Abstract
Background
Cryptococcus is a conditional pathogenic fungus causing cryptococcosis, which is one of the most serious fungal diseases faced by humans. Lateral flow immunochromatographic assay (LFA) is successfully applied to the rapid detection of cryptococcal antigens.
Methods
Studies were retrieved systematically from the Embase, PubMed, Web of Science, and Cochrane Library before July 2019. The quality of the studies was assessed by Review Manager 5.0 based on the Quality Assessment of Diagnostic Accuracy Study guidelines. The extracted data from the included studies were analyzed by Meta-DiSc 1.4. Stata 12.0 software was used to detect the publication bias.
Results
A total of 15 articles with 31 fourfold tables were adopted by inclusion and exclusion criteria. The merged sensitivity and specificity in serum were 0.98 and 0.98, respectively, and those in the cerebrospinal fluid were 0.99 and 0.99, respectively.
Conclusions
Compared to the urine and other samples, LFA in serum and cerebrospinal fluid is favorable evidence for the diagnosis of cryptococcosis with high specificity and sensitivity.
Keywords: Lateral flow immunochromatographic assay, Lateral flow assay, Cryptococcosis, Diagnostic
Background
Cryptococcosis is mainly caused by Cryptococcus, an opportunistic pathogen. Cryptococcus genus is based on C. neoformans, C. deneoformans, C. gattii, and other non-pathogenic. Those strains of serotype A or var. grubii are considered to be C. neoformans and serotype D or var. Neoformans are considered to be C. deneoformans. The strains of C. gattii consist of five species: C. gattii, C. bacillisporus, C. deuterogattii, C. tetragattii and C. decagatti i [1]. C. gattii and C. neoformans are responsible for almost all cryptococcal infections in humans [2]. Besides, people with low immunity have a high probability of being infected with Cryptococcus, for example, hunman immunodeficiency virus (HIV) patients and patients with long-term use of glucocorticoids, immunosuppressants, broad-spectrum antibiotics, and anti-tumor drugs [3, 4]. All organs of humans can be infected with Cryptococcus. Without complement and anti-Cryptococcus growth factors in cerebrospinal fluid (CSF), cryptococcal meningitis (CM) is the main clinical manifestation of the cryptococcal infection in the central nervous system [5]. In 2014, the number of cryptococcal antigen-positive people worldwide was 278,000, and the global incidence of cryptococcal meningitis was 223,100. Additionally, annual global deaths from cryptococcal meningitis were estimated at 181,100 and 135,900 deaths in sub-Saharan Africa and 15% of AIDS-related deaths are caused by cryptococcal meningitis worldwide [6]. Thus, cryptococcosis has become a serious global public health problem.
However, the cryptococcal infection is short of specificity with diverse clinical manifestations. Cryptococcosis is frequently misdiagnosed at the early stage [7]. The diagnosis of cryptococcosis relies on the cultivation of conventional fungal and bacterial culture media from biological samples (CSF, sputum and skin biopsies, etc.), cytological examination of centrifuged CSF deposits and histopathological staining of other body fluids, or using of latex agglutination, enzyme immunoassay techniques to detect cryptococcal polysaccharide capsular antigen (CrAg) which has shed in serum and CSF during infection [8]. Molecular methods, although available and extensively used for research purposes, are not used currently in routine clinical practice [7]. These methods have the advantage of high specificity, but the sensitivity is low. Moreover, these tests are time-consuming and require auxiliary equipment [9].
In 2009, Immuno-Mycologics (IMMY) invented a new cryptococcal antigen detection method, lateral flow immunoassay (LFA), for diagnosis of cryptococcal infection. LFA is a rapid diagnostic method for the quantitative or qualitative detection of analytes in complex mixtures providing results within 5–30 min [8]. LFA can detect samples without special auxiliary equipment, which can also be used for the determination of single samples and preserve the results of the test. In addition to IMMY LFA, BIOSYNEX® CryptoPS is a rapid immunochromatographic test for the semi-quantitative detection and titration of Cryptococcus capsular antigens in serum, plasma, whole blood and CSF to guide the diagnosis of cryptococcal infections, especially in cases of meningitis. Biosynex CryptoPS can detect the four serotypes of Cryptococcus, and provides results within 10 min [10]. In July 2011, the U.S. Food and Drug Administration has approved lateral flow immunoassay (LFA) (Immy, Inc., Norman, OK, USA) as a semi-quantitative tool for the rapid detection of cryptococcal capsular polysaccharide antigen in the serum or CSF [11]. The application of LFA rapid detection of Cryptococcus greatly shortens the time for the diagnosis of the disease, and also has a certain positive effect on the subsequent early treatment. Therefore, we collected relevant articles for the meta-analysis to assess LFA for the diagnostic accuracy of cryptococcosis.
Methods
Search strategy and source
Four investigators systematically searched all the articles about the Cryptococcus and LFA before July 2019 in the Embase, PubMed, Web of Science, and Cochrane Library databases. We used the keywords "cryptococcus, torula, filobasidiella" and "lateral flow immunochromatographic assay, LFA, colloidal gold immunochromatography: for advanced search. Geographical restrictions were not applied in these articles.
Study selection and screening criteria
Two investigators systematically screened all of the articles by pre-established screening criteria. The inclusion criteria were as follows: (1) Studies published in English. (2) The purpose of the study was related to LFA and cryptococcosis. (3) Studies are limited to original research. (4) Studies related to diagnostics. (5) Data can be extracted to construct fourfold tables. The exclusion criteria were as follows: (1) Duplicate studies, abstracts, conference abstracts, case reports, reviews, editorials. (2) Studies without a reference standard or a detailed number of samples. (3) Samples not from humans. (4) LFA as the reference standard.
Data extraction
In the process of carefully reading the included articles, the investigators simultaneously extracted related data from the studies, including the name of the first author, year of article, study design, geographical distribution of strains, patient population, reference standard, brand of LFA-test, sample type, true positive (TP), false positive (FP), true negative (TN), and false-negative (FN). The process of extracting data is carried out independently by the investigators, and finally, the synthesis results were compared.
Quality assessment standard
We used the Quality Assessment of Diagnostic Accuracy Study (QUADAS-2) guidelines [12] to assess the quality of included studies. Then, we analyzed the risk of bias and applicability concerns by Review Manager 5.0, including patient selection, reference standard, index test, flow, and timing. If the assessment results conflicted, the investigators reviewed the original studies, and a third investigator would intervene to achieve consensus.
Statistical analysis
We analyzed the extracted data, such as specificity, sensitivity, negative likelihood ratio (NLR), positive likelihood ratio (PLR), and diagnostic odds ratio (DOR), from the included studies using meta-DiSc 1.4 software. Also, we analyzed the summary receiver operating characteristic (SROC) curve and calculated the area under the curve (AUC). According to the sample types, these studies were analyzed by different methods. Due to the lack of adequate data on urine and other samples in the included articles, these samples were analyzed by Review Manager 5.0 software for sensitivity and specificity. Finally, publication bias was evaluated by Stata12.0 software.
Results
Search results
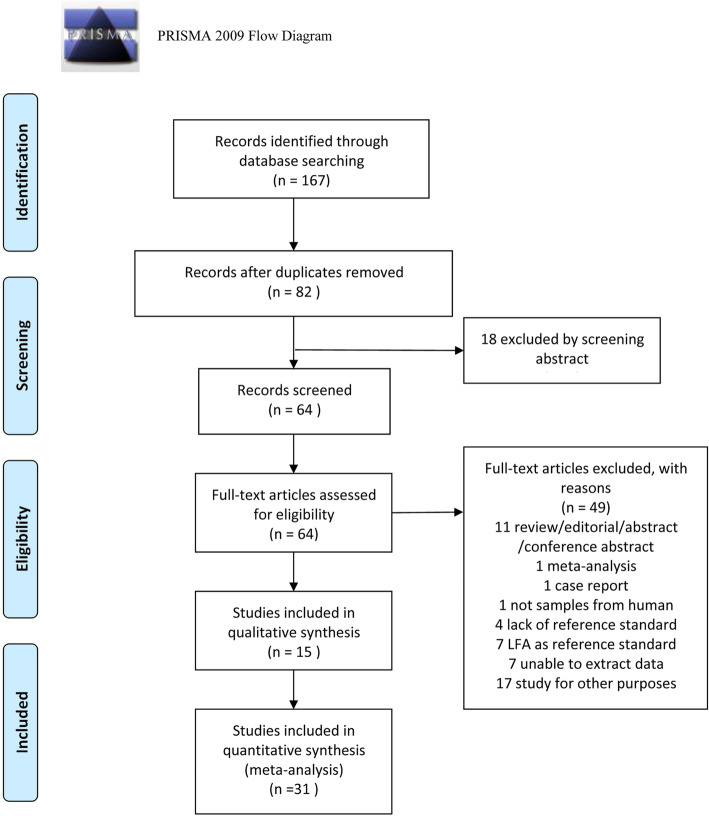
A total of 167 publications were retrieved, which decreased to 82 after excluding the duplicates. Also, 18 studies were excluded after screening the abstracts. After full-text review, we excluded 49 articles. The reasons for exclusion as shown in Fig. 1. Finally, we included 15 qualified articles [9, 11, 13–25].
Fig. 1.
Flow diagram of study identification and inclusion
Characteristics of eligible studies
Fifteen studies were published between 2011 and 2019. 13/15 articles reported data from serum samples, seven collected CSF samples, two contained urine samples, and one contained the samples of fingerprick capillary blood and whole venous blood. A total of 9312 samples were included in the meta-analysis, with an average of 620 (range 59–3447) samples. The brands of LFA-tests of total included studies were IMMY. Table 1 summarizes the characteristics of these studies.
Table 1.
Characteristics of the included studies (n = 15)
| No. | First author | Year | Geographical distribution of strains | Study design | Patient population | Sample type(s) | Sample size | Reference standard | Brand of LFA-test |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Lindsley | 2011 | Thailand | prospective | HIV | Serum Urine | 538 | EIA | IMMY |
| 2 | Binnicker | 2012 | USA | prospective& retrospective | SC | Serum | 634 | LA | IMMY |
| 3 | McMullan | 2012 | Australia | retrospective | SC | Serum | 106 | Comprehensive reference3 | IMMY |
| 4 | Escandón | 2013 | Colombia | retrospective | HIV | Serum | 421 | LA | IMMY |
| 5 | Hansen | 2013 | USA | prospective | SC | Serum CSF | 1000 | EIA | IMMY |
| 6 | Rugemalila | 2013 | Tanzania | prospective | SC | Serum | 319 | LA | IMMY |
| 7 | Boulware | 2014 | Uganda& South Africa | prospective& retrospective | HIV SM | CSF | 666 | Culture | IMMY |
| 8 | Lourens | 2014 | South Africa | prospective | HIV SM | CSF | 465 | Culture/LA | IMMY |
| 9 | Rivet-Dañon | 2015 | France | prospective& retrospective | IFI1 SC HIV | Serum CSF | 292 | LA | IMMY |
| 10 | Suwantarat | 2015 | America | retrospective &prospective | SC | Serum CSF | 1047 | EIA/Enhanced reference4 | IMMY |
| 11 | Jitmuang | 2016 | America | retrospective | HIV-N | Serum CSF | 59 | LA | IMMY |
| 12 | Cáceres | 2017 | Colombia | retrospective | CIB2 | Serum CSF | 83 | LA | IMMY |
| 13 | Frola | 2017 | Argentina | prospective | HIV | Serum | 123 | Comprehensive reference5 | IMMY |
| 14 | Temfack | 2018 | Cameroon | prospective | HIV | Serum | 186 | EIA | IMMY |
| 15 | Drain | 2019 | South Africa | prospective | HIV | VWB FCB Urine | 3447 | EIA/Combined reference6 | IMMY |
HIV hunman immunodeficiency virus, SC suspected cryptococcosis, SM suspected meningitis, HIV-N HIV-negative, CSF cerebrospinal fluid, VWB venous whole blood, FCB fingerprick capillary blood, LA latex agglutination method, EIA enzyme-linked immunoassay, LFA lateral flow assay, IMMY Immuno-Mycologics. 1:patients proven or probable invasive fungal infection other than cryptococcosis; 2:patients with or without diagnosis of cryptococcosis were randomly selected from a collection of iological samples stored in the CIB’s biobank; 3:Cryptococcosis was proven if the organism was detected by one or more of ulture, histopathology or molecular tests; 4:An enhanced reference method includes data from histopathology, cytopathology, ungal culture, and patient clinical history in addition to EIA results; 5:Pathogen identification of isolates from positive blood cultures was performed using standard microbiology methods (morphological and biochemical tests); 6:A combined reference standard for either a positive CrAg EIA or latex agglutination test
Quality assessment
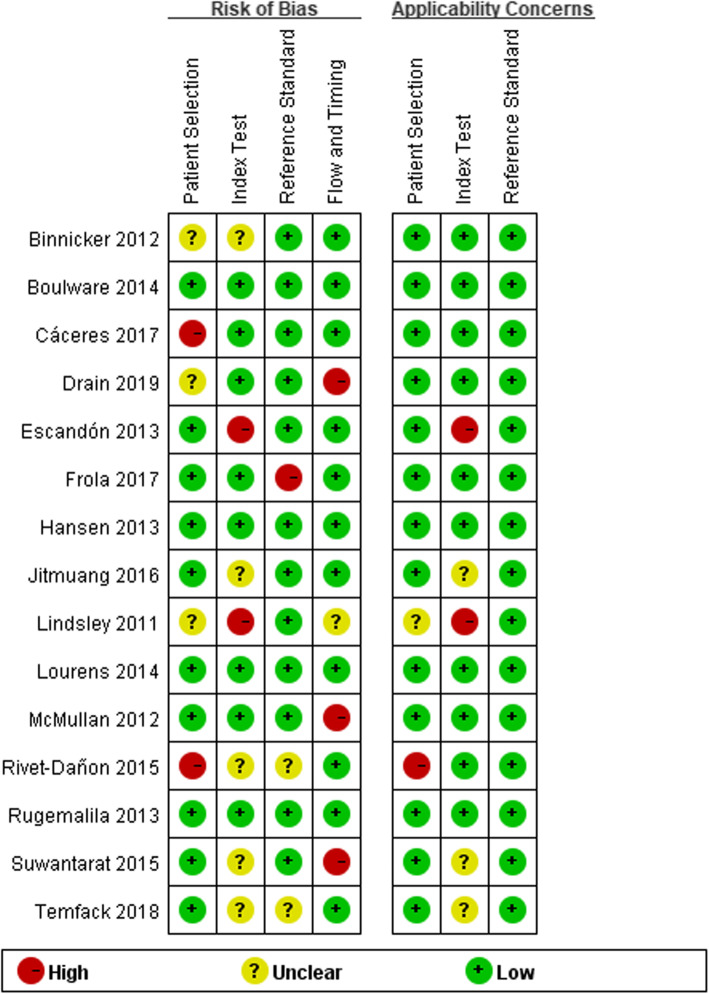
We assessed the quality of 15 articles using Review Manager 5.3. (Fig. 2).
Fig. 2.
Quality evaluation of the included studies
Data analysis
We classified the studies into different categories due to the different sample types.
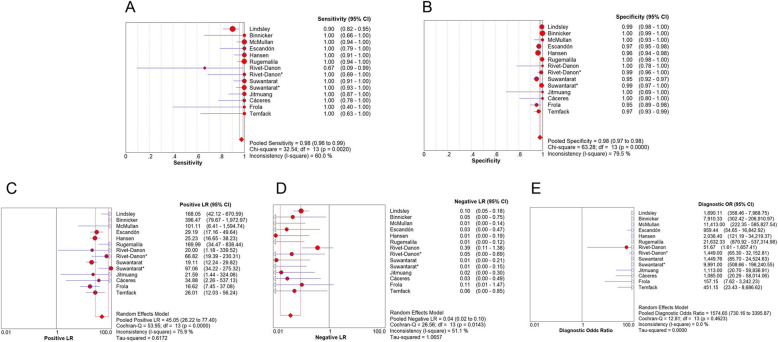
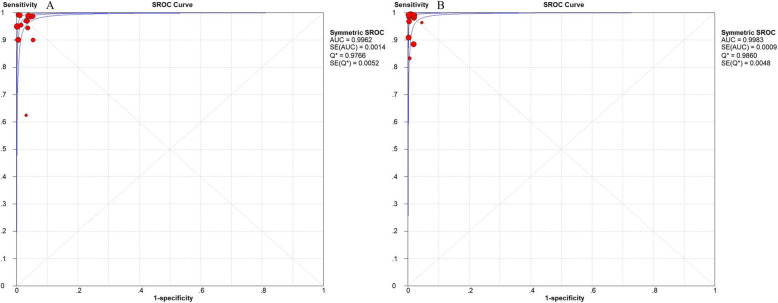
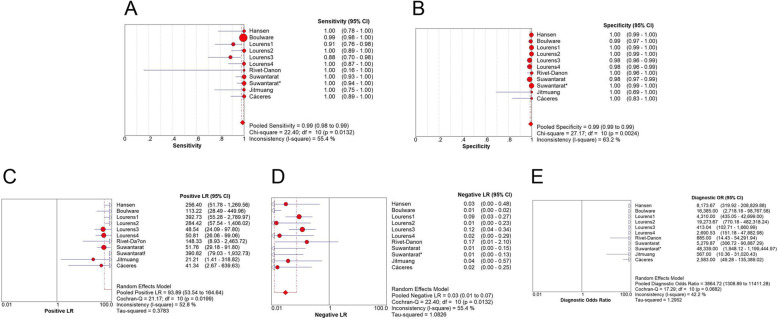
For serum specimens, the merged sensitivity and specificity values were 0.98 (95% CI: 0.96–0.99) and 0.98 (95% CI: 0.97–0.98), respectively. The average PLR of LFA in the serum was 45.05 (95% CI: 26.22–77.40) and the NLR was 0.04 (95% CI: 0.02–0.10). The merged DOR was 1574.65 (95% CI: 730.16–3395.87) and AUC was 0.9766. The results are shown in Figs. 3, 5 a.
Fig. 3.
Forest plots of a sensitivity, b specificity, c positive LR, d negative LR, e dignostic OR of LFA for the diagnosis of cryptococcosis in serum sample
Fig. 5.
Forest plots of SROC curve of the sample in a serum sample and b CSF sample
For CSF specimens, the merged sensitivity and specificity values were 0.99 (95% CI: 0.98–0.99) and 0.99 (95% CI: 0.99 to 0.99), respectively. The average PLR of LFA in CSF was 93.89 (95% CI: 53.54–164.64) and the NLR was 0.03 (95% CI: 0.01–0.07). The merged DOR was 3864.72 (95% CI: 1308.89–11,411.28) and AUC was 0.9983. The results are shown in Figs. 4, 5 b.
Fig. 4.
Forest plots of a sensitivity, b specificity, c positive LR, d negative LR, e dignostic OR of LFA for the diagnosis of cryptococcosis in CSF sample
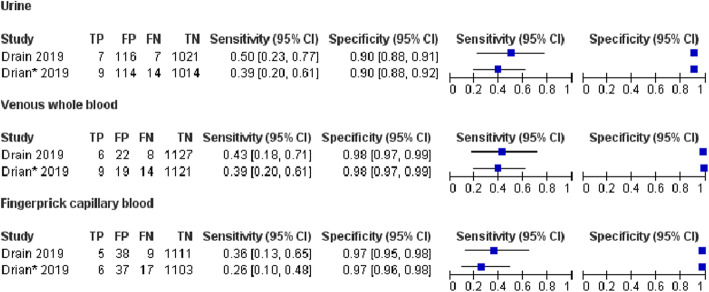
For other samples, the results of sensitivity and specificity are shown in Fig. 6.
Fig. 6.
Forest plots of the sensitivity and specificity of LFA for the diagnosis of cryptococcal infection in urine and blood
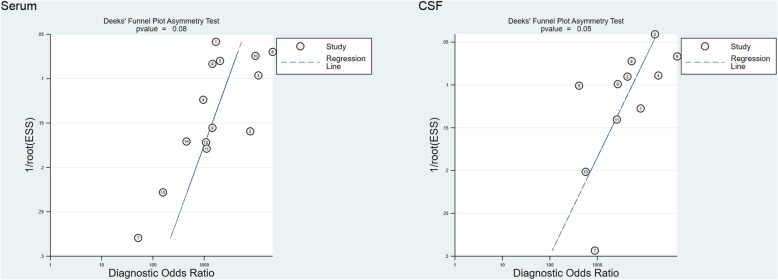
Publication bias
In this meta-analysis, the data of serum and CSF samples were tested by Stata 12.0 for publication bias. Deek’s funnel plot asymmetry test was used to assess the potential published bias in the included studies. The results of serum and CSF samples indicated that there was no obvious publication bias (Fig. 7).
Fig. 7.
Deeks’ funnel plot asymmetry test to assess publication bias in estimates of diagnostic odds ratio for LFA detection of cryptococcal infections
Discussion
Studies have shown that cryptococcosis is a disease with a relatively high mortality rate. In low- and middle-income countries, especially in sub-Saharan Africa, the mortality rate is between 26% and 63% [26]. Additionally, deaths related to cryptococcal meningitis can still reach hundreds of thousands every year [8]. Therefore, a rapid diagnosis of cryptococcal infection is necessary for patients presenting appropriate clinical symptoms. A comprehensive search with stringent screening criteria retrieved 15 articles eligible for inclusion in the study. These 15 articles encompassed 3901 serum samples, 4403 CSF samples, 1125 urine samples, 1163 venous whole blood samples, and 1163 fingerprick capillary blood samples. Moreover, the brand of LFA-tests in the included studies were IMMY, which indicated that the data we extracted would not cause great heterogeneity because of manufacturers of different brands.
The results in meta-analysis showed that the combined sensitivity of LFA in serum and CSF was 0.98 (0.96–0.99) and 0.99 (0.98–0.99); specificity was 0.98 (0.97–0.98) and 0.99 (0.99–0.99); DOR was 1574.65 (730.16–3395.87) and 2509.29 (184.18–34,187.48); SROC AUC was 0.9962 and 0.9983, respectively.
Among these indexes, the PLR of the serum and CSF was > 10, while the NLR was < 0.1. The SROC AUC of the serum and CSF was close to 1. The SROC curve was close to the upper left corner, which indicated that the area under the curve was large. Both the AUCs were > 0.9, indicating that LFA had a relatively high overall diagnostic accuracy for serum and CSF. The DOR of serum and CSF was significant, indicating that the correct diagnosis is far larger than the wrong diagnosis. In conclusion, LFA has a high degree of accuracy in the diagnosis of serum and CSF.
The current analysis of these articles revealed several factors that can explain the observed heterogeneity: the differences in the reference methods in the studies; the same reference standard was not used in the study for identification; the interpretation of the results in LFA and reference methods may cause the artificial error.
Nevertheless, the current study has some limitations. Firstly, we collected all the relevant articles. However, it was difficult to ensure that no publication was missing. Secondly, we only included the articles published in the English language, which may contribute to bias. Thirdly, our study only included the articles from inception to August 2019. The difference in the reference standard might also lead to the heterogeneity of the included studies. Finally, meta-analyses of LFA for the diagnosis of cryptococcosis, only until 2015, were included. Thus, we could comprehensively analyze the accuracy of the LFA diagnosis of the cryptococcal infection.
Conclusions
In summary, our meta-analysis indicated that LFA tested in serum and CSF has high diagnostic accuracy in the diagnosis of cryptococcal infection for high-risk patients, such as HIV-infected patients. LFA performed in urine, or other samples could be a screening tool for the early diagnosis of cryptococcal infection; however, additional studies are required for the substantiation of these results.
Acknowledgements
Not applicable.
Abbreviations
- AUC
Calculated the area under the curve
- CSF
Cerebrospinal fluid
- CM
Cryptococcal meningitis
- DOR
Diagnostic odds ratio
- FP
False positive
- FN
False negative
- HIV
Hunman immunodeficiency virus
- LFA
Lateral flow immunochromatographic assay
- NLR
Negative likelihood ratio
- PLR
Positive likelihood ratio
- QUADAS
Quality Assessment of Diagnostic Accuracy Study
- SROC
Summary receiver operating characteristic
- TP
True positive
- TN
True negative
Authors’ contributions
XG conceived and designed the experiments. LX, GL, HD and Y-XL analyzed the data and made the Tables. LX, Y-LL and JQ contributed to the production of figures by the analysis tools. All authors participated in the writing, reading, and revising of the manuscript and approved the final version of the manuscript.
Funding
No funding was used to support this study.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Idnurm A, Lin X. Rising to the challenge of multiple Cryptococcus species and the diseases they cause. Fungal Genet Biol. 2015;78:1–6. doi: 10.1016/j.fgb.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Springer DJ, Chaturvedi V. Projecting global occurrence of Cryptococcus gattii. Emer Inf Dis. 2010;16:14–20. doi: 10.3201/eid1601.090369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gibson JF, Johnston SA. Immunity to Cryptococcus neoformansand C.gattii during cryptococcosis. Fungal Genet Biol. 2015;78:76–86. doi: 10.1016/j.fgb.2014.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu J, Eastman AJ, Flaczyk A, et al. Disruption of early tumor necrosis dactor alpha signaling prevents classic alactivation of dendritic cells in lung-associated lymph nodes and development of protective immunity against cryptococcal infection. MBio. 2016;7(4):e00510–e00516. doi: 10.1128/mBio.00510-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown GD, Denning DW, Levitz SM. Tackling human fungal infections [J] Sci. 2012;336(6082):647. doi: 10.1126/science.1222236. [DOI] [PubMed] [Google Scholar]
- 6.Rajasingham R, Smith RM, Park BJ, et al. Global burden of disease of HIV-associated cryptococcal meningitis: an updated analysis. Lance Infect Dis. 2017;17(8):873–881. doi: 10.1016/S1473-3099(17)30243-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maziarz EK, Perfect JR. Cryptococcosis. Inf Dis Clin North Am. 2016;30(1):179–206. doi: 10.1016/j.idc.2015.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kozel TR, Bauman SK. CrAg lateral flow assay for cryptococcosis [J] Expert Opinion Med Diagn. 2012;6(3):245–251. doi: 10.1517/17530059.2012.681300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drain PK, Hong T, Krows M, et al. Validation of clinic-based cryptococcal antigen lateral flow assay screening in HIV-infected adults in South Africa. Sci Rep. 2019;9(1). [DOI] [PMC free article] [PubMed]
- 10.Temfack E, Kouanfack C, Mossiang L, et al. Cryptococcal antigen screening in asymptomatic HIV-infected antiretroviral Naïve patients in Cameroon and evaluation of the new semi-quantitative Biosynex CryptoPS test. Fron Micr. 2018;9:409. doi: 10.3389/fmicb.2018.00409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mcmullan BJ, Catriona H, Sorrell TC, et al. Clinical utility of the Cryptococcal antigen lateral flow assay in a diagnostic mycology laboratory [J] PLoS One. 2012;7(11):e49541. doi: 10.1371/journal.pone.0049541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lan W, Yong Z, Xian-tao Z. The QUADAS-2 tool for the quality assessment of diagnostic accuracy study:an introduction. J Hub Uni Med. 2013;32(03):201–208. [Google Scholar]
- 13.Lindsley MD, Mekha N, Baggett HC, et al. Evaluation of a newly developed lateral flow immunoassay for the diagnosis of Cryptococcosis. Clin Inf Dis. 2011;53(4):321–325. doi: 10.1093/cid/cir379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Binnicker MJ, Jespersen DJ, Bestrom JE, Rollins LO. Comparison of four assays for the detection of Cryptococcal antigen. Clin Vac Immunol. 2012;19(12):1988–1990. doi: 10.1128/CVI.00446-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Escandón, Patricia, Lizarazo J, Agudelo C I, et al. Evaluation of a rapid lateral flow immunoassay for the detection of cryptococcal antigen for the early diagnosis of cryptococcosis in HIV patients in Colombia [J]. Med Mycol, 2013, 51(7): 765–768. [DOI] [PubMed]
- 16.Hansen J, Slechta ES, Gates-Hollingsworth MA, et al. Large-scale evaluation of the Immuno-Mycologics lateral flow and enzyme-linked immunoassays for detection of Cryptococcal antigen in serum and cerebrospinal fluid. Clin Vac Immunol. 2013;20(1):52–55. doi: 10.1128/CVI.00536-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rugemalila J, Maro VP, Kapanda G, et al. Cryptococcal antigen prevalence in HIV-infected Tanzanians: a cross-sectional study and evaluation of a point-of-care lateral flow assay [J] Tropical Med Int Health. 2013;18(9):1075–1079. doi: 10.1111/tmi.12157. [DOI] [PubMed] [Google Scholar]
- 18.Boulware DR, Rolfes MA, Rajasingham R, et al. Multisite validation of Cryptococcal antigen lateral flow assay and quantification by laser thermal contrast. Emerg Inf Dis. 2014;20(1):45–53. doi: 10.3201/eid2001.130906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lourens A, Jarvis JN, Meintjes G, et al. Rapid diagnosis of Cryptococcal meningitis by use of lateral flow assay on cerebrospinal fluid samples: influence of the high-dose “hook” effect. J Clin Microbiol. 2014;52(12):4172–4175. doi: 10.1128/JCM.01683-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rivet-Dañon D, Guitard J, Grenouillet F, et al. Rapid diagnosis of cryptococcosis using an antigen detection immunochromatographic test [J] J Inf. 2015;70(5):499–503. doi: 10.1016/j.jinf.2014.12.017. [DOI] [PubMed] [Google Scholar]
- 21.Suwantarat N, Dalton JB, Lee R, et al. Large-scale clinical validation of a lateral flow immunoassay for detection of cryptococcal antigen in serum and cerebrospinal fluid specimens [J] Diagn Microbiol Inf Dis. 2015;82(1):54–56. doi: 10.1016/j.diagmicrobio.2015.01.012. [DOI] [PubMed] [Google Scholar]
- 22.Jitmuang A, Panackal AA, Williamson PR, et al. Performance of the Cryptococcal antigen lateral flow assay in non-HIV related Cryptococcosis. J Clin Microbiol. 2015;54(2):460–463. doi: 10.1128/JCM.02223-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cáceres Diego H, Alejandra Z, Tabares Ángela M, et al. Evaluation of a Cryptococcal antigen Lateral Flow Assay in serum and cerebrospinal fluid for rapid diagnosis of cryptococcosis in Colombia. Rev Do Ins Med Trop São Paulo, 2017, 59(0):e76. [DOI] [PMC free article] [PubMed]
- 24.Frola C, Guelfand L, Blugerman G, et al. Prevalence of cryptococcal infection among advanced HIV patients in Argentina using lateral flow immunoassay [J] PLoS One. 2017;12(6):e0178721. doi: 10.1371/journal.pone.0178721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elvis T, Charles K, Leonella M, et al. Cryptococcal antigen screening in asymptomatic HIV-infected antiretroviral Naïve patients in Cameroon and evaluation of the new semi-quantitative Biosynex CryptoPS test. Fron Microbiol. 2018;9:409. doi: 10.3389/fmicb.2018.00409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vidal José. Preemptive Therapy for Cryptococcal Meningitis: A Valid Strategy for Latin America?. J Fungi, 2016, 2(2):14. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.