 The search is on for effective specific inhibitors for PI3Kα mutants.
The search is on for effective specific inhibitors for PI3Kα mutants.
Abstract
The search is on for effective specific inhibitors for PI3Kα mutants. PI3Kα, a critical lipid kinase, has two subunits, catalytic and inhibitory. PIK3CA, the gene that encodes the p110α catalytic subunit is a highly mutated protein in cancer. Dysregulation of PI3Kα signalling is commonly associated with tumorigenesis and drug resistance. Despite its vast importance, only recently the FDA approved the first drug (alpelisib by Novartis) for breast cancer. A second (GDC0077), classified as PI3Kα isoform-specific, is undergoing clinical trials. Not surprisingly, these ATP-competitive drugs commonly elicit severe concentration-dependent side effects. Here we briefly review PI3Kα mutations, focus on PI3K drug repertoire and propose new, to-date unexplored PI3Kα therapeutic strategies. These include (1) an allosteric and orthosteric inhibitor combination and (2) taking advantage of allosteric rescue mutations to guide drug discovery.
Introduction
Phosphatidylinositol-3 kinases (PI3Ks) are lipid kinases that phosphorylate signaling lipid PIP2 to PIP3.1 PIP3 molecules recruit proteins bearing PIP3-binding pleckstrin homology (PH) domains such as Akt in the PI3K/Akt/mTOR signaling pathway to the plasma membrane. The PI3K pathway is among the most frequently activated in human cancers, impacting almost 50% of the malignancies.2 Class IA isoforms PI3Kα, β and δ are particularly strongly associated with cancer.3 Activating mutations in the PIK3CA gene,4 which encodes the p110α catalytic subunit of PI3Kα and suppressing mutations of tumor suppressor PTEN5,6 (a lipid phosphatase that dephosphorylates PIP3, thus opposes PI3Kα action) are the second and third most frequently mutated in cancer, making PI3Kα a vastly important target for drug discovery.7,8 However, despite considerable efforts, to date the clinical outcome of PI3K inhibitor-based treatments1 for solid tumors has been disappointing. Reasons include drug resistance such as that resulting from PTEN suppression and lack of specificity thus intolerable toxicity, making innovative drug development a major aim of the pharmaceutical industry. Structural understanding of PI3Kα activation mechanism under physiological conditions and in the presence of activating oncogenic mutations is expected to provide a better grasp of its underlying principles toward this aim. The landmark work of Peter Vogt that has uncovered mechanistic principles of PI3Kα gain-of-function mutations, including rare mutations, Roger Williams, who pointed out the premise that oncogenic mutations mimic and enhance dynamic events in natural activation of p110α, and Mario Amzel who solved the first full PI3Kα structure elucidating oncogenic mutations have been major steps in this direction.9–12
PI3Kα has two subunits, catalytic (p110) and inhibitory (p85). PIK3CA, which encodes the p110α, is frequently mutated in cancer. Dysregulation of the PI3Kα signaling pathway is commonly associated with tumorigenesis and drug resistance. Despite its vast importance, only in May 2019 the FDA approved the first drug (alpelisib by Novartis) for breast cancer.13 A second (GDC0077) is undergoing clinical trials. Although the drug has been classified as PI3K isoform-specific, severe concentration-dependent side effects are commonly observed. This is not unexpected. These drugs, like the previously developed ones, are ATP-competitive and the ATP binding site is almost identical among PI3K isoforms, and they target a major signaling pathway in the cell. Recently, it was discovered that double, triple, and multiple mutations can work conjointly,14 raising key questions, including why co-occurring mutations, why specific combinations, and how frequent can such events be in oncogenic proteins. Remarkably, alpelisib was observed to potently affect the signaling strength of particular combinations of mutations when targeting PI3Kα oncogenes, raising the question of how the drug works.
Below, we briefly discuss the mechanism of PI3Kα activation, its oncogenic mutations and their combinations. We review inhibitors developed to block it and their intrinsic off-target toxicities, and outline the challenges encountered in isoform-specific drug discovery. Finally, we propose two new possible strategies for PI3Kα specific inhibitor development. The first combines PI3Kα orthosteric and allosteric drugs; the second involves allosteric drugs at positions of rescue mutations. Drugs mimicking the allosteric effects of these mutants may rescue PI3Kα function in cancer. We further delineate determinants to accomplish these approaches.
Allostery plays a role in PI3Kα activation by RTK, but not by Ras
Recently we published in Chem. Sci. an atomistic-level mechanism of PI3Kα activation.15 In that work we delineated how, via the phosphorylated motif at the C-terminal of a receptor tyrosine kinase (RTK) PI3Kα relieves its autoinhibition by the nSH2 domain and the details of the allosteric population shifts of the PI3Kα ensemble that take place to expose the active site of this lipid kinase at the membrane. The mechanism further clarifies how these conformational changes increase the population of ATP within a catalytically required distance, permitting phosphoryl ion transfer to the signaling lipid PIP2 to produce PIP3. In addition, it provided the structural role of Ras in PI3Kα activation.16
Even though the binding of active Ras perturbs the PI3Kα structure with allosteric propagation that promotes certain conformational changes,17–21 the key question is whether this action is the dominant contribution to activation. RTK recruits PI3Kα to the membrane and promotes conformational change that relieves the autoinhibition; Ras acts to stabilize the PIP2 lipid substrate in the relatively shallow active site that accommodates it. This enhances its population, with the ‘top’ hydrophobic portion of PIP2 projecting into the plasma membrane. The allosteric conformational change promoted by Ras is unlikely to play a significant role in activation.
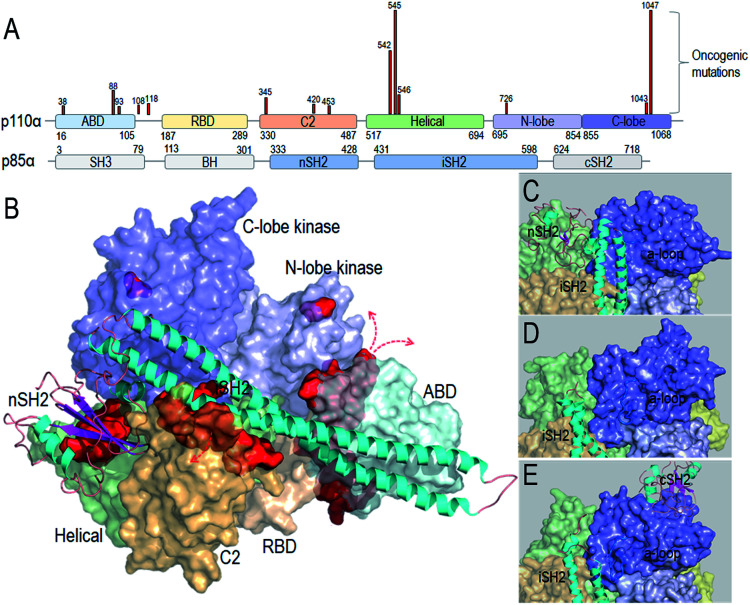
Taken together, the detailed PI3Kα activation mechanism helps in understanding how the strong cancer driver mutations activate this lipid kinase; why some other observed oncogenic mutations are weaker drivers, and why certain mutational combinations are more powerful and more potently targeted by these drugs. Hotspot mutations E542K and E545K mimic RTK's action in releasing the autoinhibition. This action promotes the allosteric conformational changes that expose PI3Kα active site at the membrane and reduce the barrier height of the transition state; the H1047R hotspot with a substitution of His by positively charged Arg mimics Ras' action. The co-acting double mutations observed by Vasan et al.14 pair a strong hotspot with one or more weak activation mutations, such as E453Q/K, E726K and M1043V/I, which either introduce charge reversal at the membrane, or altered hydrophobicity. The changes of the residue charge and hydrophobicity strengthen the activation and signaling. Most of the weak mutations are far away from both the inhibitory nSH2 in the p85α subunit and the catalytic site, confirming their allosteric roles in activation. Fig. 1 illustrates the PI3Kα structure in the autoinhibited and active states, with the location of the mutations highlighted (Fig. 1A and B). While hotspot mutations occur in the helical and kinase domains, most weak mutations are at the surfaces of the ABD and C2 domains. ABD and C2 in p110α are responsible for iSH2 interactions. The oncogenic mutations in ABD and C2 domains imply a more flexible iSH2 in activated PI3Kα. The crystal structures of PI3Kα containing the inhibitory nSH2 may represent an inactive conformation (Fig. 1C).22–24 The active conformation observed in our simulations following nSH2 release differs. Remarkably, the sampled PI3Kα active conformation resembles the crystal structure of PI3Kβ with the inhibitory nSH2 removed (PDB ID ; 2Y3A), with similar iSH2 and activation loop (a-loop) conformations (Fig. 1D and E). PI3Kβ is inhibited by both nSH2 and cSH2. The crystal structure of PI3Kβ without the nSH2 domain may represent a partially active conformation of PI3K,25 implying that PI3K isoforms may share conformational changes for activation.
Fig. 1. PI3Kα conformation and oncogenic mutations. (A) PI3Kα sequence and the frequent oncogenic mutations in p110α (data obtained from TGCA database). (B) PI3Kα structures (PDB ID 4OVV) with the oncogenic mutations highlighted by the red surfaces. The close-up of (C) the PI3Kα conformation with nSH2 in the crystal structure (PDB ID ; 4OVV), (D) the conformation of PI3Kα with nSH2 released from the simulations,15 and (E) the conformation of the PI3Kβ with nSH2 released (PDB ID ; 2Y3A).
PI3Kα activation takes place at the plasma membrane. It is mediated by interactions with the phosphorylated motif at the C-terminal of an RTK, Ras and the membrane. The activating mutations mimic the effects of these binding events. However, current inhibitors are still stymied by insufficient selectivity, the outcome of the near identity among the ATP binding sites of the PI3K isoforms. At the same time, since ATP is the principal energy currency of the cell with structurally similar cavities hosting it,26 attempts to overcome these hurdles by higher ATP-competitive drug levels encounter toxicity. This argues for allosteric drugs.27–34 Unfortunately, these efforts are yet to produce effective drugs. Notwithstanding, the encouraging successes with KRas4B, another hitherto undruggable oncogenic protein, where creative covalent allosteric drugs target KRas4BG12C, and discovery of new PI3Kα pockets,27 breed cautious optimism.
Pan PI3K and PI3Kα inhibitors
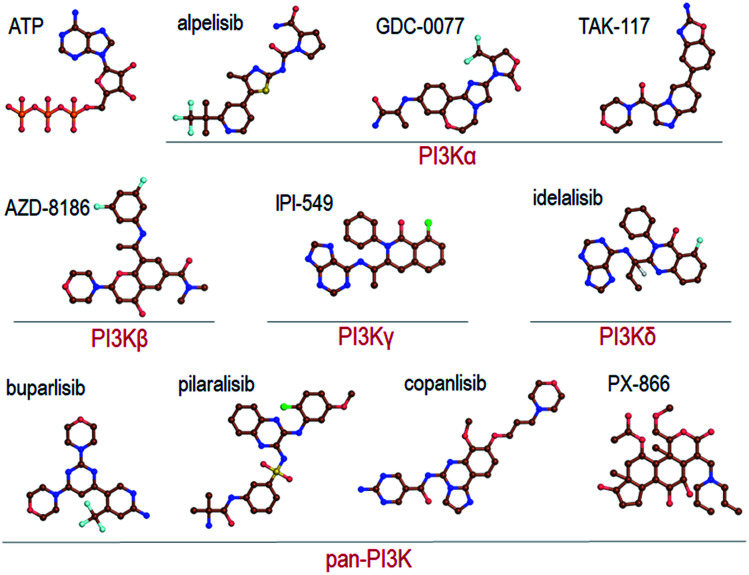
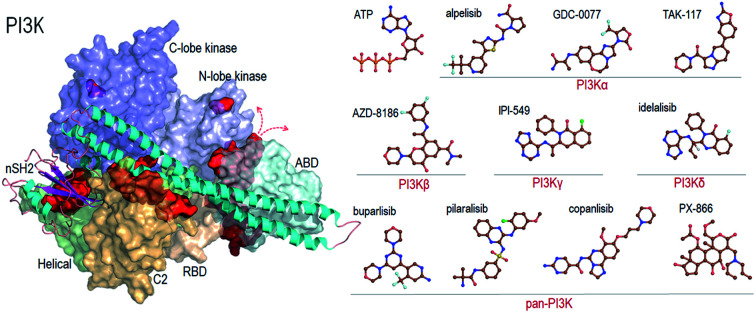
Recent reviews provide a comprehensive coverage of pan-PI3K inhibitors, PI3Kα specific inhibitors and PI3K/mTOR inhibitors, their status and the specific cancers that they target.35–38 We thus refer the readers to these. Here we provide a mere brief overview (Fig. 2).
Fig. 2. Summary of the representative pan-PI3K and PI3K isoform-specific ATP-competitive inhibitors. The molecular structures are obtained from Drugbank.
Pan-PI3K inhibitors act on all class I isoforms.38 The outcome is off-target side effects and toxicities.37 Depending on the specific targeted cancer type and its status they include buparlisib39–41 and pictilisib (GDC-0941) in breast cancer,42,43 and pilaralisib,44–47 copanlisib,48,49 PX866,50–56 CH5132799,57 and ZSTK474 and SF1126 58 in diverse cancers (Fig. 2). Their lack of isoform specificity makes them toxic albeit to different extents.37,59 Consequently, most were discontinued. Inhibitors gravitating toward isoform-specificity have lesser off-target toxicity. δ-Isoform inhibitor idelalisib is one example.37,60,61 Taselisib inhibits primarily p110β with 30-fold lower potency59 but also reduces mutant p110α levels. Alpelisib (BYL719), described as PI3Kα-specific, is another.62 Alpelisib was also prescribed in combination with the aromatase inhibitor letrozole.63 Off-target toxicities were observed. A combination with the BRAF inhibitor encorafenib and anti-EGFR monoclonal antibody cetuximab pointed to clinical efficacy and lower toxicity profiles.64 Additional combinations were tested as well, including alpelisib with inhibitors targeting PI3K pathway proteins,65 although still with side effects. As expected, at higher concentrations it accomplishes strong PI3K (80%) inhibition. TAK-117 is another PI3Kα inhibitor.66 The new PI3Kα inhibitor GDC-0077 currently undergoes clinical trials in combination with other drugs.67 Further details are provided in the reviews cited above. Exhaustive structural analyses of the active sites of all PI3Kα class I isoforms have also been reviewed and regions that can play a role in selectivity were described in detail.68 Below, we briefly overview them focusing on PI3Kα and the interactions with alpelisib.
PI3Kα ATP binding site and inhibitor interactions
We illustrate the difficulty in optimizing an ATP-competitive inhibitor specifically to the PI3Kα isoform. The ATP binding sites of class I PI3Ks are highly homologous, differing in only a few residues at the rim. The respective residues have been grouped into two regions, Regions 1 and 2.69,70 Non-conserved residues in the two regions can aid selectivity. The first non-conserved Region is near the hinge (four residues). The second less variable is at the P-loop. The chemical and conformational differences of these regions among the isoforms define their distinct interactions and have been meticulously probed with the hope that such analyses would help in improving inhibitor specificity.35 The ATP binding site in PI3K isoforms is between the two lobes of the kinase domain, which are separated by a hinge. This is where the adenine ring of ATP and the inhibitors' rings anchor. The hydrophobic residues are conserved, but not the neighboring ones. The so-called specificity pocket shows certain isoform dependent affinity differences.71 A conformational change of the P-loop upon inhibitor binding exposes it.72–74 However, the residues involved are conserved even though those around the pocket are not and may be at play in inhibitor design. There are also differences in the conformational flexibilities. Nonetheless, the opening of the specificity pocket does not appear as determining inhibitor selectivity. ATP also does not penetrate the affinity pocket, which is encircled by the conserved residues.75 Despite this, inhibitors entering the pocket have higher potency. P-loop residues play a role in PI3Kα selectivity for GDC-0941,76 which was subsequently converted into a more selective covalent drug.77 For PI3Kα the first non-conserved region appears the key. An inhibitor that extends towards Region 1 (benzoxazepine) adjusts selectivity,76,78,79 leading to GDC-0326,67 with the drug's carboxamide interacting with Gln859, and Ser854 H-bonding the drug as well.67 Additional drugs and their interactions have also been described.68
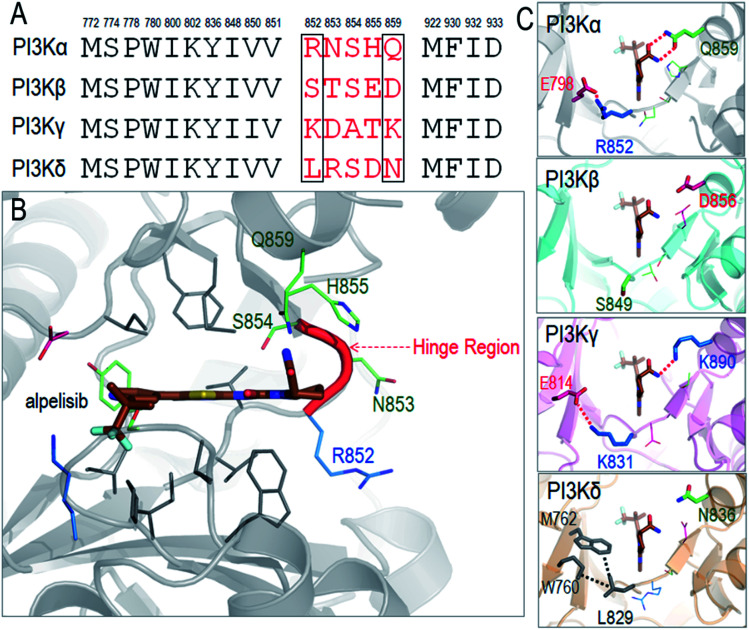
As a detailed example we describe alpelisib, the first drug to be approved by the FDA. The crystal structure of PI3Kα with alpelisib has been solved (PDB ID 4JPS). Alpelisib binds to the ATP pocket in the kinase domain, with massive contacts with the P-loop and hinge region. Due to the high sequence similarity in PI3K isoforms, most alpelisib-contacting residues in PI3Kα are conserved, with only five variable residues (852RNSH855 and Q859) in the hinge region (Fig. 3A and B). Here, we focus on two residues, R852 and Q859, since the side chains of the other three residues (NSH) are exposed to solvent and barely involve in the interactions with alpelisib. Q859 of PI3Kα forms dual hydrogen bonds with alpelisib (Fig. 3C). Its importance in isoform selectivity has been indicated and confirmed.80,81 This residue becomes shorter in PI3Kβ (D856) and PI3Kδ (N836), while the K890 in PI3Kγ is still long enough for the hydrogen bond interaction with alpelisib (Fig. 3D–F). R852 in PI3Kα forms a salt bridge with E798 in the kinase domain's N-lobe. A similar salt bridge is observed in PI3Kγ (K831-E814). L829 of PI3Kγ also establishes hydrophobic interactions with M762 and W760. However, PI3Kβ is quite different with no favorable interaction of S849. This indicates that PI3Kγ resembles more PI3Kα in those residue contacts, followed by PI3Kδ and PI3Kβ. It is interesting that the structural similarity of the hinge region relates to the selectivity of alpelisib to PI3K isoforms. The IC50 values for PI3Kα (∼5 nM) is highest, followed by PI3Kγ (∼250 nM), PI3Kδ (∼290 nM) and PI3Kβ (∼1200 nM).81 This emphasizes the significance of the hinge region in mediating the isoform selectivity.
Fig. 3. Structural insights into selectivity of alpelisib in PI3K isoforms. (A) The sequence alignment indicates the conserved and variable residues in PI3K for the interactions with alpelisib. (B) The structure of alpelisib interacting with the ATP pocket in PI3Kα (PDB ID 4JPS). (C) The structural comparison of the hinge regions in PI3K isoforms (PDB ID PI3Kβ-; 2Y3A; PI3Kγ-; 1E8X; PI3Kδ-; 4XEO). The alpelisib conformations in PI3K isoforms are modeled based on crystal structure of PI3Kα with alpelisib (PDB ID ; 4JPS).
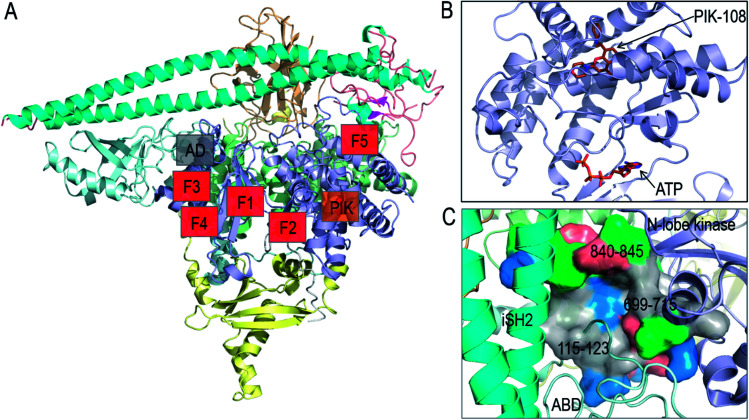
In vitro screening of oncogenic mutants has been carried out and micromolar range binders have been identified.82 However, they are not mutant-selective. A non-ATP allosteric pocket has been identified in a crystal structure of PI3Kα (PDB ; 4A55). It has been further studied by computational simulations.83 Still, currently its efficacy has been unclear, leading the authors to conclude that this binding site is unlikely to yield productive allosteric inhibitors. Fragment-based screening was also carried out to identify the allosteric inhibitor pockets with promising results27 with phosphopeptide binding at the E542K and E545K hotspots. These allosteric pockets are summarized in Fig. 4A and B. Here, we suggest another allosteric pocket. The oncogenic mutations in C2 and ABD suggest the flexible iSH2 in the activated PI3Kα, which has been observed in the simulations of wild type PI3Kα and the E545K mutant.15,84 The movement of ABD is coupled with the interacting iSH2 domain in PI3Kα.85 The more stable ABD is expected to interfere with PI3Kα activation and increase the population of the inactive PI3Kα conformation in the ensemble. By visualizing the PI3Kα structure, a pocket was identified between the ABD and the kinase domain's N-lobe. The pocket is deep with considerable hydrophobicity (Fig. 4C). The involved residues include 115–123 in the ABD–RBD linker, and 699–715 and 840–845 in the kinase domain's N-lobe. An inhibitor targeting the pocket is expected to enhance the ABD–kinase domain interface, rendering a higher stability of ABD and iSH2 to restrict the PI3Kα inactive conformation and target PI3Kα with ABD mutations.
Fig. 4. Summary of PI3Kα allosteric inhibitor pockets. (A) The full structure of PI3Kα with the allosteric pocket highlighted. The allosteric pocket for PIK-108 (PIK in the figure) is identified from the crystal structure of PI3Kα (PDB ID 4A55), the allosteric pockets F1–F5 are obtained from fragment-based screening27 and the allosteric pocket between ABD and kinase domain (AD in the figure) is proposed in the work. (B) The snapshot of PIK-108 interacting with a non-APT pocket in the C-lobe of kinase domain. The ATP is modeled based on the PI3K structure (PDB ID ; 1E8X). (C) The snapshot of the proposed allosteric pocket between ABD and N-lobe kinase domain.
Orthosteric drug design focused on the ATP binding site, with attempts to obtain highly isoform specific inhibitors. However, they encountered an almost impregnable hurdle: the near identity of the ATP binding sites across isoforms. At the same time, designing drugs to stably bind at the substrate binding site is difficult. With part of the PIP2 lipid anchored in the membrane with high avidity, the substrate binding site is shallow. The membrane-buried protein surface further constrains small molecule access. Currently available inhibitors mimicking ATP require high concentrations, resulting in toxicities. The frequent emerging deletion or mutations of PTEN, which suppresses cancer by dephosphorylating the PI3K-phosphorylated lipid PIP3 back to PIP2, which is also with no currently available inhibitors, aggravates the pharmacological scenario. PTEN is highly challenging to target: to function the inhibitor needs to activate, not suppress it. Drug-induced activation of an enzyme is difficult to accomplish.
To-date unexplored PI3Kα therapies
We propose exploring two new strategies for PI3Kα inhibition: (i) combination of allosteric and orthosteric drugs and (ii) taking advantage of rescue mutations to guide drug discovery. They can also be merged. Below, we describe the background, rationale, and map a strategy for their implementation.
(i) Allosteric plus orthosteric drugs
A few years ago, a remarkable observation was made: a Cys to Tyr mutation (C1156Y) in metastatic anaplastic lymphoma kinase (ALK) desensitized it to crizotinib, an ATP-competitive drug that was given earlier. Lorlatinib was then prescribed; however, drug resistance emerged. Sequencing indicated that an emerging allosteric mutation involving L1198F substitution conferred resistance to lorlatinib, also an ATP-competitive small-molecule inhibitor of ALK and the related tyrosine kinase ROS1. The L1198F mutation led to steric interference with lorlatinib binding. ALK L1198F has been reported as an ALK gain-of-function mutation. However, remarkably the allosteric L1198F mutation resensitized ALK to crizotinib. The patient harboring this mutation received crizotinib again, and the cancer-related symptoms abated.86 In this example L1198F is a rescue mutation. The mutation redistributed the ALK conformational ensemble, now re-populating a crizotinib binding-favored state. In line with this observation, loss of G2032R resistance mutation upon chemotherapy treatment enabled successful crizotinib rechallenge in a patient with ROS1-rearranged cancer.87 Thus, once the mutation occurred, there is no need for an allosteric drug that would mimic its action to combine with the ATP-competitive crizotinib. Had there not been such a mutation at that position, an allosteric drug mimicking it could conceivably accomplish this role, promoting a drug-accommodating conformational change at the ATP binding site. Drug resistance often emerges from conformational changes.88 Allosteric drugs can resensitize an active site in the same way that an allosteric mutation does, restoring the actions of orthosteric drugs. As we describe below, experiments in Nathaniel Gray's Lab on BCR-ABL kinase toward treatment of chronic myeloid leukemia (CML) along with subsequent work provide an outstanding example.
Imatinib, an orthosteric ATP-competitive drug, can inhibit oncogenic mutants of BCR-ABL. However, mutations often affect BCR-ABL's ATP-binding site or the activation loop can result in drug resistance. Among these, T315 substitutions that elicit steric clashes with the drug are particularly potent. By screening drug libraries the Gray team first identified an allosteric drug GNF-2 with similar antiproliferative outcome as the orthosteric drug.90 Subsequent optimization obtained the allosteric GNF-5 compound.90 Remarkably, like the L1198F rescue mutation in ALK, the GNF drug series shifted the protein ensemble toward an active site that re-sensitized BCR-ABL to imatinib and nilotinib. NMR, crystallography, mutagenesis and hydrogen exchange mass spectrometry all indicated that the drug binds to the C-terminal myristate pocket of BCR-ABL. The structure unraveled formation of considerable hydrophobic as well as vdW interactions and water-mediated H-bonds, indicating how the combination of the GNF-series allosteric drug collaborates with the ATP-competitive imatinib, overcoming the T315 drug-resistant hotspots.89,90
ABL001 (asciminib) is also a potent and selective allosteric ABL1 inhibitor that binds to the myristoyl pocket of ABL1 in a manner resembling the GNF-series. It interacts with active site residues via the same type of interactions as the GNF inhibitors, and shifts the ensemble toward the inactive kinase conformation. Each drug alone, ABL001 and the catalytic ATP-competitive inhibitor nilotinib are blocked by distinct drug resistant mutations. However, as above, their combination eradicates the tumors.91 Thus, even though mutations at the myristate site can confer resistance to ABL001, they are sensitive to nilotinib or other ATP-competitive inhibitors making their combination (or ABL001 with other ATP-competitive drugs, like imatinib or dasatinib) a viable BCR-ABL inhibition strategy.92 Combining the allosteric inhibitor asciminib with ATP-competitive ponatinib was recently found to even more powerfully suppress the emergence of mutants (Y253H and E255V) and restore efficacy against highly resistant BCR-ABL1 mutants.93 Additional examples of such combinations are available.94,95
The concept of allosteric drug working conjointly with an orthosteric drug is not new. We only need to consider that a ‘ligand’ can be an orthosteric drug, with the allosteric drug acting to modulate it.96 This powerful combinatorial concept has shown promise for the protein kinases above; but has not been tested for PI3Kα. Allosteric drugs may overcome PI3Kα drug resistance by altering the ATP binding site and resensitizing it, i.e. making it accessible to sterically blocked ATP-competitive orthosteric drugs, or by producing an altered, druggable PIP2 binding site. Cross-correlations of the motions and analysis of the conformational ensembles in long timescale molecular dynamics simulations may be one way to help identify likely targetable allosteric sites.
(ii) Rescue mutations provide a powerful concept
That mutations can rescue, or resensitize, a mutant protein to an allosteric drug has been shown for a number of diseases. As such they have been dubbed “natural gene therapy” and “nature's proof of principle that you can fix those mutations” in the case of Wiskott–Aldrich syndrome (WAS).97 Additional examples include the p.Q188R mutation in classic galactosemia98 that gives rise to a protein with lower conformational stability and lower catalytic activity99 and p53.100 Drugs mimicking the allosteric effects of these mutants may rescue function. To the best of our knowledge, in practice this however has not been followed up, likely due to the design complexity which is involved. Nonetheless, identifying them in protein structures, and their potential substitutions will provide allosteric sites and potentially certain orthosteric-drug favored conformational changes. Here we propose attempting this strategy in targeting PI3Kα.
How to a priori identify the positions of such mutations? Earlier we have taken this up for von Hippel–Lindau tumor suppressor protein (pVHL).101 To identify possible candidate residues, we used molecular dynamics simulations, seeking the least stable regions in the unbound state of the protein. This flagged the interface between the two domains. We then designed several stabilizing mutations at the interdomain interface and in the domains. Experiments showed that disease mutant Y98N at the HIF binding site destabilizes pVHL and lowers its affinity to hypoxia-inducible factor (HIF). pVHL is part of the E3 ubiquitin ligase complex that regulates the degradation of HIF. The simulations indicated that pVHL stability and binding affinity are allosterically regulated. We thus sought to allosterically stabilize pVHL and its interactions with mutations away from binding sites. The mutations were selected based on sequence alignment with a pVHL from other species (worm pVHL sequence). The worm-based substitutions stabilized pVHL, its Y98N mutant, and the complexes of pVHL with HIF, lowering its binding free energy, and the Y98N mutant with elongin C complex, thus rescuing pVHL–HIF affinity and function. The reason for choosing the worm sequence was our observation that the worm p53 is significantly more stable than the human form.102 In PI3Kα, rescuing mutations could improve the affinity of the ATP-competitive drug to its binding site without increasing its concentration, by allosterically altering its conformation. The mutations in the kinase domain's P-loop may be promising. Such a strategy may also be considered for PTEN tumor suppressor.
Alternative strategies and considerations to map and harness allosteric candidates to improve PI3Kα isoform-specific inhibitor design were also proposed.103 Key steps in successful design of allosteric drugs include identification of allosteric sites103–105 including hidden cryptic allosteric pockets106,107 shown to underlie allosteric modulator selectivity at muscarinic GPCRs,108 allosteric mutations19,109–118 and allosteric drug design.119,120 Allosteric drugs can constitute ‘anchors’ and ‘drivers’.103 The anchor docks into an allosteric pocket whose conformation is unchanged during the transition between the inactive and active states. The driver 'pulls' and/or 'pushes' protein atoms (side-chain or backbone) and triggers shifts of the receptor population to a drug binding-favored state. Based on crystal structures, characteristics of anchors and drivers have been outlined. The recent computational tools quantifying the allosteric signaling, mutation effects and their combinational actions in the signaling promote the discovery of allosteric effectors.121,122
Conclusions
To understand the behavior of biomacromolecules we need to look at the distributions of their conformational ensembles and how these are affected by their environments. Under physiological conditions PI3Kα populates the inactive state. Oncogenic mutations will shift the ensemble to populate the active state. The extent of the shift determines the strength of the mutation. The chemistry of the surface matters: the membrane surface is negatively charged, with lipid hydrophobicity playing a major role. Mutations that contribute positive charge will enhance the interactions. Mutations (orthosteric or allosteric) that weaken the interactions of p85α with p110α or between the p85α domains or strengthen the interaction with the membrane will shift the population toward the active state. Double (strong and weak) mutations can act to more fully shift the population. Drug resistance mutations can alter the ATP binding site, shifting the ensemble toward orthosteric drug-inaccessible state. Allosteric modulator drug can shift the population toward drug accessibility. Rescue mutations can identify regulatory hot spots that nature uses for functional control. Small molecule allosteric inhibitors can take aim at such hot spots in tough-to-drug targets such as kinases and phosphatases, like the SHP2.
Here we described the PI3Kα saga from the free energy landscape standpoint123 and its dynamic shifts following changes in the environment.110,124–126 Allostery is a powerful ensemble property used by nature. Harnessing it to favor an ATP-competitive drug exploits “nature's proof of principle” may be one way to target PI3Kα, the second most highly mutated protein in cancer. Here we proposed two new possible venues not exploited yet in PI3Kα drug discovery: allosteric plus orthosteric drug combinations and following mother nature rescue mutation strategy.
Conflicts of interest
No potential conflict of interest was disclosed.
Acknowledgments
This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract HHSN26120080001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This research was supported [in part] by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
References
- Carrera A. C., Anderson R. J. Cell Sci. 2019;132:jcs228395. doi: 10.1242/jcs.228395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap T. A., Bjerke L., Clarke P. A., Workman P. Curr. Opin. Pharmacol. 2015;23:98–107. doi: 10.1016/j.coph.2015.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klempner S. J., Myers A. P., Cantley L. C. Cancer Discovery. 2013;3:1345–1354. doi: 10.1158/2159-8290.CD-13-0063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- S. I. C. database, http://www.sanger.ac.uk/genetics/CGP/cosmic/.
- Wong K. K., Engelman J. A., Cantley L. C. Curr. Opin. Genet. Dev. 2010;20:87–90. doi: 10.1016/j.gde.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtney K. D., Corcoran R. B., Engelman J. A. J. Clin. Oncol. 2010;28:1075–1083. doi: 10.1200/JCO.2009.25.3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke P. A., Workman P. J. Clin. Oncol. 2012;30:331–333. doi: 10.1200/JCO.2011.38.7167. [DOI] [PubMed] [Google Scholar]
- Lawrence M. S., Stojanov P., Mermel C. H., Robinson J. T., Garraway L. A., Golub T. R., Meyerson M., Gabriel S. B., Lander E. S., Getz G. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Vogt P. K. Proc. Natl. Acad. Sci. U. S. A. 2008;105:2652–2657. doi: 10.1073/pnas.0712169105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gymnopoulos M., Elsliger M. A., Vogt P. K. Proc. Natl. Acad. Sci. U. S. A. 2007;104:5569–5574. doi: 10.1073/pnas.0701005104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke J. E., Perisic O., Masson G. R., Vadas O., Williams R. L. Proc. Natl. Acad. Sci. U. S. A. 2012;109:15259–15264. doi: 10.1073/pnas.1205508109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C. H., Mandelker D., Schmidt-Kittler O., Samuels Y., Velculescu V. E., Kinzler K. W., Vogelstein B., Gabelli S. B., Amzel L. M. Science. 2007;318:1744–1748. doi: 10.1126/science.1150799. [DOI] [PubMed] [Google Scholar]
- Markham A. Drugs. 2019;79:1249–1253. doi: 10.1007/s40265-019-01161-6. [DOI] [PubMed] [Google Scholar]
- Vasan N., Razavi P., Johnson J. L., Shao H., Shah H., Antoine A., Ladewig E., Gorelick A., Lin T. Y., Toska E., Xu G., Kazmi A., Chang M. T., Taylor B. S., Dickler M. N., Jhaveri K., Chandarlapaty S., Rabadan R., Reznik E., Smith M. L., Sebra R., Schimmoller F., Wilson T. R., Friedman L. S., Cantley L. C., Scaltriti M., Baselga J. Science. 2019;366:714–723. doi: 10.1126/science.aaw9032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M. Z., Jang H., Nussinov R. Chem. Sci. 2019;10:3671–3680. doi: 10.1039/c8sc04498h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M. Z., Jang H., Nussinov R. Phys. Chem. Chem. Phys. 2019;21:12021–12028. doi: 10.1039/c9cp00101h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussinov R., Tsai C. J., Jang H. Front. Oncol. 2019;9:1231. doi: 10.3389/fonc.2019.01231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai C. J., Nussinov R. PLoS Comput. Biol. 2014;10:e1003394. doi: 10.1371/journal.pcbi.1003394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Nussinov R. PLoS Comput. Biol. 2016;12:e1004966. doi: 10.1371/journal.pcbi.1004966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussinov R. Chem. Rev. 2016;116:6263–6266. doi: 10.1021/acs.chemrev.6b00283. [DOI] [PubMed] [Google Scholar]
- Boehr D. D., Nussinov R., Wright P. E. Nat. Chem. Biol. 2009;5:789–796. doi: 10.1038/nchembio.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M. S., Schmidt-Kittler O., Bolduc D. M., Brower E. T., Chaves-Moreira D., Allaire M., Kinzler K. W., Jennings I. G., Thompson P. E., Cole P. A., Amzel L. M., Vogelstein B., Gabelli S. B. Oncotarget. 2014;5:5198–5208. doi: 10.18632/oncotarget.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hon W. C., Berndt A., Williams R. L. Oncogene. 2012;31:3655–3666. doi: 10.1038/onc.2011.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Wjasow C., Backer J. M. J. Biol. Chem. 1998;273:30199–30203. doi: 10.1074/jbc.273.46.30199. [DOI] [PubMed] [Google Scholar]
- Zhang X. X., Vadas O., Perisic O., Anderson K. E., Clark J., Hawkins P. T., Stephens L. R., Williams R. L. Mol. Cell. 2011;41:567–578. doi: 10.1016/j.molcel.2011.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S. Y., Huang W. K., Wang Q., Shen Q. C., Li S., Nussinov R., Zhang J. Biophys. J. 2015;108:528a. [Google Scholar]
- Miller M. S., Maheshwari S., McRobb F. M., Kinzler K. W., Amzel L. M., Vogelstein B., Gabelli S. B. Bioorg. Med. Chem. 2017;25:1481–1486. doi: 10.1016/j.bmc.2017.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X., Smaill J. B., Ding K. Angew. Chem., Int. Ed. Engl. 2020;59:2–15. doi: 10.1002/anie.201914525. [DOI] [PubMed] [Google Scholar]
- Leroux A. E., Gross L. Z. F., Sacerdoti M., Biondi R. M. Adv. Exp. Med. Biol. 2019;1163:279–311. doi: 10.1007/978-981-13-8719-7_12. [DOI] [PubMed] [Google Scholar]
- Cheng X., Jiang H. Adv. Exp. Med. Biol. 2019;1163:1–23. doi: 10.1007/978-981-13-8719-7_1. [DOI] [PubMed] [Google Scholar]
- Nussinov R., Tsai C. J. Annu. Rev. Pharmacol. Toxicol. 2015;55:249–267. doi: 10.1146/annurev-pharmtox-010814-124401. [DOI] [PubMed] [Google Scholar]
- Lu S., Qiu Y., Ni D., He X., Pu J., Zhang J. Drug Discovery Today. 2020;25:177–184. doi: 10.1016/j.drudis.2019.10.006. [DOI] [PubMed] [Google Scholar]
- Zorba A., Nguyen V., Koide A., Hoemberger M., Zheng Y., Kutter S., Kim C., Koide S., Kern D. Proc. Natl. Acad. Sci. U. S. A. 2019;116:13937–13942. doi: 10.1073/pnas.1906024116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Q., Villar M. T., Artigues A., Thyfault J. P., Apte U., Zhu H., Peterson K. R., Fenton A. W. Sci. Rep. 2019;9:9031. doi: 10.1038/s41598-019-45202-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garces A. E., Stocks M. J. J. Med. Chem. 2019;62:4815–4850. doi: 10.1021/acs.jmedchem.8b01492. [DOI] [PubMed] [Google Scholar]
- Yang J., Nie J., Ma X. L., Wei Y. Q., Peng Y., Wei X. W. Mol. Cancer. 2019;18:26. doi: 10.1186/s12943-019-0954-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janku F., Yap T. A., Meric-Bernstam F. Nat. Rev. Clin. Oncol. 2018;15:273–291. doi: 10.1038/nrclinonc.2018.28. [DOI] [PubMed] [Google Scholar]
- Arafeh R., Samuels Y. Semin. Cancer Biol. 2019;59:36–49. doi: 10.1016/j.semcancer.2019.02.002. [DOI] [PubMed] [Google Scholar]
- Baselga J., Im S. A., Iwata H., Cortes J., De Laurentiis M., Jiang Z., Arteaga C. L., Jonat W., Clemons M., Ito Y., Awada A., Chia S., Jagiello-Gruszfeld A., Pistilli B., Tseng L. M., Hurvitz S., Masuda N., Takahashi M., Vuylsteke P., Hachemi S., Dharan B., Di Tomaso E., Urban P., Massacesi C., Campone M. Lancet Oncol. 2017;18:904–916. doi: 10.1016/S1470-2045(17)30376-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Leo A., Johnston S., Lee K. S., Ciruelos E., Lonning P. E., Janni W., O'Regan R., Mouret-Reynier M. A., Kalev D., Egle D., Csoszi T., Bordonaro R., Decker T., Tjan-Heijnen V. C. G., Blau S., Schirone A., Weber D., El-Hashimy M., Dharan B., Sellami D., Bachelot T. Lancet Oncol. 2018;19:87–100. doi: 10.1016/S1470-2045(17)30688-5. [DOI] [PubMed] [Google Scholar]
- Martin M., Chan A., Dirix L., O'Shaughnessy J., Hegg R., Manikhas A., Shtivelband M., Krivorotko P., Batista Lopez N., Campone M., Ruiz Borrego M., Khan Q. J., Beck J. T., Ramos Vazquez M., Urban P., Goteti S., Di Tomaso E., Massacesi C., Delaloge S. Ann. Oncol. 2017;28:313–320. doi: 10.1093/annonc/mdw562. [DOI] [PubMed] [Google Scholar]
- Krop I. E., Mayer I. A., Ganju V., Dickler M., Johnston S., Morales S., Yardley D. A., Melichar B., Forero-Torres A., Lee S. C., de Boer R., Petrakova K., Vallentin S., Perez E. A., Piccart M., Ellis M., Winer E., Gendreau S., Derynck M., Lackner M., Levy G., Qiu J., He J., Schmid P. Lancet Oncol. 2016;17:811–821. doi: 10.1016/S1470-2045(16)00106-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuylsteke P., Huizing M., Petrakova K., Roylance R., Laing R., Chan S., Abell F., Gendreau S., Rooney I., Apt D., Zhou J., Singel S., Fehrenbacher L. Ann. Oncol. 2016;27:2059–2066. doi: 10.1093/annonc/mdw320. [DOI] [PubMed] [Google Scholar]
- Tolaney S., Burris H., Gartner E., Mayer I. A., Saura C., Maurer M., Ciruelos E., Garcia A. A., Campana F., Wu B., Xu Y., Jiang J., Winer E., Krop I. Breast Canc. Res. Treat. 2015;149:151–161. doi: 10.1007/s10549-014-3248-4. [DOI] [PubMed] [Google Scholar]
- Abramson V. G., Supko J. G., Ballinger T., Cleary J. M., Hilton J. F., Tolaney S. M., Chau N. G., Cho D. C., Pearlberg J., Lager J., Shapiro G. I., Arteaga C. L. Clin. Cancer Res. 2017;23:3520–3528. doi: 10.1158/1078-0432.CCR-16-1764. [DOI] [PubMed] [Google Scholar]
- Wheler J., Mutch D., Lager J., Castell C., Liu L., Jiang J., Traynor A. M. Oncologist. 2017;22:377-e37. doi: 10.1634/theoncologist.2016-0257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soria J. C., LoRusso P., Bahleda R., Lager J., Liu L., Jiang J., Martini J. F., Mace S., Burris H. Oncologist. 2015;20:245–246. doi: 10.1634/theoncologist.2014-0449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patnaik A., Appleman L. J., Tolcher A. W., Papadopoulos K. P., Beeram M., Rasco D. W., Weiss G. J., Sachdev J. C., Chadha M., Fulk M., Ejadi S., Mountz J. M., Lotze M. T., Toledo F. G. S., Chu E., Jeffers M., Pena C., Xia C., Reif S., Genvresse I., Ramanathan R. K. Ann. Oncol. 2016;27:1928–1940. doi: 10.1093/annonc/mdw282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyling M., Santoro A., Mollica L., Leppa S., Follows G. A., Lenz G., Kim W. S., Nagler A., Panayiotidis P., Demeter J., Ozcan M., Kosinova M., Bouabdallah K., Morschhauser F., Stevens D. A., Trevarthen D., Giurescu M., Cupit L., Liu L., Kochert K., Seidel H., Pena C., Yin S. X., Hiemeyer F., Garcia-Vargas J., Childs B. H., Zinzani P. L. J. Clin. Oncol. 2017;35:3898–3905. doi: 10.1200/JCO.2017.75.4648. [DOI] [PubMed] [Google Scholar]
- Hong D. S., Bowles D. W., Falchook G. S., Messersmith W. A., George G. C., O'Bryant C. L., Vo A. C. H., Klucher K., Herbst R. S., Eckhardt S. G., Peterson S., Hausman D. F., Kurzrock R., Jimeno A. Clin. Cancer Res. 2012;18:4173–4182. doi: 10.1158/1078-0432.CCR-12-0714. [DOI] [PubMed] [Google Scholar]
- Bowles D. W., Ma W. W., Senzer N., Brahmer J. R., Adjei A. A., Davies M., Lazar A. J., Vo A., Peterson S., Walker L., Hausman D., Rudin C. M., Jimeno A. Br. J. Cancer. 2013;109:1085–1092. doi: 10.1038/bjc.2013.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yam C., Xu X. W., Davies M. A., Gimotty P. A., Morrissette J. J. D., Tetzlaff M. T., Wani K. M., Liu S. J., Deng W. L., Buckley M., Zhao J. H., Amaravadi R. K., Haas N. B., Kudchadkar R. R., Pavlick A. C., Sosman J. A., Tawbi H., Walker L., Schuchter L. M., Karakousis G. C., Gangadhar T. C. Clin. Cancer Res. 2018;24:22–32. doi: 10.1158/1078-0432.CCR-17-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimeno A., Bauman J. E., Weissman C., Adkins D., Schnadig I., Beauregard P., Bowles D. W., Spira A., Levy B., Seetharamu N., Hausman D., Walker L., Rudin C. M., Shirai K. Oral Oncol. 2015;51:383–388. doi: 10.1016/j.oraloncology.2014.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimeno A., Shirai K., Choi M., Laskin J., Kochenderfer M., Spira A., Cline-Burkhardt V., Winquist E., Hausman D., Walker L., Cohen R. B. Ann. Oncol. 2015;26:556–561. doi: 10.1093/annonc/mdu574. [DOI] [PubMed] [Google Scholar]
- Bowles D. W., Senzer N., Hausman D., Peterson S., Vo A., Walker L., Cohen R. B., Jimeno A. Invest. New Drugs. 2014;32:1197–1203. doi: 10.1007/s10637-014-0124-3. [DOI] [PubMed] [Google Scholar]
- Bowles D. W., Kochenderfer M., Cohn A., Sideris L., Nguyen N., Cline-Burkhardt V., Schnadig I., Choi M., Nabell L., Chaudhry A., Ruxer R., Ucar A., Hausman D., Walker L., Spira A., Jimeno A. Clin. Colorectal Cancer. 2016;15:337–344. doi: 10.1016/j.clcc.2016.03.004. [DOI] [PubMed] [Google Scholar]
- Blagden S., Omlin A., Josephs D., Stavraka C., Zivi A., Pinato D. J., Anthoney A., Decordova S., Swales K., Riisnaes R., Pope L., Noguchi K., Shiokawa R., Inatani M., Prince J., Jones K., Twelves C., Spicer J., Banerji U. Clin. Cancer Res. 2014;20:5908–5917. doi: 10.1158/1078-0432.CCR-14-1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevan D., Chiorean E. G., Harris W. B., Von Hoff D. D., Stejskal-Barnett A., Qi W., Anthony S. P., Younger A. E., Rensvold D. M., Cordova F., Shelton C. F., Becker M. D., Garlich J. R., Durden D. L., Ramanathan R. K. Eur. J. Cancer. 2012;48:3319–3327. doi: 10.1016/j.ejca.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janku F. Cancer Treat. Rev. 2017;59:93–101. doi: 10.1016/j.ctrv.2017.07.005. [DOI] [PubMed] [Google Scholar]
- Cheah C. Y., Fowler N. H. Blood. 2016;128:331–336. doi: 10.1182/blood-2016-02-702761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller B. W., Przepiorka D., de Claro R. A., Lee K., Nie L., Simpson N., Gudi R., Saber H., Shord S., Bullock J., Marathe D., Mehrotra N., Hsieh L. S., Ghosh D., Brown J., Kane R. C., Justice R., Kaminskas E., Farrell A. T., Pazdur R. Clin. Cancer Res. 2015;21:1525–1529. doi: 10.1158/1078-0432.CCR-14-2522. [DOI] [PubMed] [Google Scholar]
- Fritsch C., Huang A., Chatenay-Rivauday C., Schnell C., Reddy A., Liu M., Kauffmann A., Guthy D., Erdmann D., De Pover A., Furet P., Gao H., Ferretti S., Wang Y., Trappe J., Brachmann S. M., Maira S. M., Wilson C., Boehm M., Garcia-Echeverria C., Chene P., Wiesmann M., Cozens R., Lehar J., Schlegel R., Caravatti G., Hofmann F., Sellers W. R. Mol. Cancer Ther. 2014;13:1117–1129. doi: 10.1158/1535-7163.MCT-13-0865. [DOI] [PubMed] [Google Scholar]
- Mayer I. A., Abramson V. G., Formisano L., Balko J. M., Estrada M. V., Sanders M. E., Juric D., Solit D., Berger M. F., Won H. H., Li Y., Cantley L. C., Winer E., Arteaga C. L. Clin. Cancer Res. 2017;23:26–34. doi: 10.1158/1078-0432.CCR-16-0134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Geel R. M. J. M., Tabernero J., Elez E., Bendell J. C., Spreafico A., Schuler M., Yoshino T., Delord J. P., Yamada Y., Lolkema M. P., Faris J. E., Eskens F. A. L. M., Sharma S., Yaeger R., Lenz H. J., Wainberg Z. A., Avsar E., Chatterjee A., Jaeger S., Tan E., Maharry K., Demuth T., Schellens J. H. M. Cancer Discovery. 2017;7:610–619. doi: 10.1158/2159-8290.CD-16-0795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S., Shah A. N., Santa-Maria C. A., Siziopikou K., Rademaker A., Helenowski I., Cristofanilli M., Gradishar W. J. Breast Cancer Res. Treat. 2018;171:371–381. doi: 10.1007/s10549-018-4792-0. [DOI] [PubMed] [Google Scholar]
- Juric D., de Bono J. S., LoRusso P. M., Nemunaitis J., Heath E. I., Kwak E. L., Macarulla Mercade T., Geuna E., Jose de Miguel-Luken M., Patel C., Kuida K., Sankoh S., Westin E. H., Zohren F., Shou Y., Tabernero J. Clin. Cancer Res. 2017;23:5015–5023. doi: 10.1158/1078-0432.CCR-16-2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arafeh R., Samuels Y. Semin. Cancer Biol. 2019;59:36–49. doi: 10.1016/j.semcancer.2019.02.002. [DOI] [PubMed] [Google Scholar]
- Miller M. S., Thompson P. E., Gabelli S. B. Biomolecules. 2019;9:82. doi: 10.3390/biom9030082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazzetto M., Suphioglu C., Zhu J., Schmidt-Kittler O., Jennings I. G., Cranmer S. L., Jackson S. P., Kinzler K. W., Vogelstein B., Thompson P. E. Biochem. J. 2008;414:383–390. doi: 10.1042/BJ20080512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z., Amran S. I., Thompson P. E., Jennings I. G. Mol. Pharmacol. 2011;80:657–664. doi: 10.1124/mol.111.072546. [DOI] [PubMed] [Google Scholar]
- Knight Z. A., Gonzalez B., Feldman M. E., Zunder E. R., Goldenberg D. D., Williams O., Loewith R., Stokoe D., Balla A., Toth B., Balla T., Weiss W. A., Williams R. L., Shokat K. M. Cell. 2006;125:733–747. doi: 10.1016/j.cell.2006.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans C. A., Liu T., Lescarbeau A., Nair S. J., Grenier L., Pradeilles J. A., Glenadel Q., Tibbitts T., Rowley A. M., DiNitto J. P., Brophy E. E., O'Hearn E. L., Ali J. A., Winkler D. G., Goldstein S. I., O'Heam P., Martin C. M., Hoyt J. G., Soglia J. R., Cheung C., Pink M. M., Proctor J. L., Palombella V. J., Tremblay M. R., Castro A. C. ACS Med. Chem. Lett. 2016;7:862–867. doi: 10.1021/acsmedchemlett.6b00238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Certal V., Carry J. C., Halley F., Virone-Oddos A., Thompson F., Filoche-Romme B., El-Ahmad Y., Karlsson A., Charrier V., Delorme C., Rak A., Abecassis P. Y., Amara C., Vincent L., Bonnevaux H., Nicolas J. P., Mathieu M., Bertrand T., Marquette J. P., Michot N., Benard T., Perrin M. A., Lemaitre O., Guerif S., Perron S., Monget S., Gruss-Leleu F., Doerflinger G., Guizani H., Brollo M., Delbarre L., Bertin L., Richepin P., Loyau V., Garcia-Echeverria C., Lengauer C., Schio L. J. Med. Chem. 2014;57:903–920. doi: 10.1021/jm401642q. [DOI] [PubMed] [Google Scholar]
- Zheng Z. H., Miller M. S., Jennings I. G., Thompson P. E. ACS Chem. Biol. 2013;8:679–683. doi: 10.1021/cb300666s. [DOI] [PubMed] [Google Scholar]
- Berndt A., Miller S., Williams O., Le D. D., Houseman B. T., Pacold J. I., Gorrec F., Hon W. C., Liu Y., Rommel C., Gaillard P., Ruckle T., Schwarz M. K., Shokat K. M., Shaw J. P., Williams R. L. Nat. Chem. Biol. 2010;6:117–124. doi: 10.1038/nchembio.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heffron T. P., Wei B. Q., Olivero A., Staben S. T., Tsui V., Do S., Dotson J., Folkes A. J., Goldsmith P., Goldsmith R., Gunzner J., Lesnick J., Lewis C., Mathieu S., Nonomiya J., Shuttleworth S., Sutherlin D. P., Wan N. C., Wang S. M., Wiesmann C., Zhu B. Y. J. Med. Chem. 2011;54:7815–7833. doi: 10.1021/jm2007084. [DOI] [PubMed] [Google Scholar]
- Nacht M., Qiao L. X., Sheets M. P., St Martin T., Labenski M., Mazdiyasni H., Karp R., Zhu Z. D., Chaturvedi P., Bhavsar D., Niu D. Q., Westlin W., Petter R. C., Medikonda A. P., Singh J. J. Med. Chem. 2013;56:712–721. doi: 10.1021/jm3008745. [DOI] [PubMed] [Google Scholar]
- Staben S. T., Ndubaku C., Blaquiere N., Belvin M., Bull R. J., Dudley D., Edgar K., Gray D., Heald R., Heffron T. P., Jones G. E., Jones M., Kolesnikov A., Lee L., Lesnick J., Lewis C., Murray J., McLean N. J., Nonomiya J., Olivero A. G., Ord R., Pang J., Price S., Prior W. W., Rouge L., Salphati L., Sampath D., Wallin J., Wang L., Wei B., Weismann C., Wu P. Bioorg. Med. Chem. Lett. 2013;23:2606–2613. doi: 10.1016/j.bmcl.2013.02.102. [DOI] [PubMed] [Google Scholar]
- Ndubaku C. O., Heffron T. P., Staben S. T., Baumgardner M., Blaquiere N., Bradley E., Bull R., Do S., Dotson J., Dudley D., Edgar K. A., Friedman L. S., Goldsmith R., Heald R. A., Kolesnikov A., Lee L., Lewis C., Nannini M., Nonomiya J., Pang J., Price S., Prior W. W., Salphati L., Sideris S., Wallin J. J., Wang L., Wei B., Sampath D., Olivero A. G. J. Med. Chem. 2013;56:4597–4610. doi: 10.1021/jm4003632. [DOI] [PubMed] [Google Scholar]
- Zheng Z. H., Amran S. I., Zhu J. X., Schmidt-Kittler O., Kinzler K. W., Vogelstein B., Shepherd P. R., Thompson P. E., Jennings I. G. Biochem. J. 2012;444:529–535. doi: 10.1042/BJ20120499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furet P., Guagnano V., Fairhurst R. A., Imbach-Weese P., Bruce I., Knapp M., Fritsch C., Blasco F., Blanz J., Aichholz R., Hamon J., Fabbro D., Caravatti G. Bioorg. Med. Chem. Lett. 2013;23:3741–3748. doi: 10.1016/j.bmcl.2013.05.007. [DOI] [PubMed] [Google Scholar]
- Wang J., Gong G. Q., Zhou Y., Lee W. J., Buchanan C. M., Denny W. A., Rewcastle G. W., Kendall J. D., Dickson J. M. J., Flanagan J. U., Shepherd P. R., Yang D. H., Wang M. W. Acta Pharmacol. Sin. 2018;39:1816–1822. doi: 10.1038/s41401-018-0057-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gkeka P., Papafotika A., Christoforidis S., Cournia Z. J. Phys. Chem. B. 2015;119:1002–1016. doi: 10.1021/jp506423e. [DOI] [PubMed] [Google Scholar]
- Galdadas I., Gervasio F. L., Cournia Z. Chem. Sci. 2020;11:3511–3515. doi: 10.1039/c9sc05903b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leontiadou H., Galdadas I., Athanasiou C., Cournia Z. Sci. Rep. 2018;8:15544. doi: 10.1038/s41598-018-27044-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw A. T., Friboulet L., Leshchiner I., Gainor J. F., Bergqvist S., Brooun A., Burke B. J., Deng Y. L., Liu W., Dardaei L., Frias R. L., Schultz K. R., Logan J., James L. P., Smeal T., Timofeevski S., Katayama R., Iafrate A. J., Le L., McTigue M., Getz G., Johnson T. W., Engelman J. A. N. Engl. J. Med. 2016;374:54–61. doi: 10.1056/NEJMoa1508887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michels S., Scheffler M., Wagener S., Plenker D., Scheel A., Nogova L., Schultheis A., Fischer R. N., Abdulla D. S. Y., Riedel R., Bunck A., Kobe C., Baus W., Merkelbach-Bruse S., Sos M. L., Buttner R., Wolf J. JCO Precis Oncol. 2018;2:1–6. doi: 10.1200/PO.18.00121. [DOI] [PubMed] [Google Scholar]
- Rotow J., Bivona T. G. Nat. Rev. Cancer. 2017;17:637–658. doi: 10.1038/nrc.2017.84. [DOI] [PubMed] [Google Scholar]
- Adrian F. J., Ding Q., Sim T. B., Velentza A., Sloan C., Liu Y., Zhang G. B., Hur W., Ding S., Manley P., Mestan J., Fabbro D., Gray N. S. Nat. Chem. Biol. 2006;2:95–102. doi: 10.1038/nchembio760. [DOI] [PubMed] [Google Scholar]
- Zhang J. M., Adrian F. J., Jahnke W., Cowan-Jacob S. W., Li A. G., Iacob R. E., Sim T., Powers J., Dierks C., Sun F. X., Guo G. R., Ding Q., Okram B., Choi Y., Wojciechowski A., Deng X. M., Liu G. X., Fendrich G., Strauss A., Vajpai N., Grzesiek S., Tuntland T., Liu Y., Bursulaya B., Azam M., Manley P. W., Engen J. R., Daley G. Q., Warmuth M., Gray N. S. Nature. 2010;463:501–506. doi: 10.1038/nature08675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wylie A. A., Schoepfer J., Jahnke W., Cowan-Jacob S. W., Loo A., Furet P., Marzinzik A. L., Pelle X., Donovan J., Zhu W. J., Buonamici S., Hassan A. Q., Lombardo F., Iyer V., Palmer M., Berellini G., Dodd S., Thohan S., Bitter H., Branford S., Ross D. M., Hughes T. P., Petruzzelli L., Vanasse K. G., Warmuth M., Hofmann F., Keen N. J., Sellers W. R. Nature. 2017;543:733–737. doi: 10.1038/nature21702. [DOI] [PubMed] [Google Scholar]
- Schoepfer J., Jahnke W., Berellini G., Buonamici S., Cotesta S., Cowan-Jacob S. W., Dodd S., Drueckes P., Fabbro D., Gabriel T., Groell J. M., Grotzfeld R. M., Hassan A. Q., Henry C., Iyer V., Jones D., Lombardo F., Loo A., Manley P. W., Pelle X., Rummel G., Salem B., Warmuth M., Wylie A. A., Zoller T., Marzinzik A. L., Furet P. J. Med. Chem. 2018;61:8120–8135. doi: 10.1021/acs.jmedchem.8b01040. [DOI] [PubMed] [Google Scholar]
- Eide C. A., Zabriskie M. S., Stevens S. L. S., Antelope O., Vellore N. A., Than H., Schultz A. R., Clair P., Bowler A. D., Pomicter A. D., Yan D. Q., Senina A. V., Qiang W., Kelley T. W., Szankasi P., Heinrich M. C., Tyner J. W., Rea D., Cayuela J. M., Kim D. W., Tognon C. E., O'Hare T., Druker B. J., Deininger M. W. Cancer Cell. 2019;36:431–433. doi: 10.1016/j.ccell.2019.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X. Y., Yu L., Zhang Z., Ren X. M., Smaill J. B., Ding K. Med. Res. Rev. 2018;38:1550–1581. doi: 10.1002/med.21488. [DOI] [PubMed] [Google Scholar]
- To C., Jang J., Chen T., Park E., Mushajiang M., De Clercq D. J. H., Xu M., Wang S., Cameron M. D., Heppner D. E., Shin B. H., Gero T. W., Yang A., Dahlberg S. E., Wong K. K., Eck M. J., Gray N. S., Janne P. A. Cancer Discovery. 2019;9:926–943. doi: 10.1158/2159-8290.CD-18-0903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdel-Magid A. F. ACS Med. Chem. Lett. 2015;6:104–107. doi: 10.1021/ml5005365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May M. Nat. Med. 2011;17:405–407. doi: 10.1038/nm0411-405. [DOI] [PubMed] [Google Scholar]
- Filteau M., Hamel V., Pouliot M. C., Gagnon-Arsenault I., Dube A. K., Landry C. R. Mol. Syst. Biol. 2015;11:832. doi: 10.15252/msb.20156444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haskovic M., Derks B., van der Ploeg L., Trommelen J., Nyakayiru J., van Loon L. J. C., Mackinnon S., Yue W. W., Peake R. W. A., Zha L., Demirbas D., Qi W. S., Huang X. P., Berry G. T., Achten J., Bierau J., Rubio-Gozalbo M. E., Coelho A. I. Orphanet J. Rare Dis. 2018;13:212. doi: 10.1186/s13023-018-0954-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joerger A. C., Ang H. C., Veprintsev D. B., Blair C. M., Fersht A. R. J. Biol. Chem. 2005;280:16030–16037. doi: 10.1074/jbc.M500179200. [DOI] [PubMed] [Google Scholar]
- Liu J., Nussinov R. Proc. Natl. Acad. Sci. U. S. A. 2008;105:901–906. doi: 10.1073/pnas.0707401105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y. P., Ma B. Y., Levine A. J., Nussinov R. Biochemistry. 2006;45:3925–3933. doi: 10.1021/bi052242n. [DOI] [PubMed] [Google Scholar]
- Nussinov R., Tsai C. J. Trends Pharmacol. Sci. 2014;35:256–264. doi: 10.1016/j.tips.2014.03.006. [DOI] [PubMed] [Google Scholar]
- Song K., Li Q., Gao W., Lu S. Y., Shen Q. C., Liu X. Y., Wu Y. Y., Wang B. Q., Lin H. W., Chen G. Q., Zhang J. Nucleic Acids Res. 2019;47:W315–W321. doi: 10.1093/nar/gkz350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Z. W., Tee W. V., Guarnera E., Booth L., Berezovsky I. N. Nucleic Acids Res. 2019;47:D265–D270. doi: 10.1093/nar/gky1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokh D. B., Richter S., Henrich S., Czodrowski P., Rippmann F., Wade R. C. J. Chem. Inf. Model. 2013;53:1235–1252. doi: 10.1021/ci4000294. [DOI] [PubMed] [Google Scholar]
- Cimermancic P., Weinkam P., Rettenmaier T. J., Bichmann L., Keedy D. A., Woldeyes R. A., Schneidman-Duhovny D., Demerdash O. N., Mitchell J. C., Wells J. A., Fraser J. S., Sali A. J. Mol. Biol. 2016;428:709–719. doi: 10.1016/j.jmb.2016.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth S. A., Kelly B., Valant C., Michaelis J. A., Mastromihalis O., Thompson G., Venkatakrishnan A. J., Hertig S., Scammells P. J., Sexton P. M., Felder C. C., Christopoulos A., Dror R. O. Nat. Commun. 2019;10:3289. doi: 10.1038/s41467-019-11062-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayatshahi H. S., Ahuactzin E., Tao P., Wang S. Y., Liu J. J. Chem. Inf. Model. 2019;59:4691–4705. doi: 10.1021/acs.jcim.9b00447. [DOI] [PubMed] [Google Scholar]
- Guo J. J., Zhou H. X. Chem. Rev. 2016;116:6503–6515. doi: 10.1021/acs.chemrev.5b00590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarnera E., Berezovsky I. N. PLoS Comput. Biol. 2016;12:e1004678. doi: 10.1371/journal.pcbi.1004678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockless S. W., Ranganathan R. Science. 1999;286:295–299. doi: 10.1126/science.286.5438.295. [DOI] [PubMed] [Google Scholar]
- Kaya C., Armutlulu A., Ekesan S., Haliloglu T. Nucleic Acids Res. 2013;41:W249–W255. doi: 10.1093/nar/gkt284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalescky R., Liu J., Tao P. J. Phys. Chem. A. 2015;119:1689–1700. doi: 10.1021/jp5083455. [DOI] [PubMed] [Google Scholar]
- VanWart A. T., Eargle J., Luthey-Schulten Z., Amaro R. E. J. Chem. Theory Comput. 2012;8:2949–2961. doi: 10.1021/ct300377a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowerman S., Wereszczynski J. Methods Enzymol. 2016;578:429–447. doi: 10.1016/bs.mie.2016.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Sala G., Decherchi S., De Vivo M., Rocchia W. ACS Cent. Sci. 2017;3:949–960. doi: 10.1021/acscentsci.7b00211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezovsky I. N. Biochim. Biophys. Acta, Proteins Proteomics. 2013;1834:830–835. doi: 10.1016/j.bbapap.2013.01.024. [DOI] [PubMed] [Google Scholar]
- Nussinov R., Tsai C. J. Annu. Rev. Pharmacol. 2015;55:249–267. doi: 10.1146/annurev-pharmtox-010814-124401. [DOI] [PubMed] [Google Scholar]
- Huang W. K., Nussinov R., Zhang J. Methods Mol. Biol. 2017;1529:439–446. doi: 10.1007/978-1-4939-6637-0_23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Z. W., Guarnera E., Tee W. V., Berezovsky I. N. Nucleic Acids Res. 2020 doi: 10.1093/nar/gkaa338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tee W. V., Guarnera E., Berezovsky I. N. J. Mol. Biol. 2019;431:3933–3942. doi: 10.1016/j.jmb.2019.07.012. [DOI] [PubMed] [Google Scholar]
- Frauenfelder H., Sligar S. G., Wolynes P. G. Science. 1991;254:1598–1603. doi: 10.1126/science.1749933. [DOI] [PubMed] [Google Scholar]
- Ma B., Kumar S., Tsai C. J., Nussinov R. Protein Eng. 1999;12:713–720. doi: 10.1093/protein/12.9.713. [DOI] [PubMed] [Google Scholar]
- Gunasekaran K., Ma B. Y., Nussinov R. Proteins. 2004;57:433–443. doi: 10.1002/prot.20232. [DOI] [PubMed] [Google Scholar]
- Liu J., Nussinov R. Proc. Natl. Acad. Sci. U. S. A. 2017;114:7480–7482. doi: 10.1073/pnas.1709071114. [DOI] [PMC free article] [PubMed] [Google Scholar]