Abstract
One of the features distinguishing SARS-CoV-2 from its more pathogenic counterpart SARS-CoV is the presence of premature stop codons in its ORF3b gene. Here, we show that SARS-CoV-2 ORF3b is a potent interferon antagonist, suppressing the induction of type I interferon more efficiently than its SARS-CoV ortholog. Phylogenetic analyses and functional assays reveal that SARS-CoV-2-related viruses from bats and pangolins also encode truncated ORF3b gene products with strong anti-interferon activity. Furthermore, analyses of approximately 17,000 SARS-CoV-2 sequences identify a natural variant in which a longer ORF3b reading frame was reconstituted. This variant was isolated from two patients with severe disease and further increased the ability of ORF3b to suppress interferon induction. Thus, our findings not only help to explain the poor interferon response in COVID-19 patients but also describe the emergence of natural SARS-CoV-2 quasispecies with an extended ORF3b gene that may potentially affect COVID-19 pathogenesis.
Keywords: SARS-CoV-2, COVID-19, ORF3b, type I interferon, evolution
Graphical Abstract
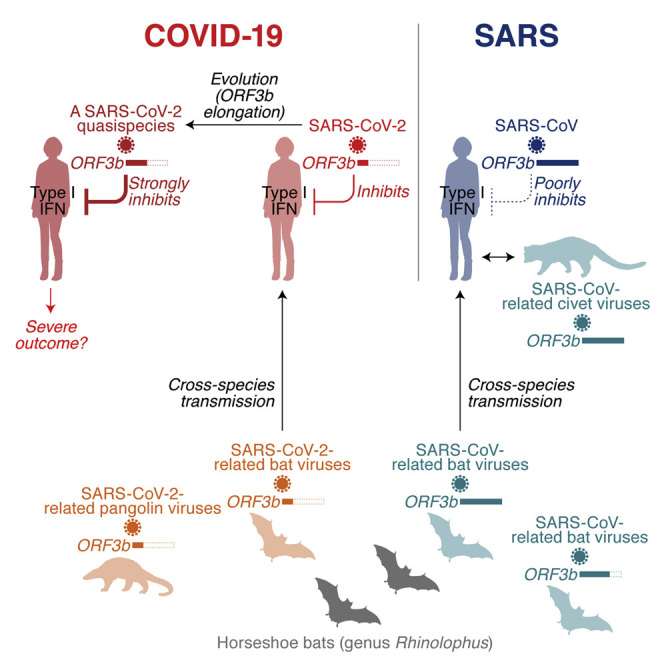
COVID-19 pathogenesis is characterized by impaired IFN responses. Konno et al. identify ORF3b proteins of SARS-CoV-2 and related animal viruses as IFN antagonists. Their anti-IFN activity depends on the C-terminal length, and a natural ORF3b variant with increased IFN-suppressive activity was isolated from two severe COVID-19 cases.
Introduction
In December 2019, an unusual outbreak of infectious pneumonia was reported in the city of Wuhan, Hubei, China. A few weeks later, a novel coronavirus (CoV) was identified as the causative agent and the disease was termed coronavirus disease 2019 (COVID-19) (Zhou et al., 2020c). Because this novel virus is phylogenetically related to severe acute respiratory syndrome (SARS) CoV (SARS-CoV), it was named SARS-CoV-2. As of July 2020, SARS-CoV-2 causes an ongoing pandemic, with more than 15 million reported cases and more than 600,000 deaths worldwide (WHO, 2020).
SARS-CoV-2 infection may be asymptomatic or result in flu-like symptoms such as fever, cough, and fatigue (Chen et al., 2020). In some cases, however, COVID-19 progresses to severe pneumonia and death (Guan et al., 2020; Hui et al., 2020; Li et al., 2020). Although it is still challenging to assess the morbidity rate of COVID-19, estimates range from 1.4 to 1.9% in China (Guan et al., 2020; Verity et al., 2020). This is substantially lower than the morbidity rate of SARS-CoV, which is about 9.6% (WHO, 2004). SARS-CoV, which frequently causes severe respiratory symptoms including fatal pneumonia, first emerged in Guangdong, China, in 2002 and was stamped out in 2004 (reviewed in Chan-Yeung and Xu, 2003; Weiss, 2020). Until then, 8,096 cases of SARS were reported in 29 countries and territories, and 774 people died (WHO, 2004). Thus, SARS-CoV is more virulent than SARS-CoV-2.
SARS-CoV-2 and SARS-CoV are phylogenetically closely related, both belonging to the family Coronaviridae, genus Betacoronavirus, and subgenus Sarbecovirus (Lam et al., 2020; Zhou et al., 2020c). Both viruses were initially transmitted from animals to humans. Thus, elucidating their zoonotic origin and phylogenetic history may help to understand genetic and phenotypic differences between SARS-CoV and SARS-CoV-2. Viruses closely related to SARS-CoV were detected in Chinese rufous horseshoe bats (Rhinolophus sinicus) (Lau et al., 2005; Li et al., 2005) and palm civets (Paguma larvata) (Wang et al., 2005). Subsequent surveillance studies identified additional clades of SARS-CoV-related viruses in various bat species (mainly of the genus Rhinolophus) (Ge et al., 2013; He et al., 2014; Hu et al., 2017a; Lau et al., 2010; Lin et al., 2017; Tang et al., 2006; Wang et al., 2017; Wu et al., 2016; Yuan et al., 2010), suggesting that zoonotic coronavirus transmission from horseshoe bats to humans led to the emergence of SARS-CoV. Similarly, SARS-CoV-2-related viruses were identified in intermediate horseshoe bats (Rhinolophus affinis) (Zhou et al., 2020c), a Malayan horseshoe bat (Rhinolophus malayanus) (Zhou et al., 2020b), and Malayan pangolins (Manis javanica) (Lam et al., 2020; Xiao et al., 2020). Although it has been suggested that the SARS-CoV-2 outbreak originated from cross-species coronavirus transmission from these mammals to humans, the exact origin remains to be determined (Andersen et al., 2020).
One prominent feature that distinguishes COVID-19 from SARS in terms of immune responses is the poor induction of a type I interferon (IFN-I) response by SARS-CoV-2 compared with SARS-CoV and influenza A virus (IAV) (Blanco-Melo et al., 2020; Hadjadj et al., 2020). Impaired IFN-I responses are associated with COVID-19 (Hadjadj et al., 2020). However, the molecular mechanisms underlying the inefficient IFN-I responses in SARS-CoV-2 infection remain unclear. In this study, we therefore aimed to characterize the viral factor or factors determining immune activation upon SARS-CoV-2 infection. We particularly focused on differences in putative viral IFN-I antagonists and revealed that the ORF3b gene products of SARS-CoV-2 and SARS-CoV differ considerably not only in their length but also in their ability to antagonize type I IFN. Furthermore, we demonstrate that the potent anti-IFN-I activity of SARS-CoV-2 ORF3b is found in related viruses from bats and pangolins. Mutational analyses revealed that the length of the C terminus determines the efficacy of IFN antagonism by ORF3b. Finally, we describe a natural SARS-CoV-2 variant with further increased ORF3b-mediated anti-IFN-I activity that emerged during the current COVID-19 pandemic.
Results
SARS-CoV-2 ORF3b Is a Potent IFN-I Antagonist
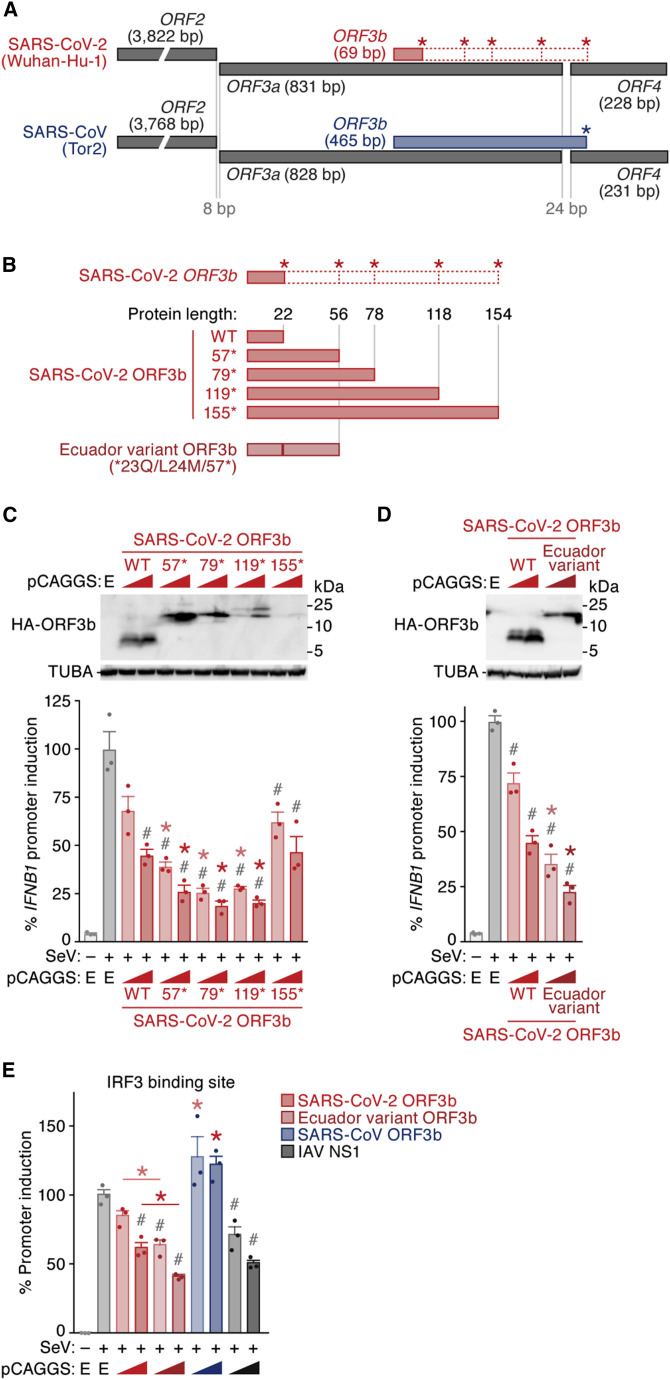
To determine virological differences between SARS-CoV-2 and SARS-CoV, we set out to compare the sequences of diverse sarbecoviruses. Consistent with recent reports (Lam et al., 2020; Zhou et al., 2020c), sarbecoviruses clustered into two groups, SARS-CoV-2-related and SARS-CoV-related viruses (Figures 1 A and S1; the sequences used are listed in Table S1). A comparison of individual viral open reading frames (ORFs) revealed that the length of ORF3b is clearly different between SARS-CoV-2 and SARS-CoV lineages, whereas the lengths of all remaining ORFs are relatively constant among sarbecoviruses (Figure 1B). More specifically, the ORF3b sequences of SARS-CoV-2 and related viruses in bats and pangolins are only 22 amino acids long (69 bp, including stop codon) and therefore considerably shorter than those of their SARS-CoV orthologs (153 amino acids on average).
Figure 1.
SARS-CoV-2 ORF3b Is a Potent IFN-I Antagonist
(A) Maximum likelihood phylogenetic tree of full-length Sarbecovirus sequences. The full-length sequences (~30,000 bp) of SARS-CoV-2 (Wuhan-Hu-1 as a representative), SARS-CoV-2-related viruses from bats (n = 4) and pangolins (n = 4), SARS-CoV (n = 190), SARS-CoV-related viruses from civets (n = 3) and bats (n = 54), and outgroup viruses (n = 2; BM48-31 and BtKY72) were analyzed. Accession number, strain name, and host of each virus are indicated for each branch. Branches including SARS-CoV (n = 190) and SARS-CoV-related viruses from civets (n = 3) were collapsed for better visualization. The uncollapsed tree is shown in Figure S1, and the sequences used are summarized in Table S1. A scale bar indicates 0.1 nucleotide substitutions per site. NA, not applicable.
(B) Comparison of the protein lengths of Sarbecovirus ORFs. The amino acid numbers of ORF1a, S (ORF2), ORF3a, ORF3b, E (ORF4), M (ORF5), ORF6, ORF7a, and N (ORF9a) of sarbecoviruses are shown. The viral sequences used correspond to those in (A). Bars indicate average values, and each dot represents one viral strain. ORFs with low similarity (e.g., ORF8 and ORF9b) were excluded from this analysis.
(C and D) Potent anti-IFN-I activity of SARS-CoV-2 ORF3b. (C) HEK293 cells were cotransfected with five amounts of plasmids expressing hemagglutinin (HA)-tagged SARS-CoV-2 ORF3b, SARS-CoV ORF3b, and IAV NS1 (50, 100, 200, 300, or 500 ng) and p125Luc, a plasmid encoding firefly luciferase under the control of the human IFNB1 promoter (500 ng). 24 h post-transfection, SeV was inoculated at MOI 10. 24 h post-infection, the cells were harvested for western blotting (top), dot blotting (middle), and luciferase assay (bottom). For western blotting, the input of cell lysate was normalized to anti-alpha-tubulin (TUBA), and one representative result of three independent experiments is shown. The band of each viral protein is indicated by a white arrowhead. kDa, kilodalton. In the luciferase assay, the value of the SeV-infected empty-vector-transfected cells was set to 100%. (D) A549 cells were electroporated with a plasmid expressing HA-tagged SARS-CoV-2 ORF3b, SARS-CoV ORF3b, or IAV NS1 (500 ng). 24 h post-transfection, SeV was inoculated at MOI 10. 24 h post-infection, the cells were harvested for western blotting (top) and real-time RT-PCR (bottom). For western blotting, the input of cell lysate was normalized to TUBA, and one representative result of three independent experiments is shown. ORF3b and NS1 were run on separate blots for better visualization. Figure S2A shows them on the same blot with high and low exposure. For qRT-PCR, the expression levels of endogenous IFNB1 and GAPDH were quantified. For the luciferase assay (C) and real-time RT-PCR (D), mean values of three independent experiments with SEM are shown, and statistically significant differences (p < 0.05) compared with the SeV-infected empty-vector-transfected cells (#) and the same amount of the SARS-CoV-2 ORF3b-transfected cells (∗) are shown. E, empty vector.
See also Figures S1 and S2 and Table S1.
Previous studies on SARS-CoV and related viruses demonstrated that at least two accessory proteins, ORF3b and ORF6, as well as the nucleocapsid (N, also known as ORF9a) have the ability to inhibit IFN-I production (Frieman et al., 2007; Hu et al., 2017b; Kopecky-Bromberg et al., 2007; Zhou et al., 2012). Because the ORF3b length is remarkably different between SARS-CoV-2 and SARS-CoV (Figure 1B), we hypothesized that the antagonistic activity of ORF3b against IFN-I also differs between these two viruses. To test this hypothesis, we monitored human IFNB1 promoter activity in the presence of ORF3b of SARS-CoV-2 (Wuhan-Hu-1) and SARS-CoV (Tor2) using a luciferase reporter assay. The IAV non-structural protein 1 (NS1) served as positive control (García-Sastre et al., 1998; Krug et al., 2003). As shown in Figure 1C, all three viral proteins dose-dependently suppressed the activation of the IFNB1 promoter upon Sendai virus (SeV) infection. The antagonistic activity of SARS-CoV-2 ORF3b was slightly but significantly higher than that of SARS-CoV ORF3b (Figure 1C, bottom). To verify this in a different experimental system, we used A549 cells, a human lung cell line, and measured the expression level of endogenous IFNB1 after SeV infection. Here, SARS-CoV-2 ORF3b, but not SARS-CoV ORF3b, significantly suppressed the induction of IFNB1 expression triggered by SeV infection (Figures 1D and S2A). Thus, our data demonstrate that SARS-CoV-2 ORF3b is a potent inhibitor of human IFN-I activation, even though it only comprises 22 amino acids.
SARS-CoV-2-Related ORF3b Proteins from Bat and Pangolin Viruses Suppress IFN-I Activation on Average More Efficiently Than Their SARS-CoV Counterparts
A phylogenetic analysis of Sarbecovirus ORF3b genes showed that the evolutionary relationship of Sarbecovirus ORF3b genes was similar to that of the full-length viral genomes (Figures 1A and 2A). Because the lengths of ORF3b proteins in SARS-CoV-2-related viruses, including those from bats and pangolins, were on average shorter than those from SARS-CoV and related viruses (Figure 1B), we next analyzed the variation of the ORF3b length in diverse sarbecoviruses. As shown in Figure 2B, most SARS-CoV-2 ORF3b proteins (16,966/16,970) had a length of 22 amino acids. Likewise, all available ORF3b proteins of SARS-CoV-2-related viruses from bats and pangolins are 22 amino acids. These observations demonstrate that the ORF3b length is highly conserved in SARS-CoV-2 and related viruses. Similarly, the length of ORF3b is highly conserved in SARS-CoV (185/190) and related viruses from civets (3/3), and almost all of them encode a 154-amino-acid ORF3b protein. In contrast, the length of ORF3b is highly variable in SARS-CoV-related bat viruses (Figure 2B; Table S2). Only 2 of the 54 ORF3b proteins of SARS-CoV-related bat viruses (3.7%) are 154 amino acids in length, whereas 50% of them express a 114-amino-acid ORF3b (Figure 2B; Table S2). These findings suggest that there was a founder effect during the cross-species transmission of SARS-CoV-like viruses from bats to humans, which resulted in the spread of a virus encoding a 154-amino-acid ORF3b protein.
Figure 2.
C-terminal Truncations Increase the IFN-Antagonistic Activity of ORF3b
(A) Maximum likelihood phylogenetic tree of Sarbecovirus ORF3b. The ORF3b sequences of SARS-CoV-2 (Wuhan-Hu-1), SARS-CoV-2-related viruses from bats (RmYN02, RaTG13, ZXC21, and ZC45) and pangolins (P5L, P1E, P4L, and Pangolin-coV), SARS-CoV (Tor2, GZ0402, GZ02, Urbani, BJ02, BJ01, BJ182-4, P3pp1, and P3pp46), SARS-CoV-related viruses from civets (civet007) and bats (Rs7327, YN2013, Rs4231, YNLF34C, Shaanxi2011, Rm1, F46, HKU3-2, GX2013, and Yunnan2011), and two outgroup viruses (BM48-31 and BtKY72) were analyzed. The ORF3b sequences of all SARS-CoV-related viruses are summarized in Table S2, and the ORF3b sequences used in this study are summarized in Table S3. Strain name and host of each virus are indicated for each branch. ∗Bootstrap value > 70%.
(B) Proportion of the ORF3b lengths in each Sarbecovirus. The distribution of different lengths of ORF3b in each viral group is summarized in pie charts. The number in parentheses (n) indicates the number of sequences used in this analysis. The numbers at the pie charts give the protein length indicated, and the numbers in bold indicate the most prevalent protein length for each viral group.
(C) Anti-IFN-I activities of different Sarbecovirus ORF3b proteins. (Top) Illustration of protein lengths of 25 Sarbecovirus ORF3b isolates used in this study. The information of the 25 Sarbecovirus ORF3b isolates is summarized in Table S3. (Middle and bottom) HEK293 cells were cotransfected with a plasmid expressing one of 25 HA-tagged Sarbecovirus ORF3b proteins (summarized in B) (100 ng) and p125Luc (500 ng). 24 h post-transfection, SeV was inoculated at MOI 10. 24 h post-infection, cells were harvested for western blotting and dot blotting (middle) and for luciferase assay (bottom). The amino acid sequences of ZXC21 and ZC45 are identical. An uncropped dot blot is shown in Figure S2B.
(D) Subcellular localization of Sarbecovirus ORF3b. Cell lysates of the HEK293 cells transfected with a plasmid expressing HA-tagged Sarbecovirus ORF3b were separated into cytosolic and nuclear fractions as described in the STAR Methods section. The percentage of ORF3b protein localized in the nucleus (top, n = 4) and a representative western blot (bottom) are shown. TUBA and anti-lamin A/C (LMNA) were used as controls for cytosolic and nuclear proteins. ORF3b proteins of HKU3-2, GX2013, and Yunnan2011 were not used in this experiment because these ORF3b proteins were only poorly expressed.
(E–G) Anti-IFN-I activity of C-terminally truncated SARS-CoV ORF3b. (E) Illustration of the ORF3b mutants of SARS-CoV (Tor2) and a SARS-CoV-related bat virus (Rs4231). NLS, nuclear localization signal of c-Myc (PAAKRVKLD). (F) Subcellular localization of the ORF3b mutants. Cell lysates of the HEK293 cells transfected with a plasmid expressing HA-tagged ORF3b mutants were separated into cytosol or nuclear fractions as described in the STAR Methods section. The percentage of ORF3b protein localized in the nucleus (top, n = 4) and a representative western blot (bottom) are shown. TUBA and LMNA were used as controls for cytosolic and nuclear proteins. (G) HEK293 cells were cotransfected with a plasmid expressing the indicated Sarbecovirus ORF3b proteins (50 or 100 ng) and p125Luc (500 ng). 24 h post-transfection, SeV was inoculated at MOI 10. 24 h post-infection, cells were harvested for western blotting (top) and luciferase assay (bottom).
(H) Subcellular localization of ORF3b and IRF3. HeLa cells were transfected with the indicated plasmids expressing HA-ORF3b and were infected with SeV as described in the STAR Methods section. Representative figures are shown. Scale bar, 10 μm. The white circles in IRF3 and DAPI panels indicate the nuclear rims of cells expressing HA-ORF3b.
For western blotting, the input of cell lysate was normalized to TUBA. One representative blot of three independent experiments is shown. For the luciferase assay, the value of the SeV-infected empty-vector-transfected cells was set to 100%. The mean values of three independent experiments with SEM are shown, and statistically significant differences (p < 0.05) compared with the SeV-infected empty-vector-transfected cells (#) are shown. In (C), red asterisks indicate statistically significant differences (p < 0.05) compared SARS-CoV-2 Wuhan-Hu-1 ORF3b-transfected cells. In (G), blue and green asterisks indicate statistically significant differences (p < 0.05) compared the same amount of either Tor2 ORF3b L115∗-transfected cells or Rs4231 ORF3b WT-transfected cells. E, empty vector.
To elucidate a potential relationship between ORF3b length and function, we compared diverse Sarbecovirus ORF3b for their ability to suppress IFN-I. For our analyses, we generated expression plasmids for ORF3b from SARS-CoV-2-related viruses from bats (RmYN02, RaTG13, and ZXC21) and a pangolin (P4L). Furthermore, we included ORF3b from nine SARS-CoV isolates (Tor2, GZ0402, GZ02, Urbani, BJ02, BJ01, BJ182-4, P3pp1, and P3pp46), a SARS-CoV-related virus from a civet (civet007), and ten SARS-CoV-related viruses from bats (Rs7327, YN2013, Rs4231, YNLF34C, Shaanxi2011, Rm1, F46, HKU3-2, GX2013, and Yunnan2011), covering essentially all length variants of this protein (Figure 2C, top; see also Tables S2 and S3). All four SARS-CoV-2-related ORF3b tested, as well as SARS-CoV-2 ORF3b (Wuhan-Hu-1), significantly suppressed human IFN-I activation (Figure 2C, bottom). In contrast, only three of the nine SARS-CoV ORF3b proteins (BJ182-4, P3pp1, and P3pp46) exhibited anti-IFN activity at the concentrations tested (Figure 2C, bottom). Intriguingly, these three SARS-CoV ORF3b proteins are C-terminally truncated and shorter than ORF3b of the reference strain SARS-CoV Tor2 (Figure 2C, top; Table S2). Similarly, only five of the ten ORF3b proteins of SARS-CoV-related viruses from bats (Rs4231, YNLF34C, Shaanxi2011, Rm1, and F46) exhibited significant anti-IFN-I effects, and all these ORF3b proteins were shorter than 114 amino acids (Figure 2C). Although three additional ORF3b proteins of SARS-CoV-related viruses from bats (HKU3-2, GX2013, and Yunnan2011) were shorter than 39 amino acids, they did not exhibit anti-IFN-I activity, most likely because of their poor expression and/or stability (Figures 2C and S2B). Altogether, these findings suggest that the C-terminal region (residues 115–154) attenuates the anti-IFN-I activity of ORF3b.
A previous study reported the presence of a nuclear localization signal (NLS) is the C terminus of SARS-CoV ORF3b that is absent from its SARS-CoV-2 ortholog (Yuan et al., 2010). Consistent with this, PSORT II Prediction (Horton and Nakai, 1997) identified amino acid residues 135–153 of SARS-CoV ORF3b as putative NLS. To address the possibility that subcellular localization of ORF3b may be associated with its anti-IFN-I activity, we performed subcellular fractionation experiments. As shown in Figure 2D, all ORF3b proteins exhibiting significant anti-IFN-I activity were mainly localized in the cytosol, whereas their inactive or poorly active counterparts were found to similar levels in both the cytosol and the nucleus. These results suggest that the presence of an NLS in the C terminus negatively interferes with the IFN-I-antagonistic activity of ORF3b. To further test this hypothesis, we generated two derivatives of SARS-CoV (Tor2) ORF3b: a C-terminally truncated version harboring a premature stop codon at position 115 (L115∗), thereby mimicking ORF3b of Rs4231 (a SARS-CoV-related bat virus), as well as a variant thereof additionally harboring the NLS of c-Myc (Ray et al., 2015) at its C terminus (L115+NLS) (Figure 2E). Furthermore, we attached the c-Myc NLS to the C terminus of Rs4231 ORF3b (Figure 2E). As expected, SARS-CoV Tor2 ORF3b L115∗, as well as Rs4231 wild type (WT), mainly localized to the cytosol, whereas the two mutants harboring the c-Myc NLS were localized to similar levels in both the cytosol and the nucleus (Figure 2F). Reporter assays showed that the SARS-CoV Tor2 L115∗ mutant exhibits significantly higher anti-IFN-I activity than WT SARS-CoV Tor2 ORF3b although both are expressed at similar levels (Figure 2G). Moreover, the anti-IFN-I activity of both Tor2 L115∗ and Rs4231 ORF3b was attenuated by the addition of an NLS (Figure 2G), suggesting that cytosolic localization of ORF3b is important to exhibit anti-IFN-I activity. Consistent with the biochemical assays (Figures 2D and 2F), immunofluorescence microscopy showed that WT SARS-CoV-2 ORF3b (Wuhan-Hu-1) and the SARS-CoV ORF3b L115∗ mutant are mainly localized in the cytosol, whereas WT SARS-CoV ORF3b (Tor2) and the Tor2 L115+NLS mutant reside in both the cytosol and the nucleus (Figure 2H). In parallel, we monitored the subcellular localization of IRF3, because this transcription factor is a key regulator of IFNB1 expression (reviewed in Park and Iwasaki, 2020) that has previously been shown to be modulated by coronavirus ORF3b orthologs (Zhou et al., 2012). Nuclear translocation of IRF3 was strongly impaired in the presence of WT SARS-CoV-2 ORF3b and the SARS-CoV ORF3b L115∗ mutant but less so by WT SARS-CoV ORF3b and its L115+NLS mutant (Figure 2H). Collectively, these findings demonstrate that the C-terminal region of SARS-CoV ORF3b attenuates its anti-IFN-I activity by impairing its ability to prevent the translocation of IRF3 into the nucleus.
A SARS-CoV ORF3b-like Sequence Is Hidden in the SARS-CoV-2 Genome
ORF3b of SARS-CoV-2 is shorter than its ortholog in SARS-CoV (Figures 1B and 2A). However, when closely inspecting the genomes of these two viruses, we noticed that the SARS-CoV-2 nucleotide sequence downstream of the stop codon of ORF3b shows a high similarity to the SARS-CoV ORF3b gene (nucleotide similarity = 79.5%; Figures 3 A and S3A). In contrast to SARS-CoV ORF3b, however, SARS-CoV-2 harbors four premature stop codons that result in the expression of a drastically shortened ORF3b protein (Figures 3A and S3A). Similar patterns were observed in SARS-CoV-2-related viruses from bats and pangolins (Figure S3B). Because the ORF3b length is closely associated with its anti-IFN-I activity (Figures 2C and 2D), we hypothesized that reversion of the premature stop codons in SARS-CoV-2 ORF3b affects its ability to inhibit human IFN-I. To test this, we generated four SARS-CoV-2 ORF3b derivatives, 57∗, 79∗, 119∗, and 155∗, lacking one to four premature stop codons (Figures 3B, top, and S3A). As shown in Figure 3C, all four derivatives inhibited human IFN-I activation in a dose-dependent manner. Consistent with the results obtained with SARS-CoV ORF3b mutants (Figure 2D), the 155∗ mutant, comprising the C-terminal region (positions 119–154), was poorly expressed and exhibited relatively low anti-IFN-I activity (Figures 3C and S2C). However, we found that the extended ORF3b derivatives, particularly 57∗, 79∗, 119∗, exhibited higher anti-IFN-I activity than WT SARS-CoV-2 ORF3b (Figure 3C). These findings confirm that the length of ORF3b determines its ability to suppress an IFN-I response. Furthermore, they show that the loss of individual ORF3b stop codons during the current SARS-CoV-2 pandemic may result in the emergence of viral variants with enhanced IFN-I-antagonistic activity.
Figure 3.
Enhanced Anti-IFN-I upon Reconstitution of the Cryptic SARS-CoV-2 ORF3b
(A) Schemes illustrating the genomic regions encoding ORF2, ORF3a, ORF3b, and ORF4 of SARS-CoV-2 and SARS-CoV. Open squares with dotted red lines indicate a cryptic ORF3b reading frame in SARS-CoV-2 that is similar to SARS-CoV ORF3b (see also Figure S3A). Asterisks indicate stop codons in the ORF3b frame.
(B) SARS-CoV-2 ORF3b derivatives characterized in this study. (Top) WT SARS-CoV-2 ORF3b and four derivatives with mutated stop codons (57∗, 79∗, 119∗, and 155∗) are shown. Asterisks indicate the stop codons in the original ORF3b frame. (Bottom) A natural ORF3b variant detected in two sequences deposited in GISAID (GISAID: EPI_ISL_422564 and EPI_ISL_422565, herein designated an Ecuador variant) are shown. The corresponding nucleotide and amino acid sequences are shown in Figure S3C.
(C) Anti-IFN-I activity of different SARS-CoV-2 ORF3b derivatives. HEK293 cells were cotransfected with two amounts of plasmids expressing the indicated HA-tagged SARS-CoV-2 ORF3b derivatives (WT, 57∗, 79∗, 119∗, and 155∗; 50 and 100 ng) and p125Luc (500 ng). 24 h post-transfection, SeV was inoculated at MOI 10. 24 h post-infection, the cells were harvested for western blotting (top) and luciferase assay (bottom).
(D) Enhanced anti-IFN-I activity of an Ecuador variant ORF3b. HEK293 cells were cotransfected with two amounts of plasmids expressing HA-tagged Ecuador variant ORF3b or parental SARS-CoV-2 ORF3b (50 and 100 ng) and p125Luc (500 ng). 24 h post-transfection, SeV was inoculated at MOI 10. 24 h post-infection, the cells were harvested for western blotting (top) and luciferase assay (bottom).
(E) Enhanced inhibition of the IRF3-mediated IFN-I activation by the Ecuador variant ORF3b. HEK293 cells were cotransfected with two amounts of plasmids expressing the indicated HA-tagged viral proteins (50 and 100 ng) and p55C1B-Luc (500 ng). 24 h post-transfection, SeV was inoculated at MOI 10. 24 h post-infection, the cells were harvested for luciferase assay.
For western blotting, the input of cell lysate was normalized to TUBA. One representative blot of three independent experiments is shown. A highly exposed blot visualizing the band of the 155∗ mutant is shown in Figure S2C. kDa, kilodalton. For the luciferase assay, the value of the SeV-infected empty-vector-transfected cells was set to 100%. The mean values of three independent experiments with SEM are shown, and statistically significant differences (p < 0.05) compared with the SeV-infected empty-vector-transfected cells (#) and the same amount of the SARS-CoV-2 ORF3b WT-transfected cells (∗) are shown. E, empty vector; NS, no significant difference.
See also Figures S2 and S3 and Table S4.
Characterization of a Natural SARS-CoV-2 ORF3b Variant with Enhanced Anti-IFN-I Activity
We then assessed the diversity of SARS-CoV-2 ORF3b during the current pandemic. A comprehensive analysis of approximately 17,000 viral sequences deposited in GISAID (https://www.gisaid.org; as of April 22, 2020) using the CoV-GLUE webtool (http://cov-glue.cvr.gla.ac.uk) revealed that the ORF3b gene is highly conserved (Figure S2D; Table S4). However, we detected two viral sequences (GISAID: EPI_ISL_422564 and EPI_ISL_422565) in which the ORF3b gene was extended because of the loss of the first premature stop codon (∗23Q) (Figures 3B, bottom, and S3C; Table S4). Table 1 summarizes the clinical information for these two patients. Both cases were part of a family cluster in Quito, Ecuador, and presented with particularly severe COVID-19 symptoms, including quick clinical deterioration and lower levels of oxygen saturation in blood. One of the patients (EPI_ISL_422565) presented elevated levels of D-dimer, a sign of hypercoagulability and inflammation in COVID-19 patients (Yu et al., 2020; Zhou et al., 2020a), and ultimately succumbed to disease. The similarity of the full-length sequences of these two viruses, which were collected from COVID-19 patients in Ecuador, is >99.6%, and the ORF3b sequences are identical. Apart from the ∗23Q mutation, the Ecuador variant harbors an L24M change compared with the SARS-CoV ORF3b-like sequence in SARS-CoV-2 (Wuhan-Hu-1 [GISAID: NC_045512.2], nucleotides 25,814–26,281) (Figure 3B, bottom; see also Figure S2D). IFNβ reporter assays revealed that the Ecuador variant ORF3b exhibits significantly higher anti-IFN-I activity than the parental SARS-CoV-2 ORF3b (Figure 3D).
Table 1.
Clinical and Laboratory Findings of Patients EPI_ISL_422564 and EPI_ISL_422565
| EPI_ISL_422564 | EPI_ISL_422565 | |
|---|---|---|
| Age | 39 | 40 |
| Gender | Male | Male |
| Fever | + | + |
| Headache | − | − |
| Cough | + | + |
| Odynophagia | + | − |
| Anosmia | − | − |
| Nausea | − | − |
| Diarrhea | − | − |
| D-dimera (ng/mL) | 326 | 4,172 |
| D-dimerb (ng/mL) | 210 | 3,630 |
| Troponinea (pg/mL) | N/A | 7.4 |
| LDH (lactate dehydrogenase)a (UI/L) | 252 | 431 |
| Leukocytesa (/μL) | 12,000 | 9,600 |
| Lymphocytesa (/μL) | 600 | 500 |
| Lymphocytesa (%) | 5 | 5.5 |
| Plateletsa (/μL) | 448,000 | 264,000 |
| Leukocytesb (/μL) | 6,500 | 8,100 |
| Lymphocytesb (/μL) | 2,100 | 800 |
| Lymphocytesb (%) | 32 | 10 |
| Plateletsb (/μL) | 330,000 | 207,000 |
| CRP (C-reactive protein)a | 154 | 109 |
| Procalcitoninea | 0.21 | 0.23 |
| Creatininea (mg/dL) | 1 | 0.89 |
| AST (aspartate transaminase) (U/L) | 20 | 45 |
| ALT (alanine transaminase) (U/L) | 35 | 62 |
| Outcome | Recovered | Deceased |
N/A, not analyzed.
Time of hospital admission.
Time of sampling.
Because we found that SARS-CoV-2 ORF3b hampers the nuclear translocation of IRF3 (Figure 2H), we investigated the ability of WT SARS-CoV-2 ORF3b, the Ecuador variant ORF3b, as well as SARS-CoV ORF3b and IAV NS1 to suppress IRF3-driven gene expression. Luciferase reporter assays revealed that the inhibitory activity of WT SARS-CoV-2 ORF3b was significantly higher than that of SARS-CoV ORF3b (Figure 3E). Importantly, the Ecuador variant ORF3b was even more effective and suppressed IRF3-driven gene expression as efficiently as IAV NS1 (Figure 3E). Altogether, these findings show that a naturally occurring SARS-CoV-2 variant, expressing an elongated ORF3b protein with enhanced anti-IFN activity, has already emerged during the current SARS-CoV-2 pandemic and more potently hampers IRF3-mediated IFN-I activation than parental SARS-CoV-2 ORF3b.
Discussion
Here, we demonstrate that SARS-CoV-2 ORF3b is a potent antagonist of human IFN-I activation. On average, ORF3b proteins from SARS-CoV-2 and related bat and pangolin viruses were more active than their SARS-CoV counterparts. Our findings may help to explain the inefficient and delayed IFN-I responses in SARS-CoV-2-infected cells, as well as COVID-19 patients (Blanco-Melo et al., 2020). Moreover, a recent study showed that impaired IFN-I responses and reduced IFN-stimulated gene expression are associated with severe COVID-19 (Hadjadj et al., 2020). This suggests that imbalanced IFN-I responses against SARS-CoV-2 infection may determine its pathogenicity and explain differences compared with SARS-CoV, and it is tempting to speculate that atypical symptoms and poor IFN-I responses in SARS-CoV-2 infection may at least to some extent be attributed to the potent anti IFN-I activity of its ORF3b.
Like SARS-CoV-2 ORF3b, its orthologs in SARS-CoV-2-related viruses from bats and pangolins efficiently antagonize IFN-I and are generally truncated because of the presence of several premature stop codons. In contrast, the anti-IFN activity of ORF3b proteins encoded by some SARS-CoV-related viruses is attenuated, most likely due to an elongated C terminus. We hypothesized that the ORF3b length variation in SARS-CoV-like viruses may be the result of recombination events. In line with this, sarbecoviruses seem to easily recombine with each other (Andersen et al., 2020; Lam et al., 2020; Zhou et al., 2020c), and some horseshoe bat species such as Rhinolophus affinis and Rhinolophus sinicus are known to harbor both SARS-CoV-2-related and SARS-CoV-related viruses (Andersen et al., 2020; Zhou et al., 2020c). Nevertheless, the phylogenetic topologies of the full-length viral genome and the ORF3b gene are similar, and we found no evidence for recombination of ORF3b between the lineages of SARS-CoV-2 and SARS-CoV. Phenotypic differences in the ability of ORF3b to suppress IFN-I responses may also be associated with the likelihood of successful zoonotic transmission of sarbecoviruses to humans, because many IFN-stimulated genes are antagonized in a species-specific manner. Although more than 50 SARS-CoV-related viruses were isolated from bats (Ge et al., 2013; He et al., 2014; Hu et al., 2017a; Lau et al., 2005, 2010; Li et al., 2005; Lin et al., 2017; Tang et al., 2006; Wang et al., 2017; Wu et al., 2016; Yuan et al., 2010), only eight viral sequences belonging to the SARS-CoV-2 lineage have been detected so far (Andersen et al., 2020; Lam et al., 2020; Xiao et al., 2020; Zhou et al., 2020b, 2020c). Thus, further investigations are needed to elucidate the dynamics of cross-species transmission events of sarbecoviruses and the evolution of the ORF3b gene.
Nevertheless, there is evidence for a founder effect on ORF3b length between SARS-CoV and its putative ancestral viruses in bats. Although ORF3b lengths are highly variable in SARS-CoV-related viruses in bats, and a length of 114 amino acids is prevailing, almost all (97.4%; 185/190) SARS-CoV ORF3b variants are 154 amino acids (Figure 2B; Table S2). These observations suggest that a virus encoding a 154-amino-acid ORF3b with poor anti-IFN-I activity was transmitted from bats to humans and resulted in the emergence of SARS-CoV in 2002.
We further show that a SARS-CoV ORF3b-like sequence is still present in the SARS-CoV-2 genome but is interrupted by premature stop codons. We demonstrate that a partial extension of SARS-CoV-2 ORF3b by reverting stop codons increases its inhibitory activity against human IFN-I. However, full reversion of all stop codons resulted in an ORF3b protein with poor anti-IFN activity. This is in line with the phenotypic difference between SARS-CoV-2 and SARS-CoV ORF3b proteins and suggests that the C terminus of ORF3b impairs its immune evasion activity.
Importantly, we also identified a naturally occurring SARS-CoV-2 ORF3b variant that expresses an elongated protein due to the loss of the first premature stop codon. This variant suppresses IFN-I even more efficiently than ORF3b of the SARS-CoV-2 reference strain. In agreement with an association of IFN suppression with disease severity (Hadjadj et al., 2020), the two patients in Ecuador harboring SARS-CoV-2 with the extended ORF3b variant were critically ill; one (GISAID: EPI_ISL_422564) was treated in an intensive care unit, and the other one (GISAID: EPI_ISL_422565) died of COVID-19 (Table 1). However, there is no direct evidence indicating that the viruses detected in these two COVID-19 patients in Ecuador are more pathogenic than the reference strain. Although we cannot tell whether this variant is associated with a different outcome in disease, it might be plausible to assume that naturally occurring length variants of ORF3b occur because of the loss of premature stop codons and potentially contribute to the emergence of more pathogenic SARS-CoV-2 variants. Thus, it will be important to continue monitoring viral sequences to see whether novel ORF3b variants emerge during the current pandemic.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| HRP-conjugated anti-HA | Roche | Cat# 12013819001; RRID: AB_390918 |
| Anti-alpha-Tubulin (TUBA) | Sigma-Aldrich | Cat# T9026; RRID: AB_477593 |
| Anti-lamin A/C (LMNA) | Cell Signaling Technology | Cat# 2032S; RRID: AB_2136278 |
| HRP-conjugated anti-mouse IgG | Cell Signaling Technology | Cat# 7076; RRID: AB_330924 |
| HRP-conjugated anti-rabbit IgG | Cell Signaling Technology | Cat# 7074S; RRID: AB_2099233 |
| FITC-conjugated anti-HA | Roche | Cat# 11988506001; RRID: AB_390916 |
| Anti-IRF3 | Abcam | Cat# ab68481; RRID: AB_11155653 |
| Alexa Fluor 546-conjugated anti-rabbit IgG | Thermo Fisher Scientific | Cat# A-11010: RRID: AB_2534077 |
| Bacterial and Virus Strains | ||
| SeV (strain Cantell, clone cCdi) | (Yoshida et al., 2018) | GenBank accession no. AB855654 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Dulbecco’s modified Eagle’s medium | Sigma-Aldrich | Cat# D6046-500ML |
| Ham’s F-12K medium | Thermo Fisher Scientific | Cat# 21127022 |
| Fetal calf serum | Sigma-Aldrich | Cat# 172012-500ML |
| Penicillin streptomycin | Sigma-Aldrich | Cat# P4333-100ML |
| L-glutamate | Thermo Fisher Scientific | Cat# 25030081 |
| PrimeSTAR GXL DNA polymerase | Takara | Cat# R050A |
| EcoRI | Takara | Cat# 1040A |
| XhoI | Takara | Cat# 1094A |
| BglII | Takara | Cat# 1021A |
| PEI Max | Polysciences | Cat# 24765-1 |
| FuGENE HD transfection reagent | Promega | Cat# E2312 |
| ProLong diamond antifade mountant with DAPI | Thermo Fisher Scientific | Cat# P36971 |
| SuperScript III reverse transcriptase | Thermo Fisher Scientific | Cat# 18080085 |
| DNase I, Amplification Grade | Thermo Fisher Scientific | Cat# 18047019 |
| Power SYBR Green PCR Master Mix | Thermo Fisher Scientific | Cat# 4367659 |
| Critical Commercial Assays | ||
| PicaGene BrillianStar-LT luciferase assay system | Toyo-b-net | Cat# BLT1000 |
| CellTiter-Glo 2.0 assay kit | Promega | Cat# G9241 |
| Nuclear/cytosol fractionation kit | BioVision | Cat# K266 |
| QIAamp RNA blood mini kit | QIAGEN | Cat# 52304 |
| Experimental Models: Cell Lines | ||
| Human: HEK293 cells | ATCC | CRL-1573 |
| Human: A549 cells | ATCC | CCL-185 |
| Oligonucleotides | ||
| Primers for plasmid construction, see Table S5 | This study | N/A |
| Oligo(dT) 12-18 primer | Thermo Fisher Scientific | Cat# 18418012 |
|
GAPDH forward primer for real-time RT-PCR: ATGGGGAAGGTGAAGGTCG |
This study | N/A |
|
GAPDH reverse primer for real-time RT-PCR: GGGTCATTGATGGCAACAATATC |
This study | N/A |
|
IFNB1 forward primer for real-time RT-PCR: AAACTCATGAGCAGTCTGCA |
This study | N/A |
|
IFNB1 reverse primer for real-time RT-PCR: AGAGGCACAGGCTAGGAGATC |
This study | N/A |
| Recombinant DNA | ||
| Plasmid: pCAGGS | (Niwa et al., 1991) | N/A |
| Sarbecovirus ORF3b, see Table S3 | This study | N/A |
| IAV A/Puerto Rico/8/34 NS1 | This study | GenBank accession no. EF467817.1 |
| Plasmid: p125Luc | (Fujita et al., 1993) | N/A |
| Plasmid: p55C1B-Luc | (Fujita et al., 1993) | N/A |
| Software and Algorithms | ||
| MEGA7 | (Kumar et al., 2016) | https://www.megasoftware.net |
| FigTree | Andrew Rambaut | http://tree.bio.ed.ac.uk/software/figtree |
| Sequencher version 5.1 | Gene Codes Corporation | N/A |
| Prism | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ |
| L-INS-i in the MAFFT version 7.453 | (Katoh and Standley, 2013) | https://mafft.cbrc.jp/alignment/software/ |
| RAxML-NG v. 0.9.0 | (Kozlov et al., 2019) | https://github.com/amkozlov/raxml-ng |
| PSORT II Prediction | (Horton and Nakai, 1997) | https://psort.hgc.jp/form2.html |
| Other | ||
| GISAID | Freunde von GISAID e.V. | https://www.gisaid.org |
| CoV-GLUE | MRC-University of Glasgow Centre for Virus Research | http://cov-glue.cvr.gla.ac.uk |
| Formaldehyde solution | FUJIFILM Wako Chemicals | Cat# 061-00416 |
| Immobilon-P PVDF 0.45-μm membrane | Merck | Cat# IPVH00010 |
| Nitrocellulose 0.20-μm membrane | Bio-Rad | Cat# 1620112 |
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Kei Sato (KeiSato@g.ecc.u-tokyo.ac.jp).
Materials Availability
All unique reagents generated in this study are listed in the Key Resources Table and available from the Lead Contact with a completed Materials Transfer Agreement.
Data and Code Availability
The published article includes all datasets generated or analyzed during this study.
Experimental Model And Subject Details
Ethics Statement
The two viral variants (GISAID accession IDs: EPI_ISL_422564 and EPI_ISL_422565) were isolated from two severely ill patients (a 39-year-old male and a 40-year-old male) treated in the Hospital General del IESS Quito Sur. Both patients belonged to the same family, and two additional family members also presented with COVID-19. All of them were hospitalized in the intensive care unit of the same hospital. Samples were collected on March 30, 2020. The main clinical and laboratory findings from both patients are summarized in Table 1. Sequencing analysis of these samples was approved by the Universidad San Francisco de Quito Bioethics Committee (CEISH) with the number P2020-022IN.
Cell Culture
HEK293 cells (a human embryonic kidney cell line; ATCC CRL-1573) and HeLa cells (a human uterus cervix cell line; ATCC CCL-2) were maintained in Dulbecco’s modified Eagle’s medium (Sigma-Aldrich) containing fetal calf serum and antibiotics. A549 cells (a human lung cell line; ATCC CCL-185) were cultured in Ham’s F-12K medium (Thermo Fisher Scientific) with 10% fetal calf serum and antibiotics.
Method Details
Viral Genomes and Phylogenetic Analyses
All viral genome sequences used in this study and the respective GenBank or GISAID (https://www.gisaid.org) accession numbers are summarized in Table S1. We first aligned the viral genomes using the L-INS-i program of MAFFT version 7.453 (Katoh and Standley, 2013). Based on the multiple sequence alignment and the gene annotation of SARS-CoV, we extracted the region of the ORF3b gene. We then constructed phylogenetic trees using the full-length genomes (Figures 1A and S1) and ORF3b genes (Figure 2A). We generated a maximum likelihood based phylogenetic tree using RAxML-NG version 0.9.0 (Kozlov et al., 2019) with a General Time Reversible model of nucleotide substitution with invariant sites and gamma distributed rate variation among sites. We visualized the tree using a FigTree software (http://tree.bio.ed.ac.uk/software/figtree).
Plasmid Construction
To construct the expression plasmids for HA-tagged Sarbecovirus ORF3b and IAV A/Puerto Rico/8/34 (H1N1 PR8; GenBank accession no. EF467817.1) NS1, pCAGGS (Niwa et al., 1991) was used as a backbone. The HA-tagged ORF of each gene (the accession numbers and sequences are listed in Table S3) and the cryptic SARS-CoV ORF3b-like sequence in SARS-CoV-2 [Wuhan-Hu-1 (GenBank accession no. NC_045512.2), nucleotides 25814-26281, see also Figure S3A) was synthesized by a gene synthesis service (Fasmac). The ORF3b derivatives were generated by PCR using PrimeSTAR GXL DNA polymerase (Takara), the synthesized ORFs as templates, and the primers listed in Table S5. The HA-tagged Ecuador variant ORF3b (GISAID accession IDs: EPI_ISL_422564 and EPI_ISL_422565, which corresponds to the S23Q/L24M mutant of SARS-CoV-2 Wuhan-Hu-1 ORF3b ∗57; see also Figure S3C) was generated by overlap extension PCR by using PrimeSTAR GXL DNA polymerase (Takara), the SARS-CoV-2 ORF3b 155∗ as the template, and the primers listed in Table S5. The obtained DNA fragments were inserted into pCAGGS via EcoRI-BglII or XhoI-BglII. Nucleotide sequences were determined by a DNA sequencing service (Fasmac), and the sequence data were analyzed by Sequencher version 5.1 software (Gene Codes Corporation). The putative NLS of SARS-CoV ORF3b was predicted using PSORT II Prediction webtool (Horton and Nakai, 1997).
Transfection, Electroporation, and SeV Infection
HEK293 cells were transfected using PEI Max (Polysciences) according to the manufacturer’s protocol. HeLa cells cultured in 6-well plates with glass coverslips were transfected with using a FuGENE HD transfection reagent (Promega) according to the manufacturer’s protocol. For luciferase reporter assay, cells were cotransfected with 500 ng of either p125Luc (expressing firefly luciferase driven by human IFNB1 promoter; kindly provided by Dr. Takashi Fujita) (Fujita et al., 1993) or p55C1B-Luc (expressing firefly luciferase driven by IRF3; kindly provided by Dr. Takashi Fujita) (Fujita et al., 1993) and the pCAGGS-based HA-tagged expression plasmid (the amounts are indicated in the figure legends). A549 cells (100,000 cells) were electroporated with 500 ng of the pCAGGS-based HA-tagged expression plasmid using a Neon transfection system (Thermo Fisher Scientific) according to the manufacturer’s protocol (1200 V; 30 ms; 2 times pulse). At 24 h post transfection, SeV (strain Cantell, clone cCdi; GenBank accession no. AB855654) (Yoshida et al., 2018) was inoculated into the transfected cells at multiplicity of infection (MOI) 10 (for HEK293 and A549 cells) or 5 (for HeLa cells).
Reporter Assay
The luciferase reporter assay was performed at 24 h post infection as described (Kobayashi et al., 2014; Konno et al., 2018; Ueda et al., 2017). Briefly, 50 μL of cell lysate was applied to a 96-well plate (Nunc), and the firefly luciferase activity was measured using a PicaGene BrillianStar-LT luciferase assay system (Toyo-b-net), and the input for the luciferase assay was normalized by using a CellTiter-Glo 2.0 assay kit (Promega) following the manufacturers’ instructions. For this assay, a 2030 ARVO X multilabel counter instrument (PerkinElmer) was used.
Subcellular Fractionation
Subcellular fractionation was performed using Nuclear/cytosol fractionation kit (Biovision) according to the manufacturer’s procedure.
Western Blotting and Dot Blotting
Transfected cells were lysed with 1x SDS sample buffer (62.5 mM Tris-HCl [pH6.8], 2% SDS, 10% glycerol, 5% 2-mercaptoethanol and 0.0025% bromophenol blue). Western blotting was performed as described (Kobayashi et al., 2014; Konno et al., 2018; Nakano et al., 2017; Yamada et al., 2018) using an HRP-conjugated rat anti-HA monoclonal antibody (clone 3F10; Roche), a mouse anti-alpha-tubulin (TUBA) monoclonal antibody (clone DM1A; Sigma-Aldrich); a rabbit anti-lamin A/C (LMNA) polyclonal antibody (Cell Signaling Technology); an HRP-conjugated horse anti-mouse IgG antibody (Cell Signaling Technology); and an HRP-conjugated goat anti-rabbit IgG antibody (Cell Signaling Technology). Dot blotting was performed using a Bio-Dot microfiltration apparatus (Bio-Rad, cat# 170-6545) according to the manufacturer’s procedure. Briefly, 100 μL of the 20 times diluted cell lysate was used for the dot blotting and the following procedure was same as western blotting. Immobilon-P PVDF 0.45-μm membranes (Merck) were used for western blotting, while nitrocellulose 0.20-μm membranes (Bio-Rad) were used for dot blotting.
Real-time RT-PCR
Cellular RNA was extracted using QIAamp RNA blood mini kit (QIAGEN) and then treated with DNase I, Amplification Grade (Thermo Fisher Scientific). cDNA was synthesized using SuperScript III reverse transcriptase (Thermo Fisher Scientific) and oligo(dT)12-18 primer (Thermo Fisher Scientific). Real-time RT-PCR was performed as previously described (Nakano et al., 2017; Yamada et al., 2018) using a Power SYBR Green PCR Master Mix (Thermo Fisher Scientific) and the primers listed in Key Resources Table. For real-time RT-PCR, a CFX Connect Real-Time PCR Detection System (Bio-Rad) was used.
Immunofluorescence Staining
Twenty-four h post transfection, transfected HeLa cells were infected with SeV at MOI 5. Twenty-four h post infection, cells were fixed with formaldehyde, permeabilized with Triton X-100, and then stained using an FITC-conjugated anti-HA antibody (clone 3F10; Roche); a rabbit anti-IRF3 polyclonal antibody (Abcam); and an Alexa 546-conjugated anti-rabbit IgG antibody (Thermo Fisher Scientific). The coverslips were mounted on glass slides using ProLong Diamond Antipode Moutant with DAPI (Thermo Fisher Scientific) and observed using an FV-1000D confocal microscope (Olympus, Japan).
CoV-GLUE
To survey the ORF3b derivatives in pandemic SARS-CoV-2 sequences, we used the viral sequences deposited in GISAID (https://www.gisaid.org) (accessed 22 April, 2020). The screening was performed using the CoV-GLUE platform (http://cov-glue.cvr.gla.ac.uk) developed by MRC-University of Glasgow Centre for Virus Research, Scotland, UK (accessed 22 April, 2020). We discarded the sequences containing undermined or mix bases in the ORF3b region, and used 16,970 sequences for the further analyses. We constructed a phylogenetic tree using RAxML-NG version 0.9.0 (Kozlov et al., 2019) with a TPM3uf substitution model (Figure S2D). We also detected the two SARS-CoV-2 sequences (GISAID accession IDs: EPI_ISL_422564 and EPI_ISL_422565, collected in Quito, Ecuador) possessing the V163T/T164N substitutions in ORF3a, which correspond to the ∗23Q/L24M/57∗ substitutions in ORF3b (see also Figure S3C).
Quantification and Statistical Analysis
Data analyses were performed using Prism 7 (GraphPad Software). The data are presented as averages ± SEM. Statistically significant differences were determined by Student’s t test. Statistical details can be found directly in the figures or in the corresponding figure legends.
Consortia
The members of USFQ-COVID19 are Sully Márquez, Belén Prado-Vivar, Juan José Guadalupe, Bernardo Gutiérrez, Manuel Jibaja, Milton Tobar, Francisco Mora, Juan Gaviria, Alejandra Garcia, Franklin Espinosa, Edison Ligña, Mauricio Espinel, Jorge Reyes, Verónica Barragán, Patricio Rojas-Silva, Gabriel Trueba, Michelle Grunauer, and Paúl Cárdenas. Affiliations for consortium members are available in Data S1.
Acknowledgments
We thank all laboratory members in Division of Systems Virology, Institute of Medical Science, the University of Tokyo, Japan and all authors who have deposited and shared genome data on GISAID. We also thank Naoko Misawa, Masayuki Horie, and Keizo Tomonaga (Institute for Life and Medical Sciences, Kyoto University, Japan) and Ryoko Kawabata (Institute of Biomedical and Health Sciences, Hiroshima University, Japan) for generous support, as well as Takashi Fujita (Institute for Life and Medical Sciences, Kyoto University, Japan) for providing p125Luc and p55C1B-Luc. We thank Kotubu Misawa for dedicated support. The super-computing resource, SHIROKANE, was provided by the Human Genome Center, the Institute of Medical Science, the University of Tokyo, Japan. This study was supported in part by the AMED Research Program on Emerging and Re-emerging Infectious Diseases (20fk0108146 to K.S., 19fk0108171 to S.N. and K.S., and 20fk0108270 to Y. Koyanagi and K.S.), AMED Research Program on HIV/AIDS (19fk0410019 to Y. Koyanagi and K.S. and 20fk0410014 to Y. Koyanagi and K.S.), JST J-RAPID (JPMJJR2007 to R.J.G. and K.S.), KAKENHI Grant-in-Aid for Scientific Research B (18H02662 to K.S.), KAKENHI Grant-in-Aid for Scientific Research on Innovative Areas (16H06429 to S.N., T.I., and K.S.; 16K21723 to S.N., T.I., and K.S.; 17H05823 to S.N.; 17H05813 to K.S.; 19H04837 to T.I.; 19H04843 to S.N.; and 19H04826 to K.S.), Fund for the Promotion of Joint International Research (Fostering Joint International Research) (18KK0447 to K.S.), JSPS Research Fellow (DC1 19J22914 to Y. Konno and DC1 19J20488 to I.K.), Takeda Science Foundation (to K.S.), Ono Medical Research Foundation (to K.S.), Ichiro Kanehara Foundation (to K.S.), Lotte Foundation (to K.S.), Mochida Memorial Foundation for Medical and Pharmaceutical Research (to K.S.), Daiichi Sankyo Foundation of Life Science (to K.S.), Sumitomo Foundation (to K.S.), Uehara Foundation (to K.S.), Joint Research Project of the Institute of Medical Science, University of Tokyo (to Y. Koyanagi), Joint Usage/Research Center of the Institute for Frontier Life and Medical Sciences, Kyoto University (to K.S.), JSPS Core-to-Core program (A. Advanced Research Networks) (to Y. Koyanagi, D.S., R.J.G., and K.S.), Canon Foundation in Europe (to D.S. and K.S.), COVID-19 research grant of the Federal Ministry of Education and Research (MWK) Baden-Württemberg (to D.S.), 2020 Tokai University School of Medicine Research Aid (to S.N.), and International Joint Research Project of the Institute of Medical Science, the University of Tokyo (2020-K3003 to R.J.G. and K.S.).
Author Contributions
Y. Konno, I.K., K.U., and T.I. performed the experiments. R.J.G. and S.N. performed molecular phylogenetic analysis. T.I., Y. Koyanagi, and D.S. prepared reagents. Y. Konno, I.K., T.I., and K.S. interpreted the results. The USFQ-COVID19 consortium provided the clinical information of the two COVID-19 patients. K.S. designed the experiments. D.S. and K.S. wrote the manuscript. All authors reviewed and proofread the manuscript.
Declaration of Interests
The authors declare no competing interests.
Published: September 22, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.108185.
Supplemental Information
References
- Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Melo D., Nilsson-Payant B.E., Liu W.-C., Uhl S., Hoagland D., Møller R., Jordan T.X., Oishi K., Panis M., Sachs D. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045.e9. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan-Yeung M., Xu R.H. SARS: epidemiology. Respirology. 2003;8(Suppl):S9–S14. doi: 10.1046/j.1440-1843.2003.00518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frieman M., Yount B., Heise M., Kopecky-Bromberg S.A., Palese P., Baric R.S. Severe acute respiratory syndrome coronavirus ORF6 antagonizes STAT1 function by sequestering nuclear import factors on the rough endoplasmic reticulum/Golgi membrane. J. Virol. 2007;81:9812–9824. doi: 10.1128/JVI.01012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita T., Nolan G.P., Liou H.C., Scott M.L., Baltimore D. The candidate proto-oncogene bcl-3 encodes a transcriptional coactivator that activates through NF-kappa B p50 homodimers. Genes Dev. 1993;7(7B):1354–1363. doi: 10.1101/gad.7.7b.1354. [DOI] [PubMed] [Google Scholar]
- García-Sastre A., Egorov A., Matassov D., Brandt S., Levy D.E., Durbin J.E., Palese P., Muster T. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology. 1998;252:324–330. doi: 10.1006/viro.1998.9508. [DOI] [PubMed] [Google Scholar]
- Ge X.Y., Li J.L., Yang X.L., Chmura A.A., Zhu G., Epstein J.H., Mazet J.K., Hu B., Zhang W., Peng C. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503:535–538. doi: 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan W.J., Ni Z.Y., Hu Y., Liang W.H., Ou C.Q., He J.X., Liu L., Shan H., Lei C.L., Hui D.S.C., China Medical Treatment Expert Group for Covid-19 Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjadj J., Yatim N., Barnabei L., Corneau A., Boussier J., Péré H., Charbit B., Bondet V., Chenevier-Gobeaux C., Breillat P. Impaired type I interferon activity and inflammatory responses in severe Covid-19 patients. Science. 2020;369:718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B., Zhang Y., Xu L., Yang W., Yang F., Feng Y., Xia L., Zhou J., Zhen W., Feng Y. Identification of diverse alphacoronaviruses and genomic characterization of a novel severe acute respiratory syndrome-like coronavirus from bats in China. J. Virol. 2014;88:7070–7082. doi: 10.1128/JVI.00631-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton P., Nakai K. Better prediction of protein cellular localization sites with the k nearest neighbors classifier. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1997;5:147–152. [PubMed] [Google Scholar]
- Hu B., Zeng L.P., Yang X.L., Ge X.Y., Zhang W., Li B., Xie J.Z., Shen X.R., Zhang Y.Z., Wang N. Discovery of a rich gene pool of bat SARS-related coronaviruses provides new insights into the origin of SARS coronavirus. PLoS Pathog. 2017;13:e1006698. doi: 10.1371/journal.ppat.1006698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Li W., Gao T., Cui Y., Jin Y., Li P., Ma Q., Liu X., Cao C. The severe acute respiratory syndrome coronavirus nucleocapsid inhibits type I interferon production by interfering with TRIM25-mediated RIG-I ubiquitination. J. Virol. 2017;91:e02143-16. doi: 10.1128/JVI.02143-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui D.S., I Azhar E., Madani T.A., Ntoumi F., Kock R., Dar O., Ippolito G., Mchugh T.D., Memish Z.A., Drosten C. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health—The latest 2019 novel coronavirus outbreak in Wuhan, China. Int. J. Infect. Dis. 2020;91:264–266. doi: 10.1016/j.ijid.2020.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T., Takeuchi J.S., Ren F., Matsuda K., Sato K., Kimura Y., Misawa N., Yoshikawa R., Nakano Y., Yamada E. Characterization of red-capped mangabey tetherin: implication for the co-evolution of primates and their lentiviruses. Sci. Rep. 2014;4:5529. doi: 10.1038/srep05529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konno Y., Nagaoka S., Kimura I., Takahashi Ueda M., Kumata R., Ito J., Nakagawa S., Kobayashi T., Koyanagi Y., Sato K. A naturally occurring feline APOBEC3 variant that loses anti-lentiviral activity by lacking two amino acid residues. J. Gen. Virol. 2018;99:704–709. doi: 10.1099/jgv.0.001046. [DOI] [PubMed] [Google Scholar]
- Kopecky-Bromberg S.A., Martínez-Sobrido L., Frieman M., Baric R.A., Palese P. Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J. Virol. 2007;81:548–557. doi: 10.1128/JVI.01782-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov A.M., Darriba D., Flouri T., Morel B., Stamatakis A. RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics. 2019;35:4453–4455. doi: 10.1093/bioinformatics/btz305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krug R.M., Yuan W., Noah D.L., Latham A.G. Intracellular warfare between human influenza viruses and human cells: the roles of the viral NS1 protein. Virology. 2003;309:181–189. doi: 10.1016/s0042-6822(03)00119-3. [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam T.T., Jia N., Zhang Y.W., Shum M.H., Jiang J.F., Zhu H.C., Tong Y.G., Shi Y.X., Ni X.B., Liao Y.S. Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature. 2020;583:282–285. doi: 10.1038/s41586-020-2169-0. [DOI] [PubMed] [Google Scholar]
- Lau S.K., Woo P.C., Li K.S., Huang Y., Tsoi H.W., Wong B.H., Wong S.S., Leung S.Y., Chan K.H., Yuen K.Y. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. USA. 2005;102:14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S.K., Li K.S., Huang Y., Shek C.T., Tse H., Wang M., Choi G.K., Xu H., Lam C.S., Guo R. Ecoepidemiology and complete genome comparison of different strains of severe acute respiratory syndrome-related Rhinolophus bat coronavirus in China reveal bats as a reservoir for acute, self-limiting infection that allows recombination events. J. Virol. 2010;84:2808–2819. doi: 10.1128/JVI.02219-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Shi Z., Yu M., Ren W., Smith C., Epstein J.H., Wang H., Crameri G., Hu Z., Zhang H. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y., Ren R., Leung K.S.M., Lau E.H.Y., Wong J.Y. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N. Engl. J. Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X.D., Wang W., Hao Z.Y., Wang Z.X., Guo W.P., Guan X.Q., Wang M.R., Wang H.W., Zhou R.H., Li M.H. Extensive diversity of coronaviruses in bats from China. Virology. 2017;507:1–10. doi: 10.1016/j.virol.2017.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano Y., Misawa N., Juarez-Fernandez G., Moriwaki M., Nakaoka S., Funo T., Yamada E., Soper A., Yoshikawa R., Ebrahimi D. HIV-1 competition experiments in humanized mice show that APOBEC3H imposes selective pressure and promotes virus adaptation. PLoS Pathog. 2017;13:e1006348. doi: 10.1371/journal.ppat.1006348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H., Yamamura K., Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- Park A., Iwasaki A. Type I and type III Interferons—induction, signaling, evasion, and application to combat COVID-19. Cell Host Microbe. 2020;27:870–878. doi: 10.1016/j.chom.2020.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray M., Tang R., Jiang Z., Rotello V.M. Quantitative tracking of protein trafficking to the nucleus using cytosolic protein delivery by nanoparticle-stabilized nanocapsules. Bioconjug. Chem. 2015;26:1004–1007. doi: 10.1021/acs.bioconjchem.5b00141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X.C., Zhang J.X., Zhang S.Y., Wang P., Fan X.H., Li L.F., Li G., Dong B.Q., Liu W., Cheung C.L. Prevalence and genetic diversity of coronaviruses in bats from China. J. Virol. 2006;80:7481–7490. doi: 10.1128/JVI.00697-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda M.T., Kurosaki Y., Izumi T., Nakano Y., Oloniniyi O.K., Yasuda J., Koyanagi Y., Sato K., Nakagawa S. Functional mutations in spike glycoprotein of Zaire ebolavirus associated with an increase in infection efficiency. Genes Cells. 2017;22:148–159. doi: 10.1111/gtc.12463. [DOI] [PubMed] [Google Scholar]
- Verity R., Okell L.C., Dorigatti I., Winskill P., Whittaker C., Imai N., Cuomo-Dannenburg G., Thompson H., Walker P.G.T., Fu H. Estimates of the severity of coronavirus disease 2019: a model-based analysis. Lancet Infect. Dis. 2020;20:669–677. doi: 10.1016/S1473-3099(20)30243-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Yan M., Xu H., Liang W., Kan B., Zheng B., Chen H., Zheng H., Xu Y., Zhang E. SARS-CoV infection in a restaurant from palm civet. Emerg. Infect. Dis. 2005;11:1860–1865. doi: 10.3201/eid1112.041293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Fu S., Cao Y., Zhang H., Feng Y., Yang W., Nie K., Ma X., Liang G. Discovery and genetic analysis of novel coronaviruses in least horseshoe bats in southwestern China. Emerg. Microbes Infect. 2017;6:e14. doi: 10.1038/emi.2016.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss S.R. Forty years with coronaviruses. J. Exp. Med. 2020;217:e20200537. doi: 10.1084/jem.20200537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . 2004. Summary of probable SARS cases with onset of illness from 1 November 2002 to 31 July 2003.https://www.who.int/csr/sars/country/table2004_04_21/en/ [Google Scholar]
- WHO . 2020. Coronavirus disease 2019.https://www.who.int/emergencies/diseases/novel-coronavirus-2019 [Google Scholar]
- Wu Z., Yang L., Ren X., Zhang J., Yang F., Zhang S., Jin Q. ORF8-related genetic evidence for Chinese horseshoe bats as the source of human severe acute respiratory syndrome coronavirus. J. Infect. Dis. 2016;213:579–583. doi: 10.1093/infdis/jiv476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao K., Zhai J., Feng Y., Zhou N., Zhang X., Zou J.-J., Li N., Guo Y., Li X., Shen X. Isolation of SARS-CoV-2-related coronavirus from Malayan pangolins. Nature. 2020;583:286–289. doi: 10.1038/s41586-020-2313-x. [DOI] [PubMed] [Google Scholar]
- Yamada E., Nakaoka S., Klein L., Reith E., Langer S., Hopfensperger K., Iwami S., Schreiber G., Kirchhoff F., Koyanagi Y. Human-specific adaptations in Vpu conferring anti-tetherin activity are critical for efficient early HIV-1 replication in vivo. Cell Host Microbe. 2018;23:110–120.e7. doi: 10.1016/j.chom.2017.12.009. [DOI] [PubMed] [Google Scholar]
- Yoshida A., Kawabata R., Honda T., Sakai K., Ami Y., Sakaguchi T., Irie T. A single amino acid substitution within the paramyxovirus Sendai virus nucleoprotein is a critical determinant for production of interferon-beta-inducing copyback-type defective interfering genomes. J. Virol. 2018;92:e02094. doi: 10.1128/JVI.02094-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B., Li X., Chen J., Ouyang M., Zhang H., Zhao X., Tang L., Luo Q., Xu M., Yang L. Evaluation of variation in D-dimer levels among COVID-19 and bacterial pneumonia: a retrospective analysis. J. Thromb. Thrombolysis. 2020 doi: 10.1007/s11239-020-02171-y. Published online June 10, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J., Hon C.C., Li Y., Wang D., Xu G., Zhang H., Zhou P., Poon L.L., Lam T.T., Leung F.C., Shi Z. Intraspecies diversity of SARS-like coronaviruses in Rhinolophus sinicus and its implications for the origin of SARS coronaviruses in humans. J. Gen. Virol. 2010;91:1058–1062. doi: 10.1099/vir.0.016378-0. [DOI] [PubMed] [Google Scholar]
- Zhou P., Li H., Wang H., Wang L.F., Shi Z. Bat severe acute respiratory syndrome-like coronavirus ORF3b homologues display different interferon antagonist activities. J. Gen. Virol. 2012;93:275–281. doi: 10.1099/vir.0.033589-0. [DOI] [PubMed] [Google Scholar]
- Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Chen X., Hu T., Li J., Song H., Liu Y., Wang P., Liu D., Yang J., Holmes E.C. A novel bat coronavirus closely related to SARS-CoV-2 contains natural insertions at the S1/S2 cleavage site of the spike protein. Curr. Biol. 2020;30:2196–2203.e3. doi: 10.1016/j.cub.2020.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The published article includes all datasets generated or analyzed during this study.