ABSTRACT
Behavioural, structural, and functional neuroimaging have implicated the hippocampus as a critical brain region in posttraumatic stress disorder (PTSD) pathogenesis. Recent work in a normative, primarily European, sample identified 15 unique genetic loci contributing to structural variability in six hippocampal subfield volumes. We explored the relevance of these loci in two samples (Mental Illness Research Education and Clinical Centre [MIRECC] and Grady; n = 290) of trauma-exposed individuals enriched for PTSD and of diverse ancestry. Four of the previous loci demonstrated nominal evidence of replication in the MIRECC dataset, primarily within non-Hispanic whites (NHW). One locus replicated in the Grady cohort, which was composed exclusively of non-Hispanic blacks (NHB). Our data supported genetic interactions with diagnosis of lifetime PTSD and genetic interactions with childhood trauma in the MIRECC sample, but not the Grady sample. Given the racial, diagnostic, and trauma-exposure differences with the original genome-wide association study (GWAS) report, we conducted a full GWAS in the MIRECC and Grady datasets. Interactions between genetic variants and lifetime PTSD or childhood trauma were interrogated for single nucleotide polymorphisms (SNPs) with evidence of main effects. Genetic associations surpassed false discovery rate (FDR)-correction within hippocampal subfields in fimbria, subiculum, cornu ammonis-1 (CA1), and hippocampal amygdala transition area (HATA). One association was replicated in the Grady cohort (rs12880795 in TUNAR with left (L)-HATA volume). The most significant association in the MIRECC dataset was between rs6906714 in LINC02571 and right (R)-fimbria volume (p = 5.99×10−8, q = 0.0056). Interestingly, the effect of rs6906714 on R-fimbria volume increased with exposure to childhood trauma (gene*environment [G*E] interaction p = 0.022). These preliminary results argue for G*E interactions between genetic loci with PTSD and childhood trauma on hippocampal phenotypes. Our results underscore the need for larger neuroimaging-genetic studies in PTSD, trauma, and ancestrally diverse populations.
KEYWORDS: PTSD, childhood trauma, genetics, hippocampus, structural MRI, hippocampal subfields
Highlights: • Gene by environment interactions of genetic loci with both PTSD and childhood trauma on hippocampal phenotypes
Las neuroimagenologia, conductual, estructural y funcional ha implicado que el hipocampo se constituye como una región cerebral critica en la patogénesis del trastorno de estrés postraumático (TEPT). Un trabajo reciente, realizado en una muestra normativa, principalmente europea, identifico 15 loci genéticos únicos que contribuyen a la variabilidad estructural de seis volúmenes de subcampos hipocampales. Exploramos la relevancia de estos loci en dos muestras enriquecidas para TEPT (del Centro de Educación y Clínica Investigación sobre Enfermedades Mentales, MIRECC por sus siglas en inglés y Grady; n=290) de individuos expuestos a trauma y de ascendencia diversa. Cuatro de los loci previos demostraron evidencia nominal de replicación en la base de datos MIRECC, principalmente en personas de raza blanca no hispánicos (NHW). Se replicó un locus en la cohorte Grady, que estaba compuesta exclusivamente por personas de raza negra no hispánicos (NHB). Nuestros datos respaldaron las interacciones genéticas con el diagnostico de TEPT a lo largo de la vida e interacciones genéticas con trauma infantil en la muestra MIRECC, pero no en la de Grady. Debido a las diferencias raciales, diagnósticas y de exposición a trauma con el reporte original del estudio de asociación del genoma completo (GWAS por sus siglas en ingles), realizamos un GWAS completo con la base de datos MIRECC y Grady. Se exploraron polimorfismos de nucleótido único (SNPs por sus siglas en ingles) en las interacciones de variantes genéticas del TEPT a lo largo de la vida y del trauma infantil, con evidencia de un efecto genético principal. Las asociaciones genéticas sobrepasaron a la corrección de tasa de falso descubrimiento (FDR) dentro de los subcampos hipocampales de la fimbria, subículo, asta de Amon-1 (CA1), y el área de transición hipocampo-amigdaliana (HATA). Una asociación se replicó en la cohorte de Grady (rs 12880795 en TUNAR con volumen izquierdo del HATA). La asociación más significativa en la base de datos de MIRECC estuvo entre rs6906714 en LINC02571 y el volumen de la fimbria derecha (p=5.99×10-8, q=0.0056). Interesantemente, el efecto rs6906714 sobre el volumen de la fimbria derecha se incrementaba con la exposición al trauma infantil (interacción gen* ambiente [G*A] p=0.022). Estos resultados preliminares orientarían a la presencia de interacciones G*A de loci genéticos con el TEPT y trauma infantil en fenotipos del hipocampo. Nuestro resultados destacan la necesidad de estudios más grandes que vinculen neuroimagenes y genética en poblaciones con TEPT, trauma, y genealogía diversa.
PALABRAS CLAVE: TEPT, trauma infantil, genética, hipocampo, Resonancia magnética estructural, subcampos hipocampales
行为性, 结构性和功能性神经影像学已将海马体视为创伤后应激障碍 (PTSD) 发病机制中的一个关键脑区。最近在一个以欧洲人为主的规范样本中的研究确定了15个导致六个海马亚区体积结构性变异的独特基因位点。我们在由高比例PTSD和多种血统的创伤暴露个体组成的两个样本 (精神疾病研究教育和临床中心[MIRECC]与Grady;n = 290) 中, 探究了这些基因位点的相关性。上述位点中四个在MIRECC数据集中展现出名义上的可重复性, 主要为非西班牙裔白人 (NHW) 。一个位点在Grady队列中重复, 此样本仅由非西班牙裔黑人 (NHB) 组成。我们的数据支持MIRECC样本 (而非Grady样本) 中诊断终身PTSD与遗传的交互作用以及童年期创伤与遗传的交互作用。考虑到与原始全基因组关联研究 (GWAS) 报告的种族, 诊断和创伤暴露差异, 我们在MIRECC和Grady数据集中进行了完整的GWAS。对于有主效应的单核苷酸多态性 (SNP) 位点, 对遗传变异与终身PTSD或童年期创伤间的交互作用进行审查。遗传关联在海马伞部, 下托, 海马角 (CA1) 亚区和海马杏仁核过渡区 (HATA) 中超过了校正错误发现率 (FDR) 。在Grady队列中重复了一种相关 (TUNAR中的rs12880795与左侧HATA体积) 。在MIRECC数据集中, 最显著的关联出现在LINC02571中的rs6906714与海马伞部右侧体积 (p = 5.99x10−8, q= 0.0056) 之间。有趣的是, rs6906714对海马伞部右侧体积的影响随童年期创伤的增加而增加 (基因*环境[G * E]交互作用 p= 0.022) 。这些初步结果证明了PTSD遗传基因位点与童年期创伤对海马表型的交互作用。我们的结果强调需要在PTSD, 创伤和多种血缘的人群中进行更大型的神经影像遗传学研究。
关键词: PTSD, 童年期创伤, 遗传学, 海马, 结构性核磁共振, 海马亚区
1. Introduction
A national survey of trauma exposure in the US indicates that nearly 90% meet criteria established in the Diagnostic and Statistical Manual (DSM)-5 (Kilpatrick et al., 2013b). Posttraumatic stress disorder (PTSD), which affects about 8% of the United States (US) population (Kessler et al., 2005b), may develop following exposure to trauma and manifests with the hallmark symptoms of hyperarousal, re-experiencing, and avoidance. The effect of PTSD on society and individual functioning is severe with a 150% increase in the chance of unemployment, 60% increase in marital instability, and higher rates of suicide than any anxiety disorder (Kessler, 2000b). Importantly, deficits in memory, including declarative memory, fragmented autobiographical, or trauma-related memories and amnesia about the details of trauma have been demonstrated in PTSD (Elzinga & Bremner, 2002b). The hippocampus has long been implicated in PTSD because of its role in memory formation and retrieval (Bremner et al., 1997), as well as the observation of lower hippocampal volume in individuals with PTSD (Logue et al., 2017b; Morey et al., 2012). In monozygotic twins who are discordant for trauma exposure, smaller hippocampal volume represents an increased risk for developing stress-related psychopathology (Gilbertson et al., 2002b). It is unclear whether lower hippocampal volume is a consequence of developing PTSD or occurs as the result of heretofore unknown genetic or biological vulnerabilities to developing PTSD. Genetic modulators of hippocampal volume have been established in non-clinical populations with genome-wide association studies (GWAS)(Hibar et al., 2015b; Stein et al., 2012; van der Meer et al., 2018b). The SNP-based heritability of hippocampal subfields is modest in normative samples (h2 = 0.14 to 0.27). By contrast, the twin-based heritability of hippocampal subfields is high (h2 = 0.38 to 0.85) (Patel et al., 2017). However, the discrepancy between the two estimates is perhaps unsurprising since twin-based estimates of heritability can be influenced by non-additive genetic factors (Yang, Zeng, Goddard, Wray, & Visscher, 2017). Overall, SNP-based heritability is lower than twin-based heritability because SNP-based heritability uses tag SNPs, which are a subset of all SNPs, and because GWAS confounds age-varying gene expression with non-genetic factors, which twin studies do not. Nonetheless, there is clearly a heritable component to hippocampal subfield volume in normative samples but the specific genetic loci associated with hippocampal subfield volume in PTSD have not been investigated.
Two GWAS from the Enhancing Neuroimaging-Genetics through Meta-Analysis (ENIGMA) Consortium has examined effects on overall hippocampal volume with significant results at the genome-wide level. Stein et al. (Stein et al., 2012) identified and replicated two quantitative trait loci (QTL) for hippocampal volumes in a large sample that included non-clinical and clinical samples with neuropsychiatric diagnoses. A large study by van der Meer and colleagues recently reported on 21,297 samples to identify 15 unique genome-wide significant loci across six hippocampal subfields, of which eight loci had not been previously linked to the hippocampus (van der Meer et al., 2018b). The SNPs mapped to genes associated with neuronal differentiation, locomotor behaviour, schizophrenia, and Alzheimer’s disease. These studies have demonstrated the ability to detect genetic associations with neuroimaging phenotypes that may be relevant to PTSD. However, as yet, there have been no published studies that have examined genetic predictors of hippocampal subfields in relation to PTSD or exposure to childhood trauma, which is an important predictor of subsequent development of PTSD (Teicher & Samson, 2014).,
In the present context, SNPs are part of genes or regulatory elements that govern protein synthesis, which in turn plays an essential role in the differentiation, metabolism, signalling, and myriad other neurobiological functions. On the other hand, behaviours secondarily emanate from carefully orchestrated actions of these neurobiological phenomena. Therefore, possible advantages of identifying neuroimaging phenotypes over diagnostic phenotypes, which consist of a constellation of symptoms or behaviours are (1) the enhanced accuracy of imaging phenotypes over a subjectively assessed diagnostic phenotype, (2) an imaging phenotype may be closer to the action of genes than the diagnostic phenotype. Subfields of the hippocampus are involved in discrete aspects of memory encoding and consolidation. For example, the dentate gyrus (DG) is important in pattern separation, a process where salient features of memories are contrasted in order to distinguish similar but discrete events (Schmidt, Marrone, & Markus, 2012; Yassa & Stark, 2011). By contrast, the entorhinal cortex (EC) and cornu ammonis subfield-3 (CA3) are crucial to pattern completion, which involves recognizing overlapping features of past events. Pattern completion is a widely investigated model of PTSD given its important implication in contextual fear-conditioning (O’Reilly & Norman, 2002; Parsons & Ressler, 2013). Preclinical animal models show that CA1 is involved in context-specific memory retrieval following extinction (Ji & Maren, 2007b). As such, the CA1 hippocampal subfield has been implicated strongly in conditioned fear and its extinction (Furini, Myskiw, & Izquierdo, 2014b). The deficits in episodic memory, contextual memory, and extinction failure suggest that CA1, CA3, and dentate subfields may play a role in developing PTSD (Chen et al., 2018).
The present study examines two samples of highly traumatized individuals. The MIRECC sample is comprised of US military veterans with roughly equal number of European- and African-Americans who experienced high levels of trauma in the military and in some cases during childhood and adolescence (Brancu et al., 2017). The Grady sample of civilians consists of African-American women in Atlanta [Georgia US] who experienced high rates of sustained trauma and interpersonal violence and in some cases during childhood and adolescence (Gillespie et al., 2009b). Based on the role of the dentate gyrus in pattern separation, CA3 in pattern completion, and CA1 in fear extinction, we hypothesized the presence of genetic variants that influence subfield volumes, which are obtained from segmentation of high-resolution structural magnetic resonance imaging (MRI), in these enriched samples of trauma-exposed individuals with and without PTSD.
2. Methods
2.1. Participants and clinical measures
2.1.1. MIRECC cohort
The MIRECC cohort consisted of 157 participants from a repository [Mid-Atlantic MIRECC, Durham NC] of Iraq and Afghanistan era military service members who contributed blood for genotyping, clinical assessment data, and MRI scans. Participants, which included 132 men and 25 women (see Table 1), were screened for inclusion/exclusion criteria based on information available in the repository. To reduce the effects of population stratification in a multi-racial sample, analyses were limited to non-Hispanic black (NHB; n = 74) and non-Hispanic white (NHW; n = 83) participants who underwent genetic and imaging components and had data available at the time of analysis. Important exclusion criteria included the presence of psychotic symptoms, high risk of suicide, contraindication to MRI, current substance abuse, neurological disorders, and age over 65 years. Participants were evaluated for PTSD, trauma exposure, and other psychiatric comorbidities (Traumatic Life Events Questionnaire, TLEQ (Kubany et al., 2000b)), combat exposure (Combat Exposure Scale, CES (Lund, Foy, Sipprelle, & Strachan, 1984b)), and depressive symptoms (Beck Depression Inventory-II, BDI-II (Beck, Steer, & Brown, 1996)). PTSD diagnosis was ascertained with the Structured Clinical Interview for DSM-IV (SCID) and confirmed with the Clinician-Administered PTSD Scale (CAPS)(Blake et al., 1995) in 152 (97%) participants and cut-off score over 36 with the Davidson Trauma Scale (DTS) (Davidson et al., 1997b) in 5 (3%) participants. Alcohol abuse and dependence were determined by the SCID. All participants provided written informed consent to participate in procedures reviewed and approved by the Institutional Review Boards at Duke University and the Durham VA Medical Centre.
Table 1.
Demographic and clinical information.
| Sample Characteristic | MIRECC (n = 157) | Grady Trauma Project (n = 133) | Group Comparison |
|---|---|---|---|
| Age, mean (SD) | 39.62 (10.03) | 38.71 (11.99) | P = 0.4815 |
| Gender, No. female (%) | 25 (15.92) | 133 (100) | P < 0.0001 |
| Race, No. Caucasian (%) | 83 (52.87) | 0 (0) | P < 0.0001 |
| Child Trauma Category 0, 1, ≥ 2 (%) | 72 (45.86), 48 (30.57), 37 (23.57) | 61 (45.86), 40 (30.08), 32 (24.06) | P = 0.9934 |
| Alcohol abuse/dependency (%) | 49 (31.61) | 42 (44.68) | P = 0.0379 |
| SCID, CAPS or PSS lifetime PTSD Diagnosis (%) | 66 (42.04) | 79 (59.40) | P = 0.0032 |
§ The Davidson Trauma Scale was used in lieu of the CAPS for 5 subjects for which CAPS was unavailable.
Abbreviations: SD = standard deviation, No = number, AUDIT = Alcohol Use Disorders Identification Test, SCID-IV = Structured Clinical Interview for DSM-IV, CAPS-IV Clinician-Administered PTSD Scaler with DSM-IV criteria.
2.1.2. Grady cohort
The Grady sample consisted of 133 participants drawn from the Grady Trauma Project (GTP), study of PTSD risk factors in participants from a low-socioeconomic status urban cohort of outpatients in general medical clinics at Grady Hospital [Atlanta, GA]. The sample was restricted to African-American women to minimize heterogeneity. Seventy-nine participants met criteria for diagnosis of PTSD, and 54 trauma-exposed controls (TC) did not. Exclusion criteria included a history of bipolar disorder, schizophrenia, current psychotic symptoms, current psychotropic medication use, contraindications to MRI scanning, neurological disorder, known structural brain abnormality, head injury with loss of consciousness, implanted metal objects, or positive urine tests for pregnancy and drug use (cocaine, marijuana, opiates, amphetamines, methamphetamines) assessed 24 to 48 hours before the MRI scan. Adult trauma history was assessed using the Traumatic Events Inventory (TEI) (Gillespie et al., 2009b). Total adult trauma exposure was measured by counting the number of trauma types (e.g. car accident, natural disaster, assault) experienced after age 17. Childhood trauma history and trauma exposure were assessed using the Childhood Trauma Questionnaire (CTQ) (Bernstein et al., 1994). Study procedures were approved by the Institutional Review Board of Emory University and the Research Oversight Committee of Grady Memorial Hospital, and all participants provided written informed consent prior to participating.
2.2. MRI acquisition
2.2.1. MIRECC sample
Images were acquired on a General Electric 3-Tesla Signa EXCITE scanner equipped with an eight-channel head coil. High-resolution T1-weighted whole-brain images using 3D-FSPGR were acquired axially with repetition time (TR) = 7.848 ms, echo time (TE) = 2.984 ms, flip angle = 12°, Inversion Time (TI) = 400 ms, field of view (FOV) = 256×256 mm, 1-mm axial slice thickness, 166 slices, 1-mm3 voxel size, 1 excitation.
2.2.2. Grady sample
Scanning took place on a 3.0 T Siemens Trio with echo-planar imaging. High-resolution T1-weighted whole-brain anatomical scans were collected using a 3D MP-RAGE sequence, TR = 2000, TE = 3.02, flip angle = 8°, TI = 900 ms, FOV = 224×256 mm, 1-mm sagittal slice thickness, 176 slices, 1-mm3 voxel size.
2.3. Hippocampal subfield volume analysis
Identical procedures were applied to the discovery and replication sample after acquisition to perform automated segmentation, labelling of subcortical volumes, and estimation of total intracranial volume (ICV) on the T1 images using the FreeSurfer image analysis suite (v. 5.3.0) and its library tool, recon-all (Fischl, 2012b). Subsequently, hippocampal subfield segmentation was performed using the FreeSurfer 6.0.0 library function, hippocampal-subfields-T1. Hippocampal subfield volumes were generated in each subject for both the left and right hemispheres on CA1, CA3, CA4, DG, fimbria, fissure, hippocampus-amygdala transition area (HATA), molecular layer, parasubiculum, presubiculum, subiculum, and tail (Figure 1). Standardized image analysis protocols for subcortical segmentation were drawn from the Consortium for Enhancing Imaging Genetics through Meta-Analysis (ENIGMA), which were previously reported in detail (Chen et al., 2018).
Figure 1.
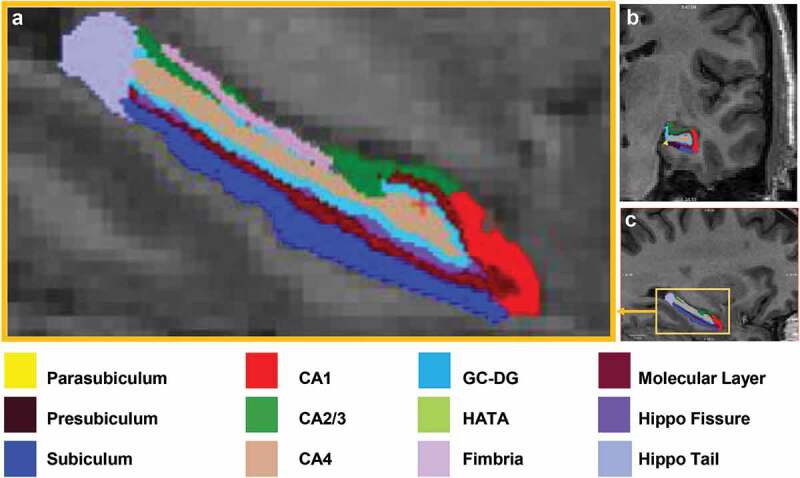
Automated Segmentation of the hippocampus into 12 subfields in each hemisphere of the brain performed with FreeSurfer v.6.0. Subfield images of CA1, CA2/3, CA4, dentate gyrus, hippocampal amygdala transition area (HATA), subiculum, tail, fissure, presubiculum, parasubiculum, molecular layer, fimbria are shown in (a) magnified sagittal plane, (b) coronal plane, and (c) sagittal plane.
2.4. Quality control
Identical visual inspection procedures were applied to all T1 images from the MIRECC (RAM, CCH) and Grady cohorts (JSS, SVR) for quality assurance purposes. We applied quality assurance for hippocampal subfield segmentations using ENIGMA-PTSD protocols (Chen et al., 2018). All participants passed both rounds of quality control. The recent emphasis on rigour and reproducibility prompted a review of the test–retest reliability hippocampal subfield segmentation. Automated segmentation for all the hippocampal subfields except the hippocampal fissure is reliable and robust quantitative phenotypes for genetic studies (Hibar et al., 2015b); therefore, we elected to remove hippocampal fissure volume from our analysis.
2.5. Genotyping
2.5.1. MIRECC sample
GWAS data were generated as part of a larger parent study of 2,312 PTSD cases and controls described previously (Ashley-Koch et al., 2015) and recapitulated here. Deoxyribonucleic acid (DNA) was extracted from whole blood using the Puregene system (Gentra Systems, Minneapolis, MN). Whole-genome genotyping data were generated in three different batches using three different platforms. A total of 2,312 samples were genotyped: 587 samples with the Illumina HumanHap650 Beadchip, 545 samples with the Illumina Human 1 M-Duo Beadchip, and 1180 samples with the Illumina Human Omni 2.5 Beadchip (Illumina, San Diego, CA). Sample classified as Centre d’Etude du Polymorphisme Humain (CEPH) and masked sample duplicates were included as controls. Each batch was analysed using the GenomeStudio software (Illumina, San Diego, CA) and subsequently passed through quality control (QC) pipelines separately. Samples were required to have a call rate >98% (n = 7 samples excluded) and no gender discrepancies (n = 7 samples excluded). Identity by state analysis was performed using the software tool PLINK (Purcell et al., 2007) to identify duplicate individuals (n = 6 samples excluded) and those related ≥50% (n = 16 samples excluded). To reduce effects of population stratification, participants who did not self-report as either NHW or NHB were removed (n = 113). Additionally, principal component analysis (PCA) was run using the smartpca program from the software package EIGENSOFT (Patterson, Price, & Reich, 2006) in order to identify remaining outliers (n = 22 excluded). Probes were required to have a call rate >97% and Hardy–Weinberg Equilibrium (HWE) p-values >10−6 in controls. Quality control was assessed separately in each batch and samples with low call rates were excluded from further analysis. A global reference panel from 1000 Genomes was used to impute missing genotypes in each batch separately; imputed SNPs with certainty <0.90 were excluded. Overlapping SNPs in the imputed NHB and NHW subsets were merged to create a final dataset comprising 2,711,511 SNPs.
2.5.2. Grady sample
Saliva collected in Oragene vials underwent DNA extraction followed by genotyping on Illumina platforms and QC measures with PLINK. Levels of heterozygosity, deviation from HWE, screening for relatedness, and low call rates were part of the QC process. DNA was extracted from saliva in Oragene collection vials (DNA Genotek, Ottawa, ON, Canada) using the DNAdvance kit (Beckman Coulter Genomics, Danvers, MA, USA), while DNA from blood was extracted using either the E.Z.N.A. Mag-Bind Blood DNA Kit (Omega Bio-Tek, Norcross, GA, USA) or ArchivePure DNA Blood Kit (5 Prime, Gaithersburg, MD, USA). Genotyping was performed using the Illumina Omni-Quad 1 M or the Omni Express BeadChip (Illumina, San Diego, CA, USA), and genotypes were called in Illumina’s GenomeStudio (Illumina). PLINK (Purcell et al., 2007) was used to perform quality control measures. Initial quality control involved removing samples with very low call rates (that is, poor quality samples with large amounts of missing data) and outside acceptable levels of heterozygosity (−0.25< Fhet>0.25), and the remaining samples were recalled in GenomeStudio (Pluzhnikov et al., 2010). Further quality control involved excluding SNPs that had a call rate <98%, a MAF<0.01 or significant deviation from Hardy–Weinberg proportions (P < 1 × 10−6 in controls and P < 1 × 10−10 in PTSD cases), and removing individuals with greater than 5% missing data (2%). We further identified and removed related individuals by using PLINK to estimate the proportion of identity by descent (IBD) for each pair of individuals. Among pairs of individuals with an IBD proportion>0.12 (indicating cousins or a closer relation), we removed the individual in each pair with the higher rate of missing genotype. Three of these variants (rs6906714, rs2553974, and rs12880795) were present in the Illumina platforms, while the others were imputed applying the PGC pipeline (Duncan et al., 2018b), using the 1000 Genomes20 phase 1 as reference, SHAPEIT for phasing and IMPUTE2 for imputation (Marchini, Howie, Myers, McVean, & Donnelly, 2007b).
2.6. Statistical analysis
Principal component analysis (PCA) was run using the smartpca program from the software package EIGENSOFT (Patterson et al., 2006) to obtain ancestry-related principal components (PCs). One principal component was necessary to account for the population variability observed in the MIRECC sample, essentially distinguishing the NHB from the NHW subjects. Linear regression was conducted using PLINK (Purcell et al., 2007) to test for association between subcortical volume and each SNP, assuming an additive genetic model. Left and right brain hemisphere volumes were analysed separately. All subcortical volumes were normally distributed except the left fimbria, which was log-transformed prior to analysis. Covariates included sex, age, one population stratification principal component, lifetime PTSD diagnosis, intracranial volume, and childhood trauma (0 = no childhood trauma; 1 = exposure to a single category of childhood trauma; 2 = exposure to two or more categories of childhood trauma, as reported from TLEQ items 12,13,15,16, and 17). In the Grady dataset, PLINK was used to prune the autosomal data in windows of 50 bp, removing 1 SNP from each pair of SNPs with r2 > 0.05 to obtain a set of roughly independent markers (~50, 000 SNPs) for use in PCA. Subjects who fell within three standard deviations of the medians of the first and second PCs were retained. Linear regression was performed in the Grady sample using the same software and model described above for the MIRECC cohort (sex was not included because all individuals in the Grady sample were female).
We interrogated the 21 genomic regions reported in van der Meer et al. (Van der Meer et al., 2018a) as both main effects and interactions with PTSD and childhood trauma in the MIRECC and Grady cohorts. Because the Van der Meer sample (Van der Meer et al., 2018a) was exclusively Caucasian, the lead SNP from that study was often not present in our cohorts because the MIRECC sample included Caucasian and African-American subjects, and the Grady sample was exclusively African American. Thus, we identified proxy SNPs that could be evaluated in our datasets, using the LDproxy tool from LDlink (Machiela & Chanock, 2015b). Proxy SNPs were chosen with the highest correlation (r2) to lead SNPs from van der Meer et al. in both the CEPH European (CEU) ancestry and Yoruba in Ibadan (YRI) parent populations. For the Caucasians, all proxy SNPs had r2 ≥ 0.83 with the reported SNP from Van der Meer (Van der Meer et al., 2018a). For African Americans, most proxy SNPs had r2 ≥ 0.63 with the reported SNP from the Norment study. Two exceptions were the SNPs rs1705614 with r2 = 0.54 and rs1337524 with r2 = 0.59. Using this approach, we were able to identify proxies for all of the van der Meer lead SNPs that were missing from the MIRECC and Grady cohorts except one SNP (rs17178006), corresponding to two significant associations in the previous study. Linear regression was then performed using the same model specified above. Finally, a conservative Bonferroni correction was applied for each analysis (19 tests, p = 0.0026).
After performing GWAS for hippocampal subfields, we used the linkage disequilibrium (LD) clumping method in PLINK (r2 threshold of 0.25 and a 500 kb window, as reported previously (Ripke et al., 2013)) to reduce genetic redundancy in this imputed dataset and to mitigate the statistical burden of multiple-comparison correction in a small sample size. The proportion of variability in the hippocampal subfield explained by the SNP in each model was calculated as the sum of squares for the SNP divided by the total sum of squares for the model. False discovery rate (FDR) q-values were generated using PROC MULTTEST in SAS version 9.4. The FDR correction was applied to the SNPs within a specific analysis but not across the 12 hippocampal subfields and two hemispheres. This was in large part due to the high correlation between the two hemispheres and subfields. However, recognizing that this multiple testing correction was somewhat permissive. We performed analyses in a second, independent dataset. Post-hoc interaction analyses were performed only for the SNPs with significant main effects. SNP*childhood trauma and SNP*lifetime PTSD interactions were investigated for associations with subfield volumes. Manhattan plots and quantile-quantile (Q-Q) plots were produced using the R package qqman (Turner, 2014) and interaction plots were generated using the R package ggplot2 (Wickham, 2016).
3. Results
Important clinical and sociodemographic information by cohort is reported in Table 1. Compared to the MIRECC cohort, the Grady cohort comprises a higher percentage of females (p < 0.0001), African-Americans (p < 0.0001), subjects reporting alcohol misuse (p = 0.0379), and lifetime PTSD (p = 0.0032). Average age and distribution of childhood trauma did not significantly differ between the two cohorts.
Our initial goal of this analysis was to evaluate the previously reported genetic associations with hippocampal subfields by van der Meer et al. (Van der Meer et al., 2018a), which was identified in a primarily normative sample, in the context of PTSD and significant trauma exposure. We evaluated a total of 19 reported genetic associations in both the MIRECC and Grady datasets. As shown in Table 2, the same SNP was generally not available in the MIRECC and Grady data sets, thus we evaluated SNPs in high LD with the reported SNP (LD proxy SNPs; see Methods). In the MIRECC dataset, we replicated four main effects, while in the Grady dataset, we replicated one of the main effects. Importantly, two of the associations in the MIRECC dataset were driven primarily by the non-Hispanic white (NHW) subset of the dataset, which ancestrally was most closely matched to the previously reported European dataset. We also explored potential interactions between the SNPs and childhood trauma and between SNPs and a lifetime diagnosis of PTSD (Supplemental Tables S1 and S2). In the interaction analysis, we identified six interactions, four with childhood trauma and two with PTSD, in the MIRECC dataset. None of these interactions were present in the Grady dataset.
Table 2.
Replication of Van der Meer associations in MIRECC and Grady.
| Right hemisphere P-values |
Left hemisphere P-values |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Structure | Chr | Lead SNP | LD proxy SNP | R2 CEU | R2 YRI | MIRECC All | MIRECC NHW | MIRECC NHB | Grady | MIRECC All | MIRECC NHW | MIRECC NHB | Grady |
| Whole hippocampus | 9 | rs7873551† | rs7858153 | 1 | 1 | 0.2692 | 0.0235 | 0.8218 | 0.7436 | 0.5698 | 0.0329 | 0.3608 | 0.8549 |
| 10 | rs12218858 | rs7099316 | 1 | 0.91 | 0.0223 | 0.1755 | 0.0723 | 0.4952 | 0.0828 | 0.2705 | 0.1874 | 0.481 | |
| 12 | rs17178139 | rs55972016 | 1 | 0.78 | 0.5221 | 0.7085 | 0.7348 | 0.1438 | 0.1032 | 0.082 | 0.3467 | 0.0029 | |
| Molecular layer | 10 | rs4962694 | rs12570348 | 1 | 1 | 0.0466 | 0.1451 | 0.1726 | 0.931 | 0.0182 | 0.1432 | 0.019 | 0.5256 |
| Hippocampal tail | 14 | rs160459 | rs221326 | 0.90 | 0.80 | 0.7344 | 0.1274 | 0.1141 | 0.5932 | 0.2305 | 0.0419 | 0.585 | 0.9763 |
Given the diagnostic and racial differences compared to the van der Meer data set, we performed a GWAS for hippocampal subfields in the MIRECC dataset and identified several SNPs associated with different hippocampal subfield volumes that survived correction for multiple testing (Table 3). The most significant associations (q < 0.01) involved R-fimbria volume that is visualized in Figure 1. The R-fimbria QQ plot is provided in Figure 2, which corresponds to a λGC (genomic control lambda) = 0.998. The SNP rs6906714, located in LINC02571 on chromosome 6 (Figure 3), was associated with R-fimbria volume (p = 5.99 × 10−8, q = 0.0056), such that for each additional G allele, R-fimbria volume increased by 15.73 mm3. An intergenic SNP, rs17012755 on chromosome 2, was also associated with R-fimbria volume (p = 6.05 × 10−8, q = 0.0056). Each additional copy of the A allele increased R-fimbria volume by 22.01 mm3. In addition to the main effects, we also identified an interaction between rs6906714 and childhood trauma affecting R-fimbria volume (p = 0.022, Figure 4(a)). As exposure to childhood trauma increased, the effect of rs6906714 genotype on R-fimbria volume became stronger. Specifically, among individuals who did not experience any childhood trauma, the association between rs6909714 genotype and R-fimbria was modest (p = 0.037; beta = 9.272 mm3). Among those who did experience childhood trauma (either 1 or 2+ categories), the association was more robust with p = 0.0002; beta = 20.19 mm3 for one category of childhood trauma and p = 0.0002; beta = 25.19 mm3 for 2+ categories. There was no appreciable difference in the effect of rs6906714 genotype on R-fimbria volume among those who experienced 2+ categories of childhood trauma compared to those who experienced a single category of childhood trauma.
Table 3.
FDR significant main effects of genetic variants on hippocampal subfield volume.
| Gene | Probe | Chr | bp | Tested allele | CEU MAF | ASW MAF | Subfield | Beta discovery | Pval discovery | FDR qval discovery | PVE discovery | Beta replica-tion | Pval replica-tion |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LINC02571 | rs6906714 | 6 | 31270038 | G | 0.071 | 0.123 | R-fimbria | 15.73 | 5.99E-08 | 0.0056 | 0.1489 | −0.517 | 0.8636 |
| rs17012755 | 2 | 76709646 | A | 0.061 | 0.098 | R-fimbria | 22.01 | 6.05E-08 | 0.0056 | 0.1479 | 1.988 | 0.6019 | |
| rs76832471 | 2 | 211570988 | G | 0.096 | 0.09 | log L-fimbria | −16.4 | 6.51E-08 | 0.0121 | 0.1462 | −2.384 | 0.5432 | |
| rs9499406 | 6 | 103816931 | C | 0.035 | 0.139 | R-subiculum | −46.38 | 8.19E-08 | 0.0151 | 0.1082 | 6.916 | 0.2368 | |
| RBBP6 | rs7196581 | 16 | 24552179 | G | 0.086 | 0.049 | L-CA1 | 53.62 | 1.51E-07 | 0.0280 | 0.0964 | −13.85 | 0.7599 |
| TUNAR | rs12880795 | 14 | 96379833 | T | 0.525 | 0.27 | L-HATA | 4.743 | 3.43E-07 | 0.0372 | 0.1206 | 4.107 | 0.0004 |
| LOC105376580 | rs2553974 | 11 | 19238380 | A† | 0.318 | 0.582 | L-HATA | 4.88 | 4.02E-07 | 0.0372 | 0.1193 | −0.4725 | 0.6168 |
| ARMC6 | rs73008928 | 19 | 19146729 | T | 0.061 | 0.016 | R-HATA | 9.609 | 2.33E-07 | 0.0424 | 0.1235 | −1.5 | 0.6048 |
| RBBP6 | rs7196581 | 16 | 24552179 | G | 0.086 | 0.049 | R-HATA | 7.939 | 4.58E-07 | 0.0424 | 0.1165 | 1.307 | 0.5551 |
| LINC01736 | rs3811492 | 1 | 230142626 | C | 0.131 | 0.451 | L-HATA | 4.866 | 8.13E-07 | 0.0498 | 0.1136 | 1.212 | 0.2411 |
*Effect size for untransformed L-fimbria.
†Tested allele in replication dataset is C.
PVE = proportion of variance in imaging phenotype explained by the SNP.
Figure 2.
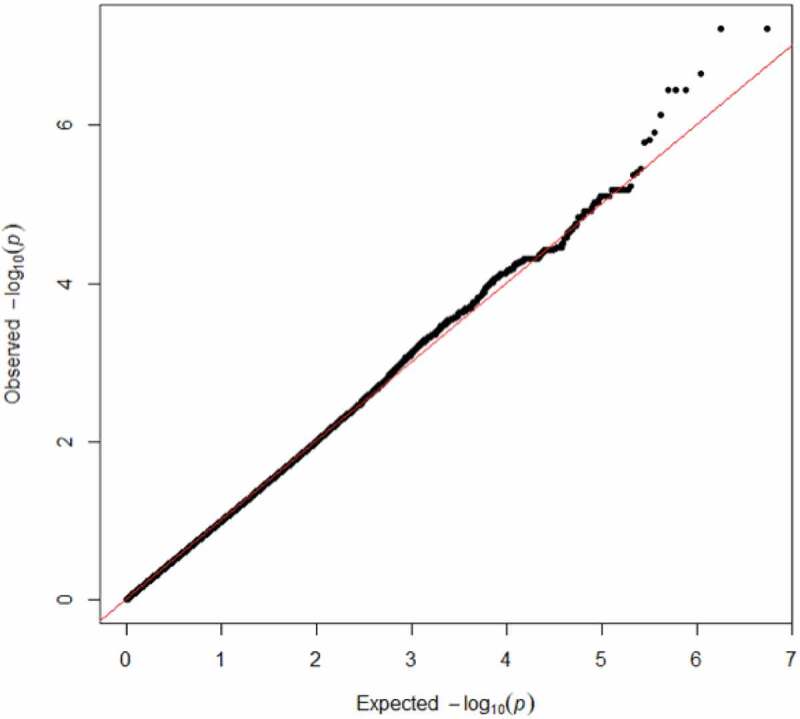
R-fimbria QQ plot, with a λGC (genomic control lambda) = 0.998.
Figure 3.
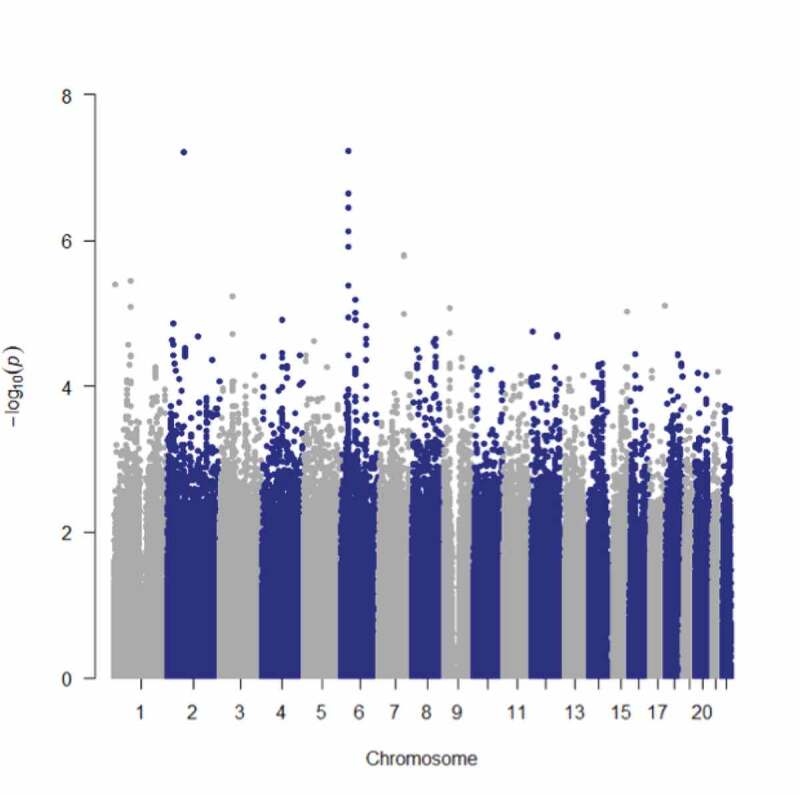
B SNP rs6906714, located in LINC02571 on chromosome 6 was associated with R-fimbria volume (p = 5.99 x 10−8, q = 0.0056). An intergenic SNP, rs17012755 on chromosome 2, was also associated with R-fimbria volume (p = 6.05 x 10−8, q = 0.0056).
Figure 4.
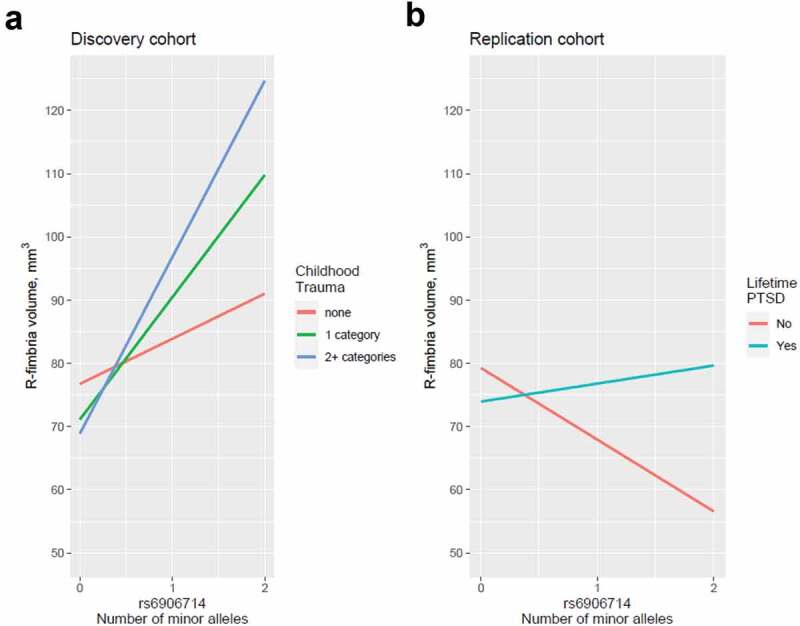
(a) Interaction between rs6906714 and childhood trauma affecting R-fimbria volume (p = 0.022). As exposure to childhood trauma increased, the effect of rs6906714 genotype on R-fimbria volume became stronger. Individuals who did not experience any childhood trauma, the association between rs6909714 genotype and R-fimbria was modest (p = 0.037; beta = 9.272 mm3). Individuals who experienced childhood trauma, the association was more robust with p = 0.0002; beta = 20.19 mm3 for 1 category of childhood trauma and p = 0.0002; beta = 25.19 mm3 for 2+ categories. There was no appreciable difference in the effect of rs6906714 genotype on R-fimbria volume among those who experienced 2+ categories of childhood trauma compared to those who experienced a single category. (b) Interaction between rs6906714 and lifetime PTSD on R-fimbria volume in the Grady cohort (p = 0.0421).
Several additional significant main effects were observed (q < 0.05) in the MIRECC cohort with different hippocampal subfields, some intergenic and some residing in genes. The SNP rs7196581 in RBBP6 was associated with two different subfields: L-CA1 (p = 1.51 x 10−7, q = 0.0280) and R-HATA (p = 4.58 × 10−7, q = 0.0424), such that for each additional G allele, L-CA1 volume increased by 53.62 mm3 and R-HATA volume increased by 7.94 mm3. Also associated with increased R-HATA volume was the T allele of rs73008928 in ARMC6 (p = 2.33 × 10−7, q = 0.0424). Three significant associations were identified with L-HATA volume: rs12880795 in TUNAR (p = 3.43 × 10−7, q = 0.0372), rs2553974 in LOC105376580 (p = 4.02 × 10−7, q = 0.0372), and rs3811492 in LINC01736 (q = 8.13 × 10−7, q = 0.0498). Finally, two intergenic SNPs were associated with L-fimbria and R-subiculum volumes: rs76832471 on chromosome 2 (p = 6.51 × 10−8, q = 0.0121) and rs9499406 on chromosome 6 (p = 8.19 × 10−8, q = 0.0151), respectively.
Aside from the significant interactions described above between rs6906714 and childhood trauma on R-fimbria volume, no other SNP displaying a significant main effect demonstrated a significant interaction with either childhood trauma or lifetime PTSD diagnosis and hippocampal subfield volume in the MIRECC cohort.
The 10 significant main effects identified in the MIRECC cohort (q < 0.05) were tested for significance in the Grady cohort. Despite significant differences in the composition of MIRECC and Grady cohorts (Table 1), we observed an association between rs12880795 genotype and L-HATA volume (p = 0.0004) in the Grady cohort. No other main effect of SNP on hippocampal subfield volume observed in the MIRECC cohort was replicated. However, we did detect a significant interaction between rs6906714 and lifetime PTSD on R-fimbria volume in the Grady cohort (p = 0.0421; Figure 4(b)). Similarly, while we found no main effect of rs17012755 on R-fimbria volume in the MIRECC cohort, we observed a significant interaction between rs17012755 and lifetime PTSD on R-fimbria volume in the Grady cohort (p = 0.0219; Figure 5).
Figure 5.
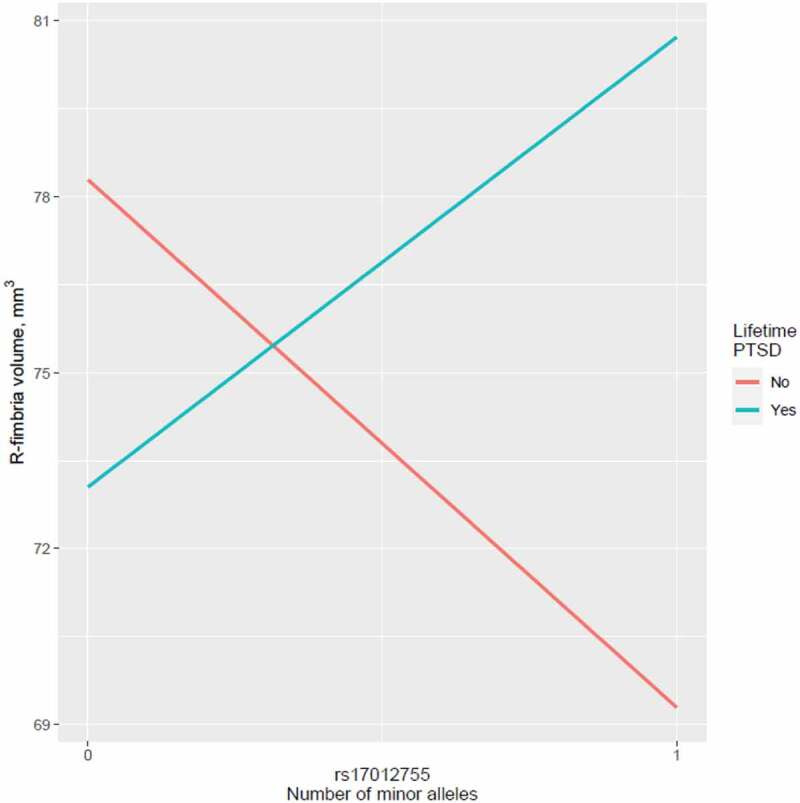
Interaction between rs17012755 and lifetime PTSD on R-fimbria volume in the Grady cohort (p = 0.0219).
4. Discussion
We performed a GWAS of multiple hippocampal subfields segmented by FreeSurfer 6.0 in trauma-exposed individuals with and without PTSD in diverse ancestry samples of US military veterans and a sample of highly traumatized civilians. We were largely unable to replicate a previously published study of hippocampal subfields in a normative sample of European ancestry individuals. This could be due to several differences between the cohorts including ancestry and exposure status. The MIRECC cohort is comprised of nearly 50% NHB individuals and the Grady cohort is 100% NHB; therefore, allele frequencies in these two cohorts are potentially quite different compared to subjects in van der Meer study et al. In addition to differences in genetic background, the subjects in MIRECC and Grady cohorts have all been exposed to trauma, and half of the individuals have been formally diagnosed with PTSD. Thus, these cohorts are more appropriate for the identification of genetic variants that are relevant to PTSD than a normative sample, despite the smaller sample size.
In our preliminary interrogation of the entire genome, several genetic markers reached genome-wide significance for specific hippocampal subfields. Two SNPs that were involved in three genome-wide significant associations lie within genes (rs73008928 in ARMC6 and rs7196581 in RBBP6). The SNP rs73008928 is particularly interesting as it has been identified as a methylation QTL and an expression QTL, according to the ARIES (Gaunt et al., 2016b) and GTEx databases (www.gtexportal.org) (Carithers et al., 2015), respectively. Specifically, rs73008928 was found to be a trans-QTL for methylation probe cg08284873 in ADARB2 using a sample of middle-aged individuals (Table 4) (Relton et al., 2015). Ribonucleic acid (RNA)-editing deaminase-2 (ADARB2) belongs to a family of genes whose other members edit RNAs encoding GluRs in the rat hippocampus (Melcher et al., 1996b). While not directly involved in RNA-editing activity, ADARB2 has been shown to inhibit the activity of the other members of this gene family, suggesting that it plays a regulatory role in RNA editing (CHEN et al., 2000). In addition to being a methylation QTL, rs73008928 is an expression QTL for SUGP2 in several tissues, as well as for TMEM161A and SLC25A42 in single tissues (Table 5). SUGP2 is a splicing factor involved in pre-mRNA processing mechanisms (Sampson & Hewitt, 2003); TMEM161A, when overexpressed, plays a role in protection against oxidative stress (Gesualdi et al., 2007b); and SLC25A42 belongs to a large family of nuclear-encoded transporters that are involved in metabolic pathways and cell functions and is widely expressed in the central nervous system (Haitina, Lindblom, Renström, & Fredriksson, 2006b; Palmieri, 2013). In summary, we have identified a SNP associated with R-HATA volume that may impact RNA editing of GluRs and regulate expression of a gene involved in oxidative stress; both are processes which have been previously implicated in the pathophysiology of PTSD (Holmes et al., 2017b; Miller, Lin, Wolf, & Miller, 2018b; Popoli, Yan, McEwen, & Sanacora, 2012).
Table 4.
Methylation QTLs in human blood from the Accessible Resource for Integrated Epigenomic Studies (ARIES) data set.
| SNP | CpG | Timepoint | Trans-QTL | p-value |
|---|---|---|---|---|
| rs2553974 | cg14704941 | Birth | N | 2.95E-09 |
| rs73008928 | cg08284873 | Middle Age | Y | 4.84E-08 |
| rs76832471 | cg07781096 | Adolescence | N | 2.21E-28 |
| rs76832471 | cg07781096 | Birth | N | 3.87E-19 |
| rs76832471 | cg07781096 | Childhood | N | 8.30E-33 |
| rs76832471 | cg07781096 | Middle Age | N | 4.74E-19 |
| rs76832471 | cg07781096 | Pregnancy | N | 2.92E-25 |
Table 5.
Expression QTLs from the Genome-Tissue Expression (GTEx) project.
| SNP | Gene Symbol | Tissue | P-Value |
|---|---|---|---|
| rs6906714 | PSORS1C1 | Thyroid | 0.0000028 |
| rs6906714 | PSORS1C2 | Thyroid | 0.000017 |
| rs6906714 | XXbac-BPG181B23.7 | Artery – Tibial | 0.000024 |
| rs6906714 | XXbac-BPG181B23.7 | Skin – Sun Exposed (Lower leg) | 0.000077 |
| rs6906714 | POU5F1 | Heart – Atrial Appendage | 0.000039 |
| rs2553974 | E2F8 | Stomach | 0.0000061 |
| rs73008928 | SUGP2 | Adipose – Subcutaneous | 1.70E-07 |
| rs73008928 | SUGP2 | Nerve – Tibial | 2.00E-07 |
| rs73008928 | SUGP2 | Artery – Aorta | 5.50E-07 |
| rs73008928 | SUGP2 | Whole Blood | 9.60E-07 |
| rs73008928 | SUGP2 | Adipose – Visceral (Omentum) | 0.0000028 |
| rs73008928 | SUGP2 | Breast – Mammary Tissue | 0.000025 |
| rs73008928 | TMEM161A | Muscle – Skeletal | 0.0000044 |
| rs73008928 | SLC25A42 | Small Intestine – Terminal Ileum | 0.000007 |
The other genome-wide significant SNP associated with hippocampal subfield volume is rs7196581 located in the RBBP6 gene. This SNP was significantly associated with both L-CA1 volume and R-HATA volume. It falls within the promoter region of RBBP6 and coincides with H3K27ac modification sites in several cell types, including neuronal progenitors, suggesting that this SNP functions as an enhancer (ENCODE-Project-Consortium, 2012b). RBBP6 is involved in apoptosis and typically considered an oncogene (Ntwasa, 2016). However, the murine ortholog is required for mouse embryogenesis and is involved in neurogenesis (Grunert, Clarke, Ahuja, Eswaran, & Nijhout, 2015b; Mukherjee et al., 2015) and thus, is a reasonable candidate gene. The CA1 subfield is highly relevant to PTSD as the CA1 has direct reciprocal projections to the medial prefrontal cortex (mPFC) that allow these regions to form a functional loop. This circuit enables interactions between cortical and subcortical areas during memory encoding and retrieval of episodic-like memories (Preston & Eichenbaum, 2013). Neurons in hippocampal CA1 code for both space and time, allowing conjoint spatial and temporal representations of experiences (Eichenbaum, 2014b). The critical role of these subfields in episodic memories makes them of particular interest in PTSD. Despite substantial research on purported episodic memory deficits in PTSD, which would implicate the functional loop, the evidence has been inconclusive (Brewin, 2014). The severity of PTSD is negatively associated with mPFC activation elicited by material that participants have subsequently forgotten (Dickie, Brunet, Akerib, & Armony, 2008b). Veterans with PTSD have lower activation in the hippocampus and amygdala when successfully encoding trauma-related stimuli into memory (Hayes et al., 2011b). Therefore, the present results that show an association of genetic markers with CA1 volume, but no interaction with PTSD diagnosis, will help build the current body of neurogenetic knowledge relevant to PTSD.
All of the other SNPs associated with hippocampal subfields in this study were intergenic, indicating there may be some underlying regulatory genomic mechanism driving the statistical associations that we observed. Of interest, three significant SNPs reside in long intervening/intergenic noncoding RNAs (lincRNAs) (rs6906714 in LINC02571, rs12880795 in TUNAR, and rs3811492 in LINC01736), which comprise the largest proportion of long non-coding RNAs (lncRNAs) in the human genome (Cabili et al., 2011). While lncRNAs do not encode proteins, they and other non-coding RNAs, such as micro-RNAs (miRNAs), have been implicated in several neuropsychiatric disorders including PTSD (Daskalakis, Provost, Hunter, & Guffanti, 2018; O’Connor, Gururajan, Dinan, Kenny, & Cryan, 2016; Schmidt, Keck, & Buell, 2015; Schouten, Aschrafi, Bielefeld, Doxakis, & Fitzsimons, 2013). lncRNAs have also been associated with changes in gene expression in animal models of PTSD. For example, lncRNAs were differentially expressed in the hippocampus of rats exposed to stress-enhanced fear learning compared to control rats (Qingzhen et al., 2016). In addition, RNA sequencing of mPFC in adult mice revealed changes in lncRNA expression after fear conditioning (Spadaro et al., 2015). The association we observed between rs12880795 in TUNAR and L-HATA volume was significantly replicated in the Grady Trauma Project cohort, providing additional confidence in this finding. These data suggest that the role of lncRNAs in PTSD should be studied further and may be useful biomarkers or therapeutic targets.
The association between rs6906714 and R-fimbria volume was the most significant finding in this study. Notably, rs6906714 is also an expression QTL for several genes including PSORS1C1 and PSORS1C2, which are psoriasis susceptibility genes that are expressed in thyroid tissue (Table 5). Genes associated with autoimmune disorders have been previously associated with PTSD (Stein et al., 2016); the overlap between stress disorders and immune/inflammatory disorders is an ongoing area of research (Song et al., 2018). We also observed an interaction of rs6906714 with childhood trauma in the MIRECC cohort, and of rs6906714 with lifetime PTSD in the Grady cohort, which is consistent with neuropsychiatric responses to stress that show increased risk of subsequent autoimmune disease (Figure 4) (Song et al., 2018). The second strongest signal was between rs17012755, an intergenic SNP on chromosome 2, and R-fimbria volume. We also observed a significant interaction between this SNP and lifetime PTSD on R-fimbria volume in the Grady (Figure 5). Interestingly, in our larger PTSD cohort of over 2000 subjects, we observed nominal evidence for the association between rs17012755 and PTSD (Ashley-Koch et al., 2015).
The fimbria is a primary source of input to the dorsal hippocampus, which is a critical element in contextual fear conditioning. Therefore, contextual fear conditioning has become a widely adopted experimental paradigm for studying PTSD (Davidson et al., 1997b; Maren, Phan, & Liberzon, 2013b). Processing of contextual memories is mediated by robust projections from hippocampal neurons to the mPFC (Jin & Maren, 2015b). Connectivity between the hippocampus and the mPFC, which involves the fimbria, is associated with mnemonic and emotion regulation deficits, which are consistent with the clinical presentation of PTSD (Maren et al., 2013b). Recent evidence suggests that the mPFC directs the retrieval of context-appropriate episodic memories in the hippocampus (Maren & Holt, 2000b; Navawongse & Eichenbaum, 2013; Preston & Eichenbaum, 2013). Electrolytic lesions to the fimbria in rats produce deficits in fear conditioning to contextual stimuli. The deficit in freezing, following dorsal hippocampus and fimbria lesions, is evident on both the conditioning day and the delayed extinction test (Maren & Fanselow, 1997b). Indeed, contextual fear deficits in rats with hippocampal damage are equivalent following lesions to either fimbria, dorsal hippocampus, or entorhinal cortex (Maren & Fanselow, 1997b). In addition to contextual fear conditioning, the fimbria plays an important role in spatial memory, which enables rats to learn the location of both a visible and hidden/submerged platform in the water maze task (Dunnett, Low, Iversen, Stenevi, & Björklund, 1982b). However, rats with lesions to the fimbria learned to swim to the visible platform but were unable to navigate to the submerged platform in the same location (McDonald & White, 1994b; Sutherland & Rudy, 1988).
The most significant effects in both cohorts involved R-fimbria volume. Deficits in contextual processing are representative of PTSD because intrusive memories and perceptions are experienced outside of the trauma context. Our finding that exposure to child trauma is a potent environmental exposure that interacts with genetic markers to influence brain structure is consistent with our recently published multi-cohort study in 1,868 subjects, which demonstrated childhood trauma exposure has a negative association with hippocampal volume but was not significant if PTSD was added as a covariate (Logue et al., 2017b). Our finding underscores the uniquely deleterious role of trauma during childhood, which is a critical neurodevelopmental time period (Morey, Haswell, Hooper, & De Bellis, 2015).
4.1. Strengths and limitations
The current work supports the value of imaging genetic studies in individuals with neuropsychiatric conditions. Our study was focused on hippocampal subfields, given their relevance to PTSD. We observed several strong associations, accounting for approximately 10–15% of the total variance for each outcome (Table 2). One of the associations was replicated in an independent dataset (Grady), yielding similar effect sizes and therefore increased confidence in this association. However, several limitations bear mention. Foremost, is the modest sample size of both the MIRECC and Grady cohorts utilized. We lacked statistical power to detect conclusive associations of main effects as well as SNP*PTSD and SNP*childhood trauma interactions given the small effect sizes typical of complex phenotypes (Button et al., 2013). As such, we restricted our investigation of interactions to only those SNPs which demonstrated a main effect with hippocampal subfield volume. The measurement invariance of PTSD is important to assess when studying G x E. An asymmetric item response theory (IRT) curves can confound interaction results we have reported with childhood trauma and with PTSD diagnosis. Palm and colleagues show the 17 DSM-IV items provide the best information in the symptom severity range that is populated by patients who fall short of meeting DSM-IV criteria for diagnosis of PTSD. This effectively means that the criteria are most effective in providing information for the subthreshold range, but then provide decreasing information throughout the severity range required to meet full criteria for PTSD. The results provided by Palm, Strong, and MacPherson (2009) are based on the National Comorbidity Study (NCS) (Kessler et al., 2005b), which used the NCS-R interview. Betemps, Smith, Baker, and Rounds-Kugler (2003) reported a similar ceiling effect with the CAPS instrument. We used the DSM-IV SCID followed by CAPS for confirmation in most subjects. Similarly, the measurement invariance of the Childhood Trauma Questionnaire has also been investigated by MacDonald et al., 2016b who report the presence of a minimization bias in an international sample of over 19,000 subjects and the administration of the Minimization-Denial subscale is essential in addressing this bias. However, the present study used the number of trauma categories, which were coded as 0, 1, 2+, rather than using the total CTQ score. Item response analysis of the childhood trauma categories is not available in the published literature to our knowledge, although coding of categories from the CTQ is used widely in the trauma literature (Dennis et al., 2019b; Logue et al., 2018b; Morey et al., 2017b).
Given the exploratory nature of our investigation, we did not impose multiple-comparison correction for the number of subfields, the number of SNP*PTSD and SNP*childhood trauma tests. In the near future, we plan to interrogate dramatically larger datasets accessed through worldwide consortia to ascertain these important interactions on a genome-wide scale (Nievergelt et al., 2018). We note that the reduced-density exploration of the genome achieved by LD-clumping may incur a penalty of false-negative associations, which otherwise might be detected by high-density GWAS. However, it is more likely that the smaller sample size of both cohorts and the differences in population structure impacted the analysis more substantially (Button et al., 2013).
One of the significant associations in the MIRECC cohort was confirmed in the Grady cohort. However, the other nine MIRECC associations were not significant in the Grady cohort. The most likely of several reasons are the differences in population structure. The MIRECC cohort had nearly equal proportions of European Americans and African Americans, while the Grady cohort was entirely African American. As such, the allele frequencies of the significant SNPs differed between the two cohorts, and in one case (rs2553974), the minor allele was reversed entirely. Other reasons for lack of replication could be cohort differences in trauma type and male/female ratio. The MIRECC cohort was comprised of military veterans while the Grady cohort was entirely civilian, which could impact the ability to detect PTSD interactions. In addition, the MIRECC sample employed three SNP chip platforms, and the Grady sample employed two SNP chip platforms for genotyping, each with varying SNP coverage and tag SNPs that were likely to introduce noise. Nonetheless, imputed data derived from tag SNPs were used for the analysis of all samples. We are encouraged that one association replicated despite differences between cohorts, which indicates this association is worthwhile examining in other trauma-exposed cohorts. Future neuroimaging genetic studies should include larger sample sizes of individuals with PTSD and other neuropsychiatric conditions, as well as evaluate the effect of childhood trauma exposure and population substructure in modulating genetic associations with neuroimaging phenotypes.
4.2. Conclusion
Variation in the volume of fimbria, CA1, hippocampal amygdala transition area, and subiculum subfields is at least partially controlled by genetic factors as well as by the interaction of genetics and environmental exposure to trauma. These findings provide new avenues for understanding neural circuits that are implicated in contextual fear conditioning and other established behavioural models of PTSD. Larger samples that include under-represented populations will be necessary to understand the role of genetic variation with respect to brain structures, in the general population, as well as in disease-enriched subpopulations. Ultimately, the promise of finding genetic determinants of PTSD is that they signal the presence of etiologic pathways for targeted therapeutic intervention. Attempts to find the disease-associated genetic variation that points to molecular mechanisms of pathogenesis has proven challenging due to the relatively distant connection between the action of SNPs and highly polygenic diagnostic phenotypes as compared to relative proximal connection with neuroimaging phenotypes, which are also polygenic (Geschwind & Flint, 2015b; McCarroll, Feng, & Hyman, 2014b). Leveraging neuroimaging phenotypes may offer a shortcut over clinical phenotypes in identifying these elusive genetic markers and relevant neurobiological pathways (Logue et al., 2015b; Nievergelt et al., 2018).
Supplementary Material
Acknowledgments
We thank the willing participation of Veterans in this research.
Funding Statement
This work was supported by the National Institutes of Health [RO1 MH111671]; VA [Mid-Atlantic MIRECC]. The views expressed in this manuscript are those of the authors and do not necessarily represent the views of the funding agencies or the federal government. This research was supported by the U.S. Department of Veterans Affairs (VA) Mid-Atlantic Mental Illness Research, Education, and Clinical Center (MIRECC) core funds. Dr. Morey also received financial support from the US Department of Veterans Affairs (VA) Office of Research and Development (5I01CX000748-01, 5I01CX000120-02). Additional financial support was provided by the National Institute for Neurological Disorders and Stroke (R01NS086885-01A1). Dr. Dedert is funded by a VA Clinical Science Research and Development (CSR&D) Career Development Award (CDA) (IK2CX000718). Dr. Naylor is funded by a VA Rehabilitation Research and Development (RR&D) CDA (1lK2RX000908). Dr. Van Voorhees is funded by a Department of Veterans Affairs Rehabilitation Research and Development Career Development Award (1K2RX001298).Dr. Kimbrel is funded by a Department of Veterans Affairs Clinical Science Research and Development Career Development Award (IK2CX000525). Dr. Beckham also received financial support from the U.S. Department of Veterans Affairs (VA) Office of Research and Development (1IK6CX001494).
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
References
- Ashley-Koch, A. E., Garrett, M. E., Gibson, J., Liu, Y., Dennis, M. F., Kimbrel, N. A., & Hauser, M. A. (2015). Genome-wide association study of posttraumatic stress disorder in a cohort of Iraq–Afghanistan era veterans. Journal of Affective Disorders, 184, 225–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck, A. T., Steer, R. A., & Brown, G. K. (eds). (1996). Manual for the Beck depression inventory-II. San Antonio, TX: Psychological Corp. [Google Scholar]
- Bernstein, D. P., Fink, L., Handelsman, L., Foote, J., Lovejoy, M., Wenzel, K., … & Ruggiero, J., et al (1994). Initial reliability and validity of a new retrospective measure of child abuse and neglect. The American Journal of Psychiatry, 151, 1132. [DOI] [PubMed] [Google Scholar]
- Betemps, E. J., Smith, R. M., Baker, D. G., & Rounds-Kugler, B. A. (2003). Measurement precision of the clinician administered PTSD scale (CAPS): A RASCH model analysis. Journal of Applied Measurement 4(1), 59–69. [PubMed] [Google Scholar]
- Blake, D. D., Weathers, F. W., Nagy, L. M., Kaloupek, D. G., Gusman, F. D., Charney, D. S., & Keane, T. M. (1995). The development of a clinician-administered PTSD scale. Journal of Traumatic Stress, 8(1), 75–90. [DOI] [PubMed] [Google Scholar]
- Brancu, M., Wagner, H. R., Morey, R. A., Beckham, J. C., Calhoun, P. S., Tupler, L. A., & Fairbank, J. A. (2017). The Post‐Deployment Mental Health (PDMH) study and repository: A multi‐site study of US Afghanistan and Iraq era veterans. International Journal of Methods in Psychiatric Research, 26. doi: 10.1002/mpr.1570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremner, J. D., Randall, P., Vermetten, E., Staib, L., Bronen, R. A., Mazure, C., & Charney, D. S. (1997). Magnetic resonance imaging-based measurement of hippocampal volume in posttraumatic stress disorder related to childhood physical and sexual abuse–a preliminary report. Biological Psychiatry, 41, 23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewin, C. R. (2014). Episodic memory, perceptual memory, and their interaction: Foundations for a theory of posttraumatic stress disorder. Psychological Bulletin, 140, 69. [DOI] [PubMed] [Google Scholar]
- Button, K. S., Ioannidis, J. P., Mokrysz, C., Nosek, B. A., Flint, J., Robinson, E. S., & Munafò, M. R. (2013). Power failure: Why small sample size undermines the reliability of neuroscience. Nature Reviews Neuroscience, 14, 365–376. [DOI] [PubMed] [Google Scholar]
- Cabili, M. N., Trapnell, C., Goff, L., Koziol, M., Tazon-Vega, B., Regev, A., & Rinn, J. L. (2011). Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes & Development, 25, 1915–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carithers, L. J., Ardlie, K., Barcus, M., Branton, P. A., Britton, A., Buia, S. A., & Moore, H. M. (2015). A novel approach to high-quality postmortem tissue procurement: The GTEx project. Biopreservation and biobanking, 13(5), 311–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C.-X., Cho, D.-S. C., Wang, Q., Lai, F., Carter, K. C., & Nishikura, K. (2000). A third member of the RNA-specific adenosine deaminase gene family, ADAR3, contains both single-and double-stranded RNA binding domains. RNA, 6, 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L. W., Sun, D., Davis, S. L., Haswell, C. C., Dennis, E. L., Swanson, C. A., … & Thompson, P. (2018). Smaller hippocampal CA-1 subfield volume in posttraumatic stress disorder. Depression & Anxiety, 5, 1–12. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daskalakis, N. P., Provost, A. C., Hunter, R. G., & Guffanti, G. (2018). Noncoding RNAs: Stress, glucocorticoids, and posttraumatic stress disorder. Biological Psychiatry, 83, 849–865. [DOI] [PubMed] [Google Scholar]
- Davidson, J. R., Book, S. W., Colket, J. T., Tupler, L. A., Roth, S., David, D., & Feldman, M. E. (1997b). Assessment of a new self-rating scale for post-traumatic stress disorder. Psychological Medicine, 27(1), 153–160. [DOI] [PubMed] [Google Scholar]
- Dennis, E. L., Disner, S. G., Fani, N., Salminen, L. E., Logue, M., Clarke, E. K., & Morey, R. A. (2019b). Altered white matter microstructural organization in posttraumatic stress disorder across 3047 adults: Results from the PGC-ENIGMA PTSD consortium. Molecular Psychiatry. doi: 10.1038/s41380-019-0631-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickie, E. W., Brunet, A., Akerib, V., & Armony, J. L. (2008b). An fMRI investigation of memory encoding in PTSD: Influence of symptom severity. Neuropsychologia, 46, 1522–1531. [DOI] [PubMed] [Google Scholar]
- Duncan, L., Ratanatharathorn, A., Aiello, A., Almli, L., Amstadter, A., Ashley-Koch, A., & Koenen, K. C. (2018b). Largest GWAS of PTSD (N= 20 070) yields genetic overlap with schizophrenia and sex differences in heritability. Molecular Psychiatry, 23(3), 666–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunnett, S. B., Low, W., Iversen, S., Stenevi, U., & Björklund, A. (1982b). Septal transplants restore maze learning in rats with fornix-fimbria lesions. Brain Research, 251, 335–348. [DOI] [PubMed] [Google Scholar]
- Eichenbaum, H. (2014b). Time cells in the hippocampus: A new dimension for mapping memories. Nature Reviews Neuroscience, 15, 732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elzinga, B. M., & Bremner, J. D. (2002b). Are the neural substrates of memory the final common pathway in posttraumatic stress disorder (PTSD). Journal of Affective Disorders, 70, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE-Project-Consortium . (2012b). An integrated encyclopedia of DNA elements in the human genome. Nature, 489, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl, B. (2012b). FreeSurfer. Neuroimage, 62, 774–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furini, C., Myskiw, J., & Izquierdo, I. (2014b). The learning of fear extinction. Neuroscience & Biobehavioral Reviews, 47, 670–683. [DOI] [PubMed] [Google Scholar]
- Gaunt, T. R., Shihab, H. A., Hemani, G., Min, J. L., Woodward, G., Lyttleto, O., & Relton, C. L. (2016b). Systematic identification of genetic influences on methylation across the human life course. Genome Biology, 17(1). doi: 10.1186/s13059-016-0926-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geschwind, D. H., & Flint, J. (2015b). Genetics and genomics of psychiatric disease. Science, 349, 1489–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesualdi, N. M., Chirico, G., Pirozzi, G., Costantino, E., Landriscina, M., & Esposito, F. (2007b). Tumor necrosis factor-associated protein 1 (TRAP-1) protects cells from oxidative stress and apoptosis. Stress, 10, 342–350. [DOI] [PubMed] [Google Scholar]
- Gilbertson, M. W., Shenton, M. E., Ciszewski, A., Kasai, K., Lasko, N. B., Orr, S. P., & Pitman, R. K. (2002b). Smaller hippocampal volume predicts pathologic vulnerability to psychological trauma. Nature Neuroscience, 5(11), 1242–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie, C. F., Bradley, B., Mercer, K., Smith, A. K., Conneely, K., Gapen, M., & Ressler, K. J. (2009b). Trauma exposure and stress-related disorders in inner city primary care patients. General Hospital Psychiatry, 31(6), 505–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunert, L. W., Clarke, J. W., Ahuja, C., Eswaran, H., & Nijhout, H. F. (2015b). A quantitative analysis of growth and size regulation in Manduca sexta: The physiological basis of variation in size and age at metamorphosis. PLoS One, 10, e0127988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haitina, T., Lindblom, J., Renström, T., & Fredriksson, R. (2006b). Fourteen novel human members of mitochondrial solute carrier family 25 (SLC25) widely expressed in the central nervous system. Genomics, 88, 779–790. [DOI] [PubMed] [Google Scholar]
- Hayes, J. P., LaBar, K. S., McCarthy, G., Selgrade, E., Nasser, J., Dolcos, F., & Morey, R. A. (2011b). Reduced hippocampal and amygdala activity predicts memory distortions for trauma reminders in combat-related PTSD. Journal of Psychiatric Research, 45(5), 660–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibar, D. P., Stein, J. L., Renteria, M. E., Arias-Vasquez, A., Desrivieres, S., Jahanshad, N., & Medland, S. E. (2015b). Common genetic variants influence human subcortical brain structures. Nature, 520(7546), 224–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes, S. E., Girgenti, M. J., Davis, M. T., Pietrzak, R. H., DellaGioia, N., Nabulsi, N., & Esterlis, I. (2017b). Altered metabotropic glutamate receptor 5 markers in PTSD: In vivo and postmortem evidence. Proceedings of the National Academy of Sciences, 114(31), 8390–8395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji, J. Z., & Maren, S. (2007b). Hippocampal involvement in contextual modulation of fear extinction. Hippocampus, 17, 749–758. [DOI] [PubMed] [Google Scholar]
- Jin, J., & Maren, S. (2015b). Prefrontal-hippocampal interactions in memory and emotion. Frontiers in Systems Neuroscience, 9, 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler, R. C. (2000b). Posttraumatic stress disorder: The burden to the individual and to society. The Journal of Clinical Psychiatry 61(Suppl. 5), 4–14. [PubMed] [Google Scholar]
- Kessler, R. C., Berglund, P., Demler, O., Jin, R., Merikangas, K. R., & Walters, E. E. (2005b). Lifetime prevalence and age-of-onset distributions of DSM-IV disorders in the national comorbidity survey replication. Archives of General Psychiatry, 62, 593–602. [DOI] [PubMed] [Google Scholar]
- Kilpatrick, D. G., Resnick, H. S., Milanak, M. E., Miller, M. W., Keyes, K. M., & Friedman, M. J. (2013b). National estimates of exposure to traumatic events and PTSD prevalence using DSM‐IV and DSM‐5 criteria. Journal of Traumatic Stress, 26, 537–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubany, E. S., Haynes, S. N., Leisen, M. B., Owens, J. A., Kaplan, A. S., Watson, S. B., Haynes, S. N., Owens, J. A., & Burns, K. (2000b). Development and preliminary validation of a brief broad-spectrum measure of trauma exposure: The traumatic life events questionnaire. Psychological Assessment, 12, 210–224. [DOI] [PubMed] [Google Scholar]
- Logue, M. W., Amstadter, A. B., Baker, D. G., Duncan, L., Koenen, K. C., Liberzon, I., & Uddin, M. (2015b). The psychiatric genomics consortium posttraumatic stress disorder workgroup: Posttraumatic stress disorder enters the age of large-scale genomic collaboration. Neuropsychopharmacology, 40(10), 2287–2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logue, M. W., van Rooij, S. J. H., Dennis, E. L., Davis, S. L., Hayes, J. P., Stevens, J. S., … & Korgaonkar, M. (2017b). Smaller hippocampal volume in posttraumatic stress disorder: A multi-site ENIGMA-PGC study. Biological Psychiatry 83(3), 244–253. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logue, M. W., van Rooij, S. J. H., Dennis, E. L., Davis, S. L., Hayes, J. P., Stevens, J. S., & Morey, R. A. (2018b). Smaller hippocampal volume in posttraumatic stress disorder: A multi-site ENIGMA-PGC study. Biological Psychiatry, 83(3), 244–253. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund, M., Foy, D., Sipprelle, C., & Strachan, A. (1984b). The combat exposure scale: A systematic assessment of trauma in the Vietnam war. Journal of Clinical Psychology, 40, 1323–1328. [DOI] [PubMed] [Google Scholar]
- MacDonald, K., Thomas, M. L., Sciolla, A. F., Schneider, B., Pappas, K., Bleijenberg, G., & Scott, J. G. (2016b). Minimization of childhood maltreatment is common and consequential: Results from a large, multinational sample using the childhood trauma questionnaire. PLoS One, 11(1), e0146058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machiela, M. J., & Chanock, S. J. (2015b). LDlink: A web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics, 31, 3555–3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchini, J., Howie, B., Myers, S., McVean, G., & Donnelly, P. (2007b). A new multipoint method for genome-wide association studies by imputation of genotypes. Nature Genetics, 39, 906–913. [DOI] [PubMed] [Google Scholar]
- Maren, S., & Fanselow, M. S. (1997b). Electrolytic lesions of the fimbria/fornix, dorsal hippocampus, or entorhinal cortex produce anterograde deficits in contextual fear conditioning in rats. Neurobiology of Learning and Memory, 67(2), 142–149. [DOI] [PubMed] [Google Scholar]
- Maren, S., & Holt, W. (2000b). The hippocampus and contextual memory retrieval in Pavlovian conditioning. Behavioural Brain Research, 110, 97–108. [DOI] [PubMed] [Google Scholar]
- Maren, S., Phan, K. L., & Liberzon, I. (2013b). The contextual brain: Implications for fear conditioning, extinction and psychopathology. Nature Reviews Neuroscience, 14(6), 417–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarroll, S. A., Feng, G., & Hyman, S. E. (2014b). Genome-scale neurogenetics: Methodology and meaning. Nature Neuroscience, 17, 756–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald, R. J., & White, N. M. (1994b). Parallel information processing in the water maze: Evidence for independent memory systems involving dorsal striatum and hippocampus. Behavioral and Neural Biology, 61, 260–270. [DOI] [PubMed] [Google Scholar]
- Melcher, T., Maas, S., Herb, A., Sprengel, R., Seeburg, P. H., & Higuchi, M. (1996b). A mammalian RNA editing enzyme. Nature, 379, 460. [DOI] [PubMed] [Google Scholar]
- Milad, M. R., Pitman, R. K., Ellis, C. B., Gold, A. L., Shin, L. M., Lasko, N. B., … & Rauch, S. L. (2009). Neurobiological basis of failure to recall extinction memory in posttraumatic stress disorder. Biological Psychiatry, 489, 1075–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, M. W., Lin, A. P., Wolf, E. J., & Miller, D. R. (2018b). Oxidative stress, inflammation, and neuroprogression in chronic PTSD. Harvard Review of Psychiatry, 26, 57–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morey, R. A., Davis, S. L., Garrett, M. E., Haswell, C. C., Marx, C. E., Beckham, J. C., & Ashley-Koch, A. E. (2017b). Genome-wide association study of subcortical brain volume in PTSD cases and trauma-exposed controls. Translational Psychiatry, 7(11), 1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morey, R. A., Gold, A. L., LaBar, K. S., Selgrade, E., Beall, S., Brown, V., Haswell, C. C., … & McCarthy, G. (2012). Amygdala volume changes in posttraumatic stress disorder in a large case-controlled veteran group. Archives of General Psychiatry, 69, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morey, R. A., Haswell, C. C., Hooper, S. R., & De Bellis, M. D. (2015). Amygdala, hippocampus, and ventral medial prefrontal cortex volumes differ in maltreated youth with and without chronic posttraumatic stress disorder. Neuropsychopharmacology 41(3), 791–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee, C., Holubowska, A., Schwedhelm-Domeyer, N., Mitkovski, M., Lee, S.-J., Kannan, M., & Stegmuller, J. (2015). Loss of the neuron-specific F-box protein FBXO41 models an ataxia-like phenotype in mice with neuronal migration defects and degeneration in the cerebellum. Journal of Neuroscience, 35(23), 8701–8717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navawongse, R., & Eichenbaum, H. (2013). Distinct pathways for rule-based retrieval and spatial mapping of memory representations in hippocampal neurons. Journal of Neuroscience, 33, 1002–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nievergelt, C. M., Ashley-Koch, A. E., Dalvie, S., Hauser, M. A., Morey, R. A., Smith, A. K., & Uddin, M. (2018). Genomic approaches to posttraumatic stress disorder: The psychiatric genomic consortium initiative. Biological Psychiatry, 83, 831–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntwasa, M. (2016). Retinoblastoma binding protein 6, another p53 monitor. Trends in Cancer, 2, 635–637. [DOI] [PubMed] [Google Scholar]
- O’Connor, R. M., Gururajan, A., Dinan, T. G., Kenny, P. J., & Cryan, J. F. (2016). All roads lead to the miRNome: MiRNAs have a central role in the molecular pathophysiology of psychiatric disorders. Trends in Pharmacological Sciences, 37, 1029–1044. [DOI] [PubMed] [Google Scholar]
- O’Reilly, R. C., & Norman, K. A. (2002). Hippocampal and neocortical contributions to memory: Advances in the complementary learning systems framework. Trends in Cognitive Sciences, 6, 505–510. [DOI] [PubMed] [Google Scholar]
- Palm, K. M., Strong, D. R., & MacPherson, L. (2009). Evaluating symptom expression as a function of a posttraumatic stress disorder severity. Journal of Anxiety Disorders, 23, 27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri, F. (2013). The mitochondrial transporter family SLC25: Identification, properties and physiopathology. Molecular Aspects of Medicine, 34, 465–484. [DOI] [PubMed] [Google Scholar]
- Parsons, R. G., & Ressler, K. J. (2013). Implications of memory modulation for post-traumatic stress and fear disorders. Nature Neuroscience, 16(2), 146–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel, S., Park, M. T. M., Devenyi, G. A., Patel, R., Masellis, M., Knight, J., & Chakravarty, M. M. (2017). Heritability of hippocampal subfield volumes using a twin and non‐twin siblings design. Human Brain Mapping, 38, 4337–4352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson, N., Price, A. L., & Reich, D. (2006). Population structure and eigenanalysis. PLoS Genetics, 2, e190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluzhnikov, A., Below, J. E., Konkashbaev, A., Tikhomirov, A., Kistner-Griffin, E., Roe, C. A., & Cox, N. J. (2010). Spoiling the whole bunch: Quality control aimed at preserving the integrity of high-throughput genotyping. The American Journal of Human Genetics, 87(1), 123–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popoli, M., Yan, Z., McEwen, B. S., & Sanacora, G. (2012). The stressed synapse: The impact of stress and glucocorticoids on glutamate transmission. Nature Reviews Neuroscience, 13, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston, A. R., & Eichenbaum, H. (2013). Interplay of hippocampus and prefrontal cortex in memory. Current Biology, 23, R764–R773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell, S., Neale, B., Todd-Brown, K., Thomas, L., Ferreira, M. A. R., Bender, D., … & Sham, P. C. (2007). PLINK: A tool set for whole-genome association and population-based linkage analyses. The American Journal of Human Genetics, 81, 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qingzhen, L., Jiehua, M., Zhiyang, Y., Hongjun, L., Chunlong, C., & Weiyan, L. (2016). Distinct hippocampal expression profiles of lncRNAs in rats exhibiting a PTSD-like syndrome. Molecular Neurobiology, 53, 2161–2168. [DOI] [PubMed] [Google Scholar]
- Relton, C. L., Gaunt, T., McArdle, W., Ho, K., Duggirala, A., Shihab, H., … Davey Smith, G. (2015). Data resource profile: Accessible resource for integrated epigenomic studies (ARIES). International Journal of Epidemiology, 44(4), 1181–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripke, S., O’Dushlaine, C., Chambert, K., Moran, J. L., Kahler, A. K., Akterin, S., … & Kim, Y. (2013). Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nature Genetics, 2(10), 1150–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson, N. D., & Hewitt, J. E. (2003). SF4 and SFRS14, two related putative splicing factors on human chromosome 19p13. 11. Gene, 305, 91–100. [DOI] [PubMed] [Google Scholar]
- Schmidt, B., Marrone, D. F., & Markus, E. J. (2012). Disambiguating the similar: The dentate gyrus and pattern separation. Behavioural Brain Research, 226, 56–65. [DOI] [PubMed] [Google Scholar]
- Schmidt, U., Keck, M. E., & Buell, D. R. (2015). miRNAs and other non-coding RNAs in posttraumatic stress disorder: A systematic review of clinical and animal studies. Journal of Psychiatric Research, 65, 1–8. [DOI] [PubMed] [Google Scholar]
- Schouten, M., Aschrafi, A., Bielefeld, P., Doxakis, E., & Fitzsimons, C. P. (2013). microRNAs and the regulation of neuronal plasticity under stress conditions. Neuroscience, 241, 188–205. [DOI] [PubMed] [Google Scholar]
- Song, H., Fang, F., Tomasson, G., Arnberg, F. K., Mataix-Cols, D., de la Cruz, L. F., & Valdimarsdóttir, U. A. (2018). Association of stress-related disorders with subsequent autoimmune disease. JAMA, 319(23), 2388–2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spadaro, P. A., Flavell, C. R., Widagdo, J., Ratnu, V. S., Troup, M., Ragan, C., & Bredy, T. W. (2015). Long noncoding RNA-directed epigenetic regulation of gene expression is associated with anxiety-like behavior in mice. Biological Psychiatry, 78(12), 848–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein, J. L., Medland, S. E., Vasquez, A. A., Hibar, D. P., Senstad, R. E., Winkler, A. M., … Thompson, P. M. (2012). Identification of common variants associated with human hippocampal and intracranial volumes. Nature Genetics, 44, 552–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein, M. B., Chen, C.-Y., Ursano, R. J., Cai, T., Gelernter, J., Heeringa, S. G., … & Nievergelt, C. M. (2016). Genome-wide association studies of posttraumatic stress disorder in 2 cohorts of US Army soldiers. JAMA Psychiatry, 44(7), 695–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland, R. J., & Rudy, J. W. (1988). Place learning in the Morris place navigation task is impaired by damage to the hippocampal formation even if the temporal demands are reduced. Psychobiology, 16, 157–163. [Google Scholar]
- Teicher, M. H., & Samson, J. A. (2014). Childhood maltreatment and psychopathology: A case for ecophenotypic variants as clinically and neurobiologically distinct subtypes. American Journal of Psychiatry 170(10), 1114–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, S. D. (2014). qqman: An R package for visualizing GWAS results using QQ and manhattan plots. bioRxiv, 005165. [Google Scholar]
- Van der Meer, D., Rokicki, J., Kaufmann, T., Córdova-Palomera, A., Moberget, T., Alnæs, D., … & Smeland, O. B. (2018a). Brain scans from 21,297 individuals reveal the genetic architecture of hippocampal subfield volumes. Molecular Psychiatry, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Meer, D., Rokicki, J., Kaufmann, T., Palomera, A. C., Moberget, T., Alnaes, D., … & Smeland, O. B. (2018b). Brain scans from 21297 individuals reveal the genetic architecture of hippocampal subfield volumes. bioRxiv, 299578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham, H. (2016). ggplot2: Elegant graphics for data analysis. New York, NY: Springer. [Google Scholar]
- Yang, J., Zeng, J., Goddard, M. E., Wray, N. R., & Visscher, P. M. (2017). Concepts, estimation and interpretation of SNP-based heritability. Nature Genetics, 49, 1304. [DOI] [PubMed] [Google Scholar]
- Yassa, M. A., & Stark, C. E. (2011). Pattern separation in the hippocampus. Trends in Neurosciences, 34, 515–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


