Abstract
Objective
Although several assays have been developed to detect SARS-CoV-2 RNA in clinical specimens, their relative performance is unknown.
Methods
The concordance between the cobas 8800 SARS-CoV-2 and a laboratory developed (LD) reverse transcriptase-polymerase chain reaction (RT-PCR) assay was assessed on 377 combined nasopharyngeal/oropharyngeal swabs in Hanks medium.
Results
The positive and negative agreement between these assays were 99.3 % (95 % CI, 97.3–99.9) and 77.1 % (95 % CI, 67.7–84.4), respectively, for an overall agreement of 93.6 % (95 % CI, 90.7–95.7) beyond random chance (kappa of 0.82, 95 % CI, 0.75−0.85). Of the 22 samples positive by cobas SARS-CoV-2 only, 9 were positive only for ORF-1 gene and had Cycle thresholds (Ct) > 35.1, 8 were positive only for the E gene with Ct > 35.5 and 5 were positive for both targets with Ct > 33.9. Samples positive only with the cobas assay were more often positive with only one gene target (77.3 %) than samples positive in both assays (16.9 %, p < 0.0001). Ct values in the cobas SARS-CoV-2 assay were significantly higher in the 279 samples testing positive in both assays (32.9 %, 95 % CI 32.3–33.6) compared to the 22 samples with discordant results (36.6 %, 95 % CI 36.2–37.1; p = 0.0009). An excellent correlation (r2 = 0.98) was obtained between Ct values of the ORF-1 and E targets in the cobas assays and a good correlation was obtained between LD RT-PCR test and cobas SARS CoV-2 ORF-1 target (r2 = 0.82).
Conclusion
Our study demonstrated an excellent concordance between a LD RT-PCR and the cobas SARS-CoV-2 tests on the 8800 platform.
Keywords: SARS-Cov-2, Cobas, Real time PCR
1. Introduction
Since December 2019, a novel coronavirus, the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerged as the cause of a severe respiratory disease and is causing a worldwide pandemic. Tests based on reverse transcription-polymerase chain reaction (RT-PCR) are the most suitable mode of rapid diagnosis of acute SARS-CoV-2 infections which is essential for patient management and contact tracing [1]. Several commercial and laboratory developed (LD) assays are available but few manufacture-independent evaluations and few comparisons between assays have been published up to now [1]. Moreover, the comparison between assays is hampered by the absence of accepted gold standard test as well as our incomplete knowledge of the natural history of SARS-CoV-2 infection. Studies evaluating the concordance between assays are thus needed at this point in the pandemic.
We evaluated the concordance between the two-target cobas SARS-CoV-2 test (Roche Molecular Diagnostics, Laval, Canada) on the fully automated cobas 8800 platform authorized by Health Canada and a laboratory-developed (LD) standardized RT-PCR test using widely used primer set and probe [2,3] in samples submitted at the diagnostic laboratory for patient care at the Centre Hospitalier de l’Université de Montréal.
2. Materials and methods
A subset of combined nasopharyngeal/oropharyngeal swabs routinely collected in Hanks media from 377 individuals between 21 April 2020 and 11 June 2020 were transported to the laboratory the same day and tested in parallel as part of routine clinical care using the cobas SARS-CoV-2 assay and the LD RT-PCR test. Testing was requested for investigation of symptomatic individuals, follow-up of positive individuals or investigation of outbreaks or contact with a positive source. According to information provided in the request form for SARS-CoV-2 detection, 121 individuals had no symptoms, 132 were symptomatic and information was not provided for 124 individuals. Both assays had been previously validated against a panel of samples with known results including negative and positive samples from the reference laboratory of the Province of Quebec, the Laboratoire de Santé Publique du Québec (LSPQ).
The validation study and publication of results was permitted without obtaining formal written consent from patients by the Directeur des Affaires Médicales Universitaires du CHUM as part of l’Urgence sanitaire de la Santé Publique du Québec, since results are published without identification of participants and as part of diagnostic purposes and assay validation.
The LD RT-PCR test developed by the LSPQ uses a standardized protocol consisting of automated RNA extraction followed by RT-PCR targeting the envelope (E) gene with widely utilized primers and probe [2]. Briefly, nucleic acid from 200 μL of specimen suspended in Hanks medium was extracted on a NucliSens® easyMAG ™ instrument (Biomérieux) using the Easy Mag extraction kit as suggested by the manufacturer’s instructions. Five of the 25 μL elution volume was amplified with real-time RT-PCR using the TaqPath 1-step multiplex no ROX mastermix and E gene specific primers at 0.4 μM and 6-carboxyfluorescein (FAM)-labeled hydrolysis probe at 0.2 μM [2]. Due to the limited availability of reagents, only one SARS-CoV-2 target was tested with the LD RT-PCR. The reaction mixture was heated at 53 °C for 10 min, at 95 °C for 2 min and then amplified for 45 cycles at 95 °C for 3 s and 60 °C for 30 s. RT-PCR was performed on a Light Cycler 480 II (Roche Molecular systems, Laval, Canada). One negative control and one positive control were included in each batch of testing. Samples were considered positive in the presence of an amplification curve below a Ct of 40. The limit of detection of the assay was 180 viral RNA copies per mL of medium (data not shown).
Prior to testing on the cobas 8800, each specimen was treated to inactivate potential infectious viruses: 400 μL of specimen in Hanks was added to 200 μL of cobas 8800 lysis buffer, followed by an incubation at room temperature for 10 min. The detection of SARS-CoV-2 RNA was then performed on the inactivated 600 μL-aliquot with the cobas 8800 instrument, as suggested by the manufacturer. A two-target RT-PCR was used on this instrument: one targeting Orf1, a non-structural region that is unique to SARS-CoV-2 coding for the RdRp activity of the virus, and the second targeting a conserved region in the structural protein envelope E gene for pan-sabercovirus detection. An internal RNA control was detected in each sample to control for amplification efficiency and RNA extraction from sample. Uracil-N-glycosylase is also included in the master mix to catalyze contaminating amplicons. A data management software in the automated apparatus assigns test results to each sample. A sample was considered positive if a positive result was obtained for the Orf1 gene only or for both Orf1 and E genes, as suggested by the manufacturer’s instructions. Samples testing positive only for the E gene were considered as SARS-CoV-2 presumptive positive. Testing was performed in 96 samples batches, including one positive control and one negative control. The Limit of detection of the assay was 23 viral RNA copies per mL of medium (data not shown). Samples with discrepant results between the cobas and LDT assays were tested, when enough sample left was available, in the Abbott RealTime SARS-CoV-2 test on the Abbott m2000 system (Abbott Molecular Inc., Des Plaines, IL), as suggested by the manufacturer [4].
In the absence of gold standard for SARS-CoV-2 RNA detection, data was analyzed using a contingency table to assess the overall, positive and negative agreement with 95 % confidence intervals (95 % CI) calculated. The level of agreement was also assessed using kappa statistics. By definition, Kappa values above 0.75 indicate excellent agreement, values between 0.40 and 0.75 indicate fair to good agreement, and values below 0.40 represent poor agreement beyond chance [5]. A McNemar’s test was applied to assess differences between matched proportions. The distributions of Ct values between assays were compared using the non-parametric Wilcoxon pair test. Linear regression analysis was performed to compare Cycle thresholds (Ct) values between gene targets and assays. Statistical analysis was done using the Statistica software (Statsoft Inc, OK, USA).
3. Results
The results of the comparison of the cobas SARS-CoV-2 and LD RT-PCR on 377 routinely collected nasopharyngeal/oropharyngeal swabs are summarized in Table 1 . The LD RT-PCR assay reported 281 samples as ‘detected’ and 96 as ‘not detected’. The cobas SARS-CoV-2 test reported 301 samples as ‘detected’ and 76 as ‘not detected’. PCR inhibition was not encountered in these specimens. The positive and negative agreement between assays between the cobas SARS-CoV-2 and the LD RT-PCR assays were 99.3 % (95 % CI, 97.3–99.9) and 77.1 % (95 % CI, 67.7–84.4), respectively. The overall percent agreement between the two tests was 93.6 %, 95 % CI 90.7–95.7. The calculated kappa coefficient of 0.82 (95 % CI, 0.75−0.85) indicated an excellent agreement between tests.
Table 1.
Detection of SARS-CoV-2 RNA in 377 nasopharyngeal-oropharyngeal samples in Hanks with cobas 8800 SARS-CoV-2 and LD RT-PCR assays.
| cobas SARS-CoV-2 results | LD RT-PCR results |
||
|---|---|---|---|
| Detected | not detected | Total | |
| Detected | 279 | 221 | 301 |
| Not detected | 2 | 74 | 76 |
| Total | 281 | 96 | 377 |
Overall agreement: 93.6 %, 95 % CI.90.7–95.7.
Kappa coefficient of 0.82, 95 % CI 0.75−0.85.
1: 8 samples were positive only with the E gene and were presumptive positive samples. The other 14 samples were positive with at least the SARS-CoV-2 Orf1 gene.
Of the 22 samples positive by cobas SARS-CoV-2 and negative by LD RT-PCR, 9 were positive only for the Orf1 gene and all had a Ct above 35.1, 5 were positive for both Orf1 and E genes and all had a Ct above 33.9 while 8 were positive only for the E gene and all had a Ct above 35.5. The later 8 samples were reported as presumptive positives as per the manufacturer’s recommendation. If these were excluded from the analysis, 74 of 88 samples were negative in the LD RT-PCR and cobas assays for a negative agreement between assays of 84.1 % (95 % CI 74.4–93.7). Moreover, samples that were found to be positive only with the cobas SARS-CoV-2 assay were more often positive for only one target [17 of the 22 (77.3 %)] than samples also positive in the LD RT-PCR assay [47 of 279 (16.9 %), p < 0.0001]. The two samples negative for both targets in the cobas SARS-CoV-2 assay yet positive in the LD RT-PCR test, had both Ct values above 35 (35.3, 35.6). Thus, discordant samples between assays all had high Ct values, suggesting a low SARS-CoV-2 load. The Ct values obtained in the cobas SARS-CoV-2 assay were significantly lower (p = 0.0009) in the 279 samples testing positive in both assays (32.9 ± 5.4 (95 % CI, 32.3–33.6), median of 34.7, range 15.4–39.1) compared to the 22 samples with discordant results between tests (36.6 ± 1.0 (95 % CI, 36.2–37.1), median of 36.5, range 35.3–38.1). Of the 22 samples positive only in the cobas SARS-CoV-2 assay, 20 could be tested in the Abbott RealTime SARS-CoV-2 test: one showed inhibition, 8 were ‘detected’ with low cycle number (CN) values between 26.7 and 31.5, and 11 were ‘not detected’. Since these latter samples were weakly reactive initially in the cobas assay, had been frozen for 6–9 weeks and thawed, the 11 negative samples in the Abbott RealTime test yet initially positive in the cobas assay were retested in the cobas assay : only one out of 10 then scored positive in the cobas test.
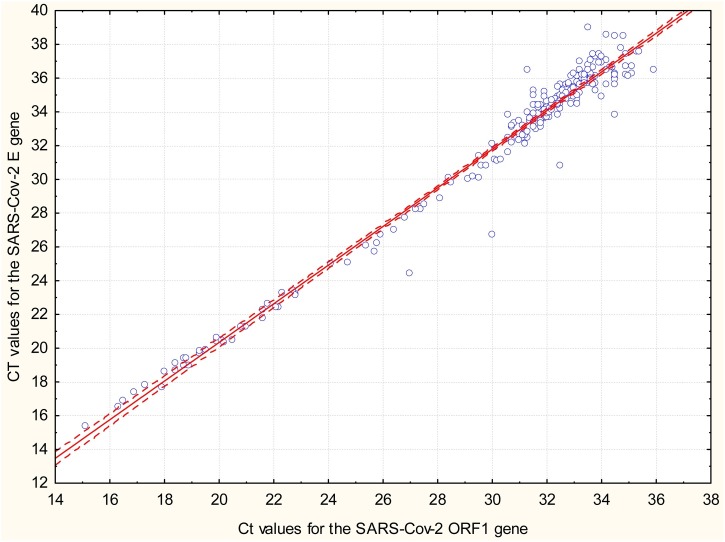
An excellent correlation (r2 = 0.98, p < 0.0001) was obtained between Ct values for SARS-CoV-2 ORF-1 target and E target in the cobas SARS-CoV-2 assays (Fig. 1 ). The correlation between Ct values obtained in the LD RT-PCR test and cobas SARS CoV-2 ORF-1 target for positive samples in both assays was good (r2 = 0.82, data not shown).
Fig. 1.
Correlation between cycle thresholds (Ct) values obtained with the cobas 8800 SARS-CoV-2 assay for target -1 Orf1 gene and target 2 –E gene (pan-sabercovirus detection) in 279 positive samples for SARS-CoV-2 virus RNA. The dotted line is the 95 % confidence internal of the regression line.
4. Discussion
Automated assays for detection of SARS-CoV-2 RNA have the advantage over LD RT-PCR protocols of requiring minimal hands-on time thus facilitating mass screening for public health and patient management purposes. Automated assays also generate consistent results between testing sites. On the other hand, LD RT-PCR tests do not rely on availability of commercial reagents and thus offer more flexibility. They also allowed laboratories to offer diagnostic services before the availability of automated platforms. Moreover, they do not require high volume of samples to be cost effective. Despite the advantages of automated platforms, few manufacture-independent evaluations of these assays have been reported and few comparisons between commercial assays and LD RT-PCR tests have been published up to now. The cobas 8800 instrument is a fully automated platform performing extraction, PCR amplification, signal detection and reporting of results. This assay has been shown to have good clinical performance in one report [3] and excellent linearity over seven logs of viral load [6]. There is currently no gold standard for the diagnosis of SARS-CoV-2 infection. Therefore, we assessed the agreement between the cobas SARS-CoV-2 test and a LD RT-PCR that had been validated with a provincial panel and is performed by several laboratories in the province of Quebec, although its clinical performance is unknown. The limit of detection of the LD RT-PCR assay is 300 SARS-CoV-2 RNA copies per mL of sample, as reported by the LSPQ (personal communication). The LD RT-PCR in the current report was using primers and probe described by Corman et al. against the E gene, reagents that have been confirmed to be sensitive in work comparing several primer-probe sets [2,7].
We found an excellent overall and positive agreement between assays. The negative agreement was lower because of samples positive in the cobas SARS-CoV-2 test but negative in the LD RT-PCR test. Samples yielding discordant results were tested in a third assay approved for SARS-CoV-2 diagnosis. Nearly half of these samples tested positive with the Abbott RealTime SARS-CoV-2 test. Retesting the Abbott RealTime SARS-CoV-2-negative sample in the cobas assay yielded negative results in all but one sample, suggesting loss of a SARS-CoV-2 RNA in the process of freezing and thawing and transportation of specimens. The reactivity of discordant samples was low compared to that of concordant samples, suggesting low SARS-CoV-2 viral load infections. None of these samples had Ct values lower than 35. Moreover, most of the discordant samples positive in the cobas 8880 but negative in the LD RT-PCR were reactive in only one out of two targets in the automated assay. These discordances may reflect differences in primer and probe sequences, detection of several gene targets in the 8800 test, assays limit of detection or sample processing differences [2].
Our study has limitations. This evaluation was conducted on samples from individual with symptoms or during follow-up of a confirmed infection. The level of agreement in individuals without symptoms at the beginning of infection was not evaluated here. The comparison was performed at a time in the pandemic with fewer individuals with low Ct results and an important proportion of specimens with high Ct values indicating low viral loads (140 (45.3 %) of 301 samples positive with cobas SARS-CoV-2 had Ct values >34.9). Because of the limited availability of reagents for the LD RT-PCR test, it was impossible to test in parallel all samples sent to our laboratory with both assays and impossible to test more than one gene target in the LD RT-PCR test. Only discordant samples were tested in a third assay, we could thus not calculate the performance of the cobas test with an expanded gold standard.
5. Conclusion
In conclusion, this study demonstrated an excellent agreement between the cobas SARS-CoV-2 test in the 8800 platform and a LD RT-PCR test, although the cobas assay detected a greater number of samples with low viral load infections. Further studies comparing different platforms in parallel, as well as using clinical information will allow establishing a gold standard and determine the true performance of automated high throughput platforms.
Source of support
This work was supported by the laboratory budget under the sanitary emergency of the Province of Quebec without funding from commercial sources.
CRediT authorship contribution statement
Catherine-Audrey Boutin: Data curation, Formal analysis, Writing - review & editing. Simon Grandjean-Lapierre: Conceptualization, Writing - review & editing. Simon Gagnon: Conceptualization, Methodology, Supervision. Annie-Claude Labbé: Formal analysis, Methodology, Review. Hugues Charest: Methodology, Review. Michel Roger: Methodology, Review. François Coutlée: Conceptualization, Formal analysis, Project administration, Writing - original draft.
Declaration of Competing Interest
The authors report no declarations of interest.
Acknowledgements
We would like to thank all the laboratory technologists who performed the real time PCR assays for this study.
References
- 1.Loeffelholz M.J., Tang Y.W. Laboratory diagnosis of emerging human coronavirus infections - the state of the art. Emerg. Microbes Infect. 2020;9(1):747–756. doi: 10.1080/22221751.2020.1745095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25(3) doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Poljak M., Korva M., Knap Gašper N., Fujs Komloš K., Sagadin M., Uršič T., et al. Clinical evaluation of the cobas SARS-CoV-2 test and a diagnostic platform switch during 48 hours in the midst of the COVID-19 pandemic. J. Clin. Microbiol. 2020;58(6) doi: 10.1128/JCM.00599-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harrington A., Cox B., Snowdon J., Bakst J., Ley E., Grajales P., et al. Comparison of Abbott ID now and Abbott m2000 methods for the detection of SARS-CoV-2 from nasopharyngeal and nasal swabs from symptomatic patients. J. Clin. Microbiol. 2020;58(8) doi: 10.1128/JCM.00798-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fleiss J.L. 2nd ed. John Wiley and Sons Inc.; New York: 1981. Statistical Methods for Rates and Proportions. 1981. [Google Scholar]
- 6.Lu X., Wang L., Sakthivel S.K., Whitaker B., Murray J., Kamili S., et al. US CDC real-time reverse transcription PCR panel for detection of severe acute respiratory syndrome coronavirus 2. Emerg. Infect. Dis. 2020;26(8) doi: 10.3201/eid2608.201246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nalla A.K., Casto A.M., Huang M.W., Perchetti G.A., Sampoleo R., Shrestha L., et al. Comparative performance of SARS-CoV-2 detection assays using seven different primer-probe sets and one assay kit. J. Clin. Microbiol. 2020;58(6) doi: 10.1128/JCM.00557-20. [DOI] [PMC free article] [PubMed] [Google Scholar]