Abstract
In this study, hypotensive peptides derived from mature flaxseed protein sequences were predicted in silico using BIOPEP-UWM with nine proteases, three each from digestive, plant and microbial sources. The physicochemical properties of 2256 ACE-inhibitory peptides and 267 renin-inhibitory peptides (including seven (7) peptides with dual inhibitory activities against both ACE and renin enzymes) were assessed in silico using the ‘Peptides’ package of R. The hypotensive peptides showed relatively low molecular weight (mol. wt.) range (132 = mol. wt. ≤ 442 Da); broad range of isoelectric point (3.61 = pI ≤ 12.50); both high (>2) and low (≤2) Boman indices, and a variety of hydrophobicity indices (hydrophilic, hydrophobic and amphipathic properties). Following this, the seven peptides with dual ACE and renin inhibitory activities were selected for molecular docking with the respective enzyme receptors. The binding energies of the seven hypotensive peptides with ACE and renin respectively ranged from −36.82 to −25.94 kJ/mol, and −33.05 to −27.61 kJ/mol; and compared well with values recorded for inhibitor drugs, captopril (−26.78 kJ/mol) and aliskiren (−34.73 kJ/mol). The seven peptides inhibited ACE through hydrogen bonds, electrostatic and hydrophobic interactions; and renin, mainly through hydrogen bonds and hydrophobic interactions. In silico prediction of adsorption, digestion, metabolism, excretion and toxicity (ADME/Tox) profile based on physicochemical properties and Lipinski's rule-of-five showed that the peptides were non-toxic and had desirable drug-like properties (flexibility, lipophilicity, molecular weight, gastrointestinal absorption, and bioavailability). This study provides insight into the molecular interactions of hypotensive peptides with their physiological targets, and the potential to develop the bioactive peptides from flaxseed proteins.
Keywords: Bioactive peptides, Hypotensive peptides, Molecular docking, Bioinformatics, Flaxseed proteins
Graphical abstract
Highlights
-
•
Flaxseed proteins were assessed in silico as source of hypotensive peptides.
-
•
Plant proteases were most suitable to release hypotensive peptides in silico.
-
•
Hypotensive peptides had molecular docking features similar to captopril and aliskiren.
-
•
In silico-derived hypotensive
peptides were non-toxic, and had drug-like properties.
1. Introduction
Increased blood pressure, especially hypertension (systolic pressure), conferring a significant cardiovascular risk, is an important public health concern. One of the blood pressure regulation systems, the renin–angiotensin system (RAS), involves two key enzymes, angiotensin-I converting enzyme (ACE) and renin (Politi et al., 2009). The first action in this system is that angiotensinogen is converted by the hydrolysis of renin into angiotensin-I (the rate-limited step), which is subsequently cleaved by ACE to generate angiotensin-II, a vasoconstrictive hormone that binds angiotensin I (AT-I) receptor to control blood pressure (He et al., 2014). The combination therapy in hypertension treatment, to simultaneously inhibit ACE and renin activities, was reported to be efficient without severe negative side effects (Oliva and Bakris, 2012).
Due to the significant importance of RAS, several ACE and renin inhibitors, such as captopril and aliskiren, respectively have already been used as antihypertensive drugs in clinical settings. However, single drugs (ACE or renin inhibitors) are not always effective and are often accompanied with several side effects such as cough and dizziness (Qian et al., 2019). Therefore, developing safer and multifunctional antihypertensive drugs from different sources is in high demand.
ACE and renin inhibitors are widely distributed in several food sources from plants and animals. Plant sources of food have attracted more attention for reasons of cost-effectiveness and sustainability (Daskaya-Dikmen et al., 2017). Therefore, a number of hypotensive peptides from plant sources have been isolated, identified or characterized from seaweed, rapeseed, hempseed, yellow field pea, and flaxseed. For example, Girgih et al. (2014) identified four hypotensive peptides with both ACE and renin inhibitory activities from hempseed (Cannabis sativa L.), and provided the inhibition kinetics and molecular docking modelling of the peptides. He et al. (2014) (He et al., 2013) purified three hypotensive rapeseed protein-derived peptides and proposed their interaction with ACE and renin using molecular docking models. Moreover, the inhibition kinetics of flaxseed (Linum usitatissimum) protein hydrolysates against ACE and renin have been reported by Udenigwe et al. (2009).
The use of conventional methods to discover and characterize novel bioactive peptides from food proteins is usually time-consuming and costly. Hence, in silico prediction and evaluation have been demonstrated to be necessary to comprehensively study the feasibility of producing bioactive peptides from various food proteins (Agyei et al., 2018, Wu et al., 2006, Udenigwe, 2014). Our previous study demonstrated that in silico proteolysis of mature flaxseed proteins with selected proteases generated structurally diverse antioxidant peptides with potential for use in developing functional foods (Ji et al., 2019). Using the diverse flaxseed proteome, the aim of this study was to evaluate the release of hypotensive flaxseed peptides using different enzymes, to predict their physicochemical properties, identify the mechanism of interactions of the dual inhibitor hypotensive peptides with both ACE and renin (docking), and the in silico drug-likeness of the multifunctional peptides.
2. Materials and methods
2.1. Preparation of hypotensive peptides
The in silico hydrolysis of mature flaxseed proteins has been described in our previous work (Ji et al., 2019). A total of 23 selected mature flaxseed protein sequences were hydrolysed in silico using the ‘enzyme(s) action’ feature of BIOPEP-UWM (http://www.uwm.edu.pl/biochemia/index.php/en/biopep). Nine (9) selected proteases were used, representative of digestive ((trypsin (E.C. 3.4.21.4), pepsin (E.C. 3.4.23.1), pancreatic elastase II (E.C. 3.4.21.71)), plant ((papain (E.C. 3.4.22.2), stem bromelain (E.C. 3.4.22.32), ficin (E.C. 3.4.4.12)) and microbial proteases (thermolysin (E.C. 3.4.24.27), subtilisin (E.C. 3.4.21.62) proteinase P1 (lactocepin; E.C. 3.4.21.96). The release of hypotensive (i.e. ACE-inhibitory and renin inhibitory) peptides from the mature flaxseed proteins was predicted using the ‘search for active fragments’ tool of the BIOPEP-UWM platform.
2.2. Physicochemical properties of hypotensive flaxseed peptides released after in silico enzymatic proteolysis
Several physicochemical properties of the in silico-derived hypotensive peptides were determined using the ‘Peptides’ package (version 2.4) in R. These properties include molecular weight (mol. wt.), peptide chain length, amino acid composition, isoelectric point (pI), Boman index, net charge, and hydrophobicity index.
2.3. Molecular docking of hypotensive peptides to enzyme receptors
Among all the identified peptides, only seven (7) had dual inhibitory activities against both ACE and renin enzymes. These 7 hypotensive dipeptides were therefore selected, together with inhibitor drugs, captopril (PDB 2X8Z; ACE inhibitor) and aliskiren (PDB 2V0Z; renin inhibitor) were selected for molecular docking studies. Molecular docking was performed using the Discovery Studio software 2019 (DS 2019) and Autodock Vina. The structures of the seven hypotensive dipeptides from flaxseed proteins were generated with DS 2019. For docking onto ACE receptor, a crystal structure of human ACE (1O8A (PDB) bound with lisinopril (an ACE inhibitor) was used. The lisinopril and water molecules were removed and polar hydrogen atoms were added prior to docking. A binding site with a radius of 13 Å and coordinates x: 40.588, y: 37.374 and z: 43.447 was created. Automated molecular docking was performed using Autodock Vina in the presence of zinc and chloride ions. For renin docking, a crystal structure of human renin 2V0Z (PDB) bound with aliskiren (a renin inhibitor), was used. The aliskiren and water molecules (except H2O-2184 and H2O-2250) were removed, and polar hydrogen atoms were added. A binding site with a radius of 12 Å and coordinates x: 7.568, y: 46.092, z: 68.842 was created, and automated molecular docking was then performed using Autodock Vina. All generated docking modes were evaluated according to affinity energy values. The dissociation constant (Ki) were also calculated using affinity energy (Galli et al., 2014), according to the following equation:
| (1) |
where R represents the gas constant and T represents the temperature. Ki was computed starting from the binding free energy values at a fixed temperature (298 K).
The DS 2019 was also utilized to view hydrogen bonds as well as hydrophilic, hydrophobic, and electrostatic interactions between residues at the ACE or renin active sites and the peptide poses.
2.4. In silico drug-likeness evaluation
In silico evaluation of drug-likeness for the seven (7) multifunctional hypotensive peptides was evaluated using the SwissADME tool (http://www.swissadme.ch/index.php# (Daina et al., 2017)), which uses the ADME (absorption, distribution, metabolism, and excretion) properties of a compounds as estimates and indicators of pharmacokinetics, and drug-likeness of a compound (Mbarik et al., 2019). In addition, ToxinPred (Gupta et al., 2013), (available at https://webs.iiitd.edu.in/raghava/toxinpred/index.html), was used for the prediction of potential toxicity of the flaxseed peptides.
3. Results and discussion
3.1. Protease sources and effect on hypotensive peptides release
The number of hypotensive peptides released from mature flaxseed proteins was enzyme dependent, as shown in Table 1. The total number of ACE-inhibiting peptides derived from plant proteases (1430) was higher than those released by digestive (169) and microbial enzymes (657). The number of renin-inhibitory peptides released by plant proteases (172) was also higher than those released by digestive (47) and microbial (48) proteases. Of all the RAS components, ACE has been the most popular target for developing antihypertensive drugs (including food-derived peptide inhibitors), especially compared to renin. Therefore, the number of ACE inhibiting peptide sequences deposited in the BIOPEP-UWM database is more than the number of peptides with other antihypertensive mechanisms. Nonetheless, the outcome of our study gives an indication of the types of enzymes that are best suited for producing hypotensive peptides from flaxseed proteins. Overall, the plant proteases were more efficient at hydrolysing flaxseed proteins and releasing peptides in silico, as evidenced by the higher degree of hydrolysis (45.5%, 43.8% and 57.13% for papain, ficin, and stem bromelain, respectively). These plant proteases are known to have broad spectra of amino acid specificity (Mótyán et al., 2013). Papain, for example cleaves at leucine, glycine, and basic amino acid residues, and especially if these residues are bonded, in the P2 position, to hydrophobic amino acids. Our previous work showed that the 23 matured proteins had high levels of basic and hydrophobic amino acids, mostly arising from oleosins and globulins, respectively (Ji et al., 2019). The presence of these unique amino acid signatures may account for the high in silico degree of hydrolysis observed in this study for the plant proteases, especially papain. The microbial proteases also produced high degree of hydrolysis (34.7%, 46.7% and 28.3% for thermolysin, lactocepin, and subtilisin, respectively) and released several hypotensive peptides, but not to the same extent as the plant proteases. The digestive enzymes gave a rather low degree of hydrolysis (9.8%, 12.0% and 14.3%, respectively for trypsin, pepsin and pancreatic elastase II). This is due to the fact that the digestive enzymes have recognition sites comprising of only a few amino acids, i.e. lysine and arginine (for trypsin), phenylalanine and leucine (for pepsin), and phenylalanine, methionine and leucine (for pancreatic elastase II). These findings demonstrate that the plant proteases are more suitable for releasing peptides with hypotensive properties from flaxseed proteins.
Table 1.
Enzyme types and the number of ACE, renin and dual inhibitory peptides realised after hydrolysis of mature flaxseed storage proteins.
| Enzyme Source | Enzyme Type | Total number of ACE-inhibitory peptides | Total number of renin-inhibitory peptides | Total number of multifunctional peptidesa |
|---|---|---|---|---|
| Digestive | Trypsin | 47 | 13 | 10 |
| Pepsin | 55 | 15 | 10 | |
| Pancreatic elastase II | 67 | 19 | 12 | |
| Plant | Papain | 583 | 65 | 22 |
| Ficin | 378 | 45 | 19 | |
| Stem bromelain | 469 | 62 | 38 | |
| Microbial | Thermolysin | 245 | 14 | 14 |
| Proteinase P1 (Lactocepin) | 171 | 13 | 6 | |
| Subtilisin | 241 | 21 | 8 | |
| Total | 2256 | 267 | 139 | |
Those with both ACE-inhibitory and renin inhibitory activities.
Moreover, all the enzymes studied were capable of releasing multifunctional peptides that had inhibitory properties against both renin and ACE. The plant proteases released 79 multifunctional peptides, whereas the digestive and microbial proteases released 32 and 28 peptides, respectively. Interestingly, stem bromelain released the greatest number of in silico-derived multifunctional peptides (38) from the flaxseed proteins. The ability of an enzyme to release peptide sequences with more than one biological property is an important and often desirable feature (Udenigwe and Mohan, 2014, Sistla, 2013). This is because the dual RAS enzyme inhibiting peptides may to able provide more pronounced reduction in blood pressure than the single RAS enzyme inhibitors (Udenigwe and Mohan, 2014), although this is yet to be clinically validated in hypertensive humans.
3.2. Physicochemical characteristics of the in silico hypotensive flaxseed peptides
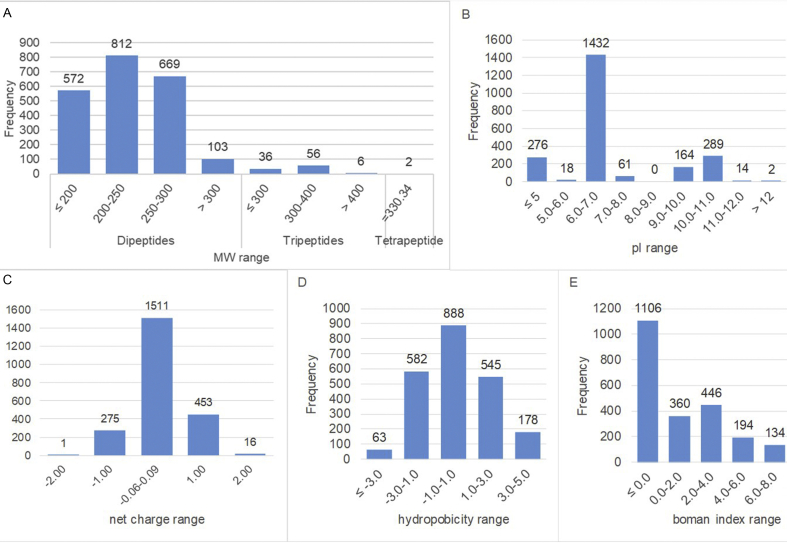
The physicochemical characteristics of in silico-hydrolysed ACE-inhibitory flaxseed peptides are presented in Fig. 1. The mol. wt. of ACE-inhibitory peptides ranged from 132 to 442 Da. The dipeptides, which were the most abundant peptides, had a mol. wt. range of 132–167 Da, while the tripeptides ranged from 233 to 442 Da. There were also two tetrapeptides of 330 Da each. As shown in Fig. 1A, dipeptides and tripeptides are most abundant in the mol. wt. range of 200–250 Da (821) and 300–400 Da (56), respectively. The pI of the ACE-inhibitory peptides ranged from 3.61 to 12.50. Out of the 2256 ACE-inhibitory peptides obtained from flaxseed proteins, 276 had pI ≤ 5 with net charges of −1.00 (275) or −2.00 (1). These peptides mostly had one of the two negatively charged amino acids (D, E) as well as other hydrophobic amino acids. There were 1511 peptides having 5 < pI ≤ 8 with net charges of about 0.00; and 453 peptides had 9 < pI ≤ 11 with net charges of 1.00, and these usually contained one of the two positively charged amino acids (R, K). Lastly, 16 peptides had pI ≥ 11, with net charges of 2.00 (Fig. 1B and C).
Fig. 1.
Distribution of physicochemical properties (A. molecular weight; B. isoelectric point, pI; C. net charge; D. hydrophobicity index; and E. Boman index) of the ACE-inhibitory peptides released from flaxseed proteins in silico.
Hydrophobicity values of the 2256 peptides ranged from −4.50 to 4.15 (Fig. 1D). There were 645 hydrophilic peptides (with hydrophobicity values of −4.50 to 1.00), 723 hydrophobic peptides (with hydrophobicity values of 1.00–4.15), and 888 amphipathic and neutral peptides (with hydrophobicity values of −1.00 to 1.00). The varying hydrophobicity of the ACE-inhibitory peptides show that they may be utilized in a wide range of food systems, such as water-soluble, lipid-soluble, or emulsion-based systems. The Boman indices (BI) of ACE-inhibitory flaxseed peptides ranged from −4.92 to 14.92 (Fig. 1E). BI of most of the peptides (1466) were less than 2. However, there were also 790 peptides with Boman index >2, which indicates higher probability of the peptides to bind to proteins and to show multifunction (Azad et al., 2011).
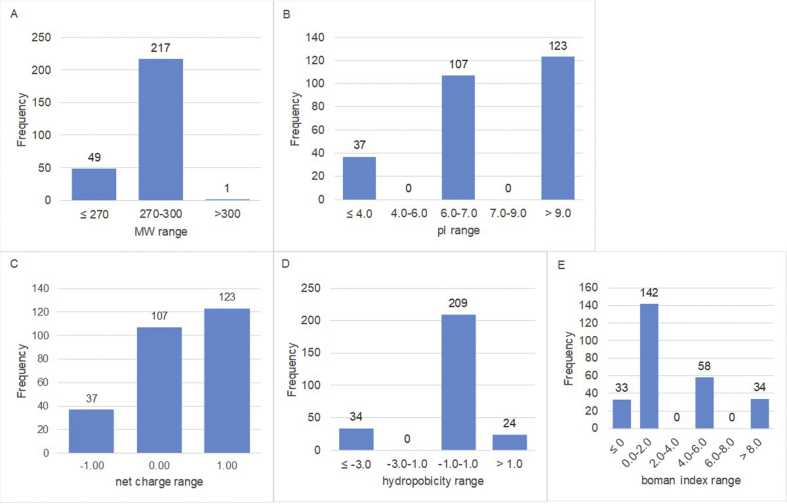
The physicochemical characteristics of in silico-hydrolysed renin-inhibitory flaxseed peptides are presented in Fig. 2. All the renin-inhibitory peptides were dipeptides having mol. wt. range of 252–317 Da. The pI of the renin-inhibitory peptides ranged from 3.85 to 10.55. Out of the 267 renin-inhibitory peptides, 37 Glu-Phe (EF) had pI of 3.85 with net charges of around −1.00. There were 107 of these peptides having 6 < pI ≤ 7 with net charges of about 0.00, and these peptides usually contained one amino acid with polar uncharged side chain (e.g. Q, T, S) and Phe (a hydrophobic amino acid). A number of the neutral peptides had only two hydrophobic amino acids (YA and LW). Furthermore, 123 of the renin-inhibitory peptides had pI greater than 9 with net charges of 1.00. Among these, KF had pI = 9.70, and IR, LR and NR had pI of ~10.55 (Fig. 2B and C). The hydrophobicity of the 267 peptides ranged from −4.00 to 1.45 (Fig. 2 D). There were 34 hydrophilic peptides (NR) with hydrophobicity values of −4.00; 24 weak hydrophobic peptides with hydrophobicity values of 1.05 or 1.45, and 209 amphipathic peptides (with hydrophobicity values from −1.00 to 1.00). Compared to ACE-inhibitory peptides, which are expected to function in both hydrophilic and hydrophobic systems, based on their physicochemical properties, the renin-inhibitory peptides may not be suitable for utilization in lipid-based systems due to their lower hydrophobicity values.
Fig. 2.
Distribution of physicochemical properties (A. molecular weight; B. isoelectric point, pI; C. net charge; D. hydrophobicity index; and E. Boman index) of the renin-inhibitory peptides released from flaxseed proteins in silico.
The Boman indices of renin-inhibitor flaxseed peptides ranged from −3.60 to 10.80. Similar to ACE-inhibitory peptides, the Boman indices of the most peptides (175 peptides) were less than 2, and 92 peptides had the Boman index of between 5.0 and 10.8 (Fig. 2E).
3.3. Molecular docking of RAS enzymes with the dual inhibitory flaxseed peptides
3.3.1. ACE
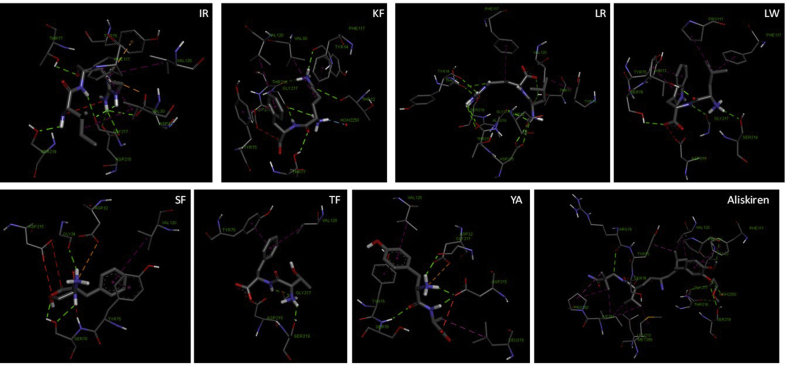
The best docking poses of the seven hypotensive flaxseed peptides and captopril interactions with the active site of ACE in the presence of Zn (II) are shown in Table 2 and Fig. 3. The results indicate affinity energies (kJ/mol) and dissociation constants (Ki) for IR, KF, LR, LW, SF, TF, YA and captopril (Table 2). The findings suggest that, compared to captopril, five flaxseed peptides (KF, LW, TF, SF, YA) have higher ACE-inhibitory properties than the other two peptides (IR, LR). These results are similar to previous docking findings with dipeptides. For example, the affinity energy of TF reported by He et al. (2014) (−31.36 kJ/mol), where the docking work was performed using CDOCKER tool of Discovery Studio 2.5, is almost the same as the result obtained in our study (−31.8 kJ/mol).
Table 2.
Predicted binding energies, computational dissociation constant and Zn(II) coordination distances of hypotensive flaxseed peptides with ACE (PDB: 1O8A).
| Ligand | Affinity energy (kJ/mol) | Ki (μM) | Zn(II) coordination |
|
|---|---|---|---|---|
| Distance (Å) | Atom | |||
| IR | −25.94 | 28.20 | – | No Zn(II) coordination |
| KF | −30.54 | 4.40 | – | No Zn(II) coordination |
| LR | −26.36 | 23.82 | – | No Zn(II) coordination |
| LW | −36.82 | 0.35 | – | No Zn(II) coordination |
| SF | −29.29 | 7.30 | 1.57 | amino group of serine |
| TF | −31.80 | 2.65 | – | No Zn(II) coordination |
| YA | −29.71 | 6.16 | 2.6 | Carboxylic acid group of alanine |
| Captopril | −26.78 | 20.12 | 2.76 | Sulfhydryl group of captopril |
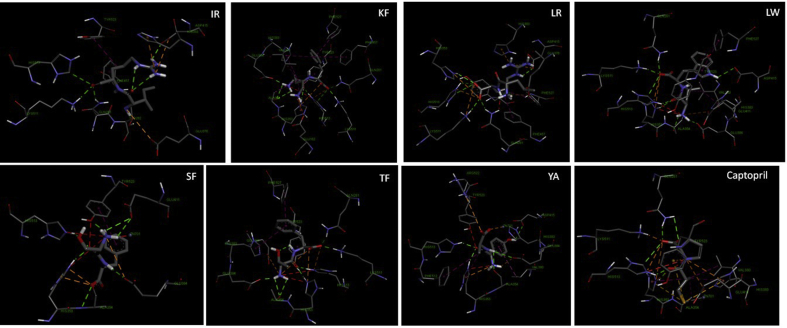
Fig. 3.
Predicted binding mode of hypotensive flaxseed peptides to ACE (PDB: 1O8A). The best scored docking pose is shown.
The best poses of each hypotensive flaxseed peptides and captopril were mainly stabilized by hydrogen bonds, hydrophobic and electrostatic interactions with the ACE catalytic site residues (Fig. 3). Four hypotensive peptides, LW, SF, TF and YA, mainly occupied the S1 and S2′ subsites via hydrogen bonds and hydrophobic interactions with Tyr523, Glu384, Ala354 residues of S1 and His353, His513, Lys511, Gln281 residues of S2′. These interactions are similar to that observed for captopril. IR mainly occupied S2′ via hydrogen bonds with His513, Lys511, Gln281. LR mainly occupied S2′ and hydrophobic pocket subsites via hydrogen bonds and electrostatic interactions with His353, His513, Lys511, and Gln281 residues of S2′ and with Phe457, Phe527 residues of the hydrophobic pocket. KF occupied S1, S1′, S2′ subsites and hydrophobic pocket via hydrophobic interactions and hydrogen bonds. Interaction was mainly with Tyr523, Glu384, and Ala354 residues of S1; Glu162 of S1′; His353, His513, Lys511, Gln281 residues of S2′; and with Phe457, Phe527 residues of the hydrophobic pocket (Table 3). The binding sites and interaction types are similar to previous studies of docking di-, or tripeptides to ACE (He et al., 2014, Jimsheena and Gowda, 2010). The hydrophobic interactions are believed to contribute to burying peptides into hydrophobic pockets of the enzyme (He et al., 2014, Jimsheena and Gowda, 2010).
Table 3.
Residues of ACE that interact with docked flaxseed peptides.
| IR | KF | LR | LW | SF | TF | YA | Captopril | |
|---|---|---|---|---|---|---|---|---|
| ALA354 | ● | ● | ● | ● | ● | ● | ||
| ARG522 | ● | |||||||
| ASP415 | ● | ● | ● | ● | ||||
| GLN281 | ● | ● | ● | ● | ● | ● | ||
| GLU162 | ● | |||||||
| GLU376 | ● | ● | ||||||
| GLU384 | ● | ● | ● | ● | ● | |||
| GLU411 | ● | ● | ● | ● | ||||
| HIS353 | ● | ● | ● | ● | ● | ● | ● | |
| HIS383 | ● | ● | ● | ● | ● | ● | ● | |
| HIS513 | ● | ● | ● | ● | ● | ● | ● | ● |
| LYS511 | ● | ● | ● | ● | ● | ● | ||
| PHE457 | ● | ● | ● | |||||
| PHE512 | ● | |||||||
| PHE527 | ● | ● | ● | ● | ||||
| THR282 | ● | |||||||
| TYR523 | ● | ● | ● | ● | ● | ● | ● | ● |
| VAL380 | ● | ● | ● | ● |
The functional importance of multiple hydrogen bonds has been reported and shown to contribute to peptide-induced inhibition of enzyme activity (He et al., 2014, Jimsheena and Gowda, 2010), and correlate with IC50 values. Wu et al. (2006) provided many di-, or tripeptides log IC50 values from different laboratories, revealing that log IC50 values of hypotensive flaxseed peptides (e.g., KF, LW, SF, TF) that form more hydrogen bonds (7–8) with ACE are significantly lower (IC50 of 2.84–2.92) than those of hypotensive flaxseed peptides (e.g., IR) that form less hydrogen bonds (4) (Table 4).
Table 4.
Hydrogen bonds observed in the best scored docking model of hypotensive flaxseed peptides with ACE.
| ACE residues in H-bonding | Number of H-bonds and their corresponding distance (Å) |
|||||||
|---|---|---|---|---|---|---|---|---|
| IR | KF | LR | LW | SF | TF | YA | Captopril | |
| ALA354:O | 2(2.74,2.68) | 1(2.33) | 2(2.54,2.68) | 2(2.56,2.20) | ||||
| ALA354:HN | 1(2.79) | |||||||
| ASP415:OD1 | 1(2.61) | |||||||
| ASP415:OD2 | 1(2.39) | 1(2.40) | 1(2.67) | |||||
| GLN281:HE21 | 1(2.96) | |||||||
| GLN281:HE22 | 1(2.06) | 1(2.07) | 1(2.16) | 1(2.22) | 1(1.93) | 1(2.11) | ||
| GLU384:OE2 | 1(1.87) | 1(2.33) | 1(2.21) | 1(1.93) | 1(2.89) | |||
| GLU411:OE1 | 2(3.11,2.98) | |||||||
| HIS353:HE2 | 1(2.15) | 1(2.69) | 1(2.16) | 1(2.08) | 1(2.01) | 1(2.11) | 1(2.13) | |
| HIS513:HE2 | 1(2.12) | 1(3.28) | 1(2.16) | 2(2.14,2.72) | 1(2.10) | 1(2.06) | 1(2.22) | |
| HIS513:CE1 | 1(3.65) | 1(3.54) | 1(3.32) | 1(3.41) | 1(3.62) | |||
| LYS511:HZ1 | 1(2.11) | 1(1.79) | 1(2.31) | 1(2.04) | 1(1.78) | |||
| TYR523:OH | 1(1.92) | |||||||
| Total | 4 | 8 | 6 | 8 | 8 | 7 | 6 | 5 |
In addition to hydrogen bonds, interaction of peptides with Zn (II) atoms is also believed to play a significant role in enzyme inhibition, and the shorter the distance between the metal and peptide, the greater the inhibition (He et al., 2014, Jimsheena and Gowda, 2010). However, as shown in Table 2, two hypotensive flaxseed peptides (SF, YA) and captopril interacted with Zn (II), although SF did not show substantially low log IC50 value (2.11, tested by only one laboratory) compared to the highly active peptides (Wu et al., 2006). To the best of our knowledge, the ACE inhibitory IC50 value of YA has not been reported. Further studies are needed to fully delineate the effect of Zn (II) atom coordination on the ACE inhibitory activity of dipeptides.
3.3.2. Renin
The molecular docking of the seven hypotensive flaxseed peptides and aliskiren within renin active sites is shown in Fig. 4. Potential affinity energies and the dissociation constants are shown in Table 5. The results suggest that the seven peptides have lower renin-inhibitory property than aliskiren. The affinity energy of TF interactions with renin reported by He et al. (2014) (−31.46 kJ/mol) is also similar to our result (−30.1 kJ/mol). LW had the best binding affinity for renin of all the flaxseed peptides (Table 5). This is because it possesses structural features (N-terminal low mol. wt. hydrophobic residue, and C-terminal residue with bulky sidechain) that were determined by chemometrics to favour renin inhibition (Udenigwe et al., 2012).
Fig. 4.
Predicted binding mode of the hypotensive flaxseed peptides to renin (PDB: 2V0Z). The best scored docking pose is shown.
Table 5.
Predicted binding energies of hypotensive flaxseed peptides with Renin (PDB: 2V0Z).
| Ligand | Affinity energy (kJ/mol) | Ki (μM) |
|---|---|---|
| IR | −27.61 | 14.35 |
| KF | −31.80 | 2.65 |
| LR | −30.12 | 5.21 |
| LW | −33.05 | 1.60 |
| SF | −28.03 | 12.12 |
| TF | −30.12 | 5.21 |
| YA | −30.12 | 5.21 |
| Aliskiren | −34.73 | 0.81 |
Similar to ACE docking results, the best poses of the seven hypotensive peptides and aliskiren interactions with renin were mainly stabilized by hydrogen bonds and hydrophobic interactions (Fig. 4, Table 6). Four hypotensive peptides, IR, KF, LW and TF, occupied four renin active pockets (S1, S3, S4, S1'). Amino acid residues Asp32, Asp215, Gly217, Phe117, Tyr75, Val30, and Val120 were used for interaction in S1 pocket; Phe117, Pro111, Ser219, and Thr12 were used for S3; Ser219, and Thr12 for S4; and Asp215, and Tyr75 for S1'. LR occupied five renin active pockets (S1, S2, S3, S4, S1') using Asp32, Asp215, Gly217, Phe117, Tyr75, and Val120 for S1; Ala218 for S2; Phe117, Ser219, and Thr12 for S3; Ser219, and Thr12 for S4; and with Asp215, and Tyr75 for S1'. SF occupied three renin active pockets (S1, S1', S2') with the aid of residues Asp32, Asp215, Tyr75, and Val120 for S1; Asp215, and Tyr75 for S1'; and Gly217 for S2'. YA occupied two renin active pockets (S1, S1') using Asp32, Asp215, Gly217, Phe117, Tyr75, and Val120 for S1; and with Asp215, and Tyr75 for S1'. Binding of aliskiren was different, compared to the flaxseed peptides, as the drug occupied five renin active pockets (S1, S3, S4, S1', S3') with Gly217, Phe117, Val30, Val120 in the S1 pocket; Phe117, and Ser219 in S3; Ser219 in S4; Leu213 and Tyr75 in S1'; and Ile291 in S2. Involvement of the active site residues in renin activity has been previously described (Sielecki et al., 1989).
Table 6.
Residues of renin that interact with the docked flaxseed peptides.
| IR | KF | LR | LW | SF | TF | YA | Aliskiren | |
|---|---|---|---|---|---|---|---|---|
| ALA218 | ● | |||||||
| ARG74 | ● | |||||||
| ASP32 | ● | ● | ● | ● | ||||
| ASP215 | ● | ● | ● | ● | ● | ● | ● | |
| GLY34 | ● | |||||||
| GLY217 | ● | ● | ● | ● | ● | ● | ● | |
| ILE291 | ● | |||||||
| LEU213 | ● | ● | ||||||
| MET289 | ● | |||||||
| PHE117 | ● | ● | ● | ● | ● | |||
| PRO111 | ● | ● | ||||||
| PRO292 | ● | |||||||
| SER76 | ● | ● | ● | ● | ||||
| SER219 | ● | ● | ● | ● | ● | |||
| THR12 | ● | ● | ||||||
| THR77 | ● | ● | ● | ● | ||||
| THR216 | ● | ● | ● | |||||
| TYR14 | ● | ● | ||||||
| TYR75 | ● | ● | ● | ● | ● | ● | ● | ● |
| VAL30 | ● | ● | ● | |||||
| VAL120 | ● | ● | ● | ● | ● | ● | ● |
As shown in Table 5, Table 7, among the docking results of seven hypotensive peptides within renin, the number of binding sites and hydrogen bonds are different but showed no significant effect on the affinity energy. For example, LR has 12 binding sites and 13 hydrogen bonds, and TF has only five binding sites and three hydrogen bonds, even though they have the same affinity energy (−30.1 kJ/mol). The reason could be that the seven peptides mainly occupied S1 and S1' sub-pocket, but did not bind well to S3 sub-pocket (0–3 binding sites). Rahuel et al. (2000) reported that high affinity inhibition of renin depends mainly on the S3 sub-pocket (Gln13, Tyr14, Val30, Tyr155, Ala218, Ser219, Ala303), that peptides have a poor ability to bind with the S3 sub-pocket, and that peptide modification to improve lipophilicity is necessary to enhance binding energy.
Table 7.
Hydrogen bonds observed in the best scored docking model of the hypotensive flaxseed peptides within renin.
| renin residues in H-bonding | Number of H-bonds and their corresponding distance (Å) |
|||||||
|---|---|---|---|---|---|---|---|---|
| IR | KF | LR | LW | SF | TF | YA | Aliskiren | |
| ALA218:O | 1(3.03) | |||||||
| ASP32:OD1 | 1(2.84) | 1(2.26) | ||||||
| ASP215:OD1 | 1(2.73) | 2(2.92,3.19) | 2(1.81,2.03) | |||||
| ASP215:OD2 | 1(2.83) | 1(3.04) | 1(3.32) | |||||
| ARG74:O | ||||||||
| GLY34:O | 1(2.23) | |||||||
| GLY217:O | 2(2.37,2.43) | 2(2.02,2.93) | 1(2.85) | 2(2.25,2.65) | 2(2.45,1.94) | 1(2.42) | 1(3.38) | |
| PRO111:O | 1(3.49) | |||||||
| SER76:HG | 1(2.69) | 1(2.60) | ||||||
| SER76:HN | 1(2.27) | |||||||
| SER76:OG | 1(2.61) | |||||||
| SER219:OG | 1(2.27) | 1(2.70) | 1(2.33) | 1(2.06) | 1(3.67) | |||
| THR12:O | 1(3.58) | 2(2.27, 3.65) | ||||||
| THR77:HG1 | 1(2.57) | 1(2.38) | 2(3.10,2.70) | 1(2.00) | ||||
| THR216:O | 1(2.37) | 1(2.53) | 1(3.51) | |||||
| TYR14:O | 1(2.37) | 1(2.60) | ||||||
| HOH2250:O | 1(2.22) | 1(3.37) | ||||||
| Total | 7 | 7 | 13 | 5 | 3 | 3 | 4 | 6 |
Additionally, Sielecki et al. (1989) reported that the residues Asp32 or Asp215 are important for renin inhibition. However, our docking models showed a conflicting result. All seven peptides had interactions with Asp215, and IR, LR, SF also had interactions with Asp32, but all these peptides had lower renin-inhibitory activities than aliskiren, which had no apparent interactions with Asp32 and Asp215. This indicates the relative importance and contributions of the active site regions to renin catalytic function.
In addition to binding within renin residues, KF and aliskiren also had a hydrogen bond with a water molecule (H2O-2250) present in the active site of renin (Fig. 4, Table 7). This water molecule and another one (H2O-2184) were reported to be important for the interaction of aliskiren (Thangapandian et al., 2011). Only one flaxseed peptide showed interaction with one water molecule; thus, it is difficult to conclude on the role of the interaction of peptides with the water molecules in renin inhibition. This factor should therefore be studied with a large number of renin-inhibiting peptides.
3.4. In silico physicochemical properties and drug-likeness
The drug-likeness of the seven multifunctional peptides were predicted using physicochemical and ADME properties calculated from SwissADME (Daina et al., 2017). SwissADME is a validated free web tool for predicting and evaluating the pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. SwissADME builds on the Lipinski's rule-of-five (Lipinski et al., 2001) and other models, while presenting a user-friendly interface for non-experts in computer-aided drug design (Daina et al., 2017).
Table 8 and Fig. 5 show the in silico physicochemical properties, drug-likeness, and pharmacokinetics of the seven multifunctional flaxseed peptides. The drug-likeness and pharmacokinetics of the peptides shared several similarities with those of the inhibitor drugs, captopril and aliskiren. As shown in Table 8, the seven multifunctional peptides did not exhibit in silico inhibition of CYP3A4, a member of the drug-metabolizing cytochrome P450 family of enzymes. This shows that the peptides may have similar metabolic profile as the ACE-inhibitors, such as captopril. ACE inhibitors are not significantly involved in cytochrome P450 interactions (Flockhart and Tanus-Santos, 2002), but some small molecule ACE inhibitors are partially metabolized by CYP3A4 (Jurima-Romet and Huang, 1993). Moreover, aliskiren showed interaction with CYP3A4 (Table 8), which is expected since CYP3A4 enzymes metabolize some renin inhibitors including aliskiren (Wal et al., 2011, Vaidyanathan et al., 2008). The role of the cytochrome P450 (CYP) family of enzymes in drug metabolism is well established (Guengerich, 2008). Consequently, the interaction, i.e. either induction or inhibition, of drug-active compounds with any of CYP isoenzymes could lead to either fast metabolism (when a CYP isozyme is induced) or bioaccumulation of drugs (when a CYP isozyme is inhibited) in the body (Zisaki et al., 2015). Both scenarios are undesirable, as the former can lead to overdosing, and the latter, toxicity. In fact, hypertension medications have been highlighted as highly prone to interaction with a variety of substances, including other drugs (β-adrenergic blocking agents, calcium channel blockers, ACE inhibitors, etc.) and food (e.g. grapefruit) compounds (Flockhart and Tanus-Santos, 2002, Zisaki et al., 2015). Food/drug-drug interaction puts patients at risk of health complications, and in silico computational approaches have been shown to be useful in predicting these interactions as part of the drug development process (Flockhart and Tanus-Santos, 2002, Zisaki et al., 2015, Singh et al., 2003).
Table 8.
In silico absorption, distribution, metabolism, excretion and toxicity (ADME/Tox) profile of the multifunctional hypotensive flaxseed peptides.
| Physicochemical properties |
Toxicity |
Lipophilicity |
Drug-likeness |
Pharmacokinetics |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| mol. wt. (g/mol) | ROTB (n) | HBA (n) | HBD (n) | ESOL Log S | ToxinPred [SVM score] | TPSA (Å) | CLogPo/w | Bioavai-lability Score | Lipinski filter | GIA | P- glycoprotein substrate | CYP3A4 inhibitor | |
| Rule | <500 | <10 | <10 | <5 | – | <140 | <5 | – | – | – | – | – | |
| IR | 287.36 | 11 | 5 | 6 | 1.54 (HS) | Non-Toxin [-0.8] | 154.32 | −0.94 | 0.55 | Yes (1) | Low | No | No |
| KF | 293.36 | 10 | 5 | 4 | 0.51 (HS) | Non-Toxin [-0.8] | 118.44 | 0.11 | 0.55 | Yes (0) | High | No | No |
| LR | 302.37 | 13 | 6 | 7 | 1.86 (HS) | Non-Toxin [-0.8] | 180.34 | −1.57 | 0.55 | Yes (1) | Low | No | No |
| LW | 334.41 | 10 | 5 | 5 | −0.18 (VS) | Non-Toxin [-0.79] | 130.47 | 0.1 | 0.55 | Yes (0) | High | Yes | No |
| SF | 252.27 | 7 | 5 | 4 | 1.14 (HS) | Non-Toxin [-0.8] | 112.65 | −0.74 | 0.55 | Yes (0) | High | No | No |
| TF | 266.29 | 7 | 5 | 4 | 0.64 (HS) | Non-Toxin [-0.8] | 112.65 | −0.35 | 0.55 | Yes (0) | High | No | No |
| YA | 252.27 | 6 | 5 | 4 | 0.94 (HS) | Non-Toxin [-0.8] | 112.65 | −0.51 | 0.55 | Yes (0) | High | No | No |
| Aliskiren | 551.76 | 20 | 7 | 4 | −4.28 (MS) | – | 146.13 | 3.47 | 0.55 | Yes (1) | Low | Yes | Yes |
| Captopril | 217.29 | 4 | 3 | 1 | −1.14 (VS) | – | 96.41 | 0.62 | 0.56 | Yes (0) | High | No | No |
Abbreviations: CLog Po/w, logarithm of compound partition coefficient between n-octanol and water; CYP3A4, Cytochrome P450 3A4; ESOL, EStimated SOLubility based on (Delaney, 2004) with classes in bracket (HS-highly soluble, VS-very soluble, MS-moderately soluble, and VS-vey soluble); GIA, gastrointestinal absorption; HBA, hydrogen bond acceptors; HBD, hydrogen bond donors; Lipinski filter (with number of violations in bracket); Kp, permeability co-efficient; mol. wt., molecular weight; n, number; ROTB, rotatable bonds; SVM, support vector machine score based on (Gupta et al., 2013); TPSA, topological polar surface area.
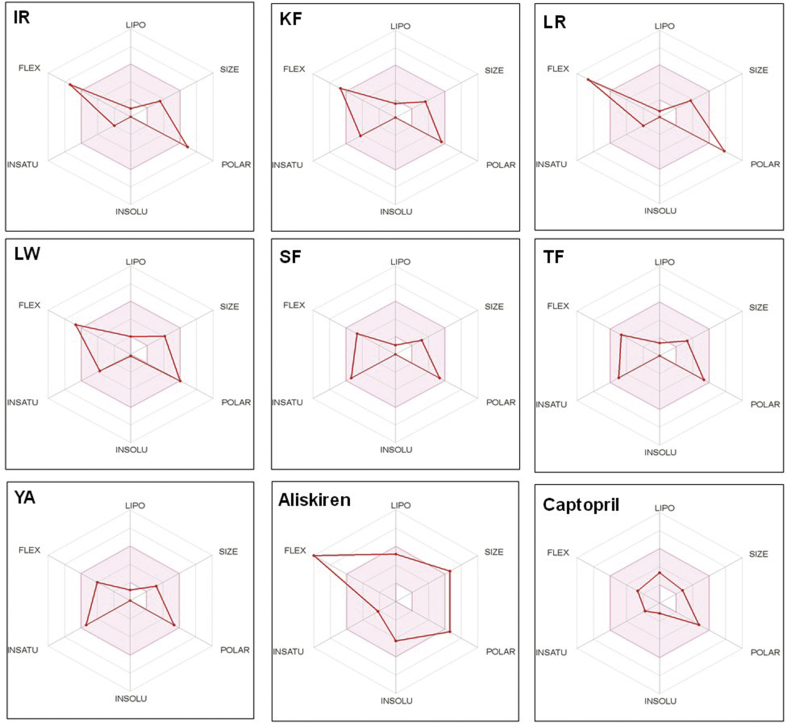
Fig. 5.
Bioavailability radar of the flaxseed peptides and inhibitor drugs (captopril and aliskiren), based on physicochemical indices ideal for oral bioavailability. LIPO, Lipophilicity: -0.7 < XLOGP3 < +5; SIZE, Molecular size: 150 g/mol < mol. wt. < 500 g/mol; POLAR, Polarity: 20 Å2 < TPSA <130 Å2; INSOLU, Insolubility: 0 < Log S (ESOL) < 6; INSATU, Insaturation: 0.25 < Fraction Csp3 < 1; FLEX, Flexibility: 0 < Number of rotatable bonds < 9. The coloured zone is the suitable physicochemical space for oral bioavailability.
Prediction of passive gastrointestinal absorption (GIA) was high for the flaxseed peptides, except for IR and LR (Table 8). The GIA prediction was based on the Brain Or IntestinaL EstimateD permeation (BOILED-Egg) model, which uses lipophilicity and polarity of molecules to predict the passive gastrointestinal absorption and blood-brain barrier permeation (Daina and Zoete, 2016). In relation to this, all the studied multifunctional flaxseed peptides showed the same bioavailability score (0.55), similar to those of aliskiren and captopril (Table 8). Aliskiren was predicted to have low GIA, which corroborates its known low bioavailability (a value of 2.6%). This is partly because aliskiren is a substrate of P-glycoprotein and thus is actively transported out of the body system (Vaidyanathan et al., 2008). The P-glycoprotein (also called permeability glycoprotein or multidrug resistance protein 1) is a cell membrane transport protein responsible for pumping xenobiotics (including drugs) out of cells, and is therefore responsible for controlling cytosolic drug accumulation (Wolking et al., 2015). Interestingly, the flaxseed peptides (except LW) were not good substrates for P-glycoprotein (see Table 8), suggesting that they are likely to have high intestinal absorption and bioavailability.
Two important properties used in predicting oral bioavailability of compounds are flexibility (FLEX, as determined by the number of rotatable bonds) and polarity (POLAR, as determined by topological polar surface area (TPSA)). Compounds with more than 10 rotatable bonds are known to have poor oral bioavailability (Veber et al., 2002), and those with low topological polar surface area (between 20 and 130 Å2) have high oral bioavailability (Mbarik et al., 2019). As shown in Fig. 5, five of the seven multifunctional flaxseed peptides had physicochemical profiles that makes them suitable for oral administration. Peptides IR and LR had FLEX and POLAR (or TPSA) values that were outside the desired range for enhanced bioavailability (see shaded regions in Fig. 5). To circumvent this issue, conjugation of suitable functional groups (e.g. fatty acids) can be used to change the polarity and flexibility of these peptides.
4. Conclusion
In this study, in silico evaluation of the release of hypotensive peptides (i.e. those possessing ACE-, or renin-inhibitory activities, or both), from flaxseed proteome, was undertaken. The plant proteases (papain, ficin, and stem bromelain), rather than microbial (thermolysin, proteinase P1, and subtilisin) and digestive (trypsin, pepsin, and pancreatic elastase II) enzymes showed superiority in releasing hypotensive peptides. Molecular docking of RAS enzymes with seven unique peptides having dual ACE and renin inhibitory activities showed binding/dissociation properties that are similar to those of captopril and aliskiren. For example, the seven peptides inhibited ACE and renin through a combination of hydrogen bonds, electrostatic interactions and hydrophobic interactions, and with bond energies that are within the range observed for captopril and aliskiren.
Moreover, in silico assessment of “medicinal chemistry friendliness” of the dual ACE and renin inhibitory peptides demonstrated that these peptides have drug-like properties on par with the standard inhibitor drugs. Physicochemical features (such as molecular weight, topological polar surface area, rotatable bonds, estimated solubility) of all peptides obeyed at least four of five rules of the Lipinksi filter (IR and LR had one violation each). Pharmacokinetics (e.g. gastrointestinal absorption, interaction with P-glycoproteins), potential toxicity, and bioavailability predictions were also favourable for all the seven hypotensive peptides. This demonstrates that flaxseed proteins have the potential to be used to generate peptides with dual functions in ACE and renin inhibition. Moreover, because they come from food proteins and are produced by enzymatic processes, these peptides may have the added advantage of being devoid of undesirable side effects when used in clinical settings.
It is widely accepted that outcomes of in silico studies involving the discovery of bioactive peptides from food proteins need experimental validation (Udenigwe, 2014). Thus, a future work by our research team will involve the synthesis of the identified ACE and renin inhibiting peptides and experimental validation of their medicinal chemistry friendliness and pharmacokinetics in vivo.
CRediT author statement
Dawei Ji: Conceptualization, Methodology, Software, Writing - original draft, writing– original draft. Min Xu: Methodology, Writing - original draft, writing– original draft, Formal analysis. Chibuike C. Udenigwe: Conceptualization, Writing - review & editing. Dominic Agyei: Conceptualization, Methodology, Writing - review & editing.
Declaration of Competing Interest
All authors declare that they have no competing interests.
References
- Agyei D., Bambarandage E., Udenigwe C.C. Reference Module in Food Science. Elsevier; 2018. The role of bioinformatics in the discovery of bioactive peptides. [DOI] [Google Scholar]
- Azad M.A., Huttunen-Hennelly H.E.K., Friedman C.R. Bioactivity and the first transmission electron microscopy immunogold studies of short de novo-designed antimicrobial peptides. Antimicrobial agents and chemotherapy. 2011;55(5):2137–2145. doi: 10.1128/AAC.01148-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daina A., Zoete V. A BOILED-egg to predict gastrointestinal absorption and brain penetration of small molecules. ChemMedChem. 2016;11(11):1117–1121. doi: 10.1002/cmdc.201600182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daina A., Michielin O., Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017;7:42717. doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daskaya-Dikmen C., Yucetepe A., Karbancioglu-Guler F., Daskaya H., Ozcelik B. Angiotensin-I-Converting enzyme (ACE)-Inhibitory peptides from plants. Nutrients. 2017;9(4) doi: 10.3390/nu9040316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney J.S. ESOL: estimating aqueous solubility directly from molecular structure. J. Chem. Inf. Comput. Sci. 2004;44(3):1000–1005. doi: 10.1021/ci034243x. [DOI] [PubMed] [Google Scholar]
- Flockhart D.A., Tanus-Santos J.E. Implications of cytochrome P450 interactions when prescribing medication for hypertension. JAMA Int. Med. 2002;162(4):405–412. doi: 10.1001/archinte.162.4.405. [DOI] [PubMed] [Google Scholar]
- Galli C.L., Sensi C., Fumagalli A., Parravicini C., Marinovich M., Eberini I. A computational approach to evaluate the androgenic affinity of iprodione, procymidone, vinclozolin and their metabolites. PloS one. 2014;9(8) doi: 10.1371/journal.pone.0104822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girgih A.T., He R., Aluko R.E. Kinetics and molecular docking studies of the inhibitions of angiotensin converting enzyme and renin activities by hemp seed (Cannabis sativa L.) peptides. J. Agric. Food Chem. 2014;62(18):4135–4144. doi: 10.1021/jf5002606. [DOI] [PubMed] [Google Scholar]
- Guengerich F.P. Cytochrome P450 and chemical toxicology. Chem. Res. Toxicol. 2008;21(1):70–83. doi: 10.1021/tx700079z. [DOI] [PubMed] [Google Scholar]
- Gupta S., Kapoor P., Chaudhary K., Gautam A., Kumar R., Open Source Drug Discovery C., Raghava G.P.S. In silico approach for predicting toxicity of peptides and proteins. PloS One. 2013;8(9) doi: 10.1371/journal.pone.0073957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He R., Malomo S.A., Alashi A., Girgih A.T., Ju X., Aluko R.E. Purification and hypotensive activity of rapeseed protein-derived renin and angiotensin converting enzyme inhibitory peptides. J. Fun. Food. 2013;5(2):781–789. [Google Scholar]
- He R., Aluko R.E., Ju X.-R. Evaluating molecular mechanism of hypotensive peptides interactions with renin and angiotensin converting enzyme. PloS one. 2014;9(3) doi: 10.1371/journal.pone.0091051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji D., Udenigwe C.C., Agyei D. Antioxidant peptides encrypted in flaxseed proteome: an in silico assessment. Food Science and Human Wellness. 2019;8(3):306–314. doi: 10.1016/j.fshw.2019.08.002. [DOI] [Google Scholar]
- Jimsheena V., Gowda L.R. Arachin derived peptides as selective angiotensin I-converting enzyme (ACE) inhibitors: structure–activity relationship. Peptides. 2010;31(6):1165–1176. doi: 10.1016/j.peptides.2010.02.022. [DOI] [PubMed] [Google Scholar]
- Jurima-Romet M., Huang H.S. Comparative cytotoxicity of angiotensin-converting enzyme inhibitors in cultured rat hepatocytes. Biochem. Pharmacol. 1993;46(12):2163–2170. doi: 10.1016/0006-2952(93)90605-v. [DOI] [PubMed] [Google Scholar]
- Lipinski C.A., Lombardo F., Dominy B.W., Feeney P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001;46(1–3):3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- Mbarik M., Poirier S.J., Doiron J., Selka A., Barnett D.A., Cormier M., Touaibia M., Surette M.E. Phenolic acid phenethylesters and their corresponding ketones: inhibition of 5-lipoxygenase and stability in human blood and HepaRG cells. Pharmacol. Res. Pers. 2019;7(5) doi: 10.1002/prp2.524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mótyán J.A., Tóth F., Tőzsér J. Research applications of proteolytic enzymes in molecular biology. Biomolecules. 2013;3(4):923–942. doi: 10.3390/biom3040923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva R.V., Bakris G.L. The Kidney in Heart Failure. Springer; 2012. Combination therapy in hypertension treatment; pp. 169–182. [Google Scholar]
- Politi A., Durdagi S., Moutevelis-Minakakis P., Kokotos G., Papadopoulos M.G., Mavromoustakos T. Application of 3D QSAR CoMFA/CoMSIA and in silico docking studies on novel renin inhibitors against cardiovascular diseases. Eur. J. Med. Chem. 2009;44(9):3703–3711. doi: 10.1016/j.ejmech.2009.03.040. [DOI] [PubMed] [Google Scholar]
- Qian B., Tian C., Huo J., Ding Z., Xu R., Zhu J., Yu L., Villarreal O.D. Design and evaluation of four novel tripeptides as potent angiotensin converting enzyme (ACE) inhibitors with anti-hypertension activity. Peptides. 2019;122:170171. doi: 10.1016/j.peptides.2019.170171. [DOI] [PubMed] [Google Scholar]
- Rahuel J., Rasetti V., Maibaum J., Rüeger H., Göschke R., Cohen N., Stutz S., Cumin F., Fuhrer W., Wood J. Structure-based drug design: the discovery of novel nonpeptide orally active inhibitors of human renin. Chem. Biol. 2000;7(7):493–504. doi: 10.1016/s1074-5521(00)00134-4. [DOI] [PubMed] [Google Scholar]
- Sielecki A.R., Hayakawa K., Fujinaga M., Murphy M., Fraser M., Muir A.K., Carilli C.T., Lewicki J.A., Baxter J.D., James M. Structure of recombinant human renin, a target for cardiovascular-active drugs, at 2.5 A resolution. Science. 1989;243(4896):1346–1351. doi: 10.1126/science.2493678. [DOI] [PubMed] [Google Scholar]
- Singh S.B., Shen L.Q., Walker M.J., Sheridan R.P. A model for predicting likely sites of CYP3A4-mediated metabolism on drug-like molecules. J. Med. Chem. 2003;46(8):1330–1336. doi: 10.1021/jm020400s. [DOI] [PubMed] [Google Scholar]
- Sistla S. Structure-activity relationships of αs-casein peptides with multifunctional biological activities. Mol. Cell. Biochem. 2013;384(1–2):29–38. doi: 10.1007/s11010-013-1778-4. [DOI] [PubMed] [Google Scholar]
- Thangapandian S., John S., Sakkiah S., Lee K.W. Potential virtual lead identification in the discovery of renin inhibitors: application of ligand and structure-based pharmacophore modeling approaches. Eur. J. Med. Chem. 2011;46(6):2469–2476. doi: 10.1016/j.ejmech.2011.03.035. [DOI] [PubMed] [Google Scholar]
- Udenigwe C.C. Bioinformatics approaches, prospects and challenges of food bioactive peptide research. Trends Food Sci. Technol. 2014;36(2):137–143. doi: 10.1016/j.tifs.2014.02.004. [DOI] [Google Scholar]
- Udenigwe C.C., Mohan A. Mechanisms of food protein-derived antihypertensive peptides other than ACE inhibition. J. Fun. Food. 2014;8:45–52. doi: 10.1016/j.jff.2014.03.002. [DOI] [Google Scholar]
- Udenigwe C.C., Lin Y.-S., Hou W.-C., Aluko R.E. Kinetics of the inhibition of renin and angiotensin I-converting enzyme by flaxseed protein hydrolysate fractions. J. Fun. Food. 2009;1(2):199–207. [Google Scholar]
- Udenigwe C.C., Li H., Aluko R.E. Quantitative structure-activity relationship modeling of renin-inhibiting dipeptides. Amino Acids. 2012;42(4):1379–1386. doi: 10.1007/s00726-011-0833-2. [DOI] [PubMed] [Google Scholar]
- Vaidyanathan S., Jarugula V., Dieterich H.A., Howard D., Dole W.P. Clinical pharmacokinetics and pharmacodynamics of aliskiren. Clin. Pharmacokinet. 2008;47(8):515–531. doi: 10.2165/00003088-200847080-00002. [DOI] [PubMed] [Google Scholar]
- Veber D.F., Johnson S.R., Cheng H.Y., Smith B.R., Ward K.W., Kopple K.D. Molecular properties that influence the oral bioavailability of drug candidates. J. Med. Chem. 2002;45(12):2615–2623. doi: 10.1021/jm020017n. [DOI] [PubMed] [Google Scholar]
- Wal P., Wal A., Rai A.K., Dixit A. Aliskiren: an orally active renin inhibitor. J. Pharm. BioAllied Sci. 2011;3(2):189–193. doi: 10.4103/0975-7406.80764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolking S., Schaeffeler E., Lerche H., Schwab M., Nies A.T. Impact of genetic polymorphisms of ABCB1 (MDR1, P-glycoprotein) on drug disposition and potential clinical implications: update of the literature. Clin. Pharmacokinet. 2015;54(7):709–735. doi: 10.1007/s40262-015-0267-1. [DOI] [PubMed] [Google Scholar]
- Wu J., Aluko R.E., Nakai S. Structural requirements of angiotensin I-converting enzyme inhibitory peptides: quantitative structure− activity relationship study of di-and tripeptides. J. Agric. Food Chem. 2006;54(3):732–738. doi: 10.1021/jf051263l. [DOI] [PubMed] [Google Scholar]
- Zisaki A., Miskovic L., Hatzimanikatis V. Antihypertensive drugs metabolism: an update to pharmacokinetic profiles and computational approaches. Curr. Pharmaceut. Des. 2015;21(6):806–822. doi: 10.2174/1381612820666141024151119. [DOI] [PMC free article] [PubMed] [Google Scholar]