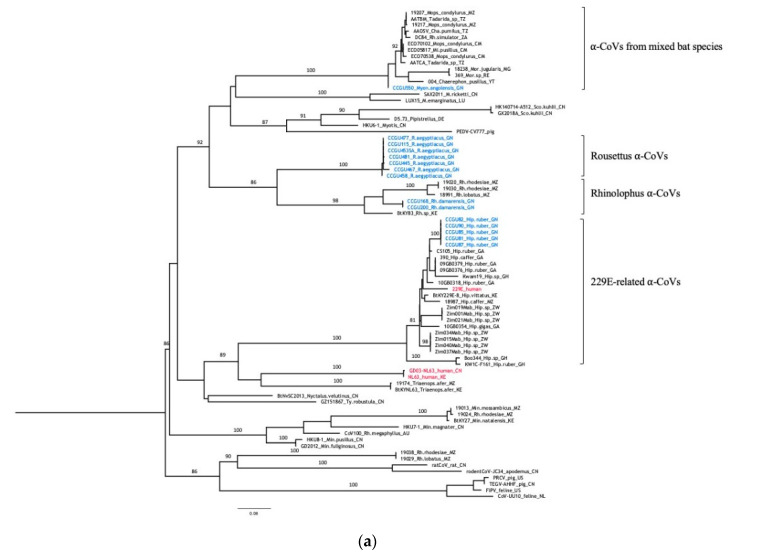
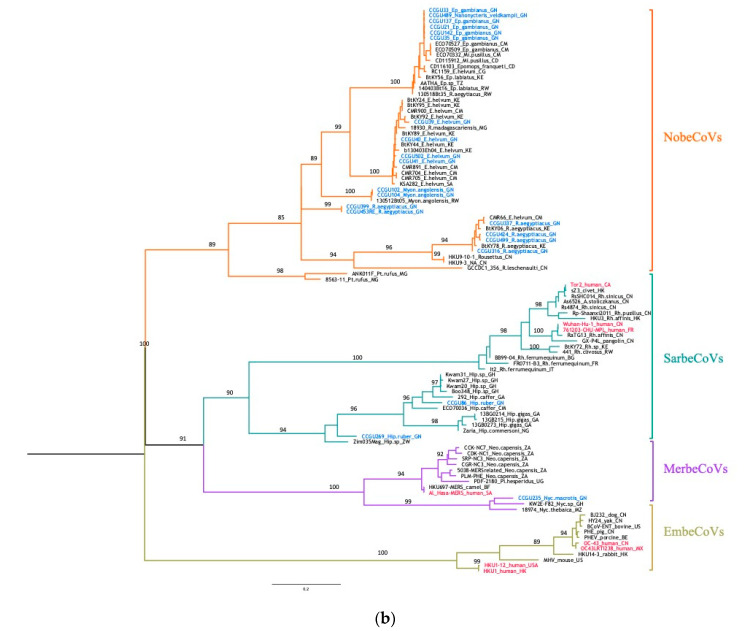
Figure 2.
Maximum likelihood (ML) consensus trees derived from coronavirus sequences in the RNA-dependent RNA-polymerase partial nucleotide sequences (374 unambiguously aligned base pairs). (a) consensus tree derived from 78 alphacoronaviruses; (b) consensus tree derived from 102 betacoronaviruses. Phylogenetic tree analysis was performed using the GTR + F + I + 4Γ nucleotide substitution model and 1000 bootstrap resampling. Sequences in blue refer to the new bat CoVs detected in this study. Human CoVs are highlighted in red. Branch supports >0.75 are indicated on the trees. Trees were generated as indicated in Material and Methods and edited with increasing nodes and midpoint rooting in FigTree. Details on reference sequences are provided in Supplementary Table S1. Abbreviations of bat genera are as follows: Ep—Epomophorus; R—Rousettus; E—Eidolon; Myon—Lissonycteris (previously Myonycteris); Mi—Micropteropus; Nyc—Nycteris; Hip—Hipposideros; Rh—Rhinolophus; Neo—Neoromicia; Pt—Pteropus; A—Aselliscus; Pi—Pipistrellus; Min—Miniopterus; Mor—Mormopterus; ChA—Chaerephon; M—Myotis; Sco—Scotophilus.