Abstract
The well-documented anticarcinogenic properties of natural polyphenolic proanthocyanidins (OPC) have been primarily attributed to their antioxidant and anti-inflammatory potency. Emerging evidence suggests that OPC may target canonical oncogenic pathways, including PI3K/AKT; however, the underlying mechanism and therapeutic potential remain elusive. Here we identify that proanthocyanidin B2 (OPC–B2) directly binds and inhibits AKT activity and downstream signalling, thereby suppressing tumour cell proliferation and metabolism in vitro and in a xenograft and diethyl-nitrosamine (DEN)-induced hepatocellular carcinoma (HCC) mouse models. We further find that OPC-B2 binds to the catalytic and regulatory PH domains to lock the protein in a closed conformation, similar to the well-studied AKT allosteric inhibitor MK-2206. Molecular docking and dynamic simulation suggest that Lys297 and Arg86 are critical sites of OPC-B2 binding; mutation of Lys297 or Arg86 to alanine completely abolishes the antitumor effects of OPC-B2 but not MK-2206. Together, our study reveals that OPC-B2 is a novel allosteric AKT inhibitor with potent anti-tumour efficacy beyond its antioxidant and anti-inflammatory properties.
Keywords: Proanthocyanidin B2 (OPC–B2), AKT, Hepatocellular carcinogenesis (HCC), Metabolic reprogramming
Graphical abstract

Highlights
-
•
OPC-B2 exerts potent anti-tumor effects in HCC.
-
•
OPC-B2 directly binds to AKT and inhibits its kinase activity.
-
•
Lys297 and Arg86 in AKT are critical sites for OPC-B2/AKT interaction.
-
•
Allosteric inhibition of AKT by OPC-2 has therapeutic potential for HCC.
1. Introduction
Proanthocyanidins are a large class of polyphenolic compounds widely found in the plants, such as grape seeds and peanut skin [38]. Proanthocyanidin-B2 (OPC–B2) consisting of two epicatechins (Fig. 1A) exhibits strong antioxidant activity owing to their phenolic hydrogen atoms, which potently intercept free radicals in the free radical chain reactions [21,30,38,45,56]. The anti-carcinogenic properties of these compounds have been well-documented and these effects have been primarily attributed to their potent antioxidant and anti-inflammatory efficacy [1,36]. In addition, emerging evidence suggests that these compounds may exert pleiotropic anticancer effects by targeting multiple pathways, including NF-κB, AMPK, Wnt β-catenin, cell apoptosis, PI3K/AKT, mTOR, and MAPK [13]. However, it remains elusive whether OPC-B2 exhibits anti-tumor effects by acting on any of these pathways as well as the therapeutic potential.
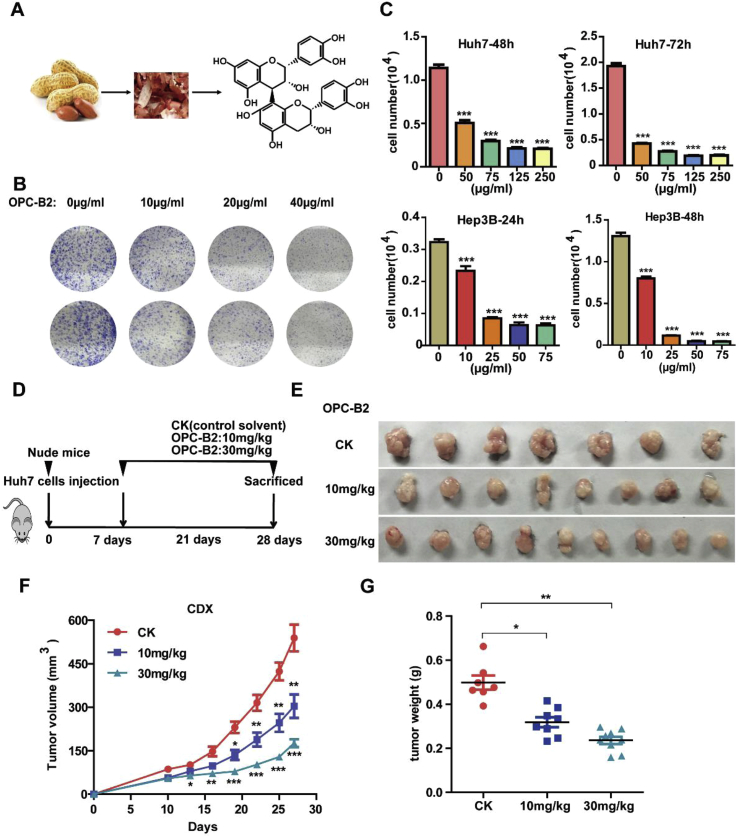
Fig. 1.
OPC-B2 inhibits proliferationof liver cancer cells and tumor growth in xenograft tumor growth models. A.OPC-B2 was extracted from peanuts skin and the purified compound was stored at 2–8 °C and protected from light. B.Huh7 cells were exposed to various concentrations of OPC-B2 for 7 days in colony formation assay. C.Cell viability of two liver cancer cell lines (Huh7 and Hep3B)treated with different concentrations of OPC-B2at 48 and 72 h. D.Schematic view ofthe experimental procedures of xenograft mouse model. Huh7 cells were subcutaneously injected into right and left flanks of nude mice (regarded as day = 0). After 7 days, micewererandomly divided into three groupsand administrated with control solvent (n = 7), 10 mg/kg (n = 8), and 30 mg/kg (n = 9),respectively, via intraperitoneal injectionevery twodays. And tumor size was measured and recorded at the same time. E.Images of nude mice tumors. F.Tumor volumesduring the period of experiment. G.Tumor weightsof nude mice. Two-tailed Student's t-test,*p < 0.05; **p < 0.01; ***p < 0.001.
As a common liver malignancy, Hepatocellular Carcinoma (HCC) represents the 4th leading cause of cancer mortality worldwide [44,47]. Although the diagnosis and treatment of HCC has been greatly improved in recent years, the 5-year survival rate of patients is far from satisfaction [55]. Several drugs are used clinically in the treatment of HCC, such as Sorafenib (first-line), Regorafenib, Cabozantinib and Ramucirumab (second-line) [33]. However, they are mainly used for advanced HCC and drug resistance is a main reason to cause chemotherapy failure in HCC. Therefore, there is an urgent need to develop safe and effective agents to treat patients with HCC [28,50].
Abnormal activation of AKT is frequently present in breast, ovarian epithelial, prostate, gastric cancers [6,18] and HCC [35,48]. In response to growth factors or cytokines, AKT is recruited to the cell membrane by phosphatidylinositol-3,4,5-triphosphate (PIP3) produced by PI3K through PH (pleckstrin homology) domain of AKT to induce a conformational change, thus mediating the assembly of PDK1 to phosphorylate AKT at Thr308. The mechanistic target of rapamycin complex 2 (mTORC2) primarily contributes to the well-characterized Ser473 phosphorylation and promotes AKT activation (Cantley; [22,39]. Once activated, AKT triggered subsequently cellular response including cell cycle progression and metabolism through phosphorylating diverse downstream proteins or inducing various genes expressions [17,57].
AKT inhibition has been explored as a therapeutic target for various cancers, including HCC. AKT inhibitors are classified as ATP-competitive inhibitors or allosteric inhibitors. ATP-competitive inhibitors target the ATP-binding pocket, whereas allosteric inhibitors bind to the PH or kinase domain of AKT to inhibit its activity [37]. Upon activation of upstream signaling by external growth factors, PH domain of AKT binds to PIP3, inducing AKT transferred to plasma membrane (“open”). Subsequently PH domain moves in and forms tight interactions with the kinase domain via polar contacts, resulting in a barrier that blocks the binding between ATP and AKT (“closed”) [20,37]. Allosteric inhibitors are generally considered to be better than ATP-competitive inhibitors as they show less toxicity, reduced side-effects, and greater specificity. As one of the most studied allosteric inhibitors of AKT, MK-2206 exhibits moderate anti-tumor efficacy in clinical studies [14,34]. However, AKT inhibitors remain challenging in clinical trials because of the side-effect of transient hyperglycemia or the E17K mutation causing AKT to be less sensitive to MK2206 [43]. Therefore, major efforts are currently under way to develop new inhibitors of AKT for cancer treatment, especially HCC.
In this study, we find that OPC-B2 extracted from peanut skin has strong anti-tumor effects in HCC through direct binding and inhibiting AKT activity, leading to cell cycle arrest and metabolic inhibition in vitro and HCC mouse models. Molecular docking and dynamic simulation suggest that Arg86 and Lys297 of AKT are critical sites for OPC-B2 binding. Experimental mutation of these two amino acids almost completely eliminates the anti-tumor efficacy of OPC-B2. Interestingly, the binding mode of OPC-B2 appears to be different from that of MK2206. Together, our study suggest that targeting the OPC-B2 binding pocket in AKT may provide a therapeutic opportunity in HCC treatment.
2. Results
2.1. OPC-B2 inhibits HCC cell proliferation and tumor growth
We isolated OPC-B2 from peanut skin and characterized its chemical structures using high resolution mass spectrometry by comparing to a commercially available standard compound (Figure S1A). To examine the effect of OPC-B2 on HCC cell proliferation, we treated Huh7 cells with various concentrations in colony formation and proliferation assays. OPC-B2 exerts strong inhibition efficacy on tumor cells proliferation and colony formation in a time and dose-dependent manner (Fig. 1B and C). Interestingly, OPC-B2 showed anti-tumor effects for several liver cancer cell lines with different oncogenic characteristics: early stage HCC cells SMMC-7721 cells, highly differentiated Hep 3B, and highly metastatic LM3 cells (Fig. 1C and Figure S1B). Although OPC-B2 showed different potencies for different cell lines, IC50 of OPC-B2 for Hep3B reached around 12.5 μg/ml (21 μM) (Figure S1C). Together, these results showed that OPC-B2 has significant inhibitory effects on HCC tumor cell proliferation in vitro.
Next, we explored the anti-tumour effects of OPC-B2 in vivo by using a xenograft tumour growth model. After the nude mice (congenital thymic defect mice) were subcutaneously injected with Huh7 cells, mice were randomly divided into three groups: control, 10 mg/kg, and 30 mg/kg. OPC-B2 was administrated intraperitoneally with two doses of 10 and 30 mg/kg every two days (Fig. 1D) and tumor sizes were recorded at the same time. After the mice were sacrificed at 28th day, we observed that OPC-B2 exhibits strong anti-tumor efficacy in a dose-dependent manner (Fig. 1E–G). Comparing with control group, OPC-B2 with 30 mg/kg treatment group had decreased tumor size, tumor volume and tumor weight up to 60% (Fig. 1E–G). Taken together, OPC-B2 exerts significant inhibitory effects on HCC cell proliferation and tumor growth in vitro and in vivo.
2.2. OPC-B2 directly binds to AKT and inhibits its kinase activity
Although multiple signal pathways have been shown to be involved in the development of HCC, such as AMPK, ERK and AKT, the aberrant activation of AKT signaling pathway plays an important role in promoting liver cancer progression through regulating cell proliferation and survival [35]. We next examined these major signaling pathways and found that OPC-B2 mainly inhibited the phosphorylation of AKT at S473 and T308 without significant impact on AMPK and ERK pathways (Figure S2A). To further investigate whether OPC-B2 inhibits tumor cell proliferation through repressing AKT activity and its downstream signaling pathway, we first transfected the Myc-tagged AKT plasmid in Huh7 cells to increase AKT expression and found that overexpression of AKT enhanced p-AKT activity and promoted cell proliferation through activating the p-AKT (S473 and T308), p-GSK3β (S9) (Fig. 2A). Conversely, inhibition of AKT activity by MK-2206 decreased the phosphorylation at S473 and T308 and attenuated tumor cell proliferation (Fig. 2B). Similarly, OPC-B2 treatment significantly inhibited cell proliferation and the phosphorylation levels of AKT at S473 and T308 in a dose-and time-dependent manner (Fig. 2C–D and Figure S2A), suggesting that OPC-B2 has similar inhibitory effect on AKT activity to MK-2206. Next, we combined OPC-B2 with MK-2206 treatment on tumor cells to test potential synergistic effects on AKT activity and cell proliferation and observed that a combination of MK-2206 with two different doses of OPC-B2 led to a significant more inhibition on cell proliferation than either of the reagents alone, suggesting a synergistic inhibition on AKT activity and cell proliferation (Figure S2B and C). To test whether OPC-B2 directly inhibits AKT activity or acts on the upstream kinases of AKT, we next examined the effects of OPC-B2 on the upstream protein kinases of AKT including PI3K [46], PDK1 [9,10], and mTORC2 [32], and found that OPC-B2 treatment did not significantly affect any of these kinase activities (Figure S2D). Taken together, all these data suggest that OPC-B2 may directly bind to AKT and inhibit its activity.
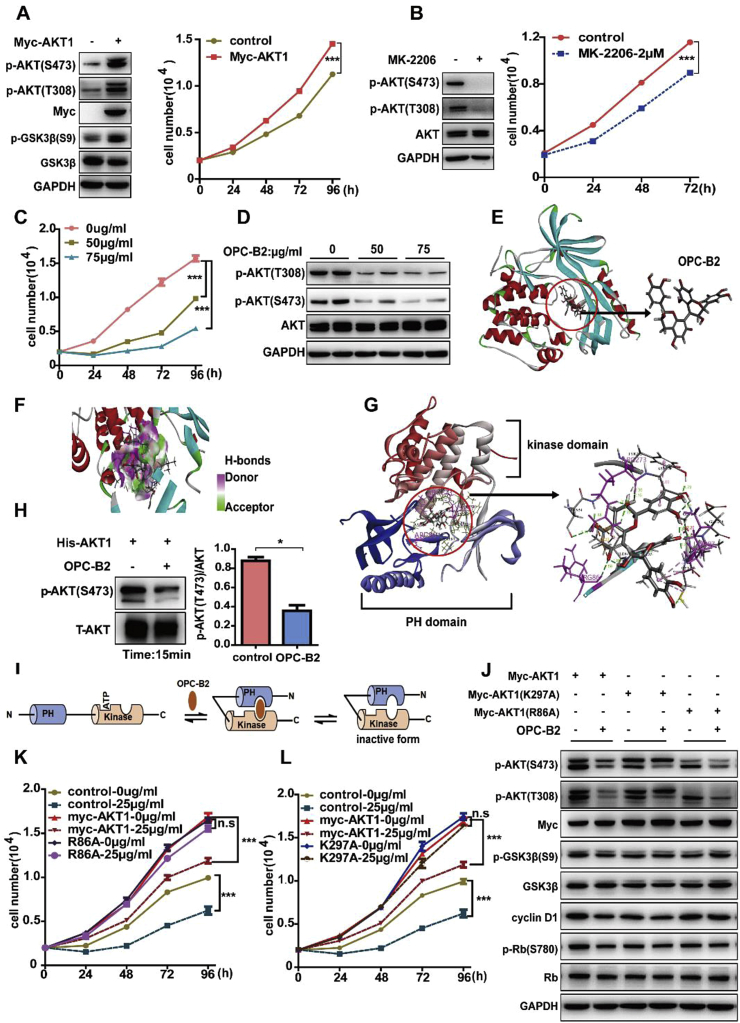
Fig. 2.
OPC-B2 directly bindsto AKT and inhibits its activity. A.Cell viability of Huh7 cells after transfected with Myc-AKT1 and immunoblots of whole cell lysates from Huh7 cells. B.Cell viability of Huh7 cells treated with MK-2206 at a dose of 2 μM and immunoblots of whole cell lysates from Huh7 cells treated with MK-2206 for 24 h. C.Cell viability of Huh7 cells at different time points treated with OPC-B2 at doses of 50 and 75 μg/ml. D.Phosphorylation levelsof AKT at serine 473 (S473) and threonine 308 (T308) after treated with OPC-B2. E.Molecular docking indicates that OPC-B2 directly binds to the AKT1. F.Hydrogen bondsbetween AKT and OPC-B2. G.Detailed interaction residues of PH and kinase domain of AKT1 between OPC-B2 and AKT1. H.Immunoblots of purified activated GST-AKT1 protein incubated with OPC-B2 (75 μg/ml) for 15min and quantitative result of p-AKT(S473). I.A schematic model ofOPC-B2 binding to AKT1 directly. J.Immunoblots of cell lysates of Huh7 cells transfected with myc-AKT1, myc-AKT1(K297A) and myc-AKT1(R86A), followed by treatment with control solvent or OPC-B2 (75 μg/ml).The upper band is the exogenously overexpressed AKT1 protein, and the lower band is the endogenous AKT1 protein. K, L.Cell number of Huh7 cells transfected with myc-AKT1, myc-AKT1(R86A) and myc-AKT1(K297A), followed by treatment with control solvent or OPC-B2 (75 μg/ml) at different time points.Two-tailed Student's t-test,*p < 0.05; **p < 0.01; ***p < 0.001.
To investigate the possibility of a direct interaction between OPC-B2 and AKT, we performed in silico docking of OPC-B2 with a full length AKT (PDB codes 3O96) using a Discovery studio software (DS 4.0) (BIOVIA, San Diego, CA, USA) [42,54]. Molecular docking results showed that OPC-B2 potentially occupies a binding pocket of AKT with several interactions with AKT (Fig. 2E–G and Figure S2E). As illustrated in Fig. 2F and supplemental Figure S2E, these potential interactions include conventional hydrogen bonding, Pi-Cation, charge-charge, and Pi-Pi T-shaped and Pi-Alkyl interactions. The molecular docking and dynamic simulation also found that OPC-B2 as a ligand could affect the activity of AKT (receptor) with a total interaction energy of around 464 kcal/mol primarily through electrostatic and Van der Waals interaction, respectively (Table S1). To provide experimental evidence that OPC-B2 directly binds and inhibits AKT activity, we performed in vitro kinase assay by incubating purified recombinant GST-AKT protein with OPC-B2 for 15 min and found that OPC-B2 inhibited nearly 50% p-AKT at S473 compared with control (Fig. 2H). Furthermore, to map the potential interaction sites of OPC-B2 with AKT, we calculated the binding energy in silico with the mutation of potential amino acids. As shown in Table S2, Arg86, Arg273 and Lys297 showed the lowest interaction energies, suggesting that three amino acids residues are most likely to be the binding sites of OPC-B2 and AKT. Based on locations of these amino acids, we speculate that OPC-B2 may bind to the PH and kinase domains of AKT to inhibit its activity through locking it in a “closed” conformation (Fig. 2I), similar to the allosteric binding of MK-2206 to AKT. Because Arg86 (R86), Arg273 (R273) and Lys297 (K297) contributed over 50% to the total interaction energy between OPC-B2 and AKT (Fig. 2G and Table S2), we predicted that these three residues may play a key role in the interaction between AKT and OPC-B2.
To support this hypothesis, we individually mutated these three amino acids (R86, R273 and K297) to alanine (Ala) and investigated the effects of these mutants on the cell proliferation and AKT activity upon OPC-B2 treatment. Interestingly, R273A mutation significantly downregulated AKT protein expression, whereas R86A and K297A did not significantly affect AKT expression and activity (Figure S2F). When cells were transfected with Myc-AKT1 (wild type), Myc-AKT1-K297A, Myc-AKT1-R86A and empty plasmids (control), cell viability significantly increased compared with control group (empty plasmids) (Figure S2G), suggesting that K297A and R86A did not significantly alter the AKT activity. Importantly, OPC-B2 treatment inhibited cell viability around 50–60% in cells transfected with Myc-AKT1 (wild type) and control group, whereas OPC-B2 had no effects on the cell proliferation in the cells transfected with Myc-AKT1-K297A or Myc-AKT1-R86A, suggesting that K297 and R86 are indeed the critical sites for OPC-B2 binding and repression of p-AKT activity (Fig. 2J-L). Surprisingly, MK-2206 still inhibited the AKT activity and cell proliferation in K297A or R86A mutant cells, suggesting that OPC-B2 and MK-2206 bind to different sites in AKT (Figure S2H and I), consistent with a synergistic effect on AKT activity and cell proliferation with MK-2206 and OPC-B2 treatment (Figure S2B, C). Notably, previous studies found that tryptophan 80 (Trp80) [54] and Aspartic acid 292 (Asp292) were vital for the binding between MK-2206 and AKT [20]. Together, these data demonstrate that OPC-B2 directly binds to AKT and inhibits its kinase activity primarily through interactions with R86 and K297, different from the binding sites of the well-studied allosteric AKT inhibitor MK2206.
2.3. OPC-B2 suppresses p-AKT activity leading to cell cycle arrest
To investigate how OPC-B2 affects AKT and its downstream signalling pathways to inhibit tumour growth, we performed a transcriptomic RNA sequencing (RNA-Seq) analysis on Huh7 cells treated with OPC-B2 (Figure S3A). We found 2430 differentially expressed genes (DEGs), among which 534 genes were significantly up-regulated and 1896 genes were down-regulated compared to control group (Figure S3B). Interestingly, from our transcriptomic RNA-Seq data, we found that there were significant changes in the expressions of cell cycle and metabolic genes (Figure S3C and D). Next, we set out to explore the effects of OPC-B2 on the cell cycle of liver cancer cells. HCC cell lines, such as Huh7 cells and SMMC-7721 cells, were treated with OPC-B2 for 24 h with indicated doses. OPC-B2 indeed cell cycle arrest at G1 phase in Huh7 (Fig. 3A) and SMMC-7721 cells (Figure S3E and F). As shown in Fig. 3B, OPC-B2 significantly inhibited AKT activity (p-AKT) and cell cycle protein cyclin D1 expression and its downstream signaling, such as p-CDK4, p-Rb (S780) (Retinoblastoma Transcriptional Corepressor) in a dose-dependent manner (Fig. 3B and Figure S3G). Furthermore, cyclin D1 knockdown clearly decreased cell proliferation (Fig. 3C and D), consistent with previous reports in which overexpression of cyclin D1 promoted HCC development by facilitating cell-cycle progression [15,53]. Notably, OPC-B2 did not significantly affect apoptosis as indicated by Annexin V and PI staining as well as the apoptosis-related protein expressions, such as Bcl2, BAX, Caspase 3 and caspase 8 (Figure S3H-I). Previous studies also reported that AKT-GSK3β is a classic signaling pathway that regulates cyclin D1 stability through ubiquitination [2,19]. Activated AKT phosphorylates the serine 9 residue of glycogen synthase kinase-3β (GSK3β) and inactivates its kinase activity on cyclin D1, which promotes cyclin D1 protein stability and cell cycles transition from G1 to S via reducing its ubiquitination/degradation [2]. Consistently, we observed that OPC-B2 inhibited p-AKT activity, leading to the downregulation of its downstream p-GSK3β activity and cyclin D1 expression (Figure S4A). To further examine whether OPC-B2 inhibited p-AKT activity causes cyclin D1 decline through ubiquitination and subsequent proteasomal degradation, we treated Huh7 cells and SMMC-7721 cells with the proteasome inhibitor MG132, in addition to OPC-B2. Proteasomal inhibition by MG132 significantly attenuated the downregulation of cyclin D1 followed by OPC-B2 treatment (Fig. 3E and F). Moreover, OPC-B2 promoted cyclin D1 degradation primarily through enhancing cyclin D1 ubiquitination and proteasomal degradation without affecting protein synthesis (Fig. 3G, and Figure S4B-F). Taken together, p-AKT activity repression by OPC-B2 causes p-GSK3β downregulation and promotes cyclin D1 instability by ubiquitination/degradation, which leads to the further suppression of cyclin D1 downstream protein CDK4 and Rb phosphorylation. The net outcome of these signaling events is the induction of tumor cell cycle arrest at G1 phase and inhibition of cell proliferation (Fig. 3H).
Fig. 3.
Inhibiting phosphorylated AKT by OPC-B2 induces cell cycle arrest at G1 phase through promoting cyclin D1 degradation and ubiquitination. A.Cell cycle distribution of Huh7 cells measured by flow cytometry after treatment with OPC-B2 at doses of 50 and 75 μg/ml. B.The protein levels of cyclin D1, phosphor-CDK4, CDK4, phosphor-Rb, and Rb in Huh7 cells upon treatment with OPC-B2 (50 and 75 μg/ml) for 24 h. C.The protein levels of cyclin D1, CDK4, phosphor-Rb (S780) and Rb ofHuh7 cells were detected by western blotting assays after knockdown cyclin D1 expression by siRNA. D.Cell viability of Huh7 cells treated with siRNA-mediated cyclin D1 protein knockdown. E.Huh7 cells were exposed to 1X PBS or OPC-B2 (75 and 125 μg/ml) for 24 h, followed by treatment with a proteasome inhibitor, MG132 (10 μM). The protein levels of cyclin D1 were detected by western blotting assays. F.Immunoblot analysis (left) and quantification (right) for cyclin D1 in SMMC-7721 cells treated withOPC-B2 (0, 25 and 50 μg/ml) for 24 h, followed by treatment with a proteasome inhibitor, MG132 (10 μM) for 6 h. G.Polyubiquitylation levels of affinity purified cyclin D1-Flag proteins transfected with flag-cyclin D, HA-UB or both plasmids after treatments with 1X PBS or OPC-B2 (75 μg/ml). H.A schematic model: OPC-B2 promotes cyclin D1 protein degradation through the ubiquitin–proteasome pathway, thereby inducing cell cycle arrest.Two-tailed Student's t-test,*p < 0.05; **p < 0.01; ***p < 0.001.
2.4. OPC-B2 inhibits AKT activity and decreases metabolic flux in glycolysis and TCA cycle
Previous studies showed that AKT activity plays a critical role in supporting tumor cell metabolism [35]. As exemplified in the classic metabolic reprogramming of cancer cells, they increase their glucose uptake and preferentially utilize glucose through aerobic glycolysis, termed Warburg effect [23,51]. Besides, the tricarboxylic acid (TCA) cycle is pivotal for oxidative phosphorylation in cells, producing ATP and the intermediates needed for macromolecule biosynthesis [4,52]. Our recent study revealed a novel mode of metabolic reprogramming of HCC due to the loss of hepatic aldolase B [31]. Emerging studies have demonstrated that not only glycolysis but also TCA cycle is a potential target for cancer therapy. As a key enzyme in the first step of glycolysis, Hexokinase 1 (HK1) catalyzes the ATP-dependent phosphorylation of glucose to yield glucose-6-phosphate, which plays a vital role in the cellular uptake and utilization of glucose. Consistently, we observed that OPC-B2 inhibited glucose uptake and HK1 expression in a dose-dependent manner (Fig. 4A–B), consistent with AKT activity on regulation of HK1 protein expressions and glucose metabolism [27,40].
Fig. 4.
Inhibiting phosphorylated AKT by OPC-B2 decreases hexokinase 1 (HK1) expression and the metabolism in glycolysis and TCA cycle. A.Glucose consumption of Huh7 cells after treatment with OPC-B2 (50 and 75 μg/ml) at different time points. B.Protein expression of HK1, the first key enzyme of glycolysis, was detected by western blotting. C.13C distribution of glycolysis and the first turn of the TCA cycle with 50% U–13C-glucose (labelled at all six carbons) and 50% glucose (unlabeled). Red circles represent 13C-labelled carbon atoms while black circles indicate unlabeled carbon atoms. D, E.Huh7 cells were treated with OPC-B2 (50 and 75 μg/ml) and incubated with 50% U–13C-glucose for 12 (D) and 24 (E) hours. Fraction of the labelled metabolites of M+3 from 13C-glucose in glycolysis and fraction of the labelled metabolites of M+2 from 13C-glucose in TCA cycle by OPC-B2 treatment for 12 and 24 h in Huh7 cells. F. Quantification of glycolysis and TCA cycle metabolites in Huh7 cells detected by GC-MS after treatment with OPC-B2 and incubation with 13C6-glucose for 24 h.Two-tailed Student's t-test,*P < 0.05, **P < 0.01, ***P < 0.001. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
To further study the effects of OPC-B2 on metabolic reprogramming of cancer cells, we used universally labelled [U–13C6] glucose as a tracer to track the cellular metabolism in glycolysis and TCA cycles upon OPC-B2 treatment (Fig. 4C). The results showed that the fraction of M+3 labelled metabolites, including pyruvate, lactate and alanine from [U–13C6]-glucose, all decreased significantly in Huh7 cells treated by OPC-B2 at different time points, demonstrating that OPC-B2 dose-dependently inhibits glycolysis through downregulation of AKT-mediated inhibition of HK1 expression (Fig. 4D and E). Similar results were observed for TCA cycles with M+2 labelled metabolites (including succinate, malate, aspartate and glutamate) from [U–13C6]-glucose upon OPC-B2 treatment (Fig. 4D and E). Quantification of intermediate metabolites and the ratio of enriched labelled carbon of metabolites from the labelled glucose were consistent with the decreased metabolic flux to glycolysis and the TCA cycles (Fig. 4F and Figure S5A and B). Taken together, all these data demonstrate that OPC-B2 suppresses p-AKT activity and further downregulates HK1 expression and activity, leading to a decreased cellular metabolism in glycolysis and TCA to inhibit cancer cell growth.
2.5. OPC-B2 inhibits AKT activity and suppresses tumorigenesis in diethyl-nitrosamine (DEN)/CCl4-induced HCC model
Next, we employed a primary HCC mouse model to further investigate whether OPC-B2 exerts anti-tumor efficacy through inhibiting AKT activity and its downstream signaling in vivo. Three-week old C57/BL6 male mice were injected with DEN once and CCl4 twice a week from 8th to 20th week. OPC-B2 (300 mg/kg), MK-2206 (40 mg/kg) [25] and control solvent were administered intragastrically every three days from 12th to 39th week before the mice were sacrificed (Fig. 5A). As expected, mice treated with OPC-B2 or MK-2206 had significantly smaller ratios of liver/body weight without significantly affecting body weights (Figure S6A and B). OPC-B2 exhibited comparable anti-tumor effects as MK-2206: tumor number and maximal tumor size were significantly decreased after OPC-B2 and MK-2206 treatment (Fig. 5B–C). Interestingly, the liver function indicators, aspartate transaminase (AST) and alanine transaminase (ALT), were also significantly improved after OPC-B2 or MK-2206 treatment (Figure S6C). Consistently, immunohistochemistry (IHC) assay found that the levels of p-AKT at Ser473, cyclin D1 and proliferation marker Ki67 expressions were all significantly decreased in mouse tumors treated with OPC-B2 or MK-2206, compared to the control (Fig. 5D and E). Similar results were verified in western blot analysis (Fig. 5F). In short, all these data clearly demonstrate that OPC-B2 acts as a potent inhibitor of AKT to suppress liver tumor growth.
Fig. 5.
OPC-B2 suppresses tumorigenesis in the DEN/CCl4-induced mouse model. A.Schematic workflow of the DEN/CCl4-induced HCC mouse model. C57BL/6 mice (3 weeks old) were injected with DEN (50 μg/g) intraperitoneally. The mice were administrated with CCl4 (0.5 ml/kg) at 8th week of age via i.p. administration for 12 weeks, and treated with control solvent, OPC-B2 (300 mg/kg) or MK-2206 (40 mg/kg) at 12th week every three days via intragastric (i.g.) administration for 27 weeks. B.Representative images of mouse livers and HE staining were shown. Red-dotted circles indicate tumors while black-dotted lines indicate the boundary of normal tissues and tumor tissues. N, normal tissue; T, tumor tissue. Scale bar: 50 μm. C.Tumor number and maximal tumor size of C57BL/6 mouse liver (n = 5/4/4). D.Immunohistochemistry analysis of Ki67, cyclin D1 and phosphor-AKT (Thr308), Scale bar: 50 μm. E.Quantification of Ki67, cyclin D1 positive cells, and quantification of phosphorylated AKT at Thr308 from C57BL/6 mice treatment with OPC-B2 or MK-2206.F.Tumors were excised and analyzed for the expression of proteins of interest by western blotting. Data are represented as mean ± SEM; p values by two-tailed unpaired t-test.Two-tailed Student's t-test,*p < 0.05; **p < 0.01; ***p < 0.001. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3. Discussion
Polyphenolic compounds, such as proanthocyanidins, have been well-documented to have potent anticancer effects primarily due to their antioxidant and anti-inflammatory properties [1]. Emerging data have also suggested that these compounds may target other oncogenic pathways, such as PI3K/AKT, but the underlying mechanism and therapeutic potential remain elusive. On the other hand, targeting hyperactive AKT signaling pathways has been explored as a potential cancer therapy but with moderate success [7,12,26]. In this study, we discover that OPC-B2 acts as a new allosteric inhibitor of AKT through directly binding to AKT and inhibiting its downstream signaling events, leading to cell cycle arrest and inhibition of glucose metabolism (Fig. 6). These anti-tumor effects were demonstrated in xenograft mouse model and in the DEN/CCl4-induced primary liver cancer mouse model.
Fig. 6.
A working model of antitumor effects of OPC-B2 through direct binding and inhibition of AKT. OPC-B2 directly binds to AKT and inhibits its phosphorylation, thereby promoting cyclin D1 degradation through ubiquitination to induce cell cycle arrest. Inhibiting AKT activity also leads to the decreased HK1 expressions and inhibition of glycolysis and TCA cycle metabolism.The net outcome of OPC-B2 treatment is the suppression oftumor growth.
AKT inhibition has emerged as a potential therapeutic strategy for multiple cancers associated with hyperactive AKT. ATP competitive or allosteric inhibitors have been explored to achieve AKT inhibition. The GSK690693 is an aminofurazan derivative and ATP-competitive inhibitor used for the treatment of acute lymphoblastic leukemia [29]. However, the clinical development was terminated because of hyperglycemia [3]. AZD5363 is a potent, selective inhibitor of the kinase activity of all three AKT isoforms and has been used as monotherapy in the breast, gastric and prostate cancer in both phase I and phase II clinical trials [5]. In addition to ATP competitive inhibitors, allosteric inhibitors of AKT have also been tested as a therapy in many types of human cancers. For example, MK-2206 was used as a monotherapy in acute myelogenous leukemia and HCC in phase II clinical trials [37]. However, the AKT constitutive membrane binding caused by the E17K mutation in some types of human cancers causes AKT to be less sensitive to MK2206 [43]. Therefore, it is of great significance to develop novel inhibitors with lower toxicity and better therapeutic efficacy for HCC therapy.
In this study, we have discovered that OPC-B2 extracted from peanut skin acts as an allosteric inhibitor of AKT, similar to MK-2206, but with different interaction amino acids. Identification of this novel mode of action may enable us to design allosteric inhibitors with diverse chemical structures. The main forces were conventional hydrogen bonding (Fig. 2F), Pi-Cation, charge-charge, and Pi-Pi T-shaped and Pi-Alkyl interaction (Figure S2E). Based on the locations of these amino acid residues, we speculate that OPC-B2 binds to the PH domain and kinase domains of AKT, forming a pocket at the interface of these two domains to lock the kinase in a closed conformation (Table S2). Besides, Lys297 and Arg86 are critical for the binding between OPC-B2 and AKT (Fig. 2G–J and Figure S2E-I). Lys297 is a key amino acid that OPC-B2 binds to AKT through Pi-bond, whereas Arg86 is important for the formation of hydrogen bond between the phenolic hydrogen atom on OPC-B2 and AKT (Figure S2E). Consistently, mutation of these two amino acids to alanine completely abolished the inhibition of OPC-B2 on AKT and the downstream signaling events leading to cell proliferation (Fig. 2K and L). Although OPC-B2 and MK-2206 showed synergistic effects on AKT inhibition, OPC-B2 appears to bind to the similar pocket to MK-2206 due to the close proximity of the critical amino acids they interact with: Trp80 [54] and Asp292 for MK-2206 while Arg86 and Lys297 for OPC-B2, respectively. Importantly, future studies are needed to examine whether OPC-B2 can be used as an adjuvant to enhance therapeutic effects on HCC treatment.
We further clarified the underlying mechanism and downstream signaling events leading to AKT inhibition by OPC-B2. Although AKT is known to regulate multiple signaling pathways associated with the pathogenesis of liver cancer, we have discovered that AKT downstream protein cyclin D1 (Fig. 3) and HK1 (Fig. 4B) are reduced by OPC-B2, which are critically important for cell cycle arrest and inhibition of glycolysis, respectively. Previous studies showed that AKT phosphorylates GSK3β to inhibit its activity, which leads to the decrease of ubiquitination level of cyclin D1 and promotes cell cycle progression and proliferation [2]. Our results have indicated that OPC-B2 can inhibit AKT activity and increase cyclin D1 ubiquitination to promote its degradation, thereby inducing tumor cell cycle arrest at G1 phase. The efficacy of OPC-B2 on cell cycle-related proteins was abolished by the mutation of AKT at Lys297 and Arg86 (Fig. 2J). In addition, HK1 is the key enzyme in glycolysis. AKT can regulate its expression and activity by upregulation of HIF1α through mTORC1 activation [24,41]. The reduction of HK1 by OPC-B2 reduced tumor cell glucose consumption (Fig. 4A), glycolysis, and TCA metabolism (Fig. 4D, E and Figure S5A, B). It is well-established that metabolic reprogramming has been regarded as a core hallmark of cancer, which sustains rapid proliferation and survival of cancer cells [8,16]. Our recent study has identified a novel metabolic reprogramming in HCC due to the loss of glycolytic enzyme aldolase B and targeting this metabolic pathway has potential therapeutic implications [31]. Taken together, our data clearly demonstrate that OPC-B2 directly inhibits AKT activity, leading to cell cycle arrest and reduced metabolic flux to glycolysis and TCA to suppress tumor growth. Nonetheless, it warrants future studies on the potential side effects of AKT inhibition by OPC-B2 on normal cell proliferations.
In summary, we have uncovered a novel mechanism underlying antitumor effects of OPC-B2 beyond its well-documented antioxidant and anti-inflammatory properties. Our work indicates that hyperactive AKT in HCC can be restrained by a direct binding of OPC-B2 to AKT. Interestingly, OPC-B2 appears to bind to the catalytically active kinase domain and regulatory PH domain to lock the protein in a closed conformation, similar to the well-studied AKT allosteric inhibitor, MK-2206. And we further found that Lys297 and Arg86 of AKT are two key amino acid residues in the binding of OPC to AKT. The inhibitory effect on AKT by OPC-B2 causes a decrease in its downstream proteins cyclin D1 and HK1, which leads to the cell cycle arrest and inhibition of tumor cell metabolisms (Fig. 6). Interestingly, however, our data show that OPC-B2 inhibited AKT activity plays an important role in liver carcinogenesis and tumor growth. Future study is needed to clarify how these two pathways may be differentially regulated by OPC-B2. Together, our study has demonstrated that OPC-B2 is an allosteric inhibitor of AKT with a great potential in clinical application for liver cancer therapy. Furthermore, this novel model of action may help to develop new allosteric inhibitors of AKT for cancer treatment.
4. Materials and methods
4.1. Reagents and antibodies
Reagents and antibodies are shown in Table S3 and S4.
4.2. Purification of OPC-B2 from peanut skin
Peanut skin powder was subject to water extraction at 45 °C with the solid/liquid ratio of 1:9 for 2.5 h, followed by a secondary water extraction of the residue of step 1 at 45 °C with solid/liquid ratio of 1:72 for 0.5 h. After filtration using 100 mesh screen, to the combined aqueous fractions was added 0.02% of Food Grade polyaluminium chloride to remove protein, heavy metal and other impurities. After high-speed centrifugation at 10000 rpm for 10 min using Plant fiber filter cloth, a clear extract was obtained. After HP20 macroporous resin adsorption and gradient elution using 10–95% ethanol water, the OPC-B2 fraction was collected in HPLC separation. After evaporation under vacuum at 65 °C, the residue was subject to spray drying with Inlet air temperature of 200 °C and outlet air temperature of 65 °C. The OPC-B2 extraction powder was obtained with yield of 12.51% and purity of 98.7%. The characterization of the final compounds was based on 1H and 13C NMR as well as high resolution MS coupled to LC by comparing with the commercially available standard from Sigma (Cat: 42157-1 MG-F).
4.3. Cell culture
All cells were purchased from Cell Bank, Chinese Academy of Science (Shanghai, China). Unless otherwise specified, all cell lines (Huh7 and Smmc-7721) were cultured in Dulbecco Minimal Essential Medium (DMEM, Hyclone) with 10%FBS (Gibco) and 1% penicillin/streptomycin (15140–142, Gibco) in a humidified incubator with 5% CO2 at 37 °C. For serum starvation of cells, after medium was removed and cells were washed with PBS, DMEM without FBS was added to cells for 12 h. Cells were treated with OPC-B2 or MK-2206 for 24 h. After stimulation, cells were washed with ice-cold PBS before cell pellets were either scraped in lysis buffer or collected for further analysis. For plasmids or siRNA transfection, flag-cyclin D1 plasmid or siRNA of cyclin D1 was transfected with Huh7 cells for 8 h, and then cells were washed with PBS and replaced fresh medium for 48 h. To determine cyclin D1 protein stability, we treated Huh7 or SMMC-7721 cells with OPC-B2 for 24 h, followed by treatment with MG132 (10μM) for 6 h. All transformed cells were tested negative for mycoplasma using an in-house PCR-based methods.
4.4. RNA-seq of Huh7 cells
Huh7 cells were administrated with control solvent or OPC-B2 (75 μg/ml) for 24 h. Cells were lysed with RNAiso reagent. The library preparation and sequencing were carried out by Beijing Genomics Institute (BGI), Shenzhen, China. We measured the expression of the transcript isoforms using RSEM and used the NOISeq method to screen differentially expressed genes (DEGs) between 2 groups. Microarray data is available here:
4.5. Cell proliferation assays
The proliferation was performed by CCK8 (OBIO Cell Counting Kit). CCK8 assay was carried out according to the standard protocol by seeding cells in a 96-well plate at a density of 2000 cells/well for 6 or 8 repeats. After 7 h, cell media were replaced with fresh media with various concentrations of test agents or control solvent and cells were proceeded to grow. Then the cell absorbance was measured at 450 nm at different time points. Cells in each well were treated by 10 μl CCK8 and 100 μl DMEM (FBS-free and penicillin-streptomycin free). Statistical significance of the difference between samples and control was obtained by Student's t-test.
4.6. Colony formation assay
Cells were seeded into 6-well plates (300 or 600 cells/well) and cultured for 7–10 days until formation of visible colonies. Cell media were changed every other day. Colonies were washed with PBS and fixed with 10% acetic acid/10% methanol for 20 min, then stained with 0.4% crystal violet in 20% ethanol for 20 min. After staining, the plates were washed and air-dried, and colony numbers were counted.
4.7. Cell cycle assay
Huh7 or Smmc-7721 cells were plated (2 × 105 cells/well) into six-well plates and subjected to indicated treatment. About 1 × 106 cells were detected using the cell cycle analysis kit (Beyotime Biotechnology, C1052). Flow cytometer (BD) was used to detect the cell cycle distribution at 450 nm. Cellular DNA content analysis and light scattering analysis were performed using FlowJo 7.6.
4.8. Apoptosis analysis
FITC Annexin V Apoptosis Detection Kit I (BD Pharmingen, 556547) was used to detect cell apoptosis. Briefly, huh7 cells were plated (2 × 105 cells/well) into six-well plates and treated with OPC-B2 for 24 h. Cells were suspended in 100 μl binding buffer, 4 μl Annexin V antibody conjugated with fluorescein isothiocyanate (FITC) and 4 μl PI. After incubation for 15 min at room temperature, shielded from light, then added another 400 μl binding buffer, the percentage of apoptotic cells was determined by flow cytometry (BD).
4.9. Western blotting
Cells were rinsed twice with ice-cold PBS and lysed in ice-cold lysis buffer (25 mM Tris pH 8.0, 150 mM NaCl, 1 mM CaCl2, 1% Triton X-100) and EDTA-free protease inhibitors (Biotool), Cell lysates were incubated on ice for 30 min. The soluble fractions of cell lysates were isolated by centrifugation at 4, 12,000 rpm for 10 min. Protein concentrations were determined using the Enhanced BCA Protein Assay Kit (Beyotime). Aliquots of proteins were boiled in 1 × loading buffer for 10 min. In western blotting experiments, samples containing 20 μg of total proteins were separated on 10% SDS-polyacrylamide gels, and transferred to Poly (Vinylidene Fluoride) (PVDF, Millipore) membranes, and incubated with primary antibodies overnight. Horseradish peroxidase-conjugated goat anti-mouse and anti-rabbit IgG were used as secondary antibodies to incubate for 1 h at room temperature. Proteins were visualized using a Chemiluminescence kit and Chemiluminescence imaging instrument (GE Image Quant LAS4000 mini).
4.10. Ubiquitination assay
For the detection of cyclin D1 ubiquitination, we transfected huh7 or Smmc-7721 cells with HA-UB and flag-cyclin D1, respectively, or transfected with both plasmids. Then fresh medium was placed into the dish 6 h post-transfection, and OPC-B2 was added to Huh7 or Smmc-7721 cells after 24 h of transfection. The cells were treated with MG132 for 6 h before protein collection. After cell lysates were quantified for protein concentration, 1–2 mg of protein was used for IP assay, while the remaining protein was used as input. Proteins were incubated with flag beads overnight at 4 °C, then affinity gel were collected by 300 g centrifuging for 4 min at 4 °C, and then affinity gel was washed five times by wash buffer (150 mM Tris-HCl, 400 mM NaCl, 0.8% Triton X-100, pH 7.4). The subsequent operations were similar to western blotting analysis.
5. Plasmids and RNA-mediated interference (RNAi)
5.1. Plasmids construction
Cyclin D1 and AKT1 expression plasmids were constructed by cloning the open reading frame of each cDNA into the multiple cloning site of pcDNA3.0 vector. Flag or myc tag was constructed through designing in one primer. The different gene fragments were amplified using their primers. pcDNA3.0 vector and each PCR fragments were both digested with their restriction enzymes EcoRI and XhoI or BamHI and XhoI or EcoRI and XhoI, and then T4 ligase was applied for ligation. Transformation was performed using DH5α competent cells. Single clones were picked and amplified to extract plasmids. All the clones were sequenced to confirm the results.
5.2. Plasmids or RNA interference
3.5 × 105 cells were seeded in a 6-well plate. After 12 h incubation, plasmid or siRNA was transfected into cells by Lipofectamine 2000 Transfection Reagent (Invitrogen), then fresh medium was replaced into the dish 6 h post-transfection and lasted for 48 h. Then cells were lysed using cell lysis buffer to perform Western Blot measurement. SiRNA sequences are showed below.
si-cyclin D1-1-F: 5′-CCUCGGUGUCCUACUUCAAdTdT-3’.
si-cyclin D1-1-R:5′-UUGAAGUAGGACACCGAGGdTdT-3’.
si-cyclin D1-2-F:5′-CCACAGAUGUGAAGUUCAUdTdT-3’.
si-cyclin D1-2-R:5′-AUGAACUUCACAUCUGUGGdTdT-3’.
5.3. In vitro AKT1 kinase assay
We performed in vitro kinase assay by incubating purified recombinant GST-AKT protein with OPC-B2 for 15 min, then detected the phosphorylation level of AKT1 at S473 by Western Blotting.
5.4. Glycose consumption assay
Huh7 cells were treated with OPC-B2 at doses of 50 and 75 μg/ml for 12, 24, 48 and 72 h. Medium was taken at different time points for glucose detection by the d-Glucose Assay Kit (R-Biopharm, 10716251035) according to the manufacturer's protocol.
5.5. Metabolic flux experiments using [U–13C6]-glucose
The metabolic flux analysis using [U–13C6]-glucose followed our previously published protocol [49]. Labelled medium was used to replace the unlabeled medium when cells grew up to ~60 % confluence, followed by treatment with OPC-B2 (50 and 75 μg/ml) for 12 h or 24 h.
Molecular docking and dynamic simulation of OPC-B2 inhibiting AKT1.
The binding of OPC-B2 to AKT1 was evaluated through the molecular docking and dynamic test using Discovery studio software (DS 4.0) (BIOVIA, San Diego, CA, USA) based on previous methods [42]. The main procedure was as follows. 1) Preparing the structure: The structure of OPC-B2 was first created using the SMALL MOLECULAR tool in Discovery studio software and then one step of optimization was employed to obtain the effective structure. The native crystal structure of AKT1 was obtained from Protein Data Bank (protein entry 3O96; resolution at 2.00 Å). 2) Defining the active site: After loading the structure of AKT1 into Discovery studio software, all water molecules and ligands were removed and CHARMs force field was applied using PREPARE PROTEIN tool in DS 4.0. Then we chose to define the active sites based on the active cavities of receptor. 10 different active sites could be produced. 3) Molecular docking: CDOCKER tools in DS 4.0 were used for the molecular docking. We first loaded the structure of OPC-B2 and AKT1 into DS 4.0 and then chose the active site with the largest diameter. The CHARMs force field was used as a default during the whole docking process. 4) Conformation Scoring: After docking process, top 10 conformation poses were generated for each ligand based on docking score. The conformation with the highest -CDocker energy (score) was selected for further analysis. 5) Dynamic simulation: The molecular dynamics simulation was performed using SIMULATION tool in the DS 4.0 with CHARMs force field. Specifically, ACE-peptide complex was first pretreated in the absence of water molecule and later a 7 Å solvation shell was added. At same time, 0.145 M NaCl was used to simulate the human environment. Two minimization cycles (steepest descent and conjugate gradient) were performed until the RMS of energy gradient was <0.1 kcal/mol·Å. The steepest descent cycle was performed with 2000 steps (time step: 0.001ps) while conjugate gradient was performed with 1000 steps (time step: 0.001ps). The SHAKE algorithm was applied throughout the MD simulation to hold all the bonds involving hydrogen atoms. The long-range electrostatic forces were treated with PME method. After minimization, the sample was gradually heated to a target temperature from 50K to 300K over an interval of 5ps. After this heating process, 5000 steps long (time step: 0.002ps) equilibration phase was applied. The production stage was performed in 50000 steps using a time step of 0.002ps using NPT canonical assembly. The decay time for the temperature coupling was 5.0 ps. 6) Conformation analysis: After the molecular dynamic simulation, 50 conformation poses were produced and we could obtain the pose with lowest total energy. Then the interaction energy of the ligand-receptor interaction was calculated and interaction sites were illustrated in 2D and 3D images. And the main interaction forces and key amino acids could be illustrated. 7) Mechanism verification: Once we got the main amino acids contributed to the interaction between ligands and AKT1, we adopted the “site-specific mutagenesis” method to confirm their key acting sites for AKT1 inhibition. We first modified the specific amino acids of AKT1 with alanine (ALA) and then used this new AKT1 to carry out the molecular docking and dynamic simulation based on the main procedures from 1) to 6). Then the structure, interaction energy and forces of the final ligand-receptor compound would be compared with the form one and we could conclude which amino acid was the key acting site for AKT1 inhibition of OPC-B2.
5.6. Mouse models
All animal experimental protocols were approved by the Institutional Animal Care and Use Committee at Shanghai Institute of Nutrition and Health (SINH), Chinese Academy of Sciences (CAS), Shanghai, China.
5.7. Xenograft mouse model
All xenograft studies of male nude mice (5 weeks old) were purchased from Shanghai Slack Laboratory Animal Corp. Ltd. We injected subcutaneously both the left and right flanks of each male BALB/c nude mouse by with 2 × 106 Huh7 cells/100 μl. Tumor size was measured every two days, and OPC-B2 was administrated at a dose of 10 mg/kg or 30 mg/kg via intraperitoneal (i.p) injection every two days. The control group received the vehicle, 1 X PBS only. The tumor volume was calculated with the equation V (in mm3) = 0.52 x length x width2. After injection for three weeks, all mice were killed and tumors were retrieved and weighed for further analysis.
5.8. DEN/CCl4-induced mouse model of primary liver cancer
DEN/CCl4-induced mouse model of primary liver cancer was established according to a previous protocol [49]. In brief, C57/BL6 mice (3 weeks old) were purchased from Shanghai Slack Laboratory Animal Corp. Ltd. DEN was injected into the mice by intraperitoneal injection at a dose of 40 μg/g. CCl4 (0.5 ml/kg, dissolved in olive oil) was administered by intraperitoneal (i.p) injection from week 8–12 for twice a week. OPC-B2 (300 mg/kg), MK-2206 (40 mg/kg) and control solvent were administered intragastrically (i.g) every three days from week 12–39 before the mice were sacrificed. Liver and plasma were harvested for further analysis.
5.9. Statistical analysis
For all experiments, including cell experiments and animal experiments, two-tailed Student's t-test was performed by GraphPad Prism 6.0. All data were reported as the means ± SD or means ± SEM from at least three independent experiments. Differences were considered statistically significant at P ≤ 0.05 and statistics significance was shown as *p < 0.05; **p < 0.01; ***p < 0.001.
Data availability
Microarray data is available here: OEP000451 (https://www.biosino.org/node/project/detail/OEP000451).
Author contributions
H.Y.Y and Y.Z.T designed this study. G.J.L., N.N.W., M.L., X.X.H., C.Z.Y. Q.C.T. performed experiments. G.J.L, Y.Z.T. and H.Y.Y., wrote the paper. X.S. perform the data analysis of RNAseq. OPC-B2 required for all experiments were provided by A.M.S and Q.W. Analysis of molecular docking and dynamic simulation of OPC-B2 inhibiting AKT1 was performed by A.M.S.
Declaration of competing interest
The authors declare no competing financial interests.
Acknowledgements
This work was financially supported by National Key R&D Program of China administered by Chinese Ministry of Science and Technology (MOST) (2016YFD0400205 and 2018YFA0800300), the National Natural Science Foundation of China (31671231 and 91857112) and Young Elite Scientist Sponsorship program by CAST (2018 QNRC001), Agricultural Science and Technology Innovation Project (CAAS-ASTIP-201X-IAPPST). We thank molecular biology/biochemistry/cell technology platform, experimental animal platform, mass spectrometry core lab, and biological sample pathology analysis platform in Shanghai Institute of Nutrition and Health, Chinese Academy of Sciences.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2020.101701.
Contributor Information
Yongzhen Tao, Email: yztao01@sibs.ac.cn.
Qiang Wang, Email: wangqiang06@caas.cn.
Huiyong Yin, Email: hyyin@sibs.ac.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Al-Ishaq R.K., Overy A.J., Büsselberg D. Phytochemicals and gastrointestinal cancer: cellular mechanisms and effects to change cancer progression. Biomolecules. 2020;10 doi: 10.3390/biom10010105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alt J.R., Cleveland J.L., Hannink M., Diehl J.A. Phosphorylation-dependent regulation of cyclin D1 nuclear export and cyclin D1-dependent cellular transformation. Genes Dev. 2000;14:3102–3114. doi: 10.1101/gad.854900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altomare D.A., Zhang L., Deng J., Di Cristofano A., Klein-Szanto A.J., Kumar R., Testa J.R. GSK690693 delays tumor onset and progression in genetically defined mouse models expressing activated Akt. Clin. Canc. Res. 2010;16:486–496. doi: 10.1158/1078-0432.CCR-09-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson N.M., Mucka P., Kern J.G., Feng H. The emerging role and targetability of the TCA cycle in cancer metabolism. Protein Cell. 2018;9:216–237. doi: 10.1007/s13238-017-0451-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Banerji U., Dean E.J., Perez-Fidalgo J.A., Batist G., Bedard P.L., You B., Westin S.N., Kabos P., Garrett M.D., Tall M. A phase I open-label study to identify a dosing regimen of the pan-AKT inhibitor AZD5363 for evaluation in solid tumors and in PIK3CA-mutated breast and gynecologic cancers. Clin. Canc. Res. 2018;24:2050–2059. doi: 10.1158/1078-0432.CCR-17-2260. [DOI] [PubMed] [Google Scholar]
- 6.Bellacosa A., Kumar C.C., Cristofano A.D., Testa J.R. 2005. Activation of AKT Kinases in Cancer: Implications for Therapeutic Targeting; pp. 29–86. [DOI] [PubMed] [Google Scholar]
- 7.Biondi R.M., Komander D., Thomas C.C., Lizcano J.M., Deak M., Alessi D.R., van Aalten D.M. High resolution crystal structure of the human PDK1 catalytic domain defines the regulatory phosphopeptide docking site. EMBO J. 2002;21:4219–4228. doi: 10.1093/emboj/cdf437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boroughs L.K., DeBerardinis R.J. Metabolic pathways promoting cancer cell survival and growth. Nat. Cell Biol. 2015;17:351–359. doi: 10.1038/ncb3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calleja V., Alcor D., Laguerre M., Park J., Vojnovic B., Hemmings B.A., Downward J., Parker P.J., Larijani B. Intramolecular and intermolecular interactions of protein kinase B define its activation in vivo. PLoS Biol. 2007;5:e95. doi: 10.1371/journal.pbio.0050095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calleja V., Laguerre M., Parker P.J., Larijani B. Role of a novel PH-kinase domain interface in PKB/Akt regulation: structural mechanism for allosteric inhibition. PLoS Biol. 2009;7:e17. doi: 10.1371/journal.pbio.1000017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cantley L.C. The phosphoinositide 3-kinase pathway. 2002;296:1655–1657. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- 13.Choy Y.Y., Fraga M., Mackenzie G.G., Waterhouse A.L., Cremonini E., Oteiza P.I. The PI3K/Akt pathway is involved in procyanidin-mediated suppression of human colorectal cancer cell growth. Mol. Carcinog. 2016;55:2196–2209. doi: 10.1002/mc.22461. [DOI] [PubMed] [Google Scholar]
- 14.Chung V., McDonough S., Philip P.A., Cardin D., Wang-Gillam A., Hui L., Tejani M.A., Seery T.E., Dy I.A., Al Baghdadi T. Effect of selumetinib and MK-2206 vs oxaliplatin and fluorouracil in patients with metastatic pancreatic cancer after prior therapy: SWOG S1115 study randomized clinical trial. JAMA Oncol. 2017;3:516–522. doi: 10.1001/jamaoncol.2016.5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deane N.G., Parker M.A., Aramandla R., Diehl L., Lee W.J., Washington M.K., Nanney L.B., Shyr Y., Beauchamp R.D. Hepatocellular carcinoma results from chronic cyclin D1 overexpression in transgenic mice. Canc. Res. 2001;61:5389–5395. [PubMed] [Google Scholar]
- 16.DelNero P., Hopkins B.D., Cantley L.C., Fischbach C. Cancer metabolism gets physical. Sci. Transl. Med. 2018;10 doi: 10.1126/scitranslmed.aaq1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deprez J., Vertommen D., Alessi D.R., Hue L., Rider M.H. Phosphorylation and activation of heart 6-phosphofructo-2-kinase by protein kinase B and other protein kinases of the insulin signaling cascades. J. Biol. Chem. 1997;272:17269–17275. doi: 10.1074/jbc.272.28.17269. [DOI] [PubMed] [Google Scholar]
- 18.Ediriweera M.K., Tennekoon K.H., Samarakoon S.R. Role of the PI3K/AKT/mTOR signaling pathway in ovarian cancer: biological and therapeutic significance. Semin. Canc. Biol. 2019;59:147–160. doi: 10.1016/j.semcancer.2019.05.012. [DOI] [PubMed] [Google Scholar]
- 19.Eo H.J., Park G.H., Jeong J.B. The involvement of cyclin D1 degradation through GSK3beta-mediated threonine-286 phosphorylation-dependent nuclear export in anti-cancer activity of mulberry root bark extracts. Phytomedicine. 2016;23:105–113. doi: 10.1016/j.phymed.2015.12.007. [DOI] [PubMed] [Google Scholar]
- 20.Fang Z., Simard J.R., Plenker D., Nguyen H.D., Phan T., Wolle P., Baumeister S., Rauh D. Discovery of inter-domain stabilizers-a novel assay system for allosteric akt inhibitors. ACS Chem. Biol. 2015;10:279–288. doi: 10.1021/cb500355c. [DOI] [PubMed] [Google Scholar]
- 21.Fracassetti D., Costa C., Moulay L., Tomas-Barberan F.A. Ellagic acid derivatives, ellagitannins, proanthocyanidins and other phenolics, vitamin C and antioxidant capacity of two powder products from camu-camu fruit (Myrciaria dubia) Food Chem. 2013;139:578–588. doi: 10.1016/j.foodchem.2013.01.121. [DOI] [PubMed] [Google Scholar]
- 22.Fruman D.A., Chiu H., Hopkins B.D., Bagrodia S., Cantley L.C., Abraham R.T. The PI3K pathway in human disease. Cell. 2017;170:605–635. doi: 10.1016/j.cell.2017.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gillies R.J., Gatenby R.A. Adaptive landscapes and emergent phenotypes: why do cancers have high glycolysis? J. Bioenerg. Biomembr. 2007;39:251–257. doi: 10.1007/s10863-007-9085-y. [DOI] [PubMed] [Google Scholar]
- 24.Gordan J.D., Thompson C.B., Simon M.C. HIF and c-Myc: sibling rivals for control of cancer cell metabolism and proliferation. Canc. Cell. 2007;12:108–113. doi: 10.1016/j.ccr.2007.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han P., Li H., Jiang X., Zhai B., Tan G., Zhao D., Qiao H., Liu B., Jiang H., Sun X. Dual inhibition of Akt and c-Met as a second-line therapy following acquired resistance to sorafenib in hepatocellular carcinoma cells. Mol. Oncol. 2017;11:320–334. doi: 10.1002/1878-0261.12039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hopkins B.D., Goncalves M.D., Cantley L.C. Insulin-PI3K signalling: an evolutionarily insulated metabolic driver of cancer. Nat. Rev. Endocrinol. 2020 doi: 10.1038/s41574-41020-40329-41579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kang M., Lee S.M., Kim W., Lee K.H., Kim D.Y. Fubp1 supports the lactate-Akt-mTOR axis through the upregulation of Hk1 and Hk2. Biochem. Biophys. Res. Commun. 2019;512:93–99. doi: 10.1016/j.bbrc.2019.03.005. [DOI] [PubMed] [Google Scholar]
- 28.Kim E., Lisby A., Ma C., Lo N., Ehmer U., Hayer K.E., Furth E.E., Viatour P. Promotion of growth factor signaling as a critical function of beta-catenin during HCC progression. Nat. Commun. 2019;10:1909. doi: 10.1038/s41467-019-09780-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levy D.S., Kahana J.A., Kumar R. AKT inhibitor, GSK690693, induces growth inhibition and apoptosis in acute lymphoblastic leukemia cell lines. Blood. 2009;113:1723–1729. doi: 10.1182/blood-2008-02-137737. [DOI] [PubMed] [Google Scholar]
- 30.Li D.L., Li X.M., Peng Z.Y., Wang B.G. Flavanol derivatives from Rhizophora stylosa and their DPPH radical scavenging activity. Molecules. 2007;12:1163–1169. doi: 10.3390/12051163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li M., He X., Guo W., Yu H., Zhang S., Wang N., Liu G., Sa R., Shen X., Jiang Y. Aldolase B suppresses hepatocellular carcinogenesis by inhibiting G6PD and pentose phosphate pathways. Nat. Canc. 2020;1:735–747. doi: 10.1038/s43018-020-0086-7. [DOI] [PubMed] [Google Scholar]
- 32.Liu P., Begley M., Michowski W., Inuzuka H., Ginzberg M., Gao D., Tsou P., Gan W., Papa A., Kim B.M. Cell-cycle-regulated activation of Akt kinase by phosphorylation at its carboxyl terminus. Nature. 2014;508:541–545. doi: 10.1038/nature13079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Llovet J.M., Montal R., Villanueva A. Randomized trials and endpoints in advanced HCC: role of PFS as a surrogate of survival. J. Hepatol. 2019;70:1262–1277. doi: 10.1016/j.jhep.2019.01.028. [DOI] [PubMed] [Google Scholar]
- 34.Ma C.X., Suman V., Goetz M.P., Northfelt D., Burkard M.E., Ademuyiwa F., Naughton M., Margenthaler J., Aft R., Gray R. A phase II trial of neoadjuvant MK-2206, an AKT inhibitor, with anastrozole in clinical stage II or III PIK3CA-mutant ER-positive and HER2-negative breast cancer. Clin. Canc. Res. 2017;23:6823–6832. doi: 10.1158/1078-0432.CCR-17-1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manning B.D., Toker A. AKT/PKB signaling: navigating the network. Cell. 2017;169:381–405. doi: 10.1016/j.cell.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nandakumar V., Singh T., Katiyar S.K. Multi-targeted prevention and therapy of cancer by proanthocyanidins. Canc. Lett. 2008;269:378–387. doi: 10.1016/j.canlet.2008.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nitulescu G.M., Margina D., Juzenas P., Peng Q., Olaru O.T., Saloustros E., Fenga C., Spandidos D., Libra M., Tsatsakis A.M. Akt inhibitors in cancer treatment: the long journey from drug discovery to clinical use (Review) Int. J. Oncol. 2016;48:869–885. doi: 10.3892/ijo.2015.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prasain J.K., Barnes S. Chapter 41 - uptake and metabolism of dietary proanthocyanidins. In: Watson R.R., Preedy V.R., Zibadi S., editors. Polyphenols in Human Health and Disease. Academic Press; San Diego: 2014. pp. 553–560. [Google Scholar]
- 39.Revathidevi S., Munirajan A.K. Akt in cancer: mediator and more. Semin. Canc. Biol. 2019 doi: 10.1016/j.semcancer.2019.06.002. [DOI] [PubMed] [Google Scholar]
- 40.Robey R.B., Hay N. Mitochondrial hexokinases, novel mediators of the antiapoptotic effects of growth factors and Akt. Oncogene. 2006;25:4683–4696. doi: 10.1038/sj.onc.1209595. [DOI] [PubMed] [Google Scholar]
- 41.Sengupta S., Peterson T.R., Sabatini D.M. Regulation of the mTOR complex 1 pathway by nutrients, growth factors, and stress. Mol. Cell. 2010;40:310–322. doi: 10.1016/j.molcel.2010.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shi A., Liu H., Liu L., Hu H., Wang Q., Adhikari B. Isolation, purification and molecular mechanism of a peanut protein-derived ACE-inhibitory peptide. PloS One. 2014;9:e111188. doi: 10.1371/journal.pone.0111188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shoji K., Oda K., Nakagawa S., Hosokawa S., Nagae G., Uehara Y., Sone K., Miyamoto Y., Hiraike H., Hiraike-Wada O. The oncogenic mutation in the pleckstrin homology domain of AKT1 in endometrial carcinomas. Br. J. Canc. 2009;101:145–148. doi: 10.1038/sj.bjc.6605109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2019. Ca - Cancer J. Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 45.Snow A.D., Castillo G.M., Nguyen B.P., Choi P.Y., Cummings J.A., Cam J., Hu Q., Lake T., Pan W., Kastin A.J. The Amazon rain forest plant Uncaria tomentosa (cat's claw) and its specific proanthocyanidin constituents are potent inhibitors and reducers of both brain plaques and tangles. Sci. Rep. 2019;9:561. doi: 10.1038/s41598-019-38645-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vanhaesebroeck B., Guillermet-Guibert J., Graupera M., Bilanges B. The emerging mechanisms of isoform-specific PI3K signalling. Nat. Rev. Mol. Cell Biol. 2010;11:329–341. doi: 10.1038/nrm2882. [DOI] [PubMed] [Google Scholar]
- 47.Villanueva A. Hepatocellular carcinoma. N. Engl. J. Med. 2019;380:1450–1462. doi: 10.1056/NEJMra1713263. [DOI] [PubMed] [Google Scholar]
- 48.Wang G., Long J., Gao Y., Zhang W., Han F., Xu C., Sun L., Yang S.C., Lan J., Hou Z. SETDB1-mediated methylation of Akt promotes its K63-linked ubiquitination and activation leading to tumorigenesis. Nat. Cell Biol. 2019;21:214–225. doi: 10.1038/s41556-018-0266-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang N., Liu H., Liu G., Li M., He X., Yin C., Tu Q., Shen X., Bai W., Wang Q. Yeast beta-D-glucan exerts antitumour activity in liver cancer through impairing autophagy and lysosomal function, promoting reactive oxygen species production and apoptosis. Redox Biol. 2020;32:101495. doi: 10.1016/j.redox.2020.101495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang W., Cheng J.W., Qin J.J., Hu B., Li X., Nijampatnam B., Velu S.E., Fan J., Yang X.R., Zhang R. MDM2-NFAT1 dual inhibitor, MA242: effective against hepatocellular carcinoma, independent of p53. Canc. Lett. 2019;459:156–167. doi: 10.1016/j.canlet.2019.114429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 52.Weinberg S.E., Chandel N.S. Targeting mitochondria metabolism for cancer therapy. Nat. Chem. Biol. 2015;11:9–15. doi: 10.1038/nchembio.1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu S.Y., Lan S.H., Wu S.R., Chiu Y.C., Lin X.Z., Su I.J., Tsai T.F., Yen C.J., Lu T.H., Liang F.W. Hepatocellular carcinoma-related cyclin D1 is selectively regulated by autophagy degradation system. Hepatology. 2018;68:141–154. doi: 10.1002/hep.29781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu W.I., Voegtli W.C., Sturgis H.L., Dizon F.P., Vigers G.P., Brandhuber B.J. Crystal structure of human AKT1 with an allosteric inhibitor reveals a new mode of kinase inhibition. PloS One. 2010;5 doi: 10.1371/journal.pone.0012913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang J.D., Roberts L.R. Hepatocellular carcinoma: a global view. Nat. Rev. Gastroenterol. Hepatol. 2010;7:448–458. doi: 10.1038/nrgastro.2010.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yin H., Xu L., Porter N.A. Free radical lipid peroxidation: mechanisms and analysis. Chem. Rev. 2011;111:5944–5972. doi: 10.1021/cr200084z. [DOI] [PubMed] [Google Scholar]
- 57.Zhang W., Patil S., Chauhan B., Guo S., Powell D.R., Le J., Klotsas A., Matika R., Xiao X., Franks R. FoxO1 regulates multiple metabolic pathways in the liver: effects on gluconeogenic, glycolytic, and lipogenic gene expression. J. Biol. Chem. 2006;281:10105–10117. doi: 10.1074/jbc.M600272200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Microarray data is available here: OEP000451 (https://www.biosino.org/node/project/detail/OEP000451).