Abstract
The HIV-1 structural polyprotein Gag drives the virus particle assembly specifically at the plasma membrane (PM). During this process, the nascent virion incorporates specific subsets of cellular lipids and host membrane proteins, in addition to viral glycoproteins and viral genomic RNA. Gag binding to the PM is regulated by cellular factors, including PM-specific phospholipid PI(4,5)P2 and tRNAs, both of which bind the highly basic region in the matrix domain of Gag. In this article, we review our current understanding of the roles played by cellular lipids and tRNAs in specific localization of HIV-1 Gag to the PM. Furthermore, we examine the effects of PM-bound Gag on the organization of the PM bilayer and discuss how the reorganization of the PM at the virus assembly site potentially contributes to the enrichment of host transmembrane proteins in the HIV-1 particle. Since some of these host transmembrane proteins alter release, attachment, or infectivity of the nascent virions, the mechanism of Gag targeting to the PM and the nature of virus assembly sites have major implications in virus spread.
Keywords: HIV-1; PI(4,5)P2; cholesterol; RNA; tetherin; CD44; CD43; PSGL-1; assembly; membrane microdomains
1. Introduction
Successful assembly of infectious virus particles is an essential step in virus spread from infected to uninfected cells. For HIV-1, the assembly process takes place at the plasma membrane (PM) in most cells, including the physiological host cell type, CD4+ T cells [1,2]. This process is driven by the viral structural protein Gag, synthesized as a precursor polyprotein Pr55Gag and comprising the following steps: 1) targeting of Gag to the site of virus assembly, 2) binding of Gag to the lipid bilayer, 3) recruitment and packaging of viral genomic RNA, 4) multimerization of Gag, 5) incorporation of viral Env glycoproteins, and 6) budding and release of nascent virus particles (not necessarily in this order for steps 1 to 5).
The HIV-1 Gag matrix (MA) domain mediates the localization and binding of Gag to the PM [3,4]. In CD4+ T cells that have a polarized morphology, Gag localizes to the PM of a rear-end protrusion known as uropod [5,6,7,8]. For Gag localization to this specific region of the PM, NC-dependent Gag multimerization is necessary, in addition to MA-mediated PM binding [5].
The assembling virus incorporates not only viral proteins and RNAs but also host PM components, i.e., lipids and host-encoded membrane proteins present at the virus assembly sites. These host proteins could modulate the production, transmission, and/or entry of progeny virions [9,10,11,12]. Therefore, further understanding of how these components are incorporated into the virus particles can help inform antiviral strategies. Notably, when HIV-1 Gag mislocalizes to intracellular membranes, virus-like particles are formed at these internal membrane sites [1,13,14,15,16,17,18,19]. Thus, subcellular Gag localization plays a major role in determining host membrane components incorporated into virions.
In this review, we will compile and examine our current understanding of the mechanism that directs Gag to the PM and the membrane environment at the site of virus assembly. We will then explore how the virus assembly site at the PM may influence the selection of host proteins into the HIV-1 particle. The major points covered in this review will be:
the interplay between PI(4,5)P2 and tRNA in mediating specific localization of Gag to the PM,
the effects of PM lipids on HIV-1 assembly, and
the potential roles played by the membrane environment in the incorporation of a subset of transmembrane proteins, which localize to uropods in T cells.
2. The Roles Played by PI(4,5)P2 in Specific Localization of Gag
Specific localization of Gag to the PM is mediated by the MA domain [1,2,4]. MA employs a bipartite signal to stably interact with lipid bilayers; an N-terminal myristate moiety and a highly basic region within its globular domain (MA-HBR) [13,15,18,20,21,22,23]. The myristate moiety is a 14-carbon fatty acid co-translationally attached to the N-terminal glycine of MA [24]. When the myristate moiety is not sequestered within the MA globular head, it mediates nonspecific hydrophobic interactions of Gag with membranes [25,26,27,28,29,30,31,32]. By itself, the myristate moiety is not sufficient to stably anchor Gag to the lipid bilayer [24]. The second determinant for the membrane binding of HIV-1 Gag is the MA-HBR (Figure 1a), a conserved patch of basic amino acids encompassing MA residues 14–31, which interacts with negatively charged lipids [23,33,34]. Earlier in vitro studies showed that HIV-1 MA can bind lipid membranes containing acidic phospholipids such as phosphatidylserine (PS) [23,35,36]. However, as described below, multiple lines of evidence indicate that MA-HBR preferentially binds to an anionic lipid that is enriched at the PM, phosphatidylinositol-(4,5)-bisphosphate [PI(4,5)P2], and that this interaction promotes specific binding of Gag to the PM [13,30,37,38,39,40,41,42,43,44,45,46,47].
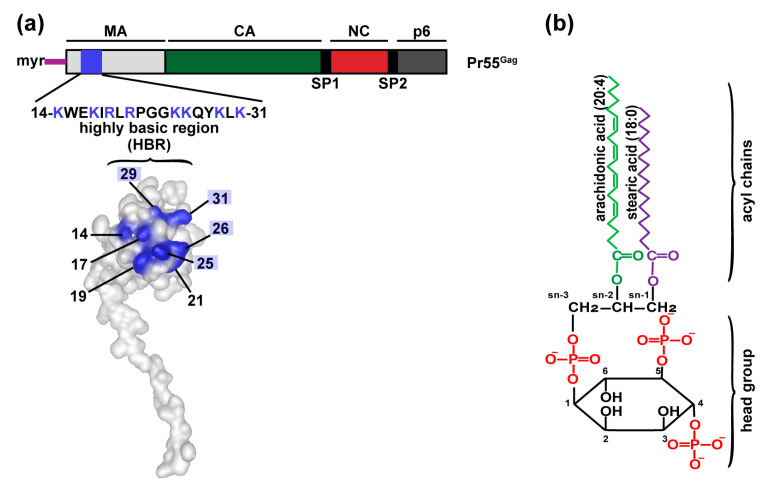
Figure 1.
Cartoons depicting HIV-1 Gag MA-HBR, and PI(4,5)P2. (a) Schematic illustration of HIV-1 Gag with the structure of HIV-1 MA (PDB accession number 2HMX) showing the basic residues of the MA-HBR in blue. The key basic residues studied in detail in our recent work [48] are indicated (in the nomenclature for MA amino acid residues used in this review, the first methionine removed upon myristylation is not counted). (b) Illustration of 38:4 PI(4,5)P2, the predominant molecular species of PI(4,5)P2 in many primary mammalian cells [49,50,51,52,53].
PI(4,5)P2 is a polyphosphoinositide bearing a glycerol backbone, with two acyl chains esterified at the sn−1 and sn−2 positions and a phosphate and myo-inositol ring the sn−3 position (Figure 1b). The myo-inositol ring in PI(4,5)P2 is phosphorylated at the fourth and the fifth positions. PI(4,5)P2 is primarily present in the inner leaflet of the PM, with minor populations distributed at other subcellular compartments [53,54,55]. Phosphatidylinositol phosphate (PIP) kinases (PIPKs), which generate PI(4,5)P2 at specific membranes, can be divided into two groups: type I (PIPKIs) and type II (PIPKIIs). Type I phosphatidylinositol 4-phosphate 5-kinases (PI4P5KI, one of PIPKIs), which preferentially phosphorylate the 5-hydroxyl group on the inositol ring of PI(4)P to produce PI(4,5)P2, are the dominant PIPKs in mammalian cells. A relatively lesser pool of PI(4,5)P2 is produced by PIPKIIs, which use PI(5)P as substrate and phosphorylate PI(5)P at the 4-hydroxyl position. Various polyphosphoinositide kinases and phosphatases act in concert to cycle PIs between different phosphorylation states. PI(4,5)P2 can be dephosphorylated by a subset of polyphosphoinositide phosphatases. The polyphosphoinositide 5-phosphatase INPP5E, also known as 5-phosphatase IV, converts PI(4,5)P2 to PI(4)P by dephosphorylating at the 5-hydroxyl position of the inositol ring [56]. Thus, spatio-temporal distribution of PI(4,5)P2 is orchestrated by the presence of PIPKs, PI(4,5)P2 phosphatases, the enzyme substrates [PI(4)P and PI(5)P], as well as PI(4,5)P2 binding/effector proteins [49,54,55,57,58,59,60,61].
When PI(4,5)P2 is depleted from the PM by the overexpression of 5-phosphatase IV or when the localization of PI(4,5)P2 is altered by inducing the formation of PI(4,5)P2-laden endosomal vesicles, the PM localization and subsequent virus-like particle (VLP) release of HIV-1 Gag is severely decreased [13,38,39,43,45,48,62,63,64], emphasizing the importance of PI(4,5)P2 in HIV-1 assembly and release. In PI(4,5)P2-depleted cells, Gag localizes promiscuously to the cytoplasmic membranes or fails to bind to any membrane and remains cytosolic. Through these studies, PI(4,5)P2 was shown to be an important cellular cofactor for HIV-1 assembly and subcellular Gag localization in multiple cell types [39,64], including the physiological host T cells [62]. A study using an inducible PI(4,5)P2 depletion system, which is based on an engineered 5-phosphatase IV, showed that even Gag multimers detach from the PM upon PI(4,5)P2 depletion [63]. Other approaches to perturb PIPs have also been used to examine the role of PI(4,5)P2 in HIV-1 assembly. For example, the expression of a constitutively active Arf6, which induces a PI(4,5)P2-enriched endosomal compartment, was observed to relocalize Gag to the induced compartment [13]. Another study showed that the inhibition of Rab27-dependent trafficking of PI4KIIα, an enzyme that catalyzes the production of PM-specific PI4P, reduced PI(4,5)P2 at the PM and suppressed Gag localization to the PM in T cells, highlighting the complexity of the PI cycle regulating HIV-1 assembly [65]. In HeLa-derived cells, knockdown of PI4P5KIα and γ, but not β, impaired the targeting of Pr55Gag to the PM and caused mislocalization of Gag to intracellular compartments [66]. Therefore, it is conceivable that HIV-1 Gag binds to a specific pool of PI(4,5)P2 in cells. Perturbation of cellular PI(4,5)P2 or other phosphoinositides has been shown to reduce particle production of other retroviruses, although some retroviruses are less sensitive to 5-phosphatase IV expression [19,39,43,64,67,68,69,70,71].
3. Determinants for Gag Binding to PI(4,5)P2
The PI(4,5)P2 headgroup is more negatively charged than a ubiquitous acidic phospholipid PS (the net charge of PI(4,5)P2, −3 or −4; PS, −1 [72]). Supporting the charge-based preference of Gag for PI(4,5)P2, a modeling study on MA-membrane interactions predicted that nonspecific electrostatic interactions between MA and PI(4,5)P2 were sufficient to significantly enhance membrane binding [34]. However, in vitro studies showed that HIV-1 Gag or MA binds more efficiently to PI(4,5)P2-containing lipid membranes than to charge-matched liposomes containing PS, suggesting that PI(4,5)P2 enhances Gag-membrane binding by specific interaction beyond mere electrostatic attraction [38,41,44,45,73,74,75,76,77]. Membrane flotation-based studies showed that PI(3,5)P2 and PI(3,4,5)P3 were also able to enhance the binding of Gag to liposomes, although the former is not as efficient [38]. Likewise, an NMR-based analysis showed enhanced binding of MA to liposomes containing PI(3,5)P2 as well, although not as efficiently as PI(4,5)P2 [78]. Therefore, both negative charge density of the inositol headgroup and distribution of the phosphate residues on the inositol ring are likely determinants for the optimal interaction between MA and PI(4,5)P2. Recent genetic and structural studies collectively suggest that HBR residues Lys 29 and Lys 31, especially Lys 31, play key roles in the interaction with the PI(4,5)P2 headgroup, although other basic residues may also be involved [41,48,78,79,80]. In addition, as we discuss later, binding of RNA to MA-HBR residues is also likely to contribute to lipid specificity.
The sn-2 position of the glycerol backbone of PI(4,5)P2 is most commonly decorated with arachidonic acid. The 1-stearoyl-2-arachidonyl (38:4) form of PI(4,5)P2 is found to be a dominant species across multiple primary mammalian cells [49,50,51,52,53] (Figure 1b). The preference of enzymes involved in the PI cycle, such as diacylglycerol kinase ε (DGKε), for substrates displaying 38:4 fatty acyl chains likely explains the enrichment of 38:4 PI(4,5)P2 [51,57]. Although an earlier structural study using water-soluble PI(4,5)P2 derivatives with short acyl chains proposed that the sn-2 acyl chain interacts with a hydrophobic cleft of the MA globular domain [30], subsequent structural and in silico studies suggest that PI(4,5)P2 acyl chains with native lengths are unlikely to directly interact with MA [78,81]. Nonetheless, the acyl chains of PI(4,5)P2 may play an indirect role in MA binding. An in vitro study using giant unilamellar vesicles (GUVs) showed that Gag prefers PI(4,5)P2 with at least one unsaturated acyl chain [82]. In the same experimental system, the pleckstrin homology domain of phospholipase Cδ1 (PHPLCδ1), which interacts with the headgroup of PI(4,5)P2, did not show a bias towards specific PI(4,5)P2 acyl chains unlike Gag [82]. In the presence of cholesterol (discussed later) or a higher concentration of PS, HIV-1 Gag binding to liposomes can be detected even in the absence of PI(4,5)P2. Under these conditions, HIV-1 Gag shows a preference for PS with unsaturated acyl chains over PS with saturated acyl chains [83], whereas RSV Gag, which lacks N-terminal myristoyl moiety, does not show such preference [84,85]. It will be intriguing to examine whether the myristoyl moiety plays a role in the observed acyl chain preference, for example, by favoring a loose packing (warranted by unsaturated acyl chains) around the acidic phospholipid that Gag binds. Lipidomics studies showed that unsaturated acyl chain species, including 38:4, were the major PI(4,5)P2 acyl chain species found in the HIV-1 particle [39,86], but this can be largely accounted for by the abundance in the PM. Currently, whether the unsaturation of acyl chains is necessary for Gag binding to PI(4,5)P2 in cells remain unclear.
4. The Balance Between MA Binding to PI(4,5)P2 and RNA
The MA-HBR has been shown to bind not only acidic lipids but also RNA [48,87,88,89,90,91,92]. Notably, in experimental systems where full-length Gag is translated in vitro using eukaryotic cell lysates and examined for binding to liposomes containing PS, RNA that is present in the cell lysates acts as a negative regulator for Gag membrane binding. Only when RNA is removed by treatment with RNase or when PI(4,5)P2 is included in the liposomes, in vitro transcribed Gag binds liposomes efficiently [38,41,45,82]. Thus, RNA bound to Gag via MA inhibits Gag binding to PS, but PI(4,5)P2 is able to overcome the negative regulation by RNA (Figure 2a).
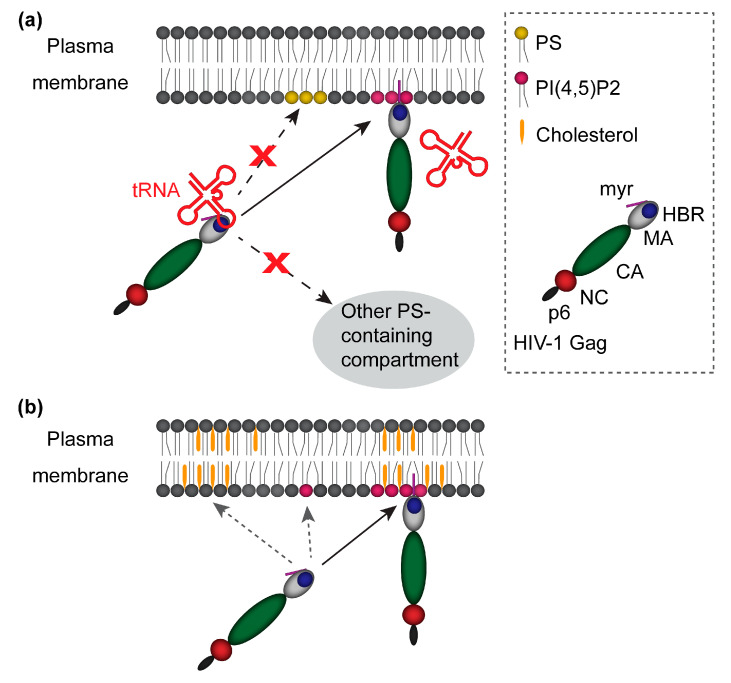
Figure 2.
Regulation of Gag binding to the plasma membrane (PM). (a) Interactions of Gag with PI(4,5)P2 and tRNA ensure specific localization of Gag to the PM. Binding of Gag to tRNA prevents localization of Gag to intracellular membranes that contain PS but not PI(4,5)P2. Interaction of Gag with PI(4,5)P2-containing membrane overcomes the block on membrane binding by tRNA, thereby recruiting Gag to the PM; (b) cholesterol-dependent membrane binding by Gag. Cholesterol increases the membrane binding of Gag, including binding to PI(4,5)P2-containing PM.
The in vitro observations of competition between RNA/nucleotide and acidic lipids [41,46] led to a proposal in which RNA binding to MA prevents MA interaction with PS, an acidic phospholipid ubiquitously distributed in cells, and thereby ensures specific binding of Gag to the PM, which contains PI(4,5)P2 (Figure 2a). In support of this model, Gag binding to RNA via MA in cells has been observed [48,89,93]. Moreover, RNA bound to MA in cells suppresses the membrane binding of Gag molecules present in the cytosol of HIV-expressing cells regardless of whether the membrane is PS-containing liposomes [93] or total cellular membranes [89]. But does RNA binding to MA prevent Gag from mislocalizing to the intracellular membranes containing PS in cells? Our comparison of HIV-1 Gag chimeras containing MA domains of various retroviruses showed that MA domains sensitive to RNA-mediated inhibition of Gag-liposome binding direct Gag chimeras to the PM, whereas Gag chimeras containing MA domains that are insensitive to RNA-mediated inhibition show promiscuous subcellular localization [43,45]. Furthermore, a strong correlation between MA-RNA binding in cells and the prevention of Gag mislocalization has been observed in our analysis of HIV-1 MA mutants [48]. This study compared Gag derivatives in which two of the eight basic amino acids in MA-HBR are substituted from Lys to Arg (KR) (preserving the total positive charge of MA-HBR) or from Lys to Thr (KT) (reducing the total positive charge) (Figure 1a). Consistent with the importance of the HBR charge in RNA binding, the KT mutants failed to bind RNA via MA efficiently in cells, unlike the KR mutants. These KT mutants bound both PM and intracellular membranes indiscriminately. In contrast, one KR mutant bound PM exclusively, consistent with the role for total MA-HBR positive charge and/or MA-bound RNA in navigating Gag specifically to the PM. Another KR mutant was predominantly cytosolic, despite retaining the total positive charge; however, when its membrane binding was enhanced through augmented Gag multimerization, it achieved specific binding to the PM without localization to other compartments. Altogether, these studies provide cell-based data supporting a model that MA binding by RNA prevents mislocalization of Gag in cells, and along with PI(4,5)P2, helps Gag localize specifically to the PM. Thus, it appears that a fine balance between MA-PI(4,5)P2 and MA-RNA binding exists and dictates the subcellular localization of Gag.
The majority of RNA that binds MA-HBR in cells has been identified as transfer RNA (tRNA) [89]. Interestingly, the affinity of MA binding to tRNA [90,94] surpasses its affinity to PI(4,5)P2 [73,78,79]. The affinity of MA to PI(4,5)P2 can be augmented when PS or cholesterol is included in the PI(4,5)P2-containing membranes [73], when MA is artificially trimerized [79], or when MA is myristoylated [73]. However, there has been no side-by-side comparison of MA binding affinity to membranes versus tRNA using identical experimental conditions and methods thus far. Of note, in the in vitro liposome binding assays in which in vitro transcribed full-length Gag is examined for liposome binding, RNase treatment drastically increases Gag binding to liposomes containing PS, but it also increases Gag binding to PI(4,5)P2-containing liposomes [41]. This observation suggests that Gag binding to PI(4,5)P2 can be susceptible to the RNA-mediated inhibition depending on the condition. Consistent with this notion, while in one in vitro study, tRNA below typical cellular concentrations suppressed binding of Gag to PC + PS but not PI(4,5)P2-containing membranes [93], in other in vitro systems where purified MA or Gag are added to lipid membranes, the addition of tRNAs at similar tRNA concentrations reduced membrane binding of MA/Gag to various degrees even when the membranes contain PI(4,5)P2 [74,76,90]. Therefore, the balance between MA binding to PI(4,5)P2 and tRNA can be affected by factors that differ among experimental systems. These factors may include the state of Gag multimerization, the presence of other lipids, the effects of divalent cations on tRNAs and membranes, and the clustering of PI(4,5)P2 (see below). Apart from the differences among in vitro systems, there are several caveats that exist for in vitro studies. One is that most tRNAs in cells may be in association with the cellular translational machinery and not free, thus the actual concentration of free tRNA in cells is difficult to assess. In addition, tRNAs shuttle between different cellular compartments in response to cellular stress [95,96], and therefore, the concentrations of tRNA in the cytoplasm may change during productive infection. Furthermore, in vitro transcribed tRNAs, which were used in some of the in vitro Gag/MA membrane binding studies, lack posttranscriptional modifications which are important for tRNA folding, structure and stability in the cell [97,98,99,100,101]. Therefore, further validation of the in vitro results regarding MA-tRNA and MA-PI(4,5)P2 associations with cell-based systems is warranted.
MA-RNA binding is mediated by the MA region containing HBR [37,48,87,88,89,90,93,102,103,104,105,106,107,108]. Therefore, it is likely that tRNA binding suppresses Gag membrane binding through inhibition of the interaction between HBR and acidic phospholipids. Interestingly, however, our previous study has shown that removal of RNA from Gag by RNase treatment increases not only Gag binding to negatively charged liposomes but also, to a lesser extent, binding to liposomes containing only a neutral lipid PC [41]. Therefore, hydrophobic interactions mediated by the myristoyl moiety may also be regulated by RNA binding to MA-HBR. A similar observation was also made in another in vitro study that used a purified Gag and yeast tRNA [76]. Of note, myristoylated MA under conditions that favor the myristate exposure has a weakened affinity to tRNA compared to when the myristate moiety is sequestered [90]. Therefore, there may be an interplay between myristoyl exposure and MA-tRNA binding. A potential mechanism may be that tRNA binding to specific MA-HBR residues prevents the exposure of the myristate moiety. However, conditions that trigger the exposure of the myristate moiety, such as Gag multimerization or a chance apposition to lipid bilayers, or the exposed myristate moiety itself may weaken tRNA binding. Obviously, these speculations need to be tested experimentally.
5. Enrichment of PI(4,5)P2 in HIV-1 Particles
The estimated amounts of PI(4,5)P2 vary depending on the type of cells. PI(4,5)P2 resides mainly in the inner leaflet of the PM at ~1–2 mol% with ~5000–20,000 molecules of PI(4,5)P2 per μm2 of PM [57]. PI(4,5)P2 is found to be enriched in the HIV-1 envelope as compared to the PM of the producer cell [39,86,109,110], and this enrichment is dependent on MA [39]. There are an estimated 8000 molecules of PI(4,5)P2 in an HIV-1 particle [86]. Total surface area for one HIV-1 particle will be 0.07 um2 assuming an average diameter of 150 nm for each virus particle. Thus, there is a ~5.6 to 22.4-fold increase in PI(4,5)P2 density on HIV-1 compared to that on the PM. A new nanodisc-based study determined that ~one PI(4,5)P2 molecule recruits one MA molecule (6 molecules of PI(4,5)P2 versus 5 molecules of MA per leaflet of the nanodisc) [79]. However, assuming an average of 2500 Gag molecules per virus particle, the ratio of PI(4,5)P2 to Gag is 3:1 in a virus particle as per the latest HIV-1 lipidomics study [86]. How does HIV-1 achieve such an enrichment of PI(4,5)P2 in its envelope?
The PM is not a homogeneous sea of lipids and proteins. Instead, the PM is a dynamic mosaic of various nanoscale or mesoscale domains (herein collectively referred to as microdomains) that have distinct lipid and protein signatures [111,112,113,114,115,116]. As for PI(4,5)P2, super-resolution microscopy studies in PC12 cells showed that PI(4,5)P2 exists in ~65–73 nm diameter clusters in the inner leaflet of the PM [117,118]. In model membranes, PI(4,5)P2 cluster formation can be driven by multivalent metal ions [119,120]. Metal ions generate PI(4,5)P2 clusters even at low PI(4,5)P2 concentrations of 0.02 mol% [120]. One can speculate that given that the MA-HBR is the established interface for the PI(4,5)P2-Gag association, the increase in PI(4,5)P2:Gag ratio in virus particle compared to nanodisc may be a result of PI(4,5)P2 clustering. In fact, a previous modeling study showed that a single basic peptide could induce the clustering of PI(4,5)P2 [121]. A combination of PI(4,5)P2 clustering and Gag multimerization is likely to lead to the incorporation of larger numbers of PI(4,5)P2 into the virus particle than what initially engaged Gag.
6. Roles Played by Cholesterol and Cholesterol-Rich Membrane Domains in HIV-1 Assembly
Another microdomain of note in HIV-1 assembly is the lipid raft, as reviewed previously [122]. Membrane rafts are highly dynamic, potentially heterogenous nanoscopic domains (10–200 nm) enriched in sterols and sphingolipids [123,124]. Segregation of these raft lipids from surrounding glycerolipid-rich regions is often correlated with the phase separation of lipid bilayers into rigid liquid-ordered (Lo) membrane and the adjacent highly fluid liquid-disordered (Ld) region [125,126]. Proteins modified with saturated acyl chains such as the palmitate moiety commonly partition into the Lo phase, which is thought to represent lipid rafts, while proteins bearing unsaturated acyl chains such as geranylgeranyl moiety do not [127,128,129,130].
Interestingly, depletion of cellular cholesterol, which disrupts lipid rafts, reduces Gag membrane binding in cells [131]. Additionally, in vitro studies have shown that cholesterol strengthens the association of Gag to liposomes in a concentration-dependent manner both in the absence and presence of PI(4,5)P2 [77,83]. Likewise, SPR on sparsely tethered bilayer lipid membranes (stBLMs) demonstrated that the inclusion of cholesterol increases the amount of myristoylated MA bound to stBLM [73]. Furthermore, an NMR-based study found that myristoylated MA exhibits significant affinity to raft-like lipids even in the presence of PI(4,5)P2 [78]. These studies strongly support the role of cholesterol in Gag membrane binding (Figure 2b).
7. Relationships Between PI(4,5)P2 and Cholesterol During HIV-1 Assembly
A substantial number of studies suggest that HIV-1 assembly takes place in the area of the PM enriched in lipid raft microdomains. The HIV-1 envelope is enriched with both cholesterol and sphingomyelin compared to producer cell membranes [39,86,109,110,132]. Consistent with this, cholesterol was apparently enriched at Gag assembly sites on the HeLa cell surface [133]. Moreover, the level of membrane order of viral envelope determined using Laurdan staining was within the range expected for Lo membranes [134]. Raft-preferring proteins are found enriched in the HIV-1 envelope [6,122,135,136,137,138]. Gag is recovered in detergent-resistant membrane (DRM) fractions [6,122,139,140,141,142,143,144,145], which are thought to contain lipid rafts, albeit DRM is unlikely to represent intact raft microdomains in cells [122,123,124]. Gag colocalizes with lipid raft markers in cells using immunofluorescence microscopy [6,122,144]. Depletion of cholesterol and/or sphingolipids in cells adversely affects normal Gag behavior in cells, virus particle production and/or infectivity [109,131,145,146,147]. Overall, the data support the role of cholesterol-enriched raft-like microdomains in HIV-1 particle production. Interestingly, however, in an in vitro study, an MA-based construct that binds PI(4,5)P2-containing GUV membrane upon induced multimerization is excluded from the Lo phase in model membranes. Instead, it colocalizes with Ld markers along with PI(4,5)P2 [75]. Thus, it is unlikely that Gag merely targets or buds through the existing raft-like domains to form virus particles, especially given the role of PI(4,5)P2 in recruiting Gag to the PM.
As discussed earlier, the majority of PI(4,5)P2 molecules in cells have an unsaturated acyl chain at its sn-2 position and are therefore less likely to partition into lipid rafts consisting of saturated lipids. This poses a question as to how HIV-1 particles can be enriched in PI(4,5)P2 and lipid raft components at the same time. It is conceivable that Gag multimerization on the membrane reorganizes the lipid landscape at the assembly sites. Consistent with this possibility, previous microscopy-based studies showed that Gag promotes colocalization of markers of lipid rafts and another distinct microdomain, tetraspanin-enriched microdomains (TEM) [148] at Gag assembly and budding sites [149,150]. Thus, Gag multimerization at the PM could lead to spatiotemporal rearrangement of membrane domains at the assembly sites, thereby allowing the distinctive HIV-1 envelope composition. Indeed, another PI(4,5)P2-binding protein, Annexin A2 (AnxA2), was found to induce the formation of microdomains rich in PI(4,5)P2, cholesterol, glycosphingolipids [151] at least in model membranes, supporting the possibility of protein-induced rearrangement of membrane microdomains.
In this regard, it is noteworthy that cholesterol facilitates the clustering of PI(4,5)P2 in model membranes in the presence [120] or the absence of multivalent cations [152]. Thus, the presence of cholesterol and Gag multimers, which are likely to bind multiple PI(4,5)P2, could prompt the formation and growth of PI(4,5)P2 nanodomains in the PM. In support of this possibility, Gag assembly on model membranes was shown to induce PI(4,5)P2 cluster formation [153]. In complex lipid mixtures, cholesterol but not sphingomyelin appeared to be co-clustered along with PI(4,5)P2 by Gag assembly [153]. Furthermore, HIV-1 Gag was found to restrict the mobility of both PI(4,5)P2 and cholesterol, but not sphingomyelin, in the PM of living CD4+ T lymphocytes by super-resolution microscopy with scanning fluorescence correlation spectroscopy [154]. Whether the microdomain reorganization mediated by Gag multimerization takes the shape of a new microdomain with an amalgamation of the raft- and nonraft-like components or whether there is a juxtaposition of two microdomains at submicroscopic scale will be interesting to determine.
8. Potential Roles Played by Membrane Environment at Virus Assembly Sites in Host Protein Incorporation into Virions
Membrane microdomain reorganization is likely to contribute to the enrichment of specific membrane proteins at the virus assembly sites. Indeed, host proteins that prefer the cholesterol-enriched raft-like microdomain cluster at the HIV-1 assembly sites [133,149] (Figure 3). However, a raft-like environment is not the sole determinant for protein recruitment to the HIV-1 assembly sites. For example, the association of the tetraspanin CD81 and some GPI-anchored proteins with the HIV assembly sites, as determined by copatching and correlative light and electron microscopy-based approaches, diminishes when Gag fails to form budding structures at the PM [149]. Furthermore, we and others have shown using super-resolution localization microscopy or live TIRF imaging that membrane curvature promotes the incorporation of tetherin to the virus assembly sites [133,155] (Figure 3a). Importantly, tetherin-mediated inhibition of virus release, which requires the incorporation of tetherin to nascent particles, is unaffected by cholesterol depletion of virus-producing cells [155]. Therefore, while tetherin possesses a GPI anchor, which is a canonical raft-targeting signal (Figure 3b), cholesterol-enriched microdomains do not appear to play a role in tetherin recruitment to nascent virions.
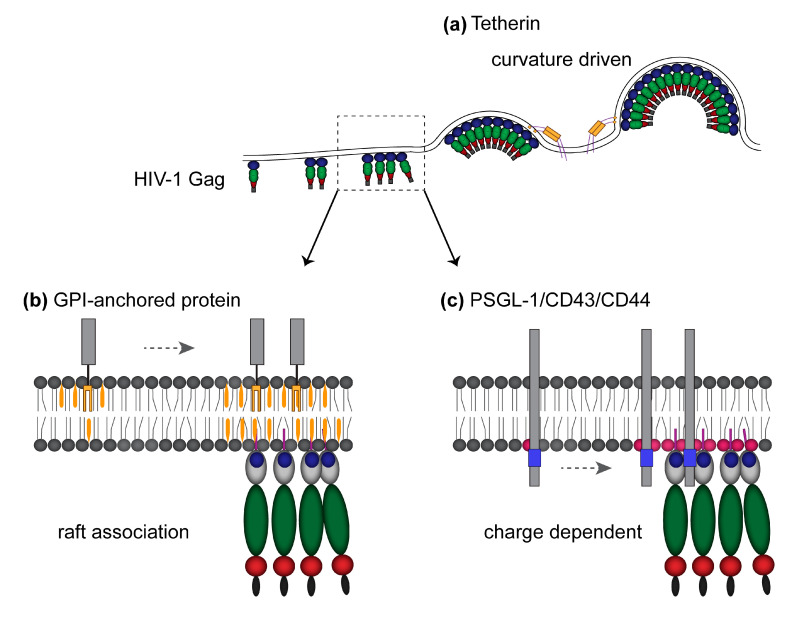
Figure 3.
Mechanisms of host protein enrichment to HIV-1 assembly sites. (a) Membrane curvature promotes the incorporation of tetherin into nascent virions. (b) Preference of cellular proteins for raft-like domains promotes their enrichment at virus assembly sites. A GPI-anchored protein is shown as an example. (c) PSGL-1, CD43, and CD44, which contain a polybasic region in their cytoplasmic tails, cocluster with Gag in an HBR-dependent manner, potentially mediated by a polyacidic entity such as clusters of acidic lipids including PI(4,5)P2.
It is conceivable that PI(4,5)P2, another lipid that is enriched at virus assembly sites, also promotes the incorporation of specific host membrane proteins. We have found that a subset of host transmembrane proteins that localize to a T cell uropod, the rear-end protrusion of polarized immune cells, copatch with each other [5] and are specifically recruited to the HIV-1 assembly sites [156]. Interestingly, coclustering between Gag and these uropod-directed proteins (PSGL-1, CD43, and CD44), determined by super-resolution localization microscopy, is dependent on the MA-HBR [156]. Gag derivatives that are engineered to bind the PM without MA-HBR failed to cocluster with PSGL-1. Moreover, the coclustering with Gag requires juxtamembrane polybasic sequences in the cytoplasmic tails of PSGL-1, CD43, and CD44. These results are consistent with the possibility that Gag multimerization facilitates the formation of microdomains enriched in acidic lipids, in particular PI(4,5)P2, which in turn recruit PSGL-1, CD43, and CD44 via electrostatic interactions (Figure 3c). In support of this possibility, a structural study suggests that the CD44 cytoplasmic tail coclusters with PI(4,5)P2 [157]. However, PSGL-1, CD43, and CD44 are also known to associate with lipid rafts [158]. Further investigation into the effects of acidic lipids and cholesterol-enriched domains on the behaviors of these transmembrane proteins is warranted, especially considering that these molecules alter HIV-1 spread significantly when incorporated into virions (see below).
9. Roles Played by PSGL-1, CD43, and CD44 in Virus Spread
PSGL-1, CD43, and CD44 are type-1 transmembrane proteins that localize to the uropod in polarized T cells [159,160,161,162,163]. While these proteins play a role in T cell migration via interactions with selectin proteins expressed by endothelial cells [164,165], PSGL-1 and CD43 can serve as a physical barrier that inhibits cell-cell interactions because the extracellular domains of PSGL-1 and CD43 are highly glycosylated with sialylated O-linked glycans and thereby, highly extended [164,165,166,167,168,169,170]. CD44 is a major receptor for hyaluronan (HA), a glycosaminoglycan, and the interactions between CD44 and HA promote cell adhesion and cell-cell contacts [171].
It has been long known that CD44 is incorporated into HIV-1 [138,172,173]. Interestingly, virion-incorporated CD44 promotes trans-infection mediated by secondary lymphoid organ stromal cells, known as fibroblastic reticular cells (FRC). HA bound to virion-incorporated CD44 interacts with CD44 on the surface of FRCs and thereby promotes virus binding to FRCs (virus capture) and subsequent trans-infection of CD4+ T cells [174]. Of note, HA present on the surface of CD44-expressing CD4+ T cells prevents direct binding and infection of CD44-containing HIV-1 [175]. Therefore, CD44 incorporation into virus particles has a pro-viral effect on virus transmission to new host T cells in the presence of lymphoid organ stromal cells.
In contrast to CD44, PSGL-1 and CD43 reduce the infectivity of progeny HIV-1 when these proteins are expressed in virus-producing cells [176,177]. We and others have found that when PSGL-1 and CD43 are incorporated into virions, these proteins inhibit HIV-1 infection via blocking of virus attachment to target cells [178,179]. Interestingly, PSGL-1 and CD43 prevent Env-independent virus-cell binding, including virus capture by FRCs. Moreover, the intact extracellular domain of PSGL-1 is required for efficient suppression of virus attachment to target cells. Therefore, virion-incorporated PSGL-1 and CD43 likely inhibit virus-cell binding regardless of molecules mediating virus attachment, perhaps because of the extended extracellular domains. However, it is also possible that PSGL-1 and CD43 on the surface of HIV-1 particles attenuate not only virus-cell binding but also subsequent steps of the HIV life cycle, such as reverse transcription, as reported by another group [176]. Regardless of the mechanism, PSGL-1 and CD43 have antiviral effects on virus entry into new host cells even though they likely share with CD44 the mechanism of recruitment to virus assembly sites and hence, virion incorporation.
Altogether, as exemplified by the uropod-directed proteins discussed above, it is likely that the mechanism by which HIV-1 determines the sites of particle assembly has an impact on the composition of virus particles, thereby affecting the fate of the nascent virions either negatively or positively.
10. Conclusions and Future Directions
Assembly of HIV-1 Gag at the PM results in the formation of infectious progeny. The virion emerging from the host cell has an envelope that, besides Env glycoprotein, contains select lipid components and host proteins that are present at the PM. These host-derived components modulate the release, transmission, or entry of HIV-1 virions. Thus, the PM of virus-producing cells plays important roles not only as the virus exit sites but also as a platform for sorting components of virus particles.
It is well established that Gag-PI(4,5)P2 interaction mediates the specific localization of Gag to the PM. Cholesterol is another cellular lipid that plays an important role in Gag membrane binding. Interestingly, even though cellular PI(4,5)P2 by virtue of its sn-2 unsaturated acyl chain is unlikely to partition into lipid raft-like microdomains, the HIV-1 envelope is enriched in PI(4,5)P2, cholesterol, and lipid raft-like components compared to the producer cell PM. Incorporation of PM proteins into the HIV-1 envelope is likely to occur in various mechanisms based on the preference for raft-like microdomains, the membrane curvature associated with virus budding, and the potential enrichment of acidic lipids such as PI(4,5)P2. Interestingly, the proteins incorporated into the viral envelope could have positive (e.g., CD44) or the negative role (e.g., tetherin, CD43, and PSGL-1) in the HIV-1 life cycle.
Given the importance of PI(4,5)P2 in HIV-1 assembly and its potential role in virus spread via incorporation of host proteins, small molecule inhibitors that prevent Gag-PI(4,5)P2 interaction could be useful as inhibitors of HIV-1 assembly. Similarly, as a shift in the balance between MA-RNA and MA-PI(4,5)P2 binding could prevent Gag binding to the PM, the use of RNA aptamers that bind MA with higher affinity than PI(4,5)P2 might prove to be an effective antiviral strategy.
Several unanswered questions pertaining to HIV-1 assembly are evident from this review. What determines the preference of HIV-1 Gag for PI(4,5)P2 with at least one unsaturated acyl chain? How does PI(4,5)P2 outcompete the negative regulation of tRNA during Gag-membrane binding? Is there a difference among tRNAs in their ability to compete with acidic lipids? Where in the cytoplasm does a tRNA bind to Gag? Does RNA serve as a chaperone for other PM-targeting proteins in the cell? How does HIV-1 Gag reorganize the host membrane microdomains at the virus assembly sites? What molecule mediates the incorporation of PSGL-1, CD43, and CD44 into the HIV-1 envelope? How are the incorporated host proteins and lipids organized on the surface of virus particles? What is the collective impact of the proteins incorporated into the virus particle? Addressing these questions will inevitably advance our understanding of both cell biology of the PM and retrovirus assembly and spread.
Acknowledgments
We thank the members of our laboratory for helpful discussions and critical reviews of the manuscript.
Author Contributions
D.T., T.M., and A.O., writing—original draft, review, and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This work is supported by NIH grants R37 AI 071727 and R21 AI 148381 (to A.O.).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Jouvenet N., Neil S.J., Bess C., Johnson M.C., Virgen C.A., Simon S.M., Bieniasz P.D. Plasma membrane is the site of productive HIV-1 particle assembly. PLoS Biol. 2006;4:e435. doi: 10.1371/journal.pbio.0040435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Finzi A., Orthwein A., Mercier J., Cohen E.A. Productive human immunodeficiency virus type 1 assembly takes place at the plasma membrane. J. Virol. 2007;81:7476–7490. doi: 10.1128/JVI.00308-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sundquist W.I., Kräusslich H.G. HIV-1 assembly, budding, and maturation. Cold Spring Harb. Perspect. Med. 2012;2:a006924. doi: 10.1101/cshperspect.a006924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Freed E.O. HIV-1 assembly, release and maturation. Nat. Rev. Microbiol. 2015;13:484–496. doi: 10.1038/nrmicro3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Llewellyn G.N., Grover J.R., Olety B., Ono A. HIV-1 Gag associates with specific uropod-directed microdomains in a manner dependent on its MA highly basic region. J. Virol. 2013;87:6441–6454. doi: 10.1128/JVI.00040-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nguyen D.H., Hildreth J.E. Evidence for budding of human immunodeficiency virus type 1 selectively from glycolipid-enriched membrane lipid rafts. J. Virol. 2000;74:3264–3272. doi: 10.1128/JVI.74.7.3264-3272.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen P., Hübner W., Spinelli M.A., Chen B.K. Predominant mode of human immunodeficiency virus transfer between T cells is mediated by sustained Env-dependent neutralization-resistant virological synapses. J. Virol. 2007;81:12582–12595. doi: 10.1128/JVI.00381-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Llewellyn G.N., Hogue I.B., Grover J.R., Ono A. Nucleocapsid promotes localization of HIV-1 gag to uropods that participate in virological synapses between T cells. PLoS Pathog. 2010;6:e1001167. doi: 10.1371/journal.ppat.1001167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramirez P.W., Sharma S., Singh R., Stoneham C.A., Vollbrecht T., Guatelli J. Plasma Membrane-Associated Restriction Factors and Their Counteraction by HIV-1 Accessory Proteins. Cells. 2019;8 doi: 10.3390/cells8091020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Firrito C., Bertelli C., Vanzo T., Chande A., Pizzato M. SERINC5 as a New Restriction Factor for Human Immunodeficiency Virus and Murine Leukemia Virus. Annu. Rev. Virol. 2018;5:323–340. doi: 10.1146/annurev-virology-092917-043308. [DOI] [PubMed] [Google Scholar]
- 11.Foster T.L., Pickering S., Neil S.J.D. Inhibiting the Ins and Outs of HIV Replication: Cell-Intrinsic Antiretroviral Restrictions at the Plasma Membrane. Front. Immunol. 2017;8:1853. doi: 10.3389/fimmu.2017.01853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burnie J., Guzzo C. The Incorporation of Host Proteins into the External HIV-1 Envelope. Viruses. 2019;11 doi: 10.3390/v11010085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ono A., Ablan S.D., Lockett S.J., Nagashima K., Freed E.O. Phosphatidylinositol (4,5) bisphosphate regulates HIV-1 Gag targeting to the plasma membrane. Proc. Natl. Acad. Sci. USA. 2004;101:14889–14894. doi: 10.1073/pnas.0405596101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joshi A., Ablan S.D., Soheilian F., Nagashima K., Freed E.O. Evidence that productive human immunodeficiency virus type 1 assembly can occur in an intracellular compartment. J. Virol. 2009;83:5375–5387. doi: 10.1128/JVI.00109-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ono A., Orenstein J.M., Freed E.O. Role of the Gag matrix domain in targeting human immunodeficiency virus type 1 assembly. J. Virol. 2000;74:2855–2866. doi: 10.1128/JVI.74.6.2855-2866.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scholz I., Still A., Dhenub T.C., Coday K., Webb M., Barklis E. Analysis of human immunodeficiency virus matrix domain replacements. Virology. 2008;371:322–335. doi: 10.1016/j.virol.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ono A., Freed E.O. Cell-type-dependent targeting of human immunodeficiency virus type 1 assembly to the plasma membrane and the multivesicular body. J. Virol. 2004;78:1552–1563. doi: 10.1128/JVI.78.3.1552-1563.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Freed E.O., Orenstein J.M., Buckler-White A.J., Martin M.A. Single amino acid changes in the human immunodeficiency virus type 1 matrix protein block virus particle production. J. Virol. 1994;68:5311–5320. doi: 10.1128/JVI.68.8.5311-5320.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saad J.S., Ablan S.D., Ghanam R.H., Kim A., Andrews K., Nagashima K., Soheilian F., Freed E.O., Summers M.F. Structure of the myristylated human immunodeficiency virus type 2 matrix protein and the role of phosphatidylinositol-(4,5)-bisphosphate in membrane targeting. J. Mol. Biol. 2008;382:434–447. doi: 10.1016/j.jmb.2008.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Göttlinger H.G., Sodroski J.G., Haseltine W.A. Role of capsid precursor processing and myristoylation in morphogenesis and infectivity of human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. USA. 1989;86:5781–5785. doi: 10.1073/pnas.86.15.5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bryant M., Ratner L. Myristoylation-dependent replication and assembly of human immunodeficiency virus 1. Proc. Natl. Acad. Sci. USA. 1990;87:523–527. doi: 10.1073/pnas.87.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yuan X., Yu X., Lee T.H., Essex M. Mutations in the N-terminal region of human immunodeficiency virus type 1 matrix protein block intracellular transport of the Gag precursor. J. Virol. 1993;67:6387–6394. doi: 10.1128/JVI.67.11.6387-6394.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou W., Parent L.J., Wills J.W., Resh M.D. Identification of a membrane-binding domain within the amino-terminal region of human immunodeficiency virus type 1 Gag protein which interacts with acidic phospholipids. J. Virol. 1994;68:2556–2569. doi: 10.1128/JVI.68.4.2556-2569.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Resh M.D. Fatty acylation of proteins: The long and the short of it. Prog. Lipid Res. 2016;63:120–131. doi: 10.1016/j.plipres.2016.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou W., Resh M.D. Differential membrane binding of the human immunodeficiency virus type 1 matrix protein. J. Virol. 1996;70:8540–8548. doi: 10.1128/JVI.70.12.8540-8548.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ono A., Freed E.O. Binding of human immunodeficiency virus type 1 Gag to membrane: role of the matrix amino terminus. J. Virol. 1999;73:4136–4144. doi: 10.1128/JVI.73.5.4136-4144.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paillart J.C., Göttlinger H.G. Opposing effects of human immunodeficiency virus type 1 matrix mutations support a myristyl switch model of gag membrane targeting. J. Virol. 1999;73:2604–2612. doi: 10.1128/JVI.73.4.2604-2612.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hermida-Matsumoto L., Resh M.D. Human immunodeficiency virus type 1 protease triggers a myristoyl switch that modulates membrane binding of Pr55(gag) and p17MA. J. Virol. 1999;73:1902–1908. doi: 10.1128/JVI.73.3.1902-1908.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang C., Loeliger E., Luncsford P., Kinde I., Beckett D., Summers M.F. Entropic switch regulates myristate exposure in the HIV-1 matrix protein. Proc. Natl. Acad. Sci. USA. 2004;101:517–522. doi: 10.1073/pnas.0305665101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saad J.S., Miller J., Tai J., Kim A., Ghanam R.H., Summers M.F. Structural basis for targeting HIV-1 Gag proteins to the plasma membrane for virus assembly. Proc. Natl. Acad. Sci. USA. 2006;103:11364–11369. doi: 10.1073/pnas.0602818103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saad J.S., Loeliger E., Luncsford P., Liriano M., Tai J., Kim A., Miller J., Joshi A., Freed E.O., Summers M.F. Point mutations in the HIV-1 matrix protein turn off the myristyl switch. J. Mol. Biol. 2007;366:574–585. doi: 10.1016/j.jmb.2006.11.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H., Dou J., Ding L., Spearman P. Myristoylation is required for human immunodeficiency virus type 1 Gag-Gag multimerization in mammalian cells. J. Virol. 2007;81:12899–12910. doi: 10.1128/JVI.01280-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hill C.P., Worthylake D., Bancroft D.P., Christensen A.M., Sundquist W.I. Crystal structures of the trimeric human immunodeficiency virus type 1 matrix protein: implications for membrane association and assembly. Proc. Natl. Acad. Sci. USA. 1996;93:3099–3104. doi: 10.1073/pnas.93.7.3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murray P.S., Li Z., Wang J., Tang C.L., Honig B., Murray D. Retroviral matrix domains share electrostatic homology: models for membrane binding function throughout the viral life cycle. Structure. 2005;13:1521–1531. doi: 10.1016/j.str.2005.07.010. [DOI] [PubMed] [Google Scholar]
- 35.Dalton A.K., Ako-Adjei D., Murray P.S., Murray D., Vogt V.M. Electrostatic interactions drive membrane association of the human immunodeficiency virus type 1 Gag MA domain. J. Virol. 2007;81:6434–6445. doi: 10.1128/JVI.02757-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ehrlich L.S., Fong S., Scarlata S., Zybarth G., Carter C. Partitioning of HIV-1 Gag and Gag-related proteins to membranes. Biochemistry. 1996;35:3933–3943. doi: 10.1021/bi952337x. [DOI] [PubMed] [Google Scholar]
- 37.Shkriabai N., Datta S.A., Zhao Z., Hess S., Rein A., Kvaratskhelia M. Interactions of HIV-1 Gag with assembly cofactors. Biochemistry. 2006;45:4077–4083. doi: 10.1021/bi052308e. [DOI] [PubMed] [Google Scholar]
- 38.Chukkapalli V., Hogue I.B., Boyko V., Hu W.S., Ono A. Interaction between the human immunodeficiency virus type 1 Gag matrix domain and phosphatidylinositol-(4,5)-bisphosphate is essential for efficient gag membrane binding. J. Virol. 2008;82:2405–2417. doi: 10.1128/JVI.01614-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chan R., Uchil P.D., Jin J., Shui G., Ott D.E., Mothes W., Wenk M.R. Retroviruses human immunodeficiency virus and murine leukemia virus are enriched in phosphoinositides. J. Virol. 2008;82:11228–11238. doi: 10.1128/JVI.00981-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alfadhli A., Barklis R.L., Barklis E. HIV-1 matrix organizes as a hexamer of trimers on membranes containing phosphatidylinositol-(4,5)-bisphosphate. Virology. 2009;387:466–472. doi: 10.1016/j.virol.2009.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chukkapalli V., Oh S.J., Ono A. Opposing mechanisms involving RNA and lipids regulate HIV-1 Gag membrane binding through the highly basic region of the matrix domain. Proc. Natl. Acad. Sci. USA. 2010;107:1600–1605. doi: 10.1073/pnas.0908661107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anraku K., Fukuda R., Takamune N., Misumi S., Okamoto Y., Otsuka M., Fujita M. Highly sensitive analysis of the interaction between HIV-1 Gag and phosphoinositide derivatives based on surface plasmon resonance. Biochemistry. 2010;49:5109–5116. doi: 10.1021/bi9019274. [DOI] [PubMed] [Google Scholar]
- 43.Inlora J., Chukkapalli V., Derse D., Ono A. Gag localization and virus-like particle release mediated by the matrix domain of human T-lymphotropic virus type 1 Gag are less dependent on phosphatidylinositol-(4,5)-bisphosphate than those mediated by the matrix domain of HIV-1 Gag. J. Virol. 2011;85:3802–3810. doi: 10.1128/JVI.02383-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chukkapalli V., Ono A. Molecular determinants that regulate plasma membrane association of HIV-1 Gag. J. Mol. Biol. 2011;410:512–524. doi: 10.1016/j.jmb.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Inlora J., Collins D.R., Trubin M.E., Chung J.Y., Ono A. Membrane binding and subcellular localization of retroviral Gag proteins are differentially regulated by MA interactions with phosphatidylinositol-(4,5)-bisphosphate and RNA. MBio. 2014;5:e02202. doi: 10.1128/mBio.02202-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alfadhli A., Still A., Barklis E. Analysis of human immunodeficiency virus type 1 matrix binding to membranes and nucleic acids. J. Virol. 2009;83:12196–12203. doi: 10.1128/JVI.01197-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alfadhli A., McNett H., Tsagli S., Bächinger H.P., Peyton D.H., Barklis E. HIV-1 matrix protein binding to RNA. J. Mol. Biol. 2011;410:653–666. doi: 10.1016/j.jmb.2011.04.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thornhill D., Olety B., Ono A. Relationships between MA-RNA Binding in Cells and Suppression of HIV-1 Gag Mislocalization to Intracellular Membranes. J. Virol. 2019;93 doi: 10.1128/JVI.00756-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.D’Souza K., Epand R.M. Enrichment of phosphatidylinositols with specific acyl chains. Biochim. Biophys. Acta. 2014;1838:1501–1508. doi: 10.1016/j.bbamem.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 50.Traynor-Kaplan A., Kruse M., Dickson E.J., Dai G., Vivas O., Yu H., Whittington D., Hille B. Fatty-acyl chain profiles of cellular phosphoinositides. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2017;1862:513–522. doi: 10.1016/j.bbalip.2017.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Epand R.M., So V., Jennings W., Khadka B., Gupta R.S., Lemaire M. Diacylglycerol Kinase-ε: Properties and Biological Roles. Front. Cell Dev. Biol. 2016;4:112. doi: 10.3389/fcell.2016.00112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barneda D., Cosulich S., Stephens L., Hawkins P. How is the acyl chain composition of phosphoinositides created and does it matter? Biochem. Soc. Trans. 2019;47:1291–1305. doi: 10.1042/BST20190205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dickson E.J., Hille B. Understanding phosphoinositides: rare, dynamic, and essential membrane phospholipids. Biochem. J. 2019;476:1–23. doi: 10.1042/BCJ20180022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sun Y., Thapa N., Hedman A.C., Anderson R.A. Phosphatidylinositol 4,5-bisphosphate: targeted production and signaling. Bioessays. 2013;35:513–522. doi: 10.1002/bies.201200171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tan X., Thapa N., Choi S., Anderson R.A. Emerging roles of PtdIns(4,5)P2--beyond the plasma membrane. J. Cell Sci. 2015;128:4047–4056. doi: 10.1242/jcs.175208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kisseleva M.V., Wilson M.P., Majerus P.W. The isolation and characterization of a cDNA encoding phospholipid-specific inositol polyphosphate 5-phosphatase. J. Biol. Chem. 2000;275:20110–20116. doi: 10.1074/jbc.M910119199. [DOI] [PubMed] [Google Scholar]
- 57.Balla T. Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiol. Rev. 2013;93:1019–1137. doi: 10.1152/physrev.00028.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Majerus P.W., York J.D. Phosphoinositide phosphatases and disease. J. Lipid Res. 2009;50:S249–254. doi: 10.1194/jlr.R800072-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gericke A., Leslie N.R., Lösche M., Ross A.H. PtdIns(4,5)P2-mediated cell signaling: emerging principles and PTEN as a paradigm for regulatory mechanism. Adv. Exp. Med. Biol. 2013;991:85–104. doi: 10.1007/978-94-007-6331-9_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hsu F., Mao Y. The structure of phosphoinositide phosphatases: Insights into substrate specificity and catalysis. Biochim. Biophys. Acta. 2015;1851:698–710. doi: 10.1016/j.bbalip.2014.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hammond G.R.V., Burke J.E. Novel roles of phosphoinositides in signaling, lipid transport, and disease. Curr. Opin. Cell Biol. 2020;63:57–67. doi: 10.1016/j.ceb.2019.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Monde K., Chukkapalli V., Ono A. Assembly and replication of HIV-1 in T cells with low levels of phosphatidylinositol-(4,5)-bisphosphate. J. Virol. 2011;85:3584–3595. doi: 10.1128/JVI.02266-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mücksch F., Laketa V., Müller B., Schultz C., Kräusslich H.G. Synchronized HIV assembly by tunable PIP. Elife. 2017;6 doi: 10.7554/eLife.25287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chan J., Dick R.A., Vogt V.M. Rous sarcoma virus gag has no specific requirement for phosphatidylinositol-(4,5)-bisphosphate for plasma membrane association in vivo or for liposome interaction in vitro. J. Virol. 2011;85:10851–10860. doi: 10.1128/JVI.00760-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gerber P.P., Cabrini M., Jancic C., Paoletti L., Banchio C., von Bilderling C., Sigaut L., Pietrasanta L.I., Duette G., Freed E.O., et al. Rab27a controls HIV-1 assembly by regulating plasma membrane levels of phosphatidylinositol 4,5-bisphosphate. J. Cell Biol. 2015;209:435–452. doi: 10.1083/jcb.201409082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gonzales B., de Rocquigny H., Beziau A., Durand S., Burlaud-Gaillard J., Lefebvre A., Krull S., Emond P., Brand D., Piver E. Type I phosphatidylinositol-4-phosphate 5-kinase α and γ play a key role in targeting HIV-1 Pr55. J. Virol. 2020 doi: 10.1128/JVI.00189-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nadaraia-Hoke S., Bann D.V., Lochmann T.L., Gudleski-O’Regan N., Parent L.J. Alterations in the MA and NC domains modulate phosphoinositide-dependent plasma membrane localization of the Rous sarcoma virus Gag protein. J. Virol. 2013;87:3609–3615. doi: 10.1128/JVI.03059-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fernandes F., Chen K., Ehrlich L.S., Jin J., Chen M.H., Medina G.N., Symons M., Montelaro R., Donaldson J., Tjandra N., et al. Phosphoinositides direct equine infectious anemia virus gag trafficking and release. Traffic. 2011;12:438–451. doi: 10.1111/j.1600-0854.2010.01153.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stansell E., Apkarian R., Haubova S., Diehl W.E., Tytler E.M., Hunter E. Basic residues in the Mason-Pfizer monkey virus gag matrix domain regulate intracellular trafficking and capsid-membrane interactions. J. Virol. 2007;81:8977–8988. doi: 10.1128/JVI.00657-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hamard-Peron E., Juillard F., Saad J.S., Roy C., Roingeard P., Summers M.F., Darlix J.L., Picart C., Muriaux D. Targeting of murine leukemia virus gag to the plasma membrane is mediated by PI(4,5)P2/PS and a polybasic region in the matrix. J. Virol. 2010;84:503–515. doi: 10.1128/JVI.01134-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ramos A.R., Ghosh S., Erneux C. The impact of phosphoinositide 5-phosphatases on phosphoinositides in cell function and human disease. J. Lipid Res. 2019;60:276–286. doi: 10.1194/jlr.R087908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McLaughlin S., Murray D. Plasma membrane phosphoinositide organization by protein electrostatics. Nature. 2005;438:605–611. doi: 10.1038/nature04398. [DOI] [PubMed] [Google Scholar]
- 73.Barros M., Heinrich F., Datta S.A.K., Rein A., Karageorgos I., Nanda H., Lösche M. Membrane Binding of HIV-1 Matrix Protein: Dependence on Bilayer Composition and Protein Lipidation. J. Virol. 2016;90:4544–4555. doi: 10.1128/JVI.02820-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Carlson L.A., Bai Y., Keane S.C., Doudna J.A., Hurley J.H. Reconstitution of selective HIV-1 RNA packaging in vitro by membrane-bound Gag assemblies. Elife. 2016;5 doi: 10.7554/eLife.14663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Keller H., Kräusslich H.G., Schwille P. Multimerizable HIV Gag derivative binds to the liquid-disordered phase in model membranes. Cell Microbiol. 2013;15:237–247. doi: 10.1111/cmi.12064. [DOI] [PubMed] [Google Scholar]
- 76.Tran R.J., Lalonde M.S., Sly K.L., Conboy J.C. Mechanistic Investigation of HIV-1 Gag Association with Lipid Membranes. J. Phys. Chem. B. 2019;123:4673–4687. doi: 10.1021/acs.jpcb.9b02655. [DOI] [PubMed] [Google Scholar]
- 77.Dick R.A., Kamynina E., Vogt V.M. Effect of multimerization on membrane association of Rous sarcoma virus and HIV-1 matrix domain proteins. J. Virol. 2013;87:13598–13608. doi: 10.1128/JVI.01659-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mercredi P.Y., Bucca N., Loeliger B., Gaines C.R., Mehta M., Bhargava P., Tedbury P.R., Charlier L., Floquet N., Muriaux D., et al. Structural and Molecular Determinants of Membrane Binding by the HIV-1 Matrix Protein. J. Mol. Biol. 2016;428:1637–1655. doi: 10.1016/j.jmb.2016.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Murphy R.E., Samal A.B., Vlach J., Mas V., Prevelige P.E., Saad J.S. Structural and biophysical characterizations of HIV-1 matrix trimer binding to lipid nanodiscs shed light on virus assembly. J. Biol. Chem. 2019;294:18600–18612. doi: 10.1074/jbc.RA119.010997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Junková P., Pleskot R., Prchal J., Sýs J., Ruml T. Differences and commonalities in plasma membrane recruitment of the two morphogenetically distinct retroviruses HIV-1 and MMTV. J. Biol. Chem. 2020;295:8819–8833. doi: 10.1074/jbc.RA119.011991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Charlier L., Louet M., Chaloin L., Fuchs P., Martinez J., Muriaux D., Favard C., Floquet N. Coarse-grained simulations of the HIV-1 matrix protein anchoring: revisiting its assembly on membrane domains. Biophys. J. 2014;106:577–585. doi: 10.1016/j.bpj.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Olety B., Veatch S.L., Ono A. Phosphatidylinositol-(4,5)-Bisphosphate Acyl Chains Differentiate Membrane Binding of HIV-1 Gag from That of the Phospholipase Cδ1 Pleckstrin Homology Domain. J. Virol. 2015;89:7861–7873. doi: 10.1128/JVI.00794-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dick R.A., Goh S.L., Feigenson G.W., Vogt V.M. HIV-1 Gag protein can sense the cholesterol and acyl chain environment in model membranes. Proc. Natl. Acad. Sci. USA. 2012;109:18761–18766. doi: 10.1073/pnas.1209408109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dick R.A., Datta S.A., Nanda H., Fang X., Wen Y., Barros M., Wang Y.X., Rein A., Vogt V.M. Hydrodynamic and Membrane Binding Properties of Purified Rous Sarcoma Virus Gag Protein. J. Virol. 2015;89:10371–10382. doi: 10.1128/JVI.01628-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wen Y., Dick R.A., Feigenson G.W., Vogt V.M. Effects of Membrane Charge and Order on Membrane Binding of the Retroviral Structural Protein Gag. J. Virol. 2016;90:9518–9532. doi: 10.1128/JVI.01102-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mücksch F., Citir M., Lüchtenborg C., Glass B., Traynor-Kaplan A., Schultz C., Brügger B., Kräusslich H.G. Quantification of phosphoinositides reveals strong enrichment of PIP. Sci. Rep. 2019;9:17661. doi: 10.1038/s41598-019-53939-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Purohit P., Dupont S., Stevenson M., Green M.R. Sequence-specific interaction between HIV-1 matrix protein and viral genomic RNA revealed by in vitro genetic selection. RNA. 2001;7:576–584. doi: 10.1017/S1355838201002023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hearps A.C., Wagstaff K.M., Piller S.C., Jans D.A. The N-terminal basic domain of the HIV-1 matrix protein does not contain a conventional nuclear localization sequence but is required for DNA binding and protein self-association. Biochemistry. 2008;47:2199–2210. doi: 10.1021/bi701360j. [DOI] [PubMed] [Google Scholar]
- 89.Kutluay S.B., Zang T., Blanco-Melo D., Powell C., Jannain D., Errando M., Bieniasz P.D. Global changes in the RNA binding specificity of HIV-1 gag regulate virion genesis. Cell. 2014;159:1096–1109. doi: 10.1016/j.cell.2014.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gaines C.R., Tkacik E., Rivera-Oven A., Somani P., Achimovich A., Alabi T., Zhu A., Getachew N., Yang A.L., McDonough M., et al. HIV-1 Matrix Protein Interactions with tRNA: Implications for Membrane Targeting. J. Mol. Biol. 2018;430:2113–2127. doi: 10.1016/j.jmb.2018.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jones C.P., Datta S.A., Rein A., Rouzina I., Musier-Forsyth K. Matrix domain modulates HIV-1 Gag’s nucleic acid chaperone activity via inositol phosphate binding. J. Virol. 2011;85:1594–1603. doi: 10.1128/JVI.01809-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kroupa T., Datta S.A.K., Rein A. Distinct Contributions of Different Domains within the HIV-1 Gag Polyprotein to Specific and Nonspecific Interactions with RNA. Viruses. 2020;12 doi: 10.3390/v12040394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chukkapalli V., Inlora J., Todd G.C., Ono A. Evidence in support of RNA-mediated inhibition of phosphatidylserine-dependent HIV-1 Gag membrane binding in cells. J. Virol. 2013;87:7155–7159. doi: 10.1128/JVI.00075-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Todd G.C., Duchon A., Inlora J., Olson E.D., Musier-Forsyth K., Ono A. Inhibition of HIV-1 Gag-membrane interactions by specific RNAs. RNA. 2017;23:395–405. doi: 10.1261/rna.058453.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Whitney M.L., Hurto R.L., Shaheen H.H., Hopper A.K. Rapid and reversible nuclear accumulation of cytoplasmic tRNA in response to nutrient availability. Mol. Biol. Cell. 2007;18:2678–2686. doi: 10.1091/mbc.e07-01-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Schwenzer H., Jühling F., Chu A., Pallett L.J., Baumert T.F., Maini M., Fassati A. Oxidative Stress Triggers Selective tRNA Retrograde Transport in Human Cells during the Integrated Stress Response. Cell Rep. 2019;26:3416–3428.e3415. doi: 10.1016/j.celrep.2019.02.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lorenz C., Lünse C.E., Mörl M. tRNA Modifications: Impact on Structure and Thermal Adaptation. Biomolecules. 2017;7 doi: 10.3390/biom7020035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Koh C.S., Sarin L.P. Transfer RNA modification and infection - Implications for pathogenicity and host responses. Biochim. Biophys. Acta Gene Regul. Mech. 2018;1861:419–432. doi: 10.1016/j.bbagrm.2018.01.015. [DOI] [PubMed] [Google Scholar]
- 99.Schimmel P. The emerging complexity of the tRNA world: mammalian tRNAs beyond protein synthesis. Nat. Rev. Mol. Cell Biol. 2018;19:45–58. doi: 10.1038/nrm.2017.77. [DOI] [PubMed] [Google Scholar]
- 100.Pan T. Modifications and functional genomics of human transfer RNA. Cell Res. 2018;28:395–404. doi: 10.1038/s41422-018-0013-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Väre V.Y., Eruysal E.R., Narendran A., Sarachan K.L., Agris P.F. Chemical and Conformational Diversity of Modified Nucleosides Affects tRNA Structure and Function. Biomolecules. 2017;7 doi: 10.3390/biom7010029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Burniston M.T., Cimarelli A., Colgan J., Curtis S.P., Luban J. Human immunodeficiency virus type 1 Gag polyprotein multimerization requires the nucleocapsid domain and RNA and is promoted by the capsid-dimer interface and the basic region of matrix protein. J. Virol. 1999;73:8527–8540. doi: 10.1128/JVI.73.10.8527-8540.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ott D.E., Coren L.V., Gagliardi T.D. Redundant roles for nucleocapsid and matrix RNA-binding sequences in human immunodeficiency virus type 1 assembly. J. Virol. 2005;79:13839–13847. doi: 10.1128/JVI.79.22.13839-13847.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cimarelli A., Luban J. Translation elongation factor 1-alpha interacts specifically with the human immunodeficiency virus type 1 Gag polyprotein. J. Virol. 1999;73:5388–5401. doi: 10.1128/JVI.73.7.5388-5401.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Webb J.A., Jones C.P., Parent L.J., Rouzina I., Musier-Forsyth K. Distinct binding interactions of HIV-1 Gag to Psi and non-Psi RNAs: implications for viral genomic RNA packaging. RNA. 2013;19:1078–1088. doi: 10.1261/rna.038869.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ramalingam D., Duclair S., Datta S.A., Ellington A., Rein A., Prasad V.R. RNA aptamers directed to human immunodeficiency virus type 1 Gag polyprotein bind to the matrix and nucleocapsid domains and inhibit virus production. J. Virol. 2011;85:305–314. doi: 10.1128/JVI.02626-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lochrie M.A., Waugh S., Pratt D.G., Clever J., Parslow T.G., Polisky B. In vitro selection of RNAs that bind to the human immunodeficiency virus type-1 gag polyprotein. Nucleic Acids Res. 1997;25:2902–2910. doi: 10.1093/nar/25.14.2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chang C.Y., Chang Y.F., Wang S.M., Tseng Y.T., Huang K.J., Wang C.T. HIV-1 matrix protein repositioning in nucleocapsid region fails to confer virus-like particle assembly. Virology. 2008;378:97–104. doi: 10.1016/j.virol.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Brügger B., Glass B., Haberkant P., Leibrecht I., Wieland F.T., Kräusslich H.G. The HIV lipidome: a raft with an unusual composition. Proc. Natl. Acad. Sci. USA. 2006;103:2641–2646. doi: 10.1073/pnas.0511136103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lorizate M., Sachsenheimer T., Glass B., Habermann A., Gerl M.J., Kräusslich H.G., Brügger B. Comparative lipidomics analysis of HIV-1 particles and their producer cell membrane in different cell lines. Cell Microbiol. 2013;15:292–304. doi: 10.1111/cmi.12101. [DOI] [PubMed] [Google Scholar]
- 111.Simons K., van Meer G. Lipid sorting in epithelial cells. Biochemistry. 1988;27:6197–6202. doi: 10.1021/bi00417a001. [DOI] [PubMed] [Google Scholar]
- 112.Simons K., Ikonen E. Functional rafts in cell membranes. Nature. 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- 113.Lingwood D., Simons K. Lipid rafts as a membrane-organizing principle. Science. 2010;327:46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- 114.Lingwood D., Kaiser H.J., Levental I., Simons K. Lipid rafts as functional heterogeneity in cell membranes. Biochem. Soc. Trans. 2009;37:955–960. doi: 10.1042/BST0370955. [DOI] [PubMed] [Google Scholar]
- 115.Kusumi A., Suzuki K.G., Kasai R.S., Ritchie K., Fujiwara T.K. Hierarchical mesoscale domain organization of the plasma membrane. Trends Biochem. Sci. 2011;36:604–615. doi: 10.1016/j.tibs.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 116.Kusumi A., Koyama-Honda I., Suzuki K. Molecular dynamics and interactions for creation of stimulation-induced stabilized rafts from small unstable steady-state rafts. Traffic. 2004;5:213–230. doi: 10.1111/j.1600-0854.2004.0178.x. [DOI] [PubMed] [Google Scholar]
- 117.van den Bogaart G., Meyenberg K., Risselada H.J., Amin H., Willig K.I., Hubrich B.E., Dier M., Hell S.W., Grubmüller H., Diederichsen U., et al. Membrane protein sequestering by ionic protein-lipid interactions. Nature. 2011;479:552–555. doi: 10.1038/nature10545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wang J., Richards D.A. Segregation of PIP2 and PIP3 into distinct nanoscale regions within the plasma membrane. Biol. Open. 2012;1:857–862. doi: 10.1242/bio.20122071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Levental I., Christian D.A., Wang Y.H., Madara J.J., Discher D.E., Janmey P.A. Calcium-dependent lateral organization in phosphatidylinositol 4,5-bisphosphate (PIP2)- and cholesterol-containing monolayers. Biochemistry. 2009;48:8241–8248. doi: 10.1021/bi9007879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wen Y., Vogt V.M., Feigenson G.W. Multivalent Cation-Bridged PI(4,5)P. Biophys. J. 2018;114:2630–2639. doi: 10.1016/j.bpj.2018.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wang J., Gambhir A., McLaughlin S., Murray D. A computational model for the electrostatic sequestration of PI(4,5)P2 by membrane-adsorbed basic peptides. Biophys. J. 2004;86:1969–1986. doi: 10.1016/S0006-3495(04)74260-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ono A., Freed E.O. Role of lipid rafts in virus replication. Adv. Virus Res. 2005;64:311–358. doi: 10.1016/S0065-3527(05)64010-9. [DOI] [PubMed] [Google Scholar]
- 123.Levental I., Levental K.R., Heberle F.A. Lipid Rafts: Controversies Resolved, Mysteries Remain. Trends Cell Biol. 2020;30:341–353. doi: 10.1016/j.tcb.2020.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Goñi F.M. "Rafts": A nickname for putative transient nanodomains. Chem. Phys. Lipids. 2019;218:34–39. doi: 10.1016/j.chemphyslip.2018.11.006. [DOI] [PubMed] [Google Scholar]
- 125.Brown D.A., London E. Structure and origin of ordered lipid domains in biological membranes. J. Membr. Biol. 1998;164:103–114. doi: 10.1007/s002329900397. [DOI] [PubMed] [Google Scholar]
- 126.Rietveld A., Simons K. The differential miscibility of lipids as the basis for the formation of functional membrane rafts. Biochim. Biophys. Acta. 1998;1376:467–479. doi: 10.1016/S0304-4157(98)00019-7. [DOI] [PubMed] [Google Scholar]
- 127.Brown D.A., London E. Structure and function of sphingolipid- and cholesterol-rich membrane rafts. J. Biol. Chem. 2000;275:17221–17224. doi: 10.1074/jbc.R000005200. [DOI] [PubMed] [Google Scholar]
- 128.Simons K., Toomre D. Lipid rafts and signal transduction. Nat. Rev. Mol. Cell Biol. 2000;1:31–39. doi: 10.1038/35036052. [DOI] [PubMed] [Google Scholar]
- 129.Melkonian K.A., Ostermeyer A.G., Chen J.Z., Roth M.G., Brown D.A. Role of lipid modifications in targeting proteins to detergent-resistant membrane rafts. Many raft proteins are acylated, while few are prenylated. J. Biol. Chem. 1999;274:3910–3917. doi: 10.1074/jbc.274.6.3910. [DOI] [PubMed] [Google Scholar]
- 130.Zacharias D.A., Violin J.D., Newton A.C., Tsien R.Y. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science. 2002;296:913–916. doi: 10.1126/science.1068539. [DOI] [PubMed] [Google Scholar]
- 131.Ono A., Waheed A.A., Freed E.O. Depletion of cellular cholesterol inhibits membrane binding and higher-order multimerization of human immunodeficiency virus type 1 Gag. Virology. 2007;360:27–35. doi: 10.1016/j.virol.2006.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Aloia R.C., Tian H., Jensen F.C. Lipid composition and fluidity of the human immunodeficiency virus envelope and host cell plasma membranes. Proc. Natl. Acad. Sci. USA. 1993;90:5181–5185. doi: 10.1073/pnas.90.11.5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sengupta P., Seo A.Y., Pasolli H.A., Song Y.E., Johnson M.C., Lippincott-Schwartz J. A lipid-based partitioning mechanism for selective incorporation of proteins into membranes of HIV particles. Nat. Cell Biol. 2019;21:452–461. doi: 10.1038/s41556-019-0300-y. [DOI] [PubMed] [Google Scholar]
- 134.Lorizate M., Brügger B., Akiyama H., Glass B., Müller B., Anderluh G., Wieland F.T., Kräusslich H.G. Probing HIV-1 membrane liquid order by Laurdan staining reveals producer cell-dependent differences. J. Biol. Chem. 2009;284:22238–22247. doi: 10.1074/jbc.M109.029256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Saifuddin M., Parker C.J., Peeples M.E., Gorny M.K., Zolla-Pazner S., Ghassemi M., Rooney I.A., Atkinson J.P., Spear G.T. Role of virion-associated glycosylphosphatidylinositol-linked proteins CD55 and CD59 in complement resistance of cell line-derived and primary isolates of HIV-1. J. Exp. Med. 1995;182:501–509. doi: 10.1084/jem.182.2.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Nakamura M., Okada H., Sasaki H., Yoshida K., Kamada M., Okada N., Terada M., Ohno T. Quantification of the CD55 and CD59, membrane inhibitors of complement on HIV-1 particles as a function of complement-mediated virolysis. Microbiol. Immunol. 1996;40:561–567. doi: 10.1111/j.1348-0421.1996.tb01109.x. [DOI] [PubMed] [Google Scholar]
- 137.Ott D.E. Cellular proteins detected in HIV-1. Rev. Med. Virol. 2008;18:159–175. doi: 10.1002/rmv.570. [DOI] [PubMed] [Google Scholar]
- 138.Chertova E., Chertov O., Coren L.V., Roser J.D., Trubey C.M., Bess J.W., Sowder R.C., Barsov E., Hood B.L., Fisher R.J., et al. Proteomic and biochemical analysis of purified human immunodeficiency virus type 1 produced from infected monocyte-derived macrophages. J. Virol. 2006;80:9039–9052. doi: 10.1128/JVI.01013-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Bhattacharya J., Repik A., Clapham P.R. Gag regulates association of human immunodeficiency virus type 1 envelope with detergent-resistant membranes. J. Virol. 2006;80:5292–5300. doi: 10.1128/JVI.01469-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Dou J., Wang J.J., Chen X., Li H., Ding L., Spearman P. Characterization of a myristoylated, monomeric HIV Gag protein. Virology. 2009;387:341–352. doi: 10.1016/j.virol.2009.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ono A., Waheed A.A., Joshi A., Freed E.O. Association of human immunodeficiency virus type 1 gag with membrane does not require highly basic sequences in the nucleocapsid: use of a novel Gag multimerization assay. J. Virol. 2005;79:14131–14140. doi: 10.1128/JVI.79.22.14131-14140.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lindwasser O.W., Resh M.D. Multimerization of human immunodeficiency virus type 1 Gag promotes its localization to barges, raft-like membrane microdomains. J. Virol. 2001;75:7913–7924. doi: 10.1128/JVI.75.17.7913-7924.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lindwasser O.W., Resh M.D. Myristoylation as a target for inhibiting HIV assembly: unsaturated fatty acids block viral budding. Proc. Natl. Acad. Sci. USA. 2002;99:13037–13042. doi: 10.1073/pnas.212409999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Holm K., Weclewicz K., Hewson R., Suomalainen M. Human immunodeficiency virus type 1 assembly and lipid rafts: Pr55(gag) associates with membrane domains that are largely resistant to Brij98 but sensitive to Triton X-100. J. Virol. 2003;77:4805–4817. doi: 10.1128/JVI.77.8.4805-4817.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ono A., Freed E.O. Plasma membrane rafts play a critical role in HIV-1 assembly and release. Proc. Natl. Acad. Sci. USA. 2001;98:13925–13930. doi: 10.1073/pnas.241320298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Gomez C.Y., Hope T.J. Mobility of human immunodeficiency virus type 1 Pr55Gag in living cells. J. Virol. 2006;80:8796–8806. doi: 10.1128/JVI.02159-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Pickl W.F., Pimentel-Muiños F.X., Seed B. Lipid rafts and pseudotyping. J. Virol. 2001;75:7175–7183. doi: 10.1128/JVI.75.15.7175-7183.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Nydegger S., Khurana S., Krementsov D.N., Foti M., Thali M. Mapping of tetraspanin-enriched microdomains that can function as gateways for HIV-1. J. Cell Biol. 2006;173:795–807. doi: 10.1083/jcb.200508165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Hogue I.B., Grover J.R., Soheilian F., Nagashima K., Ono A. Gag induces the coalescence of clustered lipid rafts and tetraspanin-enriched microdomains at HIV-1 assembly sites on the plasma membrane. J. Virol. 2011;85:9749–9766. doi: 10.1128/JVI.00743-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Krementsov D.N., Rassam P., Margeat E., Roy N.H., Schneider-Schaulies J., Milhiet P.E., Thali M. HIV-1 assembly differentially alters dynamics and partitioning of tetraspanins and raft components. Traffic. 2010;11:1401–1414. doi: 10.1111/j.1600-0854.2010.01111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Drücker P., Pejic M., Galla H.J., Gerke V. Lipid segregation and membrane budding induced by the peripheral membrane binding protein annexin A2. J. Biol. Chem. 2013;288:24764–24776. doi: 10.1074/jbc.M113.474023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Jiang Z., Redfern R.E., Isler Y., Ross A.H., Gericke A. Cholesterol stabilizes fluid phosphoinositide domains. Chem. Phys. Lipids. 2014;182:52–61. doi: 10.1016/j.chemphyslip.2014.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Yandrapalli N., Lubart Q., Tanwar H.S., Picart C., Mak J., Muriaux D., Favard C. Self assembly of HIV-1 Gag protein on lipid membranes generates PI(4,5)P. Sci. Rep. 2016;6:39332. doi: 10.1038/srep39332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Favard C., Chojnacki J., Merida P., Yandrapalli N., Mak J., Eggeling C., Muriaux D. HIV-1 Gag specifically restricts PI(4,5)P2 and cholesterol mobility in living cells creating a nanodomain platform for virus assembly. Sci. Adv. 2019;5:eaaw8651. doi: 10.1126/sciadv.aaw8651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Grover J.R., Llewellyn G.N., Soheilian F., Nagashima K., Veatch S.L., Ono A. Roles played by capsid-dependent induction of membrane curvature and Gag-ESCRT interactions in tetherin recruitment to HIV-1 assembly sites. J. Virol. 2013;87:4650–4664. doi: 10.1128/JVI.03526-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Grover J.R., Veatch S.L., Ono A. Basic motifs target PSGL-1, CD43, and CD44 to plasma membrane sites where HIV-1 assembles. J. Virol. 2015;89:454–467. doi: 10.1128/JVI.02178-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Chen X., Khajeh J.A., Ju J.H., Gupta Y.K., Stanley C.B., Do C., Heller W.T., Aggarwal A.K., Callaway D.J., Bu Z. Phosphatidylinositol 4,5-bisphosphate clusters the cell adhesion molecule CD44 and assembles a specific CD44-Ezrin heterocomplex, as revealed by small angle neutron scattering. J. Biol. Chem. 2015;290:6639–6652. doi: 10.1074/jbc.M114.589523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Shao B., Yago T., Setiadi H., Wang Y., Mehta-D’souza P., Fu J., Crocker P.R., Rodgers W., Xia L., McEver R.P. O-glycans direct selectin ligands to lipid rafts on leukocytes. Proc. Natl. Acad. Sci. USA. 2015;112:8661–8666. doi: 10.1073/pnas.1507712112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Rosenman S.J., Ganji A.A., Tedder T.F., Gallatin W.M. Syn-capping of human T lymphocyte adhesion/activation molecules and their redistribution during interaction with endothelial cells. J. Leukoc Biol. 1993;53:1–10. doi: 10.1002/jlb.53.1.1. [DOI] [PubMed] [Google Scholar]
- 160.del Pozo M.A., Sánchez-Mateos P., Nieto M., Sánchez-Madrid F. Chemokines regulate cellular polarization and adhesion receptor redistribution during lymphocyte interaction with endothelium and extracellular matrix. Involvement of cAMP signaling pathway. J. Cell Biol. 1995;131:495–508. doi: 10.1083/jcb.131.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Sánchez-Madrid F., del Pozo M.A. Leukocyte polarization in cell migration and immune interactions. EMBO J. 1999;18:501–511. doi: 10.1093/emboj/18.3.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Alonso-Lebrero J.L., Serrador J.M., Domínguez-Jiménez C., Barreiro O., Luque A., del Pozo M.A., Snapp K., Kansas G., Schwartz-Albiez R., Furthmayr H., et al. Polarization and interaction of adhesion molecules P-selectin glycoprotein ligand 1 and intercellular adhesion molecule 3 with moesin and ezrin in myeloid cells. Blood. 2000;95:2413–2419. doi: 10.1182/blood.V95.7.2413. [DOI] [PubMed] [Google Scholar]
- 163.Serrador J.M., Urzainqui A., Alonso-Lebrero J.L., Cabrero J.R., Montoya M.C., Vicente-Manzanares M., Yáñez-Mó M., Sánchez-Madrid F. A juxta-membrane amino acid sequence of P-selectin glycoprotein ligand-1 is involved in moesin binding and ezrin/radixin/moesin-directed targeting at the trailing edge of migrating lymphocytes. Eur. J. Immunol. 2002;32:1560–1566. doi: 10.1002/1521-4141(200206)32:6<1560::AID-IMMU1560>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 164.Alon R., Rossiter H., Wang X., Springer T.A., Kupper T.S. Distinct cell surface ligands mediate T lymphocyte attachment and rolling on P and E selectin under physiological flow. J. Cell Biol. 1994;127:1485–1495. doi: 10.1083/jcb.127.5.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.McEver R.P., Moore K.L., Cummings R.D. Leukocyte trafficking mediated by selectin-carbohydrate interactions. J. Biol. Chem. 1995;270:11025–11028. doi: 10.1074/jbc.270.19.11025. [DOI] [PubMed] [Google Scholar]
- 166.Carlsson S.R., Fukuda M. Isolation and characterization of leukosialin, a major sialoglycoprotein on human leukocytes. J. Biol. Chem. 1986;261:12779–12786. [PubMed] [Google Scholar]
- 167.Norgard K.E., Moore K.L., Diaz S., Stults N.L., Ushiyama S., McEver R.P., Cummings R.D., Varki A. Characterization of a specific ligand for P-selectin on myeloid cells. A minor glycoprotein with sialylated O-linked oligosaccharides. J. Biol. Chem. 1993;268:12764–12774. [PubMed] [Google Scholar]
- 168.Cyster J.G., Shotton D.M., Williams A.F. The dimensions of the T lymphocyte glycoprotein leukosialin and identification of linear protein epitopes that can be modified by glycosylation. EMBO J. 1991;10:893–902. doi: 10.1002/j.1460-2075.1991.tb08022.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Manjunath N., Correa M., Ardman M., Ardman B. Negative regulation of T-cell adhesion and activation by CD43. Nature. 1995;377:535–538. doi: 10.1038/377535a0. [DOI] [PubMed] [Google Scholar]
- 170.Matsumoto M., Miyasaka M., Hirata T. P-selectin glycoprotein ligand-1 negatively regulates T-cell immune responses. J. Immunol. 2009;183:7204–7211. doi: 10.4049/jimmunol.0902173. [DOI] [PubMed] [Google Scholar]
- 171.Aruffo A., Stamenkovic I., Melnick M., Underhill C.B., Seed B. CD44 is the principal cell surface receptor for hyaluronate. Cell. 1990;61:1303–1313. doi: 10.1016/0092-8674(90)90694-A. [DOI] [PubMed] [Google Scholar]
- 172.Bastiani L., Laal S., Kim M., Zolla-Pazner S. Host cell-dependent alterations in envelope components of human immunodeficiency virus type 1 virions. J. Virol. 1997;71:3444–3450. doi: 10.1128/JVI.71.5.3444-3450.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Orentas R.J., Hildreth J.E. Association of host cell surface adhesion receptors and other membrane proteins with HIV and SIV. AIDS Res. Hum. Retrovir. 1993;9:1157–1165. doi: 10.1089/aid.1993.9.1157. [DOI] [PubMed] [Google Scholar]
- 174.Murakami T., Kim J., Li Y., Green G.E., Shikanov A., Ono A. Secondary lymphoid organ fibroblastic reticular cells mediate trans-infection of HIV-1 via CD44-hyaluronan interactions. Nat. Commun. 2018;9:2436. doi: 10.1038/s41467-018-04846-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Li P., Fujimoto K., Bourguingnon L., Yukl S., Deeks S., Wong J.K. Exogenous and endogenous hyaluronic acid reduces HIV infection of CD4(+) T cells. Immunol. Cell Biol. 2014;92:770–780. doi: 10.1038/icb.2014.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Liu Y., Fu Y., Wang Q., Li M., Zhou Z., Dabbagh D., Fu C., Zhang H., Li S., Zhang T., et al. Proteomic profiling of HIV-1 infection of human CD4. Nat. Microbiol. 2019;4:813–825. doi: 10.1038/s41564-019-0372-2. [DOI] [PubMed] [Google Scholar]
- 177.McLaren P.J., Gawanbacht A., Pyndiah N., Krapp C., Hotter D., Kluge S.F., Götz N., Heilmann J., Mack K., Sauter D., et al. Identification of potential HIV restriction factors by combining evolutionary genomic signatures with functional analyses. Retrovirology. 2015;12:41. doi: 10.1186/s12977-015-0165-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Murakami T., Carmona N., Ono A. Virion-incorporated PSGL-1 and CD43 inhibit both cell-free infection and transinfection of HIV-1 by preventing virus-cell binding. Proc. Natl. Acad. Sci. USA. 2020;117:8055–8063. doi: 10.1073/pnas.1916055117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Fu Y., He S., Waheed A.A., Dabbagh D., Zhou Z., Trinité B., Wang Z., Yu J., Wang D., Li F., et al. PSGL-1 restricts HIV-1 infectivity by blocking virus particle attachment to target cells. Proc. Natl. Acad. Sci. USA. 2020;117:9537–9545. doi: 10.1073/pnas.1916054117. [DOI] [PMC free article] [PubMed] [Google Scholar]