Graphical abstract
Abstract
The high transmission rate and serious consequences of the unprecedented COVID-19 pandemic make it challenging and urgent to identify viral pathogens and understand their intrinsic resistance mechanisms, to pave the way for new approaches to combat severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2). Multivalent interactions are responsible for performing a broad range of biological functions in normal cells, such as cell-cell communication and adhesion. Multivalency underlies the reversibility of ligand-receptor interactions during infections. Previous studies into multivalent nanomedicines used against viruses, have revealed their ability, not only to probe the molecular processes of viral infections, but also to target pathogen-host cell binding with minimal collateral damage to normal cells. Nanomedicines are comparable in size to viruses and to cell receptor complexes (that mediate viral uptake), and can function as safe and accurate armoured vehicles to facilitate the transport of anti-viral drugs. Multivalent nanomedicines can be designed to avoid binding to extracellular serum proteins, and ultimately lead to destruction of the viruses. This brief perspective highlights the potential of innovative smart and safe multivalent nanomedicines that could target multiple viral factors involved in infections at cellular levels. For instance it is possible to target viral spike protein mediated entry pathways, as well as viral replication and cell lysis. Nanomedicine-based approaches could open new opportunities for anti-coronavirus therapies.
Introduction
Nanotechnology is a multidisciplinary field integrating diverse disciplines of engineering, chemistry, physics and medicine. Applications of nanotechnology for diagnosis, treatment, monitoring and prevention of diseases is commonly referred to as nanomedicine, and has shown the potential to make a significant impact on medical diagnosis and treatment in the near future [1]. Nanomedicine has enabled researchers to detect early biomarkers signifying the presence a disease, as well as to treat the disease in a timely fashion. Nanoparticles (NPs) are therapeutic and diagnostic agents with numerous advantages, including control over shape and size, tunable porosity, and a large surface area for drug storage and release. They also have an extremely high surface-to-volume ratio, and the ability to manipulate their dimensions and functionalise the surface with multivalent targeting moieties enables targeted delivery to diseased tissues/cells [2]. Nanomedicine has successfully been used to improve treatment in a wide range of diseases, such as cancer, cardiovascular, neurological disorders, and infectious diseases [3]. The role of nanomedicine is highly relevant to tackle the recently emerged coronavirus disease 2019 (COVID-19). The multivalent nature of nanomedicines means they have significant potential to treat COVID-19, and could provide a catalyst for novel technologies in healthcare industries.
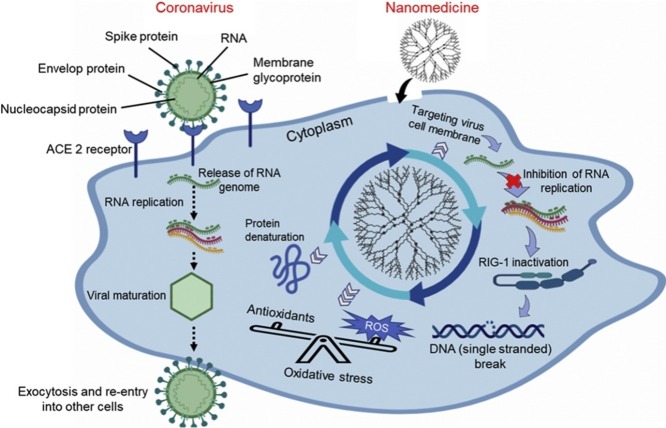
Coronaviruses (CoVs) are enveloped (covered in a lipid coat) single‐stranded positive‐sense ribonucleic acid (RNA) viruses, having a RNA genome between 26–32 kilobases, which is the longest known viral RNA genome [4,5]. CoVs have been detected in a broad range of hosts including species such as humans, mammalians and avians. In some cases they may cause serious diseases including gastrointestinal, respiratory, and central nervous system disorders [6,7]. CoVs are divided into four genera: alpha-, beta-, gamma-, and delta-CoVs. The beta-CoVs are further divided into four subgroups (A, B, C, and D). There are six strains of CoVs within the alpha and beta groups that can infect humans, including two from alpha (HCoV-229E and HCoV-NL63), two from beta A (HCoV−OC43 and HCoV-HKU1), one from beta B (severe acute respiratory syndrome CoV (SARS-CoV)) and one from beta C (Middle East respiratory syndrome CoV (MERS-CoV)) [8]. While gamma-CoVs only infect birds (avian species) and delta-CoVs infect both avians and mammalians [9]. The 21st century has witnessed an alarmingly high mortality rate of severe pulmonary diseases in humans that were associated with SARS-CoV and MERS-CoV [10]. On the 10th of January 2020, the genome sequence of the novel pathogen was found to be more similar to SARS-CoV compared to MERS-CoV and was therefore named SARS-CoV-2 [11,12]. SARS-CoV-2 belongs to the beta C subgroup with a single‐stranded positive‐sense RNA genome and was found to have 79 % genetic similarity to SARS-CoV, and 98 % similarity to bat CoV RaTG13 [13]. COVID-19 is a rapidly spreading infectious disease caused by SARS-CoV-2. The four main structural proteins encoded by CoVs include spike, envelop, matrix and nucleocapsid proteins [14].
The mechanism of viral pathogenesis associated with SARS-CoV-2 remains largely unknown. It has been shown that SARS-CoV-2 engages the same receptor used by SARS-CoV, i.e. angiotensin-converting enzyme 2 (ACE2) as a cell entry pathway. Further findings showed that the spike protein binds to ACE2 with high affinity and then facilitates viral entry into specific host cells that express high levels of ACE2 [15]. AEC2 is actively involved in the life cycle, transport of virus, replication, and regulation of viral genomes in specific host cells. On the other hand, the ACE2 receptor also plays a beneficial role in many pathophysiological processes by regulating amino acid transport thereby participating in inflammatory and fibrotic responses to other diseases, including acute lung injury and respiratory distress syndrome [16]. Briefly, ACE2 not only acts as a receptor for the entry of CoVs, but also protects living systems from severe pathological damage. ACE2 is also found on the linings of blood vessels, and in the heart, kidneys and lungs. In addition to ACE2 receptor, the cellular serine protease transmembrane protease, serine 2 (TMPRSS2) has also been found to bind to the spike protein of SARS-CoV-2 as a secondary entry pathway [17].
SARS-CoV-2 has already caused hundreds of thousands of deaths across the globe [4,5]. As we write this, the numbers are still rising worldwide. SARS-CoV2 can be rapidly transmitted from person to person. SARS-CoV-2 is zoonotic in origin, and bats have been suggested to be the primary source of transmission to humans [11]. It has also been hypothesised that SARS-CoV2 was transmitted from bats to other animals as an intermediate host [12]. The intermediate host lying in between the transmission of SARS-CoV2 from bats to humans is yet to be identified, but pangolins have been suspected. One person infected with SARS-CoV2 can pass the virus on to three other people on average [12]. In a recent study it has been found that the environmental longevity of SARS-CoV-2 is similar to that of SARS-CoV [18], so that SARS-CoV2 can remain viable and infectious on plastic or stainless steel surfaces (for up to days) and in aerosols (for up to 3 h). Currently the available molecular diagnostic tool for the detection of SARS-CoV2 is nucleic acid testing which is based on reverse transcription-polymerase chain reaction (RT-PCR) [19]. RT-PCR is widely used in virology diagnostics and involves the reverse transcription of SARS-CoV2 RNA into DNA strands using primers that amplify only specific regions of DNA strands [19]. The accuracy and sensitivity of PCR depends on a number of factors such as primer and probe design, target selection, sequencing alignment, RNA extraction and multiplexing of PCR assays (e.g., primer/buffer/reaction conditions, incubation times and conditions, and annealing temperatures) [19]. Other diagnostic methods include computed tomography (CT) and loop-mediated isothermal amplification (LAMP assay) [20]. Researchers are currently developing innovative diagnostic tools for a rapid, easy-to-apply diagnostic method to identify both active and asymptomatic SARS-CoV2 infections in a quantitative manner [20]. The design and development of innovative diagnostic methods should circumvent the clinical limitations of existing approaches such as low yields, and could focus on affinity/binding to ACE2, selectivity, half-life, stability, and immunogenicity, and biodistribution of ACE2, which may significantly improve the diagnostic accuracy.
Therapeutic interventions
The multivalency associated with SARS-CoV-2 rests on the following concepts: (a) the receptors (ACE2 and TMPRSS2) intrinsically associated with CoVs are co-localised in virus-host protein-protein networks at cellular or subcellular levels; (b) receptors and co-receptors offer an effective way towards elucidating the mechanisms of viral pathogenesis; and (c) receptors and co-receptors as multiple cellular targets offer an innovative strategy for the development of preventive and treatment modalities. There is no specifically validated therapeutic regime currently available for the treatment of COVID-19. Few Food and Drug Administration (FDA) approved drugs have been proposed as potential candidates to treat COVID-19 based on their effectiveness against other medical conditions, and against earlier strains of CoVs (such as SARS-CoV and MERS-CoV) [21,22]. One promising strategy is to use therapeutic agents, which can block the virus-host interactome (such as ACE2 and TMPRSS2) and can target the multivalent protein–protein interaction network. There are currently available drugs and inhibitors approved for other medical conditions, which could selectively target these receptors (ACE2 and TMPRSS2) [22]. To block virus entry into cells, some clinicians have proposed the idea of using small molecule inhibitors of ACE2 or TMPRSS2 to treat SARS-CoV-2 infection [23]. FDA approved Janus kinase (JAK) inhibitors could be effective in inhibiting multiple inflammatory cytokines and reducing systemic inflammation. However JAK inhibitors may also increase the risk of thrombosis and viral reactivation in a dose- and time-dependent manner [24]. For instance, baricitinib, an FDA approved JAK inhibitor for the treatment of rheumatoid arthritis, could potentially interfere with and inhibit ACE2 [25]. TMPRSS2 inhibitors such as nafamostat mesylate and camostat mesylate could also be used to block virus entry sites [26]. Oncology drugs, including sunitinib and erlotinib can inhibit and disrupt AP2-associated protein kinase 1 (AAK1), which in turn could inhibit the entry of viruses into cells, however, these drugs have been associated with severe side-effects [27]. Moreover, monoclonal antibodies (mAbs) can block the spike proteins, which in turn can inhibit membrane fusion and virus entry [28]. A few of these drugs are under clinical trials to validate their potential antiviral activity against SARS-CoV-2, and their ability to disrupt virus entry pathways. Additionally, drugs for the treatment of pulmonary fibrosis have also been proposed to treat SARS-CoV-2, such as pirfenidone [29]. The potential antiviral activities of the aforementioned drugs and HIV drugs (combination of liponavir–ritonavir) for the treatment of SARS-CoV-2 have been discussed elsewhere [30,31].
CoVs are particles about 60−140 nm in diameter, approximately 25 times smaller than an E. coli bacterial cell (−2.5 μm) [32]. One possibility is to target the multivalent interaction of viruses with cells, such as receptor-mediated endocytosis and direct fusion with the cell membrane. Based on these multivalent interactions, the effectiveness of monovalent (single target) drugs remains poor against antiviral targets at the cellular or subcellular level. On the other hand, multivalent nanomedicines could be used to treat viral infections and to regulate dysfunctional immune responses. They may be able to carry out synergistic effects to block pathogens by either interfering with the direct interaction of viruses with specific host cells, or crucial entry pathways of viruses [32,33]. Therefore, a multivalent therapeutic agent could offer tunable binding affinity towards biomolecules, ligands, receptors, co-receptors, and the cell surface, thereby degrading viral RNA and inhibiting host proteins. NPs are of a similar size to the virus and have the potential to interact with spike proteins and to bind to ACE2, which could disrupt the viral structure and interfere with viral replication [34]. In some circumstances the size of NPs can be small in comparison to drugs such as antibodies, which have larger molecular structures. Additionally, a messenger RNA (mRNA)–lipid NP-based vaccine has previously been reported to treat SARS−CoV and MERS-CoV [30]. Nanomedicine-based approaches have been extensively investigated for targeted drug/gene delivery, light-mediated therapy (photodynamic therapy and photothermal therapy) for a wide range of diseases, such as cancer, infections or neurological disorders [35,36]. Nanomedicine using biocompatible and less toxic NPs has been recognised as an enabling technology capable of providing innovative medical solutions to address unmet clinical needs. Nanomedicines have been shown to help to treat viral infections by various mechanisms, such as suppression of endocytosis in specific host cells, mitochondrial activation or inactivation, inhibition of viral genome replication, and RNA-crosslinking [37]. NPs can be tweaked, tuned and functionalised with antibodies or nanobodies to target and block pathogens before they affect living systems. NPs have also been shown to be taken up by diseased cells to a greater extent than normal cells, based on translocation mechanisms (outer wrapping, free translocation, enhanced permeability effect). The cell uptake is governed by their physiochemical characteristics, such as size, shape, and surface chemistry. Moreover their therapeutic usefulness is governed by their ease of functionalisation, and their magnetic and optical properties [38,39]. Moreover, NPs also allow improved delivery of poorly soluble drugs, and can be tailored for either active or passive targeting of specific cells or tissues. Targeting specificity leads to only minimal collateral damage to normal cells and tissue. This interplay between optical features and size cannot be tuned in the same way with other drug delivery vehicles [40].
Pristine and functionalised forms of dendritic polymers (such as polyamidoamine (PAMAM) dendrimers), silver and gold NPs, 2D-materials (such as graphene oxide nanosheets, graphene nanosheets, porous graphene), carbon nanotubes and carbon quantum dots have shown their ability to mediate multivalent inhibition of viral infection pathways. This effect could be due to synergistic targeting of receptors/co-receptors and pro-inflammatory pathways at the same time. [41]. Specifically, silver NPs have long been recognised as potential antimicrobial agents, and recently, the antiviral actions of silver NPs have also been investigated [42]. Silver ions released from silver NPs bind to the RNA viral genome and activate interferon signalling pathways in the mitochondria, which in turn activate mitochondrial proteins that may have an antiviral effect [42,43]. Villeret et al. [44] demonstrated that silver NPs could inhibit the interferon signalling pathway by stimulating the release of two clinically relevant antiviral cytokines (CCL-5 and interferon-beta) against an influenza virus. Silver NPs can also block the autophagic flux caused by the virus by disrupting the mitochondrial network. In another study, Du et al. [45] reported the antiviral activity of graphene oxide-silver NP nanocomposites against porcine reproductive and respiratory syndrome (PRRS) virus, and showed that these nanocomposites could prevent the virus from entering into specific host cells. Consequently, these nanocomposites inhibited the proliferation of the virus by enhancing the production of interferon-alpha and interferon-stimulating genes. Polymer-functionalised gold NPs have been studied as efficient antiviral agents against HIV-1, H1N1, H3N2, H5N1 and dengue virus [46,47]. Iron oxide NPs (10−15 nm) have also been developed and tested against pandemic influenza virus strains A/H1N1/Eastern India/66/PR8-H1N1 with 50 % inhibition efficacy [48]. Single walled carbon nanotubes (of diameter 0.78 nm) also showed pronounced effects against influenza A virus infection in mice as shown by reduced lung injury, lower cytokine expression, and reduction of antiviral gene expression [49].
Heparan sulfate increases the risk of virus entry, and it has also been reported to act as a mode of entry in a few viral infections, such as herpes simplex virus and dengue virus [50]. It has recently been shown that heparan sulfate is also involved in SARS-CoV-2, and can facilitate the secondary cleavage of viruses after their fusion and co-localisation with secondary receptors [50]. Graphene has shown a good viral inhibition capacity against heparan sulfate moieties, owing to its strong binding with viruses through electrostatic interactions and redox reactions [51]. Multivalent 2D polymer nanosystems have also been explored to block heparan sulfate‐dependent cell entry. Moreover, polymeric nanocomposites may be an ideal candidate to act as viral entry inhibitors owing to their ability to undergo biodegradation into small fragments, which can be cleared by the kidneys [[52], [53], [54]]. The antiviral activity of graphene oxide (functionalised with β-cyclodextrin) has been demonstrated against single-stranded RNA viruses, showing both simultaneous inactivation and inhibition of the virus [55].
Multivalent PAMAM conjugates have shown effective antiviral activity against influenza infections. For instance, Kwon et al. [56] reported that multivalent 6′-sialyllactose-polyamidoamine (6SL–PAMAM) conjugates could act as anti-influenza inhibitors by multivalent binding to a hemagglutinin trimer, and showed good inhibition of H1N1 infection with a survival rate of 75 % in mice. In another study, Stertz et al. [57] reported the synthesis of 3′-sialyllactose and 6′-sialyllactose-conjugated PAMAM dendrimers, and demonstrated these polymers could inhibit the binding of human and avian influenza virus strains to their receptors. The strains included (H1N1, H3N2 and avian strains: A/duck/England/1/1956 (EN/56), A/duck/Ukraine/1/1963 (UK/63) and A/duck/Alberta/35/1976 (AL/76)).
Some metal-based and carbon-based NPs may elicit toxicity towards surrounding healthy tissues or cells, which can occur through the release of toxic ions, proinflammatory cytokines, membrane damage and the production of reactive oxygen species [58]. One strategy to enhance the therapeutic efficacy of nanomedicines is by improving the selective and targeted accumulation of the NPs in diseased cells, thereby limiting the NP accumulation in healthy cells. The easy functionalisation and bioconjugation of NPs with targeting moieties (such as viral receptors, antibodies, peptides or proteins) can provide many exciting features, and can significantly improve the targeted delivery of NPs to diseased cells, in comparison to unmodified NPs. These approaches using functionalised NPs have been described for selective targeting and destruction of a wide range of pathogenic microorganisms [59,60]. Selective and targeted delivery of modified NPs could allow the specific identification of viral ligands and destruction of viruses at the site of infection. In order to ensure the site-specific delivery of multivalent nanomedicines to treat COVID-19, without any accumulation in non-diseased tissues, NPs must be optimised in terms of the functionalisation density of targeting moieties on the surface of the NPs, to allow their systemic administration and dosing frequency.
NPs are broadly comparable in size to viruses and their receptors, and represent a novel class of antiviral agents that can efficiently bind to ligands and disrupt receptor-host cell interactions. These multivalent NPs have several advantages in comparison to standard monovalent drugs, such as high density of binding moieties on each NP, ability to form multivalent ligand-receptor pairs, RNA hybridisation up to many fold higher, and transformation of inactive NPs into multivalent conjugates. The application of nanomedicines has a great potential in anti SARS-CoV-2 therapy by targeting entry pathways, by targeted binding to the viral genome, modulation of viral transcription, triggering the production of reactive oxygen species, or activation of AKT at a mitochondrial level. One “dichotomy” in the action of nanomedicines is related to a possible toxic effect owing to the generation of reactive oxygen species in surrounding tissues, which may induce oxidative stress [61]. Moreover, several in vivo studies have reported that different types of NPs could induce adverse side-effects including acute, sub-acute, sub-chronic damage, activation of pro-inflammatory cytokines, and delayed clearance from the body [62,63]. There are several reports suggesting that appropriate functionalisation and bioconjugation of these NPs could reduce and minimize the adverse effects and toxicity of NPs [64,65]. Nanomedicine based approaches using functionalised NPs can also be used as a photosensitiser for the photoinactivation of pathogens via the photodynamic effect. Similar nanomedicine approaches could potentially be explored for photocatalytic inactivation of viruses, as well as for photodynamic [66] and photothermal [[67], [68], [69]] therapy which can produce reactive oxygen species or photothermal heating within virus-infected host cells. Additionally, gas therapies based on the release of nitric oxide [70], hydrogen sulfide, or carbon monoxide could also be explored to complement the hypoxia produced by CoV infections. Inhibition of viral binding to the host cell surface receptors using these light-mediated or gas-releasing therapies is another possibility.
Summary
The high multivalency of the SARS-CoV-2, including viral-host interactions, the viral life cycle, and regulation of the virus by the host cellular network, highlights the importance of developing innovative multivalent antiviral nanoplatforms to specifically target cellular (sub)networks associated with SARS-CoV-2. Targeting the key indispensable steps of infection (including viral entry into specific host cells) and the multivalency of the interactions between the viral genome and host receptors could potentially be achieved by using robust multivalent nanoparticles. In this perspective, we have summarised the current status of multivalent nanomedicines, which could possibly be used as inhibitors to treat SARS-CoV-2 cell fusion by interruption of the interplay between the virus and the host, and destroying specific target sites within the viral genome. The results from different studies demonstrate that nanoparticles have the potential to target the invasive nature and high diversity of SARS-CoVs, and may potentially accelerate the leap from bench to bedside.
Author statement
Tanveer A. Tabish conceptualized and wrote the manuscript. Michael R. Hamblin supervised, reviewed and edited the manuscript.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Biographies

Tanveer A. Tabish received his PhD in 2018 from the University of Exeter, UK. His PhD research focused primarily on the development of graphene-based anti-cancer nanomedicine. He currently works as Research Fellow in Bionanophotonics at the UCL Cancer Institute, University College London and focuses on image-guided rapid, convenient and label-free disease detection. He has a mix of training/expertise in nanotechnology, photonanomedicine, free radicalmedicine, cell biology, cell biophysics, and optical imaging. He interweaves these scientific fields to develop ‘personalized’ solutions based on light-mediated therapeutic modalities that utilise nanotechnology in treating many diseases such as cancer, infections, congenital heart diseases, and Alzheimer’s disease.

Michael R Hamblin Ph.D. was Principal Investigator at the Wellman Center for Photomedicine at Massachusetts General Hospital, Associate Professor of Dermatology, Harvard Medical School, is now Visiting Professor at the University of Johannesburg, South Africa. His research interests concern photodynamic therapy and photobiomodulation. He has published >530 peer-reviewed articles, 25 textbooks including 13 SPIE proceedings. His h-index is 112 with >51,000 citations. He is Editor-in-Chief of “Photobiomodulation, Photomedicine and Laser Surgery”. Honors include Fellow of SPIE 2011, 1st Endre Mester Lifetime Achievement Award NAALT 2017, Outstanding Career Award Dose Response Society, 1st Ali Javan Award Photobiomodulation Basic Science Research from WALT 2018.
References
- 1.Lammers T., Ferrari M. The success of nanomedicine. Nano Today. 2020 doi: 10.1016/j.nantod.2020.100853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tabish T.A., Dey P., Mosca S., Salimi M., Palombo F., Matousek P., Stone N. Smart gold nanostructures for light mediated cancer theranostics: combining optical diagnostics with photothermal therapy. Adv. Sci. 2020 doi: 10.1002/advs.201903441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karimi M., Ghasemi A., Zangabad P.S., Rahighi R., Basri S.M.M., Mirshekari H., Ghosh D. Smart micro/nanoparticles in stimulus-responsive drug/gene delivery systems. Chem. Soc. Rev. 2016;45(5):1457–1501. doi: 10.1039/c5cs00798d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferretti L., Wymant C., Kendall M., Zhao L., Nurtay A., Abeler-Dörner L., Fraser C. Quantifying SARS-CoV-2 transmission suggests epidemic control with digital contact tracing. Science. 2020;368(6491) doi: 10.1126/science.abb6936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tay M.Z., Poh C.M., Rénia L., MacAry P.A., Ng L.F. The trinity of COVID-19: immunity, inflammation and intervention. Nat. Rev. Immunol. 2020:1–12. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Y., Liu Q., Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J. Med. Virol. 2020;92(4):418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Decaro N., Lorusso A. Novel human coronavirus (SARS-CoV-2): a lesson from animal coronaviruses. Vet. Microbiol. 2020 doi: 10.1016/j.vetmic.2020.108693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ashour H.M., Elkhatib W.F., Rahman M., Elshabrawy H.A. Insights into the recent 2019 novel coronavirus (SARS-CoV-2) in light of past human coronavirus outbreaks. Pathogens. 2020;9(3):186. doi: 10.3390/pathogens9030186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miłek J., Blicharz-Domańska K. Coronaviruses in avian species–review with focus on epidemiology and diagnosis in wild birds. J. Vet. Res. 2018;62(3):249–255. doi: 10.2478/jvetres-2018-0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26(4):450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.ul Qamar M.T., Alqahtani S.M., Alamri M.A., Chen L.L. Structural basis of SARS-CoV-2 3CLpro and anti-COVID-19 drug discovery from medicinal plants. J. Pharm. Anal. 2020 doi: 10.1016/j.jpha.2020.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan W.C. Nano research for COVID-19. ACS Nano. 2020 doi: 10.1021/acsnano.0c02540. [DOI] [PubMed] [Google Scholar]
- 13.Guo Y.R., Cao Q.D., Hong Z.S., Tan Y.Y., Chen S.D., Jin H.J., Yan Y. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak–an update on the status. Mil. Med. Res. 2020;7(1):1–10. doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mousavizadeh L., Ghasemi S. Genotype and phenotype of COVID-19: their roles in pathogenesis. J. Microbiol. Immunol. Infect. 2020 doi: 10.1016/j.jmii.2020.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng J. SARS-CoV-2: an emerging coronavirus that causes a global threat. Int. J. Biol. Sci. 2020;16(10):1678. doi: 10.7150/ijbs.45053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuba K., Imai Y., Ohto-Nakanishi T., Penninger J.M. Trilogy of ACE2: a peptidase in the renin–angiotensin system, a SARS receptor, and a partner for amino acid transporters. Pharmacol. Ther. 2010;128(1):119–128. doi: 10.1016/j.pharmthera.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Drucker D.J. Coronavirus infections and type 2 diabetes—shared pathways with therapeutic implications. Endocr. Rev. 2020;41(3) doi: 10.1210/endrev/bnaa011. bnaa011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Doremalen N., Bushmaker T., Morris D.H., Holbrook M.G., Gamble A., Williamson B.N., Lloyd-Smith J.O. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N. Engl. J. Med. 2020;382(16):1564–1567. doi: 10.1056/NEJMc2004973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Mulders D.G. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance. 2020;25(3) doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Udugama B., Kadhiresan P., Kozlowski H.N., Malekjahani A., Osborne M., Li V.Y., Chan W.C. Diagnosing COVID-19: the disease and tools for detection. ACS Nano. 2020 doi: 10.1021/acsnano.0c02624. [DOI] [PubMed] [Google Scholar]
- 21.Abd El-Aziz T.M., Stockand J.D. Recent progress and challenges in drug development against COVID-19 coronavirus (SARS-CoV-2)-an update on the status. Infect. Genet. Evol. 2020 doi: 10.1016/j.meegid.2020.104327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nile S.H., Nile A., Qiu J., Li L., Jia X., Kai G. COVID-19: pathogenesis, cytokine storm and therapeutic potential of interferons. Cytokine Growth Factor Rev. 2020 doi: 10.1016/j.cytogfr.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Müller M.A. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020 doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vellingiri B., Jayaramayya K., Iyer M., Narayanasamy A., Govindasamy V., Giridharan B., Rajagopalan K. COVID-19: a promising cure for the global panic. Sci. Total Environ. 2020 doi: 10.1016/j.scitotenv.2020.138277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Richardson P., Griffin I., Tucker C., Smith D., Oechsle O., Phelan A., Stebbing J. Baricitinib as potential treatment for 2019-nCoV acute respiratory disease. Lancet (London, England) 2020;395(10223):e30. doi: 10.1016/S0140-6736(20)30304-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Angelidi A.M., Belanger M.J., Mantzoros C.S. COVID-19 and diabetes mellitus: what we know, how our patients should be treated now, and what should happen next. Metabolism-Clinical Experimental. 2020 doi: 10.1016/j.metabol.2020.154245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Richardson P., Griffin I., Tucker C., Smith D., Oechsle O., Phelan A., Stebbing J. Baricitinib as potential treatment for 2019-nCoV acute respiratory disease. Lancet (London, England) 2020;395(10223):e30. doi: 10.1016/S0140-6736(20)30304-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marasco W.A., Sui J. The growth and potential of human antiviral monoclonal antibody therapeutics. Nat. Biotechnol. 2007;25(12):1421–1434. doi: 10.1038/nbt1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun P., Qie S., Liu Z., Ren J., Li K., Xi J. Clinical characteristics of hospitalized patients with SARS‐CoV‐2 infection: a single arm meta‐analysis. J. Med. Virol. 2020;92(6):612–617. doi: 10.1002/jmv.25735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu C., Zhou Q., Li Y., Garner L.V., Watkins S.P., Carter L.J., Albaiu D. Research and development on therapeutic agents and vaccines for COVID-19 and related human coronavirus diseases. ACS Cent. Sci. 2020 doi: 10.1021/acscentsci.0c00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cao B., Wang Y., Wen D., Liu W., Wang J., Fan G., Li X. A trial of lopinavir–ritonavir in adults hospitalized with severe Covid-19. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jampílek J., Kráľová K. Nanotheranostics. Springer, Cham; 2019. Nanoformulations: a valuable tool in the therapy of viral diseases attacking humans and animals; pp. 137–178. [Google Scholar]
- 33.Wu K., Chen L., Peng G., Zhou W., Pennell C.A., Mansky L.M., Li F. A virus-binding hot spot on human angiotensin-converting enzyme 2 is critical for binding of two different coronaviruses. J. Virol. 2011;85(11):5331–5337. doi: 10.1128/JVI.02274-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sportelli M.C., Izzi M., Kukushkina E.A., Hossain S.I., Picca R.A., Ditaranto N., Cioffi N. Can Nanotechnology and Materials Science Help the Fight against SARS-CoV-2? Nanomaterials. 2020;10(4):802. doi: 10.3390/nano10040802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang J., Byrne J.D., Napier M.E., DeSimone J.M. More effective nanomedicines through particle design. Small. 2011;7(14):1919–1931. doi: 10.1002/smll.201100442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tabish Tanveer A., Zhang Shaowei, Winyard Paul G. Developing the next generation of graphene-based platforms for cancer therapeutics: the potential role of reactive oxygen species. Redox Biol. 2018;15:34–40. doi: 10.1016/j.redox.2017.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McNeil S.E. Evaluation of nanomedicines: stick to the basics. Nat. Rev. Mater. 2016;1(10):1–2. [Google Scholar]
- 38.Tabish T.A., Scotton C.J., J Ferguson D.C., Lin L., der Veen A.V., Lowry S., Zhang S. Biocompatibility and toxicity of graphene quantum dots for potential application in photodynamic therapy. Nanomedicine. 2018;13(15):1923–1937. doi: 10.2217/nnm-2018-0018. [DOI] [PubMed] [Google Scholar]
- 39.McNeil S.E. Evaluation of nanomedicines: stick to the basics. Nat. Rev. Mater. 2016;1(10):1–2. [Google Scholar]
- 40.Juliano R. Nanomedicine: is the wave cresting? Nat. Rev. Drug Discov. 2013;12(3):171–172. doi: 10.1038/nrd3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Singh L., Kruger H.G., Maguire G.E., Govender T., Parboosing R. The role of nanotechnology in the treatment of viral infections. Ther. Adv. Infect. Dis. 2017;4(4):105–131. doi: 10.1177/2049936117713593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hou F., Sun L., Zheng H., Skaug B., Jiang Q.X., Chen Z.J. MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell. 2011;146(3):448–461. doi: 10.1016/j.cell.2011.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Z., Liu S., Ma J., Qu G., Wang X., Yu S., Jiang G.B. Silver nanoparticles induced RNA polymerase-silver binding and RNA transcription inhibition in erythroid progenitor cells. ACS Nano. 2013;7(5):4171–4186. doi: 10.1021/nn400594s. [DOI] [PubMed] [Google Scholar]
- 44.Villeret B., Dieu A., Straube M., Solhonne B., Miklavc P., Hamadi S., Diallo D. Silver nanoparticles impair retinoic acid-inducible gene I-mediated mitochondrial antiviral immunity by blocking the autophagic flux in lung epithelial cells. ACS Nano. 2018;12(2):1188–1202. doi: 10.1021/acsnano.7b06934. [DOI] [PubMed] [Google Scholar]
- 45.Du T., Lu J., Liu L., Dong N., Fang L., Xiao S., Han H. Antiviral activity of graphene oxide–Silver nanocomposites by preventing viral entry and activation of the antiviral innate immune response. ACS Applied Biomaterials. 2018;1(5):1286–1293. doi: 10.1021/acsabm.8b00154. [DOI] [PubMed] [Google Scholar]
- 46.Tao W., Hurst B.L., Shakya A.K., Uddin M.J., Ingrole R.S., Hernandez-Sanabria M., Gill H.S. Consensus M2e peptide conjugated to gold nanoparticles confers protection against H1N1, H3N2 and H5N1 influenza A viruses. Antiviral Res. 2017;141:62–72. doi: 10.1016/j.antiviral.2017.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang C., Zhu W., Luo Y., Wang B.Z. Gold nanoparticles conjugating recombinant influenza hemagglutinin trimers and flagellin enhanced mucosal cellular immunity. Nanomed. Nanotechnol. Biol. Med. 2018;14(4):1349–1360. doi: 10.1016/j.nano.2018.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kumar R., Nayak M., Sahoo G.C., Pandey K., Sarkar M.C., Ansari Y., Das P. Iron oxide nanoparticles based antiviral activity of H1N1 influenza A virus. J. Infect. Chemother. 2019;25(5):325–329. doi: 10.1016/j.jiac.2018.12.006. [DOI] [PubMed] [Google Scholar]
- 49.Chen H., Zheng X., Nicholas J., Humes S.T., Loeb J.C., Robinson S.E., Lednicky J.A. Single-walled carbon nanotubes modulate pulmonary immune responses and increase pandemic influenza a virus titers in mice. Virol. J. 2017;14(1):242. doi: 10.1186/s12985-017-0909-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Palmieri V., Papi M. Can graphene take part in the fight against COVID-19? Nano Today. 2020 doi: 10.1016/j.nantod.2020.100883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ziem B., Azab W., Gholami M.F., Rabe J.P., Osterrieder N., Haag R. Size-dependent inhibition of herpesvirus cellular entry by polyvalent nanoarchitectures. Nanoscale. 2017;9(11):3774–3783. doi: 10.1039/c7nr00611j. [DOI] [PubMed] [Google Scholar]
- 52.Nochi T., Yuki Y., Takahashi H., Sawada S.I., Mejima M., Kohda T., Tokuhara D. Nanogel antigenic protein-delivery system for adjuvant-free intranasal vaccines. Nat. Mater. 2010;9(7):572–578. doi: 10.1038/nmat2784. [DOI] [PubMed] [Google Scholar]
- 53.Ziem B., Thien H., Achazi K., Yue C., Stern D., Silberreis K., Rabe J.P. Highly efficient multivalent 2D nanosystems for inhibition of orthopoxvirus particles. Adv. Healthc. Mater. 2016;5(22):2922–2930. doi: 10.1002/adhm.201600812. [DOI] [PubMed] [Google Scholar]
- 54.Donskyi I.S., Azab W., Cuellar-Camacho J.L., Guday G., Lippitz A., Unger W.E., Haag R. Functionalized nanographene sheets with high antiviral activity through synergistic electrostatic and hydrophobic interactions. Nanoscale. 2019;11(34):15804–15809. doi: 10.1039/c9nr05273a. [DOI] [PubMed] [Google Scholar]
- 55.Yang X.X., Li C.M., Li Y.F., Wang J., Huang C.Z. Synergistic antiviral effect of curcumin functionalized graphene oxide against respiratory syncytial virus infection. Nanoscale. 2017;9(41):16086–16092. doi: 10.1039/c7nr06520e. [DOI] [PubMed] [Google Scholar]
- 56.Kwon S.J., Na D.H., Kwak J.H., Douaisi M., Zhang F., Park E.J., Dordick J.S. Nanostructured glycan architecture is important in the inhibition of influenza A virus infection. Nat. Nanotechnol. 2017;12(1):48. doi: 10.1038/nnano.2016.181. [DOI] [PubMed] [Google Scholar]
- 57.Günther S.C., Maier J.D., Vetter J., Podvalnyy N., Khanzhin N., Hennet T., Stertz S. Antiviral potential of 3′-sialyllactose-and 6′-sialyllactose-conjugated dendritic polymers against human and avian influenza viruses. Sci. Rep. 2020;10(1):1–9. doi: 10.1038/s41598-020-57608-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pokhrel S., Nel A.E., Mädler L. Custom-designed nanomaterial libraries for testing metal oxide toxicity. Acc. Chem. Res. 2013;46(3):632–641. doi: 10.1021/ar300032q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yuste-Calvo C., González-Gamboa I., Pacios L.F., Sánchez F., Ponz F. Structure-based multifunctionalization of flexuous elongated viral nanoparticles. ACS Omega. 2019;4(3):5019–5028. [Google Scholar]
- 60.Mout R., Moyano D.F., Rana S., Rotello V.M. Surface functionalization of nanoparticles for nanomedicine. Chem. Soc. Rev. 2012;41(7):2539–2544. doi: 10.1039/c2cs15294k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Behzadi S., Serpooshan V., Tao W., Hamaly M.A., Alkawareek M.Y., Dreaden E.C., Mahmoudi M. Cellular uptake of nanoparticles: journey inside the cell. Chem. Soc. Rev. 2017;46(14):4218–4244. doi: 10.1039/c6cs00636a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Srivastava V., Gusain D., Sharma Y.C. Critical review on the toxicity of some widely used engineered nanoparticles. Ind. Eng. Chem. Res. 2015;54(24):6209–6233. [Google Scholar]
- 63.Khlebtsov N., Dykman L. Biodistribution and toxicity of engineered gold nanoparticles: a review of in vitro and in vivo studies. Chem. Soc. Rev. 2011;40(3):1647–1671. doi: 10.1039/c0cs00018c. [DOI] [PubMed] [Google Scholar]
- 64.Sperling R.A., Parak W.J. Surface modification, functionalization and bioconjugation of colloidal inorganic nanoparticles. Philos. Trans. Math. Phys. Eng. Sci. 2010;368(1915):1333–1383. doi: 10.1098/rsta.2009.0273. [DOI] [PubMed] [Google Scholar]
- 65.Conde J., Dias J.T., Grazú V., Moros M., Baptista P.V., de la Fuente J.M. Revisiting 30 years of biofunctionalization and surface chemistry of inorganic nanoparticles for nanomedicine. Front. Chem. 2014;2:48. doi: 10.3389/fchem.2014.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kharkwal G.B., Sharma S.K., Huang Y.Y., Dai T., Hamblin M.R. Photodynamic therapy for infections: clinical applications. Lasers Surg. Med. 2011;43(7):755–767. doi: 10.1002/lsm.21080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bowman M.C., Ballard T.E., Ackerson C.J., Feldheim D.L., Margolis D.M., Melander C. Inhibition of HIV fusion with multivalent gold nanoparticles. J. Am. Chem. Soc. 2008;130(22):6896–6897. doi: 10.1021/ja710321g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vijayakumar S., Ganesan S. Gold nanoparticles as an HIV entry inhibitor. Curr. HIV Res. 2012;10(8):643–646. doi: 10.2174/157016212803901383. [DOI] [PubMed] [Google Scholar]
- 69.Gerrity D., Ryu H., Crittenden J., Abbaszadegan M. Photocatalytic inactivation of viruses using titanium dioxide nanoparticles and low-pressure UV light. J. Environ. Sci. Health, Part A. 2008;43(11):1261–1270. doi: 10.1080/10934520802177813. [DOI] [PubMed] [Google Scholar]
- 70.Lim L., Hamblin M.R. 2020. Can the Vielight X-Plus be a Therapeutic Intervention for COVID-19 Infection? [Google Scholar]