Abstract
Administration of recombinant glial cell line-derived neurotrophic factor into the putamen has been tested in preclinical and clinical studies to evaluate its neuroprotective effects on the progressive dopaminergic neuronal degeneration that characterizes Parkinson’s disease. However, intracerebral glial cell line-derived neurotrophic factor infusion is a challenging therapeutic strategy, with numerous potential technical and medical limitations. Most of these limitations could be avoided if the production of endogenous glial cell line-derived neurotrophic factor could be increased. Glial cell line-derived neurotrophic factor is naturally produced in the striatum from where it exerts a trophic action on the nigrostriatal dopaminergic pathway. Most of striatal glial cell line-derived neurotrophic factor is synthesized by a subset of GABAergic interneurons characterized by the expression of parvalbumin. We sought to identify molecular targets specific to those neurons and which are putatively associated with glial cell line-derived neurotrophic factor synthesis. To this end, the transcriptomic differences between glial cell line-derived neurotrophic factor-positive parvalbumin neurons in the striatum and parvalbumin neurons located in the nearby cortex, which do not express glial cell line-derived neurotrophic factor, were analysed. Using mouse reporter models, we have defined the genomic signature of striatal parvalbumin interneurons obtained by fluorescence-activated cell sorting followed by microarray comparison. Short-listed genes were validated by additional histological and molecular analyses. These genes code for membrane receptors (Kit, Gpr83, Tacr1, Tacr3, Mc3r), cytosolic proteins (Pde3a, Crabp1, Rarres2, Moxd1) and a transcription factor (Lhx8). We also found the proto-oncogene cKit to be highly specific of parvalbumin interneurons in the non-human primate striatum, thus highlighting a conserved expression between species and suggesting that specific genes identified in mouse parvalbumin neurons could be putative targets in the human brain. Pharmacological stimulation of four G-protein-coupled receptors enriched in the striatal parvalbumin interneurons inhibited Gdnf expression presumably by decreasing cyclic adenosine monophosphate formation. Additional experiments with pharmacological modulators of adenylyl cyclase and protein kinase A indicated that this pathway is a relevant intracellular route to induce Gdnf gene activation. This preclinical study is an important step in the ongoing development of a specific pro-endo-glial cell line-derived neurotrophic factor pharmacological strategy to treat Parkinson’s disease.
Keywords: Parkinson’s disease, pro-endo-GDNF pharmacology, parvalbumin interneurons, striatum, gene expression
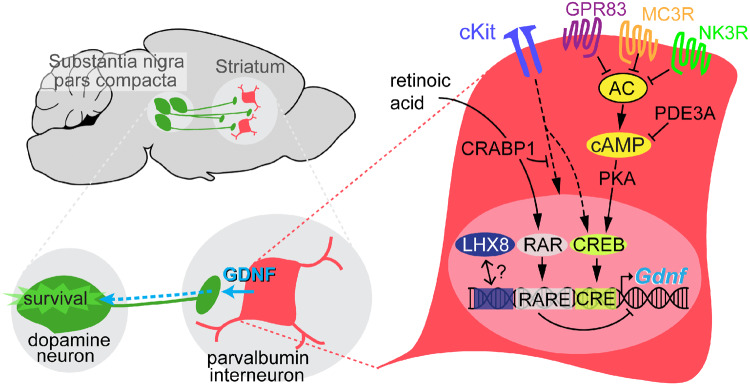
Stimulating endogenous production of glial-cell derived neurotrophic factor in the striatum is a promising therapeutic strategy to combat neuronal death in Parkinson's disease. This preclinical work provides new molecular targets and functional evidence supporting a selective pharmacological modulation of glial-cell derived neurotrophic factor by striatal parvalbumin interneurons.
Graphical Abstract
Graphical Abstract.
Introduction
Parkinson’s disease, the second most prevalent neurodegenerative disorder, is characterized by the progressive loss of mid-brain dopamine (DA) neurons (Surmeier et al., 2017). Currently, there are no therapies available to prevent damage to dopaminergic nigrostriatal neurons or promote regeneration of their axons, with existing treatments addressing symptom relief only (Pires et al., 2017).
Glial cell line-derived neurotrophic factor (GDNF) has long been known for its potent trophic action on mesencephalic DA neurons (Lin et al., 1993; Kordower and Bjorklund, 2013). GDNF binds to GDNF family receptor α1 (GFRα1), and the resulting complex activates the RET receptor tyrosine kinase and subsequent downstream signals. Both receptors are expressed by DA neurons and their selective deletion in mice is associated with progressive loss of the nigrostriatal pathway (Kramer and Liss, 2015). Numerous preclinical studies involving rodent and non-human primate models have established the efficacy of cell-derived or recombinant GDNF treatment in protecting and restoring substantia nigra pars compacta (SNpc) DA neurons (Sauer et al., 1995; Kirik et al., 2004; Gash et al., 2005; Muñoz-Manchado et al., 2013). The clinical effect of GDNF treatment delivered directly to the brain via intracerebral infusion devices has also been tested in Parkinson’s disease patients (d’Anglemont de Tassigny et al., 2015). Although safety has been shown for direct intraparenchymal injection of recombinant GDNF (Gill et al., 2003) or viral-mediated strategy (Heiss et al., 2019), clinical evidences have not fully demonstrated the efficacy of these approaches (Slevin et al., 2005; Lang et al., 2006). Recently, GDNF therapy in Parkinson’s disease has regained attention due to the publication of a novel, intermittent intraputamenal delivery strategy to optimize GDNF diffusion (Whone et al., 2019). Although the criteria for clinical improvement were not met after 40 weeks of GDNF delivery, this new approach resulted in significant progress on PET images. Indeed, there was a significant increase in 18F-DOPA putamenal uptake compared to the placebo group in patients that received GDNF (Whone et al., 2019). These data, suggesting dopaminergic sprouting in the putamen, pointed to a regenerative effect of GDNF, an outcome that had previously been observed only in a single-case report (Love et al., 2005).
The use of exogenous GDNF as restorative therapy for Parkinson’s disease is faced with serious limitations (e.g. major surgery, outpatient care or intraputamenal delivery), which highlight the importance of the further investigation to explore new options for GDNF-based neuroprotection. In this regard, new GDNF mimetic compounds, such as the brain penetrant RET receptor agonist BT13 (Mahato et al., 2020), are also being tested with a good prospect. Interestingly, GDNF is naturally produced in the adult mammalian brain, particularly in the striatum (ST) (Springer et al., 1994; Trupp et al., 1997; Mogi et al., 2001; Bäckman et al., 2006) and provides trophic signals to distantly located somas of catecholaminergic neurons by retrograde transport along the axon (Ito and Enomoto, 2016). For instance, noradrenergic neurons of the locus coeruleus depend on GDNF expressed in the septo-thalamic regions (Arenas et al., 1995; Zaman et al., 2003; Pascual et al., 2008; Enterría-Morales et al., 2020). Likewise, GDNF produced in the ST seems to promote the survival of DA neurons located in the SNpc by stimulating their broad striatal axonal network. Although the physiological effects of striatal GDNF on DA nigrostriatal neurons are a matter of debate (Kopra et al., 2015; Pascual and Lopez-Barneo, 2015; Enterría-Morales et al., 2020), it is well established that heterozygous GDNF+/− mice show accelerated impairment of the nigrostriatal pathway and decline of motor coordination with age (Boger et al., 2006). In addition, conditional deletion of the Gdnf gene in adult mice results in a progressive degeneration of the nigrostriatal dopaminergic pathway when GDNF levels are sufficiently low (Pascual et al., 2008; Enterría-Morales et al., 2020). Importantly, the main source of striatal GDNF has been identified as a subclass of GABAergic interneurons containing parvalbumin (PV), a calcium-binding protein (Hidalgo-Figueroa et al., 2012). PV neurons are uniformly distributed throughout the ST, therefore providing a homogenous trophic support to the profusely ramified axons of nigrostriatal DA neurons. Genetically, modified PV neurons producing twice the normal amount of GDNF protect DA neurons from neurotoxic insults (Kumar et al., 2015). Similarly, recent data obtained with SINEUPs, a new class of antisense long non-coding RNA targeting Gdnf mRNA, provided evidence that a 2-fold increase in GDNF is sufficient to significantly ameliorate motor deficits and prevent degeneration of DA neurons in a Parkinson’s disease neurochemical mouse model (Espinoza et al., 2020). These observations support the view that the pharmacological modulation of endogenous GDNF production in the ST, which is normally expressed at very low concentrations, could be a therapeutic approach to Parkinson’s disease complementary to other treatments. Identifying molecular targets specific to striatal PV interneurons is therefore a critical step to stimulate GDNF production.
Here, we report the differential molecular characteristics of GDNF-synthesizing striatal PV interneurons and their potential role in endogenous GDNF modulation. We performed a detailed transcriptomic analysis of two PV interneuron populations with the same embryological origin (Marín et al., 2000; Xu et al., 2004) and sharing several neurochemical and electrophysiological properties (Hu et al., 2014) but differing in their ability to express GDNF [GDNF negative in the cortex (CTX) and GDNF positive in the ST] (this work). The specific striatal PV interneuron transcriptomic profile, obtained by using fluorescence-activated cell sorting (FACS) followed by microarray comparison, was validated by additional histological and molecular analyses, with some of the studies in mice extended to striatal PV interneurons in non-human primates. Finally, we evaluated the impact on GDNF synthesis of pharmacological modulation of PV interneuron membrane receptors and the cyclic adenosine monophosphate (cAMP)-protein kinase A (PKA) intracellular pathway.
Materials and methods
Mice
Adult heterozygous GdnfLacZ mice were used for the detection of GDNF expression pattern (Sanchez et al., 1996; Hidalgo-Figueroa et al., 2012). The PV-tdTomato mice were obtained from The Jackson Laboratory (C57BL/6-Tg(Pvalb-tdTomato)15Gfng/J – JAX no. 027395), originally published in (Kaiser et al., 2016). The PV-Cre; tdTomato mice were obtained by crossing PV-Cre mice (B6; 129P2-Pvalbtm1(cre)Arbr/J - JAX Stock no. 008069) (Hippenmeyer et al., 2005) with tdTomato reporter mice (B6.Cg-Gt(ROSA)26Sortm27.1(CAGCOP4*H134R/tdTomato)Hze/J - JAX Stock no. 012567). All transgenic mice were bred in our animal facility and housed under a 12:12 h lighting schedule (lights on at 7:00 a.m.) with ad libitum access to food and water. The wild-type C57BL/6J mice for ex vivo experiments were purchased from Charles River France. Mice were sacrificed on postnatal Day 30 (P30) or at 2 months of age by thiobarbital overdose. All experiments were performed according to European Community guidelines (Council Directive 86/609/EEC) and were approved by the Ethics Committee of the ‘Virgen de la Macarena’ and “Virgen del Rocío’ hospitals (Seville, Spain) under project license no. 27-05-15-255 provided by the Government of Andalusia.
Monkeys
We analysed brain tissue sections stored in our brain bank and obtained from control monkeys (Macaca fascicularis) used in previous publications (Blesa et al., 2012; Jiménez-Sánchez et al., 2020; Monje et al., 2020). Non-human primate studies were performed according to European and Spanish guidelines (86/609/EEC and 2003/65/EC European Council Directives; and Spanish Government) and were approved by the research ethics committees of the universities of Navarra and Autónoma de Madrid.
Histology and immunohistochemistry procedures
For histological detection of GDNF expression in adult mouse coronal brain sections, we revealed β-galactosidase activity with 5-bromo-4-chloro-3-indolyl- β-D-galactopyranoside (XGal) staining. This step was followed by immunohistochemical detection of PV interneurons following the procedures detailed in the Supplementary material. To reveal PV-Cre activity and PV-tdTomato fluorescence in the PV neurons, we followed the procedures detailed in the Supplementary material (see also Enterría-Morales et al., 2020). Co-expression analyses of cKit and PV in the mouse or monkey brain were performed as indicated in the Supplementary material.
Tissue dissociation and cell preparation
Motor and somatosensory CTX or dorsal ST brain regions were dissociated into single-cell suspensions using a protocol modified from Zeisel et al. (2015). See Supplementary material for extended protocol.
Fluorescence-activated cell sorting
After dissociation of CTX and ST cells from PV-Cre; tdTomato or PV-tdTomato mice, tdTomato-positive cells were sorted using a FACS Jazz cell sorter (BD Biosciences). The tdTomato-positive cells were collected in RNAlater solution (Thermo Fisher Scientific) and immediately frozen at −80°C until RNA isolation. The yield obtained from two PV-Cre; tdTomato brains was ∼2000 cells/CTX and ∼250–1000 cells/ST. The yield from PV-tdTomato brains was ∼5000 cells/CTX and ∼2000 cells/ST. When necessary, cells from two or three sorting experiments were pooled to bring the total number to 2000 tdTomato-positive cells per replicate, which we estimated to be the minimum number required for microarray analysis (Supplementary Fig. 1C and D).
CTX and ST whole tissue preparation
As described in the ‘Tissue dissociation and cell preparation’ section in Supplementary material, mice were euthanized at P30 by thiobarbital overdose, and intracardially perfused with ice-cold oxygenated working solution (ICOWS). The brain was extracted rapidly, placed in O2-saturated ICOWS, and sectioned (thickness 300 µm) on a VT1200S vibratome (Leica). Next, brain slices were placed in oxygenated working solution at 37°C for 45 min and areas of interest (motor CTX; and dorsal ST) then dissected from each slice, snap frozen in liquid nitrogen, and stored at −80°C until used for RNA isolation.
RNA preparation, quality control and quantitation
Total RNA from cortical and striatal whole tissue samples and FACS-captured cells was isolated using the TRIzol method (Thermo Fisher) following the manufacturer’s instructions with minor modifications. Quantification and quality control were performed using a Nanodrop 2000 spectrophotometer (Thermo Fisher) for tissue RNA, or with an Agilent 2100 Bioanalyzer system for RNA obtained from FACS-captured PV cells. See Supplementary material for detailed protocol.
Microarray analysis
RNA from FACS-captured cells was amplified and labelled using Affymetrix GeneChip WT PLUS Reagent Kit (Thermo Fisher). Amplification was performed with 5 ng of total RNA as per procedures described in the WT PLUS Reagent Kit user manual. The amplified cDNA was quantified, fragmented and labelled in preparation for hybridization to an Affymetrix GeneChip Mouse Transcriptome 1.0 Array (Thermo Fisher) using 5.5 μg of single-stranded cDNA product and following protocols outlined in the user manual. Washing, staining (GeneChip Fluidics Station 450, Affymetrix) and scanning (GeneChip Scanner 3000, 10 Affymetrix) were performed as per protocols outlined in the user manual for cartridge arrays. Data were processed for gene-level background subtraction, normalization and signal summarization (SST-RMA, signal space transformation-robust multi-array average) using the BioConductor oligo package (R version 3.2.3) (Carvalho and Irizarry, 2010). To assess the overall similarity among samples, hierarchical clustering analysis of full expression profiles and principal component analysis were then performed. For hierarchical clustering analysis, the Pearson correlation metric and complete agglomeration methods were used. Differential expression analysis was performed using the R Limma (Linear Models for Microarray Analysis) package. This analysis applies a t-test empirically adjusted by a Bayes test (Huber et al., 2015). Changes in gene expression between groups were considered significant with false discovery rate (FDR) < 0.05 and fold change > 2 or < −2. Heatmap figures (hierarchical clustering) were obtained using the Transcriptome Analysis Console 3.0 Software (TAC 3.0) from Affymetrix.
Real-time quantitative RT-PCR
Real-time quantitative RT-PCR (qPCR) was run to validate the microarray results obtained from the FACS-captured PV-positive cells, as well as to compare transcriptomic differences between the PV cells and whole tissue from which they were isolated. See Supplementary Table 1 and Supplementary material for the full protocol.
RNAscope
We carried out in situ hybridization using the protocol from Advanced Cell Diagnostics (ACDbio), following the manufacturer’s instructions (RNAscope® Fluorescent Multiplex kit). This technique is used to visualize single RNA molecules in tissue samples on glass slides (Wang et al., 2012). See Supplementary material for extended protocol and analysis.
Ex vivo pharmacological experiments
We used 250 µm-thick coronal slices containing the ST (hemibrain) prepared from P30 male mice. The slices were kept in oxygenated artificial cerebrospinal fluid (aCSF) at 36°C and challenged with different pharmacological compounds for 5 or 12 h. At the end of incubation, striatal regions were dissected from each slice of the same hemibrain, placed in RNAlater and stored at −80°C until RNA extraction. Survival tests consisting of amperometric measurements of DA release in response to high potassium were performed to validate this ex vivo protocol. See Supplementary material for detailed procedures and pharmacological compounds used in this study.
In vivo stereotaxic experiments
Detailed procedures for administering stereotaxic injections into 2-month-old f male mice are available in the Supplementary material.
Statistical analysis
No statistical methods were used to predetermine sample size. For qPCR analyses, a minimum of two technical replicates were performed, with mean values used in calculations. Statistical analysis was performed using Prism software version 6.0 (GraphPad). Normality was tested with the Shapiro–Wilk test. We applied ordinary one-way analysis of variance with Tukey’s multiple comparison tests for a parametric set of data, or one-way ANOVA on ranks (Kruskal–Wallis) for non-parametric tests. A two-tailed Student’s t-test was performed for pairwise data. Data were presented as the mean ± SEM with the number (N) of experiments indicated in the figures. A value of P < 0.05 was considered statistically significant. Significance thresholds are indicated with asterisks in the figures as P < 0.05, P < 0.01, P < 0.001 and P < 0.0001, and standard errors of the mean are indicated by error bars. The statistical analysis specific to the microarray data is described in the ‘Microarray analysis’ section.
Data availability
The gene expression datasets generated in this study are available through NCBI Gene Expression Omnibus (GEO) with the series accession number (GEO: GSE100300). All other data generated or analysed during this study are included in this published article (and Supplementary material files).
Results
Differential GDNF expression in PV interneuron populations
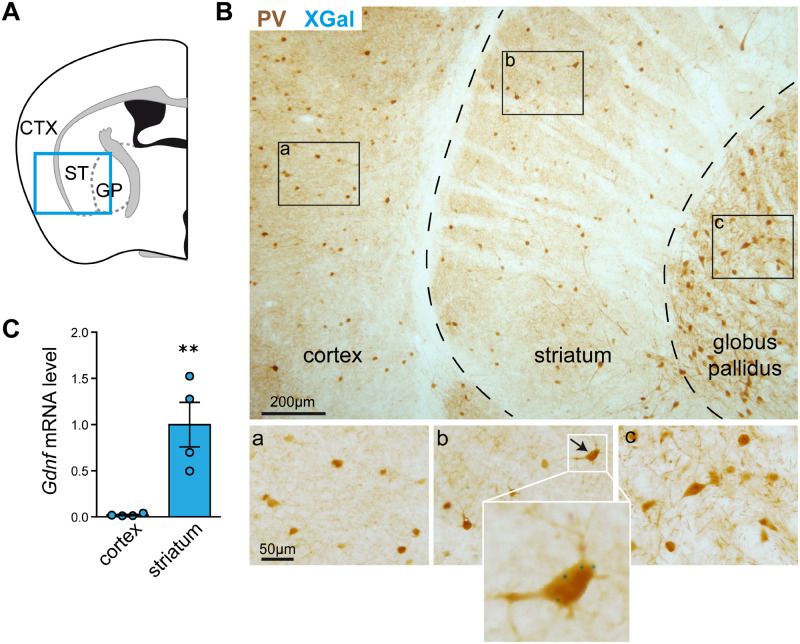
We studied neural GDNF expression by histological XGal staining of coronal brain sections from adult GdnfLacZ mice, which showed β-galactosidase activity resulting from the LacZ insert within exons 1 and 2 of the Gdnf gene. Dual XGal and PV immunohistochemical staining showed that, as reported previously (Hidalgo-Figueroa et al., 2012), GDNF is expressed in more than 95% of striatal PV-positive interneurons distributed uniformly over the ST (Fig. 1A and B). Remarkably, XGal staining, indicative of GDNF expression, was never seen in the PV neurons of the nearby somatosensory CTX or external globus pallidus (Fig. 1B). Moreover, the qPCR analysis confirmed the XGal data, as Gdnf mRNA expression levels were much higher in the ST than in the CTX of the mouse brain (P < 0.01, N = 4) (Fig. 1C). Indeed, Gdnf mRNA was barely amplified in cortical tissue. These data indicate that GDNF is selectively expressed by the striatal subclass of brain PV interneurons.
Figure 1.
Selective expression of GDNF in mouse striatal PV interneurons. (A) Schematic illustration of the CTX, ST and globus pallidus in a coronal section of the mouse brain. (B) Immunostaining of PV interneurons (brown) and selective expression of GDNF, reported by positive XGal-positive staining (blue dots), in the ST. (a), (b) and (c) are magnified fields showing details such as XGal blue dots only visible in (b). (C) Comparison of Gdnf mRNA expression (measured by qPCR) in the ST and CTX (N = 4, ** P < 0.01, two-tailed Student’s t-test).
Reporter mouse models and PV cell sorting
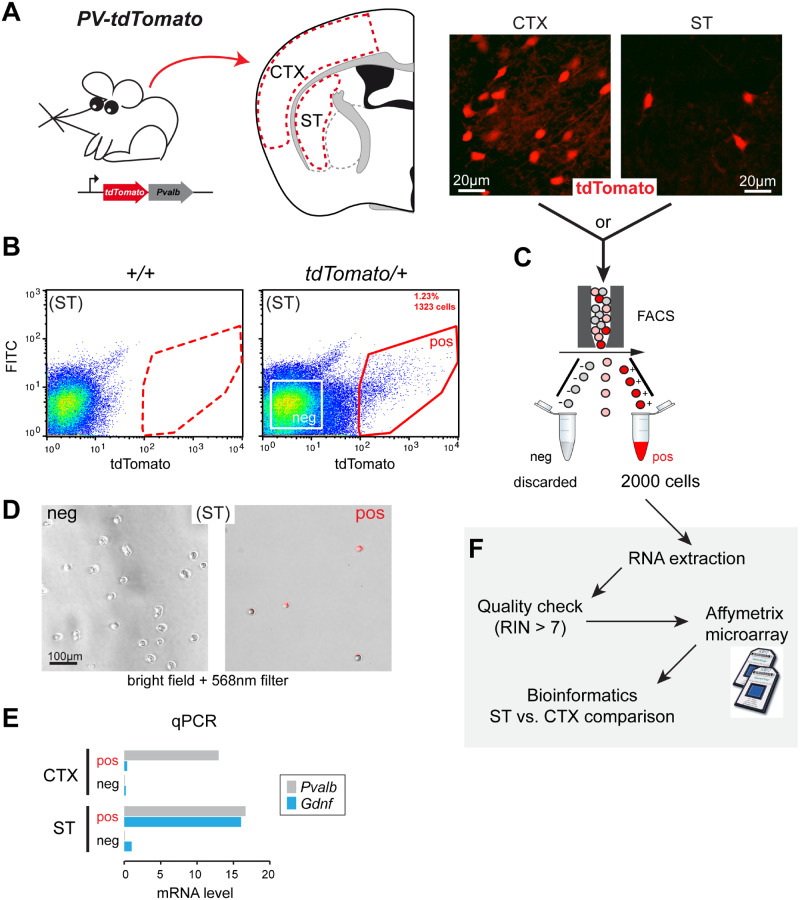
To capture PV-positive cells by FACS, we bred two mouse models expressing the red fluorescent protein variant tdTomato (Fig. 2A and Supplementary Fig. 1). We first used Cre-inducible tdTomato mice (PV-Cre; tdTomato), which harbour a loxP-flanked STOP cassette preventing the transcription of a CAG promoter-driven channelrhodopsin-2 (ChR2)/tdTomato fusion protein. Following exposure to Cre recombinase, the ChR2/tdTomato fusion protein is expressed with a predominant membrane location (Supplementary Fig. 2A). Only about half of PV interneurons are tdTomato-positive in the ST and CTX of this model due to a relatively low Cre-mediated recombination (Enterría-Morales et al., 2020). This resulted in few FACS-captured cells (see below) and, therefore, several sorting experiments were pooled to reach the critical threshold of 2000 cells required to guarantee good-quality RNA (Supplementary Fig. 1C and D).
Figure 2.
Experimental workflow for cell sorting from PV-tdTomato mice and microarray analysis. (A) Left, gene construction of PV-tdTomato mice and schematic illustration of CTX (CTX) and ST tissue samples obtained for cell dispersion. Right, histological examples of fluorescent cells (tdTomato-positive) in CTX and ST. (B) Flow cytometry dot plot graphical analysis of dispersed cells from tdTomato-negative (+/+) or positive (tdTomato/+) ST. The sorting gate (red line) was defined to separate the tdTomato-positive cells (pos) from other cells (see Supplementary Fig. 3 for extended plots). A negative gate was set to collect tdTomato-negative cells (white square). (C) 2000 tdTomato-positive cells (pos) were collected for experiment. (D) Phase contrast and fluorescence (568 nm filter) composite microphotographs of floating FACS-captured striatal cells. Left, negative gate-captured cells. Right, positive gate-captured cells. (E) Qualitative comparison of Gdnf and Pvalb mRNA expression (measured by qPCR) in tdTomato-positive and negative cells obtained from cortical and striatal cell preparations (N = 1). (F) Schematic diagram illustrating the experimental workflow after FACS capture of tdTomato-positive cells. Cells were processed for RNA extraction followed by quality check. Samples with an RNA integrity number above 7 were selected for the Affymetrix microarray procedure. Final data were analysed by bioinformatics to compare gene expression between ST and CTX parvalbumin interneurons.
The second mouse strain studied (transgenic PV-tdTomato) was free of Cre recombinase and constitutively expressed tdTomato in PV interneurons. Soluble tdTomato fluorescence was easily detected in 30 µm-thick brain cross-sections (Fig. 2A). Consistent with the original characterization of this model (Kaiser et al., 2016), we observed that most PV-immunopositive cells were tdTomato-positive in CTX and striatal regions (Supplementary Fig. 2B).
We set up a FACS protocol to specifically capture PV interneurons from striatal and cortical tissue preparations (Fig. 2B and C). In each model, dispersed cells from tdTomato-negative mice were used to detect non-specific fluorescence and to set up the gate to collect selectively tdTomato-positive cells. In a typical experiment, brain tissues were pooled from two PV-Cre; tdTomato mice and 1000–4000 (CTX) and 250–1000 (ST) tdTomato-positive cells were harvested, accounting for ∼3% and ∼0.5% of the total cell number, respectively. A typical harvest from two PV-tdTomato brains was 4000–10 000 cells from the CTX and 1000–2500 cells from the ST, representing ∼2.5% and ∼1% of the total cell number, respectively. Figure 2B shows representative FACS plots from PV; tdTomato striatal tissue; additional FACS plots for the two mouse models used are presented in Supplementary Fig. 3. To illustrate FACS selectivity, cells collected from the positive gate were red fluorescent under 568 nm excitation wavelength, whereas cells harvested from the negative gate were non-fluorescent (Fig. 2D). PV neuron enrichment was also confirmed by qPCR. Pvalb expression was high in captured tdTomato-positive cells from both the ST and CTX compared to cells harvested from the negative gate. In addition, only striatal tdTomato-positive cells exhibited high Gdnf mRNA levels (Fig. 2E). ST- and CTX-derived tdTomato-positive cells were then processed for RNA isolation and microarray analysis (Fig. 2F).
Transcriptomic differences between ST- and CTX-derived PV interneurons
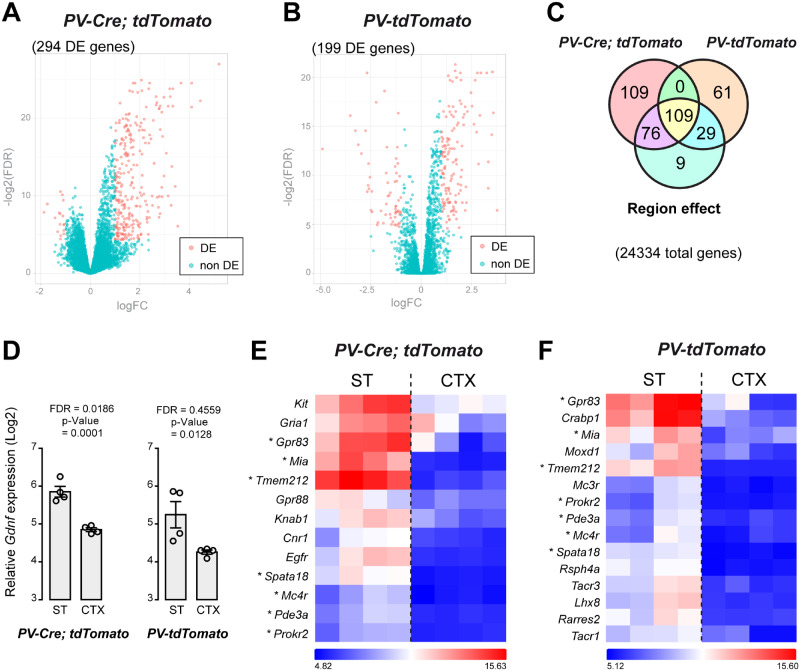
The expression profiles of 24 334 genes of FACS-captured PV neurons were compared between CTX and ST in two separate microarray experiments (corresponding to PV-Cre; tdTomato and PV-tdTomato mouse models). As illustrated by volcano plots (Fig. 3A and B), a number of differentially expressed (DE) genes between ST- and CTX-derived PV neurons were distributed in the top left (FDR < 0.05 and log2 FC < −1) and top right (FDR < 0.05 and log2 FC > 1) sections of the plots. In the PV-Cre; tdTomato model, 294 DE genes were identified (Fig. 3A and Supplementary Table 2). In contrast, 199 DE genes were identified in PV cells from PV-tdTomato mice (Fig. 3B and Supplementary Table 3). Two model-combined differential expression analyses (FDR < 0.05, log2 FC > 1 or < −1) yielded 185 DE genes specific to PV-Cre; tdTomato mice, and 90 DE genes specific to the PV-tdTomato-derived PV neurons. Finally, 109 DE genes overlapped in the two models (Fig. 3C). Although Gdnf was not DE in the initial microarray analysis, FDR and P-value indicate enrichment in striatal PV cells (Fig. 3D). Clustering of selected ST-specific DE genes showed that genes such as Gpr83, Mia, Tmem212, Spata18, Mc4r, Pde3a and Prokr2 are shared between the two reporter models (Fig. 3E and F). Other DE genes, such as Kit, were identified in PV-Cre; tdTomato mice but not in PV-tdTomato mice (Fig. 3E). On the other hand, Mc3r, Tacr1, Tacr3 and Lhx8 were DE only in striatal PV cells of PV-tdTomato mice (Fig. 3F).
Figure 3.
Microarray analysis of transcriptomes from cortical (CTX) and striatal (ST) PV interneurons. (A) Volcano plot illustrating DE genes [DE, magenta, logFC < −1 or > 1, −log2(FDR) < 0.05] compared to genes without differential expression (non-DE, green) from PV-Cre; tdTomato P30 mice. (B) Volcano plot illustrating DE genes from PV-tdTomato P30 mice. (C) Venn diagram showing ST/CTX gene expression variation present in the two set of arrays. (D) Detection of Gdnf gene transcripts (in log2 scale) in RNA samples from ST and CTX PV+ cells captured by FACS in the two reporter mouse models. (E) Hierarchical clustering of selected genes from PV-Cre; tdTomato CTX and ST (N = 4). The colour-ratio bar on the side indicates relative intensity (log2 scale) of gene up-regulation (magenta), down-regulation (black) and no change (white). (F) Hierarchical clustering of selected genes from PV-tdTomato CTX and ST. Asterisks indicate selected genes that were found in both arrays. logFC: log2-fold change, log2(FDR): log2 FDR.
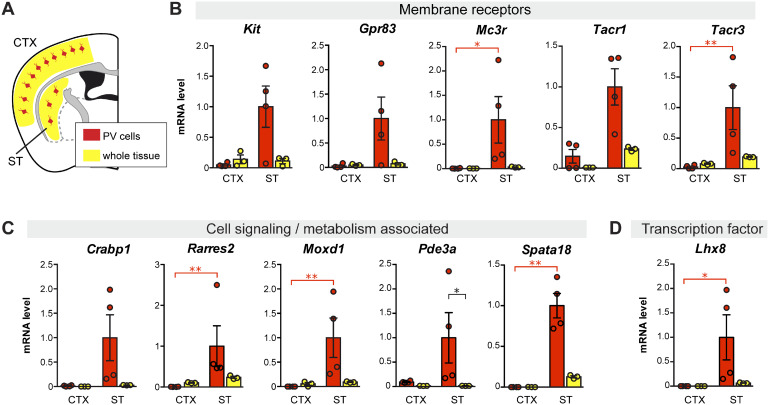
We next short-listed several DE genes obtained from the two models to carry out qPCR validation. This shortlist was based on positive ST/CTX fold change (i.e. higher expression in striatal PV interneurons) and biological interest (membrane receptors, cellular function and gene regulation). For this step, we included cDNA prepared from whole tissue samples (ST or CTX) in order to discard DE genes that although showed regional specificity was not highly expressed in PV neurons. Figure 4 shows qPCR data from eleven striatal PV-enriched genes (Kit, Gpr83, Mc3r, Tacr1, Tacr3, Crabp1, Rarres2, Moxd1, Pde3a, Spata18 and Lhx8). Other DE genes were discarded because exhibited either a low level of expression in PV neurons or high, non-PV-neuron-selective regional differences (ST versus CTX) (Supplementary Fig. 4). These genes were considered marginally selective and, therefore, less relevant for our study.
Figure 4.
Differential gene expression between parvalbumin interneurons from ST and CTX compared to tissue gene expression level in P30 mice. (A) Schematic illustration of parvalbumin interneurons (in red) within the whole CTX or ST region (yellow) in a coronal section of the mouse brain. Vertical bar graphs show cDNA level (measured by qPCR), as a measure of mRNA, in FACS-captured PV interneurons (red) and in whole tissue preparation (yellow) from ST (ST) and CTX (CTX). Gene expression for each sample was normalized to Actb mRNA. (B) Genes coding for membrane receptors. (C) Genes coding for cell signalling- and metabolism-associated proteins. (D) Gene coding for the transcription factor LHX8. N = 3 or 4, *P < 0.05, **P < 0.01, non-parametric one-way ANOVA on ranks (Kruskal–Wallis test).
Histological analysis of PV interneuron-specific gene expression
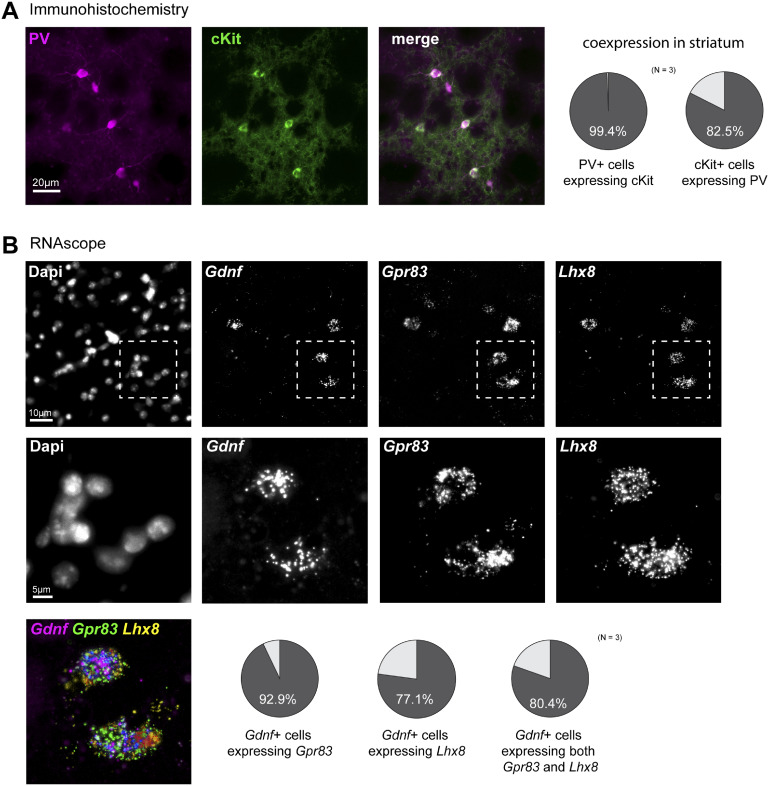
Based on microarray and qPCR data, we examined the anatomical distribution of selected PV neuron-specific genes in the mouse brain with particular attention paid to the ST. We first browsed through the Allen Mouse Brain Atlas in situ hybridization database for pattern similarity (Lein et al., 2007) and found a very similar distribution across the ST between Pvalb and some of our short-listed striatal PV neuron-specific genes (Crabp1, Lhx8, Pde3a, Tacr1, Tacr3) (Supplementary Fig. 5). We next searched for co-expression of selected genes in the mouse brain by histological methods. We chose the Kit gene for immunohistochemical analysis as highly specific antibodies for the cKit receptor are available (May et al., 2018) and determined that virtually all PV-immunopositive neurons were also cKit-positive in the mouse ST. On the other hand, only a small proportion of striatal cKit-positive cells were PV-negative (Fig. 5A;Supplementary Fig. 6A). Although cKit-positive cells were observed in the nearby somatosensory CTX, very few expressed both cKit and PV (Supplementary Fig. 6B). These histological results confirm the molecular data regarding Kit gene expression. In the absence of highly specific antibodies, we performed RNAscope®in situ hybridization analyses to examine Gpr83 and Lhx8 expression in the ST. Gpr83, Lhx8 and Gdnf mRNA were clearly co-labelled in the ST as illustrated in Fig. 5B (and Supplementary Fig. 7). These histological results support the molecular data described above.
Figure 5.
Selective expression of cKit (protein), Gpr83 and Lhx8 (mRNA) in Gdnf-positive mouse striatal parvalbumin interneurons. (A) Left, histological immunodetection of parvalbumin (PV, magenta) and cKit receptor (green) in coronal brain sections. Right, quantification of striatal PV-positive interneurons expressing cKit (circle plot on left), and cKit-positive cells expressing PV (circle plot on right) with co-expression percentages indicated (N = 3 adult mice). (B) In situ hybridization (by RNAscope method) analysis of Gdnf, Gpr83 and Lhx8 gene expression in the mouse ST. Greyscale confocal microphotographs show, from left to right: nuclei (stained by DAPI), specific probes for Gdnf, Gpr83 and Lhx8 mRNA. High magnification pictures of the fields delineated by dotted lines are displayed below the respective lower magnification images. Bottom left corner of the panel, composite image with merged labelling for Gdnf, Gpr83 and Lhx8. The circle plots indicate quantification of striatal Gdnf-positive cells expressing Gpr83 mRNA (left circle plot), Lhx8 mRNA (centre circle plot, and both Gpr83 and Lhx8 mRNAs (right circle plot) with co-expression percentages indicated (N = 3 adult mice). Additionally, 36 ± 4% of the Gpr83-positive cells were positive for Gdnf mRNA, and 86 ± 3% of Lhx8-positive cells expressed Gdnf mRNA in the ST (data not shown).
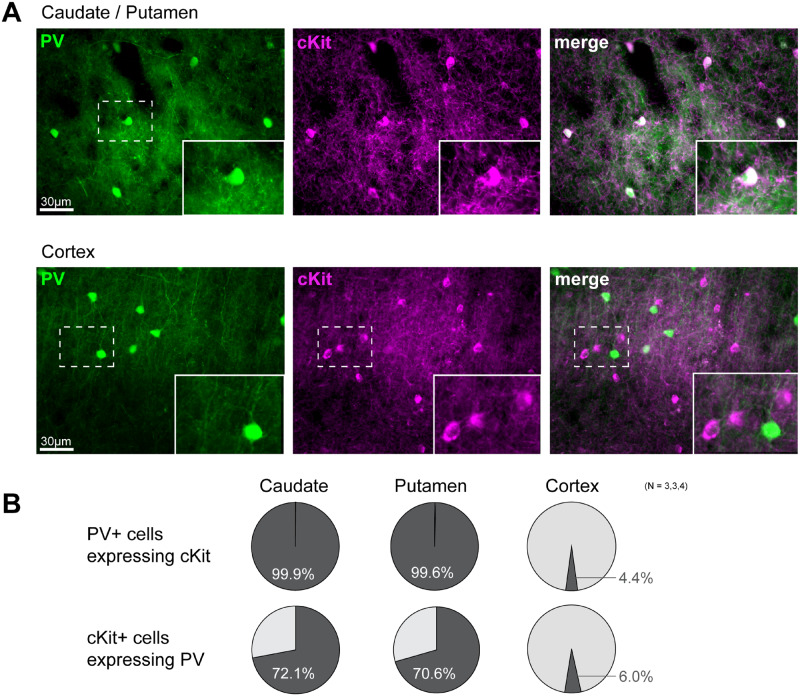
To evaluate whether the PV neuron-specific genes identified in the mouse ST might be conserved across species, we investigated the expression of cKit in the Macaca fascicularis, brain, a non-human primate model studied previously in our laboratories (Blesa et al., 2012; Jiménez-Sánchez et al., 2020; Monje et al., 2020). We stained the striatal PV-positive interneuron population and showed their resilience to 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine treatment (Supplementary Fig. 8). This observation in monkey brain supported our previous data obtained in the mouse brain showing that Gdnf-expressing striatal PV interneurons survive to 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine -induced DA depletion (Hidalgo-Figueroa et al., 2012). Dual labelling of the caudate nucleus and putamen showed a tight and selective co-expression of PV and cKit. Over 99% of PV-positive neurons appeared cKit-positive in the caudate nucleus and putamen of the Macaca fascicularis brain (Fig. 6). As previously observed in mice, cKit and PV were predominantly expressed by distinct cells in the nearby cortical region (only 4.4% of PV-positive neurons were cKit-positive). These results in the monkey are thus comparable to those obtained in the mouse brain.
Figure 6.
Selective expression of cKit in striatal parvalbumin interneurons of non-human primate brain. (A) Immunostaining of parvalbumin (PV) interneurons (green) and cKit receptor (magenta) in the putamen (top) and CTX (bottom) of the Macaca fascicularis brain. Insets show magnified images of fields demarcated by dotted lines. (B) Quantification analysis of PV-positive and cKit-positive co-labelling is presented as circle plots. Upper row, PV-positive cells expressing cKit in caudate (N = 3), putamen (N = 3) and CTX (N = 4). Lower row, cKit-positive cells expressing PV in caudate, putamen and CTX. Note the co-expression of PV and cKit in the putamen. In the CTX, PV-positive and cKit-positive cells are distinct neurons.
GDNF modulation by pharmacological activation of PV interneuron-specific G-protein-coupled receptors
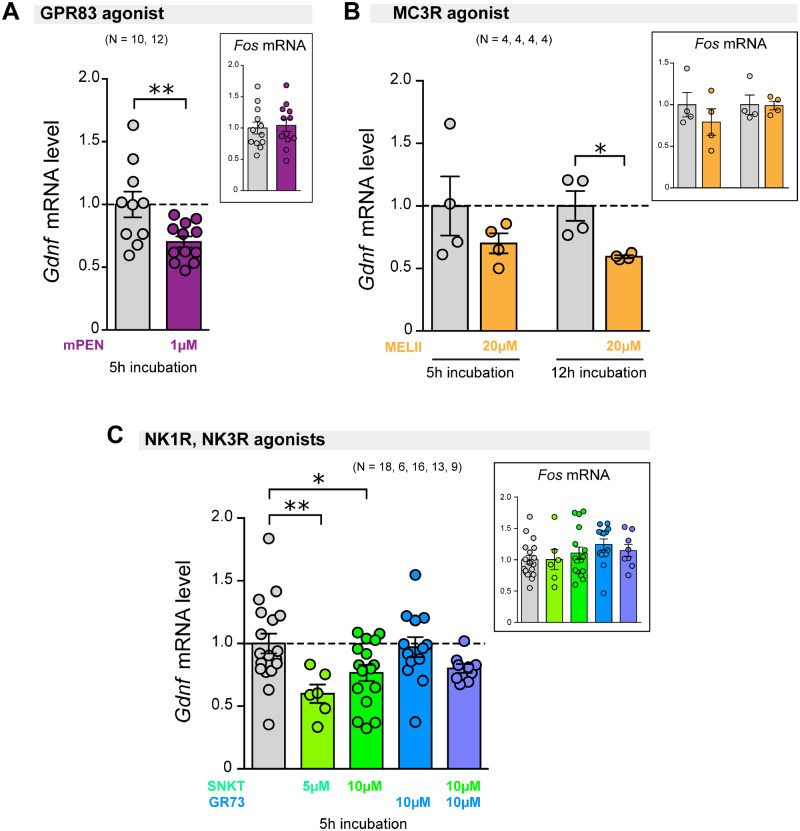
To assess whether stimulation of striatal PV interneuron-specific G-protein-coupled receptors (GPCRs) modulates GDNF expression, we tested ex vivo the effect of specific agonists of GPR83, MC3R, NK1R (from Tacr1) and NKR3 (from Tacr3). First, we showed that ex vivo slices remained viable after up to 12 h of incubation; this was done by measuring the striatal external Ca2+-dependent DA release in response to depolarization with high potassium (40 mM KCl) using an amperometric carbon-fibre electrode (Supplementary Fig. 9) (Mejías et al., 2006). The GPR83-specific agonist mPEN (Gomes et al., 2016) significantly reduced the level of Gdnf mRNA in the ST after 5 h of incubation (Fig. 7A). Likewise, incubation with the MC3R agonist Melanotan II significantly decreased Gdnf mRNA content after a long-lasting (12 h) incubation (Fig. 7B). Finally, stimulation of the tachykinin receptor NK3R, by 5 µM and 10 µM Senktide, proved to significantly inhibit Gdnf gene expression (Fig. 7C). However, stimulation of the neurokinin B receptor NK1R by its specific agonist GR73632 showed no statistically significant effect (Fig. 7C). In parallel to the actions on Gdnf modulation, we analysed the effects of the GPCR agonists on Fos gene expression, a proto-oncogene used as a marker of neuronal activity (Morgan et al., 1987; Didier et al., 2018), to rule out a non-selective pan-neuronal action of the agonists. The lack of changes in Fos expression (Fig. 7A–C) suggested that GPR83, MC3R and NK3R agonists act selectively on striatal PV interneurons to down-regulate Gdnf expression.
Figure 7.
Effect on Gdnf gene expression of pharmacological modulation of GPCRs expressed in striatal parvalbumin interneurons. Vertical bar graphs indicate Gdnf mRNA level (measured by qPCR with Hmbs gene as internal reference) in the ST from brain slices incubated for 5 or 12 h under ex vivo conditions. The level of Fos mRNA was used as a marker for pan-neuronal activation (measured by qPCR and normalized to Actb mRNA). (A) Stimulation of GPR83 by selective mPEN (1 µM) for 5 h. (B) Stimulation of Melanocortin receptor 3 (MC3R) by Melanotan II (20 µM) for 5 or 12 h. (C) Stimulation of tachykinin receptors 1 and 3 (NK1R, NK3R) for 5 h by their respective specific agonists GR73632 (GR73; 10 µM) and Senktide (10 µM). The number of experimental replicates (N) is indicated above the bar graphs. * P < 0.05, ** P < 0.01, two-tailed Student’s t-test for two sample differences, or one-way ANOVA with Tukey’s multiple comparison test were applied.
Pro-endo-GDNF effect of cyclic-AMP/PKA pathway stimulation
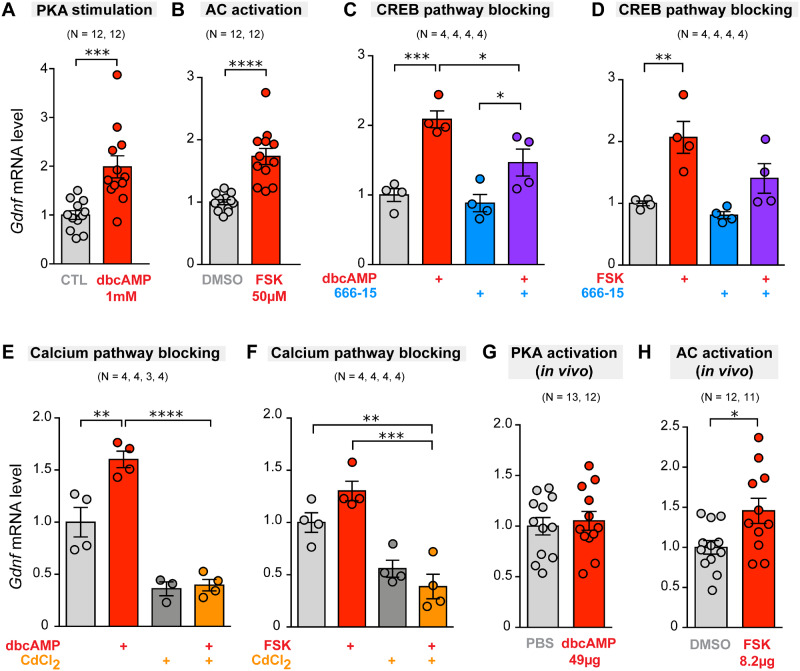
Gene ontology analysis performed on cells from PV-CRE; tdTomato and PV-tdTomato mice suggested the up-regulation of several signalling pathways in PV-positive neurons (see Supplementary Tables 4 and 5). Interestingly, gene ontology analysis of ST versus CTX gene DE from PV-tdTomato mice revealed a positive perturbation of the adenylyl cyclase (AC)/cyclic adenosine 3′,5′-monophosphate (cAMP) signalling pathway (Supplementary Table 5). Based on these data, we carried out ex vivo experiments to test the effect of pharmacological modulation of this pathway on Gdnf gene expression. We assayed the effect of dibutyryl-cAMP (dbcAMP), a cell-permeable cAMP analogue that activates cAMP-dependent protein kinases (PKA), and observed that incubation for 5 h with 1 mM dbcAMP was the optimal condition to obtain a consistent increase in striatal Gdnf mRNA levels (Fig. 8A and Supplementary Fig. 10). Similar results were obtained with direct activation of AC by forskolin (FSK, 50 µM) (Fig. 8B). The increase in Gdnf gene expression induced by dbcAMP or FSK was partially prevented by 666-15 (10 µM), a selective cAMP response element-binding protein (CREB) inhibitor (Fig. 8C and D). The effect of 666-15 was confirmed based on the expression of Ptgs2, a well-known CREB-sensitive gene coding for prostaglandin-endoperoxide synthase 2 (Xie et al., 2015) (Supplementary Fig. 11). Gdnf mRNA expression was also modulated by incubation of slices with cadmium chloride (200 µM), a blocker of voltage-gated Ca2+ channels (Ureña et al., 1994), suggesting that Ca2+ entry contributes to striatal GDNF homeostasis (Fig. 8E and F). In parallel with ex vivo experiments, we tested the effects of the intrastriatal administration of dbcAMP and FSK in anaesthetized mice. While dbcAMP had no effect on Gdnf mRNA levels (Fig. 8G), FSK evoked a significant 1.5-fold increase in Gdnf gene expression (Fig. 8H). As expected, generalized activation of the AC-cAMP-PKA pathway also induced the expression of Fos (Supplementary Fig. 10).
Figure 8.
Effect of modulation of the cyclic-AMP/protein kinase and calcium-dependent pathways on striatal Gdnf gene expression under ex vivo and in vivo conditions. Vertical bar graphs indicate Gdnf mRNA level (measured by qPCR with Hmbs or Actb genes as internal reference) in the ST after 5 h incubation under ex vivo conditions, or 4 h after stereotaxic intrastriatal administration (in vivo). (A) Ex vivo stimulation of PKA by dibutyryl-cAMP (dbcAMP; 1 mM) versus control (CTL). (B) Ex vivo stimulation of AC by FSK (50µM) versus control (DMSO vehicle) (C) Ex vivo stimulation of PKA by dbcAMP (1 mM) and effect of 666-15 (10 µM), a selective CREB inhibitor. (D) Ex vivo stimulation of AC by FSK 50µM and effect of 10 µM of 666-15. (E) Ex vivo stimulation of PKA by dbcAMP (1 mM) and effect of CdCl2 (200 µM), a blocker of voltage-gated calcium channel. (F) Ex vivo stimulation of AC by FSK (50µM) and effect of CdCl2 (200 µM). (G) In vivo stimulation of PKA with intrastriatal administration of 49 µg dbcAMP, with controls receiving PBS. (H) In vivo stimulation of AC with intrastriatal administration of 8.2 µg FSK, with controls receiving DMSO. The number of experimental replicates (N) is indicated above the bar graphs. * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001, two-tailed Student’s t-test or one-way ANOVA with Tukey’s multiple comparison test.
Discussion
The intrastriatal administration of GDNF is a potential disease-modifying therapeutic strategy for Parkinson’s disease that has been intensely studied in preclinical models as well as in patients though with variable results in the latter (Gill et al., 2003; Lang et al., 2006; Slevin et al., 2007). Recently, the design of a new protocol for GDNF delivery has raised hopes concerning its clinical efficacy (Whone et al., 2019). GDNF is produced in the mammalian brain, and adult central catecholaminergic neurons seem to rely on endogenous GDNF production for survival (Arenas et al., 1995; Pascual et al., 2008; Enterría-Morales et al., 2020). The main source of striatal GDNF in adult mice are GABAergic PV interneurons, which are densely innervated by nigrostriatal dopaminergic nerve endings that capture the GDNF necessary for the trophic maintenance of SNpc neurons (see Hidalgo-Figueroa et al., 2012 and references therein). Based on these early findings, the main goal of this work was to define a signature gene expression profile in striatal GABAergic PV interneurons, which could help to identify molecular targets for the pharmacological modulation of endogenous GDNF production.
A fundamental observation in our study was that, whereas more than 95% of mouse striatal PV neurons express GDNF, this property is completely absent in the neighbouring and more numerous neocortical (and globus pallidus) PV interneurons, despite the fact that all three neuronal classes share a similar embryological origin (Marin et al., 2000; Xu et al., 2004) and functional properties (Hu et al., 2014). This finding, which further suggests a neuroprotective role of striatal GDNF on mesencephalic dopaminergic neurons, also provides an optimal experimental model for separation of GDNF-producing (ST) and non-GDNF-producing (CTX) PV neuronal populations, thereby facilitating the study of their differential molecular properties. To this end, we performed a comparative transcriptomic analysis of tdTomato-labelled striatal versus cortical GABAergic PV cells using cDNA microarrays with posterior qPCR validation in two different transgenic mouse models. A point worth clarifying is that our analysis focused on the entire population of striatal PV interneurons, because all them express GDNF, although specific subclasses of PV striatal interneurons have been described based on their intrinsic electrical properties (Garas et al., 2016; Monteiro et al., 2018), functional inputs (Klug et al., 2018) or single-cell RNA sequencing (Muñoz-Manchado et al., 2018). Indeed, some of the PV genes reported in this last-mentioned study are coincident with the gene profile described here.
Within the several DE genes found in striatal versus cortical PV neurons, we defined 11 genes coding for potentially relevant proteins, such as membrane receptors or cell signalling and transcription factors, with high striatal PV interneuron specificity. Some differences observed in the highly expressed genes between the two reporter mouse models could be ascribed to a non-specific effect of Cre recombinase present only in the PV-Cre; tdTomato strain (Harno et al., 2013) or to a bias introduced by the distinct localization of the fluorescent signal (anchored to the plasma membrane in PV-Cre; tdTomato or free in the cytosol in PV-tdTomato) influencing the cell sorting output. However, these possibilities were ruled out given the selectivity of specific genes that was confirmed by immunocytochemistry and RNAscope in wild-type mice. The proto-oncogene Kit, which encodes a tyrosine kinase receptor, was found to be highly expressed in striatal PV interneurons in the PV-Cre; tdTomato model only. However, detailed immunohistochemical analyses in normal mice showed that this membrane-spanning receptor is actually expressed in almost 100% of striatal PV interneurons, both in mice and monkeys, and that this close association is not seen in other brain regions. In the same way, striatal Lhx8 mRNA expression, which is only seen in PV-tdTomato mice, was confirmed to be a robust property of Gdnf-positive PV interneurons of the adult normal mouse ST that is also regionally conserved in humans (Hawrylycz et al., 2012). LHX8 is a transcription factor broadly expressed in the developing mouse brain that functions as a cell fate regulator to median ganglionic eminence-derived cells (Zhao et al., 2003), which include cholinergic projection neurons and PV interneurons (Mayer et al., 2018). Embryonic deletion of the Lhx8 gene causes loss of the nucleus basalis of Meynert and a dramatic drop in the number of striatal cholinergic neurons, although PV interneurons remain present in adult Lhx8-null mice (Zhao et al., 2003).
Among the striatal PV signature genes, five code for GPCRs (GPR83, MC3R, NK1R and NK3R) and a phosphodiesterase (PDE3A), suggesting the participation of the cAMP/PKA/CREB pathway in GDNF expression. This is supported by the existence of a least three cAMP-responsive elements (CREs) in the GDNF promoter (Lamberti and Vicini, 2014) and the stimulatory effect of follicle-stimulating hormone, which acts on Gs-coupled receptors, on GDNF production by Sertoli cells (Reichert and Dattatreyamurty, 1989; Parekh et al., 2019). Indeed, we showed that direct pharmacological activation of AC or dbcAMP induced clear increases in striatal GDNF mRNA levels which were inhibited by the blockade of CREB. In contrast, stimulation of three of the four specific agonists of the GPCRs found in the PV interneurons (GPR83, MC3R and NK3R) resulted in a significant inhibition of Gdnf gene expression. It has been shown that GPR83 receptors signal via Gi (Mack et al., 2019) and that their activation with PEN (a physiological ligand) leads to a dose-dependent decrease in intracellular cAMP (Gomes et al., 2016). Therefore, it is possible that the endogenous activation of MC3R and NK3R receptors also decreases intracellular cAMP. In this manner, local neuropeptides of the tachykinin family, such as substance P, which is known to be released by striatal neurons (He et al., 2019), could provide basal inhibition of the cAMP/PKA/CREB pathway in PV cells that contributes to maintaining a low physiological production of GDNF.
Along with GPCRs, our study also suggests that the retinoic acid (RA) signalling pathway could also contribute to low basal striatal GDNF production because two RA-related genes (Crabp1 and Rarres2) appeared in the signature profile of PV neurons. CRABP1 (cellular retinoic acid-binding protein 1) is a cytoplasmic RA carrier that promotes the activation of specific RA nuclear receptors to eventually regulate the transcription of different sets of genes. However, CRABP1 is also a signalling molecule that mediates numerous rapid, non-genomic actions of RA, such as dampening growth factor sensitivity in stem cells, prevention of over-activation of calcium-calmodulin-dependent protein kinase II in heart cells, or modulation of the Raf-Ras-MEK-ERK signalling pathway (Nagpal and Wei, 2019; Park et al., 2019). Regarding the genomic actions of RA, a recent study on Sertoli cells showed that RA negatively regulates GDNF synthesis through binding of RARα to a new RA response element, DR5-RARE, on the Gdnf promoter (Saracino et al., 2020). Moreover, in some cell lines, RA induces transcriptional upregulation of retinoic acid receptor responder 2 (RARRES2), a poorly known protein that seems to act as a chemokine, adipokine or growth factor and has been associated with metabolic disorders (Helfer and Wu, 2018). However, it has been shown that RARRES2 promotes β-catenin phosphorylation and degradation (Liu-Chittenden et al., 2017). The interaction of RARRES2 with the Wnt/β-catenin canonical pathway could be of particular interest for understanding striatal GDNF modulation, as it is known that inhibition of glycogen synthetase kinase 3 prevents β-catenin degradation and leads to strong induction of GDNF in Sertoli cells (Tanwar et al., 2010). Interestingly, the Wnt/β-catenin pathway is required for embryonic differentiation and survival of mesencephalic dopaminergic neurons (Castelo-Branco et al., 2004; Joksimovic and Awatramani, 2014) and its dysregulation has been implicated in Parkinson’s disease pathogenesis (Zhang et al., 2016; Berwick et al., 2017). GDNF induction by β-catenin may also explain the functional role of Kit and Lhx8 genes that, as indicated above, are highly specific to striatal PV cells. It has been shown that cKit can directly phosphorylate tyrosine residues in β-catenin and increase its translocation to the nucleus and transcriptional activity (Kajiguchi et al., 2008; Jin et al., 2014). On the other hand, the Lhx8 promoter contains an enhancer with an evolutionarily conserved binding site for β-catenin (Landin Malt et al., 2014); transgenic LIM homeobox protein 8 (LHX8) over-expression increases β-catenin levels and its nuclear translocation (Zhou et al., 2015). Moreover, Lhx8−/− mice show a drastic down-regulation of Kit in neonatal ovaries (Choi et al., 2008). Therefore, it seems that LHX8 could induce cKit and activate the β-catenin pathway to further upregulate Lhx8 expression.
The last two specific striatal PV genes to discuss here are Spata18 and Moxd1. Spata18 codes for spermatogenesis-associated 18 homologue protein which is expressed in embryonic and adult testis (Bornstein et al., 2011). While the function of SPATA18 is poorly understood, it appears to be involved in the degradation of unhealthy mitochondria (Kitamura et al., 2011). Moxd1, which codes for DA beta hydroxylase-like monooxygenase protein, could contribute to the local inactivation of neurotransmitter in striatal dopaminergic synapses, although this function has not been demonstrated.
Taken together, our findings suggest that striatal GDNF homeostasis depends on a complex and delicate equilibrium between several signalling pathways. Strong inhibition of the cAMP/PKA/CREB pathway and the activation of RA signalling are probably responsible for a low basal level of GDNF synthesis and secretion by PV neurons. GDNF homeostasis is likely more complex and involves regulation at several non-transcriptomic levels (d’Anglemont de Tassigny et al., 2015). For instance, Gdnf mRNA degradation is highly regulated by microRNAs (Kumar et al., 2015; Xia et al., 2016; Zhang et al., 2019). However, a low basal level of expression is probably necessary to prevent over-activation of dopaminergic terminals and deleterious effects of GDNF. It is well known that local injection of GDNF produces structural aberrations in SNpc neurons (Kirik et al., 2000a, b); and that the intracerebroventricular injection of GDNF in humans induces a wide variety of side effects (Nutt et al., 2003). Overlapping with this tonic inhibition, selective activation of the cKit and Wnt/β-catenin pathways could result in a controlled production of GDNF as per physiological needs. In sum, the therapeutic potential of PV interneurons as a target is significant since they are the only cells to synthesize GDNF in the ST. These neurons remain functional, and continue producing GDNF after experimental destruction of nigrostriatal dopaminergic neurons in mice (Hidalgo-Figueroa et al., 2012) and monkeys (this work). We have shown that striatal GDNF production can be pharmacologically modulated (increased or decreased) and have also identified several pathways with druggable targets that are potentially relevant for controlling GDNF synthesis and/or release by striatal PV neurons. Development of a specific and safe pharmacology capable of modulating striatal GDNF production in man is a feasible goal that could eventually be of major therapeutic benefit for combating Parkinson’s disease.
Supplementary material
Supplementary material is available at Brain Communications online.
Supplementary Material
Acknowledgements
The authors wish to thank Dr. Patricia Ortega-Saenz for her help with the amperometric measurements, and Dr. Francisco Morón-Civanto, Mr. Juan Cordero-Varela and Ms. Maria José Castro-Pérez for their technical assistance. We also thank the IBiS animal facility staff for excellent animal care.
Funding
This research was supported by the Spanish Ministries of Science and Innovation and Health (SAF2012-39343, SAF2016-74990-R), the Carlos III Health Institute’s Miguel Servet Program (CPII17/00016, CP19/00200), the European Research Council (ERC-ADGPRJ201502629), the Fundación BBVA (Ayudas a Equipos de Investigación Científica 2015) and the Agence Nationale de la Recherche grants ANR-17-CE16-0015 and the LabEx DistAlz.
Competing interests
The authors report no competing interests. An international patent was submitted with registration number PCT/ES2018/070577 (authors: J.L.-B., X.D.T., D.E.-M. and I.L.-L.).
Glossary
- AC
adenylyl cyclase; AMP = adenosine monophosphate
- cAMP
cyclic adenosine monophosphate
- CRABP1
cellular retinoic acid-binding protein 1
- CRE
cAMP response element
- CREB
cAMP response element-binding protein
- CTX
cortex
- DA
dopamine
- DE
differentially expressed
- FACS
fluorescence-activated cell sorting; FC = fold-change
- FSK
forskolin
- GDNF
glial cell line-derived neurotrophic factor
- GO
gene ontology
- GPCR
G-protein-coupled receptor
- GPR83
G-protein-coupled receptor 83
- LHX8
LIM homeobox 8
- MC3R
melanocortin receptor 3
- NK1R
neurokinin receptor 1
- NK3R
neurokinin receptor 3
- PDE3A
phosphodiesterase 3A
- PKA
protein kinase A
- PV
parvalbumin
- RA
retinoic acid
- RARRES2
retinoic acid receptor responder 2; RET = rearranged during transfection
- SNpc
substantia nigra pars compacta
- SPATA18
spermatogenesis-associated 18
- ST
striatum
- XGal
5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside
References
- Arenas E, Trupp M, Akerud P, Ibáñez CF.. GDNF prevents degeneration and promotes the phenotype of brain noradrenergic neurons in vivo. Neuron 1995; 15: 1465–73. [DOI] [PubMed] [Google Scholar]
- Bäckman CM, Shan L, Zhang YJ, Hoffer BJ, Leonard S, Troncoso JC, et al. Gene expression patterns for GDNF and its receptors in the human putamen affected by Parkinson’s disease: a real-time PCR study. Mol Cell Endocrinol 2006; 252: 160–6. [DOI] [PubMed] [Google Scholar]
- Berwick DC Javaheri B Wetzel A Hopkinson M Nixon-Abell J Grannò S, . et al. Pathogenic LRRK2 variants are gain-of-function mutations that enhance LRRK2-mediated repression of β-catenin signaling. Mol Neurodegener 2017; 12: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blesa J, Pifl C, Sanchez-Gonzalez MA, Juri C, Garcia-Cabezas MA, Adanez R, et al. The nigrostriatal system in the presymptomatic and symptomatic stages in the MPTP monkey model: a PET, histological and biochemical study. Neurobiol Dis 2012; 48: 79–91. [DOI] [PubMed] [Google Scholar]
- Boger H, Middaugh L, Huang P, Zaman V, Smith A, Hoffer B, et al. A partial GDNF depletion leads to earlier age-related deterioration of motor function and tyrosine hydroxylase expression in the substantia nigra. Exp Neurol 2006; 202: 336–47. [DOI] [PubMed] [Google Scholar]
- Bornstein C Brosh R Molchadsky A Madar S Kogan-Sakin I Goldstein I, . et al. SPATA18, a spermatogenesis-associated gene, is a novel transcriptional target of p53 and p63. Mol Cell Biol 2011; 31: 1679–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho BS, Irizarry RA.. A framework for oligonucleotide microarray preprocessing. Bioinformatics 2010; 26: 2363–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelo-Branco G, Rawal N, Arenas E. GSK-3beta inhibition/beta-catenin stabilization in ventral midbrain precursors increases differentiation into dopamine neurons. J Cell Sci 2004; 117: 5731–7. [DOI] [PubMed] [Google Scholar]
- Choi Y, Ballow DJ, Xin Y, Rajkovic A. Lim homeobox gene, lhx8, is essential for mouse oocyte differentiation and survival. Biol Reprod 2008; 79: 442–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d’Anglemont de Tassigny X, Pascual A, López-Barneo J.. GDNF-based therapies, GDNF-producing interneurons, and trophic support of the dopaminergic nigrostriatal pathway. Implications for Parkinson’s disease. Front Neuroanat 2015; 9: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didier S, Sauve F, Domise M, Buee L, Marinangeli C, Vingtdeux V.. AMP-activated protein kinase controls immediate early genes expression following synaptic activation through the PKA/CREB pathway. Int J Mol Sci 2018; 19: 3716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enterría-Morales D, López-López I, López-Barneo J, d’Anglemont de Tassigny X.. Role of glial cell line-derived neurotrophic factor in the maintenance of adult mesencephalic catecholaminergic neurons. Mov Disord 2020; 1–12. [DOI] [PubMed] [Google Scholar]
- Espinoza S, Scarpato M, Damiani D, Managò F, Mereu M, Contestabile A, et al. SINEUP non-coding RNA targeting GDNF rescues motor deficits and neurodegeneration in a mouse model of Parkinson’s disease. Mol Ther 2020; 28: 642–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garas FN Shah RS Kormann E Doig NM Vinciati F Nakamura KC, . et al. Secretagogin expression delineates functionally-specialized populations of striatal parvalbumin-containing interneurons. Elife 2016; 5: e16088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gash DM, Zhang Z, Ai Y, Grondin R, Coffey R, Gerhardt G. A.. Trophic factor distribution predicts functional recovery in parkinsonian monkeys. Ann Neurol 2005; 58: 224–33. [DOI] [PubMed] [Google Scholar]
- Gill SS, Patel NK, Hotton GR, O'Sullivan K, McCarter R, Bunnage M, et al. Direct brain infusion of glial cell line-derived neurotrophic factor in Parkinson disease. Nat Med 2003; 9: 589–95. [DOI] [PubMed] [Google Scholar]
- Gomes I, Bobeck EN, Margolis EB, Gupta A, Sierra S, Fakira AK, et al. Identification of GPR83 as the receptor for the neuroendocrine peptide PEN. Sci Signal 2016; 9: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harno E, Cottrell EC, White A.. Metabolic pitfalls of CNS Cre-based technology. Cell Metab 2013; 18: 21–8. [DOI] [PubMed] [Google Scholar]
- Hawrylycz MJ Lein ES Guillozet-Bongaarts AL Shen EH Ng L Miller JA, . et al. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature 2012; 489: 391–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z, Liu TY, Yin YY, Song HF, Zhu XJ.. Substance P plays a critical role in synaptic transmission in striatal neurons. Biochem Biophys Res Commun 2019; 511: 369–73. [DOI] [PubMed] [Google Scholar]
- Heiss JD, Lungu C, Hammoud DA, Herscovitch P, Ehrlich DJ, Argersinger DP, et al. Trial of magnetic resonance-guided putaminal gene therapy for advanced Parkinson’s disease. Mov Disord 2019; 34: 1073–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfer G, Wu QF.. Chemerin: a multifaceted adipokine involved in metabolic disorders. J Endocrinol 2018; 238: R79–R94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidalgo-Figueroa M, Bonilla S, Gutiérrez F, Pascual A, López-Barneo J.. GDNF is predominantly expressed in the PV+ neostriatal interneuronal ensemble in normal mouse and after injury of the nigrostriatal pathway. J Neurosci 2012; 32: 864–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hippenmeyer S, Vrieseling E, Sigrist M, Portmann T, Laengle C, Ladle DR, et al. A developmental switch in the response of DRG neurons to ETS transcription factor signaling. PLoS Biol 2005; 3: e159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Gan J, Jonas P.. Interneurons. Fast-spiking, parvalbumin(+) GABAergic interneurons: from cellular design to microcircuit function. Science 2014; 345: 1255263. [DOI] [PubMed] [Google Scholar]
- Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS, et al. Orchestrating high-throughput genomic analysis with Bioconductor. Nat Methods 2015; 12: 115–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Enomoto H.. Retrograde transport of neurotrophic factor signaling: implications in neuronal development and pathogenesis. J Biochem 2016; 160: 77–85. [DOI] [PubMed] [Google Scholar]
- Jiménez-Sánchez L, Blesa J, Del Rey NL, Monje MHG, Obeso JA, Cavada C. Serotonergic innervation of the striatum in a nonhuman primate model of Parkinson's disease. Neuropharmacology 2020; 170: 107806. [DOI] [PubMed] [Google Scholar]
- Jin B, Ding K, Pan J. Ponatinib induces apoptosis in imatinib-resistant human mast cells by dephosphorylating mutant D816V KIT and silencing β-catenin signaling. Mol Cancer Ther 2014; 13: 1217–30. [DOI] [PubMed] [Google Scholar]
- Joksimovic M, Awatramani R. Wnt/β-catenin signaling in midbrain dopaminergic neuron specification and neurogenesis. J Mol Cell Biol 2014; 6: 27–33. [DOI] [PubMed] [Google Scholar]
- Kaiser T, Ting JT, Monteiro P, Feng G. Transgenic labeling of parvalbumin-expressing neurons with tdTomato. Neuroscience 2016; 321: 236–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajiguchi T, Lee S, Lee M-J, Trepel JB, Neckers L. KIT regulates tyrosine phosphorylation and nuclear localization of beta-catenin in mast cell leukemia. Leuk Res 2008; 32: 761–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirik D, Rosenblad C, Bjorklund A, Mandel RJ. Long-term rAAV-mediated gene transfer of GDNF in the rat Parkinson's model: intrastriatal but not intranigral transduction promotes functional regeneration in the lesioned nigrostriatal system. J Neurosci 2000. a; 20: 4686–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirik D, Rosenblad C, Björklund A. Preservation of a functional nigrostriatal dopamine pathway by GDNF in the intrastriatal 6-OHDA lesion model depends on the site of administration of the trophic factor. Eur J Neurosci 2000. b; 12: 3871–82. [DOI] [PubMed] [Google Scholar]
- Kirik D, Georgievska B, Björklund A. Localized striatal delivery of GDNF as a treatment for Parkinson disease. Nat Neurosci 2004; 7: 105–10. [DOI] [PubMed] [Google Scholar]
- Kitamura N Nakamura Y Miyamoto Y Miyamoto T Kabu K Yoshida M, . et al. Mieap, a p53-inducible protein, controls mitochondrial quality by repairing or eliminating unhealthy mitochondria. PLoS One 2011; 6: e16060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klug JR Engelhardt MD Cadman CN Li H Smith JB Ayala S, . et al. Differential inputs to striatal cholinergic and parvalbumin interneurons imply functional distinctions. Elife 2018; 7: e35657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopra J, Vilenius C, Grealish S, Härma M, Varendi K, Lindholm J, et al. GDNF is not required for catecholaminergic neuron survival in vivo. Nat Neurosci 2015; 18: 319–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kordower JH, Bjorklund A.. Trophic factor gene therapy for Parkinson’s disease. Mov Disord 2013; 28: 96–109. [DOI] [PubMed] [Google Scholar]
- Kramer ER, Liss B.. GDNF – ret signaling in midbrain dopaminergic neurons and its implication for Parkinson disease. FEBS Lett 2015; 589: 3760–3772. [DOI] [PubMed] [Google Scholar]
- Kumar A, Kopra J, Varendi K, Porokuokka LL, Panhelainen A, Kuure S, et al. GDNF overexpression from the native locus reveals its role in the nigrostriatal dopaminergic system function. PLOS Genet 2015; 11: e1005710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamberti D, Vicini E. Promoter analysis of the gene encoding GDNF in murine Sertoli cells. Mol Cell Endocrinol 2014; 394: 105–14. [DOI] [PubMed] [Google Scholar]
- Landin Malt A, Cesario JM, Tang Z, Brown S, Jeong J.. Identification of a face enhancer reveals direct regulation of LIM homeobox 8 (LHX8) by wingless-int (WNT)/beta-catenin signaling. J Biol Chem 2014; 289: 30289–30301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang AE, Gill S, Patel NK, Lozano A, Nutt JG, Penn R, et al. Randomized controlled trial of intraputamenal glial cell line-derived neurotrophic factor infusion in Parkinson disease. Ann Neurol 2006; 59: 459–66. [DOI] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature 2007; 445: 168–176. [DOI] [PubMed] [Google Scholar]
- Lin HF, Doherty DH, Lile JD, Bektesh S, Collins F.. GDNF: a glial cell line-derived neurotrophic factor for midbrain dopaminergic neurons. Science 1993; 260: 1130–1132. [DOI] [PubMed] [Google Scholar]
- Liu-Chittenden Y Jain M Gaskins K Wang S Merino MJ Kotian S, . et al. RARRES2 functions as a tumor suppressor by promoting β-catenin phosphorylation/degradation and inhibiting p38 phosphorylation in adrenocortical carcinoma. Oncogene 2017; 36: 3541–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love S, Plaha P, Patel N, Hotton G, Brooks D, Gill S.. Glial cell line-derived neurotrophic factor induces neuronal sprouting in human brain. Nat Med 2005; 11: 703–704. [DOI] [PubMed] [Google Scholar]
- Mack SM, Gomes I, Devi LA.. Neuropeptide PEN and its receptor GPR83: distribution, signaling, and regulation. ACS Chem Neurosci 2019; 10: 1884–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahato AK, Kopra J, Renko J-M, Visnapuu T, Korhonen I, Pulkkinen N, et al. Glial cell line-derived neurotrophic factor receptor rearranged during transfection agonist supports dopamine neurons in vitro and enhances dopamine release in vivo. Mov Disord 2020; 35: 245–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin O, Anderson SA, Rubenstein JL. Origin and molecular specification of striatal interneurons. J Neurosci 2000; 20: 6063–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May AJ, Cruz-Pacheco N, Emmerson E, Gaylord EA, Seidel K, Nathan S, et al. Diverse progenitor cells preserve salivary gland ductalarchitecture after radiation-induced damage. Development 2018; 145: dev166363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer C, Hafemeister C, Bandler RC, Machold R, Batista Brito R, Jaglin X, et al. Developmental diversification of cortical inhibitory interneurons. Nature 2018; 555: 457–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mejías R, Villadiego J, Pintado CO, Vime PJ, Gao L, Toledo-Aral JJ, et al. Neuroprotection by transgenic expression of glucose-6-phosphate dehydrogenase in dopaminergic nigrostriatal neurons of mice. J Neurosci 2006; 26: 4500–4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogi M, Togari A, Kondo T, Mizuno Y, Kogure O, Kuno S, et al. Glial cell line-derived neurotrophic factor in the substantia nigra from control and parkinsonian brains. Neurosci Lett 2001; 300: 179–81. [DOI] [PubMed] [Google Scholar]
- Monje MHG, Blesa J, García-Cabezas MÁ, Obeso JA, Cavada C. Changes in thalamic dopamine innervation in a progressive Parkinson's disease model in monkeys.Mov Disord 2020; 35: 419–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro P Barak B Zhou Y Mcrae R Rodrigues D Wickersham IR, . et al. Dichotomous parvalbumin interneuron populations in dorsolateral and dorsomedial striatum. J Physiol 2018; 596: 3695–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan JI, Cohen DR, Hempstead JL, Curran T.. Mapping patterns of c-fos expression in the central nervous system after seizure. Science 1987; 237: 192–197. [DOI] [PubMed] [Google Scholar]
- Muñoz-Manchado AB Bengtsson Gonzales C Zeisel A Munguba H Bekkouche B Skene NG, . et al. Diversity of Interneurons in the Dorsal Striatum Revealed by Single-Cell RNA Sequencing and PatchSeq. Cell Rep 2018; 24: 2179–90.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Manchado AB, Villadiego J, Suárez-Luna N, Bermejo-Navas A, Garrido-Gil P, Labandeira-García JL, et al. Neuroprotective and reparative effects of carotid body grafts in a chronic MPTP model of Parkinson’s disease. Neurobiol Aging 2013; 34: 902–15. [DOI] [PubMed] [Google Scholar]
- Nagpal I, Wei L-N. All-trans retinoic acid as a versatile cytosolic signal modulator mediated by CRABP1. Int J Mol Sci 2019; 20: 3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutt JG Burchiel KJ Comella CL Jankovic J Lang AE Laws ER, . et al. Randomized, double-blind trial of glial cell line-derived neurotrophic factor (GDNF) in PD. Neurology 2003; 60: 69–73. [DOI] [PubMed] [Google Scholar]
- Parekh PA, Garcia TX, Hofmann M-C. Regulation of GDNF expression in Sertoli cells. Reproduction 2019; 157: R95–R107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SW, Nhieu J, Persaud SD, Miller MC, Xia Y, Lin Y-W, et al. A new regulatory mechanism for Raf kinase activation, retinoic acid-bound Crabp1. Sci Rep 2019; 9: 10929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual A, Hidalgo-Figueroa M, Piruat JI, Pintado CO, Gómez-Díaz R, López-Barneo J.. Absolute requirement of GDNF for adult catecholaminergic neuron survival. Nat Neurosci 2008; 11: 755–61. [DOI] [PubMed] [Google Scholar]
- Pascual A, Lopez-Barneo J.. Reply to GDNF is not required for catecholaminergic neuron survival in vivo. Nat Neurosci 2015; 18: 322–323. [DOI] [PubMed] [Google Scholar]
- Pires AO, Teixeira FG, Mendes-Pinheiro B, Serra SC, Sousa N, Salgado AJ. Old and new challenges in Parkinson's disease therapeutics. Prog Neurobiol 2017; 156: 69–89. [DOI] [PubMed] [Google Scholar]
- Reichert LE, Dattatreyamurty B. The follicle-stimulating hormone (FSH) receptor in testis: interaction with FSH, mechanism of signal transduction, and properties of the purified receptor. Biol Reprod 1989; 40: 13–26. [DOI] [PubMed] [Google Scholar]
- Sanchez M, Silos-Santiago I, Frisen J, He B, Lira S, Barbacid M.. Renal agenesis and the absence of enteric neurons in mice lacking GDNF. Nature 1996; 382: 70–73. [DOI] [PubMed] [Google Scholar]
- Saracino R, Capponi C, Di S, Carla P, Masciarelli S, Fazi F, et al. Regulation of GDNF expression by retinoic acid in Sertoli cells. Mol Reprod Dev 2020; 87: 419–11. [DOI] [PubMed] [Google Scholar]
- Sauer H, Rosenblad C, Bjorklund A.. Glial cell line-derived neurotrophic factor but not transforming growth factor beta 3 prevents delayed degeneration of nigral dopaminergic neurons following striatal 6-hydroxydopamine lesion. Proc Natl Acad Sci USA 1995; 92: 8935–8939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slevin JT Gash DM Smith CD Gerhardt GA Kryscio R Chebrolu H, . et al. Unilateral intraputamenal glial cell line-derived neurotrophic factor in patients with Parkinson disease: response to 1 year of treatment and 1 year of withdrawal. J Neurosurg 2007; 106: 614–20. [DOI] [PubMed] [Google Scholar]
- Slevin JT, Gerhardt G. A, Smith CD, Gash DM, Kryscio R, Young B.. Improvement of bilateral motor functions in patients with Parkinson disease through the unilateral intraputaminal infusion of glial cell line-derived neurotrophic factor. J Neurosurg 2005; 102: 216–22. [DOI] [PubMed] [Google Scholar]
- Springer J, Mu X, Bergmann L, Trojanowski J.. Expression of GDNF mRNA in rat and human nervous tissue. Exp Neurol 1994; 127: 167–170. [DOI] [PubMed] [Google Scholar]
- Surmeier JD, Obeso JA, Halliday GM.. Selective neuronal vulnerability in Parkinson disease. Nat Rev Neurosci 2017; 18: 101–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanwar PS, Kaneko-Tarui T, Zhang L, Rani P, Taketo MM, Teixeira J. Constitutive WNT/beta-catenin signaling in murine Sertoli cells disrupts their differentiation and ability to support spermatogenesis. Biol Reprod 2010; 82: 422–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trupp M, Belluardo N, Funakoshi H, Ibáñez CF.. Complementary and overlapping expression of glial cell line-derived neurotrophic factor (GDNF), c-ret proto-oncogene, and GDNF receptor-alpha indicates multiple mechanisms of trophic actions in the adult rat CNS. J Neurosci 1997; 17: 3554–3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ureña J, Fernández-Chacón R, Benot AR, Alvarez De Toledo G, López-Barneo J.. Hypoxia induces voltage-dependent Ca2+ entry and quantal dopamine secretion in carotid body glomus cells. Proc Natl Acad Sci USA 1994; 91: 10208–10211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Flanagan J, Su N, Wang LC, Bui S, Nielson A, et al. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diag 2012; 14: 22–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whone A, Luz M, Boca M, Woolley M, Mooney L, Dharia S, et al. Randomized trial of intermittent intraputamenal glial cell line-derived neurotrophic factor in Parkinson’s disease. Brain 2019; 142: 512–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Z-Q, Ding D-K, Zhang N, Wang J-X, Yang H-Y, Zhang D.. MicroRNA-211 causes ganglion cell dysplasia in congenital intestinal atresia via down-regulation of glial-derived neurotrophic factor. Neurogastroenterol Motil 2016; 28: 186–195. [DOI] [PubMed] [Google Scholar]
- Xie F, Li BX, Kassenbrock A, Xue C, Wang X, Qian DZ, et al. Identification of a potent inhibitor of CREB-mediated gene transcription with efficacious in vivo anticancer activity. J Med Chem 2015; 58: 5075–5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Cobos I, De La Cruz ED, Rubenstein JL, Anderson SA.. Origins of cortical interneuron subtypes. J Neurosci 2004; 24: 2612–2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaman V, Li Z, Middaugh L, Ramamoorthy S, Rohrer B, Nelson ME, et al. The noradrenergic system of aged GDNF heterozygous mice. Cell Transplant 2003; 12: 291–303. [PubMed] [Google Scholar]
- Zeisel A, Muñoz-Manchado AB, Codeluppi S, Lönnerberg P, La Manno G, Juréus A, et al. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science (80-) 2015; 347: 1138–1142. [DOI] [PubMed] [Google Scholar]
- Zhang L Deng J Pan Q Zhan Y Fan J-B Zhang K, . et al. Targeted methylation sequencing reveals dysregulated Wnt signaling in Parkinson disease. J Genet Genomics 2016; 43: 587–92. [DOI] [PubMed] [Google Scholar]
- Zhang H, Chang Y, Zhang L, Kim S-N, Otaegi G, Zhang Z, et al. Upregulation of microRNA miR-9 is associated with microcephaly and Zika virus infection in mice. Mol Neurobiol 2019; 56: 4072–4085. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Marin O, Hermesz E, Powell A, Flames N, Palkovits M, et al. The LIM-homeobox gene Lhx8 is required for the development of many cholinergic neurons in the mouse forebrain. Proc Natl Acad Sci USA 2003; 100: 9005–9010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C Yang G Chen M Wang C He L Xiang L, . et al. Lhx8 mediated Wnt and TGFβ pathways in tooth development and regeneration. Biomaterials 2015; 63: 35–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The gene expression datasets generated in this study are available through NCBI Gene Expression Omnibus (GEO) with the series accession number (GEO: GSE100300). All other data generated or analysed during this study are included in this published article (and Supplementary material files).