Abstract
Guanine-rich DNA sequences can fold into four-stranded, noncanonical secondary structures called G-quadruplexes (G4s). G4s were initially considered a structural curiosity, but recent evidence suggests their involvement in key genome functions such as transcription, replication, genome stability, and epigenetic regulation, together with numerous connections to cancer biology. Collectively, these advances have stimulated research probing G4 mechanisms and consequent opportunities for therapeutic intervention. Here, we provide a perspective on the structure and function of G4s with an emphasis on key molecules and methodological advances that enable the study of G4 structures in human cells. We also critically examine recent mechanistic insights into G4 biology and protein interaction partners and highlight opportunities for drug discovery.
Keywords: G-quadruplex, G4, DNA, secondary structure, nucleic acids, drug discovery
Highlights
Endogenous DNA G-quadruplex (G4) structures have been detected in human cells and mapped in genomic DNA and in an endogenous chromatin context by adapting next-generation sequencing approaches, to reveal cell type- and cell state-specific G4 landscapes and a strong link of G4s with elevated transcription. Synthetic small molecules and engineered antibodies have been vital to probe G4 existence and functions in cells.
Several endogenous proteins have been found to interact with DNA G4s, including helicases, transcription factors, and epigenetic and chromatin remodellers. Detailed structural and functional studies provided novel insight into G4–protein interactions and revealed a potential involvement of G4s in a range of biological processes.
Multiple new lines of evidence suggest that G4s play a role in cancer growth and progression. More G4s are detectable in cancer cell states compared with normal state, rendering G4s highly interesting targets in drug discovery. Recent studies have started to explore the potential for synthetic lethality and global modulation of cancer gene transcription.
Beyond the DNA Double Helix
A chemist’s perspective on the function of a molecule, or a system of molecules, is typically led by a consideration of how molecular structure dictates function. The most widely recognised DNA structure is that of the classical DNA double helix [1], which defines a structural basis for the genetic code via defined base-pairing. Yet, it is evident that DNA is structurally dynamic and capable of adopting alternative secondary structures. One such class of DNA secondary structure is the four-stranded G-quadruplex (G4). Herein, we discuss some of the key scientific history that has shaped our understanding of this structural motif, its probable functions in biology, and major unanswered questions that remain to be solved.
G4 Structure
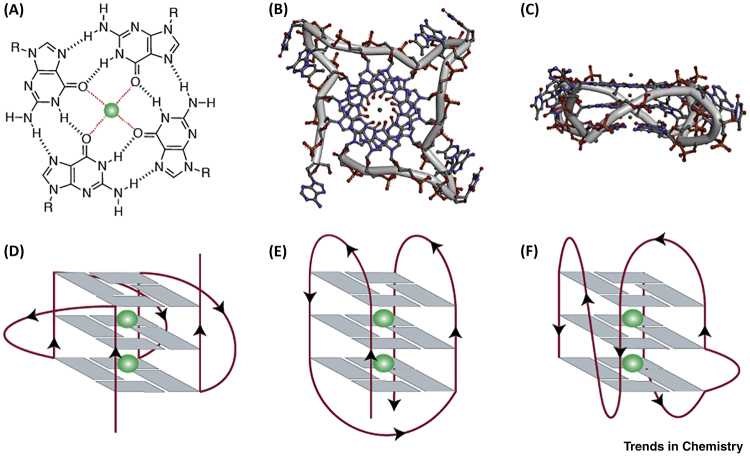
The capacity for guanylic acid derivatives to self-aggregate was noted over a century ago [2]. Some 50 years later, fibre diffraction revealed that guanylic acids form four-stranded, right-handed helices leading to a proposed model in which the strands are stabilised via Hoogsteen hydrogen-bonded guanines to form co-planar G-quartets 3, 4, 5. Subsequent biophysical studies using DNA oligonucleotides with sequences from immunoglobulin switching regions or telomeres (see Glossary) showed stable formation of G4 structures under near-physiological conditions in vitro 6, 7. Stacks of G-quartets are stabilised by cations centrally coordinated to O6 of the guanines with stabilising preference for monovalent cations in the order K+ > Na+ > Li+ (Figure 1A) [8]. G4s can be unimolecular or intermolecular and can adopt a wide diversity of topologies arising from different combinations of strand direction (Figure 1D–F), as well as length and loop composition 9, 10. Structural studies using X-ray crystallography and NMR spectroscopy have provided detailed insights into the structure of DNA G4s primarily based on the human telomeric repeat (Figure 1B,C) [11] or sequences derived from the promoter regions of certain human genes such as MYC or KIT 12, 13. Based on biophysical studies on different G4 structures, algorithms using sequence motifs such as G≥3NxG≥3NxG≥3NxG≥3 were developed and deployed to predict putative G4 structures in genomic DNA 14, 15. Early models assumed loop lengths no longer than seven and the requirement for four continuous stretches of Gs. Subsequently, several G4 structures with longer loop lengths and discontinuities in G-stretches causing bulges were observed 16, 17, leading to alternative predictive models [18]. The recent availability of large datasets on G4 formation has enabled the application of machine learning to predict G4 forming propensity [19]. Further considerations include the effects of molecular crowding [20] and DNA base modifications, such as cytosine methylation [21] and guanine oxidation [22], on the stability of G4 structures.
Figure 1.
G-Quadruplex (G4) Structures.
(A) Structure of a G-quartet formed by the Hoogsteen hydrogen-bonded guanines and central cation (coloured green) coordinated to oxygen atoms. Crystal structure of human telomeric G4s (Protein Data Bank: 1KF1): (B) top view and (C) side view; backbone is represented by grey tube and the structures are colour-coded by atoms. Schematic representation of unimolecular G4s based on the strand direction: (D) parallel, (E) anti-parallel, and (F) hybrid with a bulge.
Small Molecules that Bind G4s
Replicative immortality is a hallmark of cancer and several studies have suggested that cancer cells achieve unlimited proliferation by protecting ends of chromosomes [23]. Telomerase is a reverse transcriptase enzyme that adds repeat segments to the 3′-end of telomeric DNA and is highly expressed in most cancers 23, 24. One way to inhibit the action of telomerase was achieved using small molecules that sequester telomeric ends as stable, liganded G4 structures, which are thought to render telomeric ends inaccessible for telomerase-mediated extension [25]. An X-ray structure of the small molecule daunomycin in complex with the G4 formed by four strands of d(TGGGGT) confirmed stacking of small molecules on a terminal G-quartet [26]. Several structures based on NMR spectroscopy and X-ray crystallography of small molecule-G4 complexes have since been reported [27]. The concept of targeting telomeric G4 structures was extended to G4s located in gene promoters. A cationic porphyrin TmPyP4, which binds to G4s (but does also bind duplex DNA) in vitro was shown to inhibit transcription of oncogene MYC by a mechanism proposed to involve a G4 target in the nuclease hypersensitivity element (NHE) in the MYC promoter [28]. Since then, a variety of different G4-targeted ligands have been described to modulate the expression of genes carrying a sequence capable of forming a G4 in their respective promoters. So far, few studies have investigated transcriptional changes on a genome-wide level [29]. More carefully designed controls will be needed to assess whether a particular G4 is in fact the main biological target or if changes in target gene expression are a result of the ligand binding to other genomic (G4 or non-G4) targets. The central hypothesis would be strengthened by more explicit evidence for G4 ligands actually engaging with G4 structures in the promoters of affected genes in cells, for instance, by employing methods that enable the genome-wide mapping of ligand binding sites in native chromatin 30, 31.
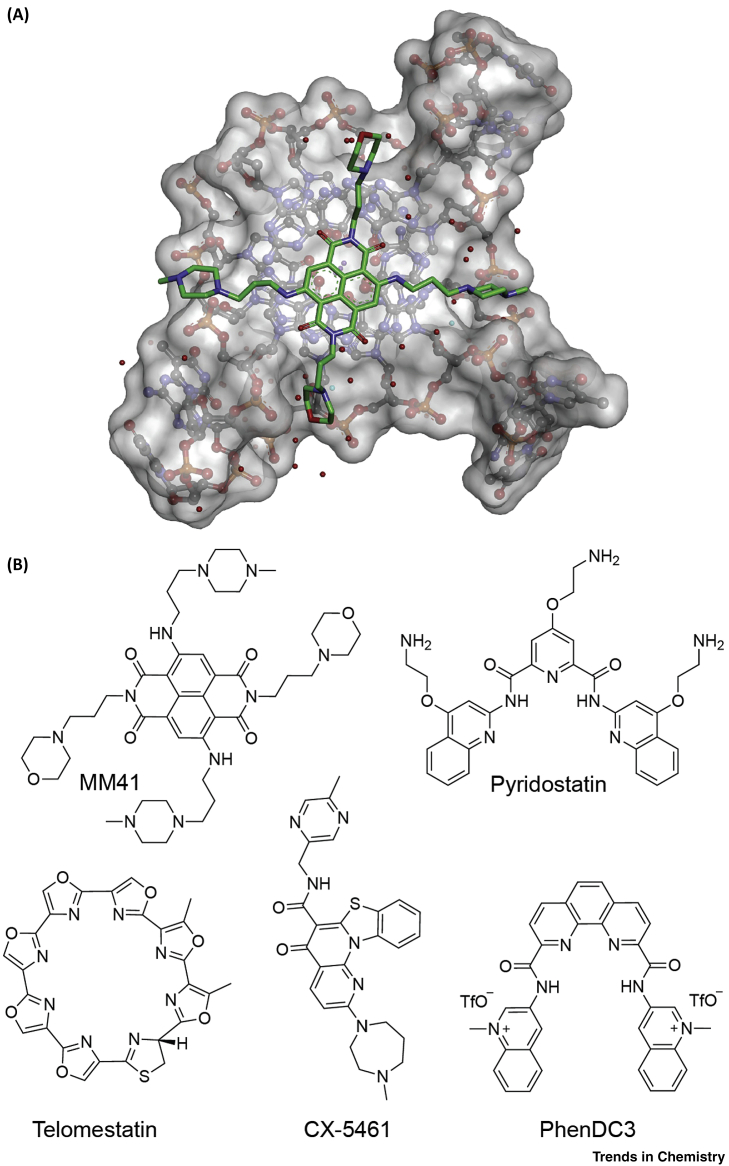
To date, around 1000 small molecules targeting G4 structures have been reported in the G-Quadruplex Ligands Database (http://www.g4ldb.org/) [32], with some examples of widely used ligands shown in Figure 2B. Small molecule G4 binders generally have an aromatic surface for π-π stacking with G-tetrads, a positive charge or basic groups to bind to loops or grooves of the G4, and steric bulk to prevent intercalation with double-stranded DNA [33]. To improve G4 binding, the aromatic ring count, positive charge, and number of hydrogen bond donors generally exceed what would be optimal for a small molecule with good pharmacokinetic properties 34, 35. Contrary to the classical perspective, it is noteworthy that a G4 ligand that lacks traditional ‘drug-like’ properties has shown significant accumulation and efficacy in tumour xenografts of human cancers [29]. The X-ray crystal structure of the G4 ligand MM41 bound to a human telomeric G4 (Figure 2A) suggests that certain structural features of G4 ligands can exploit additional interactions in the groove regions of the G4 structure [36]. Interactions with the groove and with the backbone phosphates do not require a flat aromatic structure. Therefore, compounds with reduced planarity (or high fsp3) and interactions with the grooves and/or backbone phosphates may have merit for targeting G4s. Structure–activity relationship studies on G4 ligands, controlling physicochemical properties such as planarity (fsp3), polarity (total polar surface area), lipophilicity (LogD), and rotatable bonds would enable an optimum balance between G4 binding, solubility, and permeability. In the targeting of structured RNA elements, it has previously been realised that increased planarity and strongly π-stacking compounds leads to off-target activity and that these molecules are generally difficult to improve via further medicinal chemistry [37].
Figure 2.
G-Quadruplex (G4) Ligands.
(A) Crystal structure of a naphthalene diimide, MM41 bound to an intramolecular human telomeric DNA G4, colour-coded by atoms; water molecules are shown as red spheres, MM41 carbon atoms are coloured green, surface of the G4 is coloured light grey (Protein Data Bank: 3UYH). (B) Structures of selected widely used G4 ligands.
G4 Detection and Mapping
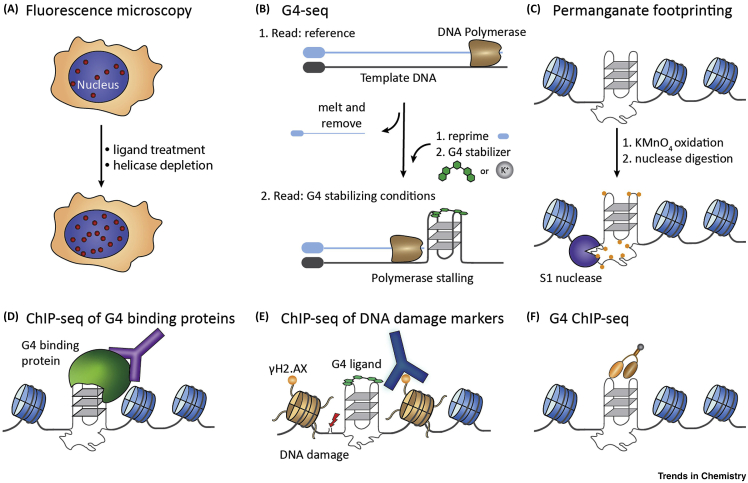
Computational algorithms have predicted over 370 000 G4 sequence motifs in the human genome, of which a general enrichment was observed in regions associated with genome regulation, such as telomeres, promoters, and 5′ untranslated regions 14, 38. G4s were first detected in vivo using a G4 structure-specific antibody to stain G4s in the telomeres of ciliates, whereby telomeric G4 structure formation was observed to be dynamically controlled through protein interactions in a cell cycle-dependent manner 39, 40. Subsequently, G4s were visualised in human cells 41, 42, 43, 44 and cancer tissue [45]. Antibodies have been used to monitor the behaviour of G4 structures in human cell lines upon ligand treatment together with depletion of the G4 resolving helicase FANCJ 41, 42, to reveal increased numbers of G4 foci staining in nuclei after pyridostatin [41] or telomestatin ligand treatment and FANCJ depletion in chicken DT40 cells [42]. Telomeric BG4 foci colocalise with human telomerase, suggesting the enzyme is recruited to G4 structures (Figure 3A) [43].
Figure 3.
Detection and Mapping of DNA G-Quadruplexes (G4s).
(A) Visualisation of G4s in fixed or live cells using structure-specific antibodies as well as labelled or intrinsically fluorescent G4 ligands. The number of detected G4 foci can be increased by small molecule treatment or helicase depletion. (B) High-throughput sequencing of G4s in human genomic DNA (G4-Seq). Two consecutive sequencing runs, under normal and G4 stabilising conditions, provide a reference map and detect G4-dependent polymerase stalling, respectively. (C) Chromatin immunoprecipitation employing antibodies against endogenous G4-binding proteins followed by next-generation sequencing (ChIP-seq). Genomic occupancy of endogenous proteins is used to infer putative G4 sites. (D) Treatment with G4 ligands induces G4-dependent DNA damage. ChIP-seq of DNA damage markers in combination with deep sequencing detects G4-associated regions. (E) Permanganate oxidation of nucleotides in transiently unwound regions traps the unpaired state, resulting in sensitivity to a single-strand specific nuclease. Computational prediction is then used to infer the type of underlying non-B-DNA structures based on sequence context. (F) G4-specific chromatin immunoprecipitation and next-generation sequencing (G4 ChIP-seq). A G4-specific antibody (e.g., BG4) is used to precipitate G4 structures directly from native chromatin and is identified by deep sequencing.
Besides antibodies, small molecules have also been used to detect G4s in cells. Early studies with radiolabelled G4 ligands also showed localisation at telomeres [46]. Subsequently, G4s were detected in human cells by treatment with alkyne functionalised G4 ligands followed by cell fixation and coupling to fluorophores via coper-catalysed azide-alkyne cycloaddition [47] or strain-promoted azide-alkyne cycloaddition [48]. Intrinsically fluorescent molecules that display different fluorescent emission or excitation maxima and fluorescent decay lifetimes upon binding to G4s have been developed for imaging live cells. The uptake of these molecules to the nucleus and a displacement by the G4 ligand pyridostatin have been monitored in living cells via fluorescence microscopy 49, 50. G4 specificity was further corroborated by colocalisation with the G4 structure-specific antibody BG4 in fixed cells [50].
The adaptation of next-generation sequencing has enabled the mapping of G4 structures in genomes. An in vitro reference map of G4s in purified human genomic DNA was obtained using differential G4-stabilising conditions (i.e., ligands or cations) to recognise G4-specific DNA polymerase stalling sites during next-generation whole genome sequencing (G4-seq, see Figure 3B) [51]. The G4-seq study identified more than 700 000 G4 sites in the human genome, exceeding some earlier predictions. Many noncanonical G4s were identified that comprised longer loops, as few as two G-tetrads or bulges caused by discontinuous G-tracts [16]. A recent study extended G4-seq to a variety of other organisms to generate G4 maps and reveal a strong potential for G4 formation in promoters that is particular to mammals (mouse, human), but mostly absent in the other organisms studied [52].
It is important to consider the effects of chromatin and all its associated proteins on G4 stability and formation, which are unaccounted for in maps generated by G4-seq or computational predictions. Efforts have been made to probe the native G4 landscape in vivo. G4 formation has been inferred from chromatin immunoprecipitation followed by high-throughput DNA sequencing (ChIP-seq) experiments that use antibodies against proteins that are known to bind G4s in vitro (Figure 3C). Enrichment of genomic regions comprising computationally predicted G4 structures was observed for α-thalassemia mental retardation X-linked (ATR-X) [53] and for the XPB and XPD helicases [54], yeast telomere binding protein RAP1-interacting factor 1 (Rif) [55], and yeast PIF1 helicase [56]. Such studies are consistent with G4 formation in vivo and suggest that the function of the respective proteins is linked to being physically associated with G4s.
The G4 stabilising ligand pyridostatin generates DNA double-strand breaks in cells. The sites of pyridostatin-induced strand breaks were determined by ChIP-sequencing of γH2.AX, a phosphorylated protein that marks stand break sites, and is found to occur predominantly at predicted G4 regions of the cellular genome (Figure 3D) [47]. In a similar approach, ChIP-seq of RAD51, which provides a more narrow ChIP-seq signal at DNA damage sites, identified ∼3000 genomic targets of the G4 ligand CX-5461 [57].
Genome-wide potassium permanganate-dependent nuclease footprinting, which identifies single-stranded, non-B DNA, was performed in mouse B cells and combined with computational analysis to discern the type and enrichment of different non-B DNA structures (Figure 3E) [58]. This approach revealed around 20 000 hypersensitivity sites featuring G4 motifs and suggested a transcription-dependent formation of the noncanonical DNA structures, when comparing resting and lipopolysaccharide-interleukin 4 activated B cells.
The BG4 antibody that binds a wide range of G4 structural types with broad selectivity and high affinity was recently used to map endogenous G4 structures in fixed chromatin from human epidermal keratinocytes (NHEKs) and from spontaneously immortalised HaCaT keratinocytes (G4 ChIP-seq, Figure 3F) [59]. In this study, ∼10 000 G4s were uncovered in precancerous HaCaT cells, while only ∼1000 were detected in the ‘normal’ counterpart NHEK cells. This represents only 1% of the sites with the capability to form G4s as observed in G4-Seq [51], suggesting a strong influence of the local chromatin context and other associated proteins on G4 formation in cells. The majority of G4s were found in nucleosome-depleted chromatin regions and were enriched in regulatory regions. In addition, G4s were particularly enriched at promoters of highly transcribed genes [59]. Mapping G4 landscapes in three other cancer cell lines revealed only moderate overlap, indicating a strong cell-type specificity [60].
In an alternative ChIP-seq approach, the G4 antibody, D1, was directly expressed in human cervical carcinoma cells as a GFP-fusion protein. In agreement with the BG4 study, a majority of the G4 signals were found at transcription start sites, in introns and intergenic regions [44]. Notably, ca. 15% of G4s were observed in exons, which were not found by BG4 ChIP-seq [59]. This may reflect different specificities of both G4 antibodies, differences in cell-type specific G4 landscapes, or competition of the D1 antibody with endogenous G4-binding proteins to reveal masked G4 structures.
Natural G4-Binding Proteins
The observation of G4s enriched at genome regulatory sites, and that G4 formation in cells is dynamic and dependent on cell type and state, suggests that the G4 landscape can be regulated by cellular proteins. Indeed, many natural proteins have been identified that interact with G4s (see G4 Interacting Proteins Database, http://bsbe.iiti.ac.in/bsbe/ipdb/) [61]. The identification of G4 associated proteins has mostly relied on affinity proteomics experiments employing RNA and DNA G4 oligomers as baits to isolate G4 interactors from cellular extracts 62, 63, 64, 65. Other approaches involve computational analysis of genomic protein binding sites to assess the enrichment of predicted G4 motifs within these binding sites 54, 66, or meta-analysis, in which shared structural features of G4-binding protein domains were compared to predict new putative G4 interactors 67, 68. In a recent study, a series of protein affinity pull-downs from nuclear lysates with oligonucleotide baits at different concentrations was combined with isobaric tandem-mass-tag labelling and mass spectrometry. This revealed apparent dissociation constants and binding profiles towards different G4 and transcription factor consensus sequences for hundreds of nuclear proteins in parallel [69].
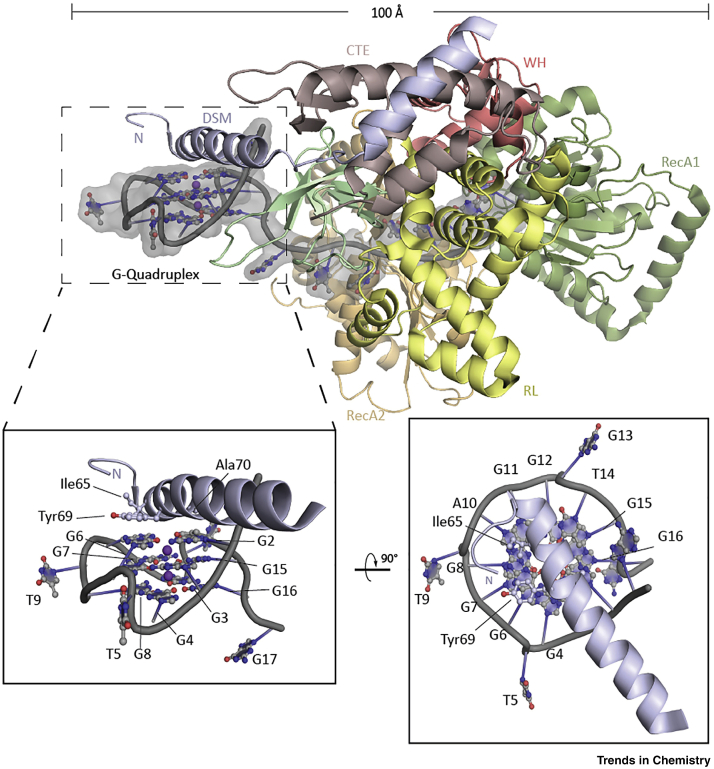
To date, over a dozen specialised helicases have been identified that target DNA G4s with up to picomolar affinity (reviewed in [70]). Recent studies have provided structural and mechanistic insights into G4 recognition and unwinding. Single-molecule imaging studies revealed a common repetitive unfolding mechanism for the specialised helicases DHX36 (also known as RHAU), Blooms (BLM), and Werner (WRN) helicases, albeit with different substrate specificity, and showed the capacity to displace G4-stabilising ligands 71, 72. A cocrystal structure of bos taurus DHX36 helicase bound to the MYC promoter G4 revealed a mode of binding in which the G4 makes contacts with a DHX36-specific motif (DSM) and the C terminal OB-fold domain (Figure 4) [73]. Strikingly, the residues of the α-helical DSM create a nonplanar hydrophobic surface that stacks on the top of a mixed quartet of the quadruplex, which forms after partial unfolding of the original G4 by the protein. The binding mode is somewhat reminiscent of that suggested for most small molecule G4 ligands, but suggesting that absolute planarity of the ligand may not be needed [73]. These findings are complemented by the crystal structure of Drosophila melanogaster DHX36, revealing a positively charged pocket that binds and destabilises the G4 [74].
Figure 4.
Crystal Structure of DHX36 Bound to the c-Myc G4 (Protein Data Bank: 5VHE).
Overall structure is shown in cartoon representation with the domain organisation of DHX36 colour-coded (top). The α-helical DHX36-specific motif (DSM) stacks on the 5′-quartet. The residues Ile65, Tyr69, and Ala70 form a nonpolar surface similar to the proposed binding mode of most small molecule G-quadruplex (G4) ligands (bottom).
In accordance with the G4 forming potential of the telomere repeat sequence, many telomere-associated proteins have been shown to interact with G4 structures [63]. For instance, POT1 [75] and RPA [76], two components of the shelterin complex, as well as the human CST complex [77], are able to bind and unwind telomeric G4 structures and assist the action of telomerase.
As G4s are strongly enriched at promoters, it is unsurprising that several transcription factors and transcriptional coactivators bind sites containing predicted G4 motifs, many of which show potential to bind or unwind G4 structures in vitro. In particular, G4 structures at promoter regions of oncogenes represent the most closely studied G4 sites. The nonmetastatic factor NM23-H2 has been reported to recognise and unwind the MYC promoter G4 [78]. Conversely, binding by nucleolin was reported to stabilise the G4 structure resulting in reduced transcription at this site [65]. Similarly, pull-down and chromatin immunoprecipitation experiments showed that Myc-associated zinc finger (MAZ) and poly(ADP-ribose) polymerase 1 (PARP-1) proteins bind the G4 structure found upstream of the KRAS transcription start site [79]. However, hnRNP A1 and MAZ were shown to bind and unfold the KRAS and HRAS promoter G4 in vitro, respectively 80, 81. Chromatin binding sites of the transcriptional helicases XPB and XPD are enriched in predicted G4 motifs. Both helicases associate with G4s in vitro, while XPD can also resolve G4 structures, suggesting that these proteins may affect transcription via a G4-associated mechanism [54].
Furthermore, various epigenetic and chromatin remodelling enzymes selectively bind DNA G4 oligomers [69]. Genomic binding sites of the chromatin remodelling protein ATR-X are enriched at GC-rich tandem repeats and CpG islands with the potential to fold G4 structures, and ATR-X loss is implicated with G4-dependent replication stress, DNA damage, and copy number alterations 53, 82. The DNA methyltransferase enzymes (DNMTs), which catalyse the formation of 5-methylcytosine at CpG dinucleotides in mammalian cells, bind to G4 structures with subnanomolar affinity in vitro 83, 84. DNMT1 shows stronger interaction with G4s compared with duplex DNA and loses enzyme activity upon G4 binding. Maps of DNMT1 chromatin binding sites and endogenous G4s in human K562 leukaemia cells identified using ChIP-seq and G4 ChIP-seq, respectively, revealed that at CpG islands most G4s occur where DNMT1 is bound, leading to a proposal that G4s regulate DNA methylation by sequestering DNMTs [84]. Conversely, DNA methylation can influence G4 topology [85] and may modulate binding by other G4-associated proteins. For instance, CpG methylation at the hTERT gene promoter was suggested to induce G4 formation, resulting in displacement of the CCCTC-binding factor (CTCF) and elevated transcription [86].
G4-protein interactions may provide a means to recruit machinery to specific parts of the genome to influence a wide range of cellular processes. Given that the G4 landscape is dynamic and dependent on the functional state of cells, proteins may be responsible for regulating G4 structural dynamics throughout the genome [60]. Approaches that can reveal the composition and dynamics of chromatin-associated protein complexes 87, 88 will be needed to uncover details of the proteins that constitute the G4 interactome, which in turn may present opportunities for small molecule modulation of these interactions.
Biological Role of G4s
The localisation of G4s at regions that regulate genome function have implicated G4s in a range of biological processes. The finding that G4-rich telomeric repeats form G4 structures [6] suggested a mechanistic link with telomerase-mediated extension of telomeres, prompting an exploration of G4 stabilising ligands that may inhibit the growth of cancer cells by interfering with telomere maintenance (reviewed in [89]). For instance, the mouse regulator of telomere elongation helicase 1 (RTEL1) was shown to maintain telomere integrity by unwinding telomeric G4s [90]. Telomeric G4s had originally been suggested to impair telomerase function [91], but might also be important for telomerase recruitment [43].
G4 structures are strongly associated with genomic and epigenetic instability, particularly when they are not efficiently regulated. The yeast Pif1 helicase was shown to prevent G4-mediated genomic instability and prevent DNA strand breaks 92, 93. Human Pif1 is recruited to DNA double-strand breaks sites to promote homologous recombination (HR) at sequences predicted to form G4s. Treatment with G4 stabilising ligands can impair Pif1 functionality, which can be rescued by PIF1 overexpression [94]. G4s have also been suggested to function as potential sensor or trapping sites of oxidative DNA damage caused by reactive oxygen species. Incorporation of 8-oxoG can affect stability of promoter G4 structures, which resulted in altered expression levels in reporter gene assays 95, 96, 97. Moreover, 8-oxoG modification was shown to disrupt the formation of telomeric DNA G4s [98] and to promote telomerase activity [99].
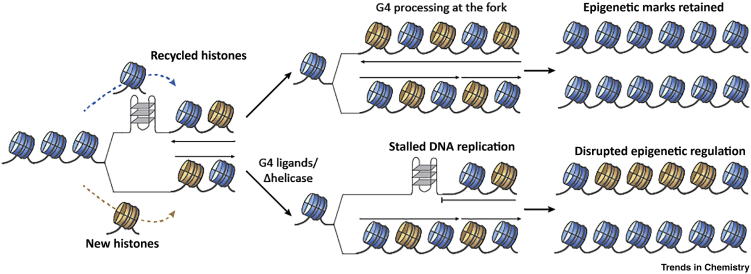
Various models have been postulated for the biological role of G4 structures during DNA replication 100, 101. G4 structures can act as an obstacle to replication fork progression when helicases are disrupted 93, 102. Introduction of G4 forming sequences at the BU-1 locus of DT40 chicken cells, resulted in pausing at the G4 sites (Figure 5). Additional replication stress due to nucleotide pool depletion decoupled the replication machinery from restoration of the parental histones, ultimately leading to altered gene expression [103]. Based on the same mechanism, small molecule stabilisation of G4 structures induced replication-dependent loss of epigenetic information showing that G4s can serve as obstacles to the replication machinery [104]. Conversely, mapping the genome-wide location of replication origins using deep-sequencing of short nascent strands in four different human cell lines uncovered enrichment of predicted G4s sequences, suggesting G4s may support initiation of DNA replication [105].
Figure 5.
G-Quadruplex (G4) Can Induce Epigenetic Reprogramming.
Unresolved G4 structures (e.g., G4 ligand treatment, impaired helicases) on the leading strand may promote uncoupling of DNA synthesis from opening of the replication fork and disrupt histone recycling. Original histone modifications (blue) are lost and replaced with new histones (brown), resulting in epigenetic reprogramming upstream of G4 sites.
Treatment with G4-stabilising small molecules resulted in reduced mRNA levels at genes that contain G4 sequences in their respective promoters, such as the proto-oncogenes MYC [28] and KRAS [106], supporting the hypothesis that G4s constitute an impediment to the progression of transcription machinery. However, G4-stabilising ligands can also lead to DNA damage and recruit associated response mechanisms 47, 57. Transcriptional changes at G4s may therefore be a result of either direct G4 stabilisation or DNA damage-mediated transcriptional repression. Aberrant function of the G4-resolving helicases WRN and BLM result in altered transcription of genes containing G4 motifs in their promoter region, corroborating a link between G4s and transcription 107, 108. Moreover, BLM-mutated cells derived from Bloom syndrome patients show high rates of sister chromatid exchange at sites of G4 motifs in transcribed genes [109]. Notably, these helicases also process duplex DNA, so not all the observed changes may be linked to G4 structures.
Where G4 structures form, the opposite DNA strand cannot form Watson-Crick base pairing. The C-rich opposite strand may be single stranded and potentially complexed with single-stranded binding proteins 110, 111. It has also been postulated that secondary structures might form on the opposite C-rich strand. Intercalated motif (i-motif) structures can be formed via stacks of intercalating hemiprotonated C-neutral C base pairs (C+:C), and are generally stabilised in slightly acidic pH [112]. Recent experiments have used an i-motif-specific antibody to image i-motifs in the nuclei of fixed human cells [113]. Furthermore, i-motif formation is cell cycle dependent, peaking at late G1 phase, whereas G4 formation was maximal during S phase [41], suggesting i-motifs and G4s may have different dependencies or even be mutually exclusive [114]. Indeed, a previously proposed model for the MYC promoter region suggested that negative superhelicity initiated at the proximal promoter travels upstream to mechanically perturb DNA structure [115] and it was suggested that a G4 and an i-motif can each be bound and stabilised by different specialised proteins to play opposite roles in the control of MYC gene transcription [116].
Approaches to map endogenous G4 structures such as permanganate footprinting, G4 ChIP-seq, and immunofluorescence with G4 antibodies have been consistent with G4 structures marking actively transcribing genes in human cells 58, 59. In addition, G4s can contribute to hypomethylation of CpG islands in promoter regions, which also contributes to elevated gene expression [84]. Further work is needed to unravel the mechanistic and molecular details of how G4s influence transcription.
Intervention and Therapeutics
G4s are associated with processes and control mechanisms that are important for the biology and growth of cancer cells, including telomere biology, transcriptional regulation of cancer-related genes, replication, and genome instability. Furthermore, G4 motifs are generally over-represented in cancer-promoting genes 51, 59 and experimental data has shown a higher presence of G4 structures in cancer states compared with normal states, which may favour G4s as molecular targets for cancer. For example, antibody staining of stomach and liver cancer tissues showed higher levels of G4 foci compared with normal tissue [45]. Also, G4 ChIP-seq detected tenfold more G4 sites in cancer-like, immortalised HaCaT cells as compared with their normal human keratinocyte counterpart 59, 60.
G4 ligands such as pyridostatin and RHPS4 cause DNA double-strand breaks in cancer cells and activate DNA repair pathways 47, 117. Cancer cells with impaired repair pathways are sensitive to G4 ligands. For example, cancer cells deficient in BRCA2, a vital component of the homologous recombination (HR) repair pathway, are sensitive to pyridostatin derivatives and RHPS4 118, 119. Similarly, knockdown of PARP1, a key regulator of the nonhomologous end joining (NHEJ) repair pathway resulted in cancer growth inhibition by RHPS4 both in cellular and xenograft models [117]. Notably, pyridostatin was also toxic to HR-deficient cells resistant to olaparib, a PARP inhibitor [119]. This result suggests that G4 ligands may be an effective therapeutic approach in HR-deficient cancers resistant to PARP inhibitors. Indeed G4 ligand, CX-5461 is currently in human clinical trials for breast cancer patients with BRCA1/2 germline aberrations [57]. Moreover, G4 ligands have also shown synergy with DNA damaging therapies in ATR-X-deficient glioma cell models [82]. G4 ligands have also been successfully used in combination with inhibitors of key proteins involved in the DNA repair pathway. Pyridostatin synergises with NU7441, an inhibitor of DNA-PK, which is an important kinase in the NHEJ pathway [118]. The combination of RHPS4 and PARP1 inhibitor GPI 15427 showed 50% reduction in growth of HT29 colon tumour-bearing mice xenografts compared with 2% and 30% with GPI 15427 and RHPS4 alone, respectively [117].
Another approach described above is to target G4-containing genes involved in cancer progression and manipulate their transcription. While the concept of regulating expression of individual genes has been explored using several small molecules 28, 106, it is rational to assume that most G4 ligands will bind to other G4s and potentially regulate expression of many genes. This multigene targeting approach can result in poly-pharmacology, affecting several pathways important for cancer progression. In support of this view, global transcriptional profiling revealed that treatment of human pancreatic ductal adenocarcinoma (PDAC) cell lines with the G4 ligand CM03 could induce global downregulation of many G4-containing genes [29]. Analysis of the downregulated genes identified G4-containing genes involved in pathways important for PDAC progression. CM03 reduced tumour growth in PDAC xenografts as well as KPC mouse models at doses that do not cause any observable toxicity. It is possible that a particular G4 ligand may result in a distinct profile of affected genes for proteins involved in several pathways, suitable for targeting certain cancers. Global transcriptional profiling of the cancer cells/tissues treated with G4 ligand may identify the affected pathways and suggest cancers best suited for efficacy studies. A better understanding of the pathways affected by G4 ligands may ultimately provide a rationale to consider combination therapy opportunities with other drugs.
Concluding Remarks
During the past two to three decades, the study of DNA G4 structures has extended from biophysical and structural work to studies in biological models and systems of increasing sophistication. Collectively, these studies have advanced the hypothesis that the G4 DNA structure is intimately linked to a number of biological functions. While there is more work to be done to unravel the mechanistic details of how and why G4 structures influence biological function (see Outstanding Questions), there appears to be rationale and merit in considering G4s as promising molecular targets for future therapeutics.
Outstanding Questions.
G4 ligands: can we develop small molecule G4 ligands with better pharmacokinetic properties that more effectively target G4s in cells? Is it possible to selectively target individual G4s or subclasses of G4s based on the molecular topology and sequence composition of the loops of G4s? Is it necessary, or desirable, to selectively target individual G4s to achieve therapeutic effects? How do we design suitable functional assays to screen for and validate direct G4 on-target effects in cells?
Detection and mapping: can we develop mapping approaches to detect G4s genome-wide in native chromatin with stranded information and at single G4 resolution? Do current antibody-based approaches provide an incomplete picture due to limited accessibility and competition with endogenous proteins? Can we develop detection methods to monitor dynamic G4-associated processes at high-resolution in living cells?
G4 landscape: under which conditions do G4s form in cells? What is the chromatin context? What are the key protein regulators that mediate G4 formation and depletion? What is the crosstalk with other epigenetic features? What happens on the opposing strand and what is the interplay with other structural phenomena (e.g., i-motifs, Z-DNA, R-loops)?
G4s and transcription: what is the exact mechanism by which G4s affect transcription? Can we control transcription efficiently by manipulating particular G4s or by interfering with particular G4 interactors?
G4s and cancer: why are G4s enriched at certain cancer genes? Are G4 signatures suitable biomarkers for diagnostic applications? What pathways are G4s involved in and are there phenotypes that make patients more tractable to G4 ligand therapies? What synthetic lethalities can potentially be exploited for combination therapies?
Alt-text: Outstanding Questions
Acknowledgements
We thank D. Tannahill for critical reading of the manuscript. The Balasubramanian laboratory is core-funded by Cancer Research UK (C14303/A17197); Cancer Research UK programme (C9681/A18618); S.B. is a Welcome Trust Senior Investigator (099232/Z/12/Z); J.S. gratefully acknowledges funding from the EU H2020 Framework Programme (H2020-MSCA-IF-2016, ID: 747297-QAPs).
Disclaimer Statement
S.B. is a founder, shareholder, and scientific advisor to Cambridge Epigenetix Ltd.
Glossary
- ChIP-seq
chromatin immunoprecipitation (ChIP) is a commonly used method to detect interactions between proteins and DNA in a native chromatin context that is based on the enrichment of DNA associated with the protein of interest. The combination with high-throughput DNA sequencing analysis (ChIP-seq) allows for genome-wide detection of protein binding sites. G4 ChIP-seq is a related technology whereby an antibody directed against the G4 structure rather than a protein is used.
- Chromatin
a complex of DNA and proteins in eukaryotic cells. Its main functions are the efficient packaging of DNA to fit into the volume of nucleus, the stabilisation and further condensation of DNA during cell division, and the control of gene expression. Euchromatin is a slightly packed chromatin, which is usually transcriptionally active, whereas heterochromatin is more condensed, inaccessible DNA that is usually transcriptionally repressed.
- DNA methyltransferases
a family of enzymes that catalyse the transfer of methyl groups to DNA. They catalyse the formation of 5-methylcytosine at CpG dinucleotides in mammalian cells.
- fsp3
number of sp3 hybridised carbons/total carbon count.
- Helicase
a class of enzymes that can bind, unpack, and remodel nucleic acids or nucleic acid protein complexes. DNA helicases play essential roles in vital cellular processes. For instance, DNA helicases unwind double-stranded into single-stranded DNA during replication or transcription so that the strand information can be copied.
- Hypersensitivity site
(nuclease) hypersensitivity sites are nucleosome-depleted regions of chromatin that are detected via their very high sensitivity to cleavage by nucleases. These regions are less densely packed to allow the binding of proteins, such as transcription factors, and often mark genomic regulatory elements.
- Next-generation sequencing
(deep sequencing) modern high-throughput sequencing technologies, such as Illumina (Solexa), Roche 454, and Ion Torrent sequencing that produce data on a genome scale in the form of millions of short sequence reads.
- 8-oxoG
8-oxoguanine is a common DNA lesion resulting from reactive oxygen species; incorporation into the DNA sequence can result in a mismatched pairing with adenine, resulting in mutations.
- Promoter
region of DNA where transcription initiates; typically located directly upstream of the transcription start site.
- Telomere
region of repetitive nucleotide sequence (TTAGGG)n at the end of each chromosome. Telomeres protect chromosomes from end-to-end fusion and prevent DNA damage and the loss of genetic information. Telomeres are typically shortened during cell division. Telomerases are enzymes that extend telomeres by addition of repeat sequences to the 3′-end, a process that is active in stem cells and cancers, but mostly absent in somatic cells.
References
- 1.Watson J.D., Crick F.H. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature. 1953;171:737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 2.Bang I. Untersuchungen über die Guanylsäure. Biochem. Z. 1910;26:293–311. (in German) [Google Scholar]
- 3.Gellert M. Helix formation by guanylic acid. Proc. Natl. Acad. Sci. U. S. A. 1962;48:2013–2018. doi: 10.1073/pnas.48.12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arnott S. Structures for polyinosinic acid and polyguanylic acid. Biochem. J. 1974;141:537–543. doi: 10.1042/bj1410537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zimmerman S.B. X-ray fiber diffraction and model-building study of polyguanylic acid and polyinosinic acid. J. Mol. Biol. 1975;92:181–192. doi: 10.1016/0022-2836(75)90222-3. [DOI] [PubMed] [Google Scholar]
- 6.Sen D., Gilbert W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature. 1988;334:364–366. doi: 10.1038/334364a0. [DOI] [PubMed] [Google Scholar]
- 7.Sundquist W.I., Klug A. Telomeric DNA dimerizes by formation of guanine tetrads between hairpin loops. Nature. 1989;342:825–829. doi: 10.1038/342825a0. [DOI] [PubMed] [Google Scholar]
- 8.Sen D., Gilbert W. A sodium-potassium switch in the formation of four-stranded G4-DNA. Nature. 1990;344:410–414. doi: 10.1038/344410a0. [DOI] [PubMed] [Google Scholar]
- 9.Burge S. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 2006;34:5402–5415. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou J. Tri-G-quadruplex: controlled assembly of a G-quadruplex structure from three G-rich strands. Angew. Chemie Int. Ed. 2012;51:11002–11005. doi: 10.1002/anie.201205390. [DOI] [PubMed] [Google Scholar]
- 11.Parkinson G.N. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002;417:876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- 12.Ambrus A. Solution structure of the biologically relevant G-quadruplex element in the human c-MYC promoter. Implications for G-quadruplex stabilization. Biochemistry. 2005;44:2048–2058. doi: 10.1021/bi048242p. [DOI] [PubMed] [Google Scholar]
- 13.Wei D. Crystal structure of a c-kit promoter quadruplex reveals the structural role of metal ions and water molecules in maintaining loop conformation. Nucleic Acids Res. 2012;40:4691–4700. doi: 10.1093/nar/gks023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huppert J.L., Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Todd A.K. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005;33:2901–2907. doi: 10.1093/nar/gki553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mukundan V.T., Phan A.T. Bulges in G-quadruplexes: broadening the definition of G-quadruplex-forming sequences. J. Am. Chem. Soc. 2013;135:5017–5028. doi: 10.1021/ja310251r. [DOI] [PubMed] [Google Scholar]
- 17.Sengar A. Structure of a (3+1) hybrid G-quadruplex in the PARP1 promoter. Nucleic Acids Res. 2018;47:1564–1572. doi: 10.1093/nar/gky1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bedrat A. Re-evaluation of G-quadruplex propensity with G4Hunter. Nucleic Acids Res. 2016;44:1746–1759. doi: 10.1093/nar/gkw006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sahakyan A.B. Machine learning model for sequence-driven DNA G-quadruplex formation. Sci. Rep. 2017;7:1–11. doi: 10.1038/s41598-017-14017-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shrestha P. Confined space facilitates G-quadruplex formation. Nat. Nanotechnol. 2017;12:582–588. doi: 10.1038/nnano.2017.29. [DOI] [PubMed] [Google Scholar]
- 21.Hardin C.C. Cytosine—cytosine+ base pairing stabilizes DNA quadruplexes and cytosine methylation greatly enhances the effect. Biochemistry. 1993;32:5870–5880. doi: 10.1021/bi00073a021. [DOI] [PubMed] [Google Scholar]
- 22.Cheong V.V. Xanthine and 8-oxoguanine in G-quadruplexes: formation of a G·G·X·O tetrad. Nucleic Acids Res. 2015;43:10506–10514. doi: 10.1093/nar/gkv826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 24.Testorelli C. Telomerase and cancer. J. Exp. Clin. Cancer Res. 2003;22:165–169. [PubMed] [Google Scholar]
- 25.Sun D. Inhibition of human telomerase by a G-quadruplex-interactive compound. J. Med. Chem. 1997;40:2113–2116. doi: 10.1021/jm970199z. [DOI] [PubMed] [Google Scholar]
- 26.Clark G.R. Structure of the first parallel DNA quadruplex-drug complex. J. Am. Chem. Soc. 2003;125:4066–4067. doi: 10.1021/ja0297988. [DOI] [PubMed] [Google Scholar]
- 27.Neidle S. Quadruplex nucleic acids as novel therapeutic targets. J. Med. Chem. 2016;59:5987–6011. doi: 10.1021/acs.jmedchem.5b01835. [DOI] [PubMed] [Google Scholar]
- 28.Siddiqui-Jain A. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. U. S. A. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marchetti C. Targeting multiple effector pathways in pancreatic ductal adenocarcinoma with a G-quadruplex-binding small molecule. J. Med. Chem. 2018;61:2500–2517. doi: 10.1021/acs.jmedchem.7b01781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anders L. Genome-wide localization of small molecules. Nat. Biotechnol. 2014;32:92–96. doi: 10.1038/nbt.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tyler D.S. Click chemistry enables preclinical evaluation of targeted epigenetic therapies. Science. 2017;356:1401. doi: 10.1126/science.aal2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Q. G4LDB: a database for discovering and studying G-quadruplex ligands. Nucleic Acids Res. 2013;41:D1115–D1123. doi: 10.1093/nar/gks1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Monchaud D., Teulade-Fichou M.P. A hitchhiker’s guide to G-quadruplex ligands. Org. Biomol. Chem. 2008;6:627–636. doi: 10.1039/b714772b. [DOI] [PubMed] [Google Scholar]
- 34.Ritchie T.J., Macdonald S.J.F. The impact of aromatic ring count on compound developability - are too many aromatic rings a liability in drug design? Drug Discov. Today. 2009;14:1011–1020. doi: 10.1016/j.drudis.2009.07.014. [DOI] [PubMed] [Google Scholar]
- 35.Shultz M.D. Two decades under the influence of the rule of five and the changing properties of approved oral drugs. J. Med. Chem. 2019;62:1701–1714. doi: 10.1021/acs.jmedchem.8b00686. [DOI] [PubMed] [Google Scholar]
- 36.Micco M. Structure-based design and evaluation of naphthalene diimide G-quadruplex ligands as telomere targeting agents in pancreatic cancer cells. J. Med. Chem. 2013;56:2959–2974. doi: 10.1021/jm301899y. [DOI] [PubMed] [Google Scholar]
- 37.Warner K.D. Principles for targeting RNA with drug-like small molecules. Nat. Rev. Drug Discov. 2018;17:547–558. doi: 10.1038/nrd.2018.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huppert J.L., Balasubramanian S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007;35:406–413. doi: 10.1093/nar/gkl1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schaffitzel C. In vitro generated antibodies specific for telomeric guanine-quadruplex DNA react with Stylonychia lemnae macronuclei. Proc. Natl. Acad. Sci. U. S. A. 2002;98:8572–8577. doi: 10.1073/pnas.141229498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paeschke K. Telomerase recruitment by the telomere end binding protein-β facilitates G-quadruplex DNA unfolding in ciliates. Nat. Struct. Mol. Biol. 2008;15:598–604. doi: 10.1038/nsmb.1422. [DOI] [PubMed] [Google Scholar]
- 41.Biffi G. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 2013;5:182–186. doi: 10.1038/nchem.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Henderson A. Detection of G-quadruplex DNA in mammalian cells. Nucleic Acids Res. 2014;42:860–869. doi: 10.1093/nar/gkt957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moye A.L. Telomeric G-quadruplexes are a substrate and site of localization for human telomerase. Nat. Commun. 2015;6:7643. doi: 10.1038/ncomms8643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu H.Y. Conformation selective antibody enables genome profiling and leads to discovery of parallel G-quadruplex in human telomeres. Cell Chem. Biol. 2016;23:1261–1270. doi: 10.1016/j.chembiol.2016.08.013. [DOI] [PubMed] [Google Scholar]
- 45.Biffi G. Elevated levels of G-quadruplex formation in human stomach and liver cancer tissues. PLoS One. 2014;9 doi: 10.1371/journal.pone.0102711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Granotier C. Preferential binding of a G-quadruplex ligand to human chromosome ends. Nucleic Acids Res. 2005;33:4182–4190. doi: 10.1093/nar/gki722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rodriguez R. Small-molecule-induced DNA damage identifies alternative DNA structures in human genes. Nat. Chem. Biol. 2012;8:301–310. doi: 10.1038/nchembio.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lefebvre J. Copper–alkyne complexation responsible for the nucleolar localization of quadruplex nucleic acid drugs labeled by click reactions. Angew. Chemie Int. Ed. 2017;56:11365–11369. doi: 10.1002/anie.201703783. [DOI] [PubMed] [Google Scholar]
- 49.Shivalingam A. The interactions between a small molecule and G-quadruplexes are visualized by fluorescence lifetime imaging microscopy. Nat. Commun. 2015;6:8178. doi: 10.1038/ncomms9178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang S. Real-time monitoring of DNA G-quadruplexes in living cells with a small-molecule fluorescent probe. Nucleic Acids Res. 2018;46:7522–7532. doi: 10.1093/nar/gky665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chambers V.S. High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat. Biotechnol. 2015;33:877–881. doi: 10.1038/nbt.3295. [DOI] [PubMed] [Google Scholar]
- 52.Marsico G. Whole genome experimental maps of DNA G-quadruplexes in multiple species. Nucleic Acids Res. 2019;1:1–13. doi: 10.1093/nar/gkz179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Law M.J. ATR-X syndrome protein targets tandem repeats and influences allele-specific expression in a size-dependent manner. Cell. 2010;143:367–378. doi: 10.1016/j.cell.2010.09.023. [DOI] [PubMed] [Google Scholar]
- 54.Gray L.T. G quadruplexes are genomewide targets of transcriptional helicases XPB and XPD. Nat. Chem. Biol. 2014;10:313–318. doi: 10.1038/nchembio.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kanoh Y. Rif1 binds to G quadruplexes and suppresses replication over long distances. Nat. Struct. Mol. Biol. 2015;22:889–897. doi: 10.1038/nsmb.3102. [DOI] [PubMed] [Google Scholar]
- 56.Paeschke K. DNA replication through G-quadruplex motifs is promoted by the Saccharomyces cerevisiae Pif1 DNA helicase. Cell. 2011;145:678–691. doi: 10.1016/j.cell.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu H. CX-5461 is a DNA G-quadruplex stabilizer with selective lethality in BRCA1/2 deficient tumours. Nat. Commun. 2017;8:14432. doi: 10.1038/ncomms14432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kouzine F. Permanganate/S1 nuclease footprinting reveals non-B DNA structures with regulatory potential across a mammalian genome. Cell Syst. 2017;4:344–356. doi: 10.1016/j.cels.2017.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hänsel-Hertsch R. G-quadruplex structures mark human regulatory chromatin. Nat. Genet. 2016;48:1267–1272. doi: 10.1038/ng.3662. [DOI] [PubMed] [Google Scholar]
- 60.Hänsel-Hertsch R. Genome-wide mapping of endogenous G-quadruplex DNA structures by chromatin immunoprecipitation and high-throughput sequencing. Nat. Protoc. 2018;13:551–564. doi: 10.1038/nprot.2017.150. [DOI] [PubMed] [Google Scholar]
- 61.Mishra S.K. G4IPDB: a database for G-quadruplex structure forming nucleic acid interacting proteins. Sci. Rep. 2016;6:38144. doi: 10.1038/srep38144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cogoi S. Structural polymorphism within a regulatory element of the human KRAS promoter: formation of G4-DNA recognized by nuclear proteins. Nucleic Acids Res. 2008;36:3765–3780. doi: 10.1093/nar/gkn120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pagano B. Identification of novel interactors of human telomeric G-quadruplex DNA. Chem. Commun. 2015;51:2964–2967. doi: 10.1039/c4cc07231f. [DOI] [PubMed] [Google Scholar]
- 64.Byrd A.K. Evidence that G-quadruplex DNA accumulates in the cytoplasm and participates in stress granule assembly in response to oxidative stress. J. Biol. Chem. 2016;291:18041–18057. doi: 10.1074/jbc.M116.718478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.González V. Identification and characterization of nucleolin as a c-myc G-quadruplex-binding protein. J. Biol. Chem. 2009;284:23622–23635. doi: 10.1074/jbc.M109.018028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Raiber E.A. A non-canonical DNA structure is a binding motif for the transcription factor SP1 in vitro. Nucleic Acids Res. 2012;40:1499–1508. doi: 10.1093/nar/gkr882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Brázda V. The amino acid composition of quadruplex binding proteins reveals a shared motif and predicts new potential quadruplex interactors. Molecules. 2018;23:2341. doi: 10.3390/molecules23092341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Huang Z.L. Identification of G-quadruplex-binding protein from the exploration of RGG motif/G-quadruplex interactions. J. Am. Chem. Soc. 2018;140:17945–17955. doi: 10.1021/jacs.8b09329. [DOI] [PubMed] [Google Scholar]
- 69.Makowski M.M. Global profiling of protein-DNA and protein-nucleosome binding affinities using quantitative mass spectrometry. Nat. Commun. 2018;9:1653. doi: 10.1038/s41467-018-04084-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sauer M., Paeschke K. G-quadruplex unwinding helicases and their function in vivo. Biochem. Soc. Trans. 2017;45:1173–1182. doi: 10.1042/BST20170097. [DOI] [PubMed] [Google Scholar]
- 71.Tippana R. Single-molecule imaging reveals a common mechanism shared by G-quadruplex–resolving helicases. Proc. Natl. Acad. Sci. U. S. A. 2016;113:8448–8453. doi: 10.1073/pnas.1603724113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Voter A.F. A guanine-flipping and sequestration mechanism for G-quadruplex unwinding by RecQ helicases. Nat. Commun. 2018;9:4201. doi: 10.1038/s41467-018-06751-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen M.C. Structural basis of G-quadruplex unfolding by the DEAH/RHA helicase DHX36. Nature. 2018;558:465–483. doi: 10.1038/s41586-018-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen W.F. Molecular mechanistic insights into Drosophila DHX36-mediated G-quadruplex unfolding: a structure-based model. Structure. 2018;26:403–415. doi: 10.1016/j.str.2018.01.008. [DOI] [PubMed] [Google Scholar]
- 75.Zaug A.J. Human POT1 disrupts telomeric G-quadruplexes allowing telomerase extension in vitro. Proc. Natl. Acad. Sci. U. S. A. 2005;102:10864–10869. doi: 10.1073/pnas.0504744102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Salas T.R. Human replication protein A unfolds telomeric G-quadruplexes. Nucleic Acids Res. 2006;34:4857–4865. doi: 10.1093/nar/gkl564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bhattacharjee A. Dynamic DNA binding, junction recognition and G4 melting activity underlie the telomeric and genome-wide roles of human CST. Nucleic Acids Res. 2017;45:12311–12324. doi: 10.1093/nar/gkx878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Thakur R.K. Metastases suppressor NM23-H2 interaction with G-quadruplex DNA within c-MYC promoter nuclease hypersensitive element induces c-MYC expression. Nucleic Acids Res. 2009;37:172–183. doi: 10.1093/nar/gkn919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cogoi S. The KRAS promoter responds to Myc-associated zinc finger and poly(ADP-ribose) polymerase 1 proteins, which recognize a critical quadruplex-forming GA-element. J. Biol. Chem. 2010;285:22003–22016. doi: 10.1074/jbc.M110.101923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Paramasivam M. Protein hnRNP A1 and its derivative Up1 unfold quadruplex DNA in the human KRAS promoter: implications for transcription. Nucleic Acids Res. 2009;37:2841–2853. doi: 10.1093/nar/gkp138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cogoi S. HRAS is silenced by two neighboring G-quadruplexes and activated by MAZ, a zinc-finger transcription factor with DNA unfolding property. Nucleic Acids Res. 2014;42:8379–8388. doi: 10.1093/nar/gku574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang Y. G-quadruplex DNA drives genomic instability and represents a targetable molecular abnormality in ATRX-deficient malignant glioma. Nat. Commun. 2019;10:943. doi: 10.1038/s41467-019-08905-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cree S.L. DNA G-quadruplexes show strong interaction with DNA methyltransferases in vitro. FEBS Lett. 2016;590:2870–2883. doi: 10.1002/1873-3468.12331. [DOI] [PubMed] [Google Scholar]
- 84.Mao S.Q. DNA G-quadruplex structures mold the DNA methylome. Nat. Struct. Mol. Biol. 2018;25:951–957. doi: 10.1038/s41594-018-0131-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tsukakoshi K. CpG methylation changes G-quadruplex structures derived from gene promoters and interaction with VEGF and SP1. Molecules. 2018;23:1–12. doi: 10.3390/molecules23040944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li P.T. Expression of the human telomerase reverse transcriptase gene is modulated by quadruplex formation in its first exon due to DNA methylation. J. Biol. Chem. 2017;292:20859–20870. doi: 10.1074/jbc.M117.808022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gupta R. DNA repair network analysis reveals shieldin as a key regulator of NHEJ and PARP inhibitor sensitivity. Cell. 2018;173:972–988. doi: 10.1016/j.cell.2018.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Papachristou E.K. A quantitative mass spectrometry-based approach to monitor the dynamics of endogenous chromatin-associated protein complexes. Nat. Commun. 2018;9:2311. doi: 10.1038/s41467-018-04619-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fouquerel E. DNA damage processing at telomeres: the ends justify the means. DNA Repair (Amst) 2016;44:159–168. doi: 10.1016/j.dnarep.2016.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Vannier J.B. RTEL1 dismantles T loops and counteracts telomeric G4-DNA to maintain telomere integrity. Cell. 2012;149:795–806. doi: 10.1016/j.cell.2012.03.030. [DOI] [PubMed] [Google Scholar]
- 91.Zahler A.M. Inhibition of telomerase by G-quartet DNA structures. Nature. 1991;350:718–720. doi: 10.1038/350718a0. [DOI] [PubMed] [Google Scholar]
- 92.Ribeyre C. The yeast Pif1 helicase prevents genomic instability caused by G-quadruplex-forming CEB1 sequences in vivo. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Paeschke K. Pif1 family helicases suppress genome instability at G-quadruplex motifs. Nature. 2013;497:458–462. doi: 10.1038/nature12149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jimeno S. The helicase PIF1 facilitates resection over sequences prone to forming G4 structures. Cell Rep. 2018;24:3262–3273. doi: 10.1016/j.celrep.2018.08.047. [DOI] [PubMed] [Google Scholar]
- 95.Fleming A.M. A role for the fifth G-track in G-quadruplex forming oncogene promoter sequences during oxidative stress: do these “spare tires” have an evolved function? ACS Cent. Sci. 2015;1:226–233. doi: 10.1021/acscentsci.5b00202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Fleming A.M. Oxidative DNA damage is epigenetic by regulating gene transcription via base excision repair. Proc. Natl. Acad. Sci. U. S. A. 2017;114:2604–2609. doi: 10.1073/pnas.1619809114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cogoi S. The regulatory G4 motif of the Kirsten ras (KRAS) gene is sensitive to guanine oxidation: implications on transcription. Nucleic Acids Res. 2018;46:661–676. doi: 10.1093/nar/gkx1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.BielskuteÌ S. Impact of oxidative lesions on the human telomeric G-quadruplex. J. Am. Chem. Soc. 2019;141:2594–2603. doi: 10.1021/jacs.8b12748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lee H.T. Molecular mechanisms by which oxidative DNA damage promotes telomerase activity. Nucleic Acids Res. 2017;45:11752–11765. doi: 10.1093/nar/gkx789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Valton A.L., Prioleau M.N. G-quadruplexes in DNA replication: a problem or a necessity? Trends Genet. 2016;32:697–706. doi: 10.1016/j.tig.2016.09.004. [DOI] [PubMed] [Google Scholar]
- 101.Lerner L.K., Sale J.E. Replication of G quadruplex DNA. Genes (Basel) 2019;10:95. doi: 10.3390/genes10020095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lopes J. G-quadruplex-induced instability during leading-strand replication. EMBO J. 2011;30:4033–4046. doi: 10.1038/emboj.2011.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Papadopoulou C. Nucleotide pool depletion induces G-quadruplex-dependent perturbation of gene expression. Cell Rep. 2015;13:2491–2503. doi: 10.1016/j.celrep.2015.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Guilbaud G. Local epigenetic reprogramming induced by G-quadruplex ligands. Nat. Chem. 2017;9:1110–1117. doi: 10.1038/nchem.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Besnard E. Unraveling cell type-specific and reprogrammable human replication origin signatures associated with G-quadruplex consensus motifs. Nat. Struct. Mol. Biol. 2012;19:837–844. doi: 10.1038/nsmb.2339. [DOI] [PubMed] [Google Scholar]
- 106.Cogoi S., Xodo L.E. G-quadruplex formation within the promoter of the KRAS proto-oncogene and its effect on transcription. Nucleic Acids Res. 2006;34:2536–2549. doi: 10.1093/nar/gkl286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Nguyen G.H. Regulation of gene expression by the BLM helicase correlates with the presence of G-quadruplex DNA motifs. Proc. Natl. Acad. Sci. U. S. A. 2014;111:9905–9910. doi: 10.1073/pnas.1404807111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Johnson J.E. Altered gene expression in the Werner and Bloom syndromes is associated with sequences having G-quadruplex forming potential. Nucleic Acids Res. 2009;38:1114–1122. doi: 10.1093/nar/gkp1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Van Wietmarschen N. BLM helicase suppresses recombination at G-quadruplex motifs in transcribed genes. Nat. Commun. 2018;9:1–12. doi: 10.1038/s41467-017-02760-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lacroix L. Identification of two human nuclear proteins that recognise the cytosine-rich strand of human telomeres in vitro. Nucleic Acids Res. 2000;28:1564–1575. doi: 10.1093/nar/28.7.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kang H.J. The transcriptional complex between the BCL2 i-motif and hnRNP LL is a molecular switch for control of gene expression that can be modulated by small molecules. J. Am. Chem. Soc. 2014;136:4172–4185. doi: 10.1021/ja4109352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Assi H.A. I-motif DNA: structural features and significance to cell biology. Nucleic Acids Res. 2018;46:8038–8056. doi: 10.1093/nar/gky735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zeraati M. I-motif DNA structures are formed in the nuclei of human cells. Nat. Chem. 2018;10:631–637. doi: 10.1038/s41557-018-0046-3. [DOI] [PubMed] [Google Scholar]
- 114.Cui Y. Mutually exclusive formation of G-quadruplex and i-motif is a general phenomenon governed by steric hindrance in duplex DNA. Biochemistry. 2016;55:2291–2299. doi: 10.1021/acs.biochem.6b00016. [DOI] [PubMed] [Google Scholar]
- 115.Kouzine F. The dynamic response of upstream DNA to transcription-generated torsional stress. Nat. Struct. Mol. Biol. 2004;11:1092–1100. doi: 10.1038/nsmb848. [DOI] [PubMed] [Google Scholar]
- 116.Sutherland C. A mechanosensor mechanism controls the G-quadruplex/i-motif molecular switch in the MYC promoter NHE III1. J. Am. Chem. Soc. 2016;138:14138–14151. doi: 10.1021/jacs.6b09196. [DOI] [PubMed] [Google Scholar]
- 117.Salvati E. PARP1 is activated at telomeres upon G4 stabilization: possible target for telomere-based therapy. Oncogene. 2010;29:6280–6293. doi: 10.1038/onc.2010.344. [DOI] [PubMed] [Google Scholar]
- 118.McLuckie K.I.E. G-quadruplex DNA as a molecular target for induced synthetic lethality in cancer cells. J. Am. Chem. Soc. 2013;135:9640–9643. doi: 10.1021/ja404868t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zimmer J. Targeting BRCA1 and BRCA2 deficiencies with G-quadruplex-interacting compounds. Mol. Cell. 2016;61:449–460. doi: 10.1016/j.molcel.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]