Highlights
-
•
Classification of normal, viral pneumonia and Covid-19 chest x-ray images.
-
•
Proposed a deep convolutional neural network model for early detection of COVID19 cases using chest x-ray images.
-
•
Trained the model on a dataset prepared by collecting x-ray images from publically available databases.
-
•
Achieved an average accuracy of 97.20% for detecting COVID-19 and an average accuracy of 96.69% for three-class classification (COVID-19 vs. normal vs. viral pneumonia). The results obtained by our proposed model are superior compared to other studies in the literature.
-
•
Promising results indicate that this model can be an interesting tool to help radiologists in the diagnosis and early detection of COVID-19 cases.
Keywords: Deep learning, Convolutional neural network, COVID-19, Coronavirus, Classification, Chest X-ray images
Abstract
The COVID-19 pandemic is an emerging respiratory infectious disease, also known as coronavirus 2019. It appears in November 2019 in Hubei province (in China), and more specifically in the city of Wuhan, then spreads in the whole world. As the number of cases increases with unprecedented speed, many parts of the world are facing a shortage of resources and testing. Faced with this problem, physicians, scientists and engineers, including specialists in Artificial Intelligence (AI), have encouraged the development of a Deep Learning model to help healthcare professionals to detect COVID-19 from chest X-ray images and to determine the severity of the infection in a very short time, with low cost. In this paper, we propose CVDNet, a Deep Convolutional Neural Network (CNN) model to classify COVID-19 infection from normal and other pneumonia cases using chest X-ray images. The proposed architecture is based on the residual neural network and it is constructed by using two parallel levels with different kernel sizes to capture local and global features of the inputs. This model is trained on a dataset publically available containing a combination of 219 COVID-19, 1341 normal and 1345 viral pneumonia chest x-ray images. The experimental results reveal that our CVDNet. These results represent a promising classification performance on a small dataset which can be further achieve better results with more training data. Overall, our CVDNet model can be an interesting tool to help radiologists in the diagnosis and early detection of COVID-19 cases.
1. Introduction
The COVID-19 pandemic is an emerging respiratory infectious disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). It was first identified in the city of Wuhan in November 2019, following the emergence of a group of patients with viral pneumonia. During the first months of 2020, COVID-19 spread worldwide through human-to-human transmission [2]. However, The COVID-19 can be responsible for several respiratory and digestive diseases in several mammals. In humans, these infections can be: Asymptomatic; Responsible for benign pathologies such as a cold or flu syndromes; Responsible for respiratory complications such as pneumonia in immunocompromised patients or infants; Responsible for severe respiratory syndrome, leading to epidemics. This virus is transmitted by: Airway; In contact with secretion and in contact with contaminated objects. As Covid-19 is spreading with unprecedented speed around the world, the healthcare system of many countries has caused overload and although definitive genetic testing for the disease is notoriously rare in many parts of the world. X-rays [1], [3], in particular, are widely used to detect COVID-19 without dedicated test kits since almost all hospitals have X-ray imaging devices. The advantage of having X-rays with the scanner is that in some cases the disease can be detected in its preliminary stages, even if the screening is still negative and its drawbacks is that an x-ray analysis requires an expert in radiology, and it takes a long time; which is precious when people are sick around the world. Therefore, it is necessary to develop an automated analysis system to detect if there are anomalies in the scans in order to save time for healthcare professionals. With the advent of data science and deep learning or machine learning algorithms, we are now able to access large amounts of data, collected from different sources of information, to detect Coronavirus from computed tomography, monitoring of the epidemic, analysis of epidemiological data, and even clinical trials in record time. Technology and artificial intelligence (AI) can significantly speed up the processing of the data needed to obtain the information, responses and recommendations to manage and combat the Covid-19 pandemic, with better than expected accuracy. Additionally, AI approaches can be useful in eliminating disadvantages such as insufficient number of available RT-PCR test kits, test costs, and waiting time of test results.
A number of artificial intelligence systems based on deep learning, in particular convolutional neural networks have been proposed and the results have been very promising in terms of accuracy in the detection of patients infected with COVID-19 using chest x-ray images. The reason for this success is that deep convolutional neural networks are not based on extracting features manually, but these algorithms automatically learn features from the data itself [4]. Deep CNN has been successfully applied in many problems such as classification of skin cancer [5], arrhythmia detection [6], classification of brain diseases [7], detection of breast cancer [8], fundus image segmentation [9], detection of pulmonary pneumonia X-ray images [10] and lung segmentation [11]. Thanks to the success of deep learning, the Canadian startup DarwinAI [12] believes that it has developed a tool that could help doctors make crucial decisions. DarwinAI has looked into this and wondered if AI could have a role to play in the fight against the Covid-19. The answer may well prove to be positive.
In this study, we review state-of-the-art models designed to identify COVID-19 infection and we propose a deep CNN model, namely CVDNet as an advanced tool to help radiologists to automatically detect COVID-19 infection using chest x-ray images and thus delay the rapid spread of this virus.
The proposed model is based on the residual neural network and it is constructed by using multi parallel levels with different kernel sizes to detect local and global features and by connecting the residuals to other levels to share information. The CVDNet is trained on 2905 chest x-ray images that is open source and publically available to classify COVID-19 cases from normal and viral pneumonia cases. Moreover, the results found from the proposed technique are compared to other studies reported in the literature.
The contributions of the paper are summarized as follows:
-
•
Building deep convolutional neural network model to accurately detect patients with COVID-19 in a very short time in order to assist in the early diagnosis.
-
•
Performing an empirical analysis of our deep model in order to classify COVID-19 disease using a chest x-ray since it is less costly than other imaging modalities.
-
•
Evaluating the performance of our model compared to other existing models.
-
•
Helping researchers continue to develop artificial intelligence techniques to fight the COVID-19 epidemic.
The rest of this paper is systematized as follows. Section 2 presents the literature review on recent developments in AI systems based on deep convolutional neural network for the detection of COVID-19. Section 3 details the design of the proposed architecture CVDNet. Section 4 describes the data set used in this study. Also, provides experimental setup and discusses the performance of our model. Section 5 draws conclusions and discusses some future directions.
2. Releted work
Motivated by the need for rapid interpretation of x-ray images, many scientists have proposed deep learning models especially convolutional neural networks to detect cases infected with COVID-19 from chest x-ray imaging. Wang et al. [12] introduced COVID-Net, a deep CNN designed for the detection of patients with Covid-19 from chest X-ray images that is an open source publically available. The dataset consists of chest radiography images that contain four classes including COVID-19 infection, Pneumonia Viral, Pneumonia bacterial and normal (non-COVID19 infection). COVID-Net achieved an overall accuracy of 83.5% for these 4-classes and an overall accuracy of 92.4% in classifying COVID-19, normal and non-COVID pneumonia cases. Hemdan et al. [13] introduced a deep learning; called COVIDX-Net to aid radiologists to automatically detect COVID-19 in chest x-ray images. This framework is based on seven deep architectures namely MobileNetV2, VGG19, InceptionV3, DenseNet201, InceptionResNetV2, ResNetV2 and Xception. Furthermore, it is validated on 50 X-ray images comprising 25 cases with COVID-19 and 25 cases without any infections. This study reveals that The VGG19 and DenseNet models have similar performance of automated COVID-19 detection with f1-scores of 0.91 and 0.89 for COVID-19 and normal, respectively, and the InceptionV3 model produce a poor classification performance with f1-scores of 0.00 for COVID-19 cases and 0.67 for normal cases. Kumar et al. [14] proposed a system based on deep Convolutional network that is developed for the detection of COVID-19 using X-ray images. This model is trained on a dataset collected from GitHub, Kaggle and Open-I repository and achieved an accuracy of 95.38% for detecting COVID-19. Ozturk et al. [15] presented a deep CNN based on DarkNet model, namely DarkCovidNet for automatic COVID-19 identification using chest X-ray images. The DarkCovidNet model is proposed to provide accurate diagnostics for multi-class classification (COVID vs. Normal vs. Pneumonia) and binary classification (COVID vs. Normal). This model is achieved a classification accuracy of 87.02% for multi-class cases and 98.08% for binary classes. Ioannis et al. [16] trained different pre- trained deep learning models on two datasets. The first is a collection of 1427 X-ray images including 504 images of normal cases, 700 images with confirmed bacterial pneumonia and 224 images with confirmed Covid-19 cases. The second is a dataset including 504 images of normal cases, 714 images with confirmed viral pneumonia and bacterial and 224 images with confirmed Covid-19 cases. Their model achieved an accuracy of 98.75% and 93.48% for two and three classes. Khan et al. [17] introduced CoroNet, a convolution neural network to detect COVID-19 using X-ray and CT scans. This model is based on Xception architecture and pre-trained on ImageNet dataset. The experimental results show that the pretrained network provides an overall accuracy of 89.6% and 95% for 4 classes (pneumonia viral vs. COVID-19 vs. Pneumonia bacterial vs. normal) and 3 classes (normal vs. COVID-19 vs. Pneumonia).Xu et al. [18] established an early screening model to differentiate COVID-19 from Influenza-A viral pneumonia and healthy cases using 618 pulmonary CT samples (i.e., 175 healthy persons, 224 patients with Influenza-A, and 219 patients with COVID-19). This model achieves a total accuracy of 86.7%. S. Wang et al. [19] proposed a deep CNN model to classify COVID-19 from viral pneumonia using 99 Chest CT images (i.e., 55 viral pneumonia and 44 COVID-19). The results of testing dataset show an overall accuracy of 73.1%, along with a sensitivity of 74.0% and a specificity of 67.0%. L. Li et al. [20] trained A ResNet50 model (COVNet) to distinguish COVID-19 from pneumonia and non-pneumonia using 4356 chest CT images (i.e., 1735 pneumonia, 1325 non-pneumonia and 1296 COVID-19). The results show that the COVNet model provides a specificity of 96%, a sensitivity of 90%, and AUC of 0.96 in classifying COVID-19. Y. Song et al. [21] introduced a system based deep learning, called DeepPneumonia to identify patients with COVID-19 from healthy people and bacterial pneumonia patients using Chest CT images. This model achieves an overall accuracy of 86.0% for (COVID-19 vs. bacterial pneumonia) classification and an overall accuracy of 94.0% for (COVID-19 vs. healthy) classification. B. Ghoshal et al. [22] trained a Bayesian Deep Learning classifier using transfer learning method to estimate model uncertainty using COVID-19 X-Ray images.
The results show that Bayesian inference improves detection accuracy of the standard VGG16 model from 85.7% to 92.9%. Zhang et al. [23] developed a deep model to identify COVID-19 infection from X-ray images. This model is trained on a dataset comprising X-ray images from 1008 non-COVID-19 pneumonia patients and 70 COVID-19 patients and achieved sensitivity of 96.0% and specificity of 70.7% along with an AUC of 95.2%.
3. The proposed CVDNet model
In this section, we present a novel deep learning based on CNN architecture, CVDNet, to detect COVID-19 infection from normal and other pneumonia cases using chest X-ray images. We first formalize the operations carried out by the network. Next, we give a detailed description of the proposed CVDNet and the training process used.
3.1. Formalization
We consider the problem of supervised learning with given dataset of NS training samples .
Let be the set of input chest x-ray data and be the set of corresponding ground truth labels, with, an image with H, W, L and C denote height, width, grayscale values and the number of channels respectively; and ti ∈ {0, 1, 2} where 0 refers to COVID-19, 1 refers to normal cases and 2 refers to viral pneumonia cases. The purpose of supervised learning is to build a classifier fw: X → Y, parameterized by a weight w. The output space Y can be different from the label space ℑ, in which case a function g: Y → ℑ is used to get the final prediction. More formally, the aim is to minimize the prediction error rate on the training set, which quantifies the difference between fw(x) and its ground truth label. The training process would be an iterative process consists of finding a set of parameters w, which minimizes the following average loss function on the training set
| (1) |
where denotes the output of the decision fw which predicts the class yi of example xi and represents a loss function of predicted label yi and ground truth label ti. We put l at the loss of cross entropy,
3.2. Network structure of CVDNet
In the previous works summarized in Table 1 , we can see that Kumar [14] suggested a deep CNN that achieves the highest accuracy (95.38%). Inspired by this work which mainly focuses on the ResNet [24], we propose to improve this network in order to more accurately distinguish COVID-19 cases from normal and viral pneumonia cases using chest x-ray images. ResNet introduced the concept of Residual Network and used skip-connections which ignore the formation of a few layers and connects directly to the output. The idea behind this residual network is that the input x passes through a series layers which we denote by F(x). This result is then added to the original input x:.
Table 1.
Representative works for Chest x-ray images based on the detection of COVID-19 infection.
| Literature | Models | Dataset | Performance |
|---|---|---|---|
| L. Wang et al. [12] | COVID-Net pre-trained with ImageNet | 5941 chest x-ray images across 2839 patient (1203 normal + 45 COVID19 + 660 non-COVID viral pneumonia + 931 bacterial pneumonia) | Accuracy of 92.4% for 2-classes and 83.5% for 4-classes |
| Hemdan et al. [13] | COVIDX-Net : based on DenseNet201, Inception v3, VGG19, MobileNet v2, Xception, InceptionResNet v2 and ResNet v2 | 50 X-ray images comprising 25 cases with COVID-19 and 25 cases without any infections | F1-scores of 0.89 for normal and 0.91 for COVID-19 |
| P. Kumar et al. [14] | Deep features from Resnet50 + SVM classifier | Dataset collected from GitHub and Kaggle comprising 25 cases with COVID-19 and 25 cases without any infections | Accuracy of 95.38% |
| Ozturk et al. [15] | DarkCovidNet | X-ray images comprising 125 with COVID-19, 500 normal and 500 Pneumonia cases | Accuracy of 87.02% for 3-class cases |
| Ioannis et al. [16] | VGG-19 | 1427 X-ray images including 504 images of normal cases, 700 images with confirmed bacterial pneumonia and 224 images with confirmed Covid-19 cases. | Accuracy of 93.48% for three classes. |
| Khan et al. [17] | CoroNet | Images collected from Kaggle repository, comprising 290 COVID-19, 1203 normal, 931 viral Pneumonia and 660 bacterial Pneumonia chest x-ray images. | Accuracy of 89.6% and 95% for 4 and 3 classes, respectively. |
| X. Xu et al. [18] | ResNet +Location Attention | 618 pulmonary CT samples (i.e., 175 healthy persons, 224 patients with Influenza-A, and 219 patients with COVID-19) | Accuracy of 86.7% |
| S. Wang et al. [19] | M-Inception | 99 Chest CT images (i.e., 55 viral pneumonia and 44 COVID-19) | Accuracy of 73.1%, along with a sensitivity of 74.0% and a specificity of 67.0% |
| L. Li et al. [20] | COVNet | 4356 chest CT images (i.e., 1735 pneumonia, 1325 non-pneumonia and 1296 COVID-19). | Specificity of 96%, sensitivity of 90%, and AUC of 0.96 |
| Y. Song et al. [21] | DeepPneumonia | Chest CT scans of 275 patients (88 patients infected with COVID-19, 101 patients infected with bacterial pneumonia, and 86 healthy) | Accuracy of 86.0% for (COVID-19 vs. bacterial pneumonia) classification and 94.0% for (COVID-19 vs. healthy) classification |
| B. Ghoshal et al. [22]. | Dropweights based Bayesian Convolutional Neural Networks | 5941 chest x-ray images across four classes (Bacterial Pneumonia: 2786, Normal: 1583, COVID-19: 68 and non-COVID-19 Viral Pneumonia: 1504). | Accuracy of 92,90% |
| J. Zhang et al. [23] | Deep CNN based on Backbone network | X-ray images from 1008 non-COVID-19 pneumonia patients and 70 COVID-19 patients | Sensitivity of 96.0% and specificity of 70.7% along with an AUC of 95.2%. |
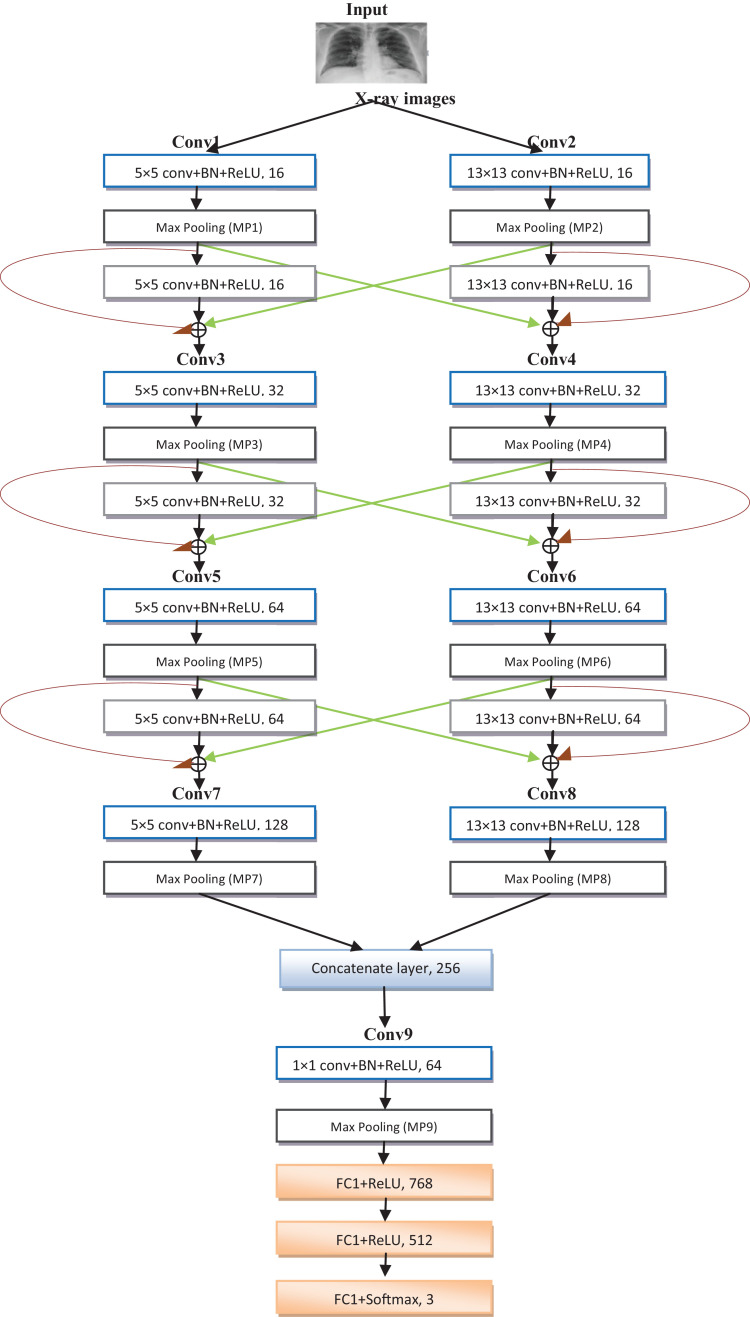
The proposed designed CNN, CVDNet, as shown in Fig. 1 consists of two parallel columns. The two columns have the same structures except for the sizes of filters. In the following, we introduce the proposed model in detail and we explain its main components such as convolutional layers, Activation unit, pooling, batch normalization, fully connected layers and the proposed block.
-
a)
Convolutional layer
Convolution is a fundamental operation for convolutional neural networks which allow extracting specific information characterizing the input. It is a mathematical operation which consists in applying a succession of filters on a set of regions of convolution of the input by a sliding window in order to produce as output a set of feature map.
- Let and be the input and output of the ith feature map in the lth. Let x 0 ∈ ℜH × W be the 2D input image and let be the kernel in the lth layer linking ith input map to jth output map. To obtain each is convoluted with the corresponding kernel, and the results are added with the bias . Finally a non linear function φ(.) is applied. Mathematically, the output of convolutional layer can be given as
(2) Where * is the convolution operation and Mj ={i | ith in the layer map linked to jth map in the lth layer}
-
a)
Activation function
Fig. 1.
The structure of the proposed CVDNet model.
After each convolution, the convolutional neural network applies a transformation to the convoluted function, in order to introduce non-linearity into the model. The ReLU function has become very popular in recent years as an activation function because it offers good performance during learning; It improves gradient convergence and it is less expensive in computation. The ReLU function is interpreted by the formula:. If the input is negative the output is 0 and if it is negative then the output is x. The output value xi (k) of a neuron i of layer k in a ReLU layer is expressed as:
| (3) |
-
a)
Pooling layer
The pooling layer consists in gradually reducing the sizes of the characteristic cards obtained during the layers of convolution while keeping the most relevant information. Thanks to this layer, the quantity of parameters and computation in the network are reduced, and this will allow controlling over-fitting. The pooling operation is characterized by a window of size hp × hp which moves with sp stride on each feature map. It is often approached by two main approaches:
-
aMax-pooling: consists of returning the maximum local value at the level of each pooling window
-
bAvg-pooling: calculates the average of the local values of each pooling window.
-
a
-
a)
Batch normalization layer
Batch normalization (BN) is a technique that greatly improves convergence during training. It consists in normalizing on average and in variance the outputs of the layers of the network.
Given a mini batch of size m, the normalized values and their linear transformations (y 1, y 2, ..., ym). The Batch normalization BN γ, β is referred to the transform BN γ, β: x 1, x 2, ..., xm → y 1, y 2, ..., ym and it is computed as:
| (4) |
where μ B and are mini-batch mean and variance, respectively, β and γ are parameters learnable via backpropagation and ɛ is a small positive number to avoid division by zero.
-
a)
Fully connected layers
In this layer, the neurons are connected with each of the neurons of the volume corresponding to its input, i.e. with each of the neurons of the previous layer. This layer outputs a vector of K dimensions, where K represents the number of classes that the network will be able to predict. This vector contains the classification probabilities for each class of an image. The fully-connected (FC) layer determines the link between the position of features in the image and a class. Indeed, the entry being the result of the previous layer, it corresponds to a feature map: the high values indicate the location of this feature in the image.
Let and be two consecutive layers and let be the weight matrix connecting them; where m(k) is the number of neurones in layet k. The output of a FC layer, x (k), is expressed as
The final fully connected layer is forwarding to a softmax function, which determines the class probabilities given the input images. The softmax function is defined as.
where is the input vector to the softmax function, the output of softmax (z)j in a range between 0 and 1 and
In chest x-ray images infected by COVID-19, Ground-glass opacities are observed, which are considered as the typical early features of COVID-19 infection. In chest x-ray images infected by COVID-19, Ground-glass opacities are observed, which is considered as the typical early features of COVID-19 infection. These features are often found in different location including in a sub-pleural and peripheral as well as they come in different sizes, shapes and quantities. For large size of Ground-glass opacities features, if using a small filter, the resulting of feature maps have few pixel containing small objet. Therefore, it does not capture holistic information. This leads to poor classification accuracy. Meanwhile, for small size of Ground-glass opacities features, if using a large filter, the receptive fields are too large. Thus, most of the information captured is irrelevant. This leads to poor classification accuracy due to loss of details. Consequently, we need to use a small filter for extracting local features, and a large filter for detecting global features. However, the sizes of the features of the input image are unpredictable and random which is not practical to construct a filter size for each scale of objects. To overcome this problem, we propose to design a structure to capture multi-scale features i.e., we applied to the input two parallel convolutions with large and small filters; each level detects a scale of features. Using these two filters in parallel is more suitable for extracting different scales of features than a single scale filter.
For the convolution, we use the concept of residual technique. The excellent performance of Residual network has been proven medical imaging tasks, which can solve the vanishing gradient problem. By connecting the residuals from the previous layers, this can also help to compensate the loss of details for local features or to obtain more holistic information for global features. However, to detect more details and more holistic information, we propose to merge the multi-scale features via residuals connections and we propose to link residual information not only at the same level but also at other levels. The merge of different scales of features makes it possible to identify several scales of objects.
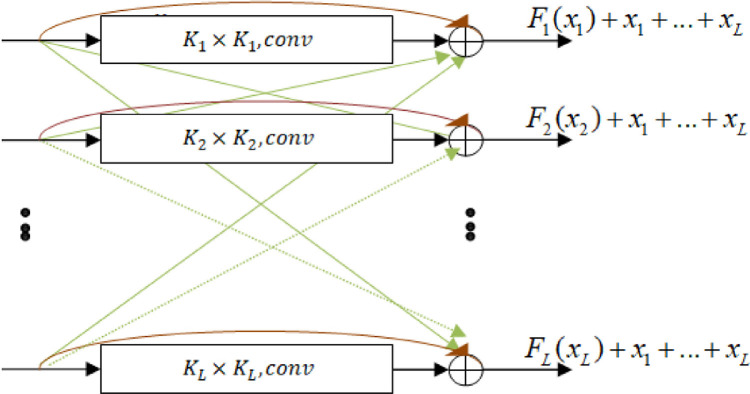
The output of the proposed block which consists of connecting a residual with all the other levels as shown in Fig. 2 is defined as:
where L is the total parallel levels, i is theith level, oi is the output of the ith level, x 1, x 2, ..., xL are the inputs of the block and F 1(x 1), F 2(x 2), ..., FL(xL) are their corresponding outputs, respectively.
Fig. 2.
Illustration of the proposed block. Inputs x are connected to the output of their layers, and added to the inputs of other levels. .
In this study, we use only two parallel levels by applying large and small filters to extract global and local features.
Based on the proposed block, we design CVDNet to classify COVID-19 infection from normal and other pneumonia cases. The proposed CVDNet model took as input chest x-ray images. Initially, the images went through a preprocessing step which consists in cropping and resizing it. Indeed, the images are resized because several letters, medical symbols and art craft are made on them as well as they are cropped because they come from different sources and consequently their sizes vary. Therefore, the size of the input images is changed to 256-by-256-by-1. After this step, the images are processed by two parallel convolutional layer having a size of 5 × 5 and 13×13 with a stride of one respectively, to produce 16 feature maps for each of them, followed by the batch normalization and ReLU layers (conv1 and conv2). The uses of conv1 and conv2 change the images dimension from 1 × 256 × 256 to16 × 251 × 251 and from 1 × 256 × 256 to 16 × 244 × 244, respectively. Features generated by conv1 and conv2 are forwarding to 2 × 2 max pooling layer (MP1 and MP2) with a stride of two to sub-sample the input images by reducing their dimensionality and therefore to provide a spatial invariance and to reduce the quantity of parameters and computation in the network. The dimensions of the resulting images after MP1 and MP2 layers will be reduced to 16 × 126 × 126 and 16 × 122 × 122. Moreover, the output of features maps is forwarding to block 1 (see Fig. 2), which contains two parallel convolutional layers with a size of 5 × 5 and 13×13. The output of each convolutional layer is added to the output of MP1 and MP2 in the same dimension. The output images dimension of this block is 16 × 126 × 126. In this block, it has no pooling layer and after each convolutional layer we normalize each batch and we apply ReLU activation function. The output of block 1 passes through 5 × 5 convolutional layer and 13 × 13 convolutional layer with a stride of 1 to generate 32 feature maps for each of them, followed by BN and ReLU function (conv3 and conv4). The uses of conv3 and conv4 change the images dimension from 16 × 126 × 126 to 32 × 122 × 122 and from 16 × 126 × 126 to 32 × 114 × 114, respectively. Features generated by conv3 and conv4 are forwarding to 2 × 2 max pooling layer (MP3 and MP4). The dimensions of the resulting images after MP3 and MP4 layers will be reduced to 32 × 61 × 61 and 32 × 57 × 57. The max pooling layer are forwarded to block 2 which is the same as the block 1 except it produced 64 feature maps for each convolutional layer. Next came again 5 × 5 and 13 × 13 parallel convolutional layers with a stride of 1, followed by 2 × 2 max pooling with a stride of 2. After this layer, block 3 is applied which is the same as the block 1 and block 2 except it produced 64 feature maps for each convolutional layer. The output of block 3 passes through 5 × 5 convolutional layer and 13 × 13 convolutional layer with a stride of 1 to generate 128 feature maps for each of them, followed by BN, ReLU function (conv7 and conv8) and max pooling (MP7 and MP8). The output of feature maps from the two parallel layers is then concatenated and passes through 1 × 1 convolutional layer (conv9) with a stride of 1in order to decrease the number of filters from 256 to 64, so the output will be reduced from 256 × 12 × 12 to 64 × 12 × 12. After this, a max pooling (MP9) is applied to reduce images dimension from 64 × 12 × 12 to 64 × 6 × 6. Finally the network had three fully connected layers; the first one has 768 feature maps each of size 1 × 1. Each of the 768 is connected to all the 2304 nodes (64 × 6 × 6) in the previous layer. The second one has 512 units is connected to all the 768 nodes and finally, there is a fully connected softmax output layer with 3 possible values. The highest value determined the predicted class. In total, the network has 5317,667 parameters. Table 2 summarizes the configuration of the proposed CVDNet architecture for COVID-19 dataset.
Table 2.
The configuration of the proposed CVDNet architecture for COVID-19 dataset.
| Layers | Filters size | Output shape (depth × height × width) |
|---|---|---|
| Input image | – | 1 × 256 × 256 |
| Conv1 | 5 × 5, stride =1 | 16 × 251 × 251 |
| MP 1 | 2 × 2, stride=2 | 16 × 126 × 126 |
| Conv2 | 13 × 13, stride =1 | 16 × 244 × 244 |
| MP 2 | 2 × 2, stride=2 | 16 × 122 × 122 |
| Block 1 | , stride=1 | 16 × 126 × 126 |
| Conv3 | 5 × 5, stride =1 | 32 × 122 × 122 |
| MP 3 | 2 × 2, stride=2 | 32 × 61 × 61 |
| Conv4 | 13 × 13, stride =1 | 32 × 114 × 114 |
| MP 4 | 2 × 2, stride=2 | 32 × 57 × 57 |
| Block 2 | , stride=1 | 32 × 61 × 61 |
| Conv5 | 5 × 5, stride =1 | 64 × 57 × 122 |
| MP 5 | 2 × 2, stride=2 | 64 × 28 × 28 |
| Conv6 | 13 × 13, stride =1 | 64 × 49 × 49 |
| MP 6 | 2 × 2, stride=2 | 64 × 24 × 24 |
| Block 3 | , stride=1 | 64 × 28 × 28 |
| Conv7 | 5 × 5, stride =1 | 128 × 24 × 24 |
| MP 7 | 2 × 2, stride=2 | 128 × 12 × 12 |
| Conv8 | 13 × 13, stride =1 | 128 × 16 × 16 |
| MP 8 | 2 × 2, stride=2 | 128 × 8 × 8 |
| Concatenation | – | 256 × 12 × 12 |
| Conv9 | 1 × 1, stride =1 | 64 × 12 × 12 |
| MP 9 | 2 × 2, stride=2 | 64 × 6 × 6 |
| Flatten | – | (1 × 2304) |
| FC1+ReLU | – | (1 × 768) |
| FC2+ReLU | – | (1 × 512) |
| FC3+Softmax | – | (1 × 3) |
3.3. Training phase
The proposed network was trained end-to-end with the adaptive moment estimation (Adam) optimizer [25] to minimize the cross-entropy loss function presented in (Eq. (1)). Adam is one of the most widely used and effective methods for gradient descent optimization. This method is based on the gradient descent with small batches and his idea is to adapt the learning rate from an estimate of the first and second moment of the gradient. The estimation of the first and second moment requires the updating of two additional variables for each parameter of the network. This type of method has the advantage of being relatively robust and makes it possible to automatically adapt the learning rate during learning for each weight. Mathematically, Adam optimizer can be defined as:
where gt is the gradient at time t, mt and vt are the first moment (the mean) and the second moment (non-centered variance) of the gradient, and is mt and vt after deviation correction respectively, the hyper-parameters β1 and β2 are used to perform execution means on the moments mt and vt respectively, wt is the model parameter value at time t, is the model parameter value of the previous moment, ɛ is a precision parameter and α is learning rate.
In practice, we set β 1, β 2, ɛ and α to 0.9, 0.999, and 0.0001. Furthermore, we initialize randomly all the hyper parameters and all weights by normal distribution with mean of 0 and standard deviation of 0.01, as well as all biases as 0.
4. Experimental setup
In this section, we describe the dataset used in this study and we detail the distribution of images to train, validation and test set; as well as we assess by different evaluation metrics the performance of our proposed model, CVDNet, to classify patients affected with COVID-19 from viral pneumonia patients and healthy persons using chest x-ray images.
4.1. Dataset description
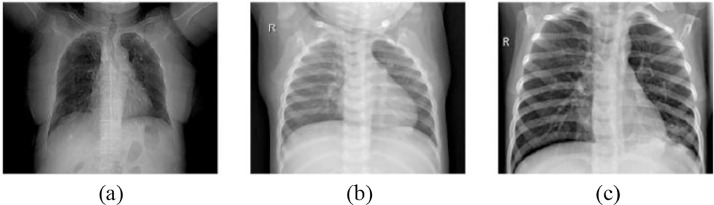
In this study, we used chest x-ray images obtained from Kaggle's COVID-19 Radiography Database [26]. This dataset was created for three different types of images classified as chest x-ray images belonging to patients infected with COVID-19, chest x-ray images of cases with viral pneumonia and Chest x-ray images of healthy parsons (Normal). Out of 2905 chest x-ray images, there are 219 COVID-19 images, 1345 viral pneumonia images and 1341 normal. Fig. 3 shows some images of this dataset and shows the difference between COVID-19, viral pneumonia and normal case images. The following findings are observed in the x-ray images of COVID-19 patients.
-
•
Ground-glass opacities (peripheral, bilateral, subpleural, multifocal, posterior, basal and medial).
-
•
Air space consolidation.
-
•
Bronchovascular thickening (in the lesion).
-
•
A crazy paving appearance. (GGOs and inter-/intra-lobular septal thickening).
-
•
Traction bronchiectasis.
Fig. 3.
Example of chest X-ray images: (a) COVID-19 chest x-ray image, (b) normal chest x-ray image, (c) viral pneumonia chest x-ray image.
Similarly, the following findings are observed in the x-ray images of pneumonia patients
-
•
Reticular opacity
-
•
Ground-glass opacities (GGO) central distribution, unilateral
-
•
Distribution more along the bronchovascular bundle
-
•
Vascular thickening
-
•
Bronchial wall thickening
The COVID-19 data is publically available on Kaggle's website and it's collected from different databases: chest x-ray images with COVID-19 were taken from Italian Society of Medical and Interventional Radiology COVID1-19 Database (SIRM) [27] and from Novel Corona Virus 2019 Dataset which is developed by Cohen et al. in Github [28], as well as from different recently published articles. The viral pneumonia and normal images were collected from Kaggle's Chest X-Ray pneumonia dataset [29].
4.2. Results and discussion
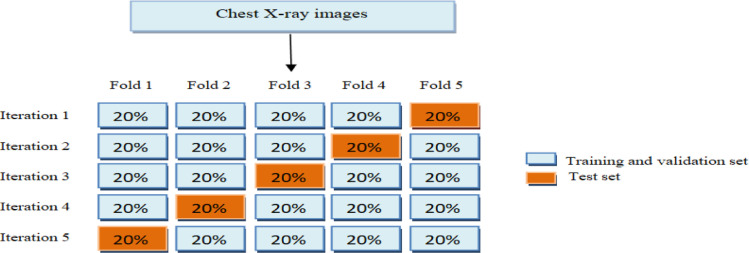
In this section we perform experiments to assess the performance of the proposed CVDNet, which is trained to classify chest x-ray images into three categories: Normal, COVID-19 and Viral pneumonia. The efficiency of the proposed model is evaluated using the five- fold cross-validation approach for (3-class) classification problem. The dataset used was divided into 5 independent and equal sets. Four out of five sets were used to train and validate our model whereas the remaining set was employed for the test (i.e. 70% of chest X-ray images are used for training, 10% for validation and 20% for testing). More details, 219 COVID-19 chest x-ray images were used, where 158 out of 219 were randomly selected for training, 18 images for validation and 43 images for testing. Moreover, 1341 normal chest x-ray images were used, where 965 out of 1341 were randomly selected for training set, 107 images for validation and remaining 269 images for testing as well as 1345 viral pneumonia chest x-ray images were used, where 969 out of 1345 were randomly selected for training, 107 images for validation and remaining 269 images for testing. The strategy of split dataset was repeated five times as illustrated in Fig. 4 and the distribution of chest x-ray images from sub-sets were summarized in Table 3 . All experiments were implemented in Tensorflow and the proposed CVDNet was trained for 20 epochs with a batch size of 8.
Fig. 4.
Schematic diagram of training, validation and test set used in the 5-fold cross-validation.
Table 3.
Details of training, validation and test set.
| Class | Number of images | Training | Validation | Test |
|---|---|---|---|---|
| COVID-19 | 219 | 158 | 18 | 43 |
| Viral pneumonia | 1345 | 969 | 107 | 269 |
| Normal | 1341 | 965 | 107 | 269 |
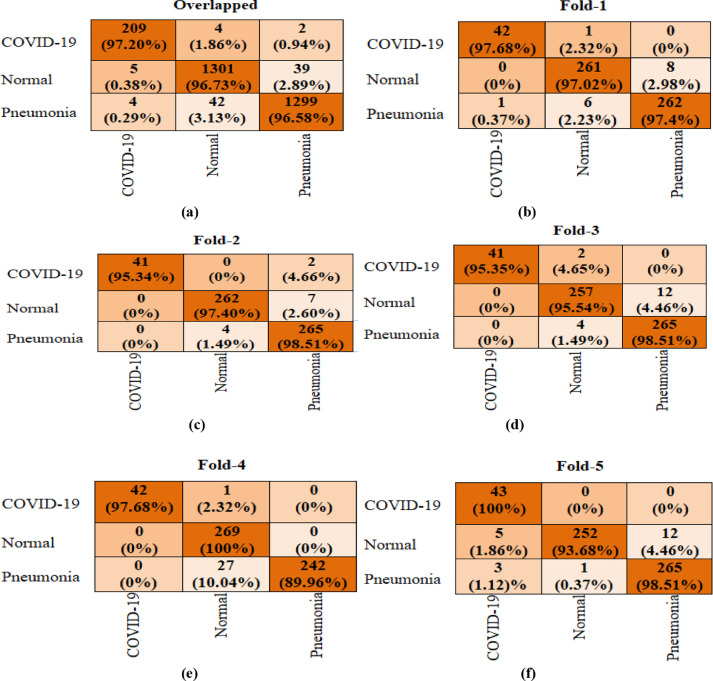
The performance of CVDNet for 3-class classification was measured for each fold using confusion matrix (CM) and top metrics given below such as precision, recall, accuracy, and F1-score. The overlapped and each separate CM are shown in Fig. 5 . Furthermore, Overall precision, recall, accuracy, and F1-score computed for each class (COVID-19, Normal and viral pneumonia) as well as for each fold are presented in Tables 4 , 5 , 6 , 7 , 8 and 9 .
Fig. 5.
Confusion matrix results of our CVDNet for 3-class Classification. (a) overlapped CM, (b) Fold-1 CM, (c) Fold-2 CM, (d) Fold-3 CM, (e) Fold-4 CM, and (f) Fold-5 CM.
Table 4.
Precision, F1-score and recall values in fold 1 for Normal, COVID-19 and pneumonia classes of the proposed CVDNet.
| Class | Precision (%) | Recall (%) | F1-score (%) |
|---|---|---|---|
| COVID-19 | 97.67 | 97.67 | 97.67 |
| Normal | 97.39 | 97.03 | 97.21 |
| Pneumonia | 97.04 | 97.40 | 97.22 |
Table 5.
Precision, F1-score and recall values in fold 2 for Normal, COVID-19 and pneumonia classes of the proposed CVDNet.
| Class | Precision (%) | Recall (%) | F1-score (%) |
|---|---|---|---|
| COVID-19 | 100 | 95.35 | 97.62 |
| Normal | 98.50 | 97.40 | 97.94 |
| Pneumonia | 96.72 | 98.51 | 97.61 |
Table 6.
Precision, F1-score and recall values in fold 3 for Normal, COVID-19 and pneumonia classes of the proposed CVDNet.
| Class | Precision (%) | Recall (%) | F1-score (%) |
|---|---|---|---|
| COVID-19 | 100 | 95.35 | 97.62 |
| Normal | 97.72 | 95.54 | 96.62 |
| Pneumonia | 95.67 | 98.51 | 97.07 |
Table 7.
Precision, F1-score and recall values in fold 4 for Normal, COVID-19 and pneumonia classes of the proposed CVDNet.
| Class | Precision (%) | Recall (%) | F1-score (%) |
|---|---|---|---|
| COVID-19 | 100 | 97.67 | 98.82 |
| Normal | 90.57 | 100 | 95.05 |
| Pneumonia | 100 | 89.96 | 94.72 |
Table 8.
Precision, F1-score and recall values in fold 5 for Normal, COVID-19 and pneumonia classes of the proposed CVDNet.
| Class | Precision (%) | Recall (%) | F1-score (%) |
|---|---|---|---|
| COVID-19 | 84.31 | 100 | 91.49 |
| Normal | 99.60 | 93.68 | 96.55 |
| Pneumonia | 95.67 | 98.51 | 97.07 |
Table 9.
Performance of the proposed CVDNet on each fold.
| Folds | Precision (%) | Accuracy (%) | Recall (%) | F1-score (%) |
|---|---|---|---|---|
| Fold 1 | 97.37 | 97.25 | 97.37 | 97.37 |
| Fold 2 | 98.40 | 97.76 | 97.09 | 97.72 |
| Fold 3 | 97.80 | 96.90 | 96.47 | 97.10 |
| Fold 4 | 96.86 | 95.18 | 95.88 | 96.20 |
| Fold 5 | 93.20 | 96.39 | 97.40 | 95.04 |
| Average | 96.72 | 96.69 | 96.84 | 96.68 |
In the following, we detail the performance of our model and we discuss the obtained results.
It can be observed from the confusion matrix reported in Fig. 5 that the proposed model has detected in fold-1 42 out of 43 patients with COVID-19 as having COVID-19, 261 out of 269 normal patients as normal patient and 262 out of 269 patients with viral pneumonia as having viral pneumonia infection; and it misclassified one patient with COVID-19 as normal patient, 8 normal patients as having viral pneumonia infection, one patient with viral pneumonia infection as having COVID-19 and 6 patients with viral pneumonia infection as normal patients. In fold-2, the proposed CVDNet has detected 41 out of 43 patients with COVID-19 as having COVID-19, 262 out of 269 normal patients as normal patient and 265 out of 269 patients with viral pneumonia as having viral pneumonia infection; and it misidentified 7 normal patients as having viral pneumonia infection and 4 patients with viral pneumonia infection as normal patients. In fold-3, CVDNet has achieved an accuracy of 95.35%, 95.54% and 98.51% to classify COVID-19, normal and viral pneumonia infection, respectively. In fold-4, CVDNet has achieved an accuracy of 97.68%, 100% and 89.96% to classify COVID-19, normal and viral pneumonia infection, respectively. In fold-5, our CVDNet has detected 43 patients with COVID-19 as having COVID-19, 252 out of 269 normal patients as normal patient and 265 out of 269 patients with viral pneumonia as having viral pneumonia infection; and it misidentified 5 normal patients as having COVID-19 infection, 12 normal patients as having viral pneumonia infection, 3 patients with viral pneumonia infection as COVID-19 infection and one patient with viral pneumonia infection as normal patients. In overall, our model achieved an average accuracy of 97.20%, 96.73% and 96.58% to classify COVID-19, normal and viral pneumonia infection, respectively.
From Table 4, it can be noted that our CVDNet has achieved in fold-1 a precision of 97.67%, 97.39% and 97.04% for COVID-19, Normal and pneumonia classes, respectively and a recall of 97.67%, 97.03% and 97.40% for COVID-19, Normal and pneumonia classes, respectively as well as a F1- score of 97.67%, 97.21% and 97.22% for COVID-19, Normal and pneumonia classes, respectively. From Table 5, it can be seen that CVDNet has achieved in fold-2 a precision of 100%, 98.50% and 96.72% for COVID-19, Normal and pneumonia classes, respectively and a recall of 95.35%, 97.40% and 98.51% for COVID-19, Normal and pneumonia classes, respectively as well as a F1- score of 97.62%, 97.94% and 97.61% for COVID-19, Normal and pneumonia classes, respectively. From Table 6, it can be observed that our model has achieved a precision of 100%, 97.72% and 95.67% for COVID-19, Normal and pneumonia classes, respectively and a recall of 95.35%, 95.54% and 98.51% for COVID-19, Normal and pneumonia classes, respectively as well as a F1- score of 95.67%, 98.51% and 97.07% for COVID-19, Normal and pneumonia classes, respectively. From Table 7, it can be observed that our model has achieved a precision of 84.31%, 99.60% and 95.67% for COVID-19, Normal and pneumonia classes, respectively and a recall of 100%, 93.68% and 98.51% for COVID-19, Normal and pneumonia classes, respectively as well as a F1- score of 91.49%, 96.55% and 97.07% for COVID-19, Normal and pneumonia classes, respectively. From Table 8, it can be observed that our model has achieved a precision of 100%, 90.57% and 100% for COVID-19, Normal and pneumonia classes, respectively and a recall of 97.67%, 100% and 89.96% for COVID-19, Normal and pneumonia classes, respectively as well as a F1-score of 98.82%, 95.05% and 94.72% for COVID-19, Normal and pneumonia classes, respectively. Therefore, the proposed CVDNet reaches an average precision, accuracy, recall and F1-score of 96.72%, 96.69, 96.84% and 96.68%, respectively for 3- class classification as shown in Table 9.
In summary, the main advantages of the proposed CVDNet are as follows:
-
-
Our model performed remarkably well in detecting COVID-19 for three class classification problem using chest x-ray images.
-
-
The model successful in classifying COVID-19 class with an accuracy of 97.20%.
-
-
It is an effective model that can assist radiologists in the diagnosis of COVID-19.
In overall, the results obtained showed that our CVDNet model outperforms the studies reported in the Table 1 in terms of accuracy for 3-class classification task (COVID-19 vs. normal vs. viral pneumonia). These results represent promising and encouraging performances in the detection of COVID-19 from chest x-ray images. This will greatly assist radiologists in overcoming load on hospitals and the medical system.
5. Conclusion and future work
In this paper, we proposed a novel deep convolutional neural network (CVDNet) model for the detection of COVID-19 cases, in order to distinguish more precisely the patients affected by COVID-19 from healthy persons and viral pneumonia patients using chest X-ray images. Our model has been trained on a small dataset of few images of various COVID-19, viral pneumonia and normal cases from publically available database. Moreover, the classification efficiency of our CVDNet was measured using k- fold cross-validation. It was observed that CVDNet achieved an average accuracy of 97.20% for detecting COVID-19 and an average accuracy of 96.69% for three-class classification (COVID-19 vs. normal vs. viral pneumonia), which exhibit superior and promising performance in classifying COVID‐19 cases. Based on these encouraging results, our CVDNet model is an interesting tool that can help clinicians to diagnose and detect COVID-19 infection in a very short time. In the future research direction, we will work to develop our CVDNet in order to detect COVID-19 cases from other lung diseases. Moreover, we intend to validate the proposed model by using more images coming from different hospitals and to address CT images for the detection of COVID-19 and to compare the obtained results with our CVDNet trained using X-ray images.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Wu F., Zhao S., Yu B. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization . Emergencies preparedness, response, disease outbreak news. World Health Organization (WHO); 2020. Pneumonia of unknown cause–China. [Google Scholar]
- 3.Zu Z.Y., Jiang M.D., Xu P.P., Chen W., Ni Q.Q., Lu G.M. Coronavirus disease 2019 (COVID-19): a perspective from China. Radiology. 2020 doi: 10.1148/radiol.2020200490. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.LeCun Y., Bengio Y., Hinton G. Deep learning. Nature. 2015;521(7553):436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 5.Esteva A., Kuprel B., Novoa R.A. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542(7639):115–118. doi: 10.1038/nature21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yoon S.H., Lee K.H. Chest radiographic and CT findings of the 2019 novel coronavirus disease (COVID-19): analysis of nine patients treated in Korea. Korean J. Radiol. 2020;21(4):494–500. doi: 10.3348/kjr.2020.0132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Talo M., Yildirim O., Baloglu U.B., Aydin G., Acharya U.R. Convolutional neural networks for multi-class brain disease detection using MRI images . Comput. Med. Imag. Graph. 2019;78 doi: 10.1016/j.compmedimag.2019.101673. [DOI] [PubMed] [Google Scholar]
- 8.Celik Y., Talo M., Yildirim O., Karabatak M., Acharya U.R. Automated invasive ductal carcinoma detection based using deep transfer learning with whole-slide images. Pattern Recognit. Lett. 2020;133:232–239. [Google Scholar]
- 9.Tan J.H., Fujita H., Sivaprasad S., Bhandary S., Rao A.K., Chua K.C., Acharya U.R. Automated segmentation of exudates, haemorrhages, microaneurysms using single convolutional neural network. Inf. Sci. 2017;420:66–76. [Google Scholar]
- 10.Rajpurkar P., Irvin J., et al. Chexnet: radiologist-level pneumonia detection on chest x-rays with deep learning, 2017 arXiv preprint
- 11.Gaal G., Maga B., Lukacs A. Attention U-net based adversarial architectures for chest x-ray lung segmentation, 2020arXiv preprint
- 12.Wang L., Lin Z.Q. and Wong A. COVID-net: a tailored deep convolutional neural network design for detection of COVID-19 cases from chest x-ray images. arXiv, Mar. 2020. [DOI] [PMC free article] [PubMed]
- 13.Hemdan E.E., Shouman M.A., Karar M.E. Covidx-net: a framework of deep learning classifiers to diagnose covid-19 in X-ray images. arXiv preprint arXiv:2003.11055. 2020 Mar 24.
- 14.Kumar P. and Kumari S. Detection of coronavirus disease (COVID-19) based on deep features. preprints.org, no. March, p. 9, Mar. 2020.
- 15.Ozturk T., Talo M., Yildirim E.A., Baloglu U.B., Yildirim O., Acharya U.R. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput Biol Med. 2020 doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ioannis D., Apostolopoulos1, T.B. COVID-19: automatic detection from x-ray images utilizing transfer learning with convolutional neural networks. 2020. [DOI] [PMC free article] [PubMed]
- 17.Khan A.I., Shah J.L., Bhat M.M. CoroNet: a deep neural network for detection and diagnosis of COVID-19 from chest X-ray images. Comput Methods Programs Biomed. 2020;196 doi: 10.1016/j.cmpb.2020.105581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu X., et al. Deep learning system to screen coronavirus disease 2019 pneumonia. arXiv, Feb. 2020.
- 19.Wang S., et al. A deep learning algorithm using CT images to screen for Corona Virus Disease (COVID-19). medRxiv, p. 2020.02.14.20023028, Apr. 2020. [DOI] [PMC free article] [PubMed]
- 20.Li L., Qin L., Xu Z., Yin Y., Wang X., Kong B. Artificial intelligence distinguishes COVID-19 from community acquired pneumonia on chest CT. Radiology. 2020 doi: 10.1148/radiol.2020200905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song Y., Zheng S., Li L., Zhang X., Zhang X., Huang, et al. Deep learning enables accurate diagnosis of novel Coronavirus (COVID-19) with CT images. MedRxiv, 2020. [DOI] [PMC free article] [PubMed]
- 22.Ghoshal B. and Tucker A. Estimating uncertainty and interpretability in deep learning for coronavirus (COVID-19) detection. arXiv:2003.10769, 2020.
- 23.Zhang J., Xie Y., Li Y., Shen C., and Xia Y. COVID-19 screening on Chest X-ray images using deep learning based anomaly detection. arXiv:2003.12338, 2020.
- 24.He K., Zhang X., Ren S., Sun J. Deep residual learning for image recognition. CVPR. 2016 [Google Scholar]
- 25.Kingma D.P., Adam Ba J.L. 3rd International Conference on Learning Representations. ICLR 2015 -Conference Track Proceedings. 2015. A method for stochastic optimization. [Google Scholar]
- 26.Rahman T., Chowdhury M.E.H, Khandakar A., Mazhar R., Kadir M.A., Mahbub Z.B., Islam K.R., Khan M.S., Iqbal A., Al‐Emadi N., Ibne Reaz M.B. 2020; COVID‐19 chest radiography database. [Online] Available: https://www.kaggle.com/tawsifurrahman/covid19‐radiography‐database.
- 27.Societa Italiana di Radiologia Medical Interventistica. 2020. COVID‐19 database. [Online]. Available: https://www.sirm.org/category/senza-categoria/covid-19/.
- 28.Monteral J.C. COVID‐chestxray database. [Online] Available: https://github.com/ieee8023/covid-chestxray-dataset.
- 29.Mooney P.Chest X‐ray images (Pneumonia). [Online] Available: https://www.kaggle.com/paultimothymooney/chest-xray-pne umonia 2018.