Abstract
There is enormous clinical value in inferring the brain regions initially atrophied in Parkinson disease for individual patients and understanding its relationship with clinical and genetic risk factors. The aim of this study is to leverage a new seed-inference algorithm demonstrated for Alzheimer’s disease to the Parkinsonian context and to cluster patients in meaningful subgroups based on these incipient atrophy patterns. Instead of testing brain regions separately as the likely initiation site for each patient, we solve an L1-penalized optimization problem that can return a more predictive heterogeneous, multi-locus seed patterns. A cluster analysis of the individual seed patterns reveals two distinct subgroups (S1 versus S2). The S1 subgroup is characterized by the involvement of the brainstem and ventral nuclei, and S2 by cortex and striatum. Post hoc analysis in features not included in the clustering shows significant differences between subgroups regarding age of onset and local transcriptional patterns of Parkinson-related genes. Top genes associated with regional microglial abundance are strongly associated with subgroup S1 but not with S2. Our results suggest two distinct aetiological mechanisms operative in Parkinson disease. The interplay between immune-related genes, lysosomal genes, microglial abundance and atrophy initiation sites may explain why the age of onset for patients in S1 is on average 4.5 years later than for those in S2. We highlight and compare the most prominently affected brain regions for both subgroups. Altogether, our findings may improve current screening strategies for early Parkinson onsetters.
Keywords: Parkinson disease, early atrophy, computational neurology, Parkinson subgroups, microglia abundance
We computationally infer the initial sites of pathology for a Parkinson cohort. These incipient patterns are highly heterogeneous and can be clustered in two unreported subgroups (S1 versus S2). Differences between subgroups extend to age-of-onset and transcriptional patterns of immune, lysosomal, autophagy and mitochondrial Parkinson’s disease-related genes.
Graphical Abstract
Graphical Abstract.
Introduction
Parkinson’s disease is a progressive neurodegenerative disorder clinically marked by tremor, bradykinesia and rigidity. The Braak staging model of Parkinson’s disease proposes that the deposition of mis-folded alpha-synuclein initiates in the brainstem sites (Stages 1 and 2), substantia nigra (SN) and limbic structures (Stages 3 and 4), finally spreading into adjoining frontal and other cortices (Stages 5 and 6) (Del Tredici et al., 2002; Braak et al., 2003). Subsequent studies improved upon the Braak system, particularly by recognizing the involvement of the olfactory bulb and the existence of two separate progression pathways for pathology: one along an initial brainstem route as proposed by Braak, and another along an initial limbic route, as proposed by later groups (Leverenz et al., 2008; Beach et al., 2009; Toledo et al., 2016; Coughlin et al., 2019). The early sites of Parkinson’s disease pathology are therefore expected to show substantial heterogeneity, a topic that has received scant attention to date. The goal of this study is to use state-of-the-art computational methods to infer the likely initiation sites of Parkinson’s disease pathology to characterize such variability.
The stereotyped, and to some extent, predictable pathological process occurring in Parkinson’s disease (Jellinger, 2009) gave rise to several computational models capable of forecasting atrophy spread in the brain (Zhou et al., 2012; Zeighami et al., 2015; Yau et al., 2018). In particular, the network diffusion model (NDM) (Raj et al., 2012, 2015) can successfully predict the trans-neuronal spread of atrophy (a commonly used radiological proxy for pathology) using brain connectivity. When applied to Parkinson’s disease, NDM is able to recapitulate radiological correlates of different stages of Braak’s Lewy pathology scheme (Pandya et al., 2019). Moreover, this computational model provides a principled way to test different brain regions as the likely initiator of pathology, which we hypothesize to vary significantly from patient to patient.
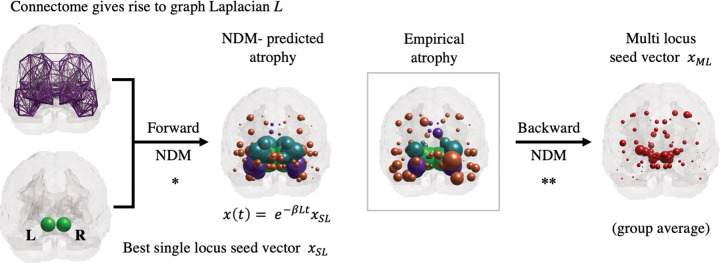
In previous studies (Freeze et al., 2019; Pandya et al., 2019), the NDM was used to rank brain regions (such as the SN) according to their likelihood of being the sole initial site with pathology (see Fig. 1, forward NDM). By excluding the possibility of multiple initiation sites, they were able to test all regions in their brain atlas via exhaustive search. To overcome this limitation and allow more nuanced initiation patterns, we will adapt the seed-inference algorithm from Torok et al. (2018) to our Parkinsonian context (see Fig. 1, backward NDM).
Figure 1.
Overview of forward and backward NDM. NDM is an established graph-theoretical model for neuropathology spread. Each node of the graph is associated with a grey matter region specified by a brain atlas. The connectome gives rise to the graph-Laplacian L. To analytically predict the value of the regional atrophy vector at a given point , one must specify the initial state of the system. It is customary in the forward NDM to test all possible single-locus seed vectors to find the one that best predicts the empirical atrophy (*). The backward NDM is a recently developed algorithm that can infer more predictive and sophisticated initial states , with multiple loci and distinct entry values (**). Brain renderings show the coronal view, with pathology loads proportional to the sphere diameters (values scaled for improved visualization). When applied to individual patients, the backward NDM gives rise to incipient patterns that are more heterogeneous than generally appreciated.
In what follows, we will show that our multi-loci seeds hide a surprising degree of intersubject heterogeneity. Specifically, that the individual subjects’ seed patterns can be clustered into two distinct groups (S1 versus S2). While only seed variables were used in the clustering analysis, the differences between subgroups extend to age-of-onset, to regional transcriptional patterns of Parkinson’s disease-related genes and to regional microglial abundance. These differences may be traced to differential expression of immune, autophagy, metabolic and microglial activity, suggesting that two divergent aetiologic processes may govern early Parkinson’s disease. Our result also highlights the value of computational backtracking Parkinson’s disease-related atrophy, since individual subject’s aetiologic heterogeneity may not be apparent from regional atrophy measurements in later stages using MRI.
Materials and methods
Participants
We test our model on a cross-sectional baseline imaging dataset from Parkinson’s Progressive Marker Initiative (PPMI) (www.ppmi-info.org/data) from 232 de novo Parkinson’s patients and 117 age-matched healthy controls (HC). Demographic and clinical features of our study are shown in Table 1.
Table 1.
Demographic and clinical features of PPMI cohort
| Parkinson’s disease | HC | |
|---|---|---|
| Size | 232 | 117 |
| Male/female | 155/77 | 74/43 |
| Age at baseline | 61.2 ± 9.1 | 59.5 ± 11.3 |
| Education (years) | 15.4 ± 2.7 | 15.7 ± 2.8 |
| MoCA | 27.3 ± 2.2 | 28.2 ± 1.2 |
| UPDRS3 | 21.9 ± 9.1.2 | n/a |
| Hoch and Yarh stage | 1.6 ± 0.5 | n/a |
| Symptom duration | 0.58 ± 0.59 | n/a |
| Handedness (L/R/A) | 17/210/5 | n/a |
Structural pre- and post-processing
For this study, we used a brain parcellation reported previously (Zeighami et al., 2015). Briefly, cortical- and basal-ganglia-related regions were included from the Hammers atlas (Hammers et al., 2003). The subthalamic nucleus, SN and red nucleus (RN) substructures were manually segmented using a high-resolution MRI template (T1-weighted ICBM152 template, resolution = 0.5 mm3), the BigBrain atlas (Amunts et al., 2013) and the brainstem anatomical atlas (Duvernoy, 1995). A subcortical atlas based on ultrahigh-field MRI confirmed the accuracy of the segmentations (Keuken et al., 2014). For the purpose of our study, we eliminated cerebellar regions and used only 78 out of 112 regions (Pandya et al., 2019).
The cross-sectional 3T high-resolution T1-weighted baseline images were denoised, normalized and corrected for non-uniformity intensity, following linear and non-linearly registration to the MNI-ICBM 152 template per the standard pipeline (Aubert-Broche et al., 2013). Regional brain atrophy was computed using deformation-based morphometry (Zeighami et al., 2015). Regional deformation-based morphometry values were extracted from the voxel-wise deformation-based morphometry maps for each subject. Parkinson’s disease atrophy was defined by computing a two tailed t-test between Parkinson’s disease and HC mean deformation-based morphometry values and was represented as a vector tPPMI = {tPPMI (i) |i ∊ [1, N]}, (N = 112). The t-statistic was converted to the natural range [0,1] using the logistic transform as followed in (Raj et al., 2015; Pandya et al., 2019). We removed 34 cerebellar regions from a previously validated brain parcellation (Zeighami et al., 2015), leaving 78 cerebral regions.
The connectome (Illinois Institute of Technology Human Brain atlas v3) was constructed from high-resolution diffusion weighted MRI data from 72 young healthy subjects. We used the anatomical connection density as the measure of connectivity (Iturria-Medina et al., 2007). This connectome is referred to as C = {ci,j}, whose elements ci,j represent the white matter pathway connectivity strength between ith and jth grey matter regions. Given that directionality is not deducible from DTI tractography, these connections are assumed to be bidirectional.
Forward NDM
The spread of proteinopathic agents over time is well captured by a dynamical system defined over a network-graph rendering of the brain, with the nodes representing grey matter structures and inter-regional connections defined as above. The forward NDM for the pathology load × at time t is given by (Raj et al., 2012):
where L is the graph-Laplacian matrix given by
The NDM has a closed-form analytical solution, , where is referred to as the seed vector or incipient atrophy pattern on which the diffusion kernel acts as a temporal and spatial blurring operator for the connectivity matrix C. In general, the unit of model’s diffusion time t is arbitrary and global diffusivity is unknown. We chose a value that would roughly span Parkinson’s progression (10–20 years), giving = 0.15. Thus, for a given initial atrophy configuration, we can use the NDM to predict the atrophy pattern at all future time points. We have previously validated the accuracy of the NDM predictions and robustness to parameter choices in multiple contexts (Raj et al., 2015; Pandya et al., 2017, 2019; Raj and Powell, 2018).
Seed-inference method (backward NDM)
The empirical atrophy z-scores are given by
where the indexes denote the i-th brain region and j-th patient, respectively; and denote mean and standard deviation calculated with respect to the HC. The scores were then normalized by a weighted logistic transform to keep values within the (0,1) range; these normalized vectors will be referred to in what follows as empirically observed atrophy vectors against which we run our inference algorithm (Raj et al., 2012; Torok et al., 2018).
The forward NDM can be used to infer the most likely pattern of disease seeding from a given vector . This inverse seed-inference process utilizes a constrained optimization algorithm with a -penalized cost function to promote sparsity while maximizing the Pearson’s correlation R between the NDM-predicted atrophy vector and . Analogous to the original formulation of the NDM, we define the two-variable function as:
Following (Torok et al., 2018), we perform the minimization over the two active variables, t and , in two steps. After determining an initial guess for the seed pattern, , and running the NDM on each region individually, we find an estimate for the that satisfies the following condition:
where is the Pearson’s correlation between the NDM-predicted atrophy using initial atrophy and the empirically observed atrophy vector. This cost function is monotonically decreasing and is minimized when the Pearson’s correlation is maximized, which is consistent with the NDM criterion. We then use this estimate of to find the that minimizes the following L1-constrained cost function:
is a tunable parameter that controls the sparsity of solutions; higher values of force more entries of the resultant vector to 0. We used the same stopping criteria as (Torok et al., 2018), and convergence under these conditions was not an issue in all runs we observed. See the original publication for further algorithmic details.
L-curves and parameter choices
The inverse-problem algorithm requires a sensible choice of the -penalty parameter (Torok et al., 2018). A priori, one does not know what this sensible choice should be, thus, it is customary to explore the parameter space within a reasonable range of values (0.1: 0.05: 1) and analyse the corresponding L-curves (see Supplementary Fig. 1). The goal is to select the ‘elbow’ of the curve, i.e. a value that provides a sensible trade-off between the mismatch of model/data—given by the term in the cost function, and the sparsity of the seed, given by the The main steps are enumerated bellow:
Infer an individualized from the observed atrophy vector b using the inverse model.
Project each into the future using the forward model to obtain the predicted atrophy vector a(t; xseed). Project the single-seed vector xsingle the same way to obtain a(t; xsingle).
Obtain the Pearson’s correlation coefficient (R-max) between (i) the pair of vectors {a(t; xseed), b}; (ii) the pair of vectors {a(t, xsingle, Subst, Nigra), b} and (iii) the pair of vectors {a(t, xsingle, Amygdala), b}, for each Parkinson's disease patient. Create histograms for R-max for all cases Supplementary Figs 1 and 2).
Supplementary Fig. 2A–C show the R-max histograms (i)–(iii) defined above in black, green and in magenta, respectively. The R-max values associated with (i) are typically higher than the ones associated with(ii) and (iii), demonstrating that inferred individualized patterns lead to significantly more predictive patterns of the patients’ atrophy vectors than a common single seed vector. This result provides strong evidence against a stereotyped/standard single seed location. We also find that (Supplementary Fig. 2C) that a single seed in the SN is more likely than a single seed in the Amygdala. Of course, if the seed-inference algorithm was giving trivial outcomes (e.g. inferred seed pattern = observed atrophy pattern) then we would erroneously obtain similar results to the above. To guard against that possibility, in Supplementary Fig. 2D, we present an R-max histogram comparing the two vectors , showing that are not obvious correlates of the observed atrophy patterns. This is consistent with the complex dynamics of disease spread in Parkinson’s disease and suggests that our seed inference is implicating a different set of regions than would be trivially predictable from the most atrophied regions.
Hierarchical clustering
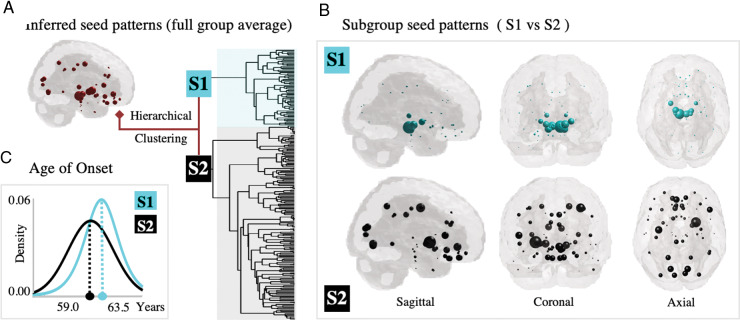
After applying our backward NDM algorithm to the baseline MRI volumetric data for all 232 Parkinson’s disease patients, we obtain a set of seed vectors given by , , …, }. For each patient, the entries of his/her seed vector represent the amount of pathology in our 78 ROI brain atlas during the incipient stage of the disease. The seed vectors in S exhibit a large amount of intersubject heterogeneity and lend themselves to exploratory data analysis and data mining. The categorization of the inferred seed vectors in two subgroups (S1 versus S2) is done via standard agglomerative hierarchical cluster tree. This unsupervised method clusters the data based on subject’s dissimilarity. We used MATLAB’s linkage function with (Ward) inner squared distance between clusters and Euclidean metric. Ward’s linkage uses the incremental sum of squares, that is, the increase in the total within-cluster sum of squares as a result of joining two clusters. The within-cluster sum of squares is defined as the sum of the squares of the distances between all objects in the cluster and the centroid of the cluster. The output of the analysis is the tree displayed in Fig. 2 is not a single set of clusters, but rather a multilevel hierarchy, where clusters at one level are joined as clusters at the next level. Thus, the classification into two subgroups (highlighted in black/cyan) follows from the algorithm, as we did not pre-specify a desired number of clusters. In what follows, we investigate if the differences in seed patterns (i.e. belonging to S1 versus S2) extend to other features not included in the clustering analysis.
Figure 2.
Analysis and clustering of Parkinson’s disease seed patterns. (A) Average inferred seed pattern for all Parkinson’s disease patients (n = 232). (B) Patients are clustered into two subgroups based on their seed patterns alone: S1 (n = 65, in cyan)—with strong involvement of brainstem and ventral nuclei, and S2 (n = 116, in black)—with involvement of cortex and striatum. (C) Post hoc analysis for variables not included in the clustering shows that the age of onset is in average 4.5 years later for S1.
Statistical analysis
Our selected HC and Parkinson’s disease cohorts passed two-sample t-test at 5% significance level for all features that may lead to substantial alterations in brain structure such as age (P-value = 0.16), sex (P-value = 0.6) and education years (P-value = 0.36). Seed patterns were inferred only for the Parkinson’s disease cohort via backward NDM and categorized as S1 or S2 via hierarchical clustering. In order to compare regional genetic differences between the subgroups, we apply t-test for 67 risk-factor genes. See the Results section and Fig. 3 for Pearson’s correlation r-coefficient and P-values for all functional classes of genes. In order to compare differences between the 72 allele frequencies, we used a chi-square test with Bonferroni multiple comparison correction. No allele survived the analysis. Finally, to compare differences in clinical scores between the subgroups, we applied t-test for all 30 features (see Supplementary Table 1). The only statistically significant differences were regarding sex and age of onset.
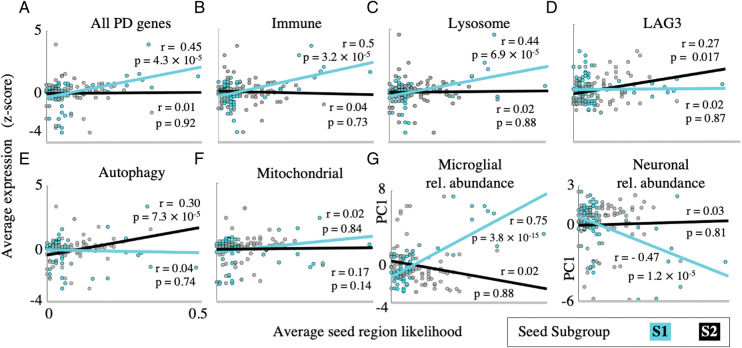
Figure 3.
Group-specific seeding patterns associated with distinct genetic predictors. Scatterplots of regional Parkinson’s disease-related gene expression versus average seed region likelihood for (A) all GWAS Parkinson’s disease risk-factor genes, (B) immune-related genes, (C) lysosomal genes, (D) autophagy-related genes, (E) mitochondrial genes and (F) LAG3. The S1 seeds are predicted by the average of all GWAS genes, immune-related and lysosomal genes, whereas S2 genes are predicted by autophagy-related genes and LAG3. (G) Scatterplots of regional microglial and neuronal abundance quantified by PC1 of high-fidelity genes, versus average seed region likelihood. Increased microglial abundance and decreased neuronal abundance predict S2 seed regions, while these measures are unrelated to S2 seed region likelihood.
Post hoc analysis for age of onset
The S1 and S2 subgroups also differ in features not included in the clustering analysis. In Fig. 2C, we fit a logistic distribution for the age of onset for patients in group S1 (in cyan, log likelihood = −226.9, mean = 63.5, var. = 68.2, and parameters mu = 63.5 and sigma = 4.5) and a normal distribution for those in group S2 (in black, log likelihood = −609.3, mean = 59.0, var. = 87.0, and parameters mu = 59.0 and sigma = 9.3). Patients in S1 are in average 4.5 years older than those in S2.
Post hoc analysis for regional genetics
In Alzheimer’s disease, it is common to use regional imaging and gene expression data as proxies for trans-neuronal and cell-autonomous processes predictors of pathology (Acosta et al., 2018). In our previous work, regional transcriptional analysis was used to show that early disease sites inferred from NDM in Parkinson’s disease patients were characterized by unusual expression of several susceptible genes, especially those responsible for aberrant synuclein processing and neuroinflammation (Freeze et al., 2019). Given that our clustering analysis revealed two distinct subgroups regarding our inferred seeds, we therefore performed a post hoc analysis to test if the differences between subgroups extend to genetic features associated with Parkinson’s disease. Regional transcriptional mapping of Parkinson’s disease-related genes was performed following (Freeze et al., 2019) using 67 putative Parkinson’s disease risk-factor genes from the recent Genome-Wide Association Study (GWAS) meta-analysis (Chang et al., 2017). The genes were mapped to the same atlas used for NDM, using data from the human Allen Brain Atlas (Hawrylycz et al., 2012). Data for the gene CDC71 could not be located. Details of transcriptional mapping on our atlas were previously described (Freeze et al., 2019). Briefly, semantic matching of expression data region to the 78-region atlas was employed except in cases of differential demarcation. In these cases, atlases were visually compared, and ABA samples were mapped to the closest corresponding grey matter region. ABA samples spanning more than one region were excluded from the analysis. For each gene, all samples across all probes and brains (n = 6 HC) with the same region were averaged and then normalized to produce a single expression value quantified as z-score. Gene samples from white matter areas were excluded from analysis. Expression data for LAG3 and the top five neuronal and microglial high-fidelity genes (Kelley et al., 2018) were mapped in the same way. Lysosomal, mitochondrial and autophagy-related were initially classified as previously described (Chang et al., 2017). Immune-related genes were initially classified according to the ImmPort database (Bhattacharya et al., 2018). Classification was manually reviewed, and additional genes were added to the relevant functional class in cases in which literature evidence supports a role in the class.
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Results
Overview of demographic and clinical information
Table 1 summarizes the demographic and clinical information for HC and Parkinson’s disease patients in our sample. All data were made publicly available by the PPMI. See the Materials and methods section for more details. Our selected HC and Parkinson’s disease cohorts passed two-sample t-test at 5% significance level for all features that may lead to substantial alterations in brain structure such as age (P-value =0.16), sex (P-value = 0.6) and education years (P-value = 0.36).
Group-level regional patterns for atrophy and seed vectors
Figure 1 shows brain renderings (coronal view) associated with the forward* and backward** NDM. The leftmost plots show the best single-locus seed vector , with non-zero entries at both hemispheres of the SN, and the connectome, that yields the graph-Laplacian . For a specified initial state (at ) and the network structure , the forward NDM analytically predicts the regional atrophy vector for all (see Materials and methods for details). The middle renderings in Fig. 1 compare the best NDM-predicted atrophy vector with the empirical atrophy vector (Parkinson’s disease group average). This step recapitulates previous NDM results for Parkinson’s disease, where all possible single-locus seed vectors were tested and ranked based on the Pearson’s correlation between and (Freeze et al., 2019; Pandya et al., 2019). The largest entries of are associated with the right Putamen (left and right Pallidum (left Precentral gyrus ( and left RN ( (see Table 2 for more details). We note that the top-10 largest entries of do not include the SN despite its association with the best single-locus seed vector.
Table 2.
Brain regions, hemispheres and relative pathology values associated with largest entries of average vectors
| Rank | Atrophy vector | Seed vector (group average) | Seed vector (S1) | Seed vector (S2) |
|---|---|---|---|---|
| 1 | Right Putamen, 0.15 | Left RN, 0.30 | Left RN, 0.49 | Right Putamen, 0.34 |
| 2 | Left Pallidum, 0.15 | Right Putamen, 0.25 | Right RN, 0.34 | Left Precentral Gyrus, 0.25 |
| 3 | Right Pallidum, 0.15 | Right Pallidum, 0.22 | Left Subthalamic Nucleus, 0.33 | Right Lingual Gyrus, 0.22 |
| 4 | Left Precentral Gyrus, 0.15 | Right SN, 0.21 | Left SN, 0.31 | Left PSGF Cortex, 0.22 |
| 5 | Left RN, 0.15 | Right RN, 0.20 | Right SN, 0.30 | Left Lingual Gyrus, 0.21 |
| 6 | Left Nucleus Accumbens, 0.14 | Left Subthalamic Nucleus, 0.20 | Left Pallidum, 0.26 | Left Inferior Frontal Gyrus, 0.20 |
| 7 | Left Putamen, 0.14 | Left SN, 0.19 | Right Subthalamic Nucleus, 0.26 | Right Cuneus, 0.19 |
| 8 | Right Thalamus, 0.14 | Left Pallidum, 0.18 | Right Thalamus, 0.20 | Left Superior Parietal Gyrus, 0.18 |
| 9 | Right Straight Gyrus, 0.14 | Left PSGF Cortex, 0.18 | Right Pallidum, 0.20 | Left Cuneus, 0.18 |
| 10 | Left Inferior Frontal Gyrus, 0.14 | Left Precentral Gyrus, 0.17 | Left Thalamus, 0.15 | Left SGF Cortex, 0.18 |
As previously stated, the set of all possible single-locus seed vectors is very limited and precludes more complex seed patterns with multiple loci and distinct entry values. We therefore apply a recent algorithm developed by our group (Torok et al., 2018) to infer the average multi-locus (ML) seed vector . See Fig. 1 rightmost plot for a brain rendering of the vector and the Materials and methods section for more details on the inference algorithm.
Table 2 shows that the largest entries of are associated with the left RN ( the right Putamen ( the right Pallidum ( and the right SN ( The main regions implicated in the multi-locus seed vector are sites where Parkinson’s disease pathology is observed (Braak et al., 2003; Huot and Parent, 2007; Hanganu et al., 2014; Lewis et al., 2016). Our result highlights a prominent role for the RN in the rostral midbrain as a potential source of the disease. This is consistent with the notion that Parkinson’s disease does not necessarily begin in the SN (Del Tredici et al., 2002; Braak et al., 2003; Lang and Obeso, 2004; Ahlskog, 2005) but could also be due to the difficulty of disambiguating fiber projections to and from SN versus those that terminate at the RN. Brain regions such as the Thalamus and the Nucleus accumbens were not among the top 10 seed locations for despite their high atrophy values. Instead, larger entries were found in the right RN and the left Pre-subgenual Frontal Cortex. See Supplementary Fig. 3 for additional brain renderings with axial and sagittal views.
S1 versus S2 subgroups for seed patterns
The outputs of our seed-inference algorithm applied to the Parkinson’s disease cohort led to incipient pathological patterns that are more heterogeneous than generally appreciated. We perform an unsupervised, hierarchical cluster analysis on the individual seed patterns (n = 232) to investigate the existence of distinct Parkinson’s disease subgroups. This technique provides not a single set of clusters, but instead, a multilevel hierarchy, where clusters at one level are joined as clusters at the next level. Figure 2 shows that without pre-specifying the number of clusters, the dendrogram splits into two large subgroups (S1 versus S2). The S1 subgroup (n = 65, in cyan) is characterized by a strong involvement of brainstem and ventral nuclei, while the S2 subgroup (n = 112, in black) is characterized by the involvement of the cortex and striatum. Table 2 shows the top 10 loci for S1 and S2. The largest entries of S1 include the left and right RN ( and , left Subthalamic Nucleus ( and left and right SN ( and . Noticeably, the right Putamen (second highest entry at the full entire group level) is not prevalent among patients in S1. Instead, it is prevalent in S2 (, along with left Precentral Gyrus (, right Lingual Gyrus ( and left Pre-Subgenual Frontal Cortex ( The brain renderings in Fig. 2 highlight the tremendous distinction between the spatial localization of these seed loci. Moreover, these regions exhibit distinct local transcriptional patterns of Parkinson’s disease-related genes, which we explore below.
Post hoc analysis
The S1 versus S2 clusters are unknown in the field as they are only apparent in the seeding patterns. We now perform a series of post hoc analyses to understand the likely genetic and aetiologic origins of these clusters, and to determine what effects, if any, the two clusters have on clinical and demographic variables in patients. The following results explore features not included in the clustering analysis. Supplementary Table 1 shows the results of student’s t-test on various clinical and demographic variables between S1 and S2 patients. None of the variables showed a significant difference save for sex ( and the age of onset ( for S1 patients, which is on average 4.5 years later than for S2 (see Fig. 2C). This is reflective of the fact that the measured atrophy patterns of the two groups is also not distinguishable (Supplementary Figs 3–5), suggesting that the patient’s atrophy profile become more similar to each other over time.
Genetic predictors of S1 versus S2 seed regions
We next test if patterns of regional genetic differences (Freeze et al., 2019) can predict the likelihood of subgroup-specific seeds (S1 versus S2). We map the regional transcript abundance of 67 Parkinson’s disease risk-factor genes (Chang et al., 2017) to the same brain atlas used in our analysis and compute the Pearson’s correlation coefficient between the average regional expression and the seed region likelihood. Figure 3A shows that the average regional expression across all Parkinson’s disease risk-factor genes predicts the seed region likelihood for the S1 group (), but not for the S2 (). The Parkinson’s disease risk-factor genes were subdivided into functional classes comprising immune-related, lysosomal, autophagy-related and mitochondrial genes. The first two classes predict the seed likelihoods for the S1 group (Fig. 3B, and Fig. 3C, ), but not for the S2 group (Fig. 3B, and Fig. 3C, ). In contrast, the expression of autophagy-related genes correlates positively with the seed region likelihood for the S2 group (Fig. 3E, ) but not for S1 (. The regional expression of mitochondrial genes does not correlate with the seed region likelihood for either group (Fig. 3F) (see Supplementary Table 2 for details).
Recent studies show that the LAG3 gene directly binds alpha-synuclein pre-formed fibrils in vitro, mediates Parkinson’s disease-like disease progression in animal models (Mao et al., 2016) and the regional expression pattern of which predict the human Parkinson’s disease atrophy pattern (Freeze et al., 2018). We performed a similar analysis for this specific gene in Fig. 3D and find that it predicts the regional seed likelihood for the S2 group (, but not for S1 . Together, these results suggest that the two patterns of inferred disease seeding may have distinct genetic and aetiologic bases.
Microglial abundance predicts S1 seed regions
Because the average region expression of immune-related genes predicts the seed region likelihood for the S1 group, we test whether the regional distribution of microglia does the same. Previous work showed that regional microglial abundance predicts the location for single-loci seeds in averaged Parkinson’s disease atrophy data (Freeze et al., 2019), and in this work, we test if this finding is group-specific. By employing high-fidelity gene expression mapping (Kelley et al., 2018), regional cell type can be derived from bulk transcriptomic data such as the Allen Brain Atlas expression data used here.
Regional microglial abundance, quantified as the first principal component of the top five high-fidelity microglial genes, is correlated with the seed region likelihood for the S1 group (Fig. 3G, but not for the S2 group (), consistent with a prominent role for immune-related pathology in S1. As a control, we also find a negative correlation between the regional neuronal abundance and seed region likelihood for the S1 group (Fig. 3G, , but no statistically significant relationship for S2 (.
S1 versus S2 differences in allelic status
We examined differences in the allelic status between the patients in the S1 and S2 clusters utilizing the template of 72 Parkinson’s disease-associated genotypic variants tabulated by the PPMI. Performing a chi-squared test to each variant yielded no statistically significant predictors after multiple hypothesis correction. Therefore, we conclude that the differences between patient Groups S1 and S2 cannot be explained by allelic status for genes that confer a risk of developing Parkinson’s disease, although a genetic basis for these two phenotypes cannot be fully ruled out.
Supplementary analyses
Supplementary Fig. 4A and B shows Parkinson’s disease subgroups for seeds and atrophy. We use MATLAB’s agglomerative hierarchical cluster tree function to categorize the seed and atrophy data into two subgroups each: S1 (65/232) versus S2 (167/232) for seeds and A1 (99/232) versus A2 (133/232) for atrophies. Notice that the majority of patients classified in S2 (100/167) are also classified in A2, but there is significant mixing between seed and atrophy subgroups. The histograms for age, MoCA scores and UPDRS3 scores exhibit significant overlap between the subgroups (see Supplementary Fig. 4C–E).
The categorization between S1 versus S2 patients does not hold at the atrophy level. Supplementary Fig. 5 shows the average predicted atrophy pattern (via NDM) of the seeds classified in S1 (cyan) and S2 (black) subgroups, i.e. and (termed AFS1 and AFS2, respectively). They are very hard to distinguish from each other (middle panels) by visual inspection and demonstrate that patient’s differences at the initiation stage of the disease diminish with progression, suggesting a process of convergence.
Discussion
In this work, we apply a novel seed-inference algorithm (Torok et al., 2018) to computationally determine the incipient pathology patterns of a Parkinson’s disease cohort (see Table 1). The method improves upon recent studies (Freeze et al., 2019; Pandya et al., 2019) by allowing the initial state of the disease to contain multiple loci instead of a single initiation site (Fig. 1, Backward NDM). The seed patterns, that were obtained indirectly from baseline MRI images, exhibit a surprising degree of intersubject variability. A cluster analysis based on the seed patterns alone reveals two very distinguished subgroups (S1 versus S2, see Fig. 2). The S1 subgroup is characterized by a high involvement of the brainstem and ventral nuclei, whereas the S2 subgroup has a strong involvement of the cortex and the striatum (see Table 2). Such clusters are only apparent in the seeding patterns and are unreported in the field. Moreover, differences between subgroups extend to other features that were not included in the clustering analysis, such as local transcriptional patterns of Parkinson’s disease-related genes and microglial abundance (Fig. 3). Together, these findings suggest two distinct early aetiological mechanisms operative in Parkinson’s disease.
Our inferred seeding patterns exhibit a high degree of variability across subjects, perhaps more than generally appreciated (Del Tredici and Braak, 2016). At the full group level (Fig. 1 and Table 2), our inverse NDM algorithm gave prominence to striatal structures that are highly affected in Parkinson’s disease such as Putamen and Nucleus Accumbens (Huot and Parent, 2007; Hanganu et al., 2014). Table 2 shows that the seeds do not merely mirror the observed atrophy patterns. Their potential relation with non-motor symptoms that appear in Parkinson’s disease, if any, merits further investigation (Shiba et al., 2000; Schuurman et al., 2002; Leentjens et al., 2003; Ross et al., 2006, 2008).
The seed patterns, when averaged across the entire Parkinson’s disease cohort, show highest seed values within the midbrain regions, in strong agreement with accepted structures involved in early stages (Braak et al., 2003). While our seeds do not contain the more caudal structures implicated in the earliest stages of the Braak scheme, there is disagreement about the extent to which detectable α-synucleopathy in those regions is necessary or sufficient for pre-determining rostral manifestation of Parkinson’s disease (Burke et al., 2008). In any case, since significant atrophy changes are known to occur in the amygdala (Harding et al., 2002) and in substructures of the basal ganglia and the SN (Davie, 2008), we can determine if our individual inferred seeds are more predictive than a single-locus seed placed at these locations.
Both the empirical atrophy and our inferred seeding patterns suggest a strong striatal involvement, but measured striatal Lewy pathology in Parkinson’s disease and dementia with Lewy bodies (DLB) is generally low (Braak et al., 2003; Jellinger and Attems, 2006). Emerging data suggest that striatal synuclein lesions are far more prevalent than previously thought (Saito et al., 2003; Braak et al., 2006; Jellinger and Attems, 2006; Mori et al., 2008). It is also possible that oligomeric or soluble synuclein might be more abundant that Lewy deposits in the striatum (Duda et al., 2002). Hence our results regarding the striatum are not surprising and might even highlight a potential role for inferring their early involvement from MRI alone.
As in Alzheimer’s disease (Torok et al., 2018), we demonstrate that the NDM predicts patient atrophy patterns using individual seeds better than a consensus seed placed at the SN or the amygdala (see Supplementary Fig. 2). In contrast to that study, however, we detected two distinct subpopulations of Parkinson’s disease patients based on their seed patterns alone (S1 versus S2). The hierarchical clustering analysis revealed, without pre-specifying the number of desired clusters, an interesting subgroup structure in the seeding patterns that were not obvious from their full group average (Figs 1 and 2 and Table 2).
The S1 subgroup is characterized by predominant involvement of the brainstem and ventral nuclei, with the RN and SN as their top seed locations (all values ≥ 0.30). The S2 subgroup is characterized by more widespread frontal and dorsal striatal cortices, with top seeds at the Putamen, Precentral Gyrus and Lingual Gyrus (all values ≥ 0.21). The most distinguishable signatures were the presence of the Thalamus for the S1 subgroup and the presence of the Lingual Gyrus for S2, a linked to letters processing, logical conditions and encoding visual memories. Finally, both left and right cuneus are among the top 10 seed regions for S2 but not for S1.
The S1 versus S2 distinctions might explain the two somewhat divergent sites implicit in Braak staging—one that involves brainstem/subcortical structures, and another, that involves predominantly cortical regions of the brain. A possible but by no means exclusive explanation of these effects may be that S1 sustains significant midbrain/BG damage due to dysfunction in the lysosomal processing of synuclein and microglia-mediated neuroinflammation. S2 is enriched for LAG3 and autophagy-related genes that may promote a different pattern of synuclein spreading in the putamen and cortical regions in the early disease state. Once synuclein reaches critical levels in SN, however, it damages the same subcortical circuits that cause the Parkinson’s disease hallmark motor symptoms. The widely distributed neocortical pattern in S2 is surprising as there is little evidence that synuclein pathology begins in the neocortex and spreads inwards. This suggests that for some patients, incipient atrophy patterns might be poorly conflated with canonical early histological patterns, which merits further exploration. However, note that the Putamen is the most prominent seed region in S2; its involvement in Parkinson’s disease is a well-established, since it is one of the first regions that suffers from a loss of dopaminergic striatal neurons. Therefore, it is possible that S2 represents an aetiologic process that begins in the Putamen and then spread quickly to the neocortex.
As noted earlier, S1 and S2 groups did not show appreciable differences in atrophy patterns or clinical scores. Interestingly, they also did not show any differences in their DaTScan striatal binding ratio, an established imaging marker of Parkinson’s disease diagnosis (see Supplementary Table 1). Hence, dopaminergic neuron loss in the striatum is similar in both groups despite marked difference in their age at onset. It is not possible to unravel the mediators of this observation in our study, but we might speculate the possibility that striatal dopaminergic loss followed by posterior-cortical cholinergic depletion—the so-called dual hypothesis—may play a role.
The identification of distinct Parkinson’s disease cohorts is not new, and previous study reported a younger disease-onset-subgroup, a tremor dominant subgroup, a non-tremor dominant subgroup and a subgroup with rapid disease progression (Szeto et al., 2015). Their cluster analysis, however, was based on numerous clinical and cognitive variables. Our study is the first to report distinct Parkinson’s disease phenotypes based solely on the initial sites of atrophy development for each patient. The differences between the subgroups, however, extended to several features not included in the categorization.
One of our most striking results is that microglial abundance is highly differential between the seeding-derived subgroups S1 versus S2 when using a healthy brain template. As in Alzheimer’s disease (Sala Frigerio et al., 2019), microglia in Parkinson’s disease may have both neurotoxic and neuroprotective effects, depending on their activation state. The list of protective effects includes the activation of multiple receptors, the release of anti-inflammatory cytokines, the initiation of neuron-microglia crosstalk, the regulation of microRNA and the modification of histone tails to suppress the expression of neurotoxic genes. Together, these actions work against inflammation-mediated injury (Le et al., 2016). It is remarkable that the first principal component of the top five high-fidelity microglial genes is correlated with the seed region likelihood for the S1 group but not for S2. While microglia may have both toxic and protective effects depending on their activation state (Joe et al., 2018), we hypothesize that in early Parkinson’s disease stages they might help suppress the neurotoxic effect of incipient synucleopathy, thus explaining why the age of onset for the S2 subgroup (regions where microglia are less abundant) is on average 4.5 years earlier than for the S1 subgroup (regions where microglia are far more abundant).
Limitations
It is challenging to measure atrophy and connectivity in the brainstem and midbrain accurately. Our 78-region parcellation, while widely used and reported, is quite large-grained, and likely encompasses substantial finer structures that are not revealed in our analysis. For instance, our imaging-derived atrophy patterns do not include the olfactory bulb, which has been reported on the basis of post-mortem neuropathology to be the most likely origination site of Parkinson’s disease. This important shortcoming is hard to address as the olfactory bulb in humans is much smaller relatively to mice (Desikan Atlas, Paxinos and Smith Brain Atlas).
Our work also inherits limitations from the forward NDM model when applied to atrophy in Parkinson’s disease (Pandya et al., 2019). The RN, for instance, plays a prominent role in the S1 subgroup, but was not reported by Braak et al. (2003) and cited as spared in later studies (Dickson, 2012). We believe that inclusion of RN as a seed region in S1 is erroneous due to tractography, and contrast and spatial resolution limitations of diffusion weighted MRI and T1-weighted MRI images. These spatial inaccuracies caused by inaccurate normalization of subjects to MNI template in atlas generation causes faulty localization of atrophy in small nuclei such as RN. Moreover, limitations are also imposed by the deformation-based morphometry methodology, which identifies iron accumulation in RN as volume changes due to T1-weighted MRI’s sensitivity to iron content. Given these limitations we had excluded RN from post hoc statistical analysis as elaborated in (Pandya et al., 2019). The dependence on atrophy as a marker of underlying pathology is clearly a limitation, and this issue was highlighted in detail in our previous work (Pandya et al., 2019). The relationship between atrophy and (synuclein) pathology in Parkinson’s disease is less well-established in comparison to Alzheimer’s, since some of the most atrophied regions are in the striatum, which does not contain high levels of Lewy deposits. One possible explanation is that atrophy could very well be trans-neuronal retrograde or anterograde from a brain region heavily affected by alpha-synuclein pathology, without much or any such pathology in the linked region. Despite these challenges, the overall success of NDM in prior work (Pandya et al., 2019) suggests that inverse seeding inference from atrophy in Parkinson’s disease is possible.
Finally, there is no ‘gold standard’ for seed-inference methods since the analyses are based on live patients at various stages of post-onset progression (Torok et al., 2018). Plausibility of results is based on (i) agreement with known pathology trends in early Parkinson’s disease; (ii) how well they are able to predict the atrophy pattern; and (iii) if they contain latent substructures that agree with other neuropathological markers. We believe that on all those counts, our results appear plausible.
Conclusion
In this work, we utilized a novel algorithm to infer the likely early sites of brain atrophy in a cohort of Parkinson patients. Images of incipient atrophy patterns are rarely available for Parkinson’s disease, and even if they were, in vivo techniques might be too coarse to properly detect them. Post-mortem studies, on another hand, typically find pathology patterns far away from their initial stages. Thus, our results provide key insights regarding the early stages of the disease. This study reveals that seed patterns are more heterogeneous than generally appreciated and can be categorized in two unreported broad subgroups (S1 versus S2). Parkinson’s disease risk-factor genes were subdivided into functional classes, which in turn, may predict the seed likelihood for each subgroup. It is clinically relevant to identify S2 patients as early as possible since their age-of-onset occurs on average 4.5 years before those in S1. Our findings highlight and compare the most prominently affected regions for both subgroups. Altogether, they may improve current screening strategies for early Parkinson’s disease onsetters.
Supplementary Material
Acknowledgments
The authors would like to thank C. Mezias and other members of the Raj Lab for their insightful comments. We also thank the Progressive Parkinson’s Markers Initiative (PPMI)—a public–private partnership—is funded by the Michael J. Fox Foundation for Parkinson’s Research and funding partners, including AbbVie, Avid, Biogen, Bristol-Myers Squibb, Covance, GE Healthcare, Genentech, GlaxoSmithKline, Lilly, Lundbeck, Merck, Meso Scale Discovery, Pfizer, Piramal, Roche, Sanofi Genzyme, Servier, Teva and UCB.
Funding
P.M. and A.R. were supported by the National Institutes of Health grant R01 EB022717. A.R., S.P., A.G. and J.T. were supported by the National Institutes of Health grant R01NS092802. B.F. was supported by Radiological Society of North America grant RR1813.
Competing interests
The authors report no competing interests.
Glossary
- HC
= healthy controls;
- NDM
= network diffusion model;
- PPMI
= Parkinson’s Progressive Marker Initiative;
- SN
= substantia nigra.
References
- Acosta D Powell F Zhao Y Raj A. Regional vulnerability in Alzheimer's: The role of cell-autonomous and transneuronal processes Alzheimers Dement 2018. pii: S1552-5260(17)33857-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahlskog JE. Challenging conventional wisdom: the etiologic role of dopamine oxidative stress in Parkinson’s disease. Mov Disord 2005; 20: 271–82. [DOI] [PubMed] [Google Scholar]
- Amunts K Lepage C Borgeat L Mohlberg H Dickscheid T Rousseau M-E, . et al. BigBrain: An ultrahigh-resolution 3D human brain model. Science 2013; 340: 1472–5. [DOI] [PubMed] [Google Scholar]
- Aubert-Broche B Fonov VS García-Lorenzo D Mouiha A Guizard N Coupé P, . et al. A new method for structural volume analysis of longitudinal brain MRI data and its application in studying the growth trajectories of anatomical brain structures in childhood. NeuroImage 2013; 82: 393–402. [DOI] [PubMed] [Google Scholar]
- Beach TG, Adler CH, Lue L, Sue LI, Bachalakuri J, Henry-Watson J, Arizona Parkinson’s Disease Consortium, et al. Unified staging system for Lewy body disorders: correlation with nigrostriatal degeneration, cognitive impairment and motor dysfunction. Acta Neuropathol 2009; 117: 613–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya S, Dunn P, Thomas CG, Smith B, Schaefer H, Chen J, et al. ImmPort, toward repurposing of open access immunological assay data for translational and clinical research. Sci Data 2018; 5: 180015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braak H, Bohl JR, Müller CM, Rüb U, de Vos RAI, Del Tredici K.. Stanley Fahn Lecture 2005: the staging procedure for the inclusion body pathology associated with sporadic Parkinson’s disease reconsidered. Mov Disord 2006; 21: 2042–51. [DOI] [PubMed] [Google Scholar]
- Braak H, Tredici KD, Rüb U, de Vos RAI, Jansen Steur ENH, Braak E.. Staging of brain pathology related to sporadic Parkinson’s disease. Neurobiol Aging 2003; 24: 197–211. [DOI] [PubMed] [Google Scholar]
- Burke RE, Dauer WT, Vonsattel JPG.. A critical evaluation of the Braak staging scheme for Parkinson’s disease. Ann Neurol 2008; 64: 485–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang D, Nalls MA, Hallgrímsdottir IB, Hunkapiller J, Van Der Brug M, Cai F, et al. International Parkinson's Disease Genomics Consortium. A meta-analysis of genome-wide association studies identifies 17 new Parkinson’s disease risk loci. Nat Genet 2017; 49: 1511–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coughlin DG, Petrovitch H, White LR, Noorigian J, Masaki KH, Ross GW, et al. Most cases with Lewy pathology in a population-based cohort adhere to the Braak progression pattern but ‘failure to fit’ is highly dependent on staging system applied. Parkinsonism Relat Disord 2019; 64: 124–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davie CA. A review of Parkinson’s disease. Br Med Bull 2008; 86: 109–27. [DOI] [PubMed] [Google Scholar]
- Del Tredici K, Braak H.. Review: sporadic Parkinson’s disease: development and distribution of α-synuclein pathology. Neuropathol Appl Neurobiol 2016; 42: 33–50. [DOI] [PubMed] [Google Scholar]
- Del Tredici K, Rüb U, de Vos RAI, Bohl JRE, Braak H.. Where does Parkinson disease pathology begin in the brain?. J Neuropathol Exp Neurol 2002; 61: 413–26. [DOI] [PubMed] [Google Scholar]
- Dickson DW. Parkinson’s disease and Parkinsonism: neuropathology. Cold Spring Harbor Perspect Med 2012; 2: a009258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duda JE, Giasson BI, Mabon ME, Lee VMY, Trojanowski JQ.. Novel antibodies to synuclein show abundant striatal pathology in Lewy body diseases. Ann Neurol 2002; 52: 205–10. [DOI] [PubMed] [Google Scholar]
- Duvernoy HM. The human brain stem and cerebellum. Vienna: Springer Vienna; 1995. [Google Scholar]
- Freeze B, Acosta D, Pandya S, Zhao Y, Raj A.. Regional expression of genes mediating trans-synaptic alpha-synuclein transfer predicts regional atrophy in Parkinson disease. Neuroimage Clin 2018; 18: 456–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeze B, Pandya S, Zeighami Y, Raj A.. Regional transcriptional pathogenesis architecture of Parkinson’s disease and network spread. Brain 2019; 142: 3072–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammers A Allom R Koepp MJ Free SL Myers R Lemieux L, . et al. Three-dimensional maximum probability atlas of the human brain, with particular reference to the temporal lobe. Hum Brain Mapp 2003; 19: 224–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanganu A, Bedetti C, Degroot C, Mejia-Constain B, Lafontaine A-L, Soland V, et al. Mild cognitive impairment is linked with faster rate of cortical thinning in patients with Parkinson’s disease longitudinally. Brain 2014; 137: 1120–9. [DOI] [PubMed] [Google Scholar]
- Harding AJ, Stimson E, Henderson JM, Halliday GM.. Clinical correlates of selective pathology in the amygdala of patients with Parkinson’s disease. Brain 2002; 125: 2431–45. [DOI] [PubMed] [Google Scholar]
- Hawrylycz MJ, Lein ES, Guillozet-Bongaarts AL, Shen EH, Ng L, Miller JA, et al. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature 2012; 489: 391–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huot P, Parent A.. Dopaminergic neurons intrinsic to the striatum. J Neurochem 2007; 101: 1441–7. [DOI] [PubMed] [Google Scholar]
- Iturria-Medina Y Canales-Rodríguez EJ Melie-García L Valdés-Hernández PA Martínez-Montes E Alemán-Gómez Y, . et al. Characterizing brain anatomical connections using diffusion weighted MRI and graph theory. NeuroImage 2007; 36: 645–60. [DOI] [PubMed] [Google Scholar]
- Jellinger KA. Formation and development of Lewy pathology: a critical update. J Neurol 2009; 256: 270–9. [DOI] [PubMed] [Google Scholar]
- Jellinger KA, Attems J.. Does striatal pathology distinguish Parkinson disease with dementia and dementia with Lewy bodies? Acta Neuropathol 2006; 112: 253–60. [DOI] [PubMed] [Google Scholar]
- Joe E-H, Choi D-J, An J, Eun J-H, Jou I, Park S.. Astrocytes, microglia, and Parkinson’s disease. Exp Neurobiol 2018; 27: 77–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley KW, Nakao-Inoue H, Molofsky AV, Oldham MC.. Variation among intact tissue samples reveals the core transcriptional features of human CNS cell classes. Nat Neurosci 2018; 21: 1171–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keuken MC Bazin P-L Crown L Hootsmans J Laufer A Müller-Axt C, . et al. Quantifying inter-individual anatomical variability in the subcortex using 7 T structural MRI. NeuroImage 2014; 94: 40–6. [DOI] [PubMed] [Google Scholar]
- Lang AE, Obeso JA.. Challenges in Parkinson’s disease: restoration of the nigrostriatal dopamine system is not enough. Lancet Neurol 2004; 3: 309–16. [DOI] [PubMed] [Google Scholar]
- Le W, Wu J, Tang Y.. Protective microglia and their regulation in Parkinson’s disease. Front Mol Neurosci 2016; 9: 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leentjens AFG, Van den Akker M, Metsemakers JFM, Lousberg R, Verhey FRJ.. Higher incidence of depression preceding the onset of Parkinson’s disease: a register study. Mov Disord 2003; 18: 414–8. [DOI] [PubMed] [Google Scholar]
- Leverenz JB, Hamilton R, Tsuang DW, Schantz A, Vavrek D, Larson EB, et al. Empiric refinement of the pathologic assessment of Lewy-related pathology in the dementia patient. Brain Pathol 2008; 18: 220–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis MM, Du G, Lee E-Y, Nasralah Z, Sterling NW, Zhang L, et al. The pattern of gray matter atrophy in Parkinson’s disease differs in cortical and subcortical regions. J Neurol 2016; 263: 68–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao X, Ou MT, Karuppagounder SS, Kam T-I, Yin X, Xiong Y, et al. Pathological α-synuclein transmission initiated by binding lymphocyte-activation gene 3. Science 2016; 353: aah3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori F, Tanji K, Zhang H, Kakita A, Takahashi H, Wakabayashi K.. α-Synuclein pathology in the neostriatum in Parkinson’s disease. Acta Neuropathol 2008; 115: 453–9. [DOI] [PubMed] [Google Scholar]
- Pandya S, Mezias C, Raj A.. Predictive model of spread of progressive supranuclear palsy using directional network diffusion. Front Neurol 2017; 8: 692. doi: 10.3389/fneur.2017.00692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandya S, Zeighami Y, Freeze B, Dadar M, Collins DL, Dagher A, et al. Predictive model of spread of Parkinson’s pathology using network diffusion. NeuroImage 2019; 192: 178–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, Kuceyeski A, Weiner M.. A network diffusion model of disease progression in dementia. Neuron 2012; 73: 1204–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, LoCastro E, Kuceyeski A, Tosun D, Relkin N, Weiner M.. Network diffusion model of progression predicts longitudinal patterns of atrophy and metabolism in Alzheimer’s disease. Cell Rep 2015; 10: 359–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, Powell F.. Models of network spread and network degeneration in brain disorders. Biol Psychiatry Cogn Neurosci Neuroimaging 2018; 3: P788–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross GW, Abbott RD, Petrovitch H, Tanner CM, Davis DG, Nelson J, et al. Association of olfactory dysfunction with incidental Lewy bodies. Mov Disord 2006; 21: 2062–7. [DOI] [PubMed] [Google Scholar]
- Ross GW, Petrovitch H, Abbott RD, Tanner CM, Popper J, Masaki K, et al. Association of olfactory dysfunction with risk for future Parkinson’s disease. Ann Neurol 2008; 63: 167–73. [DOI] [PubMed] [Google Scholar]
- Saito Y, Kawashima A, Ruberu NN, Fujiwara H, Koyama S, Sawabe M, et al. Accumulation of phosphorylated alpha-synuclein in aging human brain. J Neuropathol Exp Neurol 2003; 62: 644–54. [DOI] [PubMed] [Google Scholar]
- Sala Frigerio C, Wolfs L, Fattorelli N, Thrupp N, Voytyuk I, Schmidt I, et al. The major risk factors for Alzheimer’s disease: age, sex, and genes modulate the microglia response to Aβ plaques. Cell Rep 2019; 27: 1293–306.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuurman AG, van den Akker M, Ensinck KTJL, Metsemakers JFM, Knottnerus JA, Leentjens AFG, et al. Increased risk of Parkinson’s disease after depression: a retrospective cohort study. Neurology 2002; 58: 1501–4. [DOI] [PubMed] [Google Scholar]
- Shiba M, Bower JH, Maraganore DM, McDonnell SK, Peterson BJ, Ahlskog JE, et al. Anxiety disorders and depressive disorders preceding Parkinson’s disease: a case-control study. Mov Disord 2000; 15: 669–77. [DOI] [PubMed] [Google Scholar]
- Szeto JYY, O’Callaghan C, Shine JM, Walton CC, Mowszowski L, Naismith SL, et al. The relationships between mild cognitive impairment and phenotype in Parkinson’s disease. NPJ Parkinsons Dis 2015; 1: 15015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo JB, Gopal P, Raible K, Irwin DJ, Brettschneider J, Sedor S, et al. Pathological α-synuclein distribution in subjects with coincident Alzheimer’s and Lewy body pathology. Acta Neuropathol 2016; 131: 393–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torok J, Maia PD, Powell F, Pandya S, Raj A.. A method for inferring regional origins of neurodegeneration. Brain 2018; 141: 863–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yau Y, Zeighami Y, Baker TE, Larcher K, Vainik U, Dadar M, et al. Network connectivity determines cortical thinning in early Parkinson’s disease progression. Nat Commun 2018; 9: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeighami Y, Ulla M, Iturria-Medina Y, Dadar M, Zhang Y, Michel-Herve Larcher K, et al. Network structure of brain atrophy in de novo Parkinson’s disease. ELife 2015; 4: e08440. doi: 10.7554/eLife.08440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Gennatas ED, Kramer JH, Miller BL, Seeley WW.. Predicting regional neurodegeneration from the healthy brain functional connectome. Neuron 2012; 73: 1216–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.