Introduction
Mantle cell lymphoma (MCL) is a B-cell neoplasm characterized by the expansion of mature B cells frequently co-expressing CD5 that tend to widely spread in bone marrow, blood, lymphoid tissues and extranodal sites. The tumor cells carry the t(11;14)(q13;q32) that leads to the constitutive overexpression of cyclin D1. In spite of this common initial oncogenic event, the tumors follow a very heterogeneous biological behavior indicating that other molecular mechanisms drive the evolution of the disease. The identification of two MCL subtypes, conventional and leukemic non-nodal, with different molecular characteristics and clinical manifestations may explain, in part, the diversity of the tumor. Recent genomic and molecular studies have expanded our perspective on the cell of origin and pathogenesis of these MCL subtypes. In this review we will address new findings on the MCL pathogenesis and the two molecular subtypes that may assist in the interpretation of the clinical diversity of these tumors.
Translocation t(11;14) and overexpression of Cyclin D1
The t(11;14)(q13;q32) is considered the primary oncogenic event in MCL development, virtually present in all cases, that juxtaposes CCND1 at llql3 with the IGH regulatory region leading to a constitutive overexpression of cyclin D1. The translocation occurs at the pro/pre-B stage of differentiation during the V(D)J recombination process and is mediated by recombination activating gene (RAG) enzymes.1 The levels of cyclin D1 could be further increased by secondary rearrangements at the 3’ of the gene or point mutations in the 3’ untranslated region that create stable truncated cyclin D1 mRNAs.2 This cyclin D1 overexpression may deregulate the G1/S cell cycle transition and promote the malignant transformation of B cells. Beyond its well characterized role in cell cycle, a number of growing evidences implicate cyclin D1 in additional cellular processes including transcriptional regulation by interacting with transcription factors, chromatin-remodeling elements, and histonemodifying enzymes.3,4 Cyclin D1 may also directly participate in DNA-damage response and apoptosis regulation.5,6 Interestingly, cyclin D1 binds to a high number of active promoters and interacts with the transcription machinery leading to a global transcriptome down-modulation in neoplastic lymphoid cells,7
Variant CCND1 translocations and expression in MCL
A small subset of MCL cases show CCND1 rearrangement with IGK or IGL light chain resulting in variant translocations t(2;11)(p11;q13)8 and t(11;22)(q13;q11.2)9, respectively, that determine similar cyclin D1 dysregulation. Intriguingly, recent studies have reported a minor subset of MCL that express cyclin D1 protein and high mRNA levels but CCND1 rearrangements are not detected when evaluated by conventional cytogenetics or fluorescence in situ hybridization (FISH) using fusion or break-apart probes. Whole-genome sequencing (WGS) or FISH using custom bacterial artificial clones-labeled probes have detected that these cases carry cryptic rearrangements of IGK or IGL enhancers with CCND1 gene which may be responsible for the cyclin D1 upregulation. The clinical and pathological features of this small subgroup of patients are similar to conventional MCL, suggesting that they correspond to the same molecular entity.10,11 This finding is in line with the identification by WGS of cryptic insertions of MYC into the IGH locus in IG-MYC negative Burkitt lymphoma cases,12 or the detection of cryptic rearrangements involving MYC and BCL2 in high grade B-cell lymphomas.13
Cyclin D1 negative MCL
A particular subset of cases with the same MCL morphology and phenotype lack cyclin D1 expression and t(11;14) translocation (cyclin D1− MCL).14–16 These tumors have also similar gene expression profile, secondary chromosomal alterations, and clinical behavior as cyclin D1+ MCL, suggesting that they correspond to the same disease.15–17 These cases should be distinguished from the uncommon MCL carrying the t(11;14) in which the cyclin D1 expression is not detected by immunohistochemistry due to mutations in the C-terminal domain of CCND1 or the presence of CCND1b as the only expressed isoform that render the protein undetectable by current antibodies used in the pathology diagnoti routine.18
Initial FISH studies identified chromosomal rearrangements fusing CCND2 with IG loci (IGH, IGK, or IGL) in 55% of cyclin D1− MCL.15 However, the primary oncogenic event remained elusive in a substantial fraction of these cases. More recently, next generation sequencing (NGS) studies and FISH with custom probes have found the insertion of a small IGK region including its enhancer near CCND3 (16%) or CCND2 (7%), leading to cyclin D3 or cyclin D2 overexpression, respectively. Similar cryptic insertions involving the IGL-enhancer in the vicinity of CCND3 associated with cyclin D3 overexpression have been also detected in a subset of cyclin D1− MCL. Overall, 23% of the cyclin D1− MCL cases showed an IG light chain enhancer hijacking as initial oncogenrc event.19
SOX11: the key oncogenic factor
SOX11 is a transcription factor that plays an important oncogenic role in MCL pathogenesis through its impact in B-cell differentiation, tumor microenvironment interactions, cell cycle control and apoptosis.20,21 SOX11 is not expressed in normal lymphoid cells or other mature B cell lymphomas with the exception of 25–50% of Burkitt lymphoma, but it is highly expressed in conventional MCL, including cyclin D1− MCL.17 Hence, SOX11-nuclear staining is a useful tool in the differential diagnosis of MCL and other small B-cell neoplasias. SOX11 may contribute to MCL pathogenesis by the constitutive activation of PAX5, a master regulator of B cell development, that blocks terminal B-cell differentiation and promotes tumor growth.21 Another SOX11 direct regulated target is BCL6, an essential element for B-cell development and maintenance of follicular germinal centers (GC). SOX11 may block BCL6 expression preventing the entrance of MCL cells in the GC.20 The transgenic mouse model (EμSOX11-EGFP) developed by Kuo et al. shows hyperactivation of pBTK and other molecules of the BCR signaling pathway driving tumor development.22 Moreover, SOX11 regulates interactions of MCL cells with the microenvironment inducing angiogenesis through PDGFA23 and promoting tumor cell migration, adhesion, and cell proliferation by upregulating CXCR4 and FAK.24 Accordingly, treatment in vitro of MCL cells with FAK inhibitors could overcome ibrutinib resistance.25
In spite of the relevant role of SOX11 in MCL the mechanisms leading to its specific upregulation in this lymphoma are not well understood. Recent studies have suggested that SOX11 expression in these cells could be mediated by epigenetic mechanisms that alter the 3-dimensional configuration of chromatin bringing together a distant active enhancer region with the promoter of the gene.26
MCL molecular subtypes: distinct pathogenic pathways
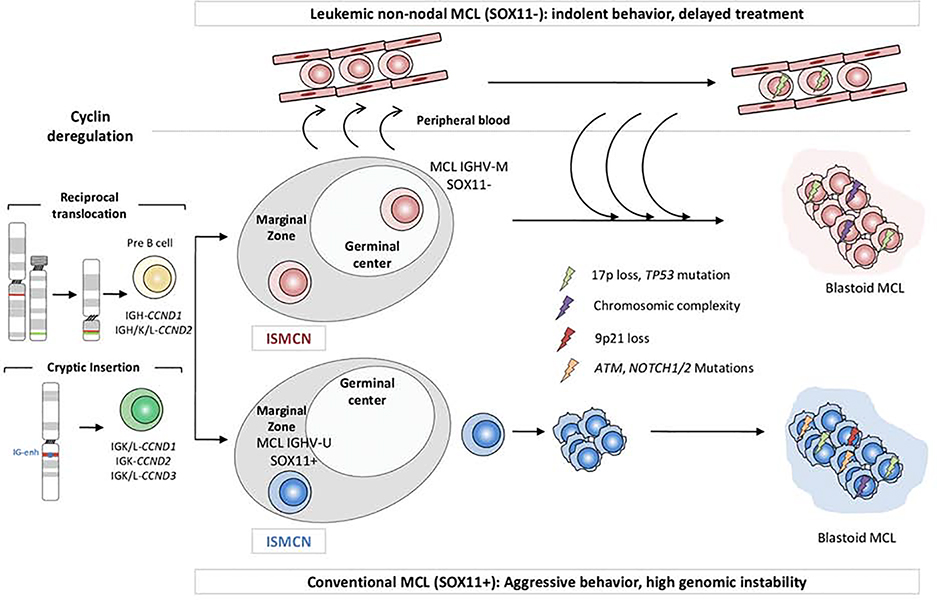
The initial translocation t(ll;14)(ql3;q32) may be followed by two distinct pathogenic pathways resulting in two subtypes of tumors with different biological behavior. The most common subtype is the conventional MCL (cMCL), that derives from a cell that does not enter into the follicular GC and carry no or a limited number of IGHV somatic mutations (Figure 1).27 The second subtype, leukemic non-nodal MCL (nnMCL), derives from a cell that has gone through the GC acquiring IGHV somatic mutations.28 These two subtypes have different cellular origin, a naive-like B-cell for cMCL and an experienced GC memory-like B cell for nnMCL. This idea is supported by the observation that both subtypes retain the DNA methylation pattern of their normal cellular counterparts.26,28
Figure 1. MCL molecular subtypes.
The naïve B-cell with cyclin deregulation may evolve into two distinct molecular subtypes with different molecular and clinicopahtological characteristics.
Although cMCL and nnMCL share similar global gene expression profiles, they differ in some genetic and molecular characteristics. cMCL cases overexpress SOX11, are genetically unstable and tend to accumulate many chromosomal alterations. Clinically, patients with cMCL usually have generalized lymphadenopathy at diagnosis and follow an aggressive clinical course.29–30 Contrarily, nnMCL present an initial indolent disease that may be stable for long periods of time. Patients have leukemic involvement with minimal lymphadenopathy and later on may develop splenomegaly.31–35 These cases might benefit of a careful observation management without negatively impacting their outcome.36–39 Although nnMCL cells initially harbor few or no chromosomal alterations besides the t(11;14), they may evolve over time acquiring TP53 mutation, 17p loss and increased genome instability that confer a dismal prognosis.34 Recently, a 16-gene assay on the NanoString platform (L-MCL16 assay) has been used to classify patients with leukemic involvement into cMCL or nnMCL subgroups and, in combination with genomic complexity and TP53 alterations, predict patients’ outcome.35
From early lesions to an overt lymphoma
The oncogenic steps from the early CCND1 rearrangement to the development of an overt lymphoma are not well known. The detection of cells carrying the t(11;14) in the peripheral blood of 8% of healthy individuals suggests that not all the cells acquiring the initial translocation will evolve into a malignant lymphoma.36 These clones may persist for long latency periods prior to evolve into a malignant neoplasm as supported by the observation of a simultaneous MCL with the same clonal origin in the recipient and donor 12 years after an allogenic bone marrow transplantation.37
Cyclin D1 positive cells are occasionally found in the inner mantle zone of reactive lymphoid follicles, a situation now known as in situ mantle cell neoplasia (ISMCN) (Figure 1). These in situ lesions are usually identified incidentally and sometimes in association with other lymphomas.38,39 Although some of these cases may affect different territories the patients have a long-term follow-up without developing progressive disease in the absence therapeutic intervention. The proportion of ISMCN that may develop an overt lymphoma is not well known but seems very low.40 However, some patients with these lesions may have already a disseminated MCL at diagnosis and therefore they should be explored carefully to rule out this situation (Figure 1).
The hallmarks of cancer in MCL
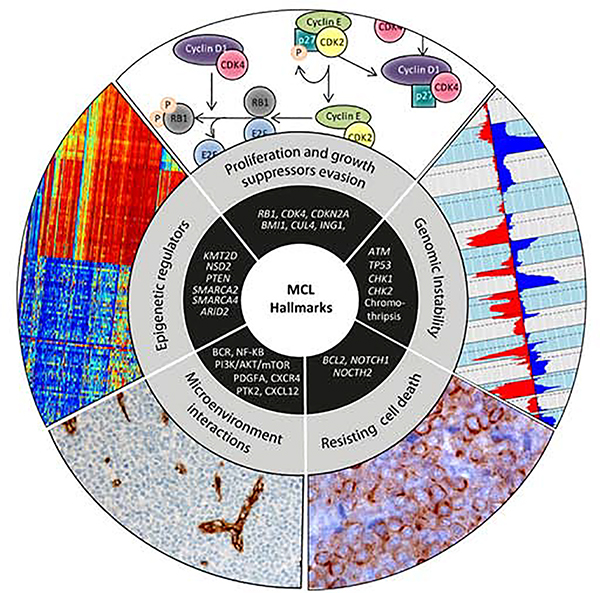
The concept of cancer hallmarks helped to dissect the malignant phenotypes that are associated with specific physiological circuits dysregulated during the oncogenic process.41 The order in which the different neoplasias acquire their hallmarks capabilities and their contribution during oncogenesis depends on the type of tumor. In MCL, the t(11;14) translocation responsible of cyclin D1 dysregulation would represent the initial acquisition of a sustained proliferative signaling. However, this event is not sufficient to explain the MCL pathogenesis. Secondary genetic alterations would reinforce the proliferation dysregulation and promote the acquisition of additional hallmarks relevant to MCL lymphomagenesis (Figure 2).
Figure 2. MCL hallmarks.
The conceptual framework encompasses many different cellular functions that transform normal cells into malignant cancer cells. All the related pathways involved in MCL pathogenesis may be globally grouped into five main hallmarks.
1. Sustaining proliferative signaling and growth suppressors evasion
An essential feature of tumor cells is their capacity for continuous proliferation together with the ability to circumvent the constrain proliferation promoted by tumor suppressors under cell stress conditions. In MCL, the initial dysregulation of cyclin D1 may promote G1/S phase transition through the binding of the cyclin to CDK4, followed by RB1 phosphorylation, and subsequent E2F release. Besides this initial alteration, secondary genetic events also impact directly in cell cycle control affecting mainly two pathways INK4A/CDK4/RB1 and ARF/MDM2/TP53. The 12ql3 amplification (20%) may led to CDK4 overexpression that would further promote cell cycle dysregulation.42 Interestingly, the inhibition of CDK4 can be a reliable mechanism to overcome the ibrutinib resistance in MCF patients.43 The 9p21 deletion (25%), involving the CDKN2A, is one of the most frequent genetic alteration in MCL. CDKN2A gene encodes for pl6 (INK4A), a cyclin dependent kinase inhibitor that specifically inhibits CDK4 and CDK6 keeping RB1 active, and for pl4 (ARF), a E3 ubiquitin-protein ligase that stabilize p53 by interacting with MDM2 preventing its degradation.42 Alternatively, few MCL cases with wild-type CDKN2A show BM11 (10ql3) amplification and overexpression which act as a transcriptional repressor of the CDKN2A locus.44 Inactivation of CDKN2A is associated with an unfavorable prognosis in MCL patients and is related to aggressive variants.45 Similarly, inactivation of RB1 by mutations and homozygous deletion, although uncommon, occurs mainly in aggressive cases.46 Other genetic alterations targeting cell cycle related genes that might confer increased proliferation rates are MYC amplification and translocation,44,47 and deletions of CUL4A and ING1 (13q34).48
The high number of genetic alterations that potentially dysregulate cell cycle in MCL underscores the relevance of this hallmark in MCL pathogenesis. Moreover, its relevance is supported by the fact that the best predictor of patient survival in MCL is a proliferation gene expression signature that may integrate the multiple genetic alterations dysregulating cell cycle and may now be reliably determined in formalin fixed paraffin embedded tissues (MCL35 proliferation assay).49,50
2. Genomic instability
Cancer cells have a tendency to accumulate genetic alterations and increased genomic instability, a hallmark of many tumors. MCL is one of the B-cell malignancies with highest degree of genomic instability. More than 90% of the MCL cases display highly altered genomes, with gains/amplifications and homozygous/heterozygous losses, as well as other non-recurrent chromosomal rearrangements. Losses in 1p, 6q, 8p, 9p, 9q, 10p, 11q, 13q, and 17p and gains in 3q, 7p, 8q, 10p, 15q and 18q are the most frequent chromosomal alterations identified in MCL.51 The high number of chromosome alterations in MCL cells is consistent with the fact that the two most common mutated genes are ATM (40–50%) and TP53 (21–45%), both involved in DNA damage response. ATM alterations, including mutations and 11q deletions, are considered early events but do not correlate with prognosis, despite being related to increased chromosomal instability52. Downregulation of CHK2 and CHK1, two critical serine-threonine kinases involved in signal transduction during DNA damage response, may constitute another mechanism to promote chromosome instability in a limited number of MCL cases.53 TP53 is frequently inactivated by point mutations and 17pl3 deletion, compromising the p53-mediated cell cycle arrest, apoptosis and senescence as response to DNA damage. TP53 alterations are identified at similar proportion in cMCL and nnMCL. Additionally, the increased number of chromosomal imbalances, TP53 mutations and global genetic instability are associated with blastoid variants and worse clinical outcome in MCL patients.35,54–56
3. Resisting cell death
The evasion of apoptotic cell death, considered a natural barrier to cancer development, is a key hallmark of cancer cells. In MCL, dysregulation of the antiapoptotic protein BCL2 mainly by amplifications or mRNA overexpression is described in 3–17% of cases. Upregulation of other proteins of the BCL-family, like BCLX, as well as occasional biallelic loss of the proapoptic BCL2L11 (2ql3) have been also observed.57 In the last decade, several reports have described recurrent gain-of-function truncating mutations in NOTCH1 (5–14%) and NOTCH2 (5%) associated with blastoid variants and dismal prognosis.58,59 NOTCH pathway is one of the most evolutionary conserved signaling cascades across species that regulates cell death but also cell proliferation and activates specific differentiation programs.60 NOTCH signaling regulates, directly or indirectly through MYC, a gene signature consisting of BCR signaling, RNA metabolism and chromatin/transcriptional modulation. Clinically, NOTCH targeted approaches may be a therapeutic option for a subset of patients.61
4. Modulation of tumor microenvironment interactions
Normal B-cell maturation involves somatic recombination and mutation of the IGHV genes that encode the antigen binding domains of the B cell receptor (BCR). The observation of restricted repertoires of IGHV genes in MCL, called BCR IG stereotypy, highlights the antigen selection and BCR signaling pathway as important hallmark for MCL pathogenesis.62 Additionally, constitutive activation of the BCR signaling pathway has a key role promoting survival and proliferation of the malignant B cells. Although the specific mechanisms involved in this deregulation are not well understood, different B-cell receptor associated kinases including tyrosine protein kinase (LYN), spleen tyrosine kinase (SYK)69, and especially Bruton’s tyrosine kinase (BTK) are considered therapeutic targets due to their constitutive activation or amplification in MCL. In this context, the oral covalent inhibitor of BTK ibrutinib, shows durable single-agent efficacy in MCL cases.63 Recently, in the MCL-0208 trial, high expression levels of a 6-gene BCR signature (AKT3, BCL2, BTK, CD79B, PIK3CD, and SYK) was associated with shorter progression-free survival and OS.64
The constitutive activation of PI3K/AKT and NF-ĸB signaling pathways also play a relevant role in MCL pathogenesis. Activated PI3K/AKT/mTOR components target a wide range of downstream processes in MCL, including angiogenesis but also cell survival, growth, and protein synthesis. Activation of downstream kinases such as SYK and PI3KCA amplification may cause activation if this pathway and can determine the therapeutic potential of small molecule inhibitors.65 The activation of the canonical NF-ĸB pathway directly correlates with increased tumor proliferation and inferior survival in MCL.66 The NF-ĸB activation can be mediated by different alterations, mainly by inactivating mutations or deletions in negative regulators as TNFAIP3/A20,67 TRAF2, BIRC3, NFKBIE, and CARD11. NFKBIE deletions are associated with poor outcome,68 meanwhile TRAF2 and BIRC3 mutations are associated with resistance to ibrutinib therapies in MCL.69 The antiapoptotic proteins BCL-2, BCL-XL, XIAP and cFLIP, are also NF-ĸB targets highly expressed in MCL.70,71
Additionally, during tumor progression the ability to sprout new blood vessels must be kept intact. Experimental studies have shown that SOX11 expression is associated with an angiogenic switch trough PDGFA activation and is characterized by increased expression of angiogenic-related signatures and vascularization.23,72 Interestingly, SOX11 directly regulates CXCR4 and PTK2 conferring a protective microenvironmentrelated signatures in SOX11+ MCL cases. CXCR4 and CXCL12 overexpression enhance FAK activation promoting MCL cell migration and adhesion facilitating the crosstalk with the stromal cells that confers survival and drug resistance to MCL cells.24
5. Epigenetic dysregulation
Epigenetic dysregulation can promote malignant cellular transformation and is considered a hallmark for a large number of different neoplasms. In the last years, the characterization of whole tumor cell methylomes confirmed the critical role that DNA methylation plays in MCL tumorigenesis and it was identified as a dynamic process prone to be altered upon neoplastic transformation. In MCL cells, an increased DNA methylation burden, defined as the number of methylation changes acquired by tumor cells, is associated with worse clinical outcome and higher number of driver mutations.26 Moreover, the genomic analysis of MCL has revealed that epigenetic modulators are recurrent altered targets. The most frequently mutated epigenetic modifier in MCL is KMT2D (17–23%), a regulator of transcriptional and posttranslational processes which mutations may have impact on the outcome of patients.55 Mutations in the catalytic domain of the NSD2 are also found in cMCL and are associated with overexpression of signatures related to proliferation and cell cycle, as well as with global chromatin methylation driving oncogenic reprogramming in other lymphoid malignancies.73,74 Recently, NSD2 has been described to mediate dimethylation of PTEN and facilitate its recruitment into DNA-damage sites contributing to the repair of DNA double strand breaks.75 Moreover, alterations in SWI-SNF chromatin remodeling complexes, including SMARCA4 mutations (5–12%) or deletions involving SMARCA2 (9p24) or ARID2 (12ql2) confer resistance to ibrutinib and venetoclax.76 Other dysregulated epigenetic modifiers are KMT2C (5–16%), BMI1 (6–12%), and TET2 (5–12%). Finally, mutations targeting regulators of transcription such as MEF2B (3–7%) or posttranscription UBR5 (7–18%) are also recurrently found in MCL.58,77
Summary
The translocation t(11;14) is the genetic hallmark of MCL although cryptic rearrangements of IG regulatory regions could be an alternative oncogenic mechanism in a minor subgroup of patients. Interestingly this IG enhancer hijacking phenomenon is found recurrently in MCL cyclin D1− cases with cyclin D2 or cyclin D3 overexpression. MCL is considered one of the most aggressive lymphomas. However, a small subset of cases may follows an initial indolent clinical course without need of treatment. The presence of IGHV somatic mutations, leukemic expression in the absence of lymphadenopathies, as well as, low number of genomic alterations are characteristics that differentiate nnMCL from cMCL cases. The dysregulation of cyclin D1, although considered the primary oncogenic event, is not enough for malignant transformation of B-cell clones. In this sense, additional somatic genetic alterations affecting genes involved in many cancer hallmarks, like cell cycle control (CDKN2A, CDK4, and RB1), DNA damage response (TP53, ATM, CDKN2A, and MYC), epigenetic modulation (KMT2D, SMARCA4, and NSD2) and NF-ĸB signaling pathways (BIRC3, NFKBIE, and TNFAIP3) among others, may impact MCL lymphomagenesis. Additionally, SOX11 seems to collaborate with cyclin D1 in MCL pathogenesis, regulating a complex transcriptional program and enhancing the aggressiveness of the tumor. The better understanding of MCL pathogenesis generated by the use of NGS technologies and the integration of multidisciplinary research, from molecular biology and pathology to the clinic, has revealed new potential management strategies for the treatment of aggressive MCL.
Key Points.
Cryptic translocations of IG enhancers near CCND1, CCND2 and CCND3 represent an alternative mechanism to the t(l1; 14) as an initial oncogenic event in MCL.
The leukemic non-nodal MCL subtype displays different clinicobiological features and better outcome than conventional MCL.
SOX11 is an oncogenic transcription factor highly expressed in conventional MCL with a relevant role in its pathogenesis, but its regulation is poorly understood.
Next generation sequencing studies have provided new insights into the molecular MCL pathogenesis and have helped to refine the MCL cancer hallmarks.
Synopsis.
Mantle cell lymphoma (MCL) is a mature B-cell neoplasm with heterogeneous clinical behavior that is molecularly characterized by the constitutive overexpression of cyclin D1 and deregulation of different signaling pathways. SOX11 expression determines an aggressive phenotype frequently associated with the accumulation of high number of chromosomal alterations and somatic gene mutations. A subset of patients with the SOX11-negative leukemic non-nodal MCL subtype follows an initial indolent clinical evolution and may not require treatment at diagnostic, although eventually may also progress to an aggressive disease. In this review, we discuss the genetic and molecular alterations with impact on the cancer hallmarks that characterize the lymphomagenesis of the two MCL subtypes.
Acknowledgments
This work was supported by research funding from Fondo de Investigaciones Sanitarias, Instituto de Salud Carlos III (PI17/01061 to S.B.), National Institute of Health “Molecular Diagnosis, Prognosis, and Therapeutic Targets in Mantle Cell Lymphoma” (P01CA229100 to E.C.); Spanish Ministerio de Ciencia, Innovación y Universidades (RTI2018-094274-B-I00 to E.C.; Generalitat de Catalunya Suport Grups de Recerca AGAUR (2017-SGR-1142 to E.C). E.C. is an Academia Researcher of the "Institució Catalana de Recerca i Estudis Avançats (ICREA)" of the Generalitat de Catalunya. This work was mainly developed at the Centre Esther Koplowitz (CEK), Barcelona, Spain. We are grateful to Sílvia Ruiz for their technical and logistic assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Alba Navarro, Institut d’Investigacions Biomèdiques August Pi i Sunyer (IDIBAPS), Barcelona, Spain; Centro de Investigación Biomédica en Red de Cáncer, Madrid, Spain.
Sílvia Beà, Institut d’Investigacions Biomèdiques August Pi i Sunyer (IDIBAPS), Barcelona, Spain; Centro de Investigación Biomédica en Red de Cáncer, Madrid, Spain; Hematopathology Unit, Hospital Clínic of Barcelona, University of Barcelona, Barcelona, Spain.
Pedro Jares, Institut d’Investigacions Biomèdiques August Pi i Sunyer (IDIBAPS), Barcelona, Spain; Centro de Investigación Biomédica en Red de Cáncer, Madrid, Spain; Hematopathology Unit, Hospital Clínic of Barcelona, University of Barcelona, Barcelona, Spain.
Elías Campo, Institut d’Investigacions Biomèdiques August Pi i Sunyer (IDIBAPS), Barcelona, Spain; Centro de Investigación Biomédica en Red de Cáncer, Madrid, Spain; Hematopathology Unit, Hospital Clínic of Barcelona, University of Barcelona, Barcelona, Spain; Hematopathology Unit, Hospital Clínic of Barcelona, Villarroel 170, 08036-Barcelona, Spain.
References
- 1.Kuppers R, La-Favera R. Mechanisms of chromosomal translocations in B cell lymphomas. Oncogene. 2001;20(40):5580–5594. [DOI] [PubMed] [Google Scholar]
- 2.Wiestner A, Tehrani M, Chiorazzi M, et al. Point mutations and genomic deletions in CCND1 create stable truncated cyclin D1 mRNAs that are associated with increased proliferation rate and shorter survival. Blood. 2007;109(11):4599–4606.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aggarwal P, Vaites LP, Kim JK, et al. Nuclear cyclin D1/CDK4 kinase regulates CUL4 expression and triggers neoplastic growth via activation of the PRMT5 methyltransferase. Cancer Cell. 2010;18(4):329–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bienvenu F, Jirawatnotai S, Elias JE, et al. Transcriptional role of cyclin D1 in development revealed by a genetic-pro teomic screen. Nature. 2010;463(7279):374–378.doi: 10.1038/nature08684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jirawatnotai S, Hu Y, Livingston DM, Sicinski P. Proteomic identification of a direct role for cyclin dl in DNA damage repair. Cancer Res. 2012;72(17):4289–4293.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beltran E, Fresquet V, Martinez-Useros J, et al. A cyclin-D1 interaction with BAX underlies its oncogenic role and potential as a therapeutic target in mantle cell lymphoma. ProcNatlAcadSciUSA. 2011;108(30):12461–12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Albero R, Enjuanes A, Demajo S, et al. Cyclin D1 overexpression induces global transcriptional downregulation in lymphoid neoplasms. J Clin Invest. 2018;128(9):4132–4147. doi: 10.1172/JCI96520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wlodarska I, Meeus P, Stul M, et al. Variant t(2;11)(p11;q13) associated with the IgK-CCND1 rearrangement is a recurrent translocation in leukemic small-cell Bnon-Hodgkin lymphoma. Leukemia. 2004;18(10): 1705–1710. [DOI] [PubMed] [Google Scholar]
- 9.Marrero WD, Cruz-Chacon A, Cabanillas F. Mantle Cell Lymphoma with t(11;22) (q1l3;q11.2) an indolent clinical variant? Leuk Lymphoma. 2018;59(10):2509–2511. doi: 10.1080/10428194.2018.1427863 [DOI] [PubMed] [Google Scholar]
- 10.Peterson JF, Baughn LB, Ketterling RP, et al. Characterization of a cryptic IGH/CCND1 rearrangement in a case of mantle cell lymphoma with negative CCND1 FISH studies. Blood Adv 2019;3(8):1298–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuster C, Martin-Garcia D, Balague O, et al. Cryptic insertions of the immunoglobulin light chain enhancer region near CCND1 in t(11;14)-negative mantle cell lymphoma. Haematologica November 2019. doi: 10.3324/haematol.2019.237073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wagener R, Bens S, Toprak UH, et al. Cryptic insertion of MYC exons 2 and 3 into the IGH locus detected by whole genome sequencing in a case of MYCnegative Burkitt lymphoma. Haematologica May 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hilton LK, Tang J, Ben-Neriah S, et al. The double-hit signature identifies double-hit diffuse large B-cell lymphoma with genetic events cryptic to FISH. Blood. 2019; 134(18): 1528–1532. doi: 10.1182/blood.2019002600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seto M. Cyclin Dl-negative mantle cell lymphoma. Blood. 2013; 121(8): 1249–1250.. doi: 10.1182/blood-2013-01-475954 [DOI] [PubMed] [Google Scholar]
- 15.Salaverria I, Royo C, Carvajal-Cuenca A, et al. CCND2 rearrangements are the most frequent genetic events in cyclin D1(−) mantle cell lymphoma. Blood. 2013; 121(8): 1394–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fu K, Weisenburger DD, Greiner TC, et al. Cyclin D1-negative mantle cell lymphoma: a clinicopathologic study based on gene expression profiling. Blood. 2005;106(13):4315–4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mozos A, Royo C, Hartmann E, et al. SOX11 expression is highly specific for mantle cell lymphoma and identifies the cyclin D1-negative subtype. Haematologica. 2009;94(11): 1555–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iaccarino I, Afify L, Aukema SM, et al. t(ll;14)-positive mantle cell lymphomas lacking cyclin D1 (CCND1) immunostaining because of a CCND1 mutation or exclusive expression of the CCND1b isoform. Haematologica. 2018;103(9):e432–e435. doi: 10.3324/haematol.2018.192435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martin-Garcia D, Navarro A, Valdes-Mas R, et al. CCND2 and CCND3 hijack immunoglobulin light-chain enhancers in cyclin D1(−) mantle cell lymphoma. Blood. 2019; 133(9):940–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Palomero J, Vegliante MC, Eguileor A, et al. SOX11 defines two different subtypes of mantle cell lymphoma through transcriptional regulation of BCL6. Leukemia. 2016;30(7): 1596–1599. [DOI] [PubMed] [Google Scholar]
- 21.Vegliante MC, Palomero J, Perez-Galan P, et al. SOX11 regulates PAX5 expression and blocks terminal B-cell differentiation in aggressive mantle cell lymphoma. Blood. 2013;121(12):2175–2185. [DOI] [PubMed] [Google Scholar]
- 22.Kuo P-Y, Jatiani SS, Rahman AH, et al. SOX11 augments BCR signaling to drive MCL-like tumor development. Blood. 2018;131(20):2247–2255. doi: 10.1182/blood-2018-02-832535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Palomero J, Vegliante MC, Rodriguez ML, et al. SOX11 promotes tumor angiogenesis through transcriptional regulation of PDGFA in mantle cell lymphoma. Blood. 2014;124(14):2235–2247. [DOI] [PubMed] [Google Scholar]
- 24.Balsas P, Palomero J, Eguileor A, et al. SOX11 promotes tumor protective microenvironment interactions through CXCR4 and FAK regulation in mantle cell lymphoma. Blood. 2017;130(4):501–513. doi: 10.1182/blood-2017-04776740 [DOI] [PubMed] [Google Scholar]
- 25.Rudelius M, Rosenfeldt MT, Leich E, et al. Inhibition of focal adhesion kinase overcomes resistance of mantle cell lymphoma to ibrutinib in the bone marrow microenvironment. Haematologica. 2018; 103(1): 116–125. doi: 10.3324/haematol.2017.177162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Queiros AC, Beekman R, Vilarrasa-Blasi R, et al. Decoding the DNA Methylome of Mantle Cell Lymphoma in the Light of the Entire B Cell Lineage. Cancer Cell. 2016;30(5):806–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jares P, Colomer D, Campo E. Molecular pathogenesis of mantle cell lymphoma. JClinlnvest. 2012;122(10):3416–3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Navarro A, Clot G, Royo C, et al. Molecular subsets of mantle cell lymphoma defined by the IGHV mutational status and SOX11 expression have distinct biologic and clinical features. Cancer Res. 2012;72(20):5307–5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fernandez V, Salamero O, Espinet B, et al. Genomic and gene expression profiling defines indolent forms of mantle cell lymphoma. Cancer Res. 2010;70(4): 1408–1418. [DOI] [PubMed] [Google Scholar]
- 30.Ek S, Dictor M, Jerkeman M, Jirstrom K, Borrebaeck CA. Nuclear expression of the non B-cell lineage Sox 11 transcription factor identifies mantle cell lymphoma. Blood. 2008;lll(2):800–805. [DOI] [PubMed] [Google Scholar]
- 31.Espinet B, Ferrer A, Bellosillo B, et al. Distinction between asymptomatic monoclonal B-cell lymphocytosis with cyclin D1 overexpression and mantle cell lymphoma: from molecular profiling to flow cytometry. ClinCancer Res. 2014;20(4): 1007–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martin P, Chadbum A, Christos P, et al. Outcome of deferred initial therapy in mantle-cell lymphoma. JClinOncol 2009;27(8): 1209–1213. [DOI] [PubMed] [Google Scholar]
- 33.Orchard J, Garand R, Davis Z, et al. A subset of t(11;14) lymphoma with mantle cell features displays mutated IgVH genes and includes patients with good prognosis, nonnodal disease. Blood. 2003;101(12):4975–4981. [DOI] [PubMed] [Google Scholar]
- 34.Royo C, Navarro A, Clot G, et al. Non-nodal type of mantle cell lymphoma is a specific biological and clinical subgroup of the disease. Leukemia. 2012;26(8):6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clot G, Jares P, Gine E, et al. A gene signature that distinguishes conventional and leukemic nonnodal mantle cell lymphoma helps predict outcome. Blood. 2018;132(4):413–422. doi: 10.1182/blood-2018-03-838136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lecluse Y, Lebailly P, Roulland S, Gac AC, Nadel B, Gauduchon P. t(l 1; 141positive clones can persist over a long period of time in the peripheral blood of healthy individuals. Leukemia. 2009;23(1476–5551 (Electronic)):1190–1193. [DOI] [PubMed] [Google Scholar]
- 37.Christian B, Zhao W, Hamadani M, et al. Mantle cell lymphoma 12 years after allogeneic bone marrow transplantation occurring simultaneously in recipient and donor. J Clin Oncol 2010;28(27):459–460. doi: 10.1200/JC0.2010.28.3077 [DOI] [PubMed] [Google Scholar]
- 38.Carvajal-Cuenca A, Sua LF, Silva NM, et al. In situ mantle cell lymphoma: Clinical implications of an incidental finding with indolent clinical behavior. Haematologica. 2012;97(2):270–278. doi: 10.3324/haematol.2011.052621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Swerdlow SH, Campo E, Pileri SA, et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood. 2016;127(20):2375–2390.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Karube K, Scarfo L, Campo E, Ghia P. Monoclonal B cell lymphocytosis and “in situ” lymphoma. Semin Cancer Biol. 2014;24:3–14. doi: 10.1016/j.semcancer.2013.08.003 [DOI] [PubMed] [Google Scholar]
- 41.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- 42.Hernandez L, Bea S, Pinyol M, et al. CDK4 and MDM2 gene alterations mainly occur in highly proliferative and aggressive mantle cell lymphomas with wild-type INK4a/ARF locus. Cancer Res. 2005;65(6):2199–2206. [DOI] [PubMed] [Google Scholar]
- 43.Chiron D, Di LM, Martin P, et al. Cell-cycle reprogramming for PI3K inhibition overrides a relapse-specific C481S BTK mutation revealed by longitudinal functional genomics in mantle cell lymphoma. Cancer Discov. 2014;4(9):1022–1035.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bea S, Ribas M, Hernandez JM, et al. Increased number of chromosomal imbalances and high-level DNA amplifications in mantle cell lymphoma are associated with blastoid variants. Blood 1999;93(12):4365–4374. [PubMed] [Google Scholar]
- 45.Hoster E, Rosenwald A, Berger F, et al. Prognostic Value of Ki-67 Index, Cytology, and Growth Pattern in Mantle-Cell Lymphoma: Results From Randomized Trials of the European Mantle Cell Lymphoma Network. JClinOncol 2016;34(1527–7755 (Electronic)): 1386–1394. [DOI] [PubMed] [Google Scholar]
- 46.Pinyol M, Bea S, Pla L, et al. Inactivation of RBI in mantle-cell lymphoma detected by nonsense-mediated mRNA decay pathway inhibition and microarray analysis. Blood. 2007;109(12):5422–5429. [DOI] [PubMed] [Google Scholar]
- 47.Hu Z, Medeiros LJ, Chen Z, et al. Mantle Cell Lymphoma With MYC Rearrangement: A Report of 17 Patients. Am J Surg Pathol 2017;41(2):216–224. doi: 10.1097/PAS.0000000000000758 [DOI] [PubMed] [Google Scholar]
- 48.Hartmann EM, Campo E, Wright G, et al. Pathway discovery in mantle cell lymphoma by integrated analysis of high-resolution gene expression and copy number profiling. Blood. 2010;116(6):953–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rosenwald A, Wright G, Wiestner A, et al. The proliferation gene expression signature is a quantitative integrator of oncogenic events that predicts survival in mantle cell lymphoma. Cancer Cell. 2003;3(2):185–197. [DOI] [PubMed] [Google Scholar]
- 50.Scott DW, Abrisqueta P, Wright GW, et al. New Molecular Assay for the Proliferation Signature in Mantle Cell Lymphoma Applicable to Formalin-Fixed Paraffin-Embedded Biopsies. JClinOncol 2017;35(15): 1668–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Salaverria I, Zettl A, Bea S, et al. Specific secondary genetic alterations in mantle cell lymphoma provide prognostic information independent of the gene expression-based proliferation signature. JClinOncol. 2007;25(10): 1216–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Camacho E, Hernandez L, Hernandez S, et al. ATM gene inactivation in mantle cell lymphoma mainly occurs by truncating mutations and missense mutations involving the phosphatidylinositol-3 kinase domain and is associated with increasing numbers of chromosomal imbalances. Blood. 2002;99(l):238–244. [DOI] [PubMed] [Google Scholar]
- 53.Tort F, Hernandez S, Bea S, et al. Checkpoint kinase 1 (CHK1) protein and mRNA expression is downregulated in aggressive variants of human lymphoid neoplasms. Leukemia. 2005;19(1): 112–117. doi: 10.1038/sj.leu.2403571 [DOI] [PubMed] [Google Scholar]
- 54.Delfau-Larue MH, Klapper W, Berger F, et al. High-dose cytarabine does not overcome the adverse prognostic value of CDKN2A and TP53 deletions in mantle cell lymphoma. Blood. 2015; 126(5):604–611. [DOI] [PubMed] [Google Scholar]
- 55.Ferrero S, Rossi D, Rinaldi A, et al. KMT2D mutations and TP53 disruptions are poor prognostic biomarkers in mantle cell lymphoma receiving high-dose therapy: a FIL study. Haematologica September 2019. doi: 10.3324/haematol.2018.214056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Eskelund CW, Dahl C, Hansen JW, et al. TP53 mutations identify younger mantle cell lymphoma patients who do not benefit from intensive chemoimmunotherapy. Blood. 2017;130(17):1903–1910. [DOI] [PubMed] [Google Scholar]
- 57.Bea S, Salaverria I, Armengol L, et al. Uniparental disomies, homozygous deletions, amplifications, and target genes in mantle cell lymphoma revealed by integrative high-resolution whole-genome profiling. Blood. 2009;113(13):3059–3069.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bea S, Valdes-Mas R, Navarro A, et al. Landscape of somatic mutations and clonal evolution in mantle cell lymphoma. ProcNatlAcadSciUSA. 2013; 110(45): 18250–18255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kridel R, Meissner B, Rogic S, et al. Whole transcriptome sequencing reveals recurrent NOTCH1 mutations in mantle cell lymphoma. Blood. 2012; 119(9): 1963–1971. [DOI] [PubMed] [Google Scholar]
- 60.Kopan R, Ilagan MXG. Thm canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137(2):216–233. doi: 10.1016/j.cell.2009.03.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Silkenstedt E, Arenas F, Colom-Sanmarti B, et al. Notchl signaling in NOTCH1mutated mantle cell lymphoma depends on Delta-Like ligand 4 and is a potential target for specific antibody therapy. J Exp Clin Cancer Res. 2019;38(1):446. doi: 10.1186/s13046-019-1458-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hadzidimitriou A, Agathangelidis A, Darzentas N, et al. Is there a role for antigen selection in mantle cell lymphoma? Immunogenetic support from a series of 807 cases. Blood. 2011; 118(11):3088–3095. [DOI] [PubMed] [Google Scholar]
- 63.Wang ML, Rule S, Martin P, et al. Targeting BTK with ibrutinib in relapsed or refractory mantle-cell lymphoma. NEnglJMed 2013;369(6):507–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bomben R, Ferrero S, D’Agaro T, et al. A B-cell receptor-related gene signature predicts survival in mantle cell lymphoma: results from the Fondazione Italiana Linfomi MCL-0208 trial. Haematologica. 2018;103(5):849–856. doi: 10.3324/haematol.2017.184325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Psyrri A, Papageorgiou S, Liakata E, et al. Phosphatidylinositol 3’-kinase catalytic subunit alpha gene amplification contributes to the pathogenesis of mantle cell lymphoma. Clin Cancer Res. 2009;15(18):5724–5732. doi: 10.1158/1078-0432.CCR-08-3215 [DOI] [PubMed] [Google Scholar]
- 66.Balaji S, Ahmed M, Lorence E, Yan F, Nomie K, Wang M. NF-kappaB signaling and its relevance to the treatment of mantle cell lymphoma. J Hematol Oncol. 2018; 11(1):83. doi: 10.1186/s13045-018-0621-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Honma K, Tsuzuki S, Nakagawa M, et al. TNFAIP3/A20 functions as a novel tumor suppressor gene in several subtypes of non-Hodgkin lymphomas. Blood. 2009; 114(12):2467–2475. doi: 10.1182/blood-2008-12-194852 [DOI] [PubMed] [Google Scholar]
- 68.Mansouri L, Noerenberg D, Young E, et al. Frequent NFKBIE deletions are associated with poor outcome in primary mediastinal B-cell lymphoma. Blood. 2016;128(23):2666–2670. [DOI] [PubMed] [Google Scholar]
- 69.Hershkovitz-Rokah O, Pulver D, Lenz G, Shpilberg O. Ibrutinib resistance in mantle cell lymphoma: clinical, molecular and treatment aspects. Br J Haematol. 2018; 181(3):306–319. doi: 10.1111/bjh.15108 [DOI] [PubMed] [Google Scholar]
- 70.Pham LV, Tamayo AT, Yoshimura LC, Lo P, Ford RJ. Inhibition of constitutive NF-kappa B activation in mantle cell lymphoma B cells leads to induction of cell cycle arrest and apoptosis. J Immunol. 2003;171(l):88–95. doi: 10.4049/jimmunol.171.1.88 [DOI] [PubMed] [Google Scholar]
- 71.Roue G, Perez-Galan P, Lopez-Guerra M, Villamor N, Campo E, Colomer D. Selective inhibition of IkappaB kinase sensitizes mantle cell lymphoma B cells to TRAIF by decreasing cellular FLIP level. J Immunol. 2007;178(3):1923–1930. doi: 10.4049/jimmunol.178.3.1923 [DOI] [PubMed] [Google Scholar]
- 72.Petrakis G, Veloza L, Clot G, et al. Increased tumour angiogenesis in SOX11positive mantle cell lymphoma. Histopathology. 2019;75(5):704–714. doi: 10.1111/his.13935 [DOI] [PubMed] [Google Scholar]
- 73.Oyer JA, Huang X, Zheng Y, et al. Point mutation E1099K in MMSET/NSD2 enhances its methyltranferase activity and leads to altered global chromatin methylation in lymphoid malignancies. Leukemia. 2014;28(1): 198–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Valdes-Mas R, Bea S, Puente DA, Fopez-Otin C, Puente XS. Estimation of copy number alterations from exome sequencing data. PLoSOne 2012;7(12):e51422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang J, Fee Y-R, Dang F, et al. PTEN Methylation by NSD2 Controls Cellular Sensitivity to DNA Damage. Cancer Discov 2019;9(9): 1306–1323. doi: 10.1158/2159-8290.CD-18-0083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Agarwal R, Chan YC, Tam CS, et al. Dynamic molecular monitoring reveals that SWI-SNF mutations mediate resistance to ibrutinib plus venetoclax in mantle cell lymphoma. NatMed. 2019;25(1): 119–129. [DOI] [PubMed] [Google Scholar]
- 77.Meissner B, Kridel R, Lim RS, et al. The E3 ubiquitin ligase UBR5 is recurrently mutated in mantle cell lymphoma. Blood. 2013;121(16):3161–3164. [DOI] [PubMed] [Google Scholar]