Abstract
We have previously shown that non-thermal dielectric-barrier discharge (DBD) plasma induces the generation of reactive oxygen species (ROS) in cells; however, the underlying mechanism has not been elucidated. This study aimed to identify the mechanisms through which DBD plasma induces the expression of NADPH oxidase (NOX) family members by epigenetic modification in human keratinocytes (HaCaT). Cell exposure to DBD plasma in 10% oxygen and 90% argon resulted in the generation of ROS, triggering oxidative stress that manifested in various forms, including lipid membrane peroxidation, DNA base modification, and protein carbonylation. DBD plasma upregulated the expression of NOX1, NOX5, and DUOX2 at the mRNA and protein levels; and siRNAs targeting NOX1, NOX5, and DUOX2 attenuated the generation of DBD plasma-induced ROS. DBD plasma upregulated the transcriptional activators TET1, MLL1, and HAT1 and downregulated the transcriptional repressors DNMT1, EZH2, and HDAC1. Additionally, DBD plasma increased the binding of transcriptional activators and decreased the binding of transcriptional repressors to the DUOX2 promoter. Methyl-specific polymerase chain reaction and bisulfite sequencing indicated that DBD plasma decreased methylation at the DUOX2 promoter. These results suggest that DBD plasma induces ROS generation by enhancing the expression of the NOX system through epigenetic DNA and histone modifications.
Keywords: Dielectric-barrier discharge plasma, NOX family, Reactive oxygen species, DNA methylation, Histone modification
Graphical abstract
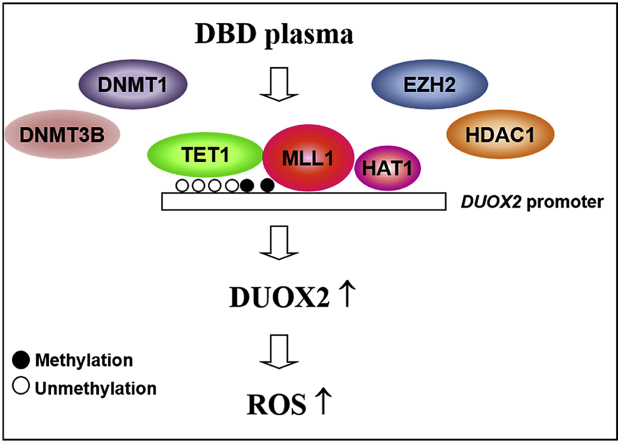
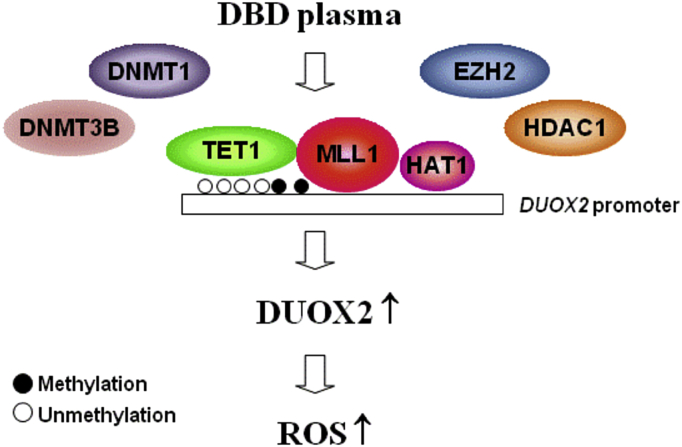
Non-thermal dielectric-barrier discharge (DBD) plasma induces ROS by enhancing the expression of the NOX system. Exposure to non-thermal, DBD plasma induces epigenetic changes in keratinocytes that prompt the competitive binding of TET1, HAT1, and MLL1 to the DUOX2 promoter, thereby displacing DNMTs, HDAC1, and EZH2, and inducing the expression of DUOX2.
1. Introduction
Plasma is composed of charged particles (negative and positive ions), free radicals, electronically excited atoms, and ultraviolet photons in gas or solution [[1], [2], [3]]. Recent studies have demonstrated the potential clinical applications of plasma in medicine, such as in dental equipment, and as therapeutics in the promotion of blood coagulation and wound healing following implant treatment, as well as for packaging in the food industry [4,5]. Additionally, plasma produces various reactive species that can be used to enhance oxidative and nitrative stresses via the generation of reactive oxygen species (ROS) and reactive nitrogen species (RNS) in various cancer cells, which eventually kill cancer cells [[6], [7], [8]]. In dermatology, plasma medicine has been used for surface modification and sterilization, and also for the treatment of infective and inflamed skin diseases [[9], [10], [11], [12]]. Plasma medicine is based on the synergistic effect of ROS, RNS, UV, ozone, and charged particles (electrons and ions) and may be helpful in the treatment of various pathological conditions [[13], [14]].
Recently, we have demonstrated that non-thermal dielectric-barrier discharge (DBD) plasma triggers complex signaling events that induce oxidative stress and endoplasmic reticulum stress by increasing ROS production [[15], [16], [17]]. Additionally, there are reports that cold, atmospheric plasma triggers H2O2 and NO2 production in cell culture media, thereby increasing intracellular ROS levels and modulating oxidative stress [[18], [19]].
Several enzymes catalyze ROS generation, including xanthine oxidase, cytochrome P450 oxidases, lipoxygenases, uncoupled nitric oxide synthase, NADPH oxidases (NOXs), and monoamine oxidases. Most of these enzymes produce ROS only after being damaged by ROS, e.g., endothelial nitric oxide synthase and xanthine oxidase. In contrast, cellular NOXs are the only known enzymes solely dedicated to ROS generation, such as superoxide or -hydrogen peroxide [20]. These enzymes are widely expressed in different tissues and organs and may play important roles in biological effects [21]. NOXs are membrane-bound proteins comprising several isoforms, including NOX1-5, DUOX1, and DUOX2, which have diverse functions. NOXs consist of two membrane-bound subunits—the catalytic subunit gp91phox and the regulatory subunit p22phox. The other regulatory components (i.e., p40phox, p48phox, p67phox, and the small GTPase Rac) are typically located in the cytoplasm. Upon stimulation, the cytosolic subunits are translocated to the membrane-bound complex to form a protein complex, thereby leading to ROS production [22].
Although the molecular mechanisms by which DBD plasma promotes the generation of ROS have been well-characterized, those causes for DBD plasma-induced intracellular ROS are currently unknown in the skin. In this study, we assessed the potential involvement of cellular NOX/DUOX in DBD plasma-induced ROS generation, focusing on its effect on epigenetic alterations in skin cells.
2. Materials and methods
2.1. Cell culture
The human keratinocyte cell line, HaCaT, was obtained from Cell Lines Service (Heidelberg, Germany). Cells were cultured in the Dulbecco's Modified Eagle Medium containing 10% heat-inactivated fetal calf serum, streptomycin (100 mg/ml), and penicillin (100 units/ml). Cells were maintained at 37 °C in a humidified incubator with an atmosphere of 5% CO2.
2.2. Non-thermal DBD plasma treatment
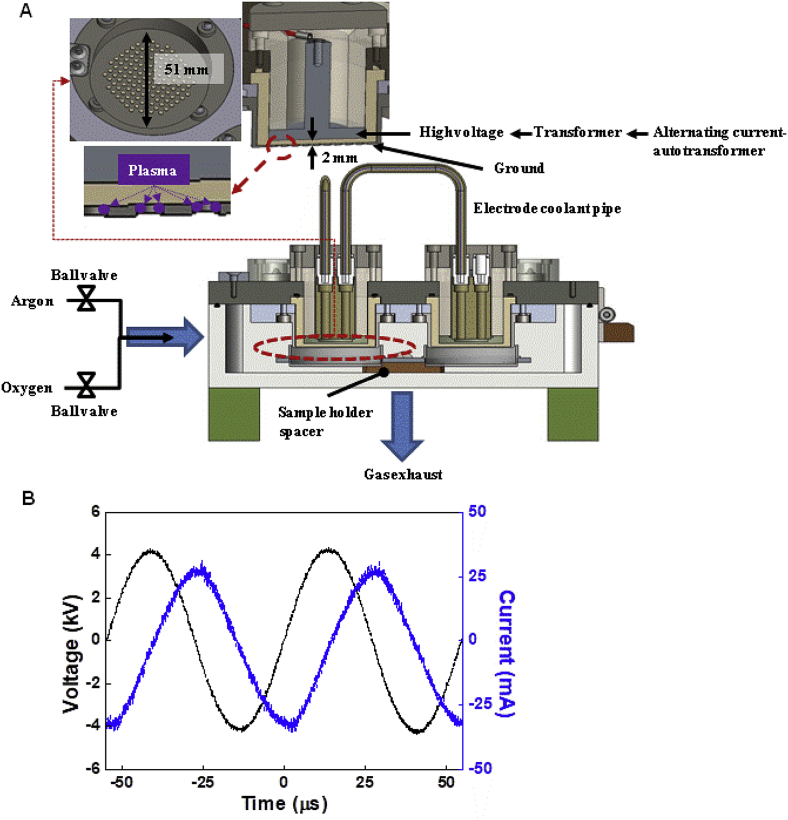
To prevent thermal damage and contamination, the experiment was conducted using non-thermal DBD plasma in a sealed chamber (Fig. 1A) [23]. With a variable alternating current autotransformer (0–220V), the voltage was amplified with a transformer (SC220–61030S, NT electronics, Seoul, Republic of Korea) and applied to the plasma electrode. The plasma electrodes were two stainless steel plates 51-mm in diameter, separated by a 2-mm thick alumina ceramic. The lower electrode had 2-mm diameter holes to generate the surface DBD. The flow rates of discharge gases, oxygen, and argon were controlled using ball-valves. The exhaust gas was discharged through a hole at the bottom of the chamber. Four samples could be placed under four identical electrodes, and the distances between the electrode and sample could be controlled using the spacer of the sample holder.
Fig. 1.
Schematic diagram of the DBD plasma device. (A) The voltage from the autotransformer was amplified in the transformer and applied to the plasma electrodes. The plasma electrode comprised two stainless steel plates 51-mm in diameter, separated by a 2-mm thick alumina ceramic. The lower electrode had 2-mm diameter holes to generate the surface dielectric-barrier discharge. (B) The voltage from the autotransformer was amplified in the transformer and applied to the plasma electrode. The amplitude and frequency of the applied sinusoidal voltage were 4.45 kV and 18.3 kHz, respectively.
The amplitude and frequency of the applied sinusoidal voltage from the transformer were 4.45 kV and 18.3 kHz, respectively (Fig. 1B). To measure the applied voltage and discharge current, a 1000:1 high voltage probe (P6015A, Tektronix, Beaverton, OR, USA) on the high voltage electrode and a current probe (110A, Pearson Electronics, Palo Alto, CA, USA) on the grounded electrode were used. Traces of the applied voltage and discharge currents were recorded with an oscilloscope (DPO4104B-L, Tektronix, Beaverton, OR, USA).
All treatments were performed in a 60-mm cell culture dish. HaCaT cells were seeded at 1.2 × 105 in a 60-mm cell culture dish and incubated for 16 h. Immediately prior to treatment, the growth media were replaced with 3 ml of fresh growth media and were placed on a cultured dish in a sample holder spacer. All cells were treated by DBD plasma for 6 and 9 min in an atmosphere of 10% oxygen and 90% argon and at 50 W input power. The 50 W input power was the power from the wall socket to the non-thermal DBD chamber and was measured using a power analyzer (HPM-300A, AD Power, Bucheon, Republic of Korea). After treatment, the cells were incubated in a humidified incubator with an atmosphere of 5% CO2 for 24 h.
2.3. Preparation of total RNA for mRNA sequencing
Total RNA was extracted using the TRI reagent® (Molecular Research Center, Cincinnati, OH, USA) and purified using the RNeasy MinElute cleanup kit (Qiagen, Valencia, CA, USA) according to the manufacturer's instructions. Sequencing libraries were prepared with 1 μg of total RNA using a TruSeq mRNA library prep kit (Illumina, Inc., San Diego, CA, USA) for library construction, which was sequenced on an Illumina NovaSeq 6000 (Illumina, Inc.) served by Macrogen Inc. (Seoul, Republic of Korea). Briefly, poly-A-containing mRNA molecules were purified using poly-T oligo attached magnetic beads. The mRNA was then thermally fragmented and converted to double-stranded cDNA. The RNA fragments were reverse transcribed into a cDNA library constructed using random hexamer primers, and paired-end sequencing (100 nucleotides) was performed using Illumina NovaSeq 6000. Two independent biological replicates were performed. To estimate expression levels and to find alternatively spliced transcripts, the RNA-Seq reads were mapped to the human genome using HISAT v2.1.0 [24], which utilizes two types of indices for alignment (a global, whole-genome index, and multiple small local indices). The reference genome sequence (hg19, Genome Reference Consortium GRCh37) and annotation data were downloaded from the UCSC website (http://genome.uscs.edu). The expression profiles were used to perform additional analyses, such as identification of differentially expressed genes (DEG). Data analysis and visualization of DEG were performed using R 3.5.2 (www.r-project.org).
2.4. Cell viability assay
Cells were seeded into 60-mm culture dishes at a density of 1.2 × 105 cells/ml, and were exposed to DBD plasma for 0, 3, 6, 9, or 12 min. After 24h incubation at 37 °C, MTT stock solution was added to each dish. Four hours later, the supernatants were aspirated. The formazan crystals in each dish were dissolved in dimethyl sulfoxide, and absorbance was read at 540 nm on a scanning, multi-well spectrophotometer (Varioskan LUX; Thermo Fisher Scientific Inc., Waltham, MA, USA).
2.5. Measurement of intracellular ROS
To determine the intracellular ROS, the 2′,7′-dichlorofluorescein diacetate (DCF-DA) assay was performed. DCF-DA is the most widely used probe for the detection of several ROS, including hydrogen peroxide, hydroxyl radical, and peroxynitrite. This probe is cell-permeable and is hydrolyzed intracellularly to the DCF2H (2′,7′-dichlorodihydrofluorescein) carboxylate anion, which is retained in the cell. Two-electron oxidation of DCF2H results in the formation of a fluorescent product, DCF, which can be monitored using several fluorescence-based techniques [25]. Cells were exposed to DBD plasma, and after 24h incubation at 37 °C, 2′,7′-dichlorofluorescein diacetate (DCF-DA) solution was added at a concentration of 5 μM. Twenty min later, the fluorescence of DCF was measured using a FACSCalibur flow cytometer (Becton Dickinson, Mountain View, CA, USA).
2.6. Measurement of intracellular H2O2
Cells were exposed to DBD plasma and after 24h incubation at 37 °C, 50 μM Amplex® red reagent and 0.1 units/ml horseradish peroxidase in phosphate buffer (Thermo Fisher Scientific, Waltham, Massachusetts, USA) were added to each well to a final volume of 100 μl, and the cells were incubated for 30 min in the dark. Fluorescence was assessed at excitation/emission values of 485 nm/580 nm in a microplate reader.
2.7. Detection of superoxide anion
Superoxide anion production was assessed with a superoxide anion assay kit (#CS1000-1 KT, Sigma-Aldrich, St. Louis MO, USA), based on the manufacturer's instructions. The principle underlying this kit-based assay exploits the oxidation of luminol by superoxide anions, which results in the formation of a chemiluminescent light. A specific, non-toxic enhancer amplifies the chemiluminescent signal. Briefly, cells were exposed to DBD plasma, and after 24 h incubation at 37 °C, cells were washed and resuspended in 100 μl of assay medium in 96-well plates. Assay buffer containing the reagents for luminol oxidation and the enhancer was added to the samples to a final volume of 200 μl. The samples were mixed and placed immediately into a luminometer, and the chemiluminescence was measured every 30 sec for 20 min.
2.8. Detection of hydroxyl radical
The hydroxyl radical detection assay was assessed using an OH580 probe (ab219931; Abcam, UK), which selectively reacts with hydroxyl radicals present in live cells. Such a reaction generates a red fluorescence signal that can be read at 540/590 nm excitation/emission. Briefly, cells were seeded in black-walled, clear-bottom 96-well microplates and incubated until confluency. Cells were washed with PBS and treated with 100 μl of the OH580 probe for 1 h at 37 °C. Next, cells were exposed to DBD plasma and incubated for 24 h at 37 °C. Next, cells were washed with PBS, and the fluorescence was read on a fluorescence plate reader.
2.9. Detection of nitric oxide (NO)
The total NO concentration was detected by measuring the concentration of nitrate and nitrite using a modified Griess reaction method. Briefly, cells were exposed to DBD plasma, and after 24h incubation at 37 °C, cells were homogenized in ice-cold nitrite assay buffer, and keep on ice for 10 min. The homogenate was centrifuged for 5 min at 10,000×g. Griess reagent (0.1% N-1-naphthylenediamine dihydrochloride and 5% H3PO4 solution; Thermo Fisher Scientific) was added to the cell supernatant at a ratio of 1:1 (v/v). After gentle mixing and 15-min incubation in the dark, the NO concentration was measured at 540 nm using a microplate reader and was compared with a nitrate standard curve.
2.10. Lipid peroxidation assay
The level of 8-isoprostane was assessed to quantify lipid peroxidation. Cells were exposed to DBD plasma, and after 24h incubation at 37 °C, conditioned media were collected, and the 8-isoprostane released into the media was measured using a commercially available enzyme immunoassay (Cayman Chemical, Ann Arbor, MI, USA) according to the manufacturer's instructions.
2.11. Analysis of 8-OHdG level
Cells were exposed to DBD plasma, and after 24h incubation at 37 °C, cellular DNA was isolated and purified using the Wizard® genomic DNA purification kit (Promega Corporation, Madison, WI, USA) and quantified using a spectrophotometer. Level of 8-hydroxy-2′-deoxyguanosine (8-OHdG), a nucleoside of 8-oxoguanine (8-oxoG) in the DNA, was quantified using the Bioxytech® 8-OHdG ELISA kit (OXIS International, Tampa, FL, USA), according to the manufacturer's instructions. The quantity of 8-OHdG was considered proportional to the 8-oxoG level.
2.12. Protein carbonyl formation
Cells were exposed to DBD plasma, and after 24h incubation at 37 °C, the cells were harvested, and the pellets were washed with PBS prior to lysis with protein carbonyl lysis buffer. The protein level in the cell lysates was quantified prior to loading the lysates into the protein carbonyl assay plate and incubating them overnight at 4 °C. The amount of protein carbonyl formation was determined using an Oxiselect™ protein carbonyl ELISA kit (Cell Biolabs, San Diego, CA, USA) according to the manufacturer's instructions.
2.13. Quantitative real-time PCR (qRT-PCR)
The qRT-PCR conditions were as follows: 10 min at 95 °C; (15 sec at 95 °C and 1 min at 60 °C) × 40 cycles; and 7 min at 72 °C. PCR amplification was carried out in a programmable thermal cycler. The qRT-PCR primers were as follows: for NOX1, forward primer 5′-GTACAAATTCCAGTGTGCAGACCAC-3′ and reverse primer 5′-ACGACAGTGGCTATAAGGTCAGAC-3′, for NOX2, forward primer 5′-GGAGTTTCAAGATGCGTGGAAACTA-3′ and reverse primer 5′-GCCAGACTCAGAGTTGGAGATGCT-3′, for NOX3, forward primer 5′-CACACCATGTTTTCATCGTCTT-3′ and reverse primer 5′- GAAGATATGGCTGGGCACTG-3′, for NOX4, forward primer 5′-GCTTACCTCCGAGGATCACA-3′ and reverse primer 5′-CGGGAGGGTGGGTATCTAA-3′, for NOX5, forward primer 5′-CCTCCTCATGTTCATCTGCTCCAGTT-3′ and reverse primer 5′-AGGAGGTAGGACAGGTGAGTCCAATA-3′, for DUOX1, forward primer 5′-TTCACGCAGCTCTGTGTCAA-3′ and reverse primer 5′-TCGGTCCTATACTAGACAGGGA-3′, for DUOX2, forward primer 5′-CCGGCAATCATCTATGGAGGT-3′ and reverse primer 5′-TTGGATGATGTCAGCCAGCC-3′, for Actin, forward primer 5′-CACCAACTGGGACGACAT-3′ and reverse primer 5′- ACAGCCTGGATAGCAACG-3'.
2.14. Western blotting
Cell lysates (40 μg protein) were electrophoresed on 10% sodium dodecyl sulfate polyacrylamide gels and transferred onto nitrocellulose membranes (Bio-Rad, Hercules, CA, USA), which were incubated with primary antibodies. Membranes were then incubated with horseradish peroxidase-conjugated secondary antibodies (Pierce, Rockland, IL, USA) and images were captured with the LAS-3000 luminescent image analyzer (Fujifilm, Tokyo, Japan). Protein bands were detected using the enhanced chemiluminescence western blotting detection kit (Amersham, Little Chalfont, Buckinghamshire, UK).
2.15. Transfection of small interfering RNAs (siRNAs)
Cells were transfected with 20 nM of control small interfering RNA (siRNA) or siRNAs against NOX1, NOX5, or DUOX2 using lipofectamine RNAiMax reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. Then, the transfected cells were exposed to DBD plasma, and after 24h incubation at 37 °C, they were analyzed by qPCR.
2.16. Immunocytochemistry
Cells plated on chamber slides were fixed with 4% paraformaldehyde for 30 min and permeabilized with PBS containing 0.1% Triton X-100 for 2.5 min. The cells were incubated in a blocking medium (PBS containing 3% bovine serum albumin) for 1 h and then probed with primary antibodies diluted in the blocking medium for 2 h. Then, the cells were probed with FITC- or Alexa 594-conjugated secondary antibody (Santa Cruz Biotechnology, Santa Cruz, CA, USA) for 1 h. Stained cells were washed with PBS and mounted on microscope slides in mounting medium containing DAPI and imaged on a Zeiss confocal microscope using LSM 510 software.
2.17. Chromatin immunoprecipitation (ChIP) and quantitative PCR (qPCR)
ChIP assays were performed with a SimpleChIP® enzymatic chromatin IP kit (Cell Signaling Technology Inc., Danvers, MA, USA) according to the manufacturer's instructions, with slight modifications. DNMT1, DNMT3B, TET1, H3K9Ac, H3K4Me3, H3K27Me3 antibodies, and normal rabbit IgG were used. DNA recovered from the immunoprecipitated complexes was subjected to qPCR. The qPCR primers were as follows: for DUOX2, forward primer 5′-GAGGACACCCTTACCGAGTG-3′ and reverse primer 5′-ACGGCGTACTGCCTTTTTAC-3'.
2.18. Quantitative methylation-specific PCR (qMSP) and bisulfite sequencing
For methylation analysis, DNA was extracted using a standard phenol-chloroform method. DNA (2 μg) was modified using bisulfite treatment with the EZ DNA methylation kit™ (Zymo Research, Orange, CA, USA), which guarantees a >99% conversion rate (non-methylated cytosine to uracil; protection of methylated cytosine residues). Promoter methylation was analyzed using methylation-specific PCR (MSP) primer pairs (designed against a region close to the putative transcription start site in the 5′ CpG island), 2 μl of bisulfite-treated DNA (template), and the JumpStart™ REDTaq® DNA polymerase (Sigma-Aldrich), as previously described [26]. For bisulfite sequencing analysis, PCR amplicons were separated by 2% agarose gel electrophoresis, purified with a gel extraction kit (Qiagen GmbH, Hilden, Germany), and cloned using the TOPO TA vector system (Invitrogen). Individual clones were isolated and purified using the NucleoSpin® plasmid isolation kit (Macherey-Nagel, Düren, Germany). Randomly selected positive clones (10–15 per sample) were sequenced using the M13F primer, and the methylation status of each CpG dinucleotide was analyzed. For the quantification of DUOX2 methylation, bisulfite-treated samples were subjected to qMSP. Methylation levels were normalized based on Alu element amplification. qPCR was performed using a CFX96TM real-time system (Bio-Rad). The primers were as follows: for DUOX2 MSP, unmethylation forward primer 5′-AGTAGTGGAATGTTGAAGTTTGTG-3′ and reverse primer 5′-ACTAACTTACCTACCCACCTACATA-3′, methylation forward primer 5′- AGTAGTGGAACGTTGAAGTTTGC-3′ and reverse primer 5′-CTAACTTACCTACCCGCCTACGTA-3′, for DUOX2 bisulfite sequencing, forward primer 5′-TTTGTTTTGGGTTTTTTAGGAGATA-3′ and reverse primer 5′-CCCCAACTTACTAACTTACCTACCC -3'.
2.19. Statistical analysis
Data represent the mean ± standard error of the mean (SEM). To analyze the differences between/among conditions, results were subjected to analysis of variance (ANOVA) followed by Tukey's test. In each case, a p-value of <0.05 was considered statistically significant.
3. Results
3.1. Identification of differentially expressed genes in cells exposed to non-thermal DBD plasma
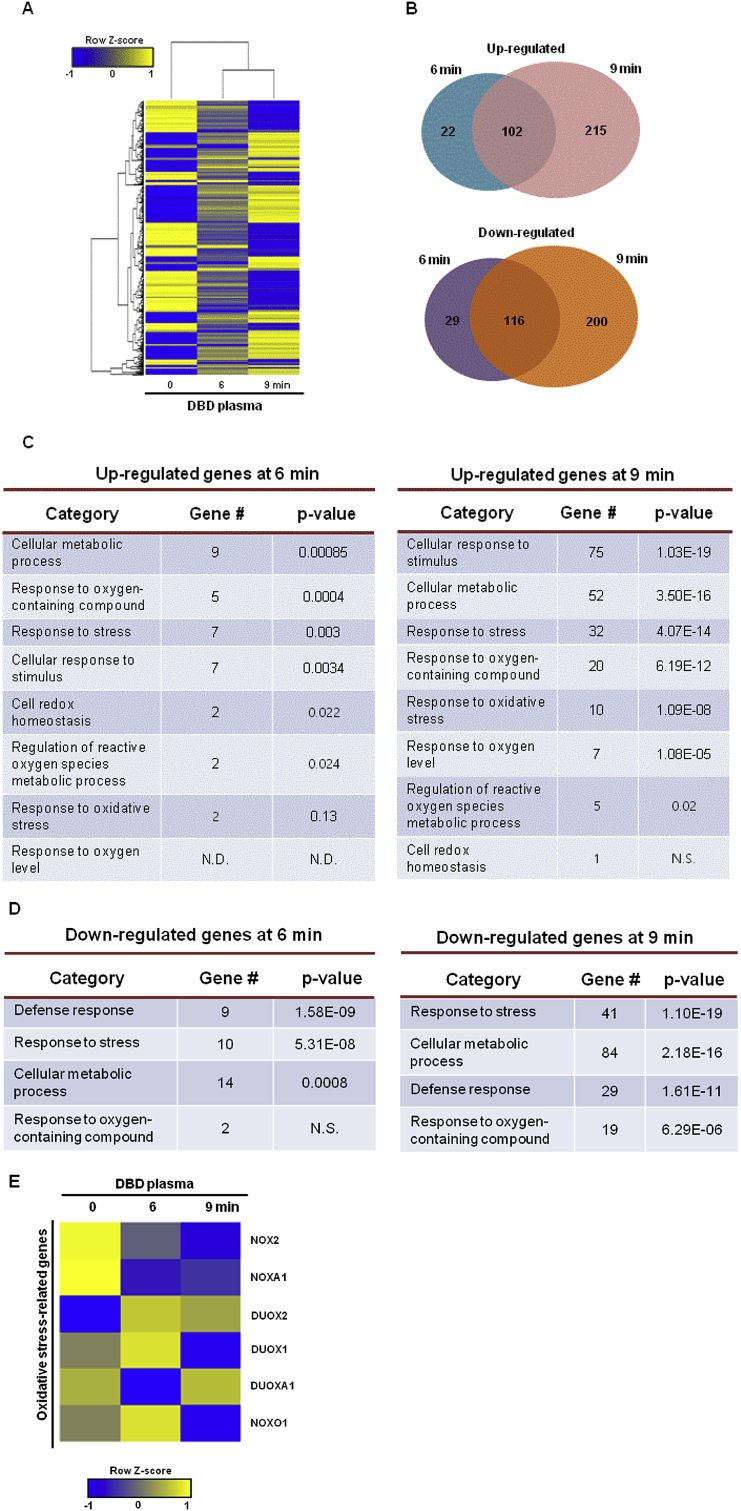
We performed whole-genome expression profiling using mRNA sequencing data to identify differentially expressed genes (DEGs) in the human keratinocyte cell line, HaCaT, exposed to non-thermal DBD plasma for 6 min and 9 min. The hierarchical clustering of a total of 684 DEGs between non-exposed cells (0 min) and cells exposed to non-thermal DBD plasma (6 min, 9 min) was shown (Fig. 2A). Moreover, the Venn diagrams showed the numbers of upregulated and downregulated genes in cells exposed to DBD plasma for 6 min and 9 min compared to those in non-exposed cells (Fig. 2B). The gene ontology (GO) classification most significantly associated with the upregulated genes in the cells exposed to plasma for 6 min overlapped with the upregulated genes in the cells exposed to plasma for 9 min, except for cell redox homeostasis and response to oxygen level (Fig. 2C). The GO classification most significantly associated with the downregulated genes in cells exposed to plasma for 6 min overlapped with the downregulated genes in cells exposed for 9 min, except for response to oxygen-containing compound (Fig. 2D). Additionally, mRNA sequencing data showed that ROS-generating enzyme-related gene DUOX2, was expressed more in cells exposed to plasma for 6 min and 9 min compared to that in non-exposed cells (Fig. 2E).
Fig. 2.
DBD plasma induces changes in biological process-associated gene expression. Genes that were differentially up- and downregulated in DBD plasma-exposed cells relative to those in non-exposed cells were identified by mRNA sequencing. (A) Hierarchical clustering of a total 684 differentially expressed genes was compared between the non-exposed keratinocytes to DBD plasma (“0” min column) and the exposed keratinocytes to DBD plasma for 6 min (“6” min column) and 9 min (“9” min column). Yellow denotes upregulated and blue denotes downregulated genes. (B) Venn diagrams display the number of genes that were upregulated (upper diagram) and downregulated (lower diagram) in keratinocytes exposed to DBD plasma for 6 or 9 min compared to those in non-exposed cells. Gene ontology (GO) analysis presented the (C) upregulated genes and (D) downregulated genes in keratinocytes exposed to DBD plasma for 6 or 9 min compared to those in non-exposed cells. (E) The heat map shows changes in the expression of NOX family-related genes in keratinocytes exposed to DBD plasma for 6 min (“6” min column) and 9 min (“9” min column) compared to keratinocytes not exposed to DBD plasma (“0” min column). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.2. Effect of non-thermal DBD plasma on ROS generation and oxidative stress
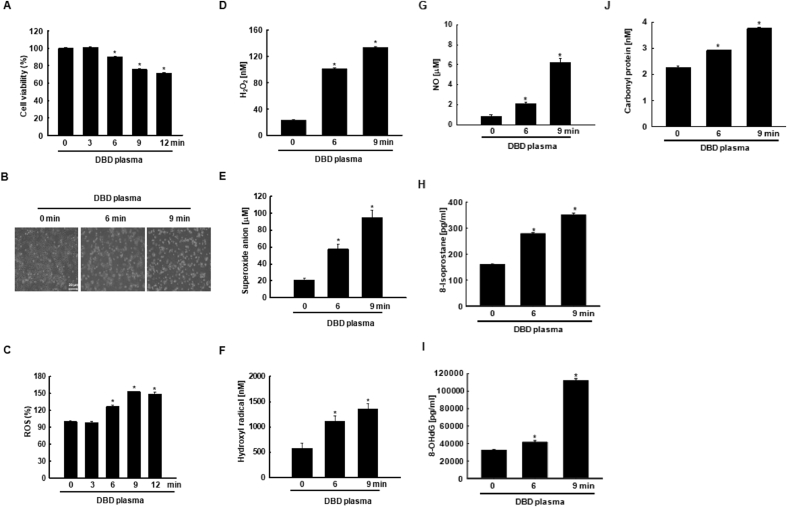
HaCaT cells exposed to non-thermal DBD plasma above 6 min were less viable than non-exposed cells (Fig. 3A–B). Exposure to DBD plasma above 6 min significantly enhanced ROS production (Fig. 3C). Additionally, cells exposed to DBD plasma for 6 min and 9 min showed a significant increase in cellular hydrogen peroxide, superoxide anion, and hydroxyl radical compared to that in non-exposed cells (Fig. 3D–F). Levels of nitric oxide (NO), an RNS, also increased in the presence of DBD plasma (Fig. 3G). Additionally, cells exposed to DBD plasma for 6 and 9 min showed a significant increase in 8-isoprostane, a marker of lipid peroxidation compared to that in non-exposed cells (Fig. 3H). 8-OHdG, a marker of DNA base modification, was present in significantly higher levels in DBD plasma-exposed cells compared to that in non-exposed cells (Fig. 3I), and the protein carbonyl content was significantly increased in cells exposed to DBD plasma compared to that in non-exposed cells (Fig. 3J).
Fig. 6.
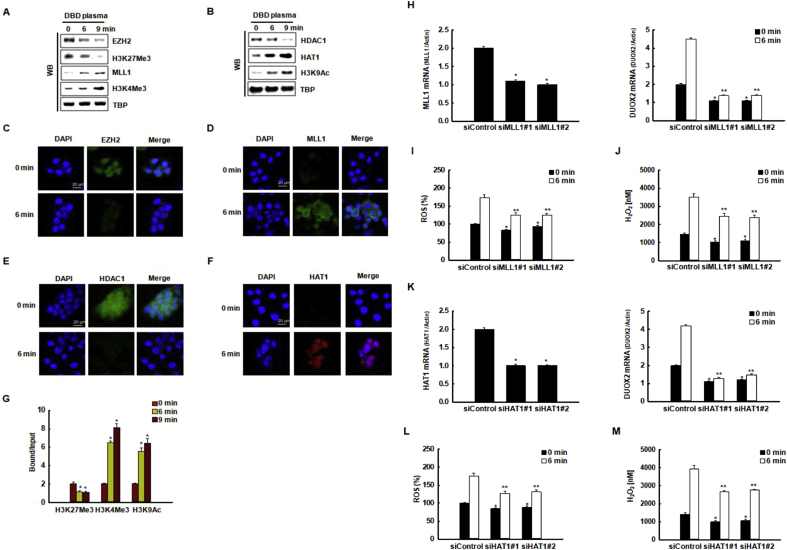
DUOX2 transcriptional upregulation by DBD plasma exposure correlates with histone modification. (A) The expression of EZH2, H3K27Me3, MLL1, and H3K4Me3, and (B) the expression of HDAC1, HAT1, and H3K9Ac in the nuclear fractions of cells were assessed by Western blot analysis. TBP used as the loading control. The nuclear localization of (C) EZH2, (D) MLL1, (E) HDAC1, and (F) HAT1 were assessed by confocal microscopy. Scale bars = 20 μm. (G) The binding to the DUOX2 promoter locus by the transcriptional repressor, H3K27Me3, and by the transcriptional activators, H3K4Me3 and H3K97Ac were assessed by chromatin immunoprecipitation coupled with quantitative PCR (ChIP-qPCR). *p < 0.05 indicates a statistically significant difference between binding of the indicated transcription factors to the DUOX2 promoter of cells exposed to DBD plasma for 6 min (“6” min) and 9 min (“9” min) and the non-exposed cells (“0” min). (H, left graph) The relative expression of MLL1 mRNA and (H, right graph) DUOX2 mRNA in cells transfected with MLL1-specific siRNAs or with control siRNAs were assessed by qRT-PCR. *p < 0.05 indicates a statistically significant difference between the expression of MLL1 mRNA in cells transfected with siMLL1 and the cells transfected with siControl. *, **p < 0.05 indicates a statistically significant difference between the expression of DUOX2 mRNA in cells transfected with siMLL1 and the cells transfected with siControl at 0 min and 6 min, respectively. (I) ROS level and (J) H2O2 concentration were assessed after reaction of DCF-DA and Amplex® red reagent, respectively. *, **p < 0.05 indicates a statistically significant difference between the expression of ROS level and H2O2 concentrations in cells transfected with siMLL1 and the cells transfected with siControl at 0 min and 6 min, respectively. (K) The relative expression of HAT1 mRNA and DUOX2 mRNA in cells transfected with HAT1-specific siRNAs or with control siRNAs were assessed by qRT-PCR. *p < 0.05 indicates a statistically significant difference between the expression of HAT1 mRNA in cells transfected with siHAT1 and the cells transfected with siControl. *, **p < 0.05 indicates a statistically significant difference between the expression of DUOX2 mRNA in cells transfected with siHAT1 and the cells transfected with siControl at 0 min and 6 min, respectively. (L) ROS level and (M) H2O2 concentration were assessed after reaction of DCF-DA and the Amplex® red reagent, respectively. *, **p < 0.05 indicates a statistically significant difference between the expression of ROS level and H2O2 concentrations in cells transfected with siHAT1 and the cells transfected with siControl at 0 and 6 min, respectively. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Fig. 3.
DBD plasma promotes ROS generation, which induces oxidative stress in keratinocytes. Cells were exposed to DBD plasma for 0, 3, 6, 9, and 12 min. After 24 h, (A) cell viability was assessed by MTT assay. *p < 0.05 indicates a statistically significant difference between the viability of DBD plasma-exposed cells (for 3, 6, 9, and 12 min) and non-exposed cells (0 min). (B) Cell morphology was assessed by an inverted microscope. Scale bars = 20 μm. (C) Cells were stained with DCF-DA, and intracellular ROS levels were detected by flow cytometry. *p < 0.05 indicates a statistically significant difference between the ROS levels in cells exposed to DBD plasma (“3, 6, 9, 12” min) and the ROS levels in non-exposed cells (“0” min). (D) H2O2 concentration was assessed using the Amplex® red reagent. *p < 0.05 indicates a statistically significant difference between the H2O2 concentration in DBD plasma-exposed cells (“6” and “9” min) and the H2O2 concentration in non-exposed cells (“0” min). (E) Superoxide anion production was assessed with a superoxide anion assay kit. *p < 0.05 indicates a statistically significant difference between the superoxide anion concentration in DBD plasma-exposed cells (“6” and “9” min) and the superoxide anion concentration in non-exposed cells (“0” min). (F) Hydroxyl radical production was assessed using the OH580 probe. *p < 0.05 indicates a statistically significant difference between the hydroxyl radical concentration in DBD plasma-exposed cells (“6” and “9” min) and the hydroxyl radical concentration in non-exposed cells (“0” min). (G) NO concentration was detected by measuring the concentration of nitrate and nitrite by a Griess reaction method. *p < 0.05 indicates a statistically significant difference between the NO concentration in DBD plasma-exposed cells (“6” and “9” min) and non-exposed cells (“0” min). (H) The lipid peroxidation was assessed by using the 8-isoprostane detection kit. *p < 0.05 indicates a statistically significant difference between the 8-isoprostane concentration in DBD plasma-exposed cells (“6” and “9” min) and the 8-isoprostane concentration in non-exposed cells (“0” min). (I) DNA damage was reflected by 8-OHdG concentration. *p < 0.05 indicates a statistically significant difference between the 8-OHdG concentration in DBD plasma-exposed cells (“6” and “9” min) and the 8-OHdG concentration in non-exposed cells (“0” min). (J) The protein oxidation was reflected by protein carbonylation. *p < 0.05 indicates a statistically significant difference between the concentrations of carbonylated proteins in DBD plasma-exposed cells (“6” and “9” min) and the concentrations of carbonylated proteins in non-exposed cells (“0” min).
3.3. Mechanism of ROS generation by non-thermal DBD plasma
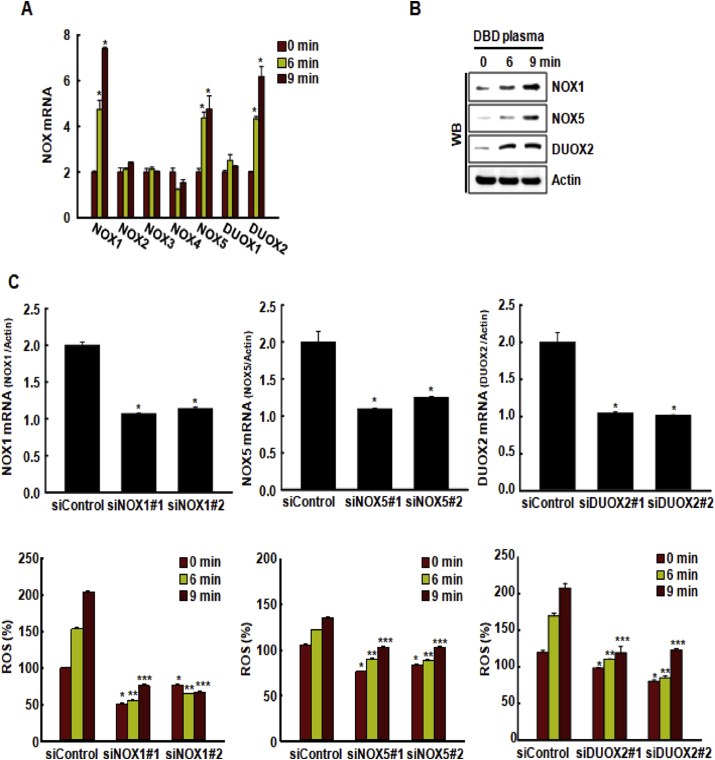
Based on the results illustrated in Fig. 2E, we speculated that DBD plasma might induce the expression of various NOX isoforms. DBD plasma exposure significantly increased the expression of NOX1, NOX5, and DUOX2 mRNA in HaCaT cells compared to that in the non-exposed cells (Fig. 4A). The effect of DBD plasma on the expression of NOX1, NOX5, and DUOX2 proteins was consistent with these findings (Fig. 4B). To assess if DBD plasma induced ROS production via NOX1, NOX5, and DUOX2, we transfected HaCaT cells with siRNAs specific to NOX1, NOX5, DUOX2, or a siRNA control. mRNA expression levels of NOX1, NOX5, and DUOX2 were significantly reduced in cells transfected with siRNAs specific to NOX1, NOX5, or DUOX2 compared to those in cells transfected with siRNA control (Fig. 4C). The plasma-enhanced generation of ROS was significantly suppressed in cells transfected with siRNAs specific to NOX1, NOX5, and DUOX2 compared to that in cells transfected with siRNA control (Fig. 4C).
Fig. 4.
DBD plasma induces ROS generation via NADPH oxidases (NOX). (A) The expression of various NOX mRNAs in cells was assessed by qRT-PCR 24 h after exposing them to DBD plasma at atmospheric pressure of 10% oxygen and 90% argon for 6 and 9 min *p < 0.05 indicates a statistically significant difference between the NOX expression by plasma-exposed cells (“6” and “9” min) and the non-exposed cells (“0” min). (B) The expression of NOX1, NOX5, or DUOX2 protein was assessed by Western blot analysis. (C) Cells were transfected with a control siRNA, or with siRNAs specific to NOX1, NOX5, or DUOX2. The transfected cells were exposed to DBD plasma for 0, 6, or 9 min, and the expression of NOX1, NOX5, and DUOX2 mRNA was assessed by qRT-PCR. *p < 0.05 indicates a statistically significant difference between the expression of NOX1, NOX5, and DUOX2 mRNA in cells transfected and siNOX1, siNOX5, or siDUOX2, respectively, and the cells transfected with control siRNAs (“siControl”). ROS level was detected by DCF-DA staining and flow cytometry. *p < 0.05 indicates a statistically significant difference between the ROS generation in cells transfected and control siRNAs (“siControl”) at 0 min; **p < 0.05 indicates a statistically significant difference between the ROS generation in cells transfected and control siRNAs (“siControl”) at 6 min; ***p < 0.05 indicates a statistically significant difference between the ROS generation in cells transfected and control siRNAs (“siControl”) at 9 min.
3.4. Epigenetic DNA regulation of DUOX2 transcription by DBD plasma in keratinocytes
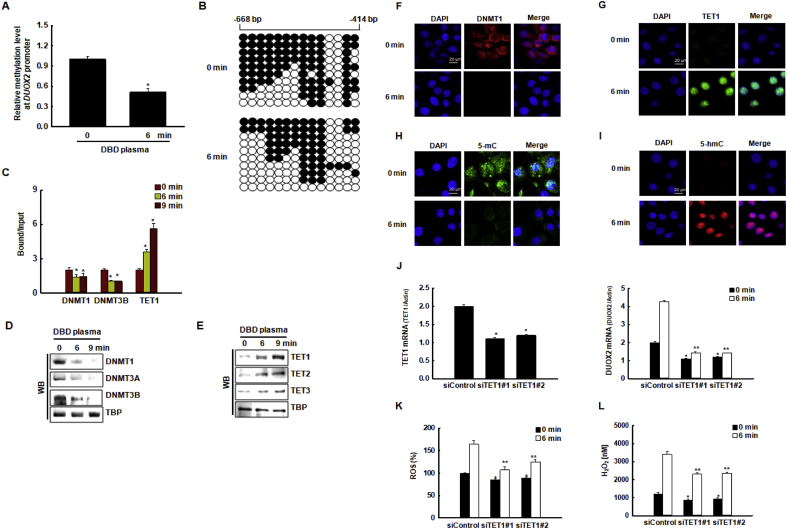
We used MethPrimer (http://www.urogene.org/methprimer/index.html) to evaluate the NOX1, NOX5, and DUOX2 gene sequences and to predict the existence of potential CpG islands, which are DNA sequences typically >100 base pairs long with a high concentration of cytosine (C) bases followed immediately by a guanine (G) base (potential methylation cites) that are often located in promoter regions of genes. Based on these criteria, we identified a sufficient number of CpG islands in the DUOX2 promoter region to analyze methylation levels; however, we were unable to detect CpG islands in the NOX1 and NOX5 promoters. We examined the methylation status of the DUOX2 promoter using qMSP and bisulfite sequencing analysis. The DNA methylation level in the DUOX2 promoter region was significantly lower in cells exposed to DBD plasma for 6 min compared to that in non-exposed cells (Fig. 5A). We then performed bisulfite sequencing analysis to examine the change in DNA methylation levels in the DUOX2 promoter region after DBD plasma exposure. Interestingly, the promoter region (from −668 to −414 bp, including regions of CpG sites of DUOX2) showed lower methylation in cells exposed to DBD plasma compared to controls (63% vs. 44%, respectively) (Fig. 5B). Next, using ChIP-qPCR analysis, we assessed whether DNA methyltransferases, DNMT1 and DNMT3B, and DNA demethylase, TET1, could directly bind to the DUOX2 promoter region (including CG sites from bp −668 to bp −414). The binding of DNMT1 and DNMT3B to the DUOX2 promoter region decreased upon DBD plasma exposure, whereas TET1 bound firmly to the DUOX2 promoter region after exposure to DBD plasma for 6 min and 9 min (Fig. 5C). These data suggest that the change in DUOX2 transcription in DBD plasma-treated cells was due to decreased DNA methylation in the promoter. Next, we measured the expression of DNA methylation-related proteins by western blotting. DNMT1, DNMT3A, and DNMT3B protein levels decreased after exposure to DBD plasma for 6 min and 9 min (Fig. 5D), whereas those of TET1, TET2, and TET3 increased (Fig. 5E). These changes in DNMT1 and TET1 protein expression detected by western blotting were confirmed by immunofluorescence (Fig. 5F–G). Concomitant with the DBD plasma-induced increase in TET1 binding to DUOX2 promoter and expression, enhanced conversion of 5-methyl cytosine (5-mC) (Fig. 5H) to 5-hydroxymethyl cytosine (5-hmC) (Fig. 5I) was observed, thereby indicating enhanced TET activity. These results suggest that these changes in DNMT1 and TET1 expression affect the transcription of ROS-associated genes in response to DBD plasma exposure. As the increase in TET expression and hypo-methylation of DUOX2 preceded the induction of DUOX2 during DBD plasma-induced ROS production, we investigated whether the expression of DUOX2 depended on TET. Transfection of HaCaT cells with TET1-specific siRNAs resulted in reduced expression of TET1 mRNA compared to that in cells transfected with control siRNAs (Fig. 5J, left graph). The knockdown of TET1 suppressed the DBD plasma-enhanced expression of DUOX2 mRNA in DBD plasma-exposed cells (Fig. 5J, right graph), thereby leading to a decrease in DBD plasma-induced ROS generation (Fig. 5K) and cellular H2O2 concentration (Fig. 5L). Thus, TET1 markedly contributes to the transcriptional activation of DUOX2, which is involved in the induction of ROS.
Fig. 5.
DBD plasma-induced transcriptional upregulation of DUOX2 correlates with DNA demethylation of the DUOX2 promoter region. (A) Relative methylation levels of the DUOX2 promoter region were normalized to the Alu element in cells exposed to DBD plasma for 0 min or 6 min *p < 0.05 indicates a statistically significant difference between DUOX2 promoter methylation in cells exposed to DBD plasma for 6 min (“6” min) and non-exposed cells (“0” min). (B) Bisulfite sequencing analysis of the DUOX2 promoter region in cells exposed to DBD plasma for 0 min or 6 min was assessed. Black circles represent methylated cytosine resides; white circles represent unmethylated cytosine residues. (C) Cells exposed to DBD plasma for 0, 6, or 9 min. The binding of DNMT1, DNMT3B, and TET1 to the DUOX2 promoter was assessed by chromatin immunoprecipitation coupled with quantitative PCR. *p < 0.05 indicates a statistically significant difference between the binding of DNMT1, DNMT3B, and TET1 to the DUOX2 promoter in DBD plasma-exposed cells (“6” and “9” min) and the non-exposed cells (“0” min), respectively. The nuclear fractions of the cells were electrophoresed, and (D) the expression of DNMT1, DNMT3A, and DNMT3B, and (E) the expression of TET1, TET2, and TET3 were detected by western blotting. TBP was used as the loading control. The nuclear localization of (F) DNMT1 and (G) TET1 was confirmed by confocal microscopy. Scale bars = 20 μm. The effect of DBD plasma on TET1 activity, i.e., the conversion of (H) 5-mC to (I) 5-hmC, was assessed by confocal microscopy. Scale bars = 20 μm. (J, left graph) The relative expression of TET1 mRNA and (J, right graph) DUOX2 mRNA in cells transfected with TET1-specific siRNAs or with control siRNAs were assessed by qRT-PCR. *p < 0.05 indicates a statistically significant difference between the expression of TET1 mRNA in cells transfected with siTET1 and cells transfected with siControl. *, **p < 0.05 indicates a statistically significant difference between the expression of DUOX2 mRNA in cells transfected with siTET1 and the cells transfected with siControl at 0 min and 6 min, respectively. (K) ROS levels were detected by DCF-DA staining and flow cytometry. *, **p < 0.05 indicates a statistically significant difference between the expression of ROS levels in cells transfected with siTET1 and the cells transfected with siControl at 0 min and 6 min, respectively. (L) H2O2 concentrations were assessed using the Amplex® red reagent. *, **p < 0.05 indicates a statistically significant difference between the expression of H2O2 concentrations in cells transfected with siTET1 and the cells transfected with siControl at 0 min and 6 min, respectively. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.5. Epigenetic histone modification of DUOX2 transcription by DBD plasma in keratinocytes
In addition to DNA methylation, we chose to investigate if epigenetic histone modifications might also be involved in modulating DBD plasma-induced DUOX2 expression. We measured the expression of the histone methyltransferases, EZH2 and MLL1, and their respective substrates, H3K27Me3 and H3K4Me3, in response to DBD plasma exposure by western blotting (Fig. 6A). The expression of EZH2, a component of the polycomb complex that has H3K27 methyltransferase activity, decreased after exposure of cells to DBD plasma, whereas the expression of MLL1, a transcriptional activator with H3K4 methyltransferase activity, increased after exposure to DBD plasma. Additionally, the expression of HDAC1, a transcriptional repressor, decreased in response to DBD plasma exposure, whereas the expression of HAT1, a transcriptional activator with H3K9 acetyltransferase activity, increased in response to DBD plasma exposure (Fig. 6B).
The decrease in EZH2 and the increase in MLL1 expression were confirmed by immunofluorescence after 6 min of DBD plasma exposure (Fig. 6C and D). Additionally, the decrease in HDAC1 and the increase in HAT1 expression were confirmed by immunofluorescence after 6 min of DBD plasma exposure (Fig. 6E and F). We then investigated if these changes in EZH2, MLL1, HDAC1, and HAT1 could affect the induction of the DUOX2 in response to DBD plasma exposure. We assessed whether the H3K27Me3, H3K4Me3, and H3K9MeAc, which are targets of EZH2, MLL1, HDAC1, and HAT1, occupied the DUOX2 locus by ChIP-qPCR analysis. We observed the binding of H3K27Me3 to the DUOX2 promoter region in the absence of DBD plasma exposure. The binding of H3K27Me3 to the DUOX2 promoter region significantly decreased in response to DBD plasma exposure (Fig. 6G). The decrease in the binding of H3K27Me3, a marker of histone transcriptional suppressor induced by DBD plasma exposure, to the DUOX2 promoter region, prompted us to investigate whether this decreased binding might be accompanied by the enhanced binding of H3K4Me3 and H3K9Ac, markers of histone transcriptional activator. We found that concomitant with the decrease in H3K27Me3 binding, the activity of H3K4Me3 and H3K9Ac sharply increased in the DUOX2 promoter region in DBD plasma-exposed cells (Fig. 6G). As the increase in MLL1 and HAT1 expression preceded the induction of DUOX2 during DBD plasma-induced ROS production, we examined whether the expression of DUOX2 depended on MLL1 and HAT1. We transfected MLL1-specific siRNAs and control siRNAs into HaCaT cells. Transfection of MLL1-specific siRNAs into these cells resulted in reduced expression of MLL1 mRNA compared to that in cells transfected with siRNA controls (Fig. 6H, left graph). The DBD plasma-induced expression of DUOX2 mRNA was decreased (Fig. 6H, right graph), resulting in a decrease of the DBD plasma-enhanced generation of ROS and decrease of the DBD plasma-enhanced concentration of cellular H2O2 in cells transfected with MLL1-specific siRNAs compared to those transfected with control siRNAs (Fig. 6I and J). Additionally, siRNA knockdown of HAT1 (Fig. 6K, left graph) suppressed the DBD plasma-induced upregulated expression of DUOX2 mRNA (Fig. 6K, right graph), reduced the DBD plasma-induced generation of ROS (Fig. 6L), and reduced the DBD plasma-induced H2O2 concentration (Fig. 6M).
4. Discussion
In this study, we investigated an intracellular ROS-generating system induced by DBD plasma and its mechanism in terms of epigenetic alteration in HaCaT human keratinocytes. Consistent with our previous reports [[15], [16], [17]], we demonstrated that exposure to DBD plasma induced the generation of ROS in HaCaT cells. The present study demonstrated that DBD plasma-induced ROS, such as superoxide anion and hydrogen peroxide, were generated via cellular NOXs. DBD plasma increased the expression of NOX1, NOX5, and DUOX2, which was confirmed by the specificity of the NOX1, NOX5, and DUOX2 siRNAs. In addition to its expression by epithelial cells, endothelial cells, and vascular smooth muscle cells, NOX1 is expressed by several other cell types [27]. NOX5 and DUOX2 are predominantly expressed in keratinocytes, thereby leading to the upregulation of calcium-dependent ROS production [[28], [29], [30]]. It has been reported that atmospheric pressure gas plasma can induce NOX2 expression and generate intracellular ROS, which promotes the programmed cell death in colorectal cancer cells by activating the ASK1-mediated apoptosis pathways [31]. However, in the present system, DBD plasma did not alter NOX2 expression, and this may be due to the difference in cell type and plasma exposure conditions.
Epigenetic regulation implies control of gene expression without accompanying genetic alteration, which remarkably can be inherited by daughter cells. There are three epigenetic events: methylation of DNA cytosine residues, structural modification of histones, and recognition of binding to the 3′-untranslated region of mRNA by microRNA. Epigenetic regulation is involved in various biological activities, including cell differentiation, inflammation, aging, and cancer [32]. Park et al. (2015) reported that cold atmospheric plasma hypermethylated and consequently downregulated oncogenes, such as ESR1, BCL2, and BNDF, whereas it hypomethylated and upregulated tumor suppressor genes such as DNAJC8, POTED, and EIF1YA [33].
In mammalian cells, DNMT1, DNMT3A, and DNMT3B are involved in the maintenance of global DNA methylation and gene-specific de novo DNA methylation. This methylation process can be reversed by DNA demethylases known as TETs. During DBD plasma exposure, TETs replaced DNMTs in the promoter region of DUOX2, thereby promoting DUOX2 transcription.
Histones undergo various chemical modifications, such as phosphorylation, acetylation, and methylation, based on the regulatory state of their corresponding gene. Methylation of histones can occur on various amino acid residues, including H3K4, H3K27, and H4K20, with one to three methyl groups. Methylation on different residues can cause completely different results in gene expression, e.g., H3K4Me3 activates the gene, whereas H3K27Me3 inactivates it. Lee et al. (2018) have reported that cold atmospheric plasma decreases the methylation level of H3K4 corresponding to oncogenes such as HSCB and PRPS1, and consequently inhibits cancer cell proliferation [34]. The present study demonstrated that in the absence of DBD plasma, EZH2 and HDAC1 binds to the DUOX2 promoter; however, when cells are exposed to DBD plasma, MLL1 and HAT1 replaces EZH2 and HDAC1 at the promoter, thereby inducing DUOX2 transcription.
Although we demonstrated that DBD plasma under an atmospheric pressure of 10% oxygen and 90% argon and 50 W input power for 6 and 9 min induce cellular NOX systems, we could not define the dose of plasma used in this study because no conclusive definition of plasma dose is available. Recently Cheng et al. (2020) proposed the equivalent total oxidation potential (ETOP), which is based on the oxidation potential of the reactive oxygen and nitrogen species (RONS), as a proxy of the plasma dose. However, considerable work is still needed to improve the ETOP, including the determination of more types of RONS, evaluation of the weight factor of each type RONS, and assessing the contribution of electrons, ions, and UV [35].
In conclusion, DBD plasma upregulates the expression of NOX1, NOX5, and DUOX2 epigenetically by regulating DNA methylation and histone modification, which leads to ROS production (Fig. 7). The current study is the first to decipher the effect of DBD plasma on epigenetic modification and can provide crucial information to comprehensively understand the epigenetic mechanism of DBD with respect to the generation of intracellular ROS.
Fig. 7.
Proposed model of intracellular ROS generation by DBD plasma. Exposure to non-thermal DBD plasma induces epigenetic changes in keratinocytes, which prompts the competitive binding of TET1, HAT1, and MLL1 to the DUOX2 promoter, thereby displacing DNMTs, HDAC1, and EZH2, and inducing the expression of DUOX2.
Author contributions
KA.K., MJ.P., and JM.Y. performed the cell experiment, analyzed the data, and wrote the manuscript. JW.H. designed, supervised, and wrote the manuscript. S.E., SY.Y., S.R., and SB.K. provided a series of experimental instructions and assistance. All authors approved the final version of the manuscript.
Declaration of competing interest
None declared.
Acknowledgment
The study was supported by the R&D Program of Plasma Advanced Technology for Agriculture and Food (Plasma Farming) via the National Fusion Research Institute of Korea (NFRI), funded by the Korea government.
References
- 1.Bernhardt T. Plasma medicine: applications of cold atmospheric pressure plasma in dermatology. Oxid. Med. Cell Longev. 2019;2019:3873928–3873938. doi: 10.1155/2019/3873928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dubuc A. Use of cold-atmospheric plasma in oncology: a concise systematic review. Ther. Adv. Med. Oncol. 2018;10 doi: 10.1177/1758835918786475. 1758835918786475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shang K., Li J., Morent R. Hybrid electric discharge plasma technologies for water decontamination: a short review. Plasma Sci. Technol. 2019;21:043001. doi: 10.1088/2058-6272/aafbc6. [DOI] [Google Scholar]
- 4.Shintani H. Inactivation of bacterial spore, endotoxin, lipid A, normal prion and abnormal prion by exposures to several sorts of gases plasma. Biocontrol Sci. 2016;21:1–12. doi: 10.4265/bio.21.1. [DOI] [PubMed] [Google Scholar]
- 5.Mohd Nasir N. Cold plasma inactivation of chronic wound bacteria. Arch. Biochem. Biophys. 2016;605:76–85. doi: 10.1016/j.abb.2016.03.033. [DOI] [PubMed] [Google Scholar]
- 6.Vandamme M. ROS implication in a new antitumor strategy based on non-thermal plasma. Int. J. Canc. 2012;130:2185–2194. doi: 10.1002/ijc.26252. [DOI] [PubMed] [Google Scholar]
- 7.Ratovitski E.A. Anti-cancer therapies of 21st century: novel approach to treat human cancers using cold atmospheric plasma. Plasma Process. Polym. 2014;11:1128–1137. doi: 10.1002/ppap.201400071. [DOI] [Google Scholar]
- 8.Li Y., Kang M.H., Uhm H.S., Lee G.J., Choi E.H., Han I. Effects of atmospheric-pressure non-thermal bio-compatible plasma and plasma activated nitric oxide water on cervical cancer cells. Sci. Rep. 2017;7:45781. doi: 10.1038/srep45781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng K.Y. Wound healing in streptozotocin-induced diabetic rats using atmospheric-pressure argon plasma jet. Sci. Rep. 2018;8:12214–12229. doi: 10.1038/s41598-018-30597-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heinlin J. Plasma medicine: possible applications in dermatology. J. Dtsch. Dermatol. Ges. 2010;8:968–976. doi: 10.1111/j.1610-0387.2010.07495.x. [DOI] [PubMed] [Google Scholar]
- 11.Potter M.J., Harrison R., Ramsden A., Bryan B., Andrews P., Gault D. Facial acne and fine lines: transforming patient outcomes with plasma skin regeneration. Ann. Plast. Surg. 2007;58:608–613. doi: 10.1097/01.sap.0000252481.84134.fe. [DOI] [PubMed] [Google Scholar]
- 12.Lu X., Keidar M., Laroussi M., Choi E., Szili E.J., Ostrikov K. Transcutaneous plasma stress: from soft-matter models to living tissues. Mater. Sci. Eng. R Rep. 2019;138:36–59. doi: 10.1016/j.mser.2019.04.002. [DOI] [Google Scholar]
- 13.Haertel B. Non-thermal atmospheric-pressure plasma possible application in wound healing. Biomol. Ther. 2014;22:477–490. doi: 10.4062/biomolther.2014.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu X., Naidis G.V., Laroussi M., Reuter S., Graves D.B., Ostrikov K. Reactive species in non-equilibrium atmospheric-pressure plasmas: generation, transport, and biological effects. Phys. Rep. 2016;630:1–84. doi: 10.1016/j.physrep.2016.03.003. [DOI] [Google Scholar]
- 15.Kim K.C. Non-thermal dielectric-barrier discharge plasma damages human keratinocytes by inducing oxidative stress. Int. J. Mol. Med. 2016;37:29–38. doi: 10.3892/ijmm.2015.2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim K.C. Exposure of keratinocytes to non-thermal dielectric barrier discharge plasma increases the level of 8-oxoguanine via inhibition of its repair enzyme. Mol. Med. Rep. 2017;16:6870–6875. doi: 10.3892/mmr.2017.7454. [DOI] [PubMed] [Google Scholar]
- 17.Ruwan Kumara M.H. Non-thermal gas plasma-induced endoplasmic reticulum stress mediates apoptosis in human colon cancer cells. Oncol. Rep. 2016;36:2268–2274. doi: 10.3892/or.2016.5038. [DOI] [PubMed] [Google Scholar]
- 18.Girard P.M. Synergistic effect of H2O2 and NO2 in cell death induced by cold atmospheric He plasma. Sci. Rep. 2016;6:29098–29115. doi: 10.1038/srep29098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Turrini E. Cold atmospheric plasma induces apoptosis and oxidative stress pathway regulation in T-lymphoblastoid leukemia cells. Oxid. Med. Cell Longev. 2017;2017:4271065–4271078. doi: 10.1155/2017/4271065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Magnani F. Crystal structures and atomic model of NADPH oxidase. Proc. Natl. Acad. Sci. U.S.A. 2017;114:6764–6769. doi: 10.1073/pnas.1702293114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Altenhöfer S. Evolution of NADPH oxidase inhibitors: selectivity and mechanisms for target engagement. Antioxid. Redox Signal. 2015;23:406–427. doi: 10.1089/ars.2013.5814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Panday A. NADPH oxidases: an overview from structure to innate immunity-associated pathologies. Cell. Mol. Immunol. 2015;12:5–23. doi: 10.1038/cmi.2014.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brandenburg R. Dielectric barrier discharges: progress on plasma sources and on the understanding of regimes and single filaments. Plasma Sources Sci. Technol. 2017;26:053001. doi: 10.1088/1361-6595/aa6426/meta. [DOI] [Google Scholar]
- 24.Kim D., Langmead B., Salzberg S.L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y.F., Dai M.H., Yuan Z.H. Methods for the detection of reactive oxygen species. Anal. Methods. 2018;10:4625–4638. doi: 10.1039/C8AY01339J. [DOI] [Google Scholar]
- 26.Herman J.G. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc. Natl. Acad. Sci. U.S.A. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fu X.J. NADPH oxidase 1 and its derived reactive oxygen species mediated tissue injury and repair. Oxid. Med. Cell Longev. 2014;2014:282854–282864. doi: 10.1155/2014/282854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Choi H. Sphingosylphosphorylcholine down-regulates filaggrin gene transcription through NOX5-based NADPH oxidase and cyclooxygenase-2 in human keratinocytes. Biochem. Pharmacol. 2010;80:95–103. doi: 10.1016/j.bcp.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 29.Benedyk M. HaCaT keratinocytes overexpressing the S100 proteins S100A8 and S100A9 show increased NADPH oxidase and NF-kappaB activities. J. Invest. Dermatol. 2007;127:2001–2011. doi: 10.1038/sj.jid.5700820. [DOI] [PubMed] [Google Scholar]
- 30.Ko E. Dual oxidase 2 is essential for house dust mite-induced pro-inflammatory cytokine production in human keratinocytes. Exp. Dermatol. 2015;24:936–941. doi: 10.1111/exd.12808. [DOI] [PubMed] [Google Scholar]
- 31.Ishaq M., Evans M.D., Ostrikov K.K. Atmospheric pressure gas plasma-induced colorectal cancer cell death is mediated by Nox2-ASK1 apoptosis pathways and oxidative stress is mitigated by Srx-Nrf2 anti-oxidant system. Biochim. Biophys. Acta. 2014;1843:2827–2837. doi: 10.1016/j.bbamcr.2014.08.011. [DOI] [PubMed] [Google Scholar]
- 32.Zhang W. The ageing epigenome and its rejuvenation. Nat. Rev. Mol. Cell Biol. 2020;21:137–150. doi: 10.1038/s41580-019-0204-5. [DOI] [PubMed] [Google Scholar]
- 33.Park S.B. Differential epigenetic effects of atmospheric cold plasma on MCF-7 and MDA-MB-231 breast cancer cells. PloS One. 2015;10:0129931–0129947. doi: 10.1371/journal.pone.0129931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee S. ChIP-seq analysis reveals alteration of H3K4 trimethylation occupancy in cancer-related genes by cold atmospheric plasma. Free Radic. Biol. Med. 2018;126:133–141. doi: 10.1016/j.freeradbiomed.2018.08.004. [DOI] [PubMed] [Google Scholar]
- 35.Cheng H., Xu J.X., Li X., Liu D.W., Lu X.P. On the dose of plasma medicine: equivalent total oxidation potential (ETOP) Phys. Plasmas. 2020;27:063514. doi: 10.1063/5.0008881. [DOI] [Google Scholar]