Highlights
-
•
miRNAs are important epigenetic regulators of placental gene and protein expression.
-
•
miRNAs play a role in placenta developmental processes such as trophoblast invasion.
-
•
Placental miRNAs are susceptible to exposure to environmental chemicals.
-
•
miRNAs are involved in the pathogenesis of pregnancy-related diseases.
Keywords: microRNA, Placenta, Epigenetic, Prenatal, Maternal
Abstract
MicroRNAs (miRNAs) are epigenetic modifiers that play an important role in the regulation of the expression of genes across the genome. miRNAs are expressed in the placenta as well as other organs, and are involved in several biological processes including the regulation of trophoblast differentiation, migration, invasion, proliferation, apoptosis, angiogenesis and cellular metabolism. Related to their role in disease process, miRNAs have been shown to be differentially expressed between normal placentas and placentas obtained from women with pregnancy/health complications such as preeclampsia, gestational diabetes mellitus, and obesity. This dysregulation indicates that miRNAs in the placenta likely play important roles in the pathogenesis of diseases during pregnancy. Furthermore, miRNAs in the placenta are susceptible to altered expression in relation to exposure to environmental toxicants. With relevance to the placenta, the dysregulation of miRNAs in both placenta and blood has been associated with maternal exposures to several toxicants. In this review, we provide a summary of miRNAs that have been assessed in the context of human pregnancy-related diseases and in relation to exposure to environmental toxicants in the placenta. Where data are available, miRNAs are discussed in their context as biomarkers of exposure and/or disease, with comparisons made across-tissue types, and conservation across studies detailed.
1. Introduction
The placenta is a critical organ that has multiple roles including oxygen balance, metabolite transfer between fetus and mother, hormone production and acts as a selective barrier protecting the fetus [1]. As a mediator between maternal and fetal compartments, the placenta has been shown to trigger robust responses to changes in the maternal environment. For example, depending on the particular insult, the placenta may elicit a variety of responses ranging from changes in the placenta morphology, physiology and epigenome to potentially affect fetal development and in some instances having later life consequences in the offspring [[2], [3], [4]]. There is increasing awareness of the modifications to the epigenome in relation to environmental stimuli. These epigenetic changes serve as a biological mechanism by which placental gene expression is altered. Such changes in gene expression often lead to downstream effects contributing to pregnancy complications that may have a deleterious impact on the developing fetus.
MicroRNA (miRNA)s are epigenetic modifiers that are important for placental development and contribute to the pathogenesis of pregnancy-related diseases. Specifically, miRNAs in the placenta have been implicated in pregnancy complications such as pregnancy loss, preeclampsia, intrauterine growth restriction, gestational diabetes mellitus (GDM), obesity, and preterm birth [5]. Furthermore, placenta miRNAs have been used to differentiate between normal pregnancies and disease related to pregnancies, thus underscoring their roles in the pathogenesis of these diseases [5]. Additionally, miRNAs in the placenta, as well as placental-specific miRNAs in maternal circulation, are susceptible to changes in the maternal milieu including environmental exposure to toxicants [5]. Thus, miRNAs may serve as biomarkers to xenobiotic exposures as well as related clinical syndromes.
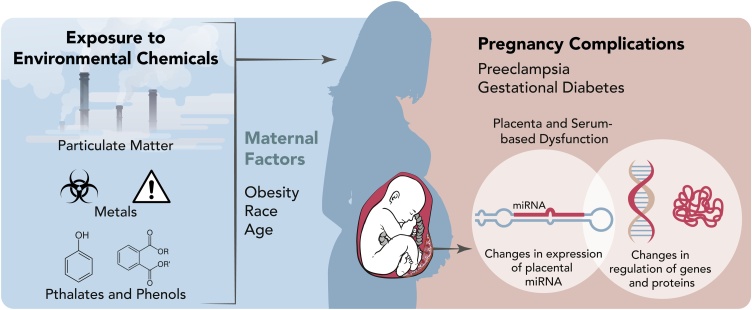
In this review, we provide a summary of literature highlighting associations among miRNA expression in the placenta, pregnancy-related complications and/or environmental toxicants (Fig. 1). The role of miRNAs that influence placental miRNA expression, and their potential effect on pregnancy and fetal development are discussed.
Fig. 1.
Various environmental factors are associated with pregnancy compications including preeclampsia and gestational diabetes. Evidence supports a role for miRNAs in the placenta as biomarkers of exposure and/or effect that may be impacted by obesity, race and age.
2. The role of miRNAs in the placenta
miRNAs are important regulators of the genome and influence cell function in tissues/organs throughout the body. miRNAs are endogenous, single-stranded, short non-coding RNA sequences of approximately 22 nucleotides that regulate gene expression [6]. It is estimated that more than 2000 miRNAs exist in the human genome and are key regulators of development, cell identity and diseases [7]. miRNAs may repress gene translation by binding to complementary sequences of specific mRNAs resulting in the degradation of target genes or translational blockade resulting in gene silencing [8,9].
In the placenta, miRNAs are essential for many physiological processes including developmental timing, apoptosis, proliferation, angiogenesis, stem cell differentiation, neurogenesis, and immune responses [[10], [11], [12], [13]]. Moreover, biological molecules important for miRNA biogenesis, such as Argonaute 2 (Ago2), Drosha, Exportin 5, Dicer, and DP103, have all been identified in placental trophoblast cells, indicating that active miRNA biogenesis exists in the placenta [14,15]. There are more than 600 miRNAs that are expressed in the placenta. Of the miRNAs expressed in the placenta, the chromosome 19 miRNA cluster (C19MC) is termed placental-specific because this group of miRNAs is predominantly expressed in the placenta [[16], [17], [18], [19], [20], [21]]. C19MC is the largest described miRNA gene cluster in the human genome to date, spanning about 100 kb on chromosome 19q13.4 and consisting of about 54 miRNAs [16,22]. The C19MC family is found exclusively in primates [23]. Interestingly, C19MC is regulated by genomic imprinting with only the paternally-inherited allele being expressed [23]. The fact that the C19MC cluster expression is controlled by the paternally-inherited allele suggests that there may be increased susceptibility in relation to paternal exposure to environmental chemicals. Thus, as this is often understudied, future research should examine the effects of both maternal and paternal exposures to contaminants.
Studies of the expression profile of the C19MC miRNAs in term human placentae have reported that the majority of the human trophoblasts express C19MC miRNAs at a higher level compared to all other miRNAs in the placenta. The most highly expressed C19MC miRNAs are miR-517a, miR517b, miR-516b, miR-525−5p, miR-512−3p, and miR-515−3p [24]. One intriguing aspect of these placental-specific miRNAs is their ability to be secreted in maternal circulation during gestation. For example, miR-526b, miR-517a, miR-517c, miR-518b, miR-515−3p, miR-520 h, miR-525, miR-520miR-526a, miR-516−5p are placental-specific miRNAs, are highly expressed in maternal circulation during gestation but significantly decreased at the end of pregnancy [5,25]. Placental-specific miRNAs released into maternal circulation are stabilized by binding to argonaute proteins or stored in exosomes [26]. These placental-derived miRNAs dynamically adapt to the feto-maternal milieu [27], making them potential biomarkers for pregnancy-associated complications such as preeclampsia, gestational diabetes, obesity, ectopic pregnancies, fetal growth restriction, recurrent pregnancy loss, and preterm birth [28,29] This underscores a role for miRNAs in the pathogenesis of these pregnancy-associated diseases [5].
In addition to the C19MC, another large cluster of miRNAs have been found in the placenta, though this group of miRNAs is not placental-specific. Embedded within the maternally imprinted DLK-DIO3 domain and located on the human chromosomal site 14q32, the miRNA cluster on chromosome 14 (C14MC) is conserved in mammals and has been shown to play important roles in placentation, brain development, and prenatal growth [30,31]. The C14MC cluster is grouped into two genomic regions: the miR-127/miR-136 cluster and the miR-379/miR-410 cluster [32]. The miR-127/miR-136 cluster regulates fetal capillaries in the labyrinth zone during placentation [32] and the miR-379/miR-410 promotes proliferation, migration and invasion in human trophoblast cell line HTR8/SVneo [33].
In the placenta, miRNAs play important roles during placental development. Specifically, they are involved in trophoblast differentiation, migration, invasion, vasculogenesis and cellular metabolism [34,35]. For example, miR-21, an oncogene known to promote cell proliferation and migration by inhibiting PTEN in ovarian epithelia carcinomas has been shown to play similar roles in the placenta [36]. Overexpression of miR-21 in the third trimester placenta cell line (TCL-1) results in a 50 % reduction of PTEN, suggesting that miR-21 specifically targets PTEN in the placenta [36]. Interestingly, downregulation of miR-21 in placental cells results in decreased invasion of the maternal decidua, decreased migration, and decreased growth, suggesting that miR-21 is involved in trophoblast cell migration and invasion [37].
In relation to gestation, miRNA expression patterns in the placenta change from the first trimester to the third trimester. For example, a comparison between placentas isolated from the first and third trimester highlighted a total of 208 miRNA transcripts that were differentially expressed [38]. Interestingly, distinct cluster expression profiles showed that miRNAs which regulate angiogenic and proliferative genes were highly expressed in the first trimester, whereas miRNAs that promoted cell differentiation and suppressed proliferation were highly expressed in the third trimester, highlighting the important roles of miRNA in normal placental development [38]. Notably, the chromosome 19 miRNA cluster increases expression across gestation from the first trimester to the third trimester, while the chromosome 14 miRNA cluster decreases expression across gestation [38,39].
miRNA expression patterns differ not only across gestation but are also heterogenous within the placenta. Specifically, in the human placenta, the miRNA expression pattern in the basal plate is different from the expression pattern in the chorionic plates [40]. Similar to humans, this heterogeneity of placenta miRNAs has also been observed in pigs where miRNA expression varies between the maternal and fetal sides of the placenta [41]. The spatiotemporal expression of miRNA in the placenta underscores the important role of miRNAs during placentation adding to the complexity of the placental miRNA landscape.
Beyond the placenta, miRNAs have been detected in biological fluids including blood, breast milk, amniotic fluid, saliva, and urine [42]. These extracellular miRNAs are incredibly resistant to nucleases and can be aberrantly expressed during diseases and pregnancy. For this reason, extracellular miRNAs, particularly those in circulation, have promising potential to be used as diagnostic biomarkers. However, the origin and biological function of these circulatory miRNAs remain largely unknown and are subject to several hypotheses. Since perturbations in blood cell counts and hemolysis have been shown to significantly disrupt circulatory miRNAs, it is thought that blood cells secrete miRNAs into the serum [43]. In contrast, several studies have suggested that other tissues including the liver, muscle, brain, and heart may contribute to circulatory miRNAs [[44], [45], [46], [47], [48]]. During pregnancy, placental-specific miRNAs have been detected in maternal blood, and these miRNAs disappear after birth. These data suggest that the placenta also contributes to the production of circulatory miRNAs [5].
3. Pregnancy-related complications
Studies that detail miRNAs that display altered expression in relation to pregnancy complications are detailed below (Table 1).
Table 1.
miRNAs in the placenta and plasma that are responsive to pregnancy-related complications.
| Pregnancy-related complication | Target Tissue(s) | Sample Size (N) | # miRNAs Measured | Differentially Expressed miRNAs | Methods |
Author (Year) | ||
|---|---|---|---|---|---|---|---|---|
| Extraction | miRNA Quantification | Normalization | ||||||
| Preeclampsia | Placenta, plasma | N = 40 (20 PE, 20 C) | 1 | miR-195a | mirVana miRNA Isolation Kit (Ambion) | qRT-PCR; targeted analysis | U6 | (Gunel, Kamali et al. 2020) |
| Placenta, plasma, HTR8/SVneo Cells | N = 53 (20 PE, 33 C) | 184 | miR-195a, miR-223, miR-218, miR-17, miR-18a, miR-19b1, miR-92a1, miR-379, miR-411, miR-210b, miR-30a-3p, miR-518b, miR-524, miR-17−3p, miR-151, miR-193b | TRIzol Reagent (Invitrogen) and PEG method | miRNA microarray; qRT-PCR | U6 | (Xu, Zhao et al. 2014) | |
| Placenta, plasma | Placenta N = 20 (13 PE, 7 C) Plasma N = 16 (8 PE, 8 C) |
4 | miR-210b | TRIzol Reagent (Invitrogen) | qRT-PCR | hsa-miR-103a | (Biró, Fóthi et al. 2019) | |
| Placenta | N = 34 (23 PE, 11 C) | 34 | miR-210b, miR-152, miR-411, miR-377, miR-518b, miR-18a, miR-363, miR-542−3p | TRIzol Reagent (Invitrogen) | miRNA microarray; qRT-PCR | U6 | (Zhu, Han et al. 2009) | |
| Placenta | N = 18 | 3 | miR-21c, miR-122, miR-155 | mirVana miRNA Isolation Kit (Ambion) | TaqMan qRT-PCR | RNU44 | (Lasabová, Vazan et al. 2015) | |
| Placenta | N = 21 | 866 | miRNA-451, miR-720 | TRIzol Reagent (Invitrogen) | miRNA microarray; TaqMan qRT-PCR | RNU24 | (Vidal, Ramão et al. 2018) | |
| Placenta, HTR8/SVneo Cells | N = 32 (15 PE, 17 C) | 1 | miR-195a | TRIzol Reagent (Invitrogen) | qRT-PCR; targeted analysis | U6 | (Bai, Yang et al. 2012) | |
| HTR8/SVneo Cells | 1 | miR-195a | TRIzol Reagent (Invitrogen) | qRT-PCR; targeted analysis | U6 | (Wu, Wang et al. 2016) | ||
| Placenta, plasma | N = 70 (34 PE, 36 C) | 7 | miR-532−5p, miR -423−5p, miR -127−3p, miR -539−5p, miR -519a-3p, miR -6295p, let-7c-5p | Placenta: miRNeasy Micro Kit (Qiagen); Plasma: Serum Plasma kit (Qiagen) | miRNA deep sequencing; qRT-PCR | Placenta: SNORD68; Plasma: cel-miR-39 | (Timofeeva, Gusar et al. 2018) | |
| Fetal Growth | Placenta | N = 107 | 6 | miR-16d, miR-21c | mirVana miRNA Isolation Kit (Ambion) | TaqMan qRT-PCR | RNU44 | (Maccani, Padbury et al. 2011) |
| Gestational Diabetes Mellitus | Placenta | N = 16 (8 GDM, 8 C) | 32 | miR-138−5p | mirVana PARIS Kit (Ambion) | miRNA sequencing; qRT-PCR | U6 | (Ding, Guo et al. 2018) |
| Serum | N = 48 (24 GDM, 24 C) | 73 | miR-132, miR-29a, miR-222f | TRIzol Reagent (Invitrogen), miRNeasy Mini kit (Qiagen) | TaqMan Low Density Array, qRT-PCR | cel-miR-39 | (Zhao, Dong et al. 2011) | |
| Placenta | N = 20 (10 GDM, 10 C) | 29 | miR-508−3p, miR-27a, miR-9, miR-137, miR-92a, miR-33a, miR-30d, miR-362−5p, miR-502−5p | RecoverAllTM Total Nucleic Acid Isolation Kit (Ambion) | miRNA microarray; qRT-PCR | U6 | (Li, Song et al. 2015) | |
| Placenta | N = 70 (20 preOB, 25 gestOB, 25 C) | 723 | miR-100, miR-1269, miR-1285, miR-181, miR-185, miR-214, miR-296, miR-487 | mRNeasy mini kit (Qiagan) | TaqMan Low Density Array; qRT-PCR | Pool A: miR-523, miR-532, miR-425p Pool B: miR-30e-3p, miR-519b-3p, miR-520d-3p |
(Carreras-Badosa, Bonmatí et al. 2017) | |
| Obesity | Placenta | N = 211 | 7 | miR-20a, miR-34a, miR-222f, miR-146ae, miR-210b | miRNeasy kit (Qiagan) | qRT-PCR | RNU6 | (Tsamou, Martens et al. 2017) |
Reproduced miRNAs across studies: a miR-195, b miR-210, c miR-21, d miR-16, e miR-146a, f miR-222.
C = control. PE = preeclampsia. GDM = Gestational Diabetes Mellitus. preOB = pre-gestational obesity. gestOB = gestational obesity.
3.1. Preeclampsia
Preeclampsia is a multisystem disorder that is associated with hypertension and proteinuria after 20 weeks of gestation [49]. Preeclampsia complicates 3–8 % of all pregnancies and is the most common cause of neonatal morbidity and perinatal mortality world-wide [[49], [50], [51]]. Although the exact etiology of preeclampsia is not understood, miRNAs are suspected to play an essential role in the pathological processes associated with preeclampsia. Preeclampsia is related to a placentation defect which is characterized by impaired cytotrophoblast differentiation during their invasion into the spiral arteries of the uterus. This causes a restriction of placental blood flow, resulting in hypoxia. Thus, hypoxia, necrosis, and trophoblast apoptosis are all part of the molecular pathogenesis of preeclampsia [52]. miRNA-210 is a hypoxia-sensitive miRNA found to be overexpressed in preeclamptic placentas [53]. The high levels of miR-210 in preeclamptic placentas also correlate with increased exosome encapsulation of miR-210 and Ago-bound miR-210 in circulation, which may contribute to the etiology of preeclampsia [26]. It is worth noting that miR-210 expression is altered in cardiovascular diseases such as myocardial infarction and atherosclerosis [54], therefore these diseases may also contribute to the overall increase in miR-210 in circulation.
miRNAs play a critical role in regulating placental development. Some specific miRNAs in the placenta that regulate normal placental physiology are miR-141 and miR-519d-3p [[55], [56], [57]]. These miRNAs are responsible for regulating trophoblast cell proliferation, invasion, migration and intercellular communication [58]. Another important placental miRNA is miR-451, which is suggested to be a tumor suppressor involved in the regulation of biological processes, such as cell proliferation, migration and treatment responses in certain cancer cells [59]. The following miRNAs play an important role in possibly explaining the pathogenesis of preeclampsia: miR-21, miR-122, miR-26a-5p, miR‐103a-3p, and miR-145−5p, miR-499a-5p, miR-195, and miR-210 [49,60,61]. Specifically, miR-195 is associated with abnormal placental growth mechanisms such as impaired cellular proliferation, inadequate trophoblastic invasion and apoptosis [60].
Preeclamptic placental samples have been compared to samples obtained from healthy women. A difference in the expression of miR-21 and miR-122 was observed in the placental cells [61]. The results showed that apoptosis-associated miRNAs (miR-21 and miR-122) were significantly (p < 0.001) overexpressed in preeclamptic placentas [61]. Increased expression of these microRNAs may potentially downregulate several mRNA targets including those involved in trophoblast proliferation contributing to the etiology of preeclampsia [61].
In a recent study, circulating miRNAs from both maternal plasma and placentas from 20 preeclamptic patients and 10 normotensive pregnancy were analyzed [60]. miR-195 was downregulated by a 3.83-fold in preeclamptic placentas compared to normotensive placentas [60] suggesting a plausible role for miRNAs in the placental pathology of preeclampsia. Bioinformatic analysis predicted that miR-195 targets ActRIIA, which is the type II receptor for Activin A and Nodal. A functional role for miR-195 in the placenta was investigated using in vitro human trophoblast cells, HTR8/SVneo cells. Here it was observed that miR-195 represses ActRIIA and promotes trophoblast invasion [62]. Thus, decreased expression of miR-195, as seen in preeclamptic placentas, may contribute to the etiology through the Activin/Nodal signaling pathway in the human placenta [62,63].
Another study utilized miRNA deep sequencing and quantitative RT-PCR to compare miRNA expression profiles in the placentas and maternal serum of healthy women and women with early- to late-onset of preeclampsia [64]. The results indicated a significant decrease in the expression of miR-532−5p, miR-423−5p, miR-127−3p, miR-539−5p, miR-519a-3p, and miR-629−5p and miR-let-7c-5p and an increase in the expression of miR-423−5p, miR-519a-3p, and miR-629−5p and miR-let-7c-5p in maternal plasma [64].
Of note, variation in the expression of the preeclampsia-associated miRNAs reviewed here may be due to differences in cohort characteristics, diagnostic criteria, sampling technique, and/or miRNA profiling platforms [65]. Nonetheless, a systemic review of miRNA expression that compared 58 studies to identify the most miRNAs associated with preeclampsia pathogenesis reported that miR-210, miR-223, and miR-126 are strongly associated with the etiological domains of hypoxia, immune cell regulation, and angiogenesis [65]. Furthermore, miR-515, belonging to the C19MC family and aberrantly expressed in preeclampsia, is strongly associated with trophoblast invasion [65]. Overall, the differential expression of miRNA in placentas and serum of preeclamptic patients suggest that they potentially contribute to the etiology of the disease and can be useful predictive biomarkers for diagnosing preeclampsia.
3.2. Gestational diabetes
Gestational diabetes mellitus (GDM), the most common medical complication of pregnancy, is defined as glucose intolerance with onset or first recognition during pregnancy. Affecting between 6–7 % of pregnant women, gestational diabetes occurs when a woman’s pancreatic function is unable to overcome the diabetogenic environment of pregnancy [66]. Gestational diabetes is characterized by high glucose levels in the mother and can lead to maternal and neonatal adverse outcomes [67,68].
A comparison in the expression of miRNAs in placental tissues from GDM and normal controls revealed nine miRNAs that were differentially expressed. Specifically, miR-508−3p was upregulated and miR-27A, miR-9, miR-137, miR-92a, miR-30d, miR-362−5p, and miR-502p were downregulated. These miRNAs are known to control genes involved in key pathways such as epidermal growth factor receptor (EGFR), phosphoinositide 3-kinase (PI3K), and protein kinase B (AKT) [69]. To further confirm that these pathways were altered in the placentas of GDM patients, western blot and RT-PCR analysis were performed in terminal placentas of both normal (N = 5) and GDM (N = 5) subjects. mRNA levels of PIK3CD and PIK3R3 were significantly increased in expression (p < 0.01) whereas mRNA expression levels of EGFR, PIK3CA, PIK3CB, PIK3CG did not differ between the two groups [69]. Additionally, EGFR, PIK3CG and p-AKT protein levels were upregulated, and PIKFyve was significantly decreased in GDM tissues. Moreover, these pathways play important roles in both placenta and fetal development.
Additionally, RNAseq analysis of 16 placentas from 8 GDM patients and 8 controls resulted in the identification of 32 miRNAs that are differentially expressed between GDM patients and controls. Of these, miR-138−5p was shown to target the 3′ untranslated region of the transducing β-like protein 1 (TBL1X) gene and significantly inhibit the migration and proliferation of trophoblasts [70].
In addition to placental miRNAs, circulating miRNAs that are not placental-specific have been shown to be differentially expressed in maternal serum from the early second trimester (16–19 weeks) in GDM patients and controls [71]. Here, miR-132, miR-29a and miR-222 were significantly decreased in the serum of GDM patients compared to controls of similar gestational age [71]. Although the role of these miRNAs in the placenta are not known, knockdown of miR-29a in human hepatocytes cells (HepG2) resulted in significant induction of insulin-induced gene 1 (Insig 1), which activates genes involved in cholesterol, fatty acid metabolism and glucose homeostasis. Overall, these studies suggest that miRNAs may play an important role in gene regulation of glucose homeostasis and may contribute to the pathogenesis of GDM. Thus, they can be useful biomarkers that can be used in diagnosing GDM.
4. Obesity
Obesity is defined as an extremely high body weight to height ratio [72]. Obesity affects over a third of the population, and is expected to affect approximately 38 % of the world population by 2038 [72]. Excess body weight is not only associated with a variety of chronic diseases, including disability, depression, type 2 diabetes and cardiovascular disease, but can also result in mortality. During fetal development, there is a critical period when exposure to extrauterine stressors can have major effects on the growth and development of the fetus [73]. Additionally, there is an association between maternal obesity and expression of miRNAs in the placenta [73].
In obese mothers, the intrauterine environment is disturbed as obesity causes a proinflammatory milieu and a number of metabolic disruptions within the placenta [73]. Obesity is associated with hypertensive disorders, gestational diabetes and thromboembolic events; therefore, obesity affects the outcome of pregnancy [73]. Additionally, maternal obesity affects the fetus by resulting in large-for-gestational age and intrauterine growth-restricted infants, which may result in obesity, cardiovascular disease and diabetes later in life [73]. A few studies have suggested that miRNAs may contribute to the pathogenesis of pregnancy-related obesity. For example, studies of the placental miRNA profiles of women who were obese/overweight pre-pregnancy have established the relationship between altered expression of placental miRNAs and maternal obesity [73]. The study recruited a total of 70 pregnant women with either pre-gestational obesity (preOB), gestational obesity (gestOB), or normal weight (control) based on their body mass index (BMI) [73]. Eight miRNAs were identified to display differential expression in placentas from obese women versus women with normal weight, namely: miR-100, miR-1269, miR-1285, miR-181, miR-185, miR-214, miR-23, miR-296, and miR-487. Among them, miR-100, miR-185, and miR-487 were significantly decreased in gestOB women compared with the control group. Additionally, miR-1269, miR-1285, miR-181, miR-214 and miR 296 were significantly reduced between preOB and gestOB compared with control (p < 0.05). Other dysregulated miRNAs (miR-100, miR-1285, and miR-487) were associated with metabolic parameters such as BMI, homeostatic model assessment of insulin resistance (HOMA-IR), and C-peptide. These were also found to be predictors of neonates with lower birthweight, increased postnatal weight gain and type-2 diabetes. Therefore, these results indicate that the placenta may respond to the maternal obesogenic environment by altering the expression of critical miRNAs involved in growth parameters of the fetus and the maternal metabolic parameters.
Maternal pre-pregnancy BMI has also been associated with the altered expression of placental miRNAs [74]. Specifically, seven miRNAs are known for their role in adipogenesis. Placental miR-20a, miR-34A and miR-222 expression is downregulated in relation to higher maternal pre-pregnancy BMI. Interestingly, the expression of the dysregulated miRNAs is modified by the sex of the offspring such that the aberrant miRNAs are only observed in placentas of newborn females and not males [74]. These data suggest that miRNAs in the placenta may have a sexual dimorphic response to pre-pregnancy BMI.
5. Environmental toxicants
Studies that detail miRNAs that display altered expression in relation to environmental toxicant exposure are detailed below (Table 2).
Table 2.
miRNAs in the placenta that are responsive to exposure to environmental toxicants.
| Extrinsic Factors | Target Tissue(s) | Sample Size (N) | # of miRNAs Measured | Differentially Expressed miRNAs | Methods |
Author (year) | ||
|---|---|---|---|---|---|---|---|---|
| Extraction | miRNA quantification | Normalization | ||||||
| Toxic Metals | Placenta, JEG-3 cells | N = 32 | 861 | miR-26a, miR-155 | AllPrep DNA/RNA/miRNA Univerisal Kit (Qiagen) | Human miRNA Oligo microarray (Agilent); qRT-PCR | miRNA U6 | (Brooks, Martin et al. 2016) |
| Phthalates | Placenta | N = 179 | 29 | miR-185, miR-142−3p, miR-15a-5p | mirVANA RNA Isolation Kit (Ambion) | PCR array (Qiagen); qRT-PCR | SNORD61, SNORD95 | (LaRocca, Binder et al. 2016) |
| Phenols | Placenta | N = 80 | 1349 | miR-146ae | TRIzol Reagent (Invitrogen) | Human miRNA Microarray (Agilent); qRT-PCR | RNU6B | (De Felice, Manfellotto et al. 2015) |
| Placenta | N = 179 | 29 | miR-185, miR-142−3p, miR-15a-5p | mirVANA RNA Isolation Kit (Ambion) | PCR array (Qiagen); qRT-PCR | SNORD61, SNORD95 | (LaRocca, Binder et al. 2016) | |
| Particulate Matter | Placenta | N = 210 | 6 | miR-16d, miR-20a, miR-21c, miR-146ae, miR-222f | miRNeasy mini kit (Qiagen) | Taqman miRNA qRT-PCR assay | RNU6 | (Tsamou, Vrijens et al. 2018) |
| Placenta | N = 25 | 4 | miR-16d, miR-21c, miR-146ae | miRVANA miRNA Isolation Kit (Ambion) | Taqman miRNA qRT-PCR Assay | RNU44 | (Maccani, Avissar-Whiting et al. 2010) | |
Reproduced miRNAs across studies: c miR-21, d miR-16, e miR-146a.
5.1. Phthalates
Phthalates are endocrine-disrupting chemicals regularly used in plastics to increase their flexibility and durability [75]. Perinatal exposure to phthalates has been associated with adverse health outcomes, including poor semen quality, pregnancy complications, cardiovascular diseases, and cognitive and behavioral outcomes in children [[76], [77], [78], [79], [80]]. Studies have investigated the relationship between miRNAs and phthalates in connection with pregnancy. Associations have been observed between first trimester urine concentrations of eight phenols and 11 phthalates and the expression of 29 candidate placental miRNAs using qRT-PCR [81]. The miRNAs included in the study were shown to be differentially expressed in placentas exposed to phthalates and phenols in a pilot study conducted by the researchers. Additive phthalate concentrations displayed a negative correlation with miR-185 expression with monoethyl phthalate showing the strongest suppression of miR-185. Ten of the 29 miRNAs studied showed significantly altered expression in the presence of at least one phthalate [81], suggesting that prenatal phthalate exposure alters miRNA expression in the placenta. Interestingly, five of the ten miRNAs found to be differentially expressed were also associated with preeclampsia [[82], [83], [84], [85], [86]]. In particular, monobenzyl phthalate (MBzP) was associated with altered expression of miR-20a-5p [81]. miR-185 and miR-20a-5p have both been shown to have altered expression in preeclamptic mothers [81]. miR-20a-5p expression is altered in preeclamptic placentas through mechanisms that inhibit invasion and proliferation of trophoblast cells [84,87].
In another study, blood and urine from women pregnant with uncomplicated, healthy twins were analyzed for the correlation between prenatal phthalate exposure and miRNA profiles in extracellular vesicles (EV-miRNA) in maternal blood [88]. The researchers evaluated 20 placental-derived EV-miRNAs circulating in maternal plasma in relation to 13 phthalates and eight other chemicals used in plastics. Only MBzP displayed a positive association with miR-518 [88]. MBzP is a metabolite formed from butyl benzyl phthalate [89], and seems to have a potent effect on placental miRNA expression, specifically those implicated in preeclampsia. These results have been replicated by Zhong et al. and LaRocca et al. [81,88]. Altered expression of miR-518 has been associated with preeclamptic pregnancies, low fetal birth weight, and fetal growth restriction [5,84,90,91]. Thus, miR-518 could be potentially used as biomarker for adverse pregnancy outcomes.
In addition to epidemiologic studies, in vitro models using placental cell lines have been employed to further elucidate the effects of phthalates on placental cells. More specifically, the effects of mono-(2-ethylhexyl) phthalate (MEHP) exposure and its role in apoptosis in HTR-8/SVneo placental cells in vitro have been studied. Exposure to MEHP increased miR-16 expression in a time- and dose- dependent manner, while inducing apoptosis. The results showed that miR-16 could be induced after only four hours of exposure, and was significantly upregulated with increasing concentrations of MEHP [92]. miR-16 has been shown to regulate cell cycle genes, including cyclin D1 (CCND1), cyclin E1 (CCNE1), and CDK1, to induce G0/G1 arrest; it has also been implicated in playing a role in tumor growth [[93], [94], [95]]. However, this result was not seen in the epidemiological study previously described; MEHP was significantly associated with altered miR-128, miR-200c-3p, miR-143−3p, and miR-19a-3p [81].
The results may not have been replicated due to the fact that the two studies utilized two different experimental models. Meruvu et al. explored the effects of phthalates through an in vitro model, which may not fully represent the complex interactions that occur in system-wide model [92]. On the other hand, LaRocca et al. implemented a population-based study, where it is more difficult to control for potential confounders that can influence miRNA expression [81].
Based on the evidence, it is possible that perinatal phthalate exposure is associated with the onset of preeclampsia through alterations in miRNA expression. Specifically, miR-20a-5p, miR-518, and miR-16 have all been linked to preeclampsia [81,88,92]. However, the results are still conflicting. MEHP was found to significantly alter miR-16 expression in human placental cells in vitro [92], however, this change was not observed in human populations [81,88]. In an epidemiologic study, MEHP was associated with altered expressions of miR-128, miR-200c-3p, miR-143−3p, and miR-19a-3p [81]. The discrepancies in results could be attributed to the differences in experimental models. MBzP’s effect on miRNAs also displayed differences in the observed findings of miRNA change. One found that MBzP was associated with altered miR-20a-5p [81], but another found an association with miR-518 [88]. The issue with reproducibility between human population studies may also be related to inconsistencies between the candidate miRNAs and phthalate metabolites analyzed. Nevertheless, across the studies, phthalate exposure was significantly associated with altered miRNAs in preeclamptic mothers.
5.2. Phenols
Phenols are a group of endocrine-disrupting chemicals used in the manufacturing of nylon and other synthetic fibers, pesticides, disinfectants, and antiseptic products [77]. Bisphenol A (BPA) is one of the most commonly studied phenolic compounds due to its detrimental effects on the human body. Significant evidence suggests that prenatal exposure to BPA and its analogs can affect neurobehavior [77,[96], [97], [98], [99], [100]] and metabolic disorders (e.g. obesity and diabetes) in children [101,102]. In a genome-wide analysis of placental miRNAs, the expression of 1349 miRNAs from placental samples derived from mothers living in polluted areas were compared to placental samples from mothers living in nonpolluted areas using miRNA microarray technology. The researchers found that 34 miRNAs had at least a significant 2.5-fold difference. High concentrations of BPA in the placenta were related to significantly increased miR-146a expression. miR-146a targeted genes associated with 19 significant biological functions, including cell differentiation and proliferation, cancer, neural disease, signal transduction, and enzyme function [103].
Several phenols in maternal blood were analyzed in relation to 20 placenta-driven circulating EV-miRNAs: 2,4-dichlorophenol (24-DCP), 2,5-dichlorophenol (25-DCP), benzophenone-3 (BP-3), BPA, bisphenol F, bisphenol S, butyl paraben (B-PB), ethyl paraben (E-PB), methyl paraben (M-PB), Propyl paraben (P-PB), and triclosan (TCS). Increases in 24-DCP, 25-DCP, TCS concentrations were associated with decreased in miR-543 expression. M-PB was positively correlated to miR-518e, and negatively correlated with miR-373−3p and miR-543. E-PB and P-PB were negatively correlated with miR-543 [88]. These differentially expressed miRNAs associated with phenol exposure are known to play important roles during pregnancy. For example, miR-543 and miR-518e are typically upregulated during the first trimester of pregnancy, and have a vital role in cell differentiation and immune regulation [38,39,104]. miR-373 is a part of miR-371 cluster that is known to have a role in stem cell signature, oncogenesis, and immune suppression [38].
An association has been reported between urinary phenol levels and placental miRNAs. [81]. There was a significant association between additive phenols and miR-142 and miR-15a-5p. An increase in additive phenol concentration was linked to an increase in miR-142 expression. Interestingly, there was an inverse relationship between urinary additive phenol concentrations and placental miR-15a-5p expression. Additionally, 16 of the 29 candidate miRNAs explored were significantly associated with at least one phenol compound. BPA was associated with a total of six miRNAs: miR-185, miR-16−5p, miR-15b-5p, miR-25−3p, miR-92a-3p, and miR-30a-5p. Five miRNAs were differentially expressed with exposure to benzophenone-3 (BP-3): miR-106b-5p, miR-20a-5p, miR-17−5p, miR-19a-3p, and miR-142.3p [81]. miR-19a-3p and miR-20a-5p are a part of the miR-17−92 cluster, which plays a role in immune tolerance, proliferation, and oncogenic, angiogenic, and antiapoptotic characteristics [38]. Additionally, miR-25 is involved in modulating TGFβ signaling, and oncogenic and antiapoptotic activities [38]. miR-185, miR-19a-3p, miR-25−3p, miR-92a-3p, and miR-20a-5p are also typically upregulated in the first trimester [38]. Interestingly, the results of this study were not reproduced by Zhong et al. likely due to differences in technique [88,81].
BPA exposure to placental cells lines (HTR-8/SVneo, 3A, and TCL-1) revealed that 25 miRNAs had significantly altered expression in 3A cells, while 60 miRNAs were differentially expressed in HTR-8/SVneo cells. Between the 3A cells and HTR-8/SVneo cells, there was an overlap of 21 miRNA, of which miR-146a had a positive association with BPA concentration [105]. miR-146a has been shown to target genes involved in inflammation, oxidation, and apoptosis [106]. miR-146a is also associated with risk for preeclampsia, preterm birth, and unexplained recurrent spontaneous abortion [39,[105], [106], [107], [108]].
The relationship between perinatal BPA exposure and miR-146 has been reproduced in both human populations and in vitro studies [81,103,105]. As mentioned earlier, miR-146 has been associated with increased risk for preeclampsia, preterm birth, and unexplained recurrent spontaneous abortion [39,[105], [106], [107], [108]]. Subsequently, there is evidence to suggest a relationship between prenatal BPA exposure and preeclampsia [[109], [110], [111]]. It is possible that BPA exposure during pregnancy can lead to preeclampsia by modulating the expression of miR-146.
5.3. Metals
Toxic metals such as lead (Pb), cadmium (Cd), inorganic arsenic (iAs), and mercury (Hg) are abundant in the environment and exposure can lead to various detrimental health effects, including neurodevelopmental deficits [107,108,112,113], metabolic diseases [107], adverse birth outcomes [80,107,108,[112], [113], [114], [115], [116], [117], [118]], and endocrine disruption [[119], [120], [121]]. There is a growing body of literature examining the relationship between exposure to toxic metals and miRNA expression in the placenta or blood. For example, prenatal iAs exposure has been associated with the altered expression of miRNAs in newborn cord blood [122]. Specifically, a total of 12 cord blood-derived miRNAs were identified to have increased expression in relation to iAs in maternal urine. These 12 miRNAs identified were let-7a, miR-107, miR-126, miR-16, miR-17, miR-195, miR-20a, miR-20b, miR-26b, miR-454, miR-96, and miR-98. Pathway analysis of these miRNAs showed association to cancer, reproductive system disease, inflammatory disease/response, and connective tissue disorders amongst many others [122].
Conversely, other results have demonstrated that there is no association between iAs levels in the placenta and miRNA expression [69]. However, high levels of Hg are associated with reduced expression of many miRNAs, including miR-151−5p, miR-10a, miR-193b, miR-1975, and a large number of let-7 miRNAs (let-7a, let-7b, let-7c, let-7d, let-7 g, and let-7i). High levels of Pb are associated with decreased expression of let-7f, miR-146a, miR-10a, and miR-431 and linked with increased expression of miR-651. The let-7 miRNA family is highly conserved across species, and plays a major role in cell differentiation, cell development, and inflammation [123,124]. Additionally, high concentrations of Cd are positively correlated to miR-1537 expression [69]. Placenta trophoblast cells treated with Cd display dysregulation of miRNAs that specifically targets transforming growth factor beta (TGF-β) pathway [125]. The TGF-β pathway mediates trophoblast migration and invasion [126], and it is altered in preeclamptic placentas. Interestingly, Cd-responsive miRNAs, including miR-26a and miR-155, have been implicated in preeclampsia [125], suggesting a plausible link between Cd exposure and impaired placenta development. Several of the identified miRNAs associated with toxic metal exposures are known to target genes that play significant roles in maintaining homeostasis in the body. For example, miR-431, associated with Pb exposure, has been found to inhibit trophoblast cell migration and is also implicated in preeclampsia [127]. miR-193b associated with Hg exposure is aberrantly expressed in preeclamptic placentas. Furthermore, the overexpression of the miR-193b represses trophoblasts cell HT4−8/SVneo invasion, migration, and growth [128].
It is difficult to assess the reproducibility of the studies’ results due to the limited number of studies available. Of all the metals investigated, the effects of Cd have been explored in two studies, however the results were not replicated possibly due to differences in experimental models. One study used placental trophoblast cells, while the other explored the relationship through clinically-derived samples. Overall, these studies suggest that placenta miRNAs are responsive to toxic metal exposures and may mediate placenta abnormalities associated with exposures or pregnancy related disease pathogenesis.
5.4. Air toxics
The effects of air pollution on placental miRNAs remain vastly understudied. There is evidence that exposure to air pollutants during the prenatal period is associated with detrimental health outcomes including preterm birth, altered fetal growth and birth weight, and behavioral issues in children [129,130]. Some studies have found that prenatal exposure to tobacco smoke has a direct impact on brain function, cognition and behavior [[131], [132], [133]]. While the mechanisms underlying the association between particulate matter exposure and adverse birth effect is largely understudied, miRNAs may be potential biological molecules mediating this association.
The expression of four miRNAs (miR-16, miR-21, miR-146a, and miR-182) known to be implicated in growth and developmental processes were differentially expressed between placentas of mothers who smoked during pregnancy and those who did not. The qRT-PCR results showed a downregulation of miR-16, miR-21, and miR-146a in association with maternal cigarette smoking [134]. Recent studies describe miR-21 as oncogenic with the capability to target and alter key regulators of proliferation, invasion, migration, and the cell cycle [37]. To further identify the components of cigarette smoke responsible for alterations to miRNA expression in the placenta, three placental cell lines (3A cells, HTR-8/SVneo cells, and TCL-1 cells) were exposed to nicotine and benzo(a)pyrene, two chemicals commonly found in cigarettes. TCL-1 cells exposed to nicotine showed a dose-dependent downregulation of miR-146a but not miR-16 or miR-21. Similar to nicotine, benzo(a) pyrene downregulated miR-146a across all doses and showed no differential expression of miR-16 or miR-21 [134]. In silico analyses have predicted TNF-receptor associated factor 6 (TRAF6) as a target for miR-146a. TRAF6 plays an important role in the NFkB signaling pathways, however its role in the placenta is not clear [135]. The NFkB pathway is known to mediate placental growth and has been implicated in preeclampsia and preterm birth [136]. Overall, the results from this study suggest that miR-146a is particularly sensitive to chemicals and that smoking may trigger a cascade of downstream effects through dysregulation of miRNA expression in the placenta.
Further support for the impact of prenatal tobacco smoke on miRNA expression circulating in maternal and cord blood comes from a study where the researchers analyzed maternal tobacco exposure on urine metabolites and indoor volatile organic compounds (VOCs) concentrations [137]. The researchers found that maternal tobacco smoke exposure was associated with increased concentrations of benzene in the subjects’ homes as well as increased urinary cotinine concentrations. When looking at the effects of maternal tobacco smoke exposure on miRNA expression, high concentrations of benzene and toluene in homes and maternal urinary cotinine were associated with increased miR-223 expression in maternal blood during pregnancy [137]. Urinary cotinine levels were also related to increased miR-223 expression in cord blood. Interestingly, several studies have also noted altered expression of miR-223 in preeclamptic placentas [85,[138], [139], [140], [141]]. A recent study demonstrated that miR-223 targets STAT3 to promote trophoblast survival and invasion. miR-155 expression in cord blood is associated with decreased concentrations of S-benzylmercapturic acid (a metabolite of toluene) in maternal urine. However, there was no relationship observed between maternal exposure to tobacco smoke and maternal miR-155 expression [137]. This particular miRNA regulates inflammatory activity, proliferation, migration, and invasion of trophoblasts, as well as differentiation and regeneration of syncytiotrophoblasts [142]. miR-155 is also linked to preeclampsia by potentially downregulating cysteine-rich protein 61 (CYR61) [143].
Expression of six miRNAs (miR-16, miR-20a, miR-21, miR-34a, miR-146a, and miR-222) have been investigated in response to maternal exposure to air pollution. The air pollutants, particulate matter (PM2.5) and nitrogen dioxide (NO2), were averaged for each trimester of pregnancy [144]. There were positive associations between first trimester PM2.5 exposure and the expression of miR-20a and miR-21. PM2.5 during the second trimester was inversely linked to miR-16, miR-20a, miR-21, miR-146a, and miR-222 expressions [144]. miR-222 targets cyclin-dependent kinase to regulate endometrial stromal cell differentiation [58]. miR-146a expression was inversely associated with third trimester exposure to PM2.5. NO2 exposure had a positive correlation with miR-21 expression during the first trimester, and inverse associations with expressions of miR-20a, miR-21, and miR-146a during the second trimester. All miRNA expressions were identified at term [144].
Together, these studies suggest that air toxics exposure during the prenatal period may alter miRNA expression and potentially elicit downstream effects that can contribute to the pathogenesis of pregnancy-related disorders by disrupting biological processes such as trophoblast proliferation and invasion.
6. Conclusions
miRNAs are unique epigenetic regulators that play a critical role in gene regulation with a key role in the placenta. miRNAs are abundantly expressed in the placenta, are regulated across gestation, and are involved in a plethora of placental biological processes. From the literature, it is clear that several pathways which are under miRNA control may be critical for contaminant-induced effects as well as pregnancy-associated complications. These includes trophoblast cell migration, differentiation, invasion, proliferation, and angiogenesis. There is now mounting evidence suggesting that miRNAs in the placenta as well as maternal serum can be aberrantly expressed in relation to exposure to environmental contaminants and pregnancy-related complications. Unlike other epigenetic markers, placental-specific miRNAs can be secreted into maternal circulation, where they are highly stable and can be used as important biomarkers for diagnosing pregnancy-related disorders. In spite of the essential role of miRNAs in the human genome, the understanding of precisely how they contribute to normal placentation to pathogenesis in the context of environmental toxicants is not known. The studies summarized here highlight a number of miRNA candidates that are implicated either in pregnancy complications and/or in relation to exposure to environmental toxicants. Identification of their gene targets in the placenta will improve the understanding of their role in disease pathogenesis and as responder to exposure to environmental contaminants.
Three major epigenetic processes are active in the placenta namely: DNA methylation, histone acetylation and miRNAs [35,[145], [146], [147]]. These epigenetic processes are closely intertwined as DNA methylation can regulate miRNA expression and vice versa [148]. Similarly, miRNAs have been shown to regulate histone acetylation by targeting histone-modifying enzymes [149]. Interestingly, sex differences in several epigenetic modifications have been identified in the placenta. For example, X-linked H3K4me3 demethylase is expressed higher in the female placenta, whereas expression of the Y-linked gene for H3K27me3 is expressed higher in male placentas [150,151]. Sex differences in DNA methylation patterns have also been observed in human placenta [152]. Moreover, the placenta has been shown to respond to environmental exposures in a sexually-dimorphic manner [153]. These differences suggest that sensitivity or resistance to maternal insults, such as exposure to environmental contaminants, affect epigenetic control of gene expression in the placenta and potentially influence sex-differences. Yet, very little is known about how infant sex contributes to miRNA expression patterns in the placenta or maternal circulation. In some of the studies reviewed here, sex of the offspring was not factored as a potential modifier of miRNAs. Future studies should determine whether the sex of the fetus modifiesmiRNA expression in both the placenta and circulation as it relates to pregnancy-associated diseases or environmental exposures. Furthermore, mechanistic studies involving knock-out mouse models, next-generation sequencing and in vitro models are needed to enhance our understanding of the role of miRNAs and how they contribute to the pathogenesis of pregnancy-related diseases and responses to environmental chemicals.
When interpreting the studies reviewed here, caution must be taken due to the following five limitations. First, differences in subject characteristics and sample size can contribute to the noted discrepancies across studies. For example, with regards to preeclampsia, the studies reviewed here have sample sizes ranging from 6−40. Sample size has been shown to influence biomarker discovery [154]. In studies with a small sample size, the statistical power is significantly reduced, leading to the potential detection of many false positives, which may not be replicated in studies with large sizes. Implementation of power calculations in large, high throughput experiments is necessary to minimize discrepancies [154]. Second, differences in study design, definition of inclusion and exclusion criteria, and differences in cohort characteristics across studies makes it difficult to compare studies which can also contribute to the discrepancies [155]. To circumvent these limitations, a large prospective study in a well-defined population may be necessary to further elucidate the clinical relevance of the identified miRNAs. Third, because miRNAs in the placenta are expressed in a spatiotemporal manner, differences in sampling methodologies can potentially contribute to the lack of reproducibility across studies. During early gestation, miRNAs involved in the regulation of cell proliferation and immune suppression are highly expressed. In late gestation of term placentas, miRNAs that that are involved in tumor suppressor and hematopoietic activities, such as those in the let-7 family, miR-34 cluster or miR-29 cluster are highly expressed [38]. Furthermore, miRNA expression is not uniform in the placenta. Particularly, some miRNAs in the C19MC clusters tend to be expressed higher in the villous trophoblast than in the extravillous trophoblast [156]. Because placental miRNAs are spatiotemporally expressed, it is important to adjust for gestational age when comparing miRNAs across studies. Additionally, standardized methodologies for tissue sampling will be necessary to improve rigor and reproducibility. Fourth, many computational algorithms used in identifying miRNA gene targets are often based on identifying phylogenetically-conserved matches of miRNA 5′end (i.e., the seed region consisting of about 2–7 nucleotides of the miRNA) to the target site [157]. However, sequences may be conserved for other reasons, while being incidentally complementary to miRNAs. This can potentially lead an overestimate of targets and false predictions. Algorithms which incorporate additional characterization and do not rely solely on sequence homology may be necessary for precise miRNA target precision. Finally, it is worth noting that the trends in the relationships either showing concordant or discordant expression patterns between circulating miRNAs and tissue-specific miRNAs vary according to tissue-type and miRNA. For example, the same miRNAs were identified to display expression in placenta and maternal serum, yet the magnitude was different with 10-fold higher expression in the placenta than in maternal serum [158]. Also, in studies of other tissues besides the placenta, similar miRNA expression patterns have been observed between tissue and plasma. For example, miRNA expression in matched tissue and plasma samples from colorectal patients and controls both showed upregulation of miR-187, miR-675 and miR-3591−3 P in both [159]. Still, other studies have highlighted discrepancies and found that when comparing miRNAs from placentas and maternal plasma of normotensive and preeclamptic patients, miR-195 was significantly downregulated in the placenta of preeclamptics but was undetected in the plasma samples [60]. Together, these studies suggest that there is no absolutely predictive correlation between placental-specific or circulating miRNAs.
There are three major takeaways from the review. First, miRNAs often display differential expression depending on the tissue studied. Second, miRNAs may serve as biomarkers of disease and/or environmental contaminant exposure. Third, some miRNA results are reproduced across studies. Related to the first concept, the studies highlighted here show that miRNAs often display differential expression depending on the tissue studied. For example, studies have identified that even in the same assessment for a pregnancy-associated complication, miRNA expression may be different depending on the tissue studied. For instance, there was variation in detected miRNAs in preeclamptic mothers based on whether the tissue examined was placenta or blood. While the placenta may secrete miRNAs into maternal circulation, other tissues also contribute to circulating miRNAs. This can contribute to placental-specific and circulating miRNAs being vastly different. Finally, Due to the lack of correlation between tissue-specific miRNAs and circulatory miRNAs, and the fact that multiple tissues secrete miRNAs into circulation during endogenous and pathological conditions, it is difficult to directly link dysregulated plasma miRNAs to pregnancy complications directly. In studies where certain miRNAs in maternal circulation have been associated with either environmental contaminants or pregnancy-related complications, several tissues including the liver or pancreas rather than the placenta may have secreted these miRNAs into maternal circulation though it is not clear which tissue makes the major contribution. Related to the second concept, miRNAs can be used as biomarkers. Several of the studies reviewed here have indicated miR-210 as a potential biomarker for preeclampsia because it is dysregulated in circulation of preeclamptic patients, yet miR-210 is strongly linked to the hypoxia pathway and has also been associated with several diseases including cancers and cardiovascular diseases. miR-210 plasma levels have been shown to fluctuate in pathological conditions associated with liver disease. Specifically, miR-210 has been shown to be elevated in response to hypoxia-induced liver inflammation [48]. In the event where a preeclamptic patient also has a cardiovascular disease, it will be difficult to identify which tissue contributes the majority of miR-210 in circulation. Thus, the disease or the environmental exposure may not be directly responsible for the dysregulated miRNA in circulation. For circulating miRNAs to have public health relevance, it will be necessary for future studies to investigate whether other pathological conditions are reflected in maternal circulation. Additional studies are needed to establish a correlation between tissue-specific miRNAs in circulation and their overall role in pregnancy. Related to the third concept, there were specific miRNAs that were reproduced across studies. This includes pregnancy-related complications (Table 1) and miRNAs that are differentially expressed in relation to exposure to environmental contaminants (Table 2). For example, miR-210 and miR-195 were observed to be differentially expressed across several studies focused on preeclampsia. Similarly, miR-21, miR-16, miR-146a and miR-222 were found to be differentially expressed in pregnancies exposed to particulate matter.
In conclusion, changes in the expression of miRNAs in the placenta and in maternal plasma have been observed in relation to exposure to environmental chemicals and their dysregulation is likely involved in the development of pregnancy-related complications. While technologies over the past decade have advanced the knowledge of the roles of miRNAs during pregnancy, there is still much that is unknown. This is, in part, due to a limited number of studies that have adequately explored the impact of pregnancy-related complications and prenatal environmental exposures on miRNAs. Additionally, the lack of standardized methodologies has resulted in discrepancies across studies, leaving knowledge gaps as it relates to the role of miRNAs during pregnancy. Moving forward, it will be necessary to validate candidate biomarkers and examine their clinical utility in large scale retrospective and prospective studies that adjust for race, gestational age, lifestyle factors (such as smoking status), and other pathologies as these factors have been shown to influence miRNA expression. Additionally, integrative modeling approaches which combine miRNA assessment with mRNA expression measurements will enhance the understanding of the potential impact of miRNA expression changes on biological processes.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
We would like to acknowledge funding from the National Institutes of Health (NIH) (R01HD092374; T32ES007126).
References
- 1.Gude N.M. Growth and function of the normal human placenta. Thromb. Res. 2004;114(5–6):397–407. doi: 10.1016/j.thromres.2004.06.038. [DOI] [PubMed] [Google Scholar]
- 2.Gabory A. Maternal diets trigger sex-specific divergent trajectories of gene expression and epigenetic systems in mouse placenta. PLoS One. 2012;7(11):e47986. doi: 10.1371/journal.pone.0047986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Green B.B. Epigenome-wide assessment of DNA methylation in the placenta and arsenic exposure in the New Hampshire birth cohort study (USA) Environ. Health Perspect. 2016;124(8):1253–1260. doi: 10.1289/ehp.1510437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amankwah K.S., Kaufmann R.C. Ultrastructure of human placenta: effects of maternal drinking. Gynecol. Obstet. Invest. 1984;18(6):311–316. doi: 10.1159/000299099. [DOI] [PubMed] [Google Scholar]
- 5.Kotlabova K., Doucha J., Hromadnikova I. Placental-specific microRNA in maternal circulation--identification of appropriate pregnancy-associated microRNAs with diagnostic potential. J. Reprod. Immunol. 2011;89(2):185–191. doi: 10.1016/j.jri.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 6.O’Brien J. Overview of MicroRNA biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. (Lausanne) 2018;9:402. doi: 10.3389/fendo.2018.00402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hammond S.M. An overview of microRNAs. Adv. Drug Deliv. Rev. 2015;87:3–14. doi: 10.1016/j.addr.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Catalanotto C., Cogoni C., Zardo G. MicroRNA in control of gene expression: an overview of nuclear functions. Int. J. Mol. Sci. 2016;17(10) doi: 10.3390/ijms17101712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wahid F. MicroRNAs: synthesis, mechanism, function, and recent clinical trials. Biochim. Biophys. Acta. 2010;1803(11):1231–1243. doi: 10.1016/j.bbamcr.2010.06.013. [DOI] [PubMed] [Google Scholar]
- 10.Williams A.E. Functional aspects of animal microRNAs. Cell. Mol. Life Sci. 2008;65(4):545–562. doi: 10.1007/s00018-007-7355-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang H. Expression profile of circular RNAs in placentas of women with gestational diabetes mellitus. Endocr. J. 2019;66(5):431–441. doi: 10.1507/endocrj.EJ18-0291. [DOI] [PubMed] [Google Scholar]
- 12.Inoue K. The rodent-specific MicroRNA cluster within the Sfmbt2 gene is imprinted and essential for placental development. Cell Rep. 2017;19(5):949–956. doi: 10.1016/j.celrep.2017.04.018. [DOI] [PubMed] [Google Scholar]
- 13.Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 14.Rezaei M. The effect of the placental DROSHA rs10719 and rs6877842 polymorphisms on PE susceptibility and mRNA expression. J. Hum. Hypertens. 2019;33(7):552–558. doi: 10.1038/s41371-018-0156-9. [DOI] [PubMed] [Google Scholar]
- 15.Hansen T.B. Argonaute-associated short introns are a novel class of gene regulators. Nat. Commun. 2016;7:11538. doi: 10.1038/ncomms11538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bentwich I. Identification of hundreds of conserved and nonconserved human microRNAs. Nat. Genet. 2005;37(7):766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- 17.Morin R.D. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 2008;18(4):610–621. doi: 10.1101/gr.7179508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morin R. Profiling the HeLa S3 transcriptome using randomly primed cDNA and massively parallel short-read sequencing. Biotechniques. 2008;45(1):81–94. doi: 10.2144/000112900. [DOI] [PubMed] [Google Scholar]
- 19.Li M. Frequent amplification of a chr19q13.41 microRNA polycistron in aggressive primitive neuroectodermal brain tumors. Cancer Cell. 2009;16(6):533–546. doi: 10.1016/j.ccr.2009.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rippe V. The two stem cell microRNA gene clusters C19MC and miR-371-3 are activated by specific chromosomal rearrangements in a subgroup of thyroid adenomas. PLoS One. 2010;5(3):e9485. doi: 10.1371/journal.pone.0009485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liang Y. Characterization of microRNA expression profiles in normal human tissues. BMC Genomics. 2007;8:166. doi: 10.1186/1471-2164-8-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang R., Wang Y.Q., Su B. Molecular evolution of a primate-specific microRNA family. Mol. Biol. Evol. 2008;25(7):1493–1502. doi: 10.1093/molbev/msn094. [DOI] [PubMed] [Google Scholar]
- 23.Noguer-Dance M. The primate-specific microRNA gene cluster (C19MC) is imprinted in the placenta. Hum. Mol. Genet. 2010;19(18):3566–3582. doi: 10.1093/hmg/ddq272. [DOI] [PubMed] [Google Scholar]
- 24.Donker R.B. The expression profile of C19MC microRNAs in primary human trophoblast cells and exosomes. Mol. Hum. Reprod. 2012;18(8):417–424. doi: 10.1093/molehr/gas013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miura K. Identification of pregnancy-associated microRNAs in maternal plasma. Clin. Chem. 2010;56(11):1767–1771. doi: 10.1373/clinchem.2010.147660. [DOI] [PubMed] [Google Scholar]
- 26.Biro O. Circulating exosomal and Argonaute-bound microRNAs in preeclampsia. Gene. 2019;692:138–144. doi: 10.1016/j.gene.2019.01.012. [DOI] [PubMed] [Google Scholar]
- 27.Sabapatha A., Gercel-Taylor C., Taylor D.D. Specific isolation of placenta-derived exosomes from the circulation of pregnant women and their immunoregulatory consequences. Am. J. Reprod. Immunol. 2006;56(5–6):345–355. doi: 10.1111/j.1600-0897.2006.00435.x. [DOI] [PubMed] [Google Scholar]
- 28.Zhao Z., Moley K.H., Gronowski A.M. Diagnostic potential for miRNAs as biomarkers for pregnancy-specific diseases. Clin. Biochem. 2013;46(10–11):953–960. doi: 10.1016/j.clinbiochem.2013.01.026. [DOI] [PubMed] [Google Scholar]
- 29.Hosseini M.K. MicroRNA expression profiling in placenta and maternal plasma in early pregnancy loss. Mol. Med. Rep. 2018;17(4):4941–4952. doi: 10.3892/mmr.2018.8530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumar A. Identification of miR-379/miR-656 (C14MC) cluster downregulation and associated epigenetic and transcription regulatory mechanism in oligodendrogliomas. J. Neurooncol. 2018;139(1):23–31. doi: 10.1007/s11060-018-2840-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.da Rocha S.T. Genomic imprinting at the mammalian Dlk1-Dio3 domain. Trends Genet. 2008;24(6):306–316. doi: 10.1016/j.tig.2008.03.011. [DOI] [PubMed] [Google Scholar]
- 32.Malnou E.C. Imprinted microRNA gene clusters in the evolution, development, and functions of mammalian placenta. Front. Genet. 2018;9:706. doi: 10.3389/fgene.2018.00706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fu G. MicroRNA-376c impairs transforming growth factor-beta and nodal signaling to promote trophoblast cell proliferation and invasion. Hypertension. 2013;61(4):864–872. doi: 10.1161/HYPERTENSIONAHA.111.203489. [DOI] [PubMed] [Google Scholar]
- 34.Hayder H. MicroRNAs: crucial regulators of placental development. Reproduction. 2018;155(6):R259–R271. doi: 10.1530/REP-17-0603. [DOI] [PubMed] [Google Scholar]
- 35.Mouillet J.F. MicroRNAs in placental health and disease. Am. J. Obstet. Gynecol. 2015;213(4 Suppl):S163–72. doi: 10.1016/j.ajog.2015.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lou Y. MicroRNA-21 promotes the cell proliferation, invasion and migration abilities in ovarian epithelial carcinomas through inhibiting the expression of PTEN protein. Int. J. Mol. Med. 2010;26(6):819–827. doi: 10.3892/ijmm_00000530. [DOI] [PubMed] [Google Scholar]
- 37.Maccani M.A., Padbury J.F., Marsit C.J. miR-16 and miR-21 expression in the placenta is associated with fetal growth. PLoS One. 2011;6(6):e21210. doi: 10.1371/journal.pone.0021210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gu Y. Differential miRNA expression profiles between the first and third trimester human placentas. Am. J. Physiol. Endocrinol. Metab. 2013;304(8):E836–43. doi: 10.1152/ajpendo.00660.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morales-Prieto D.M. MicroRNA expression profiles of trophoblastic cells. Placenta. 2012;33(9):725–734. doi: 10.1016/j.placenta.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 40.Xu P. Variations of microRNAs in human placentas and plasma from preeclamptic pregnancy. Hypertension. 2014;63(6):1276–1284. doi: 10.1161/HYPERTENSIONAHA.113.02647. [DOI] [PubMed] [Google Scholar]
- 41.Wessels J.M. The microRNAome of pregnancy: deciphering miRNA networks at the maternal-fetal interface. PLoS One. 2013;8(11):e72264. doi: 10.1371/journal.pone.0072264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Turchinovich A., Weiz L., Burwinkel B. Extracellular miRNAs: the mystery of their origin and function. Trends Biochem. Sci. 2012;37(11):460–465. doi: 10.1016/j.tibs.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 43.Kirschner M.B. The impact of hemolysis on cell-free microRNA biomarkers. Front. Genet. 2013;4:94. doi: 10.3389/fgene.2013.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Y. Plasma microRNA-122 as a biomarker for viral-, alcohol-, and chemical-related hepatic diseases. Clin. Chem. 2010;56(12):1830–1838. doi: 10.1373/clinchem.2010.147850. [DOI] [PubMed] [Google Scholar]
- 45.Lewis A.P., Jopling C.L. Regulation and biological function of the liver-specific miR-122. Biochem. Soc. Trans. 2010;38(6):1553–1557. doi: 10.1042/BST0381553. [DOI] [PubMed] [Google Scholar]
- 46.Corsten M.F. Circulating microRNA-208b and microRNA-499 reflect myocardial damage in cardiovascular disease. Circ. Cardiovasc. Genet. 2010;3(6):499–506. doi: 10.1161/CIRCGENETICS.110.957415. [DOI] [PubMed] [Google Scholar]
- 47.Laterza O.F. Plasma MicroRNAs as sensitive and specific biomarkers of tissue injury. Clin. Chem. 2009;55(11):1977–1983. doi: 10.1373/clinchem.2009.131797. [DOI] [PubMed] [Google Scholar]
- 48.Li J. Circulating miR-210 and miR-22 combined with ALT predict the virological response to interferon-alpha therapy of CHB patients. Sci. Rep. 2017;7(1):15658. doi: 10.1038/s41598-017-15594-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lv Y. Roles of microRNAs in preeclampsia. J. Cell. Physiol. 2019;234(2):1052–1061. doi: 10.1002/jcp.27291. [DOI] [PubMed] [Google Scholar]
- 50.Duley L. The global impact of pre-eclampsia and eclampsia. Semin. Perinatol. 2009;33(3):130–137. doi: 10.1053/j.semperi.2009.02.010. [DOI] [PubMed] [Google Scholar]
- 51.Uzan J. Pre-eclampsia: pathophysiology, diagnosis, and management. Vasc. Health Risk Manag. 2011;7:467–474. doi: 10.2147/VHRM.S20181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Soleymanlou N. Molecular evidence of placental hypoxia in preeclampsia. J. Clin. Endocrinol. Metab. 2005;90(7):4299–4308. doi: 10.1210/jc.2005-0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang X., Le Q.T., Giaccia A.J. MiR-210--micromanager of the hypoxia pathway. Trends Mol. Med. 2010;16(5):230–237. doi: 10.1016/j.molmed.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chan Y.C. miR-210: the master hypoxamir. Microcirculation. 2012;19(3):215–223. doi: 10.1111/j.1549-8719.2011.00154.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cao G. MicroRNA regulation of transthyretin in trophoblast biofunction and preeclampsia. Arch. Biochem. Biophys. 2019;676:108129. doi: 10.1016/j.abb.2019.108129. [DOI] [PubMed] [Google Scholar]
- 56.Steinfeld I. miRNA target enrichment analysis reveals directly active miRNAs in health and disease. Nucleic Acids Res. 2013;41(3):e45. doi: 10.1093/nar/gks1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xie L., Sadovsky Y. The function of miR-519d in cell migration, invasion, and proliferation suggests a role in early placentation. Placenta. 2016;48:34–37. doi: 10.1016/j.placenta.2016.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cai M., Kolluru G.K., Ahmed A. Small molecule, big prospects: MicroRNA in pregnancy and its complications. J. Pregnancy. 2017;2017:6972732. doi: 10.1155/2017/6972732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vidal D.O. Highly expressed placental miRNAs control key biological processes in human cancer cell lines. Oncotarget. 2018;9(34):23554–23563. doi: 10.18632/oncotarget.25264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gunel T. Regulatory effect of miR-195 in the placental dysfunction of preeclampsia. J. Matern. Fetal. Neonatal. Med. 2020;33(6):901–908. doi: 10.1080/14767058.2018.1508439. [DOI] [PubMed] [Google Scholar]
- 61.Lasabova Z. Overexpression of miR-21 and miR-122 in preeclamptic placentas. Neuro Endocrinol. Lett. 2015;36(7):695–699. [PubMed] [Google Scholar]
- 62.Bai Y. Downregulated miR-195 detected in preeclamptic placenta affects trophoblast cell invasion via modulating ActRIIA expression. PLoS One. 2012;7(6):e38875. doi: 10.1371/journal.pone.0038875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu H. MiR-195 participates in the placental disorder of preeclampsia via targeting activin receptor type-2B in trophoblastic cells. J. Hypertens. 2016;34(7):1371–1379. doi: 10.1097/HJH.0000000000000948. [DOI] [PubMed] [Google Scholar]
- 64.Timofeeva A.V. Identification of potential early biomarkers of preeclampsia. Placenta. 2018;61:61–71. doi: 10.1016/j.placenta.2017.11.011. [DOI] [PubMed] [Google Scholar]
- 65.Sheikh A.M. Systematic review of Micro-RNA expression in pre-eclampsia identifies a number of common pathways associated with the disease. PLoS One. 2016;11(8):e0160808. doi: 10.1371/journal.pone.0160808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gilmartin A.B., Ural S.H., Repke J.T. Gestational diabetes mellitus. Rev. Obstet. Gynecol. 2008;1(3):129–134. [PMC free article] [PubMed] [Google Scholar]
- 67.Alfadhli E.M. Gestational diabetes mellitus. Saudi Med. J. 2015;36(4):399–406. doi: 10.15537/smj.2015.4.10307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Buchanan T.A., Xiang A.H., Page K.A. Gestational diabetes mellitus: risks and management during and after pregnancy. Nat. Rev. Endocrinol. 2012;8(11):639–649. doi: 10.1038/nrendo.2012.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li J. A MicroRNA signature in gestational diabetes mellitus associated with risk of Macrosomia. Cell. Physiol. Biochem. 2015;37(1):243–252. doi: 10.1159/000430349. [DOI] [PubMed] [Google Scholar]
- 70.Ding R. Integrated transcriptome sequencing analysis reveals role of miR-138-5p/ TBL1X in placenta from gestational diabetes mellitus. Cell. Physiol. Biochem. 2018;51(2):630–646. doi: 10.1159/000495319. [DOI] [PubMed] [Google Scholar]
- 71.Zhao C. Early second-trimester serum miRNA profiling predicts gestational diabetes mellitus. PLoS One. 2011;6(8):e23925. doi: 10.1371/journal.pone.0023925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hruby A., Hu F.B. The Epidemiology of Obesity: A Big Picture. Pharmacoeconomics. 2015;33(7):673–689. doi: 10.1007/s40273-014-0243-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Carreras-Badosa G. Dysregulation of Placental miRNA in Maternal Obesity Is Associated With Pre- and Postnatal Growth. J. Clin. Endocrinol. Metab. 2017;102(7):2584–2594. doi: 10.1210/jc.2017-00089. [DOI] [PubMed] [Google Scholar]
- 74.Tsamou M. Mother’s pre-pregnancy BMI and placental candidate miRNAs: findings from the ENVIRONAGE birth cohort. Sci. Rep. 2017;7(1):5548. doi: 10.1038/s41598-017-04026-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.National Biomonitoring Program. Phthalates. 2017 04/07/2020 [cited 2020 01/17/2020]; Available from: https://www.cdc.gov/biomonitoring/Phthalates_FactSheet.html.
- 76.Ejaredar M. Phthalate exposure and childrens neurodevelopment: a systematic review. Environ. Res. 2015;142:51–60. doi: 10.1016/j.envres.2015.06.014. [DOI] [PubMed] [Google Scholar]
- 77.Agency for Toxic Substances and Disease Registry. Phenol. 2011 03/03/2011 [cited 2020 01/17/2020]; Available from: https://www.atsdr.cdc.gov/substances/toxsubstance.asp?toxid=27.
- 78.Mariana M. The effects of phthalates in the cardiovascular and reproductive systems: a review. Environ. Int. 2016;94:758–776. doi: 10.1016/j.envint.2016.07.004. [DOI] [PubMed] [Google Scholar]
- 79.Meeker J.D., Sathyanarayana S., Swan S.H. Phthalates and other additives in plastics: human exposure and associated health outcomes. Philos. Trans. R. Soc. Lond., B, Biol. Sci. 2009;364(1526):2097–2113. doi: 10.1098/rstb.2008.0268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zlatnik M.G. Endocrine-disrupting chemicals and reproductive health. J. Midwifery Womens Health. 2016;61(4):442–455. doi: 10.1111/jmwh.12500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.LaRocca J. First-trimester urine concentrations of phthalate metabolites and phenols and placenta miRNA expression in a cohort of U.S. Women. Environ. Health Perspect. 2016;124(3):380–387. doi: 10.1289/ehp.1408409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ding G.C. MicroRNA-128a-induced apoptosis in HTR-8/SVneo trophoblast cells contributes to pre-eclampsia. Biomed. Pharmacother. 2016;81:63–70. doi: 10.1016/j.biopha.2016.03.040. [DOI] [PubMed] [Google Scholar]
- 83.Shen L. Placenta-associated serum exosomal miR155 derived from patients with preeclampsia inhibits eNOS expression in human umbilical vein endothelial cells. Int. J. Mol. Med. 2018;41(3):1731–1739. doi: 10.3892/ijmm.2018.3367. [DOI] [PubMed] [Google Scholar]
- 84.Wang Y. Aberrantly up-regulated miR-20a in pre-eclampsic placenta compromised the proliferative and invasive behaviors of trophoblast cells by targeting forkhead box protein A1. Int. J. Biol. Sci. 2014;10(9):973–982. doi: 10.7150/ijbs.9088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xu P. Variations of MicroRNAs in Human Placentas and Plasma From Preeclamptic Pregnancy. Hypertension. 2014;63(6):1276–1284. doi: 10.1161/HYPERTENSIONAHA.113.02647. [DOI] [PubMed] [Google Scholar]
- 86.Zhang X. MiR-200c regulates apoptosis of placental trophoblasts in preeclampsia rats through Wnt/beta-catenin signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2019;23(17):7209–7216. doi: 10.26355/eurrev_201909_18822. [DOI] [PubMed] [Google Scholar]
- 87.Ishibashi O. Hydroxysteroid (17-beta) dehydrogenase 1 is dysregulated by miR-210 and miR-518c that are aberrantly expressed in preeclamptic placentas: a novel marker for predicting preeclampsia. Hypertension. 2012;59(2):265–273. doi: 10.1161/HYPERTENSIONAHA.111.180232. [DOI] [PubMed] [Google Scholar]
- 88.Zhong J. Maternal phthalate and personal care products exposure alters extracellular placental miRNA profile in twin pregnancies. Reprod. Sci. 2019;26(2):289–294. doi: 10.1177/1933719118770550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ema M. Decreased anogenital distance and increased incidence of undescended testes in fetuses of rats given monobenzyl phthalate, a major metabolite of butyl benzyl phthalate. Reprod. Toxicol. 2003;17(4):407–412. doi: 10.1016/s0890-6238(03)00037-6. [DOI] [PubMed] [Google Scholar]
- 90.Mouillet J.F. The levels of hypoxia-regulated microRNAs in plasma of pregnant women with fetal growth restriction. Placenta. 2010;31(9):781–784. doi: 10.1016/j.placenta.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yang S. Deregulated microRNA species in the plasma and placenta of patients with preeclampsia. Mol. Med. Rep. 2015;12(1):527–534. doi: 10.3892/mmr.2015.3414. [DOI] [PubMed] [Google Scholar]
- 92.Meruvu S. Mono-(2-ethylhexyl) phthalate induces apoptosis through miR-16 in human first trimester placental cell line HTR-8/SVneo. Toxicol. In Vitro. 2016;31:35–42. doi: 10.1016/j.tiv.2015.11.010. [DOI] [PubMed] [Google Scholar]
- 93.Bottoni A. miR-15a and miR-16-1 down-regulation in pituitary adenomas. J. Cell. Physiol. 2005;204(1):280–285. doi: 10.1002/jcp.20282. [DOI] [PubMed] [Google Scholar]
- 94.Calin G.A. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99(24):15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Liu Q. miR-16 family induces cell cycle arrest by regulating multiple cell cycle genes. Nucleic Acids Res. 2008;36(16):5391–5404. doi: 10.1093/nar/gkn522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Braun J.M. Impact of early-life bisphenol A exposure on behavior and executive function in children. Pediatrics. 2011;128(5):873–882. doi: 10.1542/peds.2011-1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Braun J.M. Prenatal bisphenol A exposure and early childhood behavior. Environ. Health Perspect. 2009;117(12):1945–1952. doi: 10.1289/ehp.0900979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Evans S.F. Prenatal bisphenol A exposure and maternally reported behavior in boys and girls. Neurotoxicology. 2014;45:91–99. doi: 10.1016/j.neuro.2014.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Harley K.G. Prenatal and early childhood bisphenol A concentrations and behavior in school-aged children. Environ. Res. 2013;126:43–50. doi: 10.1016/j.envres.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Perera F. Prenatal bisphenol a exposure and child behavior in an inner-city cohort. Environ. Health Perspect. 2012;120(8):1190–1194. doi: 10.1289/ehp.1104492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Shankar A., Teppala S. Relationship between urinary bisphenol A levels and diabetes mellitus. J. Clin. Endocrinol. Metab. 2011;96(12):3822–3826. doi: 10.1210/jc.2011-1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wang T. Urinary bisphenol A (BPA) concentration associates with obesity and insulin resistance. J. Clin. Endocrinol. Metab. 2012;97(2):E223–7. doi: 10.1210/jc.2011-1989. [DOI] [PubMed] [Google Scholar]
- 103.De Felice B. Genome-wide microRNA expression profiling in placentas from pregnant women exposed to BPA. BMC Med. Genomics. 2015;8:56. doi: 10.1186/s12920-015-0131-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hiura H. Genome-wide microRNA expression profiling in placentae from frozen-thawed blastocyst transfer. Clin. Epigenetics. 2017;9:79. doi: 10.1186/s13148-017-0379-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Avissar-Whiting M. Bisphenol A exposure leads to specific microRNA alterations in placental cells. Reprod. Toxicol. 2010;29(4):401–406. doi: 10.1016/j.reprotox.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhong X. Toll-like 4 receptor /NFkappaB inflammatory/miR-146a pathway contributes to the ART-correlated preterm birth outcome. Oncotarget. 2016;7(45):72475–72485. doi: 10.18632/oncotarget.11987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Shapiro G.D. Exposure to phthalates, bisphenol A and metals in pregnancy and the association with impaired glucose tolerance and gestational diabetes mellitus: the MIREC study. Environ. Int. 2015;83:63–71. doi: 10.1016/j.envint.2015.05.016. [DOI] [PubMed] [Google Scholar]
- 108.Wai K.M. Prenatal heavy metal exposure and adverse birth outcomes in Myanmar: a birth-cohort study. Int. J. Environ. Res. Public Health. 2017;14(11) doi: 10.3390/ijerph14111339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Cantonwine D.E. Urinary concentrations of bisphenol a and phthalate metabolites measured during pregnancy and risk of preeclampsia. Environ. Health Perspect. 2016;124(10):1651–1655. doi: 10.1289/EHP188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Leclerc F., Dubois M.F., Aris A. Maternal, placental and fetal exposure to bisphenol A in women with and without preeclampsia. Hypertens. Pregnancy. 2014;33(3):341–348. doi: 10.3109/10641955.2014.892607. [DOI] [PubMed] [Google Scholar]
- 111.Ye Y. Maternal serum bisphenol A levels and risk of pre-eclampsia: a nested case-control study. Eur. J. Public Health. 2017;27(6):1102–1107. doi: 10.1093/eurpub/ckx148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Al-Saleh I. Birth outcome measures and maternal exposure to heavy metals (lead, cadmium and mercury) in Saudi Arabian population. Int. J. Hyg. Environ. Health. 2014;217(2–3):205–218. doi: 10.1016/j.ijheh.2013.04.009. [DOI] [PubMed] [Google Scholar]
- 113.Sun H. The effects of prenatal exposure to low-level cadmium, lead and selenium on birth outcomes. Chemosphere. 2014;108:33–39. doi: 10.1016/j.chemosphere.2014.02.080. [DOI] [PubMed] [Google Scholar]
- 114.Yang J. Maternal urinary cadmium concentrations in relation to preterm birth in the Healthy Baby Cohort Study in China. Environ. Int. 2016;94:300–306. doi: 10.1016/j.envint.2016.06.003. [DOI] [PubMed] [Google Scholar]
- 115.Shirai S. Maternal exposure to low-level heavy metals during pregnancy and birth size. J. Environ. Sci. Health A. Tox. Subst. Environ. Eng. 2010;45(11):1468–1474. doi: 10.1080/10934529.2010.500942. [DOI] [PubMed] [Google Scholar]
- 116.Kippler M. Environmental exposure to arsenic and cadmium during pregnancy and fetal size: a longitudinal study in rural Bangladesh. Reprod. Toxicol. 2012;34(4):504–511. doi: 10.1016/j.reprotox.2012.08.002. [DOI] [PubMed] [Google Scholar]
- 117.Rahman A. Arsenic exposure and risk of spontaneous abortion, stillbirth, and infant mortality. Epidemiology. 2010;21(6):797–804. doi: 10.1097/EDE.0b013e3181f56a0d. [DOI] [PubMed] [Google Scholar]
- 118.Laine J.E. Maternal arsenic exposure, arsenic methylation efficiency, and birth outcomes in the Biomarkers of Exposure to ARsenic (BEAR) pregnancy cohort in Mexico. Environ. Health Perspect. 2015;123(2):186–192. doi: 10.1289/ehp.1307476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Henson M.C., Chedrese P.J. Endocrine disruption by cadmium, a common environmental toxicant with paradoxical effects on reproduction. Exp. Biol. Med. (Maywood) 2004;229(5):383–392. doi: 10.1177/153537020422900506. [DOI] [PubMed] [Google Scholar]
- 120.Stasenko S. Metals in human placenta: focus on the effects of cadmium on steroid hormones and leptin. J. Appl. Toxicol. 2010;30(3):242–253. doi: 10.1002/jat.1490. [DOI] [PubMed] [Google Scholar]
- 121.Yang K. Cadmium reduces 11 beta-hydroxysteroid dehydrogenase type 2 activity and expression in human placental trophoblast cells. Am. J. Physiol. Endocrinol. Metab. 2006;290(1):E135–E142. doi: 10.1152/ajpendo.00356.2005. [DOI] [PubMed] [Google Scholar]
- 122.Rager J.E. Prenatal arsenic exposure and the epigenome: altered microRNAs associated with innate and adaptive immune signaling in newborn cord blood. Environ. Mol. Mutagen. 2014;55(3):196–208. doi: 10.1002/em.21842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Chan H.W. The expression of the let-7 miRNAs and Lin28 signalling pathway in human term gestational tissues. Placenta. 2013;34(5):443–448. doi: 10.1016/j.placenta.2013.02.008. [DOI] [PubMed] [Google Scholar]
- 124.Roush S., Slack F.J. The let-7 family of microRNAs. Trends Cell Biol. 2008;18(10):505–516. doi: 10.1016/j.tcb.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 125.Brooks S.A. miRNAs as common regulators of the transforming growth factor (TGF)-β pathway in the preeclamptic placenta and cadmium-treated trophoblasts: links between the environment, the epigenome and preeclampsia. Food Chem. Toxicol. 2016;98:50–57. doi: 10.1016/j.fct.2016.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Caniggia I. Inhibition of TGF-beta 3 restores the invasive capability of extravillous trophoblasts in preeclamptic pregnancies. J. Clin. Invest. 1999;103(12):1641–1650. doi: 10.1172/JCI6380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Yang X., Meng T. MicroRNA-431 affects trophoblast migration and invasion by targeting ZEB1 in preeclampsia. Gene. 2019;683:225–232. doi: 10.1016/j.gene.2018.10.015. [DOI] [PubMed] [Google Scholar]
- 128.Zhou X. The aberrantly expressed miR-193b-3p contributes to preeclampsia through regulating transforming growth factor-β signaling. Sci. Rep. 2016;6:19910. doi: 10.1038/srep19910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yorifuji T. Prenatal exposure to outdoor air pollution and child behavioral problems at school age in Japan. Environ. Int. 2017;99:192–198. doi: 10.1016/j.envint.2016.11.016. [DOI] [PubMed] [Google Scholar]
- 130.van den Hooven E.H. Air pollution exposure during pregnancy, ultrasound measures of fetal growth, and adverse birth outcomes: a prospective cohort study. Environ. Health Perspect. 2012;120(1):150–156. doi: 10.1289/ehp.1003316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Cornelius M.D., Day N.L. Developmental consequences of prenatal tobacco exposure. Curr. Opin. Neurol. 2009;22(2):121–125. doi: 10.1097/WCO.0b013e328326f6dc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Espy K.A. Prenatal tobacco exposure: developmental outcomes in the neonatal period. Dev. Psychol. 2011;47(1):153–156. doi: 10.1037/a0020724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lin Q. Prenatal exposure to environmental tobacco smoke and hyperactivity behavior in Chinese young children. Int. J. Environ. Res. Public Health. 2017;14(10):1132. doi: 10.3390/ijerph14101132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Maccani M.A. Maternal cigarette smoking during pregnancy is associated with downregulation of miR-16, miR-21, and miR-146a in the placenta. Epigenetics. 2010;5(7):583–589. doi: 10.4161/epi.5.7.12762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Funakoshi-Tago M. TRAF6 negatively regulates TNFalpha-induced NF-kappaB activation. Cytokine. 2009;45(2):72–79. doi: 10.1016/j.cyto.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 136.Lappas M. Nuclear factor-kappaB mediates placental growth factor induced pro-labour mediators in human placenta. Mol. Hum. Reprod. 2012;18(7):354–361. doi: 10.1093/molehr/gas007. [DOI] [PubMed] [Google Scholar]
- 137.Herberth G. Maternal and cord blood miR-223 expression associates with prenatal tobacco smoke exposure and low regulatory T-cell numbers. J. Allergy Clin. Immunol. 2014;133(2):543–550. doi: 10.1016/j.jaci.2013.06.036. e4. [DOI] [PubMed] [Google Scholar]
- 138.Choi S.Y. MicroRNA expression profiles in placenta with severe preeclampsia using a PNA-based microarray. Placenta. 2013;34(9):799–804. doi: 10.1016/j.placenta.2013.06.006. [DOI] [PubMed] [Google Scholar]
- 139.Vashukova E.S. Placental microRNA expression in pregnancies complicated by superimposed pre‑eclampsia on chronic hypertension. Mol. Med. Rep. 2016;14(1):22–32. doi: 10.3892/mmr.2016.5268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Weedon-Fekjær M.S. Placental miR-1301 is dysregulated in early-onset preeclampsia and inversely correlated with maternal circulating leptin. Placenta. 2014;35(9):709–717. doi: 10.1016/j.placenta.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 141.Zhu X.-m. Differential expression profile of microRNAs in human placentas from preeclamptic pregnancies vs normal pregnancies. Am. J. Obstet. Gynecol. 2009;200(6):661. doi: 10.1016/j.ajog.2008.12.045. e1-661.e7. [DOI] [PubMed] [Google Scholar]
- 142.Nizyaeva N.V. Expression of MicroRNA-146a and MicroRNA-155 in placental villi in early- and late-onset preeclampsia. Bull. Exp. Biol. Med. 2017;163(3):394–399. doi: 10.1007/s10517-017-3812-0. [DOI] [PubMed] [Google Scholar]
- 143.Bounds K.R. MicroRNAs: new players in the pathobiology of preeclampsia. Front. Cardiovasc. Med. 2017;4:60. doi: 10.3389/fcvm.2017.00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Tsamou M. Air pollution-induced placental epigenetic alterations in early life: a candidate miRNA approach. Epigenetics. 2018;13(2):135–146. doi: 10.1080/15592294.2016.1155012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Acevedo N. Histone acetylation of immune regulatory genes in human placenta in association with maternal intake of olive oil and fish consumption. Int. J. Mol. Sci. 2019;20(5) doi: 10.3390/ijms20051060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Harb H. Epigenetic modifications in placenta are associated with the child’s sensitization to allergens. Biomed Res. Int. 2019;2019:1315257. doi: 10.1155/2019/1315257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Robinson W.P., Price E.M. The human placental methylome. Cold Spring Harb. Perspect. Med. 2015;5(5):a023044. doi: 10.1101/cshperspect.a023044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Wang S., Wu W., Claret F.X. Mutual regulation of microRNAs and DNA methylation in human cancers. Epigenetics. 2017;12(3):187–197. doi: 10.1080/15592294.2016.1273308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kim H.S., Shen Q., Nam S.W. Histone deacetylases and their regulatory MicroRNAs in hepatocarcinogenesis. J. Korean Med. Sci. 2015;30(10):1375–1380. doi: 10.3346/jkms.2015.30.10.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Howerton C.L. O-GlcNAc transferase (OGT) as a placental biomarker of maternal stress and reprogramming of CNS gene transcription in development. Proc Natl Acad Sci U S A. 2013;110(13):5169–5174. doi: 10.1073/pnas.1300065110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Nugent B.M., Bale T.L. The omniscient placenta: metabolic and epigenetic regulation of fetal programming. Front. Neuroendocrinol. 2015;39:28–37. doi: 10.1016/j.yfrne.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Bommarito P.A., Martin E., Fry R.C. Effects of prenatal exposure to endocrine disruptors and toxic metals on the fetal epigenome. Epigenomics. 2017;9(3):333–350. doi: 10.2217/epi-2016-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Addo K.A. Acetaminophen use during pregnancy and DNA methylation in the placenta of the extremely low gestational age newborn (ELGAN) cohort. Environ. Epigenet. 2019;5(2):dvz010. doi: 10.1093/eep/dvz010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Kok M.G.M. Small sample sizes in high-throughput miRNA screens: a common pitfall for the identification of miRNA biomarkers. Biomol. Detect. Quantif. 2018;15:1–5. doi: 10.1016/j.bdq.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Tiberio P. Challenges in using circulating miRNAs as cancer biomarkers. Biomed Res. Int. 2015;2015:731479. doi: 10.1155/2015/731479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Xie L. C19MC microRNAs regulate the migration of human trophoblasts. Endocrinology. 2014;155(12):4975–4985. doi: 10.1210/en.2014-1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Pinzon N. microRNA target prediction programs predict many false positives. Genome Res. 2017;27(2):234–245. doi: 10.1101/gr.205146.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Chim S.S. Detection and characterization of placental microRNAs in maternal plasma. Clin. Chem. 2008;54(3):482–490. doi: 10.1373/clinchem.2007.097972. [DOI] [PubMed] [Google Scholar]
- 159.Nagy Z.B. Comparison of circulating miRNAs expression alterations in matched tissue and plasma samples during colorectal cancer progression. Pathol. Oncol. Res. 2019;25(1):97–105. doi: 10.1007/s12253-017-0308-1. [DOI] [PubMed] [Google Scholar]