Abstract
Coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) has rapidly become a global pandemic. Although great efforts have been made to develop effective therapeutic interventions, only the nucleotide analog remdesivir was approved for emergency use against COVID-19. Remdesivir targets the RNA-dependent RNA polymerase (RdRp), an essential enzyme for viral RNA replication and a promising drug target for COVID-19. Recently, several structures of RdRp in complex with substrate RNA and remdesivir were reported, providing insights into the mechanisms of RNA recognition by RdRp. These structures also reveal the mechanism of RdRp inhibition by nucleotide inhibitors and offer a molecular template for the development of RdRp-targeting drugs. This review discusses the recognition mechanism of RNA and nucleotide inhibitor by RdRp, and its implication in drug discovery.
Keywords: COVID-19, SARS-CoV-2, RNA-dependent RNA polymerase, Structure, RNA recognition, Remdesivir
1. Introduction
Coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) was initially emerged in December 2019 [1,2]. The exponential growth of infections and death cases makes COVID-19 a worldwide health threat, and the World Health Organization (WHO) declares it a global pandemic in March 2020. There have been over 20 million infections and 800,000 deaths in 188 countries as of August 2020 (GISAID database). Genome sequence analysis has revealed that SARS-CoV-2 belongs to the coronaviridae family. Its sequence is closely related to the severe acute respiratory syndrome coronavirus (SARS-CoV) and several betacoronavirus of bat and pangolin origins [[3], [4], [5]]. SARS-CoV-2 is related to two highly pathogenic human coronaviruses, SARS-CoV and Middle East respiratory syndrome coronavirus (MERS-CoV), which have caused more than 8000 and 2500 confirmed cases, with approximately 10% and 36% mortality rates, respectively [6]. Compared to these two coronaviruses, SARS-CoV-2 exhibits a higher human-to-human transmission rate and lower mortality (∼3.5%). A more potent binding affinity of the spike protein of SARS-CoV-2 to angiotensin-converting enzyme 2 (ACE2) from host cells than other coronaviruses may explain for its much higher transmission rate [[7], [8], [9], [10], [11]]. Recently, a SARS-CoV-2 variant harboring the spike protein D614G mutation was identified, which is associated with the higher viral loads in patients [12]. Although scientists have responded promptly to the emergency and endeavored to discover effective treatments, remdesivir is the only drug authorized for emergency use against COVID-19 on May 1, 2020. In total, 27 vaccines for COVID-19 are under development in clinical trials (Clarivate Analytics Integrity database), and no vaccine has been approved so far. More effective antiviral treatments are urgently needed for the treatment and control of COVID-19.
Remdesivir, an approved drug against the Ebola virus, targets the RNA-dependent RNA polymerase (RdRp) and inhibits the synthesis of viral RNA [[13], [14], [15]]. As a critical enzyme in the virus life cycle, RdRp is referred to as an attractive target for the treatment of COVID-19. Recent progress on the structural studies of RdRp complexes provides promising insights into the mechanism of substrate RNA recognition by RdRp [[16], [17], [18], [19]]. Moreover, these structures also clarified the inhibition mechanism of RdRp by nucleotide analog remdesivir [17,18], thus providing a structural template for developing efficient antiviral drugs against COVID-19.
2. Organization of SARS-CoV-2 genome
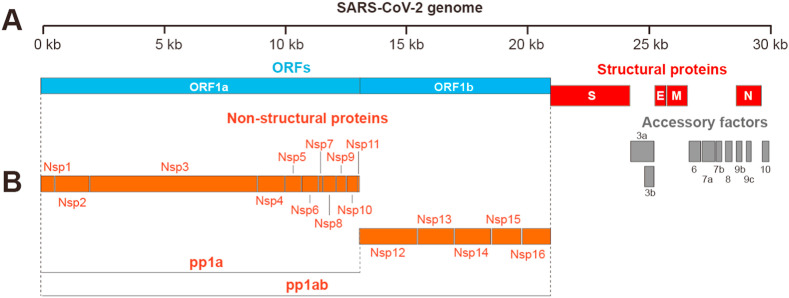
SARS-CoV-2 is a positive-strand RNA virus. Its genome is approximately 30 kb in size and encodes 14 open reading frames (ORFs). The 5′-ORF-1a/1b occupies 67% of SARS-CoV-2 genome, and encode polyprotein 1a (pp1a) and polyprotein 1 ab (pp1ab) precursor polyproteins, which were further cleavage into 16 non-structural proteins (nsp1-16) [2]. These non-structural proteins constitute multiple enzymes required for the virus life cycle, including papain-like protease (nsp3), chymotrypsin-like main protease (3CL protease, nsp5), RdRp (nsp12), helicase (nsp13), and exoribonuclease (nsp14) [20]. Other nsp proteins are also reported to be involved in SARS-CoV-2 replications or host immune system regulation. Nsp 1 suppresses host translation machinery by binding to the ribosome [[21], [22]]. Nsp9 is involved in viral genomic RNA reproduction and virulence [23]. Nsp15 is a endoribonuclease which processes viral RNA to evade detection by host defense system [24]. Nsp16, a methyltransferase in complex with nsp10, is capable of capping viral mRNA transcripts for efficient translation and to evade immune surveillance [25]. The four structural proteins——spike (S), envelope (E), membrane (M), and nucleocapsid (N) proteins occupy ∼33% of the virus genome at C-terminus, and play a crucial role in viral structure integrity, as in the case of spike protein which mediated the entry of SARS-CoV-2 into the host. The 3′ end of the genome also contains nine putative ORFs for accessory factors (Fig. 1 ) [20].
Fig. 1.
The genome of SARS-CoV-2. (A) The organization of SARS-CoV-2 genome. (B) The polyproteins (pp1a and pp1ab) are cleaved into 16 non-structural proteins (nsp1-16).
3. The overall structure of the RdRp complex
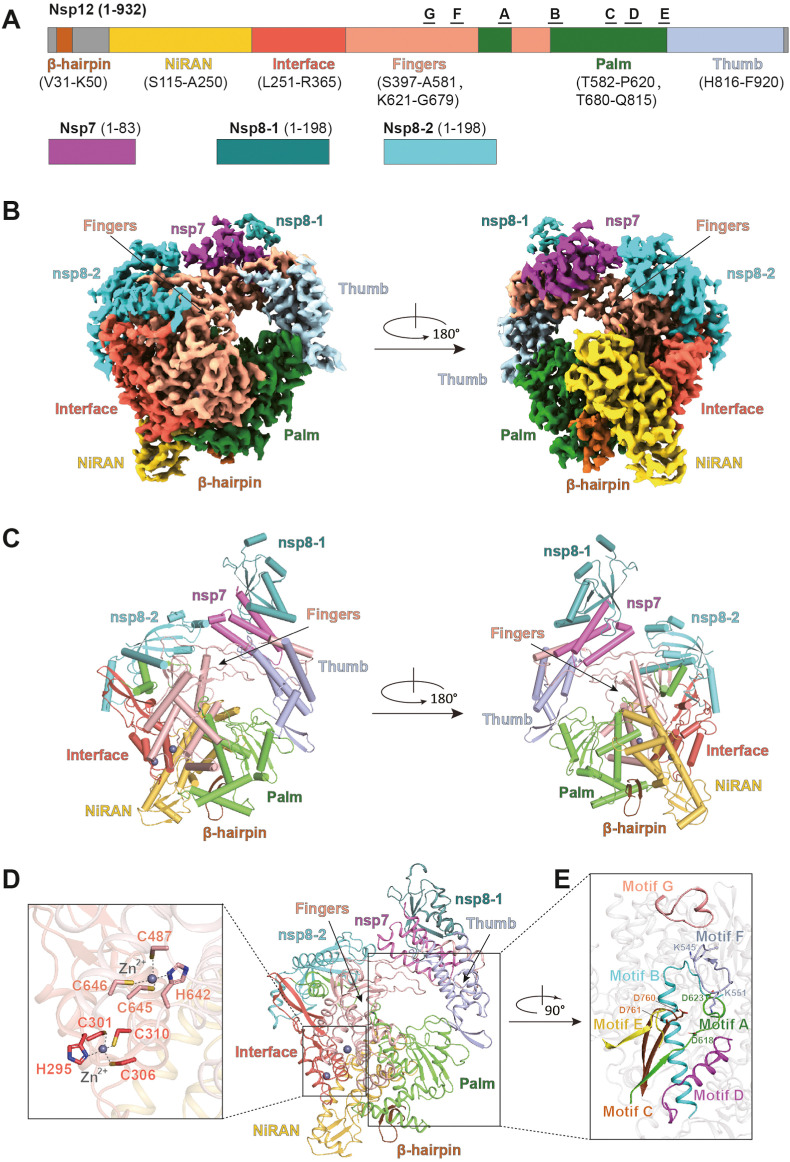
The RNA-dependent RNA polymerase (RdRp), also known as nsp12, is a core component of the virus replication and transcription complex and handles the replication and transcription of viral RNA [26]. Nsp12 exhibits significant polymerase activities with the assistance of other cofactors, nsp7 and nsp8, while nsp12 itself shows limited or no catalytic activity [26,27]. Thus, nsp12-nsp7-nsp8 is defined as the minimal core component for virus RNA replication. The structure of the SARS-CoV-2 RdRp complex consists of an nsp12 core catalytic unit, an nsp7-nsp8 (nsp8-1) heterodimer, and an additional nsp8 subunit (nsp8-2).
The overall conformation of RdRp resembles the SARS-CoV RdRp [27]. The N-terminal portion of nsp12 contains a β-hairpin (residues V31 to K50) and a nidovirus-specific extension domain (NiRAN, residues S115 to A250). The β-hairpin is sandwiched by the palm subdomain in RdRp core and NiRAN, a configuration not observed in the SARS-CoV RdRp structure. The C-terminal catalytic domain of nsp12 (residues L366 to F920) connects to NiRAN through an interface subdomain (residues L251 to R365) (Fig. 2 A). Recently, an RdRp mutation 14408C > T was identified in Europe, which is associated with an increased mutation rate compared to viral genomes from Asia through an unknown mechanism [28]. The resulting mutation P323L located at the interface domain of RdRp, which is far from the active catalytic site and may exert its effects through altered interaction with other components of the replication-transcription complex or with the RNA template [29]. The C-terminal catalytic domain of nsp12 adopts a canonical cupped right-handed configuration of all viral RdRp, composed of the finger, palm, and thumb subdomains. The finger subdomain (residues S397 to A581 and K621 to G679) forms a closed-ring structure with the thumb subdomain (residues H816 to F920) (Fig. 2A, B and C). The similarly closed conformation can also be observed in other positive-strand RNA viruses [30,31]. Additionally, similar to the SARS-CoV-RdRp, two zinc ions bind to the conserved metal binding motifs constituted by H295–C301–C306–C310 and C487–H642–C645–C646 respectively (Fig. 2D) [27].
Fig. 2.
Structure of the SARS-CoV-2 apo RdRp complex. (A) The schematic diagram for the domain organization of the RdRp complex, containing nsp12, nsp7, and two copies of nsp8 (nsp8-1 and nsp8-2). The polymerase motifs A to G in the catalytic site are highlighted. The β-hairpin is indicated. (B–C) Two views of the cryo-EM map (B) and structure (C) of SARS-CoV-2 apo RdRp complex (PDB code: 7BV1). (D) The conserved zinc-binding motifs. The zinc-binding residues are shown as sticks. The subdomains and components of the RdRp complex are colored as follows: β-hairpin, chocolate; NiRAN, gold; Interface, tomato; Fingers, salmon; Palm, green; Thumb, light blue; nsp7, magenta; nsp8-1, dark cyan; nsp8-2, cyan. (E) The active site of the RdRp complex. The conserved seven motifs A-G are highlighted as indicated colors. The conserved residues, K545 and R555 in motif F, D618 and D623 in motif A, as well as D760 and D761 in motif C, are shown as sticks. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
The SARS-CoV-2 nsp7-nsp8 heterodimer shows a conserved structural similarity to that of SARS-CoV [27,32]. The nsp7-nsp8 heterodimer binds above the thumb subdomain and stabilizes the thumb-finger interface. Nsp7 makes a major contribution to the binding of the heterodimer to nsp12, while nsp8 only contacts a few residues from nsp12. The other copy of nsp8 (nsp8-2) sitting atop the finger subdomain and form additional interactions with the interaction subdomain (Fig. 2B and C) [17].
4. The active site of SARS-CoV-2 RdRp
The active site of the SARS-CoV-2 RdRp is formed by seven conserved catalytic motifs, motifs A to G. Five of these motifs (A-E) are located within the palm subdomain, while the other two (F and G) reside in the finger subdomain. Motif A (residues T611 to M626) houses the catalytic motif DX2-4D, in which the first aspartic acid D618 is invariant in most viral polymerases, including the hepatitis C virus and poliovirus [33,34]. The flexible loop in Motif B (residues G678 to T710) serves as a hinge to undergo conformation arrangement associate with template RNA and substrate binding [35]. Motif C (residues F753 to N767) contains the catalytic motif SDD (residues S759 to D761), which is essential for binding the metal ion [36,37]. The recent structures of RdRp also confirm that the D760 and D761 are involved in the coordination of two magnesium ions at the catalytic center [17]. These conserved aspartic acids from catalytic motif DX2-4D and SDD motifs are involved in the regulation of catalytic activity. Motif F (residues L544 to V557) interacts with the phosphate group of incoming NTP. Structurally, the side chains of K545 and R555 contact with the +1 base to direct the incoming NTP to the correct position for catalysis. Motif G (residues D499 to L514) interacts with the template strand and may direct the RNA template to the active catalytic site (Fig. 2E).
5. The structural basis for RNA recognition by SARS-CoV-2 RdRp
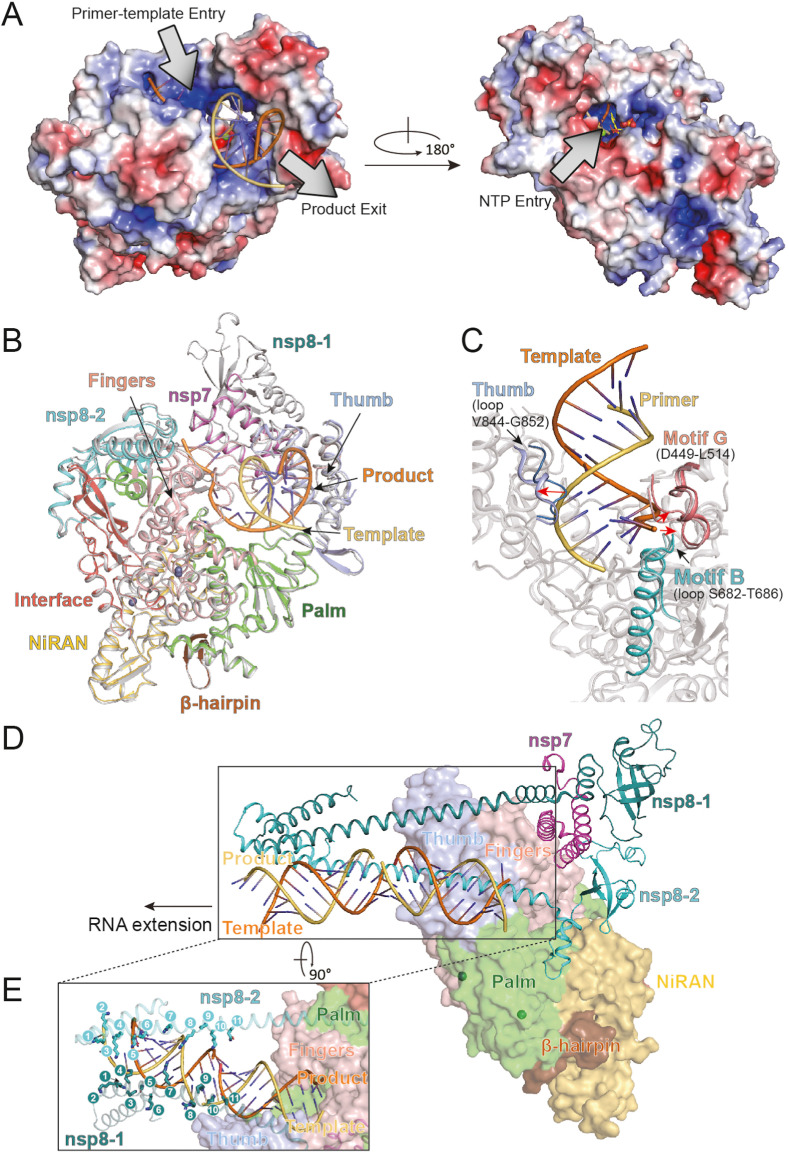
The recent structures of SARS-CoV-2 RdRp in complex with double-stranded RNA helix reveal the recognition mechanism of substrate RNA by SARS-CoV-2 RdRp [17,18]. Like other RNA polymerases, the primer-template entry, NTP entry, and the nascent strand exit routes are positively charged and solvent accessible, providing a preferable environment for RNA binding (Fig. 3 A). The primer-template RNA is embraced by the finger-palm-thumb subdomains (Fig. 3B). A total of 41 residues from nsp12 direct contact with helical RNA, of which 26 residues to the template strand and 15 to the primer strand. No interactions are observed between base pairs of RNA and residues from nsp12, providing a structural basis for RNA sequence-independent binding of RdRp. Additionally, most of these interactions are mediated by the phosphate-ribose backbone, especially the 2′-OH groups, which offers a structural explanation for RdRp to distinguish RNA from DNA [17].
Fig. 3.
The structural basis for RNA recognition by SARS-CoV-2 RdRp. (A) Two surface views of the RdRp with the electrostatic potential (Red, negative; blue, positive). The primer-template entry, product exit, and nucleotide (NTP) entry routes are highlighted. (B) Superposition of structures of apo (PDB codes: 7BV1) and RNA bound RdRp complex (PDB codes: 7BV2). The subdomains of the RNA bound RdRp complex are colored as indicated, while that of the apo RdRp complex are shown in gray. (C) Structural rearrangements of RdRp active site after RNA binding. The loops in motif B, motif G, and the thumb subdomain in RNA bound RdRp structure are colored in cyan, salmon, and light blue, respectively, while corresponding loops in apo RdRp structure are indicated in pale cyan, light salmon, and gray. The directions of red arrows indicate the shift of the structural fragments from apo RdRp toward that from RNA bound RdRp complex. (D–E) The “sliding poles”-like conformation of the N-terminal extension of nsp8 stabilizes the long exiting helical RNA (PDB codes: 6YYT). The subdomains of the RdRp complex, and the template and product strands are colored as indicated. The solid circled numbers 1 to 11 indicate the positively charged residues at the N-terminal of nsp8, including K36, K37, K39, K40, N43, K46, R51, R57, K58, and Q65, respectively (E). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Interestingly, the structure of complexed nsp12 is almost identical to nsp12 in apo RdRp, with an RMSD of 0.5 Å [17], coinciding with the high processivity of the viral RNA polymerase, which does not need to consume extra energy for conformation changes in the active site during the replication cycle (Fig. 3B). Although nearly identical conformation between the apo and RNA bound nsp12 structures, several subtle structural rearrangements can still be observed (Fig. 3C). Compared to the structure of apo RdRp complex, the loop in motif G (residues D449 to L514) moves outward by 2.2–3.0 Å (as measured by nsp12 residue S501), and loop (residues S682 to T686) in motif B by 1.7–2.0 Å (as measured by nsp12 residue A685) [17,18]. The loop connecting the first and second helices of the thumb subdomain also shifts outward by 6.0 Å (as measured by nsp residue I847) [17]. These conformational changes produce a more open cleft of nsp12 in response to accommodate the template-primer RNA.
A recent structure of RdRp-RNA complex represents a long template-product helical RNA strands exiting from the active core of RdRp, which is not observed in other RdRp-RNA complex [38]. The two copies of nsp8 exhibit long helical extensions at their N-terminus fragments, and serve as platforms for coordinating the exiting RNA backbones, forming positively charged “sliding poles” (Fig. 3D) [38]. These “sliding poles” are stabilized by interactions formed between the positively charged residues at the extended N-terminal of nsp8 and bases in RNA backbones (Fig. 3E) and reported to account for the known processivity of the RdRp, which is required for replicating the long coronavirus genomes [39]. In this structure of RdRp in complex with the protruding RNA, the prominent nsp8-2 extends up to 28 base pairs away from the active site, which is a unique conformation not observed in structures of other RNA viruses [33,34] and SARS-CoV-2 RdRp-RNA complexes [17,19]. The N-terminal ends of two copies of nsp8 direct to different orientations, which synergistically prevent premature dissociation of helical RNA from RdRp (Fig. 3D and E). Similarly, the structure of helicase-RdRp-RNA complex also exhibits a similar nsp8 conformation [40], while structure of pre-translocated SARS-CoV-2 RdRp-RNA complex exhibits two conformations of nsp8-2 with a ∼45° rotation at its N-terminus, one of which structurally overlaps with the “sliding poles”-like conformation of nsp8-2 [18]. The alanine mutation of K58, which located in the nsp8 extension, is lethal to the virus, supporting the model of “sliding poles” [26]. Collectively, these findings indicate that the N-terminal conformation of nsp8 is flexible in solution and dominates the “sliding pole”-like state during RNA elongation.
6. Mechanism of RdRp inhibition by remdesivir
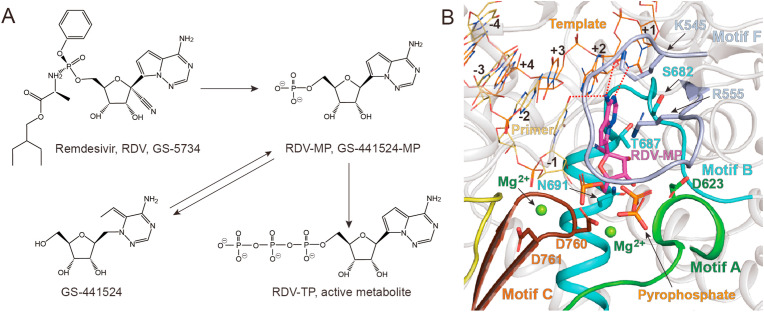
Although the sequence identity of nsp12 across the RNA viruses is low, the polymerase active site is structurally highly conserved, suggesting that RdRp inhibitors may serve as a potential broad-spectrum antiviral drug against RNA viruses. Remdesivir (RDV, GS-5734) is a 1′-cyano-substituted adenosine nucleotide analog prodrug, which is originally developed as a treatment for the Ebola virus by inhibiting RNA synthesis. It is metabolized into its active form (RDV-MP, GS-441524-MP), which has a monophosphate moiety to enhance intracellular metabolism into its active triphosphate metabolite (RDV-TP) (Fig. 4 A) [41].
Fig. 4.
Inhibition mechanism of RdRp by remdesivir. (A) Chemical structure of remdesivir and its cellular metabolic pathway. (B) The binding mode of remdesivir in the active catalytic site of SARS-CoV-2 RdRp (PDB code: 7BV2). The covalently bound remdesivir in the monophosphate form (RDV-MP, colored in magenta), two magnesium ions (green), and pyrophosphate (orange) are shown. Residues interacted with RDV-MP in motif A (green), motif B (cyan), motif C (chocolate), and motif F (light blue) are shown as sticks. Polar interactions formed between RDV-MP and primer strand (yellow orange), as well as template strand (orange) are highlighted as red dashed lines. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Like many nucleotide analog inhibitors, RDV-TP competes with the incorporation of nucleotide counterparts and inhibits transcription of viral RNA. The structure of the RdRp-RNA-RDV complex reveals a molecular mechanism for transcription inhibition by remdesivir [17]. Only one RDV-MP molecule incorporates into the primer strand at +1 position and forms base-stacking interaction with bases in primer strand and two hydrogen bonds with the uridine base from the template strand. RDV-MP also interacts with residues K545 and R555 in motif F. Near RDV-MP are two magnesium ions and pyrophosphate, whose densities are missing in other structures of SARS-CoV-2 RdRp-RNA complexes. The pyrophosphate may block the entry of NTP to the active site by occupying the entrance of nucleotides. Two magnesium ions contact with phosphate diester backbone and form polar interactions with two conserved aspartic acids in motif C (Fig. 4B).
In contrast to other classic chain terminators, the featured inhibition mechanism of remdesivir is a delayed chain termination of nascent viral RNA at i+3 position [42]. When the RNA synthesis process to the i+3 position, the incorporated RTP will be at −3 position. The molecular simulation analysis shows that the 1′- cyano substituent of incorporated RDV sterically clashed with the side chain of S861, a residue faces with −3 position, and probably causes significant distortion of the position of RNA, hampering the translocation of RNA to the −4 position [42]. The S861A mutant abates the chain termination reaction, supporting this steric clash hypothesis for the delayed chain termination [18].
7. Opportunity for targeting RdRp
The RdRp is critical for the replication of viral RNA, and also a promising drug target for COVID-19 treatment. Firstly, like other proteins of SARS-CoV-2, RdRp lacks the closed-related host cell counterparts. Thus, targeting RdRp may circumvent the off-target side effects. Secondly, compared to the spike protein and other virus surface proteins, the active catalytic motifs of the RdRp are highly conserved among RNA viruses, making RdRp an attractive antiviral drug target for a broad-spectrum of viruses. Several nucleoside analog inhibitors have shown inhibitory activities against a broad spectrum of RNA viruses [43]. Finally, repurposing of the RdRp-targeted drugs remains a promising strategy for COVID-19 treatment. The drugs developed for other viruses, such as the anti-influenza drug favipiravir [44], anti-hepatitis C virus drug sofosbuvir, and the broad-spectrum antiviral drug ribavirin, are being evaluated in clinical trials for COVID-19 treatment (Clarivate Analytics Integrity database and ClinicalTrials.gov).
Remdesivir has been approved for emergency use against COVID-19. Although clinical treatment efficacy is limited, it validated the utility of RdRp inhibitors for the treatment of COVID-19. A variety of antiviral drugs and natural products were suggested as lead candidates against COVID-19 through homolog model-based virtual screening and molecular docking [[45], [46], [47]]. Recent progresses on structure determination of the RdRp-RNA bound to remdesivir provide a structural template to improve the accuracy of virtual screening. Additionally, a combination of the RdRp inhibitors with antiviral drugs targeting other virus proteins and immunomodulators may provide a promising strategy for COVID-19 therapeutics.
8. Conclusion
The replication and transcription of SARS-CoV-2 is a spatiotemporally regulated multi-step process, which is mediated by the replication-transcription complex composed by primarily the virus-encoded non-structural proteins. The field of structural studies on the RTC is rapidly developed, as witnessed by the copious amount of the recent RdRp complex structures [[16], [17], [18], [19],38]. Recently, a structure of SARS-CoV-2 holo-RdRp-RNA in complex with two molecules of the nsp13 helicase was released, providing a structural basis for putative nsp13 helicase functions during viral genome replication and transcription [40]. The structures of SARS-CoV-2 RdRp and lessons learned from other coronaviruses have shed light on the mechanisms of RNA binding and inhibitor recognition of RdRp. However, the structural organization and the regulation mechanism of the replication-transcription complex have not been thoroughly studied. More high-resolution structures with diverse nsps organizations are essential to understand how these cofactors conduct the orchestra of RNA processing enzyme to achieve the replication and transcription of virus RNA. Moreover, the complete picture of intricate RNA processing will also provide new candidate targets for designing drugs against COVID-19.
The structures of RdRp in complex with RNA helical strands and remdesivir have provided the structure details of RdRp inhibition by remdesivir and a rational basis for designing nucleotide analog inhibitors. More structures of RdRp bound to specific inhibitors with a diversity of chemical structures, especially the non-nucleotide inhibitors, are awaiting to provide a comprehensive insight into mechanisms of these inhibitors bound to RdRp and a more accuracy structural model for the development of drugs targeting RdRp.
Funding sources
This work is partially supported by the National Key R&D Programs of China 2018YFA0507002; Shanghai Municipal Science and Technology Major Project 2019SHZDZX02; CAS Strategic Priority Research Program XDB08020303 to H.E.X.; the National Natural Science Foundation of China, 31770796; the National Science & Technology Major Project “Key New Drug Creation and Manufacturing Program” 2018ZX09711002 to Y.J.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W., China Novel Coronavirus I., Research T. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., Yuan M.L., Zhang Y.L., Dai F.H., Liu Y., Wang Q.M., Zheng J.J., Xu L., Holmes E.C., Zhang Y.Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lam T.T., Jia N., Zhang Y.W., Shum M.H., Jiang J.F., Zhu H.C., Tong Y.G., Shi Y.X., Ni X.B., Liao Y.S., Li W.J., Jiang B.G., Wei W., Yuan T.T., Zheng K., Cui X.M., Li J., Pei G.Q., Qiang X., Cheung W.Y., Li L.F., Sun F.F., Qin S., Huang J.C., Leung G.M., Holmes E.C., Hu Y.L., Guan Y., Cao W.C. Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature. 2020;583:282–285. doi: 10.1038/s41586-020-2169-0. [DOI] [PubMed] [Google Scholar]
- 5.Zhang T., Wu Q., Zhang Z. Probable pangolin origin of SARS-CoV-2 associated with the COVID-19 outbreak. Curr. Biol. 2020;30:1346–1351. doi: 10.1016/j.cub.2020.03.022. e1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu W., Chen C.E.Z., Gorshkov K., Xu M., Lo D.A.C., Zheng W. RNA-dependent RNA polymerase as a target for COVID-19 drug discovery. Slas Discov. 2020 doi: 10.1177/2472555220942123. Artn 2472555220942123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yan R.H., Zhang Y.Y., Li Y.N., Xia L., Guo Y.Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wrapp D., Wang N.S., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lan J., Ge J.W., Yu J.F., Shan S.S., Zhou H., Fan S.L., Zhang Q., Shi X.L., Wang Q.S., Zhang L.Q., Wang X.Q. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020 doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 10.Shang J., Ye G., Shi K., Wan Y.S., Luo C.M., Aihara H., Geng Q.B., Auerbach A., Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020 doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., Hastie K.M., Parker M.D., Partridge D.G., Evans C.M., Freeman T.M., de Silva T.I., Sheffield C.-G.G., McDanal C., Perez L.G., Tang H., Moon-Walker A., Whelan S.P., LaBranche C.C., Saphire E.O., Montefiori D.C. Tracking changes in SARS-CoV-2 spike: Evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020 doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holshue M.L., DeBolt C., Lindquist S., Lofy K.H., Wiesman J., Bruce H., Spitters C., Ericson K., Wilkerson S., Tural A., Diaz G., Cohn A., Fox L., Patel A., Gerber S.I., Kim L., Tong S., Lu X., Lindstrom S., Pallansch M.A., Weldon W.C., Biggs H.M., Uyeki T.M., Pillai S.K., Washington State -nCo V.C.I.T. First case of 2019 novel coronavirus in the United States. N. Engl. J. Med. 2020;382:929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sheahan T.P., Sims A.C., Graham R.L., Menachery V.D., Gralinski L.E., Case J.B., Leist S.R., Pyrc K., Feng J.Y., Trantcheva I., Bannister R., Park Y., Babusis D., Clarke M.O., Mackman R.L., Spahn J.E., Palmiotti C.A., Siegel D., Ray A.S., Cihlar T., Jordan R., Denison M.R., Baric R.S. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci. Transl. Med. 2017;9 doi: 10.1126/scitranslmed.aal3653. ARTN eaal3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao Y., Yan L., Huang Y., Liu F., Zhao Y., Cao L., Wang T., Sun Q., Ming Z., Zhang L., Ge J., Zheng L., Zhang Y., Wang H., Zhu Y., Zhu C., Hu T., Hua T., Zhang B., Yang X., Li J., Yang H., Liu Z., Xu W., Guddat L.W., Wang Q., Lou Z., Rao Z. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020;368:779–782. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yin W., Mao C., Luan X., Shen D.D., Shen Q., Su H., Wang X., Zhou F., Zhao W., Gao M., Chang S., Xie Y.C., Tian G., Jiang H.W., Tao S.C., Shen J., Jiang Y., Jiang H., Xu Y., Zhang S., Zhang Y., Xu H.E. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. 2020;368:1499–1504. doi: 10.1126/science.abc1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Q., Wu J., Wang H., Gao Y., Liu Q., Mu A., Ji W., Yan L., Zhu Y., Zhu C., Fang X., Yang X., Huang Y., Gao H., Liu F., Ge J., Sun Q., Yang X., Xu W., Liu Z., Yang H., Lou Z., Jiang B., Guddat L.W., Gong P., Rao Z. Structural basis for RNA replication by the SARS-CoV-2 polymerase. Cell. 2020;182:417–428. doi: 10.1016/j.cell.2020.05.034. e413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peng Q., Peng R., Yuan B., Zhao J., Wang M., Wang X., Wang Q., Sun Y., Fan Z., Qi J., Gao G.F., Shi Y. Structural and biochemical characterization of the nsp12-nsp7-nsp8 core polymerase complex from SARS-CoV-2. Cell Rep. 2020;31:107774. doi: 10.1016/j.celrep.2020.107774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O’Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., Tummino T.A., Huttenhain R., Kaake R.M., Richards A.L., Tutuncuoglu B., Foussard H., Batra J., Haas K., Modak M., Kim M., Haas P., Polacco B.J., Braberg H., Fabius J.M., Eckhardt M., Soucheray M., Bennett M.J., Cakir M., McGregor M.J., Li Q., Meyer B., Roesch F., Vallet T., Mac Kain A., Miorin L., Moreno E., Naing Z.Z.C., Zhou Y., Peng S., Shi Y., Zhang Z., Shen W., Kirby I.T., Melnyk J.E., Chorba J.S., Lou K., Dai S.A., Barrio-Hernandez I., Memon D., Hernandez-Armenta C., Lyu J., Mathy C.J.P., Perica T., Pilla K.B., Ganesan S.J., Saltzberg D.J., Rakesh R., Liu X., Rosenthal S.B., Calviello L., Venkataramanan S., Liboy-Lugo J., Lin Y., Huang X.P., Liu Y., Wankowicz S.A., Bohn M., Safari M., Ugur F.S., Koh C., Savar N.S., Tran Q.D., Shengjuler D., Fletcher S.J., O’Neal M.C., Cai Y., Chang J.C.J., Broadhurst D.J., Klippsten S., Sharp P.P., Wenzell N.A., Kuzuoglu-Ozturk D., Wang H.Y., Trenker R., Young J.M., Cavero D.A., Hiatt J., Roth T.L., Rathore U., Subramanian A., Noack J., Hubert M., Stroud R.M., Frankel A.D., Rosenberg O.S., Verba K.A., Agard D.A., Ott M., Emerman M., Jura N., von Zastrow M., Verdin E., Ashworth A., Schwartz O., d’Enfert C., Mukherjee S., Jacobson M., Malik H.S., Fujimori D.G., Ideker T., Craik C.S., Floor S.N., Fraser J.S., Gross J.D., Sali A., Roth B.L., Ruggero D., Taunton J., Kortemme T., Beltrao P., Vignuzzi M., Garcia-Sastre A., Shokat K.M., Shoichet B.K., Krogan N.J. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thoms M., Buschauer R., Ameismeier M., Koepke L., Denk T., Hirschenberger M., Kratzat H., Hayn M., Mackens-Kiani T., Cheng J., Straub J.H., Sparrer K.M.J., Beckmann R. Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2. Science. 2020;369(6508):1249e1255. doi: 10.1126/science.abc8665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yuan S., Peng L., Park J.J., Hu Y., Devarkar S.C., Dong M.B., Wu S., Chen S., Lomakin I., Xiong Y. 2020. Nonstructural Protein 1 of SARS-CoV-2 Is a Potent Pathogenicity Factor Redirecting Host Protein Synthesis Machinery toward Viral RNA; p. 243451. 2020.2008.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Littler D.R., Gully S., Colson R.N., Rossjohn J. Crystal structure of the SARS-CoV- 2 non-structural protein 9, Nsp9. iScience. 2020;23(7):101258. doi: 10.1016/j.isci.2020.101258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pillon M.C., Frazier M.N., Dillard L.B., Williams J.G., Kocaman S., Krahn J.M., Perera L., Hayne C.K., Gordon J., Stewart Z.D., Sobhany M., Deterding L.J., Hsu A.L., Dandey V.P., Borgnia M.J., Stanley R.E. 2020. Cryo-EM Structures of the SARS-CoV-2 Endoribonuclease Nsp15; p. 244863. 2020.2008.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosas-Lemus M., Minasov G., Shuvalova L., Inniss N.L., Kiryukhina O., Wiersum G., Kim Y., Jedrzejczak R., Maltseva N.I., Endres M., Jaroszewski L., Godzik A., Joachimiak A., Satchell K.J.F. 2020. The Crystal Structure of Nsp10-Nsp16 Heterodimer from SARS-CoV-2 in Complex with S-Adenosylmethionine. 2020.2004.2017.047498. [DOI] [Google Scholar]
- 26.Subissi L., Posthuma C.C., Collet A., Zevenhoven-Dobbe J.C., Gorbalenya A.E., Decroly E., Snijder E.J., Canard B., Imbert I. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc. Natl. Acad. Sci. U. S. A. 2014;111:E3900–E3909. doi: 10.1073/pnas.1323705111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kirchdoerfer R.N., Ward A.B. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat. Commun. 2019;10:2342. doi: 10.1038/s41467-019-10280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pachetti M., Marini B., Benedetti F., Giudici F., Mauro E., Storici P., Masciovecchio C., Angeletti S., Ciccozzi M., Gallo R.C., Zella D., Ippodrino R. Emerging SARS-CoV-2 mutation hot spots include a novel RNA-dependent-RNA polymerase variant. J. Transl. Med. 2020;18:179. doi: 10.1186/s12967-020-02344-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eskier D., Karakulah G., Suner A., Oktay Y. RdRp mutations are associated with SARS-CoV-2 genome evolution. PeerJ. 2020;8:e9587. doi: 10.7717/peerj.9587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Godoy A.S., Lima G.M., Oliveira K.I., Torres N.U., Maluf F.V., Guido R.V., Oliva G. Crystal structure of Zika virus NS5 RNA-dependent RNA polymerase. Nat. Commun. 2017;8:14764. doi: 10.1038/ncomms14764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gerlach P., Malet H., Cusack S., Reguera J. Structural insights into bunyavirus replication and its regulation by the vRNA promoter. Cell. 2015;161:1267–1279. doi: 10.1016/j.cell.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhai Y., Sun F., Li X., Pang H., Xu X., Bartlam M., Rao Z. Insights into SARS-CoV transcription and replication from the structure of the nsp7-nsp8 hexadecamer. Nat. Struct. Mol. Biol. 2005;12:980–986. doi: 10.1038/nsmb999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Appleby T.C., Perry J.K., Murakami E., Barauskas O., Feng J., Cho A., Fox D., 3rd, Wetmore D.R., McGrath M.E., Ray A.S., Sofia M.J., Swaminathan S., Edwards T.E. Viral replication. Structural basis for RNA replication by the hepatitis C virus polymerase. Science. 2015;347:771–775. doi: 10.1126/science.1259210. [DOI] [PubMed] [Google Scholar]
- 34.Gong P., Peersen O.B. Structural basis for active site closure by the poliovirus RNA-dependent RNA polymerase. Proc. Natl. Acad. Sci. U. S. A. 2010;107:22505–22510. doi: 10.1073/pnas.1007626107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garriga D., Ferrer-Orta C., Querol-Audi J., Oliva B., Verdaguer N. Role of motif B loop in allosteric regulation of RNA-dependent RNA polymerization activity. J. Mol. Biol. 2013;425:2279–2287. doi: 10.1016/j.jmb.2013.03.034. [DOI] [PubMed] [Google Scholar]
- 36.Vazquez A.L., Alonso J.M., Parra F. Mutation analysis of the GDD sequence motif of a calicivirus RNA-dependent RNA polymerase. J. Virol. 2000;74:3888–3891. doi: 10.1128/jvi.74.8.3888-3891.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Y., Xiao M., Chen J., Zhang W., Luo J., Bao K., Nie M., Chen J., Li B. Mutational analysis of the GDD sequence motif of classical swine fever virus RNA-dependent RNA polymerases. Virus Gene. 2007;34:63–65. doi: 10.1007/s11262-006-0001-z. [DOI] [PubMed] [Google Scholar]
- 38.Hillen H.S., Kokic G., Farnung L., Dienemann C., Tegunov D., Cramer P. Structure of replicating SARS-CoV-2 polymerase. Nature. 2020;584:154–156. doi: 10.1038/s41586-020-2368-8. [DOI] [PubMed] [Google Scholar]
- 39.Posthuma C.C., Te Velthuis A.J.W., Snijder E.J. Nidovirus RNA polymerases: Complex enzymes handling exceptional RNA genomes. Virus Res. 2017;234:58–73. doi: 10.1016/j.virusres.2017.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen J., Malone B., Llewellyn E., Grasso M., Shelton P.M.M., Olinares P.D.B., Maruthi K., Eng E.T., Vatandaslar H., Chait B.T., Kapoor T.M., Darst S.A., Campbell E.A. Structural basis for helicase-polymerase coupling in the SARS-CoV-2 replication-transcription complex. Cell. 2020 doi: 10.1016/j.cell.2020.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cardile A.P., Warren T.K., Martins K.A., Reisler R.B., Bavari S. Will there be a cure for Ebola? Annu. Rev. Pharmacol. 2017;57:329–348. doi: 10.1146/annurev-pharmtox-010716-105055. [DOI] [PubMed] [Google Scholar]
- 42.Gordon C.J., Tchesnokov E.P., Woolner E., Perry J.K., Feng J.Y., Porter D.P., Gotte M. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J. Biol. Chem. 2020;295:6785–6797. doi: 10.1074/jbc.RA120.013679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Campagnola G., Gong P., Peersen O.B. High-throughput screening identification of poliovirus RNA-dependent RNA polymerase inhibitors. Antivir. Res. 2011;91:241–251. doi: 10.1016/j.antiviral.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Du Y.X., Chen X.P. Favipiravir: Pharmacokinetics and concerns about clinical trials for 2019-nCoV infection. Clin. Pharmacol. Ther. 2020;108:242–247. doi: 10.1002/cpt.1844. [DOI] [PubMed] [Google Scholar]
- 45.Wu C.R., Liu Y., Yang Y.Y., Zhang P., Zhong W., Wang Y.L., Wang Q.Q., Xu Y., Li M.X., Li X.Z., Zheng M.Z., Chen L.X., Li H. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharmacol. Sin. B. 2020;10:766–788. doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Naik B., Gupta N., Ojha R., Singh S., Prajapati V.K., Prusty D. High throughput virtual screening reveals SARS-CoV-2 multi-target binding natural compounds to lead instant therapy for COVID-19 treatment. Int. J. Biol. Macromol. 2020;160:1–17. doi: 10.1016/j.ijbiomac.2020.05.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Elfiky A.A. SARS-CoV-2 RNA dependent RNA polymerase (RdRp) targeting: an in silico perspective. J. Biomol. Struct. Dyn. 2020:1–9. doi: 10.1080/07391102.2020.1761882. [DOI] [PMC free article] [PubMed] [Google Scholar]