins9010036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang X., Bai Y., Huang H., Tu T., Wang Y., Wang Y., Luo H., Yao B., Su X. Degradation of aflatoxin B1 and zearalenone by bacterial and fungal laccases in presence of structurally defined chemicals and complex natural mediators. Toxins. 2019;11:609. doi: 10.3390/toxins11100609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alberts J., Gelderblom W., Botha A., van Zyl W. Degradation of aflatoxin B1 by fungal laccase enzymes. Int. J. Food Microbiol. 2009;135:47–52. doi: 10.1016/j.ijfoodmicro.2009.07.022. [DOI] [PubMed] [Google Scholar]
- 22.Loi M., Fanelli F., Zucca P., Liuzzi V.C., Quintieri L., Cimmarusti M.T., Monaci L., Haidukowski E.M., Logrieco A.F., Sanjust E., et al. Aflatoxin B1 and M1 Degradation by Lac2 from Pleurotus pulmonarius and Redox Mediators. Toxins. 2016;8:245. doi: 10.3390/toxins8090245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cao H., Liu D., Mo X., Xie C., Yao D. A fungal enzyme with the ability of aflatoxin B1 conversion: Purification and ESI-MS/MS identification. Microbiol. Res. 2011;166:475–483. doi: 10.1016/j.micres.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 24.Wu Y.-Z., Lu F.-P., Jiang H.-L., Tan C.-P., Yao D.-S., Xie C.-F., Liu D.-L., Chu M.-Q. The furofuran-ring selectivity, hydrogen peroxide-production and low Km value are the three elements for highly effective detoxification of aflatoxin oxidase. Food Chem. Toxicol. 2015;76:125–131. doi: 10.1016/j.fct.2014.12.004. [DOI] [PubMed] [Google Scholar]
- 25.Giardina P., Faraco V., Pezzella C., Piscitelli A., Vanhulle S., Sannia G. Laccases: A never-ending story. Cell. Mol. Life Sci. 2010;67:369–385. doi: 10.1007/s00018-009-0169-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zeinvand-Lorestani H., Sabzevari O., Setayesh N., Amini M., Nili-Ahmadabadi A., Faramarzi M.A. Comparative study of in vitro prooxidative properties and genotoxicity induced by aflatoxin B1 and its laccase-mediated detoxification products. Chemosphere. 2015;135:1–6. doi: 10.1016/j.chemosphere.2015.03.036. [DOI] [PubMed] [Google Scholar]
- 27.Dellafiora L., Galaverna G., Reverberi M., Dall’Asta C. Degradation of Aflatoxins by Means of Laccases from Trametes versicolor: An in Silico Insight. Toxins. 2017;9:17. doi: 10.3390/toxins9010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Y., Mao H., Hu C., Tron T., Lin J., Wang J., Sun B. Molecular docking studies and in vitro degradation of four aflatoxins (AFB1, AFB2, AFG1, and AFG2) by a recombinant laccase from Saccharomyces cerevisiae. J. Food Sci. 2020;85:1353–1360. doi: 10.1111/1750-3841.15106. [DOI] [PubMed] [Google Scholar]
- 29.Loi M., Fanelli F., Cimmarusti M.T., Mirabelli V., Haidukowski E.M., Logrieco A.F., Caliandro R., Mulè G. In vitro single and combined mycotoxins degradation by Ery4 laccase from Pleurotus eryngii and redox mediators. Food Control. 2018;90:401–406. doi: 10.1016/j.foodcont.2018.02.032. [DOI] [Google Scholar]
- 30.Guo Y., Qin X., Tang Y., Ma Q., Zhang J., Zhao L. CotA laccase, a novel aflatoxin oxidase from Bacillus licheniformis, transforms aflatoxin B1 to aflatoxin Q1 and epi-aflatoxin Q1. Food Chem. 2020;325:126877. doi: 10.1016/j.foodchem.2020.126877. [DOI] [PubMed] [Google Scholar]
- 31.Yang J., Lin Q., Ng T.B., Ye X., Lin J. Purification and Characterization of a Novel Laccase from Cerrena sp. HYB07 with Dye Decolorizing Ability. PLoS ONE. 2014;9:e110834. doi: 10.1371/journal.pone.0110834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rogalski J., Janusz G. Purification of Extracellular Laccase from Cerrena unicolor. Prep. Biochem. Biotechnol. 2010;40:242–255. doi: 10.1080/10826068.2010.488967. [DOI] [PubMed] [Google Scholar]
- 33.Bekker E.G., Petrova S.D., Ermolova O.V., Elisashvili V.I., Sinitsyn A.P. Extraction, purification and some properties of laccase from Cerrena unicolor. Biokhimiya. 1990;55:2019–2024. [Google Scholar]
- 34.Michniewicz A., Ullrich R., Ledakowicz S., Hofrichter M. The white-rot fungus Cerrena unicolor strain 137 produces two laccase isoforms with different physico-chemical and catalytic properties. Appl. Microbiol. Biotechnol. 2006;69:682–688. doi: 10.1007/s00253-005-0015-9. [DOI] [PubMed] [Google Scholar]
- 35.Janusz G., Mazur A., Checinska Sielaff A., Małek W., Rogalski J., Ohga S. Cloning and characterization of a laccase gene from biotechnologically important basidiomycete Cerrena Unicolor. J. Fac. Agric. Kyushu Univ. 2012;57:41–49. [Google Scholar]
- 36.Kachlishvili E., Metreveli E., Elisashvili V. Modulation of Cerrena unicolor laccase and manganese peroxidase production. SpringerPlus. 2014;3:1–7. doi: 10.1186/2193-1801-3-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pawlik A., Mazur A., Wielbo J., Koper P., Żebracki K., Kubik-Komar A., Janusz G. RNA Sequencing Reveals Differential Gene Expression of Cerrena Unicolor in Response to Variable Lighting Conditions. Int. J. Mol. Sci. 2019;20:290. doi: 10.3390/ijms20020290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang J., Ng T.B., Lin J., Ye X. A novel laccase from basidiomycete Cerrena sp.: Cloning, heterologous expression, and characterization. Int. J. Biol. Macromol. 2015;77:344–349. doi: 10.1016/j.ijbiomac.2015.03.028. [DOI] [PubMed] [Google Scholar]
- 39.Zhang J., Sun L., Zhang H., Wang S., Zhang X., Geng A. A novel homodimer laccase from Cerrena unicolor BBP6: Purification, characterization, and potential in dye decolorization and denim bleaching. PLoS ONE. 2018;13:e0202440. doi: 10.1371/journal.pone.0202440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen S.-C., Wu P.-H., Su Y.-C., Wen T.-N., Wei Y.-S., Wang N.-C., Hsu C.-A., Wang A.H.-J., Shyur L.-F. Biochemical characterization of a novel laccase from the basidiomycete fungus Cerrena sp. WR1. Protein Eng. Des. Sel. 2012;25:761–769. doi: 10.1093/protein/gzs082. [DOI] [PubMed] [Google Scholar]
- 41.Wang S.-S., Ning Y.-J., Wang S.-N., Zhang J., Zhang X., Chen Q.-J. Purification, characterization, and cloning of an extracellular laccase with potent dye decolorizing ability from white rot fungus Cerrena unicolor GSM-01. Int. J. Biol. Macromol. 2017;95:920–927. doi: 10.1016/j.ijbiomac.2016.10.079. [DOI] [PubMed] [Google Scholar]
- 42.Zhu M., Zhang G., Meng L., Wang H., Gao K., Ng T.B. Purification and Characterization of a White Laccase with Pronounced Dye Decolorizing Ability and HIV-1 Reverse Transcriptase Inhibitory Activity from Lepista nuda. Molecules. 2016;21:415. doi: 10.3390/molecules21040415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nicolás-Vázquez M.I., Méndez-Albores A., Moreno-Martínez E., Miranda R., Castro M. Role of Lactone Ring in Structural, Electronic, and Reactivity Properties of Aflatoxin B1: A Theoretical Study. Arch. Environ. Contam. Toxicol. 2010;59:393–406. doi: 10.1007/s00244-010-9501-x. [DOI] [PubMed] [Google Scholar]
- 44.Raters M., Matissek R. Thermal stability of aflatoxin B1 and ochratoxin A. Mycotoxin Res. 2008;24:130–134. doi: 10.1007/BF03032339. [DOI] [PubMed] [Google Scholar]
- 45.Doyle M.P., Marth E.H. Aflatoxin is degraded at different temperatures and pH values by mycella of Aspergillus parasiticus. Eur. J. Appl. Microbiol. Biotechnol. 1978;6:95–100. doi: 10.1007/BF00500860. [DOI] [Google Scholar]
- 46.Kiermeier F., Ruffer L. Veränderung von Aflatoxin B1 in Alkalischer Lösung. Z. Lebensm. Unters. Forch. 1974;155:129–141. doi: 10.1007/BF01141061. [DOI] [Google Scholar]
- 47.Baiocco P., Barreca A.M., Fabbrini M., Galli C., Gentili P. Promoting laccase activity towards non-phenolic substrates: A mechanistic investigation with some laccase-mediator systems. Org. Biomol. Chem. 2003;1:191–197. doi: 10.1039/B208951C. [DOI] [PubMed] [Google Scholar]
- 48.Loi M., Renaud J.B., Rosini E., Pollegioni L., Vignali E., Haidukowski M., Sumarah M.W., Logrieco A.F., Mulè G. Enzymatic transformation of aflatoxin B1 by Rh_DypB peroxidase and characterization of the reaction products. Chemosphere. 2020;250:126296. doi: 10.1016/j.chemosphere.2020.126296. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.