Abstract
Background/Aim: Prostate cancer (PCa) is the second-most commonly occurring cancer among men, worldwide. Although the mechanisms associated with the progression of castration-resistant prostate cancer (CRPC) have been widely studied, the mechanism associated with more distant metastases from the bone remains unknown. This study aimed to characterize potential pathogenic kinases associated with highly metastatic PCa, that may regulate phosphorylation in extensively involved and diverse signaling pathways that are associated with the development of various cancers. Materials and Methods: A mass spectrometry (MS)-based comparative phosphoproteome strategy was utilized to identify differentially expressed kinases between the highly aggressive PCa cell-lines PC-3 and PC-3M. Results: Among 2,968 phosphorylation sites in PCa cells, 151 differently expressed phosphoproteins were identified. Seven motifs: -SP-, -SxxE-, -PxS-, -PxSP-, -SxxK-, -SPxK-, and -SxxxxxP- were found to be highly expressed in PC-3M cells. Based on these motifs, the kinases p21-activated kinase (PAK)2, Ste20-like kinase (SLK), mammalian Ste20-like kinase (MST)4, mitogen-activated kinase kinase (MAP2K)2, and A-Raf proto-oncogene serine/threonine kinase (ARAF) were up-regulated in PC-3M cells. Conclusion: PAK2, SLK, MST4, MAP2K2, and ARAF are kinases that are potentially associated with the progression of increased migration in PC-3M cells and may represent molecule regulators or drug targets for highly metastatic PCa therapy.
Keywords: Metastatic prostate cancer, proteomics, phosphorylation, kinases
Prostate cancer (PCa) is the second-most commonly occurring cancer among men, worldwide (1,2). According to the latest report by the American Cancer Society, in 2020, 191,930 new cases of PCa will be diagnosed and 33,330 individuals will die from PCa, in the United States (3). Unlike other types of cancers, PCa grows very slowly and may not cause symptoms for years, until the cancer cells metastasize out of the prostate, developing to advanced PCa (4,5). This characteristic has limited most cases of PCa being recognized and treated during the early stages, and prostate-specific antigen technology began to be applied to the detection of primary-stage PCa in 1994 (6,7).
Because PCa is an androgen-dependent disease (8), androgen deprivation therapy is the standard therapy for the initial stages of advanced PCa; however, treatment can eventually result in the development of castration-resistant PCa (CRPC) (9). Androgen receptor (AR) inhibition remains a key factor in CRPC therapies. Metastatic CRPC (mCRPC) develops when PCa cells spread to other parts of the body, where they can grow and spread, even under medical treatment (10). According to the “seed and soil” theory, proposed by Paget (11), PCa spreads in the bloodstream as “seeds”, which settle in specific organs, which represent “soil” (12). Although organs, such as the liver, lungs, and brain, are common targets of PCa colonization, PCa spreads most predominantly to the bone (13). The transforming growth factor (TGF)-β signaling pathway, bone morphogenetic protein (BMP), and the receptor activator of nuclear factor ĸB (NF-ĸB) (RANK)/RANK ligand (RANKL)/osteoprotegerin (OPG) system are known to be important factors in the formation of bone metastases (14-16). However, the metastasis process to distant organ such as liver or lung metastasis remains largely unknown (17,18).
Protein phosphorylation is the most common post-translational modification observed in proteins (19,20). Dynamic protein phosphorylation plays a vital role in the activation and deactivation of enzymes and receptors, which are phosphorylated and dephosphorylated by kinases and phosphatases, respectively (21). Protein kinases represent a large category containing various enzymes that catalyze protein phosphorylation (22). Kinase inhibitors have played increasingly prominent roles in the treatment of cancer and other diseases (23). The phosphoinositol-3 kinase (PI3K)/protein kinase b (AKT)/NF-ĸB/BMP-2-Smad (24), phosphatase and tensin homolog (PTEN)/PI3K/AKT (25), and PI3K/AKT/mammalian target of rapamycin (mTOR) signaling pathways regulate tumor cell metastasis and invasion in PCa (26). However, the roles played by kinases in the progression of highly metastatic PCa remains underexplored.
In this study, we performed a global, comparative phosphorylation analysis on the PCa cell lines PC-3 and PC-3M. The PC-3 cell line was established in 1979, from a bone metastasis of a grade IV prostate cancer (27), whereas PC-3M cells are a metastasis-derived variant of PC-3 cells, which present an increased metastatic capacity and have been found to metastasize to the liver (28). This study aimed to characterize differences in kinase expression cancer during PCa that are associated with the development of increased metastatic capacity. This study is distinct from previous investigations, due to the following aspects: (i) this study represents the first study of global phosphorylation in PC-3 and PC-3M cells, using quantitative proteomics approaches, and (ii) this study focused on the relationships between kinase expression and the transformation into increasingly metastatic PCa cells. Our results provide a phosphoproteome database for the future study of PCa and suggested that increased kinase expression in PC-3M may be used to diagnose PCa or represent potential drug targets for the prevention of PCa progression to a highly metastatic form.
Materials and Methods
Prostate cancer cell culture. The PCa cell lines PC-3 and PC-3M were purchased from Korea Cell Line Bank (Seoul, Republic of Korea). PC-3M cells were cultured in RPMI 1640 medium (K0R0) (Thermo Fisher Scientific, Waltham, MA, USA), containing 10% fetal bovine serum (HyClone Laboratories, Inc. Logan, UT, USA) and 1% penicillin-streptomycin (Thermo Fisher Scientific). PC-3 cells were cultured in a stable isotope labeling with amino acids in cell culture (SILAC) RPMI 1640 media containing “medium” (K4R6) lysine and arginine isotopes (Cambridge Isotope Laboratory, Andover, MA, USA). The cell culture incubator was maintained at 5% CO2 and 37˚C. Cells were subcultures at 70-80% confluence, using 0.05% trypsin-ethylenediaminetetraacetic acid (EDTA) (Thermo Fisher Scientific) to detach the cells for subculture.
Protein extraction. PCa cells were washed in ice-cold phosphate-buffered saline (PBS, 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 1.8 mM KH2PO4, pH 7.4). After PBS buffer was removed, cells were lysed in ice-cold RIPA buffer (Thermos Fisher Scientific), containing 1% protease inhibitor and 1% phosphatase inhibitor (Thermo Fisher Scientific). Cells were lysed by sonication, and the lysates were centrifuged at 16,000 × g, at 4˚C for 10 min, to remove cellular debris. Clarified lysis supernatants were then transferred to new sample tubes. The protein concentrations were determined using a bicinchoninic acid (BCA) protein assay kit (Thermo Fisher Scientific). Protein lysate was maintained at –80˚C until use.
Western blotting analysis. To denature proteins for western blotting analysis, 100 μg PCa cell lysate was mixed with 4× sodium dodecyl sulfate (SDS) sample buffer [200 mM Tris-Cl, pH 6.8, 400 mM dithiothreitol (DTT), 8% SDS, 0.4% bromophenol blue, and 40% glycerol]. Distilled water was used to dilute the mixture to 1× SDS. Protein was denatured by heating at 95˚C, for 5 min. Denatured protein was allowed to cool to room temperature (RT) and then loaded at 10 μg per/well in a 10% SDS-polyacrylamide electrophoresis (SDS-PAGE) gel. The SDS-PAGE gel was run at 120 V, at RT. The protein was then electro-transferred from the gel to a polyvinylidene fluoride (PVDF) membrane (Roche, Basel, Switzerland), by wet transfer. The transfer was performed at a voltage of 90 V, for 120 min, in an ice-cold environment. After transfer, the membrane was blocked with 5% (v/v) bovine serum albumin (BSA, GenDEPOT, Barker, TX, USA), in 1× Tris-buffered saline containing Tween-20 (TBST, 137 mM sodium chloride and 20 mM Tris, with 0.1% Tween-20), at RT for 2 h.
The membrane was then incubated with the following primary antibodies: anti-phosphoserine and anti-phosphohistidine (Abcam, Cambridge, MA, USA) and anti-phosphothreonine and anti-phosphotyrosine (Cell Signaling Technology, Beverly, MA, USA). Antibodies were diluted at 1:1,000 (v/v), in 5% (v/v) BSA, and the membrane was incubated with gentle shaking, at 4˚C overnight. Membranes were then washed in TBST buffer for 10 min, 3 times, followed by incubation with horseradish peroxidase (HRP)-conjugated anti-mouse or anti-rabbit antibody (Cell Signaling Technology, Danvers, MA, USA), at RT for 90 min. The membrane was then washed in TBST for 10 min, 3 times. For visualization, the membranes were dampened using 1 ml ECL reagent (GE Healthcare, Chicago, IL, USA), and exposure was performed using the Image Quant LAS 4000 mini (GE Healthcare, Chicago, IL, USA).
Protein in-solution digestion. Prior to in-solution digestion, protein lysates from SILAC-labeled PC-3 (K4R6) and PC-3M (K0R0) cells were mixed at equal protein amounts. A total of 5 mg mixed protein lysate was utilized for the in-solution digestion (29). Protein lysate was reduced in 5 mM DTT, at 56˚C for 30 min, and alkylated in 15 mM iodoacetamide (IAA), at RT for 45 min. The DTT and IAA solutions were dissolved in 25 mM ammonium bicarbonate (ABC) buffer. To purify proteins, a 4-fold lysate volume of –20˚C cold acetone was added to the lysate. Protein was precipitated at –20˚C for 4 h. The protein lysate-acetone mixture was then centrifuged at 16,000 × g, at 4˚C for 15 min. The supernatant was discarded, and the pellet was re-dissolved in 100 mM ABC buffer. To perform trypsin digestion, sequencing-grade modified trypsin (Promega Corporation, Madison, WI, USA) was added, at 1:50 (w/w), and incubated at 37˚C, for 12 h. Trypsin digestion was quenched by adding 10% (v/v) trifluoroacetic acid (TFA), at a final concentration of 1% (v/v) TFA. To remove any undigested proteins, the protein mixture was centrifuged, at 16,000 × g, at 4˚C for 10 min. After centrifugation, the digested peptides were soluble in the supernatant, whereas undigested proteins remained in the pellet. The supernatant was then transferred to a new tube and the peptide concentration was measured by a quantitative, colorimetric peptide assay kit (Thermo Fisher Scientific). The peptide solution was then dried using a speed vacuum, without heating. The dried peptides were maintained at –80˚C until use.
Phosphorylated peptides, enriched by the TiO2 column. Phosphorylated peptides were enriched using a TiO2 Phosphopeptide Enrichment Kit (Thermo Fisher Scientific), following the manufacturer’s instructions. One milligram of peptides was applied to one TiO2 tip. Eluted peptides were fractionated, using a high-pH, reversed-phase, peptide fractionation kit (Thermo Fisher Scientific) and desalted using styrene-divinylbenzene (SDB) and graphite carbon tips (GL Science, Tokyo, Japan). Eluted, desalted peptides were dried in a speed vacuum, without heating. The dried peptides were maintained at −80˚C until use.
LC-MS analysis. Prior to MS analysis, the peptides were desalted using 5 μg Zip-tip (Millipore, Milford, MA, USA), according to the manufacturer’s instructions. The desalted peptide sample was dissolved in 2.5 μl solvent A (solvent A consisted of 2% acetonitrile and 0.1% formic acid, and solvent B consisted of 98% acetonitrile and 0.1% formic acid). The peptide sample was separated using a nano-LC (Eksigent, Dublin, CA, USA), equipped with an auto-sampler system. A C12 column (Proteo C12, 4 μm beads, 90-Å pore size, Phenomenex, Torrance, CA, USA) was used to connect the LC with the MS. The C12 column was packed with capillary tubing (length: 10 cm, OD: 75 μm, ID: 150 μm, Polymicro Technologies, Molex, Lisle, IL, USA). A 75-min gradient nano-LC was employed to analyze phosphorylated peptides, and the flow rate was set to 300 nl/min. The procedure began with 6% solvent B and end with 21% solvent B. For global proteomics analysis, a 3-23% solvent B gradient was performed over 60 min, with a flow rate set to 300 nl/min. For the peptide analysis, the nano-LC was coupled with the LTQ Orbitrap Velos MS (Thermo Fisher Scientific), in positive-ion mode. MS data were acquired in the data-dependent mode. The electrospray voltage was set to 1.8 kV. The precursor ion scans, ranging from m/z 300-1,800, were acquired at a resolution of 60,000, and the automatic gain control (AGC) target value was 1.0×106 for the MS scan. Tandem MS was obtained using the collision-induced dissociation mode. The data-dependent mode results, which produced 20 of the most abundant ions from full scans, were fragmented in high-energy collisional dissociation mode, with 35% normalized collision energy. The maximum IT was 100 ms for a full scan and 60 ms for MS2. The mass data are available via ProteomeXchange, with the identifier PXD016282.
Database search. MS raw files were searched in MaxQuant (version 1.5, http://www.maxquant.org/) against a Homo sapiens database (30). For the global proteomic peptide search, the following parameter settings were used: oxidation and acetyl (protein N-term) were chosen as variable modifications, Trypsin/P was chosen as digest enzymes, protein false discovery ratio (FDR) was set at 0.01, and Lys4 and Arg6 were chosen as the isotope labels. For the phosphorylated peptide analysis data search, phosphorylation (SYTH) was added as an option in the variable modifications. A Homo sapiens proteome database was downloaded from UniProt (https://www.uniprot.org/proteomes/UP000005640) (31). For result filtration, “contamination +” and “reverse +” were removed from the results file, and proteins with “scores” below 40 and phosphorylation “localization prob” below 70% were filtered from the results.
Bioinformatics analysis. Gene functional annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway mapping were performed using the database for annotation, visualization and integrated discovery (DAVID, version 6.8, https://david.ncifcrf.gov/), according to a previously published protocol (32). A –log10 value of the p-value from Fisher’s exact test was used as an enrichment factor, and the top five most enriched GO terms were used in the figure. Gene ontology biological process (GOBP), gene ontology cell component (GOCC), gene ontology molecular function (GOMF), InterPro, and KEGG pathways were analyzed in this study.
To characterize the phosphorylation patterns, motif analysis of the phosphorylated peptides was performed, using Motif-All algorithm (33). The pre-aligned sequences had 15-amino-acid lengths, which were used for the foreground format. Phospho-modified amino acids, including serine, threonine, tyrosine, and histidine, were arranged as the central characters. The p-value threshold was set to 10–6. The sequence logo was visualized through web plogo (https://plogo.uconn.edu/), after inputting pre-aligned sequences. The background was set to “human”, and a sequence logo was generated, according to the detailed methods in a previously published research article (34). Site-specific kinase-substrate relationships were predicted using the software package iGPS (Version 1.0) (35). Using a previously described analysis procedure, the protein phosphorylation network (PPN) among these activated kinases was visualization using PNC 1.0.
The protein-protein interaction (PPI) network was retrieved from the web resource STRING (Version 10.5 https://string-db.org/) (36). During STRING analysis, the UniProt accession IDs were inputted as the queries, Homo sapiens was chosen as the organism, and the “minimum required interaction score” was set to 0.9, which indicated the highest confidence. To simplify the display, “disconnected nodes” were hidden from the network.
Results
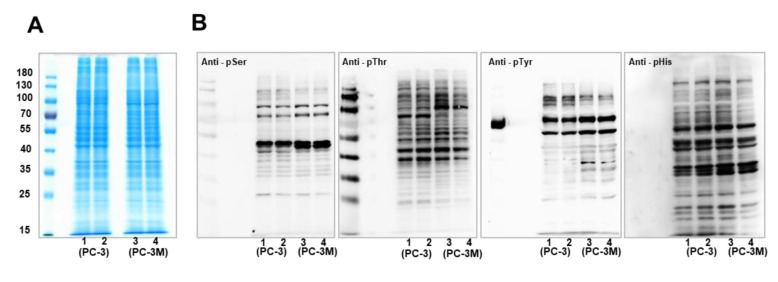
Proteomics screening of phosphorylation in PCa cells. To eliminate protein expression level effects on the comparison of phosphorylation abundance, protein expression levels in PCa cells were profiled by SDS-PAGE analysis (Figure 1A), which indicated similar protein expression levels between whole-cell lysates derived from PC-3 and PC-3M cells. Then, the phosphorylation levels in PCa cells were explored by western blotting analysis (Figure 1B). Phosphorylation on the amino acids serine (Ser), threonine (Thr), tyrosine (Tyr), and histidine (His) was detected using relevant antibodies. Western blot results showed different phosphorylation expression levels between PC-3 and PC-3M cells. The expression levels of pSer and pThr in PC-3 cells were relatively higher than in PC-3M cells, whereas pTyr exhibited elevated levels in PC-3 cells for proteins larger than 100 kDa, compared to PC-3M cells. Variations in pHis levels between these two PCa cell lines were not obvious.
Figure 1. Screening of phosphorylation expression patterns in PCa cells by western blotting analysis. (A) Protein expression profiling in PC-3 and PC-3M cells by western blotting analysis. Ten micrograms of protein were loaded into lanes 1-4. Lanes 1-2: duplicate loading of protein extracts from PC-3 cells, lanes 3-4: duplication loading from PC-3M cells. (B) Phosphorylation validation in PC-3 and PC-3M cells, by western blotting analysis. Phosphorylation patterns for the amino acids serine (Ser), threonine (Thr), tyrosine (Tyr), and histidine (His) were detected using corresponding antibodies.
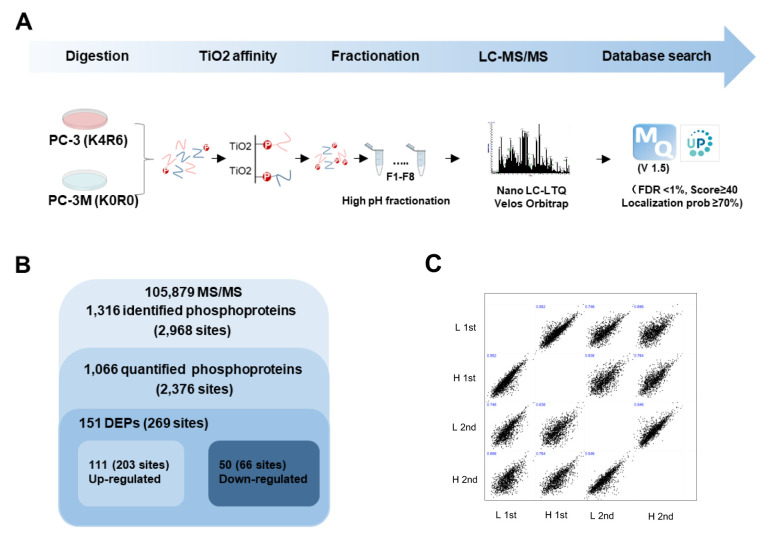
The proteomics strategy applied to the global profiling of phosphorylation in PCa cells is depicted in a flow chart (Figure 2A). SILAC PC-3 (K4R6) and PC-3M (K0R0) cells were lysed and mixed at equal protein amounts. After in-solution digestion, phosphorylated peptides were enriched by TiO2 bead-embedded tips. Eluted peptides were fractionated by high-pH, reversed-phased chromatography, based on hydrophobicity. The phosphorylated peptides were then chromatographically separated by a nano-LC system and analyzed using an LTQ-Orbitrap Velos. The generated MS spectra were searched using MaxQuant against a Homo sapiens database (version 1.5). A total number of 105,879 MS/MS spectra were generated from the MS raw data. MS raw data are available via ProteomeXchange, with the identifier PXD016282. Data were filtered using a FDR <1%, score ≥40, and localization prob ≥70. As a result, 2,968 non-redundant phosphorylation sites in 1,316 proteins were identified (Figure 2B). Among them, 2,376 phosphorylation sites in 1,066 phosphoproteins were quantified. Pearson’s correlation coefficient analysis was used to evaluate the coefficiency among technical duplicates was shown in (Figure 2C). The R-values reached as high as 0.74, which indicated that the replicates were highly correlated. Totally, 151 differently expressed phosphoproteins (DEPs) were quantified between PC-3 and PC-3M cells, including 111 up-regulated and 50 down-regulated phosphoproteins in PC-3M cells compared with PC-3 cells (under 0.5: down-regulated, over 2: up-regulated).
Figure 2. MS-based quantitative proteomic profiling of phosphorylation in PCa. (A) Experimental scheme for quantitative proteomic analysis. PC- 3 (K4R6) and PC-3M (K0R0) were lysed and mixed at equal protein amounts, and tryptic digestion was performed. Phosphorylated peptides were affinity purified using a TiO2 column, and eluted peptides were injected into the nano-LC-MS/MS system. MS spectra were searched in MaxQuant (version 1.5). (B) Overview of the frequency of differentially expressed phosphorylation from proteomics analysis. (C) Correlation of technically replication of MS data.
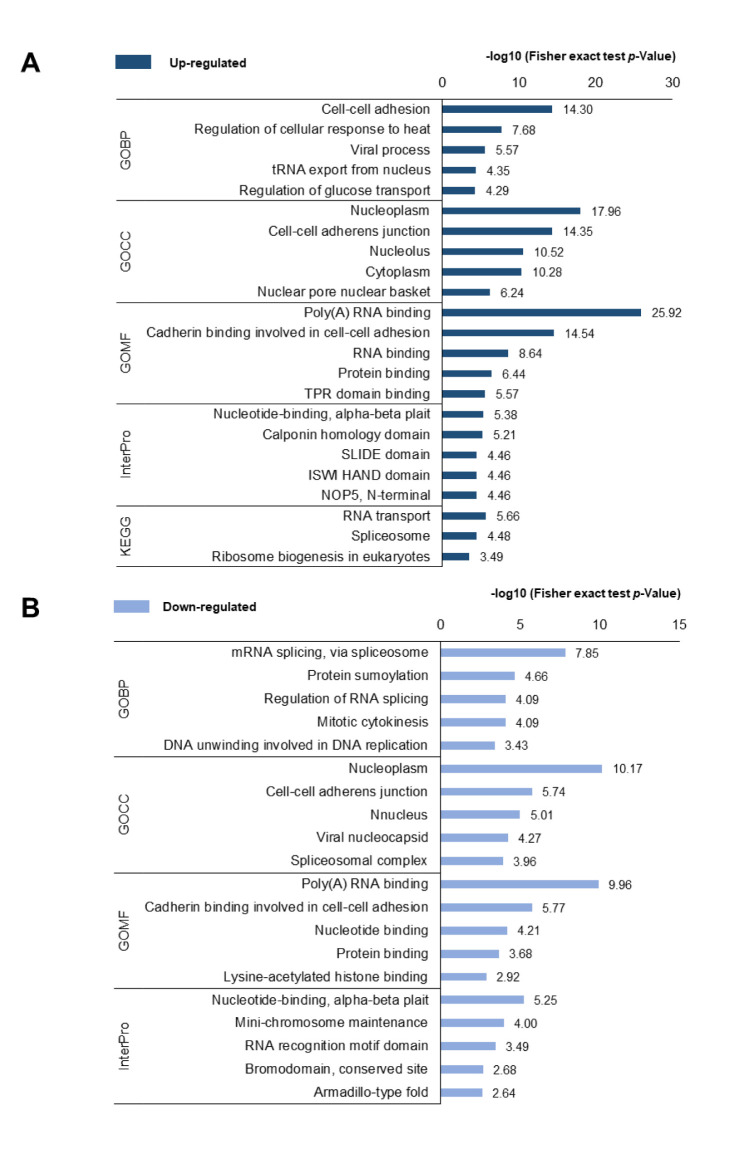
Functional annotation of differently expressed phosphoproteins. To investigate the functional differences in DEPs between PC-3 and PC-3M cells, functional annotation of the DEPs was performed using DAVID (Figure 3). The GOBP results showed that, up-regulated proteins were predominantly enriched in the process of “cell-cell adhesion”, followed by “regulation of cellular response to heat”, whereas down-regulated proteins were associated with “mRNA splicing, via spliceosome” and “protein sumoylation” processes. The misregulation of cell-cell adhesion in cancer cells has been shown to increase invasion into surrounding stromal tissues, resulting in the formation of advanced cancer (37). GOCC analysis indicated that the DEPs localized to similar cellular components, including the “nucleoplasm” and “cell-cell adherens junctions”. In the GOMF analysis, the functions of “poly(A) RNA binding”, “cadherin binding involved in cell-cell adhesion”, and “protein binding” increased among DEPs. In the InterPro analysis, nucleotide-binding was highly enriched among the DEPs, and “calponin homology domain” and “SIDE domain” were enriched among the up-regulated phosphoproteins, whereas “mini-chromosome maintenance” and “RNA recognition motif domain” were enriched among the down-regulated phosphoproteins. In the KEGG pathway analysis, up-regulated phosphoproteins were primarily associated with “RNA” transport, “Spliceosome”, and “ribosome biogenesis in eukaryotes”. These functional annotation results indicated that transcripts associated with the mRNA process were highly activated in highly metastatic PCa cells.
Figure 3. DAVID-generated GO enrichment and KEGG pathway analysis of DEPs. (A) Up-regulated phosphoproteins and (B) down-regulated phosphoproteins. The –log of Fisher’s exact test was used to represent the enrichment index. GOBP: Gene Ontology Biological Process, CC: cellular component, MF: molecular function, KEGG: Kyoto Encyclopedia of Genes and Genomes.
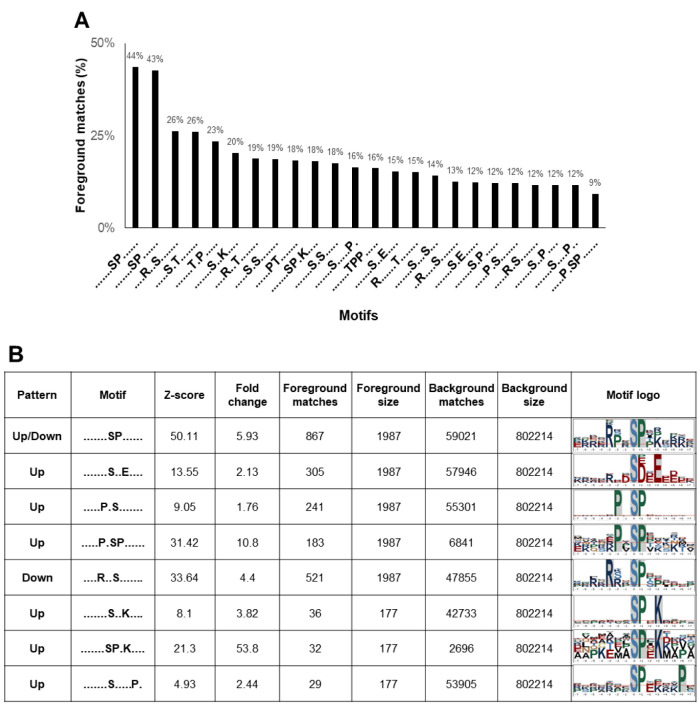
Phosphorylation motifs in highly metastatic PCa cells. The motifs that surround phosphorylation sites are recognized to be specific substrates for different kinases. To characterize the motifs identified in PC-3 and PC-3M cells, for further kinase prediction analyses, all phosphorylated peptides and DEPs were analyzed using the Motif-All algorithm. The percentage of foreground matches was used as a motif-enrichment index. As a result, 24 motifs, including 18 pSer and 6 pThr motifs, were significantly enriched among our identified phosphosites (Figure 4A). Among the 24 extensively enriched motifs, 8 pSer motifs were differentially expressed between PC-3 and PC-3M cells (Figure 4B). Seven motifs, -SxxE-, -PxS-, -PxSP-, -SxxK-, -SPxK-, -SxxxxxP- and -SP-, were enriched among the up-regulated phosphoproteins in PC-3M cells, whereas the motifs -RxxS- and -SP- were enriched in down-regulated phosphoproteins (The motif -SP- was enriched among both up- and down-regulated phosphosites). In addition, -SxxxxxP- was identified as a novel motif, in our study. Motif logos were visualized by the sequence logo tool from plogo (Figure 4B).
Figure 4. Differentially expressed motifs in PCa. (A) Twenty-four extensively enriched motifs were identified among the phosphosites, including 18 pSer and 6 pThr motifs. Percentage of foreground matches was used as the enrichment index. (B) Differentially expressed motifs between PC-3 and PC-3M cells. Seven motifs -SxxE-, -PxS-, -PxSP-, -SxxK-, -SPxK-, -SxxxxxP- and -SP-, were up-regulated in PC-3M cells. The motifs -RxxS- and - SP- were down-regulated. (Motif -SP- was enriched among both up- and down-regulated phosphosites).
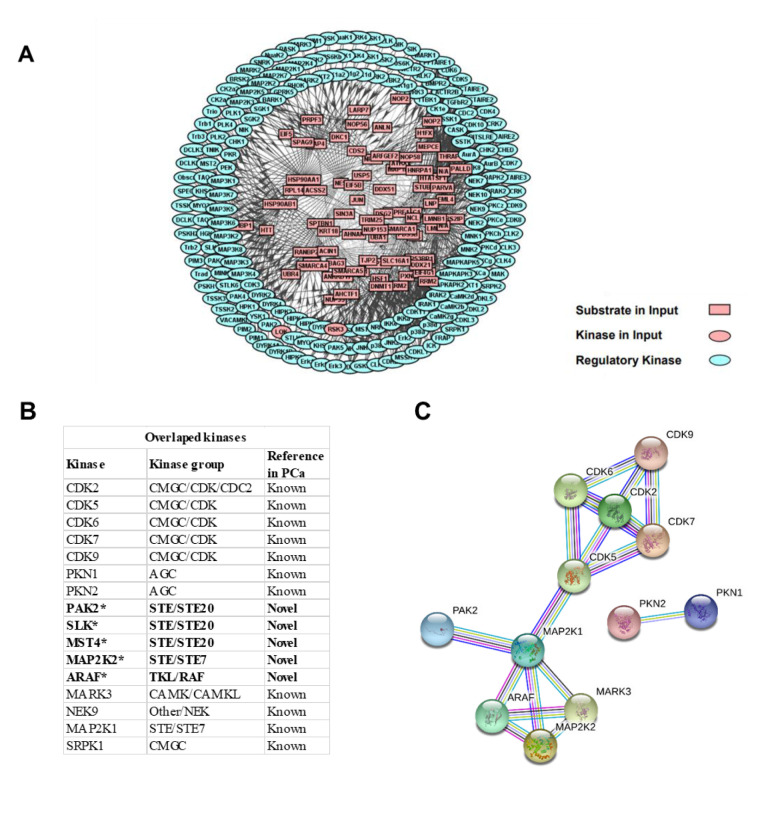
Kinases in highly metastatic PCa. To investigate the potential regulatory role played by kinases in the highly metastatic characteristics of PC-3M cell, site-specific kinase and substrate prediction analyses were performed, using the identified DEP phosphosites, through iGPS. As a result, 195 kinases were predicted to regulate the up-regulated phosphosites. Furthermore, a PPN was used to interpret connections between the kinase and specific substrates. A protein network calculator (PNC) was utilized to build the PPNs for the two kinase groups. As a result, 195 kinases (predicted from up-regulated phosphosites) displayed intensive interactions with various substrates (Figure 5A).
Figure 5. Protein–-protein network (PPN) of the upstream kinases in highly metastatic PCa cell. (A) A total of 195 kinase-substrate interactions were predicted from the up-regulated phosphosites. (B) A total of 16 overlapping kinases between phosphosite-predicated and the global proteome experiment were detected. Five kinases (*) are identified as novel kinases regulators during the progression to highly metastatic PCa. (C) PPN of the 16 overlapping kinases. PPN was mapped through the STRING database, with a high confidence value of 0.9.
To verify our findings and obtain a more reliable result, we co-analyzed the identified kinases using global phosphoproteome and proteome strategies. The co-analysis results revealed that 16 kinases predicted to interact with the identified phosphosites originating from our global proteome analysis (Figure 5B). The results suggested that the CMGC/CDK and AGC kinase groups extensively interacted with the DEPs. Five kinases, p21-activated kinase (PAK)2, Ste20-like kinase (SLK), mammalian Ste20-like kinase (MST)4, mitogen-activated kinase kinase (MAP2K)2, and A-Raf proto-oncogene serine/threonine kinase (ARAF) were firstly found to be up-regulated during the progression to PC-3M. PPI networks for these overlapping kinases were visualized using STRING (Figure 5C), which demonstrated that these kinases presented extensive interactions with each other.
Discussion
This study integrated two kinds of high-throughput MS-based datasets: global proteome and phosphoproteome. The results demonstrated that several known and novel kinases are associated with the highly metastatic PCa cell line, PC-3M (Figure 5B). First, we identified several kinases that are known regulators of PCa. For example, the cyclin-dependent kinases (CDKs), including CDK2, CDK5, CDK6, CDK7, and CDK9, were found to be up-regulated in PC-3M cells. CDKs are known to regulate cell-cycle phases and the overexpression of CDKs can induce cell proliferation (38,39). A previous study showed that inhibition of CDK2 suppressed PCa metastases by inactivating PI3K/Akt signaling (40). CDK5, CDK6, CDK7, and CDK9 can suppress CRPC through AR inactivation (41-44). In addition, CDK inhibitors are currently being evaluated in clinical trials (45). Second, serine/threonine-protein kinase N1 (PKN1) and PKN2 have been reported to contribute to the motility of PCa cells, and PKN1 overexpression was observed during clinical CRPC progression (46-48). PKN1 and PKN2 have been suggested to be potential PCa treatment targets. Our results are consistent with previous reports, which suggested that the present study’s strategy and results are reliable.
In addition, we identified several novel kinase molecules that were associated with the increased migration mechanisms of PC-3M cells. For example, PAK2 was up-regulated in PC-3M cells in our study, and the inhibition of PAK2 was able to inhibit the growth of androgen-independent PC3 xenografts (49). SLK is involved in cell-cycle progression through the G2/M phase (50,51) and is also required for migration in fibroblasts (52,53). SLK is also essential for ErbB2-driven breast cancer cell motility (54). According to these experiments, the elevation of SLK in PC-3M cells suggested that this kinase may increase PCa cell migration through an unknown pathway.
In addition, MST4 is another Ste20 kinase that is known to play a role in PCa progression through signal transduction pathways in the LNCaP, DU 145, and PC-3 cell lines (55). MST4 is not expressed in all benign prostatic hyperplasia cases but is expressed in 38.7% of PCa (56). In our study, MST4 was also found to be up-regulated in PC-3M cells compared to PC-3 cells. These results suggest a positive correlation between MST4 and PCa.
The dual-specificity MAP2K2 can activate both extracellular signal-related kinase (ERK)1 and ERK2 MAPKs (57,58). MAPKs are also known to phosphorylate substrates with the motif -PSP- (59). The increased expression of the motif -PSP- was observed in PC-3M cells (Figure 4B), which indirectly indicated that MAPKs might be activated by MAP2K2. Therefore, MAP2K2 is expected to represent a candidate regulator for cancer cell migration. In addition, ARAF has also been reported to promote the MAPK pathway activation and cell migration (60,61).
In this study, we characterized the global phosphorylation pattern in PC-3M cells, that are highly metastatic PCa cells, to explore the secondary metastatic regulation of PCa from bone to more distant organs, such as the liver and lung. We identified 2,968 phosphorylation sites and 151 DEPs, and bioinformatics analysis was conducted. We identified 5 novel kinases, PAK2, SLK, MST4, MAP2K2, and ARAF, that are up-regulated in PC-3M compared to PC-3 cells, which may represent candidates for therapeutic targets against the secondary metastasis of PCa. The present study was limited by a low abundance of identified phosphoproteins, which may result in some potential kinase regulators being overlooked. For example, it was known that tyrosine phosphorylation acts as a central regulator in cell signaling. Relative abundance of pSer:pThr:pTyr (1800:200:1) has been estimated in vertebrate cells. Therefore, characterization of tyrosine phosphorylation is a more challenging task. More recently, this deficiency could be improved by using multiple phosphorylation enrichment kits, specific antibodies affinity beads, or increasing the length of the LC column to improve the peptide sequence coverage.
Taken together, the current analysis indicated that the up-regulated kinase: PAK2, SLK, MST4, MAP2K2, and ARAF are concerns to the activation of PC-3M metastasis pathway. Although the current study was investigated in the proteomics field, our result could provide kinase information relating to the prostate cancerization study.
Conflicts of Interest
The Authors declare no conflicts of interest.
Authors’ Contributions
Conceptualization: Gao, Y., Lee, S. and Lee, J.N.; Methodology and analysis: Gao, Y. and Lee, S.; Resources: Ha, Y., Kwon, T.G. and Lee, J. N.; Data Curation: Gao, Y. Cho, Y. and Lee, S.; Writing: Gao, Y., Lee, S. and Lee, J.N.; Funding Acquisition: Ha, Y., Kwon, T.G., Lee, S and Lee, J.N.
Acknowledgements
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea and funded by the Korean government (2016R1C1B1011180, 2018R1D1A1A02043591, 2018R1C1B5040264, 2019R1H1A1079839).
Availability of Data and Materials
The mass spectrometry raw data are available via ProteomeXchange with identifier PXD016282.
References
- 1.Ferlay J, Steliarova-Foucher E, Lortet-Tieulent J, Rosso S, Coebergh JW, Comber H, Forman D, Bray F. Cancer incidence and mortality patterns in europe: Estimates for 40 countries in 2012. Eur J Cancer. 2013;49(6):1374–1403. doi: 10.1016/j.ejca.2012.12.027. [DOI] [PubMed] [Google Scholar]
- 2.Center MM, Jemal A, Lortet-Tieulent J, Ward E, Ferlay J, Brawley O, Bray F. International variation in prostate cancer incidence and mortality rates. Eur Urol. 2012;61(6):1079–1092. doi: 10.1016/j.eururo.2012.02.054. [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 4.Teo MY, Rathkopf DE, Kantoff P. Treatment of advanced prostate cancer. Annu Rev Med. 2019;70:479–499. doi: 10.1146/annurev-med-051517-011947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stockle M, Junker K, Fornara P. Low-risk prostate cancer prior to or after kidney transplantation. Eur Urol Focus. 2018;4(2):148–152. doi: 10.1016/j.euf.2018.07.003. [DOI] [PubMed] [Google Scholar]
- 6.Pezaro C, Woo HH, Davis ID. Prostate cancer: measuring psa. Intern Med J. 2014;44(5):433–440. doi: 10.1111/imj.12407. [DOI] [PubMed] [Google Scholar]
- 7.Saini S. Psa and beyond: alternative prostate cancer biomarkers. Cell Oncol (Dordr) 2016;39(2):97–106. doi: 10.1007/s13402-016-0268-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huggins C, Hodges CV. Studies on prostatic cancer. I. The effect of castration, of estrogen and androgen injection on serum phosphatases in metastatic carcinoma of the prostate. CA Cancer J Clin. 1972;22(4):232–240. doi: 10.3322/canjclin.22.4.232. [DOI] [PubMed] [Google Scholar]
- 9.Hoang DT, Iczkowski KA, Kilari D, See W, Nevalainen MT. Androgen receptor-dependent and -independent mechanisms driving prostate cancer progression: Opportunities for therapeutic targeting from multiple angles. Oncotarget. 2017;8(2):3724–3745. doi: 10.18632/oncotarget.12554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vlaeminck-Guillem V. When prostate cancer circulates in the bloodstream. Diagnostics (Basel) 2015;5(4):428–474. doi: 10.3390/diagnostics5040428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paget S. The distribution of secondary growths in cancer of the breast. 1889. Cancer Metastasis Rev. 1989;8(2):98–101. doi: 10.1016/S0140-6736(00)49915-0. [DOI] [PubMed] [Google Scholar]
- 12.Wong SK, Mohamad NV, Giaze TR, Chin KY, Mohamed N, Ima-Nirwana S. Prostate cancer and bone metastases: The underlying mechanisms. Int J Mol Sci. 2019;20(10) doi: 10.3390/ijms20102587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Sousa M. Microenvironment to a lymphoid cell is nothing more than interaction with its neighbours. Adv Exp Med Biol. 1976;66:165–169. doi: 10.1007/978-1-4613-4355-4_25. [DOI] [PubMed] [Google Scholar]
- 14.Cao Z, Kyprianou N. Mechanisms navigating the tgf-beta pathway in prostate cancer. Asian J Urol. 2015;2(1):11–18. doi: 10.1016/j.ajur.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu X, Jin EJ, Cheng X, Feng S, Shang X, Deng P, Jiang S, Chang Q, Rahmy S, Chaudhary S, Lu X, Zhao R, Wang YA, DePinho RA. Opposing roles of tgfbeta and bmp signaling in prostate cancer development. Genes Dev. 2017;31(23-24):2337–2342. doi: 10.1101/gad.307116.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Christoph F, Konig F, Lebentrau S, Jandrig B, Krause H, Strenziok R, Schostak M. Rankl/rank/opg cytokine receptor system: Mrna expression pattern in bph, primary and metastatic prostate cancer disease. World J Urol. 2018;36(2):187–192. doi: 10.1007/s00345-017-2145-y. [DOI] [PubMed] [Google Scholar]
- 17.Cotogno PM, Ranasinghe LK, Ledet EM, Lewis BE, Sartor O. Laboratory-based biomarkers and liver metastases in metastatic castration-resistant prostate cancer. Oncologist. 2018;23(7):791–797. doi: 10.1634/theoncologist.2017-0564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zang L, Ma M, Hu J, Qiu H, Huang B, Chu T. The effects of lung and prostate cancer bone metastasis on serum osteoprotegerin levels: A meta-analysis. Sci Rep. 2015;5:18324. doi: 10.1038/srep18324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li X, Wilmanns M, Thornton J, Kohn M. Elucidating human phosphatase-substrate networks. Sci Signal. 2013;6(275):rs10. doi: 10.1126/scisignal.2003203. [DOI] [PubMed] [Google Scholar]
- 20.Sacco F, Perfetto L, Castagnoli L, Cesareni G. The human phosphatase interactome: An intricate family portrait. FEBS Lett. 2012;586(17):2732–2739. doi: 10.1016/j.febslet.2012.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ardito F, Giuliani M, Perrone D, Troiano G, Lo Muzio L. The crucial role of protein phosphorylation in cell signaling and its use as targeted therapy (review) Int J Mol Med. 2017;40(2):271–280. doi: 10.3892/ijmm.2017.3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cicenas J, Zalyte E, Bairoch A, Gaudet P. Kinases and cancer. Cancers (Basel) 2018;10(3) doi: 10.3390/cancers10030063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gross S, Rahal R, Stransky N, Lengauer C, Hoeflich KP. Targeting cancer with kinase inhibitors. J Clin Invest. 2015;125(5):1780–1789. doi: 10.1172/JCI76094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Graham TR, Odero-Marah VA, Chung LW, Agrawal KC, Davis R, Abdel-Mageed AB. Pi3k/akt-dependent transcriptional regulation and activation of bmp-2-smad signaling by nf-kappab in metastatic prostate cancer cells. Prostate. 2009;69(2):168–180. doi: 10.1002/pros.20870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mediwala SN, Sun H, Szafran AT, Hartig SM, Sonpavde G, Hayes TG, Thiagarajan P, Mancini MA, Marcelli M. The activity of the androgen receptor variant ar-v7 is regulated by foxo1 in a pten-pi3k-akt-dependent way. Prostate. 2013;73(3):267–277. doi: 10.1002/pros.22566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen H, Zhou L, Wu X, Li R, Wen J, Sha J, Wen X. The pi3k/akt pathway in the pathogenesis of prostate cancer. Front Biosci (Landmark Ed) 2016;21:1084–1091. doi: 10.2741/4443. [DOI] [PubMed] [Google Scholar]
- 27.Kaighn ME, Narayan KS, Ohnuki Y, Lechner JF, Jones LW. Establishment and characterization of a human prostatic carcinoma cell line (pc-3) Invest Urol. 1979;17(1):16–23. [PubMed] [Google Scholar]
- 28.Pettaway CA, Pathak S, Greene G, Ramirez E, Wilson MR, Killion JJ, Fidler IJ. Selection of highly metastatic variants of different human prostatic carcinomas using orthotopic implantation in nude mice. Clin Cancer Res. 1996;2(9):1627–1636. [PubMed] [Google Scholar]
- 29.Kim SC, Chen Y, Mirza S, Xu Y, Lee J, Liu P, Zhao Y. A clean, more efficient method for in-solution digestion of protein mixtures without detergent or urea. J Proteome Res. 2006;5(12):3446–3452. doi: 10.1021/pr0603396. [DOI] [PubMed] [Google Scholar]
- 30.Tyanova S, Temu T, Cox J. The maxquant computational platform for mass spectrometry-based shotgun proteomics. Nat Protoc. 2016;11(12):2301–2319. doi: 10.1038/nprot.2016.136. [DOI] [PubMed] [Google Scholar]
- 31.Pundir S, Martin MJ, O’Donovan C, UniProt Consortium Uniprot tools. Curr Protoc Bioinformatics. 2016;53:1.29.1–1.29.15. doi: 10.1002/0471250953.bi0129s53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using david bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 33.He Z, Yang C, Guo G, Li N, Yu W. Motif-all: Discovering all phosphorylation motifs. BMC Bioinformatics. 2011;12(Suppl 1):S22. doi: 10.1186/1471-2105-12-S1-S22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O’Shea JP, Chou MF, Quader SA, Ryan JK, Church GM, Schwartz D. Plogo: A probabilistic approach to visualizing sequence motifs. Nat Methods. 2013;10(12):1211–1212. doi: 10.1038/nmeth.2646. [DOI] [PubMed] [Google Scholar]
- 35.Song C, Ye M, Liu Z, Cheng H, Jiang X, Han G, Songyang Z, Tan Y, Wang H, Ren J, Xue Y, Zou H. Systematic analysis of protein phosphorylation networks from phosphoproteomic data. Mol Cell Proteomics. 2012;11(10):1070–1083. doi: 10.1074/mcp.M111.012625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P, Jensen LJ, von Mering C. String v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43(Database issue):D447–452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vasioukhin V. Adherens junctions and cancer. Subcell Biochem. 2012;60:379–414. doi: 10.1007/978-94-007-4186-7_16. [DOI] [PubMed] [Google Scholar]
- 38.Sanchez I, Dynlacht BD. New insights into cyclins, cdks, and cell cycle control. Semin Cell Dev Biol. 2005;16(3):311–321. doi: 10.1016/j.semcdb.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 39.Malumbres M, Barbacid M. Cell cycle, cdks and cancer: A changing paradigm. Nat Rev Cancer. 2009;9(3):153–166. doi: 10.1038/nrc2602. [DOI] [PubMed] [Google Scholar]
- 40.Yin X, Yu J, Zhou Y, Wang C, Jiao Z, Qian Z, Sun H, Chen B. Identification of cdk2 as a novel target in treatment of prostate cancer. Future Oncol. 2018;14(8):709–718. doi: 10.2217/fon-2017-0561. [DOI] [PubMed] [Google Scholar]
- 41.Maddox AM, Freireich EJ, Keating MJ, Frasier-Scott KF, Haddox MK. Alterations in human circulating and bone marrow mononuclear cell polyamine levels in hematologic malignancies as a consequence of difluoromethylornithine administration. Invest New Drugs. 1988;6(2):125–134. doi: 10.1007/bf00195371. [DOI] [PubMed] [Google Scholar]
- 42.Lim JT, Mansukhani M, Weinstein IB. Cyclin-dependent kinase 6 associates with the androgen receptor and enhances its transcriptional activity in prostate cancer cells. Proc Natl Acad Sci USA. 2005;102(14):5156–5161. doi: 10.1073/pnas.0501203102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rasool RU, Natesan R, Deng Q, Aras S, Lal P, Sander Effron S, Mitchell-Velasquez E, Posimo JM, Carskadon S, Baca SC, Pomerantz MM, Siddiqui J, Schwartz LE, Lee DJ, Palanisamy N, Narla G, Den RB, Freedman ML, Brady DC, Asangani IA. Cdk7 inhibition suppresses castration-resistant prostate cancer through med1 inactivation. Cancer Discov. 2019;9(11):1538–1555. doi: 10.1158/2159-8290.CD-19-0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rahaman MH, Kumarasiri M, Mekonnen LB, Yu M, Diab S, Albrecht H, Milne RW, Wang S. Targeting cdk9: A promising therapeutic opportunity in prostate cancer. Endocr Relat Cancer. 2016;23(12):T211–T226. doi: 10.1530/ERC-16-0299. [DOI] [PubMed] [Google Scholar]
- 45.Rizzolio F, Tuccinardi T, Caligiuri I, Lucchetti C, Giordano A. Cdk inhibitors: from the bench to clinical trials. Curr Drug Targets. 2010;11(3):279–290. doi: 10.2174/138945010790711978. [DOI] [PubMed] [Google Scholar]
- 46.Yang CS, Melhuish TA, Spencer A, Ni L, Hao Y, Jividen K, Harris TE, Snow C, Frierson HF Jr., Wotton D, Paschal BM. The protein kinase c super-family member pkn is regulated by mtor and influences differentiation during prostate cancer progression. Prostate. 2017;77(15):1452–1467. doi: 10.1002/pros.23400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Venkadakrishnan VB, DePriest AD, Kumari S, Senapati D, Ben-Salem S, Su Y, Mudduluru G, Hu Q, Cortes E, Pop E, Mohler JL, Azabdaftari G, Attwood K, Shah RB, Jamieson C, Dehm SM, Magi-Galluzzi C, Klein E, Sharifi N, Liu S, Heemers HV. Protein kinase n1 control of androgen-responsive serum response factor action provides rationale for novel prostate cancer treatment strategy. Oncogene. 2019;38(23):4496–4511. doi: 10.1038/s41388-019-0732-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.O’Sullivan AG, Mulvaney EP, Hyland PB, Kinsella BT. Protein kinase c-related kinase 1 and 2 play an essential role in thromboxane-mediated neoplastic responses in prostate cancer. Oncotarget. 2015;6(28):26437–26456. doi: 10.18632/oncotarget.4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jiang N, Hjorth-Jensen K, Hekmat O, Iglesias-Gato D, Kruse T, Wang C, Wei W, Ke B, Yan B, Niu Y, Olsen JV, Flores-Morales A. In vivo quantitative phosphoproteomic profiling identifies novel regulators of castration-resistant prostate cancer growth. Oncogene. 2015;34(21):2764–2776. doi: 10.1038/onc.2014.206. [DOI] [PubMed] [Google Scholar]
- 50.O’Reilly PG, Wagner S, Franks DJ, Cailliau K, Browaeys E, Dissous C, Sabourin LA. The ste20-like kinase slk is required for cell cycle progression through g2. J Biol Chem. 2005;280(51):42383–42390. doi: 10.1074/jbc.M510763200. [DOI] [PubMed] [Google Scholar]
- 51.Wang K, Hong RL, Lu JB, Wang DL. Ste20-like kinase is upregulated in glioma and induces glioma invasion. Neoplasma. 2018;65(2):185–191. doi: 10.4149/neo_2018_170318N193. [DOI] [PubMed] [Google Scholar]
- 52.Wagner S, Storbeck CJ, Roovers K, Chaar ZY, Kolodziej P, McKay M, Sabourin LA. Fak/src-family dependent activation of the ste20-like kinase slk is required for microtubule-dependent focal adhesion turnover and cell migration. PLoS One. 2008;3(4):e1868. doi: 10.1371/journal.pone.0001868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wagner SM, Sabourin LA. A novel role for the ste20 kinase slk in adhesion signaling and cell migration. Cell Adh Migr. 2009;3(2):182–184. doi: 10.4161/cam.3.2.7229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roovers K, Wagner S, Storbeck CJ, O’Reilly P, Lo V, Northey JJ, Chmielecki J, Muller WJ, Siegel PM, Sabourin LA. The ste20-like kinase slk is required for erbb2-driven breast cancer cell motility. Oncogene. 2009;28(31):2839–2848. doi: 10.1038/onc.2009.146. [DOI] [PubMed] [Google Scholar]
- 55.Sung V, Luo W, Qian D, Lee I, Jallal B, Gishizky M. The ste20 kinase mst4 plays a role in prostate cancer progression. Cancer Res. 2003;63(12):3356–3363. [PubMed] [Google Scholar]
- 56.Zhang H, Ma X, Peng S, Nan X, Zhao H. Differential expression of mst4, stk25 and pdcd10 between benign prostatic hyperplasia and prostate cancer. Int J Clin Exp Pathol. 2014;7(11):8105–8111. [PMC free article] [PubMed] [Google Scholar]
- 57.Lavoie H, Sahmi M, Maisonneuve P, Marullo SA, Thevakumaran N, Jin T, Kurinov I, Sicheri F, Therrien M. Mek drives braf activation through allosteric control of ksr proteins. Nature. 2018;554(7693):549–553. doi: 10.1038/nature25478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Seger R, Seger D, Reszka AA, Munar ES, Eldar-Finkelman H, Dobrowolska G, Jensen AM, Campbell JS, Fischer EH, Krebs EG. Overexpression of mitogen-activated protein kinase kinase (mapkk) and its mutants in nih 3t3 cells. Evidence that mapkk involvement in cellular proliferation is regulated by phosphorylation of serine residues in its kinase subdomains vii and viii. J Biol Chem. 1994;269(41):25699–25709. [PubMed] [Google Scholar]
- 59.Wang ZK, Wang J, Liu J, Ying SH, Peng XJ, Feng MG. Proteomic and phosphoproteomic insights into a signaling hub role for cdc14 in asexual development and multiple stress responses in beauveria bassiana. PLoS One. 2016;11(4):e0153007. doi: 10.1371/journal.pone.0153007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wei H, Xu L, Yu M, Zhang L, Wang H, Wei X, Ruan Y. Monocillin ii inhibits human breast cancer growth partially by inhibiting mapk pathways and cdk2 thr160 phosphorylation. Chembiochem. 2012;13(3):465–475. doi: 10.1002/cbic.201100558. [DOI] [PubMed] [Google Scholar]
- 61.Mooz J, Oberoi-Khanuja TK, Harms GS, Wang W, Jaiswal BS, Seshagiri S, Tikkanen R, Rajalingam K. Dimerization of the kinase araf promotes mapk pathway activation and cell migration. Sci Signal. 2014;7(337):ra73. doi: 10.1126/scisignal.2005484. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The mass spectrometry raw data are available via ProteomeXchange with identifier PXD016282.