Abstract
Risk assessment in cutaneous melanoma (CM) patients is one of the major challenges in the effective treatment of CM patients. Traditionally, clinico-pathological features such as Breslow thickness, American Joint Committee on Cancer (AJCC) tumor staging, etc. are utilized for this purpose. However, due to advancements in technology, most of the upcoming risk prediction methods are gene-expression profile (GEP) based. In this study, we have tried to develop new GEP and clinico-pathological features-based biomarkers and assessed their prognostic strength in contrast to existing prognostic methods. We developed risk prediction models using the expression of the genes associated with different cancer-related pathways and got a maximum hazard ratio (HR) of 2.52 with p-value ~10−8 for the apoptotic pathway. Another model, based on combination of apoptotic and notch pathway genes boosted the HR to 2.57. Next, we developed models based on individual clinical features and got a maximum HR of 2.45 with p-value ~10−6 for Breslow thickness. We also developed models using the best features of clinical as well as gene-expression data and obtained a maximum HR of 3.19 with p-value ~10−9. Finally, we developed a new ensemble method using clinical variables only and got a maximum HR of 6.40 with p-value ~10−15. Based on this method, a web-based service and an android application named ‘CMcrpred’ is available at (https://webs.iiitd.edu.in/raghava/cmcrpred/) and Google Play Store respectively to facilitate scientific community. This study reveals that our new ensemble method based on only clinico-pathological features overperforms methods based on GEP based profiles as well as currently used AJCC staging. It also highlights the need to explore the full potential of clinical variables for prognostication of cancer patients.
Keywords: Bioinformatics, Cancer research, Genetics, Oncology, Cancer, Prognosis, Melanoma, Survival analysis, Skin, Risk prediction
Bioinformatics; Cancer research; Genetics; Oncology; Cancer; Prognosis; Melanoma; Survival analysis; Skin; Risk prediction.
1. Introduction
Cutaneous melanoma (CM), which accounts for less than 5% of all skin cancers, has been reported to be the most aggressive and fatal form of skin cancer [1]. Melanoma also referred to as malignant melanoma or cutaneous melanoma, arises due to genetic mutations in the melanocytes. Melanocytes are the cells that produce the pigment melanin (the pigment responsible for skin colour) and are located in the skin, inner ear, eye, vaginal epithelium, leptomeninges, heart and bones [2, 3, 4, 5]. In the last 30 years, the number of deaths has increased drastically due to melanoma. According to the estimation by The American Cancer Society, in 2019, about 96480 new melanomas would have been diagnosed (57220 in men and 39260 in women) in the US alone, out of which around 7230 people were expected to die (about 4740 men and 2490 women) [6].
Cutaneous melanomas are curable when diagnosed in the earlier stage (~98% survival rate) but once metastasised, they are tough to cure (~25% survival rate); thus, early identification of melanoma has been deemed crucial for the patient's survival [7, 8]. This uncertainty between patient's death or survival is directly affected by the choice of the therapy given to the patients, which in turn depends on the features that were used for prognostication. According to the present AJCC cancer staging criterion for melanoma, features derived from biopsies and/or blood samples, are primarily used for this purpose in CM patients [9]. There are also demographic factors like ethnicity of a patient, which has been associated with Melanoma occurrence [10]. While features like Breslow thickness, Ulceration status, Metastatic staging etc. can be clinically observed and monitored, underlying molecular factors such as alteration of protein expression and genetic mutations are difficult to incorporate during clinical observation of a patient, due to a huge plethora of genes/proteins to account for. Thus, the problem for risk prediction with the help of minimal but relevant features in addition to the current clinico-pathological features remains a major challenge. Identification of these features would help in deciding better therapeutic strategies and bridge the uncertainty gap that exists at present.
Over the past decades, protein and gene-based biomarkers have provided valuable prognostic information about patient outcome and aided in the design of therapy. Several of these gene/protein based biomarkers have been utilized in the case of prognosis and risk prediction for various diseases such as breast cancer, colon cancer, prostate cancer, lung cancer, leukemia, and melanoma [11, 12, 13, 14]. Along with these biomarkers and due to the modern techniques used for purifying RNA from formalin-fixed paraffin-embedded (FFPE) tumour samples, there have also been efforts to identify/develop multiple protein and multiple genes-based biomarkers associated with prognostication of SNL (sentinel lymph node status), locoregional recurrences, distant metastases, and survival [15]. In the case of melanoma, several protein candidates have been significantly associated with prognostication, including lactate dehydrogenase (LDH), C-reactive protein and S100B [16, 17, 18, 19]. Out of these, only LDH has been included in the AJCC staging system for categorizing metastasis. However, it is shown to function well only in Stage IV patients. NCOA3, SPP1, and RGS1 is another known example of multiple protein-based biomarker [20] that has been shown to be a significant predictor of SLN status and disease-specific survival as compared to other clinical features. Though validated [21], this 3-protein marker was also not included in the AJCC staging criteria. Amongst single and multiple-gene expression profile (GEP) based markers, TRPM1 expression [22], NRAS mutation status [23], BRAF mutation status [24], circulating miRNA biomarkers [25], DecisionDx-Melanoma (31 GEP) [26, 27, 28, 29], Melagenix (9 GEP), ITLP group [30] and 53-gene immune GEP [31] are a few mainstream examples in the scientific community. The prognostic marker Melagenix (9 GEP) was able to differentiate high and low risk patients on the basis of overall survival, DecisionDx-Melanoma segregated patients on the basis of RFS (relapse free survival), DMFS (distant metastasis-free survival) and MSS (microsatellite instability) while models such as 53-gene immune GEP and ITLP group are prediction models for metastasis progression and SLN positivity respectively. While almost all of these models compare their results with the clinical factors (mainly by employing multivariate Cox-PH analysis), only a few of these models incorporate the combined effect of clinical factors with gene expression. However, unlike other cancers, none of the independent gene based or GEP based prognostic biomarkers for melanoma have been included in the AJCC staging system so far.
In the present study, we make a systematic attempt to identify/develop new prognostic markers based on the overall survival (OS) of CM patients to predict risk. For this purpose, we utilized the TCGA-SKCM RNAseq expression dataset to build machine learning-based regression (MLR) and prognostic index (PI) models based on GEP and clinical factors. GEP based models were constructed based on genes involved in different cancer related pathways, most of which have been related to melanoma carcinogenesis in the past while others have been attributed a role in other cancers. Models were also developed using best genes associated with survival as obtained via rfSRC feature selection method. Combinations of GEP models and clinical features were also analyzed. Briefly, we present here a comparative assessment between these various models and show that a new model based on clinico-pathological features alone outperforms GEP based models.
2. Materials and Methods
2.1. Dataset and pre-processing
The original dataset consisted of RNAseq expression values for 458 Skin Cutaneous Melanoma (SKCM) patients that was obtained from The Cancer Genome Atlas (TCGA) using TCGA Assembler 2 [32]. Out of which, information about OS time and censoring was available for 449 patients. Thus, the final dataset was reduced to 449 samples constituting RNAseq values for 20530 genes. Following the approach similar to [33], genes with ‘NA’ or zero expression data for more than 50% of the samples were removed. Table 1 presents a summary of clinicopathological features in the final TCGA-SKCM cohort. Further, the final dataset was normalized by quantile normalization method, which has been extensively used in the past for similar studies [34, 35, 36].
Table 1.
Summary of TCGA-SKCM cohort used in this study.
| Factor | Value | Percentage |
|---|---|---|
|
Age (at diagnosis), in years | ||
| Mean | 58.006 | |
| Median | 58 | |
| Range |
15–90 |
|
|
Gender | ||
| Male | 280 | 62.36 |
| Female |
169 |
37.63 |
|
Ethnicity | ||
| Non-Hispanic/Non-Latino | 426 | 94.88 |
| Hispanic/Latino | 10 | 2.23 |
| Unknown |
13 |
2.90 |
|
Race | ||
| White | 426 | 94.88 |
| Black/African American | 1 | 0.22 |
| Asian | 12 | 2.67 |
| Unknown |
10 |
2.23 |
|
Known primary melanoma tumor | ||
| Yes | 402 | 89.53 |
| No |
47 |
10.47 |
|
Location of primary melanoma | ||
| Head and Neck | 35 | 7.80 |
| Trunk | 155 | 34.52 |
| Extremity | 194 | 43.21 |
| Other | 13 | 2.90 |
| Unknown |
52 |
11.58 |
|
Breslow thickness (mm) | ||
| Mean | 5.49 | |
| Median | 3 | |
| Unknown |
105 |
23.39 |
|
Clark level | ||
| I–III | 98 | 21.83 |
| IV–V | 211 | 46.99 |
| Unknown |
140 |
31.18 |
|
Ulceration status | ||
| Yes | 159 | 35.41 |
| No | 141 | 31.40 |
| Unknown |
149 |
33.18 |
|
Mitotic rate | ||
| <1 | 19 | 4.23 |
| ≥ 1 | 151 | 33.63 |
| Unknown |
279 |
62.14 |
|
T stage | ||
| Tis | 7 | 1.56 |
| T0 | 23 | 5.12 |
| T1 | 10 | 2.23 |
| T1a | 22 | 4.90 |
| T1b | 10 | 2.23 |
| T2 | 30 | 6.68 |
| T2a | 31 | 6.90 |
| T2b | 15 | 3.34 |
| T3 | 14 | 3.12 |
| T3a | 37 | 8.24 |
| T3b | 37 | 8.24 |
| T4 | 15 | 3.34 |
| T4a | 23 | 5.12 |
| T4b | 104 | 23.16 |
| TX | 44 | 9.80 |
| Unknown |
27 |
6.01 |
|
N stage | ||
| N0 | 223 | 49.67 |
| N1 | 16 | 3.56 |
| N1a | 18 | 4.01 |
| N1b | 38 | 8.46 |
| N2 | 6 | 1.34 |
| N2a | 13 | 2.90 |
| N2b | 20 | 4.45 |
| N2c | 9 | 2.00 |
| N3 | 53 | 11.80 |
| NX | 34 | 7.57 |
| Unknown |
19 |
4.23 |
|
M stage | ||
| M0 | 400 | 89.09 |
| M1 | 5 | 1.11 |
| M1a | 4 | 0.89 |
| M1b | 5 | 1.11 |
| M1c | 9 | 2.00 |
| Unknown |
26 |
5.79 |
|
Pathological Stage | ||
| Stage 0 | 6 | 1.34 |
| Stage I | 30 | 6.68 |
| Stage IA | 18 | 4.01 |
| Stage IB | 29 | 6.46 |
| Stage II | 30 | 6.68 |
| Stage IIA | 17 | 3.79 |
| Stage IIB | 27 | 6.01 |
| Stage IIC | 58 | 12.92 |
| Stage III | 40 | 8.91 |
| Stage III A | 15 | 3.34 |
| Stage III B | 45 | 10.02 |
| Stage III C | 66 | 14.70 |
| Stage IV | 22 | 4.90 |
| I/II NOS | 10 | 2.23 |
| Unknown |
36 |
8.02 |
|
Anatomic site | ||
| Primary Tumor | 91 | 20.27 |
| Regional Lymph Node | 218 | 48.55 |
| Regional Cutaneous or Subcutaneous tissue | 73 | 16.26 |
| Distant Metastasis | 64 | 14.25 |
| Unknown |
3 |
0.67 |
|
Overall Survival (months) | ||
| Mean | 52.96 | |
| Median | 29.5 | |
| Range |
0–362.33 |
|
|
Vital Status | ||
| Alive | 295 | 65.70 |
| Dead | 154 | 34.30 |
2.2. Survival analysis
Hazard ratios were computed to predict the risks of death associated with high-risk and low-risk groups based on overall survival time of patients. These were stratified on the basis of mean and median values of various factors, using the univariate unadjusted Cox-Proportional Hazard (Cox-PH) regression models. Kaplan-Meier (KM) plots were used to compare survival curves of high risk and low risk groups. Survival analyses on these datasets were performed using ‘survival’ and ‘survminer’ packages (V.2.42–6) in R (V.3.4.4, The R Foundation). Statistical significance between the survival curves was estimated using log-rank tests. Wald tests were performed to estimate the significance of the explanatory variables used for HR calculations. Concordance index (C) provided the strength of predictive ability of the model [37, 38, 39]. p-values less than 0.05 were considered as significant.
2.3. Prognostic genes
Cox-Proportional Hazard models were used to find the genes that are related to CM patient survival. A cutoff of HR > 1.2 and p-value<0.05 was used. Univariate Cox-PH analysis, at median expression cutoffs, revealed a total of 1343 good prognostic marker (GPM) genes and 1294 bad prognostic marker (BPM) genes. GPM genes are defined as genes whose expression is positively correlated with patient OS time and vice-versa for BPM. For a GPM gene, patients with GPM gene expression < median (GPM gene expression) are at high risk and for a BPM gene, patients with BPM gene expression > median (GPM gene expression) are at high risk. Supplementary S1 TableA shows the survival associated parameters such as HR, p-value, C, and Cox regression coefficient-Beta (β) corresponding to each gene. The distribution of the GPM and BPM genes based on Hazard Ratio (HR > 1.2 and p < 0.05) is represented in Supplementary S2 Figure S1.
2.4. Machine learning based regression (MLR) models
Regression models from ‘caret’ package (V.3.4.4, The R Foundation) were implemented to fit the gene expression values against the OS time. Various regressors such as Support vector machine (SVR), Decision-tree (DT), Random-forest (RF), K-nearest neighbors (KNN), Ridge, Lasso and Elastic-Net were used. The fitting and test evaluations were carried using a five-fold cross-validation scheme. Combination of all five evaluated test datasets (predicted OS) was then used to classify the actual patient survival time (OS) at mean and median cutoffs to estimate HR, CI and p-values. Hyperparameter optimization and regularization was achieved using the in-built function ‘expand.grid’.
2.5. Five-fold cross-validation
The dataset is shuffled randomly and divided into 5 subsets. After these groups are prepared, an iterative process begins. During each iteration, a unique group is taken as a test dataset and combination of remaining groups as a training dataset. Model is fitted on the training dataset and evaluated on the test dataset. Model's performance is evaluated using standard parameters viz. RMSE (root mean squared error) and MAE (mean absolute error). The process is repeated five times and each sample is processed once as a testing data point and four times as training data point. Five-fold cross-validation has been successfully implemented in previous studies [40, 41, 42, 43, 44, 45, 46].
2.6. Prognostic index (PI)
As implemented in [33, 47, 48], PI for a set of n genes was evaluated as:
where β represents regression coefficient obtained for a gene g from a univariate Cox-PH model. PI for different set of genes was used for stratifying risk groups and standard metrics such as HR, p-value etc. were estimated.
3. Results
3.1. Multiple gene expression profile (GEP) based risk prediction
3.1.1. Models based on genes associated with cancer pathways
Several signaling pathways have been attributed a role in cancer progression and development in the past. We collected the list of 11 cancer-related pathways and the genes associated with those pathways from a recent study [49]. These are namely Apoptosis, MYC, NRF2, NOTCH, P53, WNT, HIPPO, CELL CYCLE, PI3K-AKT, RAS and TGF-BETA pathways. Amongst these pathways, many have been associated with melanoma tumorigenesis. Table 2 shows the PMIDs of the studies which have explored the role of these pathways in CM progression and/or development. Table 2 also shows the original gene-count in the pathway, number of GPM and number of BPM genes as a result of univariate analysis. Combined gene count is the sum of GPM and BPM genes. Supplementary S2 Figure S2 shows the upset plot representing gene overlaps between different pathways in the original gene set. MLR and PI based models were built using the GPM, BPM and combined (i.e. GPM + BPM) genes in these pathways, as explained in the following sections. The information regarding genes involved in original gene set and the three filtered gene sets is provided in Supplementary S1 TableB.
Table 2.
Genes related to pathways associated with cancer. PMIDs of the studies that relate to role of the pathway in melanoma and gene-count before and after univariate Cox-PH analysis is provided.
| S. no. | Pathway | PMID | No. of Genes | No. of GPM | No. of BPM | Combined |
|---|---|---|---|---|---|---|
| 1 | NRF2 | 27344172, 18353146 | 481 | 27 | 26 | 53 |
| 2 | P53 | 32377702, 31374895 | 201 | 17 | 16 | 33 |
| 3 | Apoptosis | 32687246, 32645331 | 161 | 29 | 4 | 33 |
| 4 | WNT | 32659938, 32073511 | 151 | 7 | 9 | 16 |
| 5 | CELL-CYCLE | - | 128 | 4 | 17 | 21 |
| 6 | PI3K-AKT | 32626712, 32558531 | 105 | 18 | 11 | 29 |
| 7 | TGF-β | 31667872, 31599708 | 86 | 3 | 1 | 4 |
| 8 | NOTCH | 30569717, 30941830 | 47 | 3 | 4 | 7 |
| 9 | MYC | 32283126 | 25 | 2 | 2 | 4 |
| 10 | RAS | 32605090, 32568870 | 23 | 2 | 1 | 3 |
| 11 | HIPPO | 32407182, 32561850 | 22 | 1 | 2 | 3 |
Machine learning based regression (MLR) models: MLR models were built using GPM, BPM and combined gene sets for each pathway. Various regressors were used to predict OS time by employing the expression data for the genes in the corresponding gene set (Materials and Methods). Similar models have also been used in the past with protein concentrations as independent variables [45].
Table 3 provides detailed results for each gene-set and the corresponding best model. The Lasso regression model that predicts the survival time on the basis of the combination of GPM and BPM genes (7 genes) of the NOTCH pathway shows the best results, where patients with pred OS median (pred OS) are at 2.34 times higher risk than patients with pred OS > median (pred OS) with a p-value of the order of 10−7 and C = 0.62. Second best results are seen in the case of the RF model built using GPM genes of the apoptotic pathway (29 genes), where HR is 2.24 with a significant p-value (~10−6) and C = 0.59, at median cutoff. However, it should be noted that the combination of the apoptotic GPM geneset and NOTCH combined geneset (36 genes) improves the risk stratification further with HR = 2.54 and p~10−8. For mean cutoff, see Supplementary S1 TableC.
Table 3.
MLR models for risk stratification. The table shows the best models for each pathway and corresponding gene set used. Patients with predicted OS less than or equal to the median cutoff are at higher risk than patients with predicted OS more than cutoff.
| S. no. | Pathway | Gene set | Regressor | HR | p-val | C |
|---|---|---|---|---|---|---|
| 1 | NRF2 | Combined | Ridge | 2.17 | 3 × 10−6 | 0.60 |
| 2 | P53 | Combined | RF | 2.04 | 1 × 10−5 | 0.59 |
| 3∗ | Apoptosis | GPM | RF | 2.24 | 1.2x10−6 | 0.59 |
| 4 | WNT | Combined | RF | 2.22 | 2 × 10−6 | 0.59 |
| 5 | CELL-CYCLE | Combined | RF | 1.76 | 5.7 × 10−3 | 0.59 |
| 6 | PI3K-AKT | Combined | RF | 1.90 | 8 × 10−5 | 0.60 |
| 7 | TGF-β | Combined | KNN | 1.73 | 9 × 10−4 | 0.58 |
| 8 | NOTCH | Combined | Lasso | 2.34 | 3 × 10−7 | 0.62 |
| 9 | MYC | GPM | Lasso | 1.68 | 1.7 × 10−3 | 0.57 |
| 10 | RAS | Combined | Lasso | 1.79 | 3.8 × 10−4 | 0.58 |
| 11 | HIPPO | Combined | KNN | 1.11 | 0.51 | 0.52 |
| 12∗ | Apoptosis + NOTCH | GPM + Combined | RF | 2.54 | 2.3x10−8 | 0.61 |
∗Boldface indicates the best results. Hyperparameters for above models are provided in Supplementary S1 TableC.
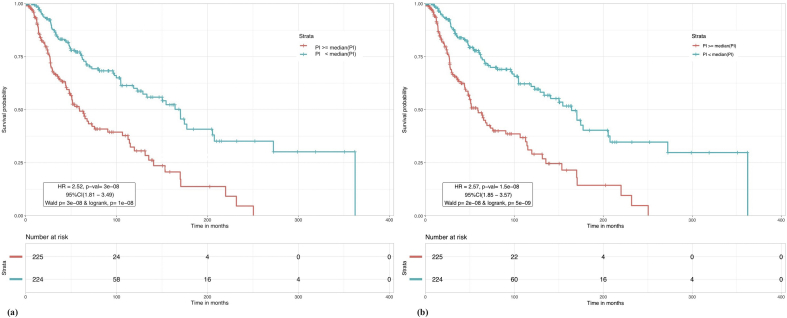
Prognostic Index (PI) based models: PI was evaluated for the three types of gene-sets corresponding to each pathway. Based on PI, the patients were stratified into high and low risk groups such that patients with PI median (PI) were at higher risk than patients with PI < median (PI) (similarly with mean in Supplementary S1 TableD). Results for each pathway are shown in Table 4. As evident from the results, PI for apoptotic GPM genes improved the risk prediction over the models developed previously. High and low risk patients show a 2.52 fold difference in survival when stratified using the PI for apoptotic GPM genes at median cutoff, with a p-value ~10−8 and C value of 0.62. 10-year survival rate for high risk patients in this case is around 30% whereas low risk patients have around 60% survival probability. Figure 1a shows the Kaplan Meier plot for median cutoff. The PI model built using the combination of apoptotic GPM genes and NOTCH GPM and BPM genes, is again seen to enhance the risk stratification with HR = 2.57 though p-value and C remain of the same order as apoptotic GPM geneset alone. KM plot for this model is shown in Figure 1b.
Table 4.
Prognostic index (PI) based risk stratification. The table shows the results for each pathway and corresponding gene set used. Patients with PI less than the median cutoff are at lower risk than patients with PI greater than cutoff.
| S. no. | Pathway | Gene set | HR | p-val | C |
|---|---|---|---|---|---|
| 1 | NRF2 | GPM | 1.87 | 1.2 × 10−4 | 0.58 |
| 2 | P53 | Combined | 2.20 | 1.5 × 10−6 | 0.61 |
| 3∗ | Apoptosis | GPM | 2.52 | 3.2x10−8 | 0.62 |
| 4 | WNT | GPM | 1.97 | 3.6 × 10−5 | 0.59 |
| 5 | CELL-CYCLE | GPM | 1.48 | 1.6 × 10−2 | 0.57 |
| 6 | PI3K-AKT | GPM | 1.82 | 2.4 × 10−4 | 0.58 |
| 7 | TGF-β | BPM | 1.48 | 1.6 × 10−2 | 0.53 |
| 8 | NOTCH | Combined | 2.26 | 9.4 × 10−7 | 0.60 |
| 9 | MYC | BPM | 1.67 | 1.8 × 10−3 | 0.57 |
| 10 | RAS | BPM | 1.79 | 4.5 × 10−4 | 0.56 |
| 11 | HIPPO | Combined | 1.67 | 1.9 × 10−3 | 0.55 |
| 12∗ | Apoptosis + NOTCH | GPM + Combined | 2.57 | 1.5x10−8 | 0.62 |
∗Boldface indicates the best results.
Figure 1.
Kaplan Meier plot for risk stratification of CM patients. (a) Based on prognostic index of apoptotic genes. Based on GPM gene set, patients with PI ≥ median (PI) are at a greater risk than patients with PI < median (PI) with HR = 2.52 and p-val = 3 × 10−8. (b) Based on prognostic index of apoptotic GPM and NOTCH combined genes. Patients with PI ≥ median (PI) are at a greater risk than patients with PI < median (PI) with HR = 2.57 and p-val = 1.5 × 10−8.
3.2. Models based on complete set of genes
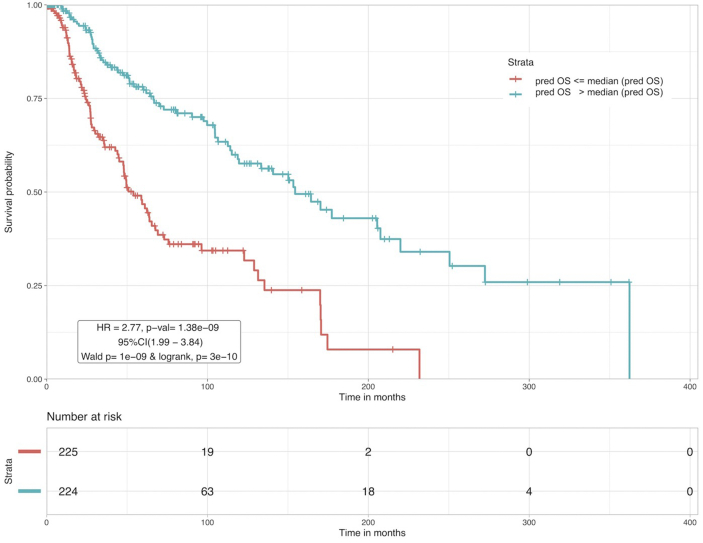
In addition to developing models for pathway-specific gene sets, we also developed similar models for the total GPM (1343), BPM (1294) and combined gene set (2637). Feature selection was performed to obtain the most important genes using random survival forests-variable hunting (rfSRC) for 100 iterations, on each of these three gene sets. rfSRC resulted in 58 GPM genes, 52 BPM genes and 129 combined genes (Supplementary S1 TableE). This feature selection method has been extensively used in similar survival-based studies in the past [47, 50, 51, 52]. First, MLR models were built and predicted OS was used to stratify high and low risk patients, similar to the pathway-specific analysis before. Subsequently, prognostic index based stratification was performed using the selected GPM, BPM and combined genes. Results are provided in Supplementary S2 Table S1. The SVR regression model based on 52 BPM genes performs the best out of all these models with highest HR value of 2.77, lowest p-value ~ 10−9 and concordance index of 0.63. A comparison of apoptotic genes based PI models, NOTCH genes based regression models, apoptosis and NOTCH genes combined models with the current 52 total BPM based SVR model shows that HR, p-value and concordance have improved. KM plot corresponding to this model is shown in Figure 2.
Figure 2.
CM patients were stratified based on the predicted survival time (pred OS) by the 52 BPM based SVR model. Patients with pred OS ≤ median (pred OS) are at higher risk than the patients with pred OS > median (pred OS).
3.3. Using clinico-pathological features for prognostication
3.3.1. Univariate analysis shows Breslow thickness as the most significant feature
In order to see whether the models developed earlier in this study perform better than the previously established prognostic markers, patients were stratified using clinical factors such as AJCC pathological staging, age, TNM staging, Breslow thickness, Gender and Ulceration status. These have been reported to be associated with risk in CM patients in the past [53, 54, 55, 56]. Table 5 shows these results.
Table 5.
Risk estimation in CM patients using clinical factors. N represents no. of samples for which data was available.
| Factor | Strata | N | HR | p value |
|---|---|---|---|---|
| Age | >63y vs 63y | 449 | 1.83 | 4 × 10−4 |
| continuous | 449 | 1.02 | 1.9 × 10−6 | |
| AJCC 6th ed. | Stage III, IV vs I,II | 138 | 1.60 | 0.071 |
| AJCC 7th ed. | Stage III, IV vs I,II | 215 | 2.26 | 0.025 |
| N staging | N1, N2, N3 vs N0 | 396 | 1.82 | 9 × 10−4 |
| T staging | T2, T3, T4 vs Tis, T1 | 378 | 1.68 | 4.8 × 10−2 |
| M staging | M1 vs M0 | 423 | 1.90 | 9.9 × 10−2 |
| Breslow thickness | >3mm vs 3mm | 342 | 2.45 | 3 × 10−6 |
| continuous | 342 | 1.03 | 10−4 | |
| Gender | Male vs Female | 449 | 1.20 | 0.277 |
| Ulceration status | Yes vs No | 300 | 2.06 | 5 × 10−4 |
Though our results agree with the previously reported observations, such as patients with age greater than 63 years [57], males [58], patients with metastasized tumors, stage III/IV patients etc. are at higher risk and thus show a high HR value, some of them are either insignificant (p > 0.05) or have a low HR/high p-value except Breslow thickness. Patients with Breslow thickness more than 3mm (median) were found to be at 2.45 times higher risk (p-value = 3 × 10−6) than compared to the patients with a lesser Breslow thickness. Nevertheless, both apoptotic GPM based PI model and total BPM based SVR model outperform the individual clinical factors and AJCC staging based risk stratification.
3.3.2. GEP and clinical features based combinatorial models
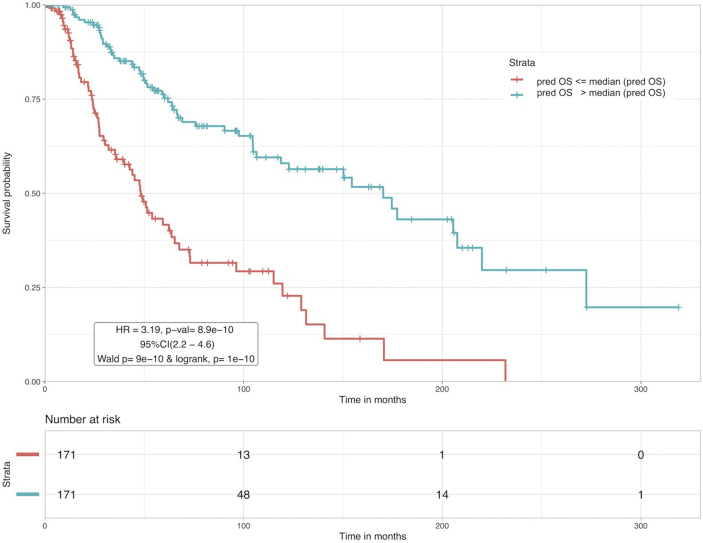
In one of our previous studies [59], Human Leukocyte Antigen (HLA) alleles-based features were used in combination with clinical information, for the prognostication of melanoma patients. The study used a dataset of 401 patients for developing ML based models. Here, we follow a similar strategy with a bigger dataset (449 patients) and additional features (TNM staging, Ulceration status), to combine clinical-features with the apoptotic GPM set, NOTCH combined set and total BPM set to see whether performance increases. New MLR models and PI based models were built with the combinations and patients were classified based on pred OS and PI in the same way as earlier. Best results based on combination with ‘Breslow Thickness’ are shown in Supplementary S2 Table S2. Clearly, the predicted OS obtained from the SVR model based on a combination of 52 total BPM genes and Breslow thickness boosted the performance even further as compared to using BPM genes alone. An HR value of 3.19 with a p-value ~ 10−10 and C value of 0.65 was achieved at median cutoff, which is the best amongst other combinatorial models as well as previous models. At mean cutoff, HR was 3.19, p-value ~ 9.16 × 10−10 and C = 0.65 (not shown in the table). KM plot illustrating the survival rates associated with high/low risk patients corresponding to this model at median cutoff is shown in Figure 3. Gene-enrichment analysis was performed on this set of 52 genes and the results are shown in Supplementary S1 TableF. A pathway enrichment corresponding to the associated proteins showed that the proteins KRT4, KRT13, KRT27 and SPRR3 are involved in the cornification process, which has a direct association with risk of skin cancer [60, 61].
Figure 3.
Kaplan Meier plots for risk stratification of CM patients based on SVR model with combination of 52 BPM and Breslow thickness as features. Patients with Pred OS median (Pred OS) are at a greater risk than patients with Pred OS > median (Pred OS) with HR = 3.19 and p-val = 8.9 × 10−10.
3.3.3. Clinico-pathological information-based ensemble model as a superior prognostic marker
We devised a new ensemble model for integrating the prognostic potential of important clinical features. Here, entries corresponding to each clinical feature were allotted a risk point (r) as r = 1, 0 or -1 based on the risk group (high risk: r = 1, low risk: r = -1, unavailable: r = 0), according to Table 5. For example, in the feature ‘Breslow Thickness’, entries that were >3mm (high-risk) were given an r = 1, entries that were 3mm (low-risk) were given r = -1 and missing/unavailable entries were labelled with r = 0. This method ensured fixed length vectors, each of equal dimensions i.e. 449. Various linear combinations comprising of two or more features were evaluated and the best results were achieved with the combination of Breslow thickness, N staging, M staging and Ulceration status. We termed this combination as Risk Grade (RG) where RG for a patient is defined as:
RG = r (Breslow thickness) +r (N staging) +r (M staging) + r (Ulceration status)
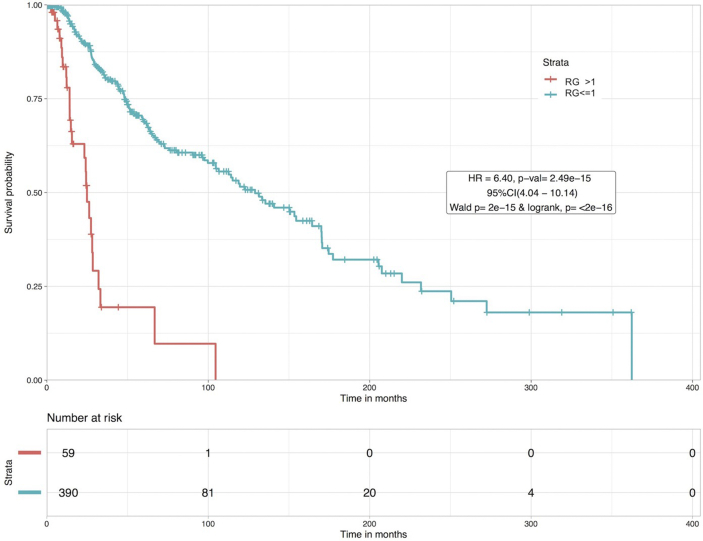
The hazard ratio for RG was 6.40 with a p-value of 2.49 × 10−15. Patients with RG > 1 were at higher risk than patients that had an RG 1, as represented by the KM plot in Figure 4. 10-year survival rate for high risk patients is seen to fall to zero whereas patients in low risk group have a 50% survival probability. Results for other top combinations are shown in Supplementary S2 Table S3. It is important to note the inferior risk stratification achieved using the AJCC 7th edition staging (Table 5) in contrast to RG. It should also be noted that even when the patients with data available for all four clinical features were taken (~259 patients), RG was able to stratify high (RG > 1) and low risk (RG1) patients with a significant HR of 4.04 (95%CI 2.09–7.79) with a p-value of 3 × 10−5 (Wald test p-val = 3 × 10−5, logrank test p-val = 6 × 10−6), which is still superior than AJCC staging. High risk patients in this case had a 10-year survival probability of zero as compared to a 40% survival chance for low risk patients.
Figure 4.
Kaplan Meier plot for risk stratification of CM patients based on Risk Grade (RG). Patients with RG > 1 are at a greater risk than patients with RG ≤ 1 with HR = 6.40 and p-val = 2.49 × 10−15.
3.3.4. Comparative validation of ‘Risk Grade’
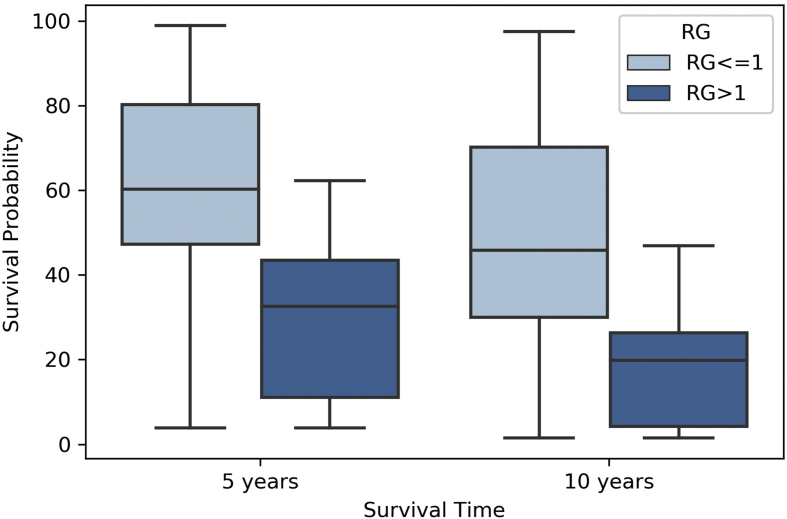
We performed a comparative assessment of the strength of RG as prognostic marker by employing a popular melanoma survival prediction model [62]. To do this, we used our dataset's features as input to the web-server “AJCC individualised melanoma patients outcome prediction tool” for prediction of 5 and 10-year survival probabilities of the patient samples in our dataset. The web-server required a total of seven input features i.e (i) whether patient had localized melanoma or regional melanoma (ii) tumour-thickness (iii) age (iv) tumor burden (v) lesion site (vi) number of nodes and (vii) ulceration status. After the queries were executed, it was able to predict survival rates for 162 patients out of 449 patients in our dataset. A pre-computed RG score based on the ensemble model for these 162 patients was used to classify these patients. Figure 5 shows the web-server predicted survival rates between two risk groups (RG > 1 or High Risk vs RG<=1 or Low Risk) in the form of a boxplot. The high risk group (n = 46) had a ~30% 5-year and ~20% 10-year mean survival rate as compared to the low risk group (n = 116) with a ~65% 5-year and ~55% 10-year mean survival rate. Apart from the risk groups segregation, the strength of RG can also be implied from the minimal number of required input features i.e four features as compared to seven features in the “AJCC individualised melanoma patients outcome prediction tool” for survival prediction.
Figure 5.
Boxplot representing the distinct segregation of risk groups by RG on the basis of 5- and 10- year survival outcomes predicted by “AJCC individualised melanoma patients outcome prediction tool”. A total of 162 predictions were made using the tool out of which 116 were low-risk patients (RG<=1) and 46 were high-risk (RG > 1).
4. Web-server and mobile application
A web-server and an android application named “CMcrpred” were developed and are freely available at (http://webs.iiitd.edu.in/raghava/cmcrpred/) and Google Play Store. Users can predict the survival outcome and risk of a melanoma patient using these services. The web-server provides a detailed prediction (graphical and tabular output) about the survival outcome of a patient belonging to a specific RG. On the other hand, the android application has been kept less detailed and more user friendly for on-the-go usage by clinicians and/or patients. The web server has been designed by using a responsive HTML template for adjustment to the browsing device. Thus, our web server is compatible with a wide range of devices, including desktops, tablets, and smartphones.
5. Discussion
So far, the primary methods for the prognosis of melanoma patients include patient classification on the basis of various clinico-pathological features — following which the therapy choice is made for the majority of cases. However, alleged inadequate assessment of individual tumor prognosis by clinicopathological variables led to the need for molecular biomarkers that provide sufficient predictive value to allow personal treatment in the case of cancer. Molecular biomarkers such as S100 levels in pembrolizumab treated patients with metastatic melanoma [63], higher levels of serum tryptase levels in deeper melanomas and melanomas with ulceration [64], serum levels of vitamin D in regards to shield site melanoma vs non-shield site melanoma [65] are examples of a few recent studies. Another widely emerging class is GEP based biomarkers and combinations of these biomarkers. These genomic biomarkers are directly related to the intricacies of cancer biology and could adjunct the phenotypic staging system. To that endeavor, many studies in the past have suggested prognostic markers that could enhance the risk management in melanoma patients notably [22, 23, 24, 25, 26, 27, 28, 29]. While there has been a major interest in this area, none of the GEP based prognostic markers have been included in the AJCC staging system as of yet.
In this study, we were able to present new models for risk assessment in melanoma patients, based on OS time. We first analyzed various cancer pathway associated genes and showed that a PI constructed with the GEP of 29 apoptotic pathway linked genes could be used for risk prediction in melanoma. These genes mainly encode for proteins that are directly related to promoting cell death and are under-expressed in tumor cells. Examples include MCL1, XIAP, FAS, FASLG, Caspases [3, 7, 8], which are key players in the intrinsic and extrinsic apoptotic pathway [66, 67]. The PI for these 29 genes was able to significantly discriminate between high and low-risk patients by a 2.5 hazard ratio. Another model employed the genes involved in both Apoptosis and NOTCH pathways and increased the HR, though only by a marginal amount. The strong biological roles of these pathways in the general case of cancer are well established [68, 69, 70, 71]. Further, a popular feature selection method, rfSRC was used to filter out important survival associated genes, which were then used to make MLR and PI-based models. We show that OS time predicted by using GEP of a set of novel 52 genes was able to stratify high and low-risk patients with a higher HR (2.77) than 29-apoptotic genes based PI. The PI-based on these 52 genes also performed better than the 29-apoptotic genes based PI. By integration of this 52 GEP and Breslow thickness in an SVR based regression model, we were able to enhance the risk classification further (HR = 3.19).
Further, to assess the comparative strength of the clinico-pathological features against the GEP based prognostication, we developed models with combinations of two or more clinical features. To achieve this, we employed a simple categorization method, which helped to avoid data loss and consequently guaranteed a bigger dataset. “Risk Grade (RG)” built on the information from N stage, M stage, Breslow thickness, and Ulceration status of a patient was shown to stratify high and low-risk patients with an HR value almost two-fold (HR = 6.40) of the 52 GEP and Breslow thickness-based model. Though, the efficacy of RG was also shown for a subset of 259 patients where data for every clinical feature was present. The strength of this risk segregation scheme can be compared with the current AJCC staging. While in the conventional AJCC staging criteria, T stage encompasses the information about Breslow thickness and Ulceration, here we constructed RG with the explicit use of both of these features. It is also necessary to point out that even though we have shown the performance of RG in comparison to AJCC 7th and 6th editions, it is bound to perform equally well when compared with 8th edition, since the major change in AJCC 8th edition was within the substages (primarily in stage III sub staging) [16]. Briefly, as compared to using any of the GEP based methods, the number of required prognostic features was reduced to a minimum, with a huge improvement in risk stratification. Although these models are based on features that are already involved in the AJCC staging system, an integration of these features, such as the one done in this study, has been missing in the previous studies. We exhibit the strength of using the clinico-pathological features over the molecular biomarkers by means of combinatorial models. We also show by making a comparative assessment with an existing survival prediction tool that our model which employs information about only four clinical features displays both risk segregation and survival prediction ability. Clearly, there needs to be more development and integration of prognostic biomarkers, such as the one presented here, for therapeutic decision-making in melanoma. To promote this concept further and facilitate the scientific as well as clinical community, we have also developed a web-server (http://webs.iiitd.edu.in/raghava/cmcrpred) and an android application “CMcrpred”, based on this model.
6. Limitations of the study
The in-silico analysis presented here was performed by employing a recent RNAseq dataset obtained from TCGA which contained clinical as well as bio-specimen information. To the best of our knowledge, only publicly accessible free dataset which contains all the clinical features (i.e Breslow Thickness, N staging, M staging and Ulceration status) that are required as input to our model, is the TCGA-SKCM cohort. Though, we have used cross-validation techniques and comparative analysis with another prediction tool to assess the risk prediction models, an independent validation on external dataset is crucial for the application of the model in a clinical setting. Absence of such an external validation dataset and constraints within TCGA dataset are major limitations of this study and thus demand future efforts.
Declarations
Author contribution statement
Chakit Arora: Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Dilraj Kaur: Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Anjali Lathwal: Contributed reagents, materials, analysis tools or data.
Gajendra P.S. Raghava: Conceived and designed the experiments; Analyzed and interpreted the data; Wrote the paper.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
The Authors are thankful to funding agencies Indraprastha Institute of Information Technology, New Delhi and University Grants Commission, for fellowship and support. Authors are also thankful to Pranay Raj Anand (Postgraduate student, IIIT-Delhi) for his support in developing the “CMcrpred” android application.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Ossio R., Roldan-Marin R., Martinez-Said H., Adams D.J., Robles-Espinoza C.D. Melanoma: a global perspective. Nat. Rev. Canc. 2017;17:393–394. doi: 10.1038/nrc.2017.43. [DOI] [PubMed] [Google Scholar]
- 2.Mintz B. Clonal basis of mammalian differentiation. Symp. Soc. Exp. Biol. 1971;25:345–370. https://www.ncbi.nlm.nih.gov/pubmed/4940552 Available at: [PubMed] [Google Scholar]
- 3.Markert C.L., Silvers W.K. The effects of genotype and cell environment on melanoblast differentiation in the house mouse. Genetics. 1956;41:429–450. doi: 10.1093/genetics/41.3.429. https://www.ncbi.nlm.nih.gov/pubmed/17247639 Available at: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Theriault L.L., Hurley L.S. Ultrastructure of developing melanosomes in C57 black and pallid mice. Dev. Biol. 1970;23:261–275. doi: 10.1016/0012-1606(70)90098-9. [DOI] [PubMed] [Google Scholar]
- 5.Barden H., Levine S. Histochemical observations on rodent brain melanin. Brain Res. Bull. 1983;10:847–851. doi: 10.1016/0361-9230(83)90218-6. [DOI] [PubMed] [Google Scholar]
- 6.Siegel R.L., Miller K.D., Jemal A. Cancer statistics. CA Cancer J. Clin. 2019;69:7–34. doi: 10.3322/caac.21551. (2019) [DOI] [PubMed] [Google Scholar]
- 7.Buttner P., Garbe C., Bertz J., Burg G., d’Hoedt B., Drepper H., Guggenmoos-Holzmann I., Lechner W., Lippold A., Orfanos C.E. Primary cutaneous melanoma. Optimized cutoff points of tumor thickness and importance of Clark’s level for prognostic classification. Cancer. 1995;75:2499–2506. doi: 10.1002/1097-0142(19950515)75:10<2499::aid-cncr2820751016>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 8.Bristow I.R., de Berker D.A., Acland K.M., Turner R.J., Bowling J. Clinical guidelines for the recognition of melanoma of the foot and nail unit. J. Foot Ankle Res. 2010;3:25. doi: 10.1186/1757-1146-3-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Balch C.M., Gershenwald J.E., Soong S.J., Thompson J.F., Atkins M.B., Byrd D.R., Buzaid A.C., Cochran A.J., Coit D.G., Ding S. Final version of 2009 AJCC melanoma staging and classification. J. Clin. Oncol. 2009;27:6199–6206. doi: 10.1200/JCO.2009.23.4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Linos E., Swetter S.M., Cockburn M.G., Colditz G.A., Clarke C.A. Increasing burden of melanoma in the United States. J. Invest. Dermatol. 2009;129:1666–1674. doi: 10.1038/jid.2008.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mehta S., Shelling A., Muthukaruppan A., Lasham A., Blenkiron C., Laking G., Print C. Predictive and prognostic molecular markers for cancer medicine. Ther. Adv. Med. Oncol. 2010;2:125–148. doi: 10.1177/1758834009360519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee J.K., Chan A.T. Molecular prognostic and predictive markers in colorectal cancer: current status. Curr. Colorectal. Canc. Rep. 2011;7:136–144. doi: 10.1007/s11888-011-0091-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crijns A.P.G., Duiker E.W., de Jong S., Willemse P.H.B., van der Zee A.G.J., de Vries E.G.E. Molecular prognostic markers in ovarian cancer: toward patient-tailored therapy. Int. J. Gynecol. Canc. 2006;16(Suppl 1):152–165. doi: 10.1111/j.1525-1438.2006.00503.x. [DOI] [PubMed] [Google Scholar]
- 14.Kutomi G., Mizuguchi T., Satomi F., Maeda H., Shima H., Kimura Y., Hirata K. Current status of the prognostic molecular biomarkers in breast cancer: a systematic review. Oncol. Lett. 2017;13:1491–1498. doi: 10.3892/ol.2017.5609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hyams D.M., Cook R.W., Buzaid A.C. Identification of risk in cutaneous melanoma patients: prognostic and predictive markers. J. Surg. Oncol. 2019;119:175–186. doi: 10.1002/jso.25319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gershenwald J.E., Scolyer R.A., Hess K.R., Sondak V.K., Long G.V., Ross M.I., Lazar A.J., Faries M.B., Kirkwood J.M., McArthur G.A. Melanoma staging: evidence-based changes in the American Joint Committee on Cancer eighth edition cancer staging manual. CA Cancer J. Clin. 2017;67:472–492. doi: 10.3322/caac.21409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deichmann M., Kahle B., Moser K., Wacker J., Wust K. Diagnosing melanoma patients entering American Joint Committee on Cancer stage IV, C-reactive protein in serum is superior to lactate dehydrogenase. Br. J. Canc. 2004;91:699–702. doi: 10.1038/sj.bjc.6602043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weide B., Elsasser M., Buttner P., Pflugfelder A., Leiter U., Eigentler T.K., Bauer J., Witte M., Meier F., Garbe C. Serum markers lactate dehydrogenase and S100B predict independently disease outcome in melanoma patients with distant metastasis. Br. J. Canc. 2012;107:422–428. doi: 10.1038/bjc.2012.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wevers K.P., Kruijff S., Speijers M.J., Bastiaannet E., Muller Kobold A.C., Hoekstra H.J. S-100B: a stronger prognostic biomarker than LDH in stage IIIB-C melanoma. Ann. Surg Oncol. 2013;20:2772–2779. doi: 10.1245/s10434-013-2949-y. [DOI] [PubMed] [Google Scholar]
- 20.Kashani-Sabet M., Venna S., Nosrati M., Rangel J., Sucker A., Egberts F., Baehner F.L., Simko J., Leong S.P., Haqq C. A multimarker prognostic assay for primary cutaneous melanoma. Clin. Canc. Res. 2009;15:6987–6992. doi: 10.1158/1078-0432.CCR-09-1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kashani-Sabet M., Nosrati M., Miller J.R., 3rd, Sagebiel R.W., Leong S.P.L., Lesniak A., Tong S., Lee S.J., Kirkwood J.M. Prospective validation of molecular prognostic markers in cutaneous melanoma: a correlative analysis of E1690. Clin. Canc. Res. 2017;23:6888–6892. doi: 10.1158/1078-0432.CCR-17-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brozyna A.A., Guo H., Yang S.E., Cornelius L., Linette G., Murphy M., Sheehan C., Ross J., Slominski A., Carlson J.A. TRPM1 (melastatin) expression is an independent predictor of overall survival in clinical AJCC stage I and II melanoma patients. J. Cutan. Pathol. 2017;44:328–337. doi: 10.1111/cup.12872. [DOI] [PubMed] [Google Scholar]
- 23.Johnson D.B., Lovly C.M., Flavin M., Panageas K.S., Ayers G.D., Zhao Z., Iams W.T., Colgan M., DeNoble S., Terry C.R. Impact of NRAS mutations for patients with advanced melanoma treated with immune therapies. Canc. Immunol. Res. 2015;3:288–295. doi: 10.1158/2326-6066.CIR-14-0207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Long G.V., Hauschild A., Santinami M., Atkinson V., Mandala M., Chiarion-Sileni V., Larkin J., Nyakas M., Dutriaux C., Haydon A. Adjuvant dabrafenib plus trametinib in stage III BRAF-mutated melanoma. N. Engl. J. Med. 2017;377:1813–1823. doi: 10.1056/NEJMoa1708539. [DOI] [PubMed] [Google Scholar]
- 25.Mumford S.L., Towler B.P., Pashler A.L., Gilleard O., Martin Y., Newbury S.F. Circulating MicroRNA biomarkers in melanoma: tools and challenges in personalised medicine. Biomolecules. 2018:8. doi: 10.3390/biom8020021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cook R.W., Middlebrook B., Wilkinson J., Covington K.R., Oelschlager K., Monzon F.A., Stone J.F. Analytic validity of DecisionDx-Melanoma, a gene expression profile test for determining metastatic risk in melanoma patients. Diagn. Pathol. 2018;13:13. doi: 10.1186/s13000-018-0690-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Greenhaw B.N., Zitelli J.A., Brodland D.G. Estimation of prognosis in invasive cutaneous melanoma: an independent study of the accuracy of a gene expression profile test. Dermatol. Surg. 2018;44:1494–1500. doi: 10.1097/DSS.0000000000001588. [DOI] [PubMed] [Google Scholar]
- 28.Hsueh E.C., DeBloom J.R., Lee J., Sussman J.J., Covington K.R., Middlebrook B., Johnson C., Cook R.W., Slingluff C.L., Jr., McMasters K.M. Interim analysis of survival in a prospective, multi-center registry cohort of cutaneous melanoma tested with a prognostic 31-gene expression profile test. J. Hematol. Oncol. 2017;10:152. doi: 10.1186/s13045-017-0520-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gastman B.R., Gerami P., Kurley S.J., Cook R.W., Leachman S., Vetto J.T. Identification of patients at risk of metastasis using a prognostic 31-gene expression profile in subpopulations of melanoma patients with favorable outcomes by standard criteria. J. Am. Acad. Dermatol. 2019;80:149–157 e4. doi: 10.1016/j.jaad.2018.07.028. [DOI] [PubMed] [Google Scholar]
- 30.Meves A., Nikolova E., Heim J.B., Squirewell E.J., Cappel M.A., Pittelkow M.R., Otley C.C., Behrendt N., Saunte D.M., Lock-Andersen J. Tumor cell adhesion as a risk factor for sentinel lymph node metastasis in primary cutaneous melanoma. J. Clin. Oncol. 2015;33:2509–2515. doi: 10.1200/JCO.2014.60.7002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sivendran S., Chang R., Pham L., Phelps R.G., Harcharik S.T., Hall L.D., Bernardo S.G., Moskalenko M.M., Sivendran M., Fu Y. Dissection of immune gene networks in primary melanoma tumors critical for antitumor surveillance of patients with stage II-III resectable disease. J. Invest. Dermatol. 2014;134:2202–2211. doi: 10.1038/jid.2014.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wei L., Jin Z., Yang S., Xu Y., Zhu Y., Ji Y. TCGA-assembler 2: software pipeline for retrieval and processing of TCGA/CPTAC data. Bioinformatics. 2018;34:1615–1617. doi: 10.1093/bioinformatics/btx812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y., Ren F., Chen P., Liu S., Song Z., Ma X. Identification of a six-gene signature with prognostic value for patients with endometrial carcinoma. Cancer Med. 2018;7:5632–5642. doi: 10.1002/cam4.1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mandelboum S., Manber Z., Elroy-Stein O., Elkon R. Recurrent functional misinterpretation of RNA-seq data caused by sample-specific gene length bias. PLoS Biol. 2019;17 doi: 10.1371/journal.pbio.3000481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.He X., Xu H., Zhao W., Zhan M., Li Y., Liu H., Tan L., Lu L. POPDC3 is a potential biomarker for prognosis and radioresistance in patients with head and neck squamous cell carcinoma. Oncol. Lett. 2019;18:5468–5480. doi: 10.3892/ol.2019.10888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Akter S., Xu D., Nagel S.C., Bromfield J.J., Pelch K., Wilshire G.B., Joshi T. Machine learning classifiers for endometriosis using transcriptomics and methylomics data. Front. Genet. 2019;10:766. doi: 10.3389/fgene.2019.00766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van der Net J.B., Janssens A.C., Defesche J.C., Kastelein J.J., Sijbrands E.J., Steyerberg E.W. Usefulness of genetic polymorphisms and conventional risk factors to predict coronary heart disease in patients with familial hypercholesterolemia. Am. J. Cardiol. 2009;103:375–380. doi: 10.1016/j.amjcard.2008.09.093. [DOI] [PubMed] [Google Scholar]
- 38.Dyrskjot L., Reinert T., Algaba F., Christensen E., Nieboer D., Hermann G.G., Mogensen K., Beukers W., Marquez M., Segersten U. Prognostic impact of a 12-gene progression score in non-muscle-invasive bladder cancer: a prospective multicentre validation study. Eur. Urol. 2017;72:461–469. doi: 10.1016/j.eururo.2017.05.040. [DOI] [PubMed] [Google Scholar]
- 39.Chaudhary K., Poirion O.B., Lu L., Garmire L.X. Deep learning-based multi-omics integration robustly predicts survival in liver cancer. Clin. Canc. Res. 2018;24:1248–1259. doi: 10.1158/1078-0432.CCR-17-0853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chauhan J.S., Mishra N.K., Raghava G.P.S. Prediction of GTP interacting residues, dipeptides and tripeptides in a protein from its evolutionary information. BMC Bioinf. 2010;11:301. doi: 10.1186/1471-2105-11-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Singh H., Kumar R., Singh S., Chaudhary K., Gautam A., Raghava G.P. Prediction of anticancer molecules using hybrid model developed on molecules screened against NCI-60 cancer cell lines. BMC Canc. 2016;16:77. doi: 10.1186/s12885-016-2082-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Singh H., Singh S., Singla D., Agarwal S.M., Raghava G.P. QSAR based model for discriminating EGFR inhibitors and non-inhibitors using Random forest. Biol. Direct. 2015;10:10. doi: 10.1186/s13062-015-0046-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nagpal G., Usmani S.S., Dhanda S.K., Kaur H., Singh S., Sharma M., Raghava G.P.S. Computer-aided designing of immunosuppressive peptides based on IL-10 inducing potential. Sci. Rep. 2017;7:42851. doi: 10.1038/srep42851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Agrawal P., Kumar S., Singh A., Raghava G.P.S., Singh I.K. NeuroPIpred: a tool to predict, design and scan insect neuropeptides. Sci. Rep. 2019;9:5129. doi: 10.1038/s41598-019-41538-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lathwal A., Arora C., Raghava G.P.S. Prediction of risk scores for colorectal cancer patients from the concentration of proteins involved in mitochondrial apoptotic pathway. PloS One. 2019;14 doi: 10.1371/journal.pone.0217527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kaur D., Arora C., Raghava G.P.S. A hybrid model for predicting pattern recognition receptors using evolutionary information. Front. Immunol. 2020;11:71. doi: 10.3389/fimmu.2020.00071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li P., Ren H., Zhang Y., Zhou Z. Fifteen-gene expression based model predicts the survival of clear cell renal cell carcinoma. Med. 2018;97 doi: 10.1097/MD.0000000000011839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lathwal A., Kumar R., Arora C., Raghava G.P.S. Identification of prognostic biomarkers for major subtypes of non-small-cell lung cancer using genomic and clinical data. J. Canc. Res. Clin. Oncol. 2020 doi: 10.1007/s00432-020-03318-3. [DOI] [PubMed] [Google Scholar]
- 49.Sanchez-Vega F., Mina M., Armenia J., Chatila W.K., Luna A., La K.C., Dimitriadoy S., Liu D.L., Kantheti H.S., Saghafinia S. Oncogenic signaling pathways in the cancer Genome Atlas. Cell. 2018;173:321–337 e10. doi: 10.1016/j.cell.2018.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu Y., Yang N., Peng X., Liu G., Zhong H., Liu L. One-lincRNA and five-mRNA based signature for prognosis of multiple myeloma patients undergoing proteasome inhibitors therapy. Biomed. Pharmacother. 2019;118:109254. doi: 10.1016/j.biopha.2019.109254. [DOI] [PubMed] [Google Scholar]
- 51.Zhang Z.L., Zhao L.J., Xu L., Chai L., Wang F., Xu Y.P., Zhou S.H., Fu Y. Transcriptomic model-based lncRNAs and mRNAs serve as independent prognostic indicators in head and neck squamous cell carcinoma. Oncol. Lett. 2019;17:5536–5544. doi: 10.3892/ol.2019.10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang J., Xu M., Gao H., Guo J.C., Guo Y.L., Zou M., Wu X.F. Two protein-coding genes act as a novel clinical signature to predict prognosis in patients with ovarian serous cystadenocarcinoma. Oncol. Lett. 2018;15:3669–3675. doi: 10.3892/ol.2018.7778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Marghoob A.A., Koenig K., Bittencourt F.V., Kopf A.W., Bart R.S. Vol. 88. Cancer; 2000. pp. 589–595.https://www.ncbi.nlm.nih.gov/pubmed/10649252 (Breslow Thickness and clark Level in Melanoma: Support for Including Level in Pathology Reports and in American Joint Committee on Cancer Staging). Available at: [PubMed] [Google Scholar]
- 54.Cherobin A., Wainstein A.J.A., Colosimo E.A., Goulart E.M.A., Bittencourt F.V. Prognostic factors for metastasis in cutaneous melanoma. An. Bras. Dermatol. 2018;93:19–26. doi: 10.1590/abd1806-4841.20184779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chao C., Martin R.C., 2nd, Ross M.I., Reintgen D.S., Edwards M.J., Noyes R.D., Hagendoorn L.J., Stromberg A.J., McMasters K.M. Correlation between prognostic factors and increasing age in melanoma. Ann. Surg Oncol. 2004;11:259–264. doi: 10.1245/aso.2004.04.015. [DOI] [PubMed] [Google Scholar]
- 56.Morton D.L., Wanek L., Nizze J.A., Elashoff R.M., Wong J.H. Improved long-term survival after lymphadenectomy of melanoma metastatic to regional nodes. Analysis of prognostic factors in 1134 patients from the John Wayne Cancer Clinic. Ann. Surg. 1991;214:491–501. doi: 10.1097/00000658-199110000-00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Watts C.G., Madronio C., Morton R.L., Goumas C., Armstrong B.K., Curtin A., Menzies S.W., Mann G.J., Thompson J.F., Cust A.E. Clinical features associated with individuals at higher risk of melanoma: a population-based study. JAMA Dermatol. 2017;153:23–29. doi: 10.1001/jamadermatol.2016.3327. [DOI] [PubMed] [Google Scholar]
- 58.Smalley K.S. Why do women with melanoma do better than men? Elife. 2018:7. doi: 10.7554/eLife.33511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dhall A., Patiyal S., Kaur H., Bhalla S., Arora C., Raghava G.P.S. Computing skin cutaneous melanoma outcome from the HLA-alleles and clinical characteristics. Front. Genet. 2020;11:221. doi: 10.3389/fgene.2020.00221. https://www.frontiersin.org/article/10.3389/fgene.2020.00221 Available at: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Eckhart L., Lippens S., Tschachler E., Declercq W. Cell death by cornification. Biochim. Biophys. Acta. 2013;1833:3471–3480. doi: 10.1016/j.bbamcr.2013.06.010. [DOI] [PubMed] [Google Scholar]
- 61.Zhou Y., Cheng Y., Feng D., Ling B., Liu P. Induction the cornification of squamous cancerous cells to eliminate tumor cells by promotion cell differentiation and stratum. Med. Hypotheses. 2011;77:763–764. doi: 10.1016/j.mehy.2011.07.032. [DOI] [PubMed] [Google Scholar]
- 62.Soong S., Ding S., Coit D., Balch C.M., Gershenwald J.E., Thompson J.F., Gimotty P. Predicting survival outcome of localized melanoma: an electronic prediction tool based on the AJCC Melanoma Database. Ann. Surg Oncol. 2010;17:2006–2014. doi: 10.1245/s10434-010-1050-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Simetic L., Blazicevic K., Medugorac K., Golcic M., Herceg D. Relative change in S100 as a biomarker of survival in patients with metastatic melanoma treated with pembrolizumab. Anticancer Res. 2020;40:2157–2163. doi: 10.21873/anticanres.14175. [DOI] [PubMed] [Google Scholar]
- 64.Paolino G., Moliterni E., Lopez T., Cardone M., Macri G., Garelli V., Richetta A.G., Didona D., Bottoni U., Calvieri S. Serum tryptase levels in melanoma patients: first results of clinicopathological features. Melanoma Res. 2016;26:207–208. doi: 10.1097/CMR.0000000000000231. [DOI] [PubMed] [Google Scholar]
- 65.Paolino G., Moliterni E., Didona D., Garelli V., Corsetti P., Lopez T., Richetta A.G., Cantisani C., Bottoni U., Calvieri S. Clinicopathological features, vitamin D serological levels and prognosis in cutaneous melanoma of shield-sites: an update. Med. Oncol. 2015;32:451. doi: 10.1007/s12032-014-0451-4. [DOI] [PubMed] [Google Scholar]
- 66.Elmore S. Apoptosis: a review of programmed cell death. Toxicol. Pathol. 2007;35:495–516. doi: 10.1080/01926230701320337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pfeffer C.M., Singh A.T.K. Apoptosis: a target for anticancer therapy. Int. J. Mol. Sci. 2018;19 doi: 10.3390/ijms19020448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sever R., Brugge J.S. Signal transduction in cancer. Cold Spring. Harb. Perspect Med. 2015:5. doi: 10.1101/cshperspect.a006098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Debatin K.M. Apoptosis pathways in cancer and cancer therapy. Cancer Immunol. Immunother. 2004;53:153–159. doi: 10.1007/s00262-003-0474-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mandinova A., Lee S.W. The p53 pathway as a target in cancer therapeutics: obstacles and promise. Sci. Transl. Med. 2011;3:64rv1. doi: 10.1126/scitranslmed.3001366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Han Y. Analysis of the role of the Hippo pathway in cancer. J. Transl. Med. 2019;17:116. doi: 10.1186/s12967-019-1869-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.