ABSTRACT
Brucellosis caused by Brucella melitensis is considered to be one of the most important zoonotic diseases in China. In this study, Conventional bio-typing, MLVA (multiple locus variable-number tandem repeat analysis), and WGS (whole-genome sequencing)-SNP (single nucleotide polymorphism) were used to study the genetic similarity of B. melitensis in northern and southern China and analyze its relationship with worldwide lineages. Currently, the distribution of species/biovars of B. melitensis has obviously changed, and B. melitensis has become the dominant species in southern regions of China. Strains from the southern had a common geographic origin with strains from the northern. Many MLVA-16 events were shared in the genotypes of the southern and northern strains, suggest that genotypic movement occurred from north to south. Based on WGS-SNP analysis, strains from different provinces were closely related and may have descended from one common ancestor, suggests that the southern strains originated from northern China. These data indicate that B. melitensis is a latent “travel bacterium” that spread and expanded from North China to South China. Moreover, B. melitensis strains from China are also genetically related to strains from other Asian regions (Kazakhstan, Russia, Mongolia, and India). The movement of infected sheep and their products requires control.
KEYWORDS: Brucella melitensis, species/biovars, genetic relatedness, MLVA, WGS-SNP, China
Introduction
Brucellosis, a common zoonotic disease globally, is caused by bacteria of the genus Brucella [1], which are nonmotile, gram-negative α-proteobacteria that are facultative intracellular pathogens [2]. Human brucellosis is largely dependent on animal reservoirs and through direct contact with infected animals or consumption of contaminated animal products [3]. At present, Brucella. melitensis, Brucella. abortus, and Brucella. suis remain the main causes for human and animal brucellosis worldwide [4]. Chronic infections with severe complications in humans are a major public health problem [5,6]. The brucellosis epidemiological situation remains complex, and there are serious epidemics in many low-income countries, including in the Mediterranean region, South and Central America, Africa, Asia, the Arabian Peninsula, the Indian subcontinent, Eastern Europe, the Middle East, and China [7]. Despite its low mortality rates, brucellosis is a very important public health problem in rural and pasturing areas in China [8]. Brucellosis has reemerged since 1995, and human brucellosis has been reported in all mainland provinces, a total of 513,034 brucellosis cases were recorded from 1955 to 2014, of which 99.3% were in northern China [9]. Animal husbandry suffers great economic losses from brucellosis due to reduced productivity, the culling of livestock, and costs of associated control measures [10,11]. The incidence of human brucellosis in southern China increased in 2005 and 2014 and the affected area expanded from northern to southern coastal and southwestern areas [9,12]. B. melitensis is responsible for the vast majority of brucellosis in humans and animals in China [13], but the genetic relatedness of B. melitensis within China and its relationship to strains in other world areas is unknown. Investigation of species and genotype distributions, genetic relatedness, and molecular epidemiology of the main circulating strains is essential for understanding the epidemiology of human brucellosis, managing disease outbreaks and for establishing efficient prevention and control programmes [14].
Multiple-locus variable-number tandem-repeat analysis (MLVA) has high power to discriminate closely related strains and can be used for tracing infections [15], achieves result largely in agreement with WGS-SNP-based typing [16]. In addition, its low cost and fast results allow its use as a routine first-line assay [17]. Ma et al. [18] reported that the Brucella strain in Qinghai was different from strains in other regions of the world, possibly owing to the unique geography, such as the high altitude, of the QTP. Extensive genotype-sharing events between isolates obtained from humans and animals showed that yaks, sheep, and blue sheep were important zoonotic reservoirs of brucellosis that caused human infections. Liu et al. [13] reported that human brucellosis in Ulanqab, Inner Mongolia (China) occurred as a multipoint outbreak epidemic caused by multiple common sources of infection. Many shared MLVA-16 genotypes were observed among isolates from different regions of Ulanqab and from other provinces of China. This suggests that infected animal movement between different regions is not controlled. Consequently, an investigation of the genetic relatedness, molecular epidemiology, and potential transmission route of B. melitensis from humans and animals over the whole country is needed. The purpose of this study was to determine the distribution profiles, genetic relatedness, and potential transmission pattern of 1,382 B. melitensis collected from 29 different regions from humans and animals at the whole-country scale.
Materials and methods
Ethics statement
This study was carried out according to the principles of the Declaration of Helsinki. This study is a retrospective investigation of historical strain collections using molecular typing methods, and the research protocol was approved by the Ethics Committees of the National Institute for Communicable Disease Control and Prevention and the Chinese Center for Disease Control and Prevention. All strains from humans were collected as a part of a standard clinical investigation of patients with suspected brucellosis. The patients were anonymized. All strains from animals were obtained during related research on animal brucellosis. The majority Brucella strains (human and animals) used in this study were collected from published academic articles found on PubMed and Chinese life science databases (e.g. WanFang data and CNKI) and MLVA bank (http://microbesgenotyping.i2bc.paris-saclay.fr/databases).
Clinical strains characterization
A total of 1382 B. melitensis (385 in animals and 997 in humans) were collected from patients and animals from 1955 to 2018 in 29 provinces of China. Fewer B. melitensis strains from Anhui Provinces and Tibet have been reported but MLVA genotyping of these strains has not yet completed. Because of this, strains from these regions were excluded from this study. All strains were isolated and identified according to standard bacteriology approaches [19]. Biotypes were assigned by conventional identification methods, and all strains were gram negative, agglutinated with polyvalent brucellosis serum, had oxidase and catalase activity, did not produce H2S, synthesized urease, and were capable of growing in atmospheric conditions. Both AMOS-PCR [20] and ladder PCR [4] were applied to verify the results from bio-typing assays.
DNA preparation, genotyping, and data analysis
Bacterial cultures were scrapped from the surfaces of solid agar medium. DNA was isolated using the QIAamp DNA Mini Kit (Qiagen, United States) according to the manufacturer’s instructions. The MLVA-16 assay was performed as previously described [21,22]. Briefly, 16 loci were divided into three panels: panel 1 (also called MLVA8), panel 2A, and panel 2B. The combination of panels MLVA8 and 2A was called MLVA11, while the combination of all three panels (16 loci) was designated MLVA16. The MLVA11 panel allows for tracing the geographic origin of strains analyzed, while the panel 2B loci are highly discriminatory and their combination with MLVA11 was used in tracking local outbreaks. PCR was used to determine the number of repeats from a sample, and its products were purified and directly sequenced using an ABI Prism Big Dye Terminator. Size analysis of VNTR repeats was performed using GeneMapper 4.1 (Applied Biosystems). Dendrograms from strains analyzed (Table S1) were constructed using BioNumerics 5.0 (Applied Maths, Sint-Martens-Latem, Belgium) based on the categorical coefficient and unweighted pair group method using arithmetic averages (UPGMA). Minimum spanning trees (MST) were constructed based on MLVA-11 (Table S2) and MLVA-16 (Table S3) data using BioNumerics 7.6 to investigate the geographic origin and genetic relatedness of strains. Phylogenetic analysis of representative strains (Table S4) was performed based on WGS-SNP using the maximum parsimony method [17], B. abortus bv.1 str. 9–941 used as the outgroup strain. Microsoft Excel 2016 (Microsoft, Redmond, WA, USA) was used for data processing, and ArcGis 10.5 (ESRI, Redlands, CA, USA) was applied to display analysis results.
Results
Distribution characteristics of species biotypes of Chinese B. melitensis strains
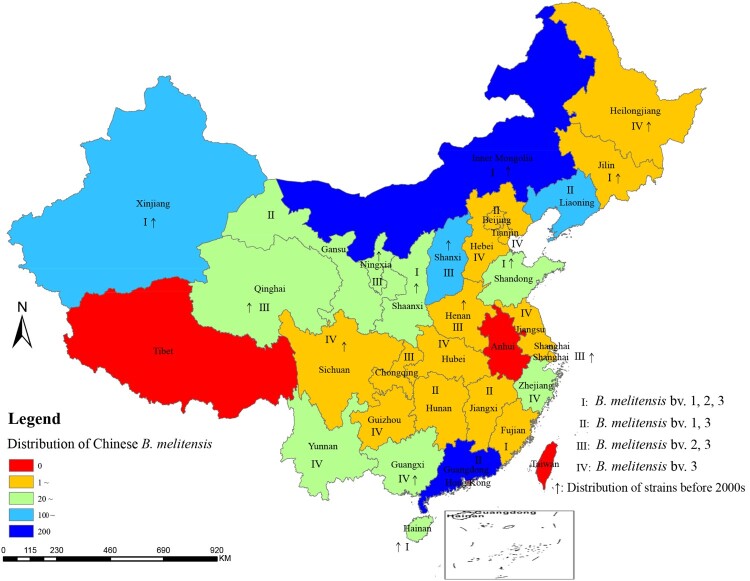
A total of 1382 B. melitensis strains were collected from 29 provinces (including autonomous regions and cities); with the exceptions of Tibet and Anhui, all mainland provinces had obtained B. melitensis strains. Among them, 977 were obtained from human blood samples and 385 strains were recovered from animal samples: 291 in Inner Mongolia, 200 in Guangdong, 165 in Liaoning, 113 in Xinjiang, 107 in Shanxi, 91 in Qinghai, 54 in Shandong, 52 in Ningxia, and other regions contained 3–44 strains as shown in Table 1. A total of 35.5% of the strains were from Inner Mongolia and Guangdong Provinces; the former is a historical area for brucellosis, and the latter is an emerging region for brucellosis. 1382 B. melitensis samples were divided into four distinct epidemic areas: seven provinces in which all three B. melitensis biovars (1, 2, and 3) were collected (I); six provinces in which B. melitensis biovars bv. 1 and 3 were found (II); six provinces in which B. melitensis bv. 2 and 3 were found (III); and ten provinces in which B. melitensis bv. 3 was obtained (IV) (Figure 1). B. melitensis strains were found in 14 provinces in China before 2000, while strains were found in 29 provinces in 2018 (Figure 1). These data demonstrated that B. melitensis has expanded its distribution to all of mainland China.
Table 1. Location, numbers, percentages (%), species, and hosts of 1382 B. melitensis isolates in 29 provinces.
| Province | No. | % | Species-biovar | Host |
|---|---|---|---|---|
| Hubei | 3 | 0.22 | B. melitensis bv. 3 | Human |
| Tianjin | 3 | 0.22 | B. melitensis bv. 3 | Human |
| Beijing | 4 | 0.29 | B. melitensis bv. 1, 3 | Human |
| Shanghai | 4 | 0.29 | B. melitensis bv. 2, 3 | Human, Sheep, Cattle |
| Chongqing | 4 | 0.29 | B. melitensis bv.2, 3 | Human, Sheep |
| Guizhou | 5 | 0.36 | B. melitensis bv. 3 | Human, Goat |
| Hunan | 5 | 0.36 | B. melitensis bv. 1, 3 | Human |
| Sichuan | 7 | 0.51 | B. melitensis bv. 3 | Human Sheep, Cattle, Yak |
| Heilongjiang | 8 | 0.58 | B. melitensis bv. 3 | Human, Sheep |
| Jilin | 8 | 0.58 | B. melitensis bv. 1, 2, 3 | Human, Sheep, Cattle, Deer |
| Henan | 12 | 0.87 | B. melitensis bv. 2, 3 | Human, Sheep |
| Jiangxi | 14 | 1.01 | B. melitensis bv. 1, 3 | Human |
| Jiangsu | 17 | 1.23 | B. melitensis bv. 3 | Human |
| Fujian | 18 | 1.30 | B. melitensis bv. 1, 2, 3 | Human |
| Hebei | 19 | 1.37 | B. melitensis bv. 3 | Human, Sheep |
| Yunnan | 20 | 1.45 | B. melitensis bv. 3 | Human |
| Guangxi | 22 | 1.59 | B. melitensis bv. 3 | Human |
| Gansu | 26 | 1.88 | B. melitensis bv. 1, 3 | Sheep |
| Shaanxi | 28 | 2.03 | B. melitensis bv. 1, 2, 3 | Human |
| Zhejiang | 38 | 2.75 | B. melitensis bv. 3 | Human, Goat |
| Hainan | 44 | 3.18 | B. melitensis bv. 1, 2, 3 | Human, Sheep |
| Ningxia | 52 | 3.76 | B. melitensis bv. 2, 3 | Human, Sheep, Goat |
| Shandong | 54 | 3.91 | B. melitensis bv. 1, 2, 3 | Human, Sheep |
| Qinghai | 91 | 6.58 | B. melitensis bv. 2, 3 | Human, Sheep, Cattle, Blue sheep, Yak, Pseudois nayaur, Tibetan gazelle |
| Shanxi | 107 | 7.74 | B. melitensis bv. 2, 3 | Human, Sheep, Cattle |
| Xinjiang | 113 | 8.18 | B. melitensis bv. 1, 2, 3 | Human, Sheep, Cattle, Goat, Yak |
| Liaoning | 165 | 11.94 | B. melitensis bv. 1, 3 | Human |
| Guangdong | 200 | 14.47 | B. melitensis bv. 1, 3 | Human |
| Inner Mongolia | 291 | 21.06 | B. melitensis bv. 1, 2, 3 | Human, Sheep, Cattle, Camel |
Figure 1.
Geographic distribution of B. melitensis samples in China.
Note: the map of this study does not represent the true borders of administrative regions of China.
Geographic origins of Chinese B. melitensis strains based on MLVA-11
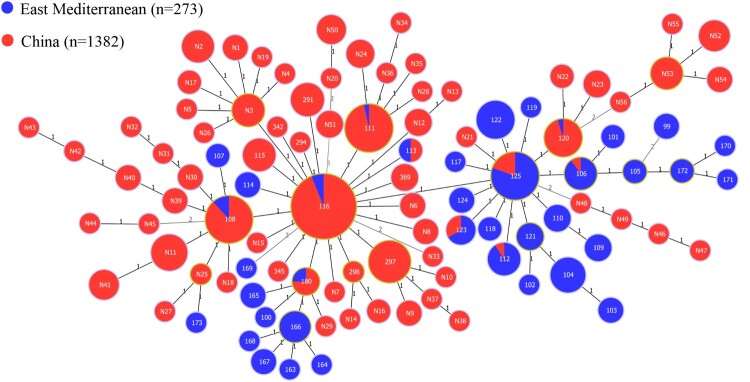
1382 B. melitensis strains yielded 71 MLVA-11 genotypes, including 53 new MLVA-11 genotypes and 18 known genotypes, of which 69 belonged to the East Mediterranean lineage, and two new (N50 and N20) genotypes were of the Americas lineage (Figure 2). Seven MLVA-11 genotypes (116, 111,108, 297, N11, N24, and N3) made up the predominant circulating genotypes, of which 69% (951/1382) were genotype 116, which was shared by strains from 28 different provinces in northern and southern China (S. Figure 1). These southern provinces had fewer B. melitensis before 2000 (S. Figure 1), suggesting that there has been continuous expansion from northern to southern regions. These dominant MLVA-11 genotypes were shared by strains from 5 to 28 distinct provinces (S. Figure 2), of which 89 strains were MLVA-11 genotype 111, accounting for 6.4% (89/1382) and distributed in eleven provinces; 76 strains were MLVA-11 genotype 108, accounting for 5.5% (76/1382) and distributed in seven regions; 39 strains were MLVA-11 genotype 297, accounting for 2.8% (39/1382) and shared by strains from ten provinces; 20 strains were MLVA-11 genotype 120, accounting for 1.4% (20/1382) and shared by strains from five regions, dominating in southern regions including Fujian, Hainan, Yunnan, and Guangxi provinces. The distributed range in the remaining genotypes was limited. All predominant MLVA-11 genotypes were shared by strains from different epidemic periods of brucellosis, including 1950–1970, 1980–2000, and 2001–2018 (S. Figure 3). Moreover, 14 circulating MLVA-11 genotypes were also shared by strains from north and south China (S. Figure 4).
Figure 2.
Minimum spanning tree for B. melitensis using MLVA-11 data with Chinese isolates (red) and East Mediterranean isolates (blue).
Note: numbers in lines show the values of locus variants and numbers in nodes represent MLVA-11 genotypes.
Epidemiological characteristics of animal and human brucellosis
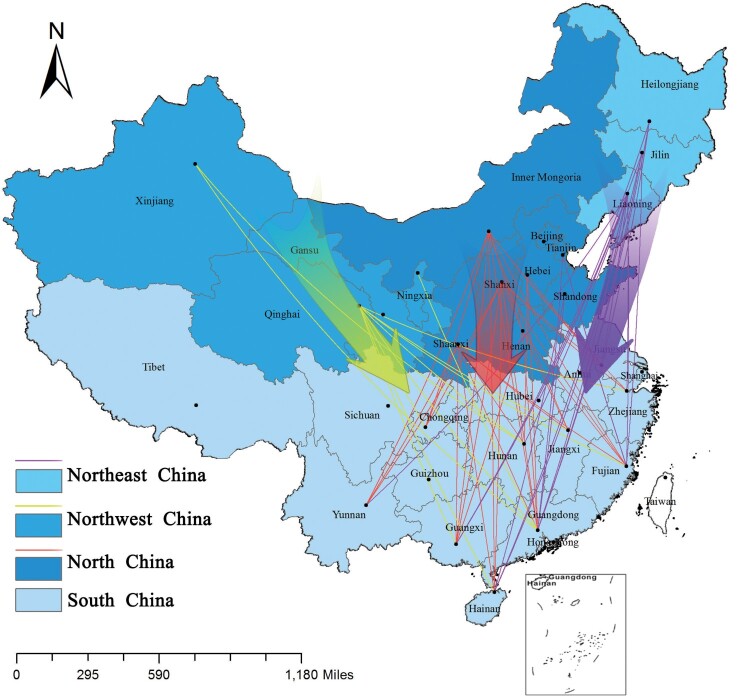
Based on the MLVA-16 genotype, 385 animal B. melitensis strains were sorted into 157 genotypes, of which 91 were shared genotypes in that each genotype present in 2–17 strains, and the cluster rate of strains was 82.3% (317/385). Among the 91 shared genotypes, 24 shared genotypes were present in 103 strains from two to four different provinces (S. Figure 5) (Table S5), accounting for 32.5% (103/317); the other shared genotypes were all from strains from the same provinces. A total of 977 human B. melitensis strains were divided into 391 MLVA-16 genotypes, of which 158 shared genotypes were present in 744 strains, and the cluster rate of these strains was 76.2% (744/977); 88 genotypes were in 268 strains from two to eight different provinces (Table S5), accounting for 36.0% (268/744); and the remaining 233 strains represented single genotypes, with each being an independent strain, accounting for 24% (233/977). The strains from the southern provinces had more similar MLVA-16 genotypes with strains from northern regions, the latter being a historical area of animal and human brucellosis (Figure 3). In particular, three shared genotypes were present in strains from both southern and northern provinces, including Jilin, Qinghai, Guangdong, Inner Mongolia, Guangxi, Fujian, Liaoning, and Shaanxi; Inner Mongolia, Qinghai, Henan, Guangdong, Shanxi, Shaanxi, and Guangxi; and Inner Mongolia, Jilin, Shanxi, Liaoning, Shandong, Guangdong, and Hainan (S. Figure 6). The other 70 shared genotypes were present in strains from the same provinces, accounting for 64% (476/744). Meanwhile, completely identical MLVA-16 genotypes were shared by strains from the three different epidemic periods (S. Figure 7). Moreover, many shared genotypes were observed among strains from livestock, humans, and wild animals (S. Figure 8).
Figure 3.
Transmission pattern of B. melitensis isolates from humans.
Note: the map of this study does not represent the true borders of administrative regions of China.
Genetic relatedness of B. melitensis strains on a global scale
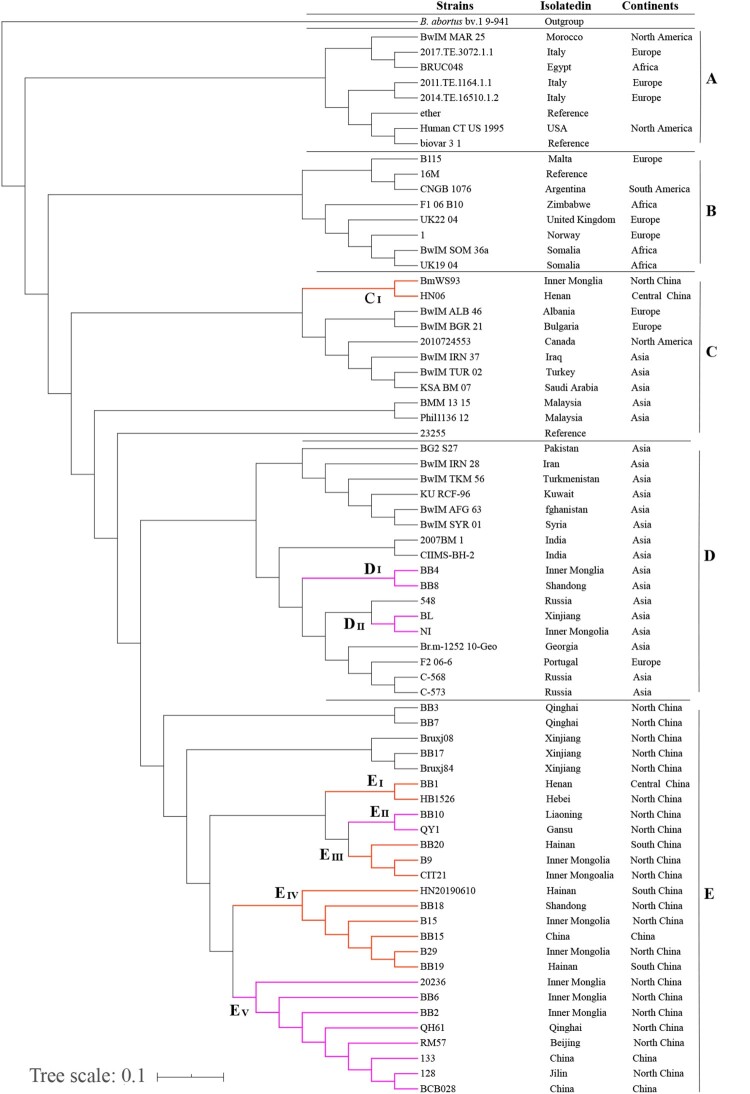
To reveal the genetic links of Chinese strains with those in the rest of the world, the genetic relatedness among 3480 B. melitensis strains on a global level were compared using MST based on MLVA-16 data. MST analysis showed that these strains sort into three groups (G I ∼ III) (Figure 4). Many shared genotypes were observed in strains from this study with strains from Kazakhstan, Turkey, and Mongolia (Figure 4, G I); these regions are important states along the silk road with close geographies. However, there were significant genetic differences between strains from China and Italy, France, and Peru (Figure 4, G II and III). Phylogenetic analysis of strains was performed based on WGS-SNP of 74 B. melitensis strains from NCBI GenBank. Phylogenetic analysis based on whole-genome SNPs and geographical distribution of the isolates revealed spatial clustering of the B. melitensis isolates were divided into five clades (A-E) (Figure 5). The Mediterranean strains, identified as clade A, occupied the basal node of the phylogenetic tree. The majority of the Chinese B. melitensis strains clustered into clade E and represented the Asia lineage (Figure 5). Strains obtained from southern and central regions had close relationships with strains from northern regions, China, including the Inner Mongolian Autonomous Region, Xinjiang, and Shandong Provinces (Figure 5 C (I), D (I and II), E (I - V)), which are ongoing epidemic regions of animal and human brucellosis. However, B. melitensis strains from Chinese provinces had a close genetic relationship to strains from other Asian regions including Russia, India, and Georgia, which are traditional epidemic regions of brucellosis (Figure 5(C and D)).
Figure 4.
Minimum spanning tree (MST) was constructed using MLVA-16 data on a global scale.
Figure 5.
Phylogenetic tree of B. melitensis strains based on WGS-SNP over all of global. (Clades coloured with orange that representation strains from South were closely related to North and Central regions, Clades coloured with purple showed that there were closely related among strains from North regions.)
Discussion
Brucellosis is the most common zoonotic disease and it is mainly caused by B. melitensis infection (biovars 1 and 3). Brucellosis poses a threat to both animals and humans [23]. A previous study showed that 84.5% of the Brucella strains isolated from humans with brucellosis in China were B. melitensis [24]. B. melitensis strains are now widely distributed throughout China and the distribution of pathogenic species of brucellosis in China has obviously changed. Since the 1950s, B. melitensis was most common in the grassland areas of northern China, where sheep and goats are the main livestock [24]. B. melitensis strains were found in 14 provinces in China before 2000. Now, B. melitensis strains occur in 29 provinces (autonomous regions and cities) and B. melitensis is the dominant circulating species in southern regions of China [25,26]. This supports our hypothesis that B. melitensis has spread and expanded from northern to southern China. Brucellosis occurred earlier in the north than in the south and due to the introduction of northern sheep and other species, human brucellosis is increasing in the southern provinces [12]. Since 2010, human brucellosis has occurred or reappeared in all provinces in southern China [27]. These data confirm that the geographic distribution of the disease has also evolved, with the movement of B. melitensis strains to the south.
In this study, 69% (951/1382) of the strains were genotype 116 and belonged to the East Mediterranean lineage. Genotype 116 is shared by strains from 28 different provinces in northern and southern China, revealing that these strains had a common geographic origin. Genotype 116 is responsible for the vast majority of Brucella infections in humans and animals [28] and is predominant in many countries, accounting for more than 77% of cases in Portugal and Kazakhstan, 37% in Spain, 16% in Turkey, and 10% in France [29]. Many shared MLVA-16 genotypes were observed among strains from northern and southern regions, and three brucellosis epidemic periods. This finding coincides with species distributions and geographic origin profiles of strains in this study. These data suggest that B. melitensis spreads and expands continually and has moved from northern to southern, China [30], while affected area now covers all of mainland, China [9].
Brucellosis is associated with large-scale farming and trading of sheep and goats. The inventory of sheep is correlated with the presence of brucellosis cases in mainland China [31]. The non-regulated animal trade has an important impact on the dissemination of brucellosis. Infected sheep and their products that have not been quarantined in the north before import to the south may be the main cause for the increasing incidence of human brucellosis in southern China [27]. Introduced infected sheep from northern regions may have led to a brucellosis outbreak epidemic in Zhejiang Province, China [32]. The increasing demand for meat and expanding animal husbandry in southern China have also increased the infection risk for brucellosis due to occupational exposure [33]. Brucellosis is an emerging disease and there is unfamiliarity with its infection in most provinces in southern China. Therefore, the occupational protections used in populations are relatively few. Control of brucellosis in southern China requires strict restrictions on the inter-provincial movement of infected animals. Livestock serologically negative for brucellosis can be allowed to move without restriction. Livestock that test positive for brucellosis, or those with unknown disease status, should only be allowed to move if they have a negative serological test issued up to 30 days before movement [34]. An improved culling policy is also needed. Sick animals cannot be properly disposed because of unacceptable compensation funds for farmers and this leads to persistence of the source of infection [35]. There should be improved capability in immunization, quarantine, diagnosis, and treatment in animal disease control and prevention organizations to meet the increasing need for disease control. Lastly, improved awareness of the need for personal protection for those working with animals or animal products is needed. Individuals should wear gloves and other appropriate protective clothing.
Many shared genotypes were observed among strains from different hosts. These data showed a potential transmission pattern for B. melitensis in China and demonstrate direct or indirect transmission among livestock and wild animals, eventually infecting humans. Previous reports have shown that wild animals are a significant brucellosis reservoir for livestock and humans [36,37]. To better understand the epidemiological characteristics of brucellosis in China, studies on brucellosis epidemics in wild animals at the country level are needed.
MST analysis showed that the strains studied here had a common geographic origin and a close relationship with strains from Kazakhstan, Mongolia and Turkey [15]. The most MLVA-16 shared genotypes were found for strains from China and Kazakhstan. These regions are geographically close and have historically exchanged livestock. Animals were a common form of payment in ancient commerce, and the modern version of this practice has promoted disease transmission [38].
Based on WGS-SNP analysis, the Mediterranean strains, identified as clade A, occupied the basal node of the phylogenetic tree. This indicates that B. melitensis may have originated in the Mediterranean regions. Brucellosis may have been identified in the late Roman era; however, the disease was first described by Sir David Bruce, Hughes, and Zammit while working in Malta [39]. Clade E comprised 26 out of the 74 B. melitensis strains used in the study. It represented the largest B. melitensis genotype, with isolates collected from diverse locations of China. It exhibited a ladder-like phylogram, suggesting a possible single introduction of these genotype strains into China [40]. WGS-SNP analysis showed close relationships among strains from southern provinces, central, and many northern regions, indicating that B. melitensis strains from the southern region originated from northern regions. This conclusion is consistent with previous reports that the sources of infection of human brucellosis in southern regions (Guangxi, Hunan and Hainan Provinces) originated in northern provinces, including Inner Mongolia [25,26,41]. However, B. melitensis strains from China have a genetic relationship to strains from Asian regions including Russia, India, and Georgia, which are traditional epidemic regions of brucellosis. Although few B. melitensis isolates were from Russia, they are genetically similar to Chinese strains [42], and these strains had high homogeneity [43]. India harbours the largest ruminant populations and there are high seroprevalence estimates of brucellosis in livestock and humans [44]. Importantly, the absence of a clear differentiation according to territorial affiliation between these regions indicates the frequent penetration of the B. melitensis strains from one country to another [43], and active trade based on the ancient Silk Road, Tea Horse Road and Trans-Eurasia exchange among these nearby regions could have promoted this process. Brucellosis has a “knows no borders” character, making it challenging to monitor and control [45]. A positive response to a “National brucellosis control plan (2016–2020)” is needed. It is also necessary to enforce animal controls, vaccinate all susceptible animals, and increase the level of education and awareness among people, especially regarding contact with infected animals and consumption of contaminated milk and other byproducts.
This study has several limitations. First, there was considerable variability in the number of strains collected among different regions and periods. In some high-endemic areas of brucellosis, Jilin and Heilongjiang had fewer strains, affecting the study conclusions. Second, data on the distribution of prevalence of animal brucellosis, sheep population mobility, and strains obtained from animals (sheep) from southern provinces were lacking. Further animal studies including seroprevalence and strain distributions are essential. Genomic data of strains from China in the Gene bank were limited and additional phylogenetic analysis of more Chinese B. melitensis is warranted.
Conclusion
Although a national brucellosis control programme in China has been ongoing for many years (2009–2020), disease prevalence has not obviously declined. This indicates a need to reformulate the existing management strategies. At present, B. melitensis is the predominant species in southern China. Sheep and animal products are traded frequently between northern and southern regions and this promotes the spread and expansion of B. melitensis strains. Because sheep play a large role of the spread of brucellosis, surveillance and disease countermeasures for sheep populations should be a priority. Quarantine and inspections of sheep transfer and trade are urgently needed. Brucellosis is a disease that has spread across extensive regions and provinces and is transmitted by food, air, and soil. It occurs in areas dominated by traditional hygiene practices and these areas have limited access to health services. We encourage government organizations (veterinary and health authorities) to play a greater role in disease management. The active participation of livestock producers as well as industry partners is also essential.
Supplementary Material
Acknowledgements
We are grateful to the staff in the Branch of Brucellosis Control and Prevention, and to the Centers for Disease Control and Prevention of each province for assistance with the strains identified and collected. We are also grateful to Buyun Cui from the China CDC for experimental guidance. LZG and ZZZ performed strain identification and collected, ZX and LZG performed genotyping and cluster analysis and drafted the manuscript; GZW and WM conducted epidemiological investigations and data analysis; ZX and MSY prepared the DNA samples; LZG participated in the design of the study and critically reviewed the manuscript; ZX and LZJ participated in the design of the study and managed the project. All authors read and approved the final version of the manuscript.
Glossary
Abbreviations: MLVA: multiple locus variable-number tandem repeat analysis; WGS: whole genome sequencing; SNP: single nucleotide polymorphism; MST: minimum spanning tree
Funding Statement
This study was supported by National Key Research and Development Project (No. 2019YFC1200705), the China Mega-project for Infectious Disease (Nos. 2017ZX10303401, 2018ZX10734401, and 2018ZX10734404), and the Natural Science Foundation of Inner Mongolia Autonomous Region of China (No. 2018MS08004). The funding agencies had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Ethics approval and consent to participate
This study focused on genotyping Brucella isolates using a modern molecular approach and did not involve animal work or the collection of patient information. This study was approved by and adhered to the rules of the Ethics Committee of the National Institute for Communicable Disease Control and Prevention at the Chinese Center for Disease Control and Prevention.
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- 1.Charypkhan D, Sultanov AA, Ivanov NP, et al. . Economic and health burden of brucellosis in Kazakhstan. Zoonoses Public Health. 2019 Aug;66(5):487–494. [DOI] [PubMed] [Google Scholar]
- 2.Pappas G, Papadimitriou P, Akritidis N, et al. . The new global map of human brucellosis. Lancet Infect Dis. 2006 Feb;6(2):91–99. [DOI] [PubMed] [Google Scholar]
- 3.O'Brien MP, Beja-Pereira A, Anderson N, et al. . Brucellosis transmission between wildlife and livestock in the greater Yellowstone ecosystem: inferences from DNA genotyping. J Wildl Dis. 2017 Apr;53(2):339–343. [DOI] [PubMed] [Google Scholar]
- 4.Kumar S, Tuteja U, Sarika K, et al. . Rapid multiplex PCR assay for the simultaneous detection of the Brucella genus, B. abortus, B. melitensis, and B. suis. J Microbiol Biotechnol. 2011 Jan;21(1):89–92. [DOI] [PubMed] [Google Scholar]
- 5.Galinska EM, Zagorski J.. Brucellosis in humans–etiology, diagnostics, clinical forms. Ann Agric Environ Med. 2013;20(2):233–238. [PubMed] [Google Scholar]
- 6.Facciola A, Palamara MAR, D'Andrea G, et al. . Brucellosis is a public health problem in southern Italy: Burden and epidemiological trend of human and animal disease. J Infect Public Health. 2018 Nov–Dec;11(6):861–866. [DOI] [PubMed] [Google Scholar]
- 7.Garofolo G, Di Giannatale E, Platone I, et al. . Origins and global context of Brucella abortus in Italy. BMC Microbiol. 2017 Feb 2;17(1):28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li K, Zhang L, Shahzad M, et al. . Increasing incidence and changing epidemiology of brucellosis in China (2004–2016). Travel Med Infect Dis. 2019 Jul 30: 101464. [DOI] [PubMed] [Google Scholar]
- 9.Lai S, Zhou H, Xiong W, et al. . Changing epidemiology of human brucellosis, China, 1955–2014. Emerging Infect Dis. 2017 Feb;23(2):184–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zeng JY, Robertson ID, Ji QM, et al. . Evaluation of the economic impact of brucellosis in domestic yaks of Tibet. Transbound Emerg Dis. 2019 Jan;66(1):476–487. [DOI] [PubMed] [Google Scholar]
- 11.Li MT, Sun GQ, Zhang WY, et al. . Model-based evaluation of strategies to control brucellosis in China. Int J Environ Res Public Health. 2017 Mar 12;14(3):295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kong W. Brucellosis infection increasing in southern China. Eur J Intern Med. 2018 May;51:e16–e18. [DOI] [PubMed] [Google Scholar]
- 13.Liu ZG, Di DD, Wang M, et al. . MLVA genotyping characteristics of human brucella melitensis isolated from Ulanqab of inner Mongolia, China. Front Microbiol. 2017;8:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanogo M, Fretin D, Thys E, et al. . Exploring the diversity of field strains of brucella abortus biovar 3 isolated in West Africa. Front Microbiol. 2017;8:1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shevtsov A, Ramanculov E, Shevtsova E, et al. . Genetic diversity of Brucella abortus and Brucella melitensis in Kazakhstan using MLVA-16. Infect Genet Evol. 2015 Aug;34:173–180. [DOI] [PubMed] [Google Scholar]
- 16.Sun M, Jing Z, Di D, et al. . Multiple locus variable-number tandem-repeat and single-nucleotide polymorphism-based Brucella typing reveals multiple lineages in Brucella melitensis currently endemic in China. Front Vet Sci. 2017;4:215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ledwaba MB, Gomo C, Lekota KE, et al. . Molecular characterization of Brucella species from Zimbabwe. PLoS Negl Trop Dis. 2019 May;13(5):e0007311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma JY, Wang H, Zhang XF, et al. . MLVA and MLST typing of Brucella from Qinghai, China. Infect Dis Poverty. 2016 Apr 13;5:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smirnova EA, Vasin AV, Sandybaev NT, et al. . Current methods of human and animal brucellosis diagnostics. Adv Infect Dis. 2013;3(3):177–184. [Google Scholar]
- 20.Matope G, Bhebhe E, Muma JB, et al. . Characterization of some Brucella species from Zimbabwe by biochemical profiling and AMOS-PCR. BMC Res Notes. 2009 Dec 22;2:261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Le Fleche P, Jacques I, Grayon M, et al. . Evaluation and selection of tandem repeat loci for a Brucella MLVA typing assay. BMC Microbiol. 2006 Feb 9;6:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Al Dahouk S, Fleche PL, Nockler K, et al. . Evaluation of Brucella MLVA typing for human brucellosis. J Microbiol Methods. 2007 Apr;69(1):137–145. [DOI] [PubMed] [Google Scholar]
- 23.Grilló MJ, Blasco JM, Gorvel JP, et al. . What have we learned from brucellosis in the mouse model? Vet Res. 2012 Apr 13;43:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deqiu S, Donglou X, Jiming Y.. Epidemiology and control of brucellosis in China. Vet Microbiol. 2002 Dec 20;90(1–4):165–182. [DOI] [PubMed] [Google Scholar]
- 25.Li Z, Wang XM, Zhu X, et al. . Molecular characteristics of Brucella isolates collected from humans in Hainan Province, China. Front Microbiol. 2020;11:452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu ZG, Wang M, Zhao HY, et al. . Investigation of the molecular characteristics of Brucella isolates from Guangxi Province, China. BMC Microbiol. 2019 12 16;19(1):292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang H, Zhang S, Wang T, et al. . Epidemiological characteristics and spatiotemporal trend analysis of human brucellosis in China, 1950–2018. Int J Environ Res Public Health. 2020 Mar 31;17(7):2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ferreira AC, Chambel L, Tenreiro T, et al. . MLVA16 typing of Portuguese human and animal Brucella melitensis and Brucella abortus isolates. PLoS ONE. 2012;7(8):e42514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shevtsova E, Vergnaud G, Shevtsov A, et al. . Genetic diversity of Brucella melitensis in Kazakhstan in relation to world-wide diversity. Front Microbiol. 2019;10:1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhong Z, Yu S, Wang X, et al. . Human brucellosis in the people's Republic of China during 2005–2010. Int J Infect Dis. 2013 May;17(5):e289–e292. [DOI] [PubMed] [Google Scholar]
- 31.Peng C, Li YJ, Huang DS, et al. . Spatial-temporal distribution of human brucellosis in mainland China from 2004 to 2017 and an analysis of social and environmental factors. Environ Health Prev Med. 2020 Jan 2;25(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang H, Xu WM, Zhu KJ, et al. . Molecular investigation of infection sources and transmission chains of brucellosis in Zhejiang, China. Emerg Microbes Infect. 2020 Apr 14;9(1):889–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu T, Perrings C, Kinzig A, et al. . Economic growth, urbanization, globalization, and the risks of emerging infectious diseases in China: a review. Ambio. 2017 Feb;46(1):18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fevre EM, Bronsvoort BM, Hamilton KA, et al. . Animal movements and the spread of infectious diseases. Trends Microbiol. 2006 Mar;14(3):125–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Godfroid J. Brucellosis in livestock and wildlife: zoonotic diseases without pandemic potential in need of innovative one health approaches. Arch Public Health. 2017;75:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bhandi S, Pfukenyi DM, Matope G, et al. . Brucellosis and chlamydiosis seroprevalence in goats at livestock-wildlife interface areas of Zimbabwe. Onderstepoort J Vet Res. 2019 Aug 22;86(1):e1–e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kamath PL, Foster JT, Drees KP, et al. . Genomics reveals historic and contemporary transmission dynamics of a bacterial disease among wildlife and livestock. Nat Commun. 2016 May 11;7:11448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shevtsova E, Shevtsov A, Mukanov K, et al. . Epidemiology of brucellosis and genetic diversity of Brucella abortus in Kazakhstan. PLoS ONE. 2016;11(12):e0167496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khan MZ, Zahoor M.. An overview of brucellosis in Cattle and humans, and its serological and molecular diagnosis in control strategies. Trop Med Infect Dis. 2018 Jun 14;3(2):65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tan KK, Tan YC, Chang LY, et al. . Full genome SNP-based phylogenetic analysis reveals the origin and global spread of Brucella melitensis. BMC Genomics. 2015 Feb 18;16:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu ZG, Wang M, Zhan ZF, et al. . Epidemiology of human brucellosis and source of Brucella isolates in Hunan province. Zhonghua Liu Xing Bing Xue Za Zhi. 2019 Sep 10;40(9):1150–1154. [DOI] [PubMed] [Google Scholar]
- 42.Daugaliyeva capital AC, Sultanov A, Usserbayev B, et al. . Genotyping of Brucella melitensis and Brucella abortus strains in Kazakhstan using MLVA-15. Infect Genet Evol. 2018 Mar;58:135–144. [DOI] [PubMed] [Google Scholar]
- 43.Pisarenko SV, Kovalev DA, Volynkina AS, et al. . Global evolution and phylogeography of Brucella melitensis strains. BMC Genomics. 2018 May 10;19(1):353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Singh BB, Khatkar MS, Aulakh RS, et al. . Estimation of the health and economic burden of human brucellosis in India. Prev Vet Med. 2018 Jun 1;154:148–155. [DOI] [PubMed] [Google Scholar]
- 45.Pappas G. The changing Brucella ecology: novel reservoirs, new threats. Int J Antimicrob Agents. 2010 Nov;36(Suppl 1):S8–S11. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files.