Dear Editor
The coronavirus disease-2019 (COVID-19) has become a global pandemic affecting over 192 countries [1]. It is caused by the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), which enters cells via interactions with angiotensin converting enzyme II (ACE2) receptors that line the respiratory tract, esophagus, ileum and colon, thereby providing a potential means for fecal-oral transmission [2]. Concurrent gastrointestinal (GI) complaints have been reported in up to 61% of COVID-19 patients [3], and early reports from Asian populations suggested that hospitalized COVID-19 patients with concurrent GI symptoms require more antibiotics and have worse coagulopathy compared to those without GI symptoms [4].
We undertook a mortality analysis of COVID-19 patients hospitalized at our New York City (NYC) hospital at the height of the pandemic to determine whether or not the presence of gastrointestinal symptoms was associated with patient survival to discharge. Our primary endpoint was survival to discharge with and without the presence of GI symptoms upon admission, which included nausea, vomiting, hematemesis, abdominal pain, diarrhea, and hematochezia.
We performed a retrospective review of the electronic health records (EHR) of all adults aged 18 years and above who have tested positive on nasal swabs for SARS-CoV-2 polymerase chain reaction (PCR) and required hospital admission. We identified patients with assistance from our institutional information technology (IT) department and utilization of the international classification of diseases code 10th Revision (ICD-10) U07.1. Data abstraction from the EHR was uniform for all patients. We collected demographic data, vital status, and clinical history, among other variables, listed in Table 1 . Our study period was from our institution's first positive SARS-CoV-2 PCR test on 10 March 2020 through 13 April 2020, with follow-up to 30 April 2020. We obtained approval from our hospital's Institutional Review Board (IRB-1591128–1). We excluded patients ≤ 17 years of age, those still hospitalized at time of analysis, those who tested positive for COVID-19 but who did not require hospitalization, and persons under investigation for COVID-19 who tested negative despite having typical COVID-19 symptoms.
Table 1.
Baseline characteristics.
| Variables | Number | Percent | |
|---|---|---|---|
| Survived to Discharge | Yes | 497 | [67.7] |
| No | 237 | [32.3] | |
| Sex | Women | 355 | [48.4] |
| Men | 379 | [51.6] | |
| Any GI Symptoms on Admission | Yes | 231 | [31.5] |
| No | 503 | [68.5] | |
| Nausea | Yes | 109 | [14.9] |
| No | 625 | [85.1] | |
| Vomiting | Yes | 62 | [8.45] |
| No | 672 | [91.6] | |
| Abdominal Pain | Yes | 68 | [9.26] |
| No | 666 | [90.7] | |
| Diarrhea | Yes | 149 | [20.3] |
| No | 585 | [79.7] | |
| Hematemesis | Yes | 4 | [0.545] |
| No | 730 | [99.5] | |
| Hematochezia | Yes | 12 | [1.63] |
| No | 722 | [98.4] | |
| Underlying Hypertension | Yes | 494 | [67.3] |
| No | 240 | [32.7] | |
| Underlying Chronic Renal Insufficiency | Yes | 174 | [23.7] |
| No | 560 | [76.3] | |
| Underlying History of Coronary Artery Disease | Yes | 281 | [38.3] |
| No | 452 | [61.7] | |
| Underlying History of Diabetes Mellitus | Yes | 319 | [43.5] |
| No | 415 | [56.5] | |
| Underlying Chronic Lung Disease | Yes | 138 | [18.8] |
| No | 596 | [81.2] | |
| Underlying Autoimmune Disease | Yes | 74 | [10.1] |
| No | 660 | [89.9] | |
| Underlying Chronic Liver Disease | Yes | 26 | [3.54] |
| No | 708 | [96.5] | |
| Active Neoplastic Disease | Yes | 27 | [3.7] |
| No | 703 | [96.3] | |
We used Chi-square tests to compare the survival percentages between those with and without GI symptoms. T-tests were used to examine continuous variables and their relationship to survival. We utilized multivariable Cox proportional hazard regressions to calculate hazard ratios (HR) for mortality for patients with and without GI symptoms after adjusting for significant variables identified with univariable analysis. A two-tailed p-value <0.05 was considered statistically significant.
To corroborate our findings, we also used predictive analysis, which is a technique that allows direct estimation of relevant probabilities, in this case survival to discharge. This technique provided direct probabilities of survival to discharge based upon the demographic and clinical variables of interest. The predictive models are presented in plots that depict the probability of surviving at each age level therefore there are no confidence bounds or p-value involved. The models were then expanded to incorporate those with and without GI complaints on admission. Since predictive analysis gives direct probabilities, there are no confidence bounds or p-values involved. All analyses were carried out in R version 3.6.3 (R Foundation for Statistical Computing, Vienna, Austria) using the rstanarm version 2.18.2, and models fit using default priors and 10,000 iterations per chain, 4 chains per model until convergence was reached.
During the study period of 3/10/20 through 4/13/20, over 3000 patients tested positive for SARS-Cov-2 at our institution, of whom 865 were adults who required hospitalization. We excluded 131 patients who remained hospitalized at the end of the study period, thus leaving 734 patients in our analysis. Our cohort included 355 women (48.4%), 372 Blacks (50.7%), 214 whites (29.2%), and 92 Hispanics (12.5%). The mean age for the cohort was 66.1 years ± 15.6 (median 68 years, range 18–99 years). Our cohort's baseline characteristics are summarized in Table 1.
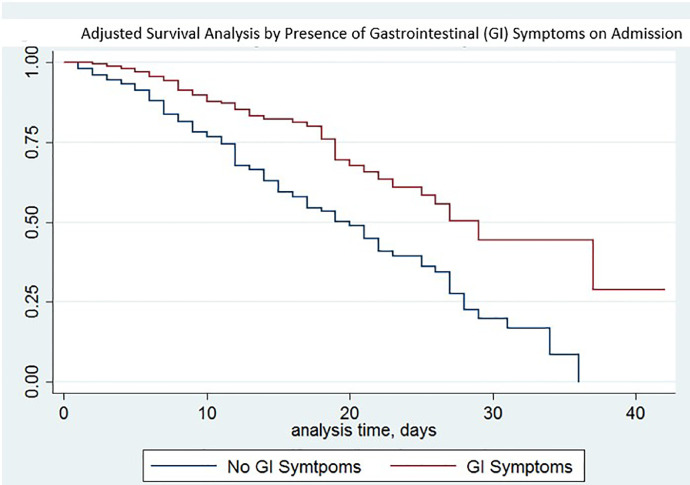
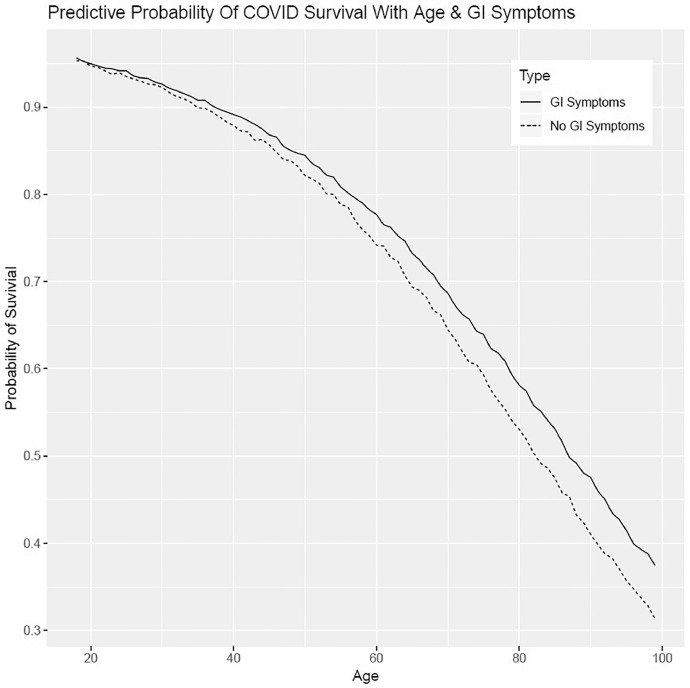
A total of 231 patients (31.5%) were documented to have GI symptoms on admission. The presence of GI symptoms was significantly associated with survival to discharge (169/231, 73.2%) when compared to those without GI symptoms (328/503, 65.2%; p = 0.04). This finding persisted after multivariable analyses accounting for the presence of underlying coronary artery disease (CAD), diabetes mellitus (DM), hypertension (HTN), and chronic kidney disease (CKD), with hazard ratio for in-hospital mortality for COVID-19 patients with GI symptoms 0.66 (95% CI, 0.49–0.89; p = 0.01). There were no statistically significant differences in mortality based on individual GI symptoms, specifically nausea, vomiting, abdominal pain, or diarrhea. Our predictive models found that lack of GI symptoms portended lower survival at all ages within our cohort. Our findings are shown in Table 2 . In-hospital mortality based upon our multivariable Cox proportions and presence of GI symptoms are presented in a Kaplan-Meier graph and predictive model in Fig. 1, Fig. 2 .
Table 2.
Predictors of in-hospital mortality.
| Variables | Survived to Discharge? |
P-Value (Chi-Squared Analysis) | Probability of Survival (Predictive Analysis) | ||
|---|---|---|---|---|---|
| Yes | No | ||||
| Total | 497 | 237 | N/A | N/A | |
| Any GI Symptoms | Yes | 169 | 62 | 0.04 | 0.75 |
| No | 328 | 175 | 0.65 | ||
| Underlying Hypertension | Yes | 317 | 177 | 0.004 | 0.64 |
| No | 180 | 60 | 0.75 | ||
| Underlying Chronic Renal Insufficiency | Yes | 104 | 70 | 0.013 | 0.60 |
| No | 393 | 167 | 0.72 | ||
| Underlying History of Coronary Artery Disease | Yes | 156 | 125 | <0.001 | 0.55 |
| No | 340 | 112 | 0.75 | ||
| Underlying History of Diabetes Mellitus | Yes | 198 | 121 | 0.005 | 0.63 |
| No | 299 | 116 | 0.72 | ||
| Underlying Chronic Lung Disease | Yes | 87 | 51 | 0.23 | 0.63 |
| No | 410 | 186 | 0.70 | ||
Fig. 1.
Adjusted survival analysis by presence of gastrointestinal (GI) symptoms on admission
Fig. 2.
Predictive probability of COVID survival with age & GI symptoms
The presence of GI symptoms was associated with reduced in-hospital mortality.
In our cohort. This association may hint at a crucial characteristic about SARS-CoV-2 and its route of infectivity. We surmise that SARS-CoV-2 may be transmitted via fecal-oral route in addition to the well-established respiratory droplet route, and this fecal-oral transmission may lead to a milder form of COVID-19. Researchers have detected live SARS-CoV-2 in saliva via viral culture as well as in stool samples [5]. Nobel's group found that gastrointestinal symptoms increased likelihood of positive SARS-CoV-2 testing by 70% [6]. Wei, et al. found that diarrhea was associated with detection of SARS-CoV-2 in stool despite negative nose swab testing [7], a finding later corroborated in a pooled meta-analysis from Hong Kong [3]. More recently, Lamers, et al. have demonstrated that SARS-CoV-2 readily infects human small intestinal organoids and that the intestinal epithelium supports viral replication [8]. These findings suggest that, despite negative respiratory testing, SARS-CoV-2 shedding may still be present via the GI tract and can potentially act as a nidus for asymptomatic spread and may provide a milder disease course.
Our findings are dissimilar to what Wan, et al. found, specifically that diarrhea was associated with higher in-hospital mortality, but their study limited GI symptoms of interest to diarrhea, and their symptomatic sample size was both smaller–49 patients—and included only 6 deaths [9]. In contrast, our study included both upper and lower GI complaints and our univariable analysis revealed a numerical increase in survival to discharge among our 149 patients with diarrhea compared to those without (74% vs 66% respectively, p = 0.09). Virologic studies have also revealed that early transmission of COVID-19 in NYC originated in Europe, not in Asia, which could account for differences in disease severity from those reported in China [10].
Despite our best efforts to incorporate multivariable analysis, the retrospective design allows for selection bias and potential confounders. We limited our focus to those requiring hospitalization. We did not obtain prognostic index scores such as Acute Physiology and Chronic Health Evaluation II (APACHE II) due to inconsistent documentation at the onset of the pandemic. We did not analyze endoscopic outcomes in those requiring emergent procedures and we did not account for the various medications taken prior or during the hospitalization. These may bias our sample because we could not exclude the mortality risk of hospitalized patients associated with the aforementioned variables.
We have a sizable U.S.-based cohort from when NYC was the pandemic's epicenter that nonetheless highlights a potential role for the digestive system in COVID-19 transmission and hospitalization outcomes. We look forward to studies that will further elucidate the fecal-oral transmission route of SARS-CoV-2.
Author Contributions
Intellectual genesis: BDR, DSJ; Data extraction and cleaning: BDR, NK, KC, EB, DSJ; Statistical consultation and formal data analysis: WMB; Data interpretation: BDR, DSJ; Manuscript writing: BDR, WMB, DSJ; Critical manuscript review and editing: BDR, DSJ. All authors approved the final draft as submitted.
Financial Support
None
Declaration of Competing Interest
None
References
- 1.John Hopkins University & Medicine Coronavirus Resource Centerhttps://coronavirus.jhu.edu/data/racial-data-transparencyAccessed May 2020.
- 2.Gu J., Han B., Wang J. COVID-19: gastrointestinal manifestations and potential fecal-oral transmission. Gastroenterology. 2020 doi: 10.1053/j.gastro.2020.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheung K.S., Hung I.F., Chan P.P., et al. Gastrointestinal manifestations of SARS-CoV-2 infection and virus load in fecal samples from the Hong Kong Cohort and systematic review and meta-analysis. Gastroenterology. 2020 doi: 10.1053/j.gastro.2020.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pan L., Mu M., Yang P., et al. Clinical characteristics of COVID-19 patients with digestive symptoms in Hubei, China: a descriptive, cross-sectional, multicenter study. Am J Gastroenterol. 2020 doi: 10.14309/ajg.0000000000000620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.To K.K., Tsang O.T., Chik-Yan Yip C., et al. Consistent detection of 2019 novel coronavirus in saliva. Clin Infect Dis: Off Publ Infect Dis Soc Am. 2020 doi: 10.1093/cid/ciaa149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nobel Y.R., Phipps M., Zucker J., et al. Gastrointestinal Symptoms and COVID-19: case-control study from the United States. Gastroenterology. 2020 doi: 10.1053/j.gastro.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wei X.S., Wang X., Niu Y.R., et al. Diarrhea is associated with prolonged symptoms and viral carriage in COVID-19. Clin Gastroenterol Hepatol: Off Clin Pract J Am Gastroenterol Assoc. 2020 doi: 10.1016/j.cgh.2020.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lamers M.M., Beumer J., van der Vaart J., et al. SARS-CoV-2 productively infects human gut enterocytes. Science. 2020 doi: 10.1126/science.abc1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wan Y., Li J., Shen L., et al. Enteric involvement in hospitalised patients with COVID-19 outside Wuhan. Lancet Gastroenterol Hepatol. 2020 doi: 10.1016/S2468-1253(20)30118-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gonzalez-Reiche A.S., Hernandez M.M., Sullivan M.J., et al. Introductions and early spread of SARS-CoV-2 in the New York City area. Science (New York, NY) 2020 doi: 10.1126/science.abc1917. [DOI] [PMC free article] [PubMed] [Google Scholar]