Abstract
Factors contributing to development of complex regional pain syndrome (CRPS) are not fully understood. This study examined possible epigenetic mechanisms that may contribute to CRPS after traumatic injury. DNA methylation profiles were compared between individuals developing CRPS (n = 9) and those developing non-CRPS neuropathic pain (n = 38) after undergoing amputation following military trauma. Linear Models for Microarray (LIMMA) analyses revealed 48 differentially methylated cytosine-phosphate-guanine dinucleotide (CpG) sites between groups (unadjusted P’s < 0.005), with the top gene COL11A1 meeting Bonferroni-adjusted P < 0.05. The second largest differential methylation was observed for the HLA-DRB6 gene, an immune-related gene linked previously to CRPS in a small gene expression study. For all but 7 of the significant CpG sites, the CRPS group was hypomethylated. Numerous functional Gene Ontology-Biological Process categories were significantly enriched (false discovery rate-adjusted q value <0.15), including multiple immune-related categories (eg, activation of immune response, immune system development, regulation of immune system processes, and antigen processing and presentation). Differentially methylated genes were more highly connected in human protein–protein networks than expected by chance (P < 0.05), supporting the biological relevance of the findings. Results were validated in an independent sample linking a DNA biobank with electronic health records (n = 126 CRPS phenotype, n = 19,768 non-CRPS chronic pain phenotype). Analyses using PrediXcan methodology indicated differences in the genetically determined component of gene expression in 7 of 48 genes identified in methylation analyses (P’s < 0.02). Results suggest that immune- and inflammatory-related factors might confer risk of developing CRPS after traumatic injury. Validation findings demonstrate the potential of using electronic health records linked to DNA for genomic studies of CRPS.
Keywords: Complex regional pain syndrome, CRPS, Methylation, Genetic, Immune, Inflammatory, Chronic pain, Risk
1. Introduction
Complex regional pain syndrome (CRPS) is a chronic pain condition characterized by allodynia and hyperalgesia, skin temperature and color changes, edema, and trophic changes.7,8 Its pathophysiological mechanisms are only incompletely understood, but available data suggest that neuropathic, central and autonomic nervous system, inflammatory, and immune mechanisms all are involved.7,8 Complex regional pain syndrome is a somewhat heterogeneous diagnosis, with several distinct phenotypes9,10,15; multiple mechanisms may contribute to different degrees across patients and over time.8,10
Familial aggregation of CRPS cases have led to suspicions of a heritable component of CRPS risk.20,21,35 Supporting this, prior work has suggested possible genetic risk factors for CRPS. Most frequently reported are genetic differences in the human leukocyte antigen (HLA) system, the system underlying the adaptive immune response.42 Differences have been noted in a number of studies in the frequency of several HLA alleles between patients with CRPS and controls,40,67-69 with replication observed for HLA-DQ8 effects.22,70
Beyond putative genetic drivers of susceptibility, epigenetic modifications provide an additional path for transmitting CRPS risk. There have been multiple epigenetic processes identified, with one of the most widely studied being DNA methylation.14,65 Methylation of the DNA sequence, particularly at cytosine-phosphate-guanine dinucleotide (CpG) sites, is known to be affected by both genetic and environmental factors (eg, diet, smoking, and trauma), and suppresses transcriptional activity.38,65 To the best of our knowledge, no prior studies have evaluated associations between CRPS and DNA methylation profiles. However, limited animal and human work support the potential importance of altered gene expression in CRPS.
Reviews of animal research conclude that altered gene expression in dorsal root ganglion neurons could enhance inflammatory responses, contributing to CRPS risk.71,72 Human work regarding gene regulatory effects on CRPS risk is quite limited. One study described altered expression of 18 micro-RNAs, which are involved in posttranscriptional regulation of gene expression, in patients with CRPS compared to controls.52 A more recent study conducting genome-wide gene expression profiling of blood samples from 4 patients with CRPS and 5 pain-free controls revealed 80 genes that were differentially expressed.37 A reanalysis of these data using Linear Models for Microarray analysis revealed 257 differentially expressed genes in patients with CRPS.62 In both studies, 3 of the largest gene expression differences were for genes in the HLA system.37,62
No published studies have reported associations between CRPS and DNA methylation status, and no gene expression studies have compared patients with CRPS to other pain patients. The latter may be important because any gene expression differences between patients with CRPS and controls might reflect chronic pain status rather than CRPS per se. The current study therefore sought to evaluate whether presence of CRPS (vs non-CRPS neuropathic pain) after traumatic injury is associated with differential DNA methylation profiles. We further examined whether genes showing significant methylation differences in the primary sample could be independently validated (in terms of the genetically determined component of gene expression)26 in a large deidentified clinical sample.25,57
2. Method
2.1. Primary methylation study
2.1.1. Design
Patients were all enrolled in the Veterans Integrated Pain Evaluation Research (VIPER) study, a case-control study that included recent traumatic amputees from Operation Enduring Freedom/Operation Iraqi Freedom (OEF/OIF) who were enrolled 3 to 18 months after amputation. All subjects were enrolled between November 2011 and July 2013.
2.1.2. Subjects
All study procedures were approved by the institutional review board of Walter Reed National Military Medical Center (WRNMMC). Subjects included 9 CRPS and 38 non-CRPS neuropathic pain patients, all of whom had experienced military trauma and had subsequently undergone posttraumatic amputation surgery. Subjects were included if they were a military health care system beneficiary aged 18 years or older and undergoing treatment at WRNMMC with a diagnosis of postinjury amputation of all or part of one limb performed 3 to 18 months previously. Patients were excluded if they were afflicted with severe traumatic brain injury, significant cognitive deficits, substantial hearing loss, spinal cord injury with permanent or persistent deficits, ongoing tissue damage that might cause pain, infection, heterotrophic ossification, poorly fitting prosthesis, or hip disarticulation.
Patients were eligible for this study if they reported clinically significant residual limb pain at the time of evaluation, operationally defined as an average pain score over the past week of ≥3/10 on a numeric rating scale (NRS). Those patients with clinically significant pain were further adjudicated into pain subtypes (see Ref. 11 for full details). For use in the adjudication process, all patients underwent a history and physical examination to assess signs and symptoms used in CRPS diagnosis. Per the diagnostic algorithm in the larger study, patients were initially classified into phantom or residual limb pain. Subsequently, the residual limb pain group was further classified as somatic vs neuropathic through use of the Self-Report Leeds Assessment of Neuropathic Symptoms and Signs (S-LANSS), based on the standard cutoff of ≥12 on the S-LANSS.4 Finally, patients identified with neuropathic pain qualities were further characterized into diagnostic groups, including CRPS (based on the Budapest clinical criteria).33 The non-CRPS neuropathic pain subgroup consisted of those patients meeting criteria for neuropathic pain who did not meet the Budapest criteria for CRPS. The sample reported in the current work included all CRPS and non-CRPS neuropathic pain patients as defined above in the VIPER sample who had DNA methylation assay data available. The final sample was 98% male and 89% white, with a mean age of 26.9 years.
2.1.3. Procedures
After providing written informed consent, patients completed 0 to 10 NRS ratings of their pain intensity, with NRS anchors being 0 = “no pain” and 100 = “worst possible pain.” Blood samples were also obtained from each patient at the time of enrollment for subsequent analysis. For plasma preparation, 6 mL of blood was collected in EDTA-containing K2 tubes and inverted to mix. Tubes were then spun at 3,000g for 20 minutes at 4°C. Plasma fraction was collected with a pipette and aliquoted into 1.5-mL cryovials and stored at −20°C for 24 hours and subsequently at −80°C until assays were conducted.
2.2. DNA methylation assays
DNA was extracted from whole blood and then sent to the Molecular Genomics Shared Resource of the Duke Molecular Physiology Institute for methylation analysis. As recommended by Illumina, we used the Zymo EZ DNA Methylation kit (Zymo Research, Irvine, CA) for bisulfite conversion of DNA before running the Illumina Methylation 450 k protocol. Before beginning the main portion of the protocol, the CT Conversion Reagent was prepared. Then all steps were completed until step 2 where reagent was instead vortexed constantly for 10 minutes instead of frequently. The protocol was then continued starting with 500 ng of good-quality DNA and proceeding with the manufacturer’s protocol using the alternative thermocycler conditions for Infinium Methylation assay in downstream applications for steps 4 and 5 of the Zymo EZ DNA Methylation protocol. These alternative conditions dictate instead of an incubation of 50°C for 16 hours, the thermocycler program is as follows: (95°C for 30 seconds, 50°C for 60 minutes) × 16 cycles, then 4°C hold.
To use the Illumina Methylation 450 k kit (Illumina, San Diego, CA), 4 μL of bisulfite-converted DNA was used and the manufacturer’s protocol for Infinium HD Methylation was followed. Briefly, the samples were denatured and amplified overnight for 20 to 24 hours. Fragmentation, precipitation, and resuspension of the samples followed overnight incubation. After resuspension, samples were then hybridized to the Illumina Infinium Methylation 450 k BeadChip for 16 to 24 hours. Finally, the BeadChips were washed to remove any unhybridized DNA and then labeled with nucleotides to extend the primers to the DNA sample. Following the Infinium HD Methylation protocol, the BeadChips were imaged using the Illumina iScan system (Illumina).
2.3. Data analysis
2.3.1. Differential methylation analysis
We conducted Linear Models for Microarray (LIMMA)56 analyses to identify differentially methylated CpG sites. LIMMA estimates the log-fold change (logFC) for each CpG site.
Let j be a given CpG site. LIMMA’s Bayesian model assumes an inverse χ2 prior for the unknown variance with mean and degrees of freedom d0:
| (1) |
For statistical inference, the approach uses a modified t-statistic, in which the posterior variance is substituted for the regular variance in the classical t-statistic:
| (2) |
Here, dj is the residual degrees of freedom for the jth CpG site. Note that the variance estimates are “moderated” using a shared value from the Bayesian prior Equation 1, reducing the possibility of false positives that may arise from underestimation of the variance. The modified t-statistic has higher degrees of freedom, d0 + dj, in comparison with the ordinary t-statistic.
Most CpG sites were not variably methylated across the individuals. Hence, we filtered out those CpG sites that were not sufficiently variable. Only those with variance >0.002 were analyzed for this report.
2.3.2. Protein–protein interactions
We generated a protein–protein interaction (PPI) subnetwork from the genes annotated to the differentially methylated CpG sites using DAPPLE.58 A direct interaction between 2 proteins, represented as nodes, from a database of high-confidence in vitro direct interactions (InWeb)46 is represented by an “edge” (line) between the corresponding annotated genes. We tested the significance of the centrality index, defined as the average number of direct connections to a tested gene. The significance was evaluated as the proportion of within-degree node-label permutations (n = 1000) with a permutation statistic greater than or equal to the centrality index value in the actual (nonpermuted) data.
2.3.3. Enrichment for known pathways and functional annotations
Using DAVID,17 we performed gene ontology analysis of the top 48 genes from the differential methylation analysis to determine which genes belong to known pathways, gene sets, and functional annotations. We used an enrichment analysis approach using the enrichGO function of the Cluster Profiler R package (http://bioconductor.org/packages/release/bioc/html/clusterProfiler.html). This analysis generated a list of pathways and functional annotations with significant enrichment, as indicated by a false discovery rate-adjusted (FDR) q value <0.15 for the genes with differentially methylated sites. The number of annotated genes for significant categories is provided in Supplementary Figure 1 (available at http://links.lww.com/PAIN/A818) and Supplementary Figure 2 (available at http://links.lww.com/PAIN/A819).
2.4. Validation study
2.4.1. Design
Results in the primary discovery sample were validated using a case-control design in a DNA biobank linked to an electronic health records (EHR) database.
2.4.2. Sample
The validation sample was drawn from a large pool of clinical patients seen at Vanderbilt University Medical Center since 2002 who had DNA samples available in BioVU, the Vanderbilt biobank of deidentified DNA samples obtained for research purposes from discarded blood.55,57 BioVU DNA samples were linked in a deidentified manner to pain-relevant phenotypes previously derived based on informatics algorithms applied to ICD-9 codes in the Synthetic Derivative, the Vanderbilt deidentified electronic medical records database.19 The current study compared patients with a CRPS phenotype (n = 126) to patients with a non-CRPS chronic pain phenotype (n = 19,768). In the CRPS sample, 69% were female, vs only 50% being female in the non-CRPS sample. This predominance of female patients in the CRPS validation sample is consistent with the broader CRPS population.8 Mean age in the 2 validation samples was 62.6 (±14.18) and 64.4 (±20.7), respectively.
2.4.3. Procedures
As a means of validating epigenetic effects observed in methylation analyses, we tested for the contribution of differential gene expression to CRPS using the PrediXcan methodology.25,26 PrediXcan estimates the genetically determined component of gene expression and evaluates this component for association with the target phenotype (ie, CRPS). Using the weights derived from the gene expression imputation model31 and the number of effect alleles Xij at the variant predictor j, the genetically determined component of gene expression is calculated as the additive effect:
The imputation model was generated using Elastic Net. A significant association between the genetically determined component of gene expression and the CRPS phenotype (in this case, based on logistic regression) suggests a causal direction of effect (from gene expression to phenotype) because the germline genetic profile (on which the genetic component of gene expression is based) is not influenced by the CRPS phenotype.
2.4.4. Data analysis
We performed logistic regression modeling CRPS vs non-CRPS chronic pain status using the genetically determined component of gene expression as the independent variable. We tested in the independent BioVU validation sample the 48 genes found to be significant in the differential methylation discovery analysis.
3. Results
3.1. Differential methylation between complex regional pain syndrome and non–complex regional pain syndrome neuropathic pain patients
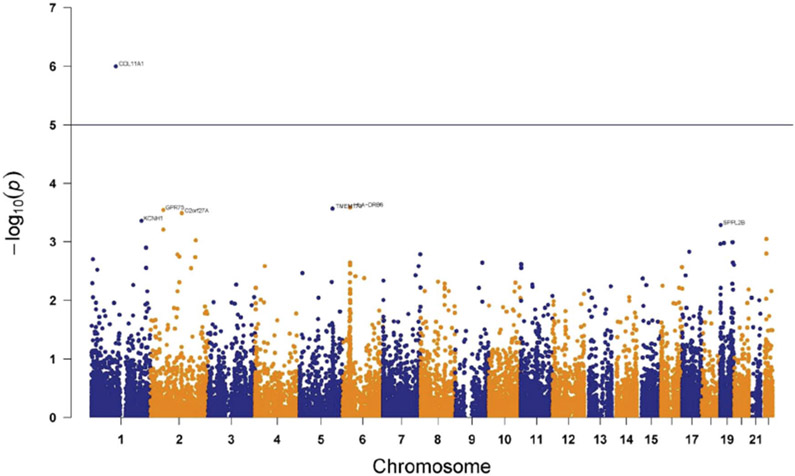
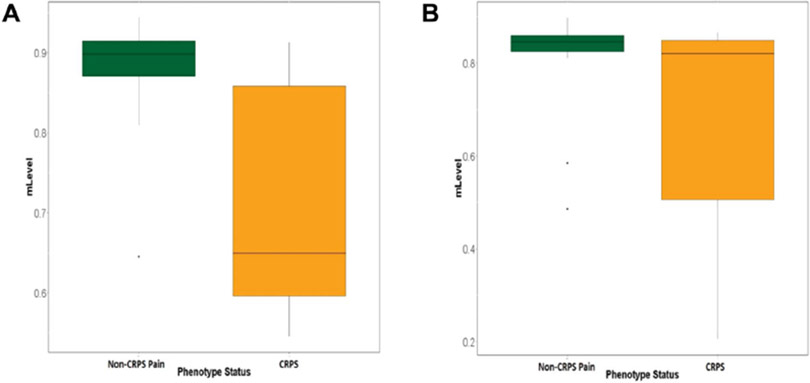
Overall methylation levels by individual subject are provided in Supplementary Figure 3 (available at http://links.lww.com/PAIN/A820). We compared patients meeting vs not meeting diagnostic criteria for CRPS to test whether presence of CRPS after traumatic injury is associated with differential methylation profiles. LIMMA analyses revealed 48 CpG sites differentially methylated (P < 0.005, uncorrected) between CRPS and non-CRPS groups (Table 1). For all but 7 CpG sites, the CRPS group was hypomethylated relative to the non-CRPS neuropathic pain group. A Manhattan plot summarizing differential methylation at all CpG sites highlights genomic regions in which this epigenetic mechanism may contribute to differences between the groups (Fig. 1). One CpG site (cg17820060, COL11A1, P = 9.98 × 10−7) was differentially methylated at a Bonferroni-corrected P < 0.05 level. Additional CpG sites at P < 5 × 10−4 were noted for the following genes: HLA-DRB6, TMEM173, GPR75, C2orf27A, and KCNH1. Box and whisker plots for methylation differences across CRPS and non-CRPS groups for the top 2 genes identified (COL11A1 and HLA-DRB6) are provided in Figures 2A and B, respectively. In both cases, hypomethylation was observed in the CRPS group, with wide variability in individual methylation levels noted, possibly reflecting the suspected heterogeneity in contributions of various CRPS mechanisms across individual patients.
Table 1.
Cytosine-phosphate-guanine dinucleotide sites (and associated genes) showing differential methylation at an uncorrected P < 0.005 level between CRPS and non-CRPS chronic pain patients.
| CpG site | Gene | Log FC | t value | P |
|---|---|---|---|---|
| cg17820060_chr1:103343469 | COL11A1 | −0.166219474 | −5.623121047 | 9.98E-07 |
| cg15591384_chr6:32525960 | HLA-DRB6 | −0.176122632 | −3.955685059 | 0.000256198 |
| cg00070715_chr5:138862796 | TMEM173 | −0.08300769 | −3.939373128 | 0.000269618 |
| cg01876338_chr2:54087008 | GPR75 | −0.065696053 | −3.922495243 | 0.000284217 |
| cg17854650_chr2:132481613 | C2orf27A | −0.062572719 | −3.882752019 | 0.000321678 |
| cg09518270_chr1:211308115 | KCNH1 | −0.237310731 | −3.785369214 | 0.000434713 |
| cg23803868_chr19:2340235 | SPPL2B | −0.412810322 | −3.730180616 | 0.000514858 |
| cg14832904_chr2:54087028 | GPR75 | −0.068758392 | −3.671198847 | 0.000616167 |
| cg20571420_chr22:25160308 | TOP1P2; PIWIL3 | −0.056746842 | −3.54875351 | 0.000890954 |
| cg10560079_chr2:191398806 | TMEM194B | 0.057021579 | 3.531565975 | 0.000937857 |
| cg04375683_chr19:52692431 | PPP2R1A | −0.058455731 | −3.506453217 | 0.001010652 |
| cg10226967_chr19:16606648 | C19orf44; CALR3 | −0.055366199 | −3.496832101 | 0.001039946 |
| cg15975960_chr19:1035450 | CNN2 | −0.183742719 | −3.482898424 | 0.001083811 |
| cg04610742_chr1:230513809 | PGBD5 | −0.205357602 | −3.434243521 | 0.001251244 |
| cg25115829_chr17:29057586 | SUZ12P | −0.053363596 | −3.380452852 | 0.00146496 |
| cg13627197_chr22:25160378 | TOP1P2; PIWIL3 | −0.059896462 | −3.354150242 | 0.001581692 |
| cg23565757_chr7:157646605 | PTPRN2 | −0.080209854 | −3.344032445 | 0.001628906 |
| cg02675527_chr2:114030824 | PAX8 | 0.119200439 | 3.341899694 | 0.001639028 |
| cg03287111_chr2:121625634 | GLI2 | −0.051736842 | −3.314028541 | 0.001776912 |
| cg23206520_chr2:189158094 | GULP1 | −0.062407164 | −3.30687498 | 0.001814038 |
| cg26858423_chr1:6149477 | KCNAB2 | 0.084953041 | 3.277223241 | 0.001975912 |
| cg04573316_chr19:54216680 | MIR519D | −0.063664591 | −3.230689232 | 0.002257805 |
| cg22451923_chr6:32370879 | BTNL2 | −0.085751053 | −3.230266241 | 0.002260534 |
| cg23964386_chr9:114362249 | PTGR1 | −0.067227339 | −3.229947345 | 0.002262593 |
| cg01122627_chr11:1474386 | BRSK2 | −0.079350234 | −3.210120217 | 0.002394169 |
| cg03848831_chr11:2397685 | CD81 | −0.091655 | −3.206372603 | 0.002419837 |
| cg16929496_chr19:58952249 | ZNF132 | −0.054303684 | −3.200189318 | 0.002462757 |
| cg26805579_chr6:32372962 | BTNL2 | −0.083768012 | −3.195461202 | 0.00249606 |
| cg03870448_chr4:41214687 | APBB2 | −0.055146374 | −3.183598257 | 0.0025815 |
| cg02027878_chr7:150711138 | ATG9B; NOS3 | −0.049219181 | −3.182685168 | 0.002588189 |
| cg10330187_chr16:88666383 | ZC3H18 | −0.063355117 | −3.169273341 | 0.002688349 |
| cg24474852_chr1:230850050 | AGT | −0.07147886 | −3.158041045 | 0.002775033 |
| cg09701145_chr11:2019436 | MIR675; H19 | 0.049746433 | 3.157926055 | 0.002775934 |
| cg19846314_chr2:171680113 | GAD1 | 0.052606901 | 3.153575381 | 0.002810223 |
| cg18323236_chr1:24743029 | NIPAL3 | −0.066710292 | −3.131413078 | 0.002991171 |
| cg15975890_chr5:11903145 | CTNND2 | −0.05833652 | −3.084777665 | 0.003408353 |
| cg18584440_chr6:32370815 | BTNL2 | −0.084480673 | −3.079664007 | 0.003457285 |
| cg24212267_chr7:137801780 | AKR1D1 | −0.051366959 | −3.054308822 | 0.003709806 |
| cg15214448_chr17:13506230 | HS3ST3A1 | 0.058285497 | 3.052583402 | 0.003727604 |
| cg14989959_chr6:54711471 | FAM83B | 0.056408596 | 3.041369712 | 0.003845243 |
| cg18683523_chr6:91297516 | MAP3K7 | −0.057792047 | −3.01326307 | 0.00415557 |
| cg22835630_chr15:25434030 | SNORD115 | −0.071314181 | −3.007297156 | 0.004224389 |
| cg07065756_chr7:2119340 | MAD1L1 | −0.045774766 | −2.97828233 | 0.004574559 |
| cg16320788_chr8:73794096 | KCNB2 | −0.05781769 | −2.963333609 | 0.004765386 |
| cg17275074_chr5:135701422 | TRPC7 | −0.058669094 | −2.958925012 | 0.004823067 |
| cg02844892_chr6:31370412 | MICA | −0.046332544 | −2.957764382 | 0.004838361 |
| cg13713922_chr2:121625577 | GLI2 | −0.050111784 | −2.954838104 | 0.004877122 |
| cg12580156_chr10:112588051 | RBM20 | −0.04473193 | −2.948632141 | 0.004960286 |
Negative logFC and t values indicate lower methylation in the CRPS group.
CpG, cytosine-phosphate-guanine dinucleotide; CRPS, complex regional pain syndrome.
Figure 1.
Manhattan plot for differential methylation across all CpG sites evaluated. Any gene above the horizontal line indicates Bonferroni-corrected P < 0.05. One gene COL11A1 (nominal P = 9.98E-07) passed this threshold. CpG, cytosine-phosphate-guanine dinucleotide.
Figure 2.
Box and whisker plots displaying source of differential methylation for the top 2 identified genes, COL11A1 (A) and HLA-DRB6 (B).
To address potential pain confounders, we considered whether the 2 patient groups also differed in the primary clinical feature of chronic pain intensity based on responses on the NRS pain intensity rating scale used to determine study eligibility. Comparisons of mean limb pain intensity ratings across the 2 groups revealed that the CRPS group (4.7 ± 1.41) and the non-CRPS neuropathic pain group (4.5 ± 2.00) did not significantly differ [t(45) = −0.283, P = 0.78].
3.2. Functional analysis of differentially methylated cytosine-phosphate-guanine dinucleotide sites
Using the Gene Ontology (GO) classification system,27 we tested whether CpG sites showing differential methylation in patients with CRPS exhibited patterns of functional enrichment. This functional enrichment analysis revealed numerous GO-Biological Process (GO-BP) categories that were significantly enriched for differentially methylated genes at FDR (q-value) <0.15.60 As indicated in Supplementary Figure 1 (available at http://links.lww.com/PAIN/A818), a notable subset of these enriched GO-BP categories involved immune function, including leukocyte-mediated cell toxicity, regulation of cell killing and negative regulation of cell killing, immune effector process, immune response and activation of immune response, production of molecular mediator of immune response, immune system development, regulation of immune system processes, negative and positive regulation of immune system process, antigen processing and presentation, and leukocyte activation.
Supplementary Figure 2 (available at http://links.lww.com/PAIN/A819) portrays the number of differentially methylated genes by GO-Molecular Function (GO-MF) categories. The 5 GO-MF categories with the largest number of differentially methylated genes were protein binding, organic cyclic compound binding, ion binding, heterocyclic compound binding, and antigen binding. This latter immune-related GO-MF category supports the pervasive immune-relevant findings in GO-BP enrichment analyses.
3.3. Protein–protein interactions for differentially methylated genes in patients with complex regional pain syndrome
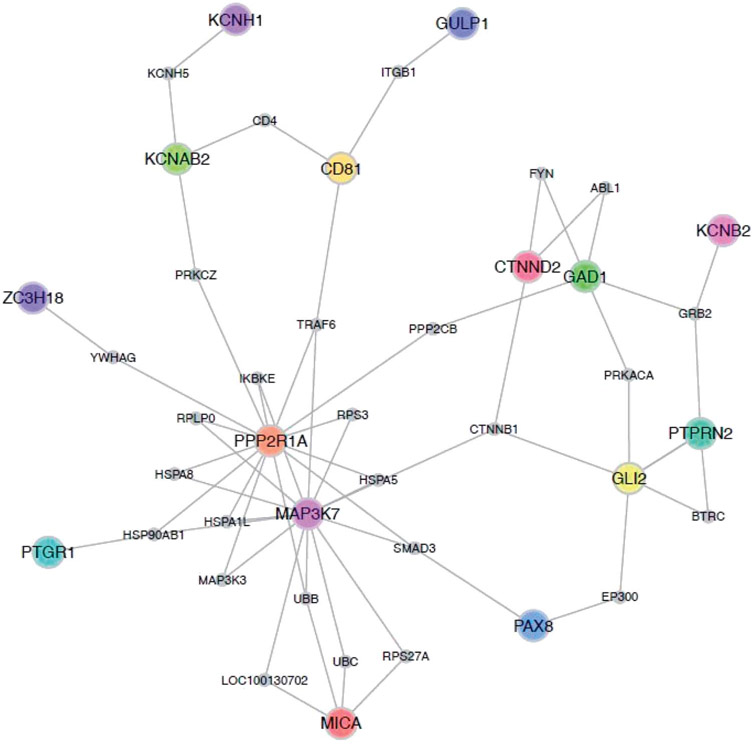
We examined whether the 48 differentially methylated CpG sites reflected known PPIs. Among the genes annotated to the differentially methylated CpG sites, we found genes that assemble in PPI subnetworks (Fig. 3). Using a within-degree node-label permutation, we found that the differentially methylated genes were more highly connected in human PPI networks than expected by chance (P = 0.03), indicating that the genes are more likely to be essential genes.39
Figure 3.
Protein–protein interactions (PPIs) among the genes annotated to the differentially methylated CpG sites. Nodes represent proteins associated with the annotated genes displaying significant differential methylation in primary analyses. Known direct interactions between proteins are represented as an edge between the annotated genes. CpG, cytosine-phosphate-guanine dinucleotide.
3.4. Independent validation
We sought to validate primary findings in a large deidentified clinical sample of patients who had provided DNA that was subjected to genome-wide level (GWAS) genetic assays.18 Prior work by one of the authors (E.R.G.) had developed methods for using reference transcriptome data to impute the genetically determined component of gene expression from GWAS genetic information alone (PrediXcan).26 Applying the PrediXcan approach, we expected that compared to patients with a non-CRPS chronic pain phenotype, patients displaying a CRPS phenotype would show evidence for differential genetically determined gene expression in similar genes as revealed in primary analyses.
Results (Table 2) indicated differential gene expression for the CRPS phenotype relative to the non-CRPS chronic pain phenotype for 7 of the 48 genes identified as loci for differential methylation in the primary sample. These 7 genes were PTGR1, HS3ST3A1, FAM83B, CD81, GPR75, APBB2, AGT, and BTNL2. Taken together, these results demonstrate that the genes identified as showing CRPS-related differential methylation in the primary analyses had some predictive power in an independent patient data set in which the genetic component of gene expression was estimated using the germline genetic profile.
Table 2.
Genes displaying CRPS-related differential methylation profiles in the primary sample that showed additional support (defined as P < 0.05) in the BioVU validation sample in terms of the genetic component of gene expression.
| Gene | Beta | Validation P |
|---|---|---|
| PTGR1 | −0.728786 | 1.28E-03 |
| HS3ST3A1 | 1.53113 | 1.64E-02 |
| FAM83B | 0.620845 | 2.41E-02 |
| CD81 | −2.49833 | 2.43E-02 |
| GPR75 | 3.61601 | 2.86E-02 |
| APBB2 | 0.550918 | 3.87E-02 |
| AGT | −0.441226 | 4.70E-02 |
| BTNL2 | 2.24399 | 1.39E-02 |
Beta is the beta-weight observed in the logistic regression analysis for each gene indicated.
CRPS, complex regional pain syndrome.
4. Discussion
To evaluate epigenetic mechanisms that may be associated with CRPS after injury, we compared DNA methylation profiles in individuals with chronic pain who met Budapest diagnostic criteria for CRPS to those not meeting CRPS criteria. We identified 48 CpG sites that were (nominally) differentially methylated between the 2 groups. Of the 48, one met methylome-wide significance (COL11A1). Functional analyses revealed numerous GO-BP categories that were significantly enriched in the CRPS group at FDR (q-value) <0.15.60 A substantial portion of these functionally enriched categories were related to immune function. It is notable that differences in methylation profiles and functional enrichment patterns were observed despite both groups reporting similar pain intensity. Thus, differential methylation was not related simply to presence of chronic pain, but rather specifically to clinical features reflected in the diagnostic criteria defining the CRPS group (eg, edema, skin temperature changes). Validation sample findings regarding the genetically determined component of gene expression suggest that altered expression of implicated genes may confer risk of CRPS rather than simply being a consequence of CRPS.
Interpretation of these findings is best considered in context of clinical and experimental data regarding CRPS mechanisms. Research has increasingly highlighted a likely role for immune mechanisms in CRPS. Several studies suggest that autoimmune mechanisms are relevant to CRPS24,28 with, for example, over one-third of CRPS patients across 3 studies exhibiting higher levels of antineuronal antibodies than population norms.6,23,43,44 Moreover, a passive transfer model has demonstrated that serum IgG from patients with CRPS can induce CRPS in animals after tissue injury.30,63 A key role for inflammatory mechanisms has also been suggested. Compared to both healthy controls and non-CRPS pain patients, CRPS patients display significantly elevated levels of proinflammatory cytokines (eg, TNF-alpha, IL-1 beta).2,49,66,74,75 A study using informatics-based network analysis34 suggests that such proinflammatory changes may derive in part from upstream alterations in nfKappaB, an inducible transcription factor involved in immune regulation and inflammation,64 a finding supported in a preclinical model of CRPS.16
In light of evidence for immune and inflammatory mechanism in CRPS, several aspects of the current findings are notable. One of the top 2 CpG sites most strongly associated with CRPS was in HLA-DRB6, an HLA class II pseudogene. This gene has previously exhibited differential gene expression between patients with CRPS and pain-free controls.37 Class II HLA genes are involved in immune regulation, and are expressed by B cells, activated T cells, and dendritic cells.42 HLA-DRB6 is hypomethylated in rheumatoid arthritis, a painful autoimmune condition,32 a pattern similar to patients with CRPS in the current study. Also, among the 48 CpG sites exhibiting differential methylation were genes including MICA, TMEM173, and BTNL2, all of which were hypomethylated in patients with CRPS. MICA is an HLA class I gene, expressed by somatic cells rather than specifically in immune cells.42 Variations in the MICA gene have been linked with other painful (Sjogren syndrome)12 and nonpainful automimmune conditions (cutaneous lupus erythematosus).45 TMEM173 produces the stimulator of interferon genes (STING) protein, a regulator of innate immune responses that also is involved in autoimmune diseases48,50 and promotes inflammation.78 BTNL2, a gene identified both in the primary methylation sample and in the validation sample, is an HLA class II-related gene that has been associated with painful autoimmune conditions due to its being in strong linkage disequilibrium with HLA-DQB1 and HLA-DRB1 haplotypes.53 Interestingly, work in CRPS suggests that both the HLA-DQB1 and HLA-DRB1 genes are differentially expressed (downregulated) in patients with CRPS compared to pain-free controls.37,62
The current findings are also relevant in terms of inflammatory CRPS mechanisms. Among the 48 genes displaying differential methylation in CRPS were the COL11A1 and MAP3K7 genes. These genes are part of an nfKappaB-related gene network.64,73,76 Activation of the nfKappaB pathway triggers cytokine release, leading to inflammation.34 Inflammatory clinical features (eg, edema and erythema) and elevated proinflammatory cytokine levels are both characteristic of CRPS, particularly in its early stages.5,10,47 Differential methylation was also noted in the GPR75 gene, another gene validated in terms of the genetic component of gene expression in our large informatics sample. Impaired GPR75 function has been linked to neuroinflammation.36 Oxidative stress is also linked to inflammation,3 and current findings of differential methylation of PTGR1 in patients with CRPS may be relevant in this regard. PTGR1 codes for prostaglandin reductase-1, which is involved in catabolism of eicosanoids and lipid peroxidation, and is part of the nuclear factor E2-related factor 2 (NRF2) pathway.59 A potential link between PTGR1 and CRPS was also validated in the informatics sample in the current study. The NRF2 pathway controls expression of genes enhancing cellular antioxidant capacity.51
Preclinical CRPS models13,41 and meta-analysis of human studies1 suggest that CRPS risk after injury may be mitigated by administration of antioxidants, consistent with a possible role for oxidative stress in CRPS.
The largest differential methylation effect in the current work was for the COL11A1 gene. COL11A1 is involved in collagen formation, and notably, the only prior gene expression work in CRPS found that one of the top differentially expressed genes was also a collagen-related gene, MMP9.37 There is no prior evidence of a specific role for collagen-related factors in CRPS, although we might speculate that given the role of collagen in skin formation,54 altered expression of genes such as COL11A1 and MMP9 might contribute to altered skin growth often characteristic of CRPS.7,8 In addition, it may be relevant that both the COL11A1 gene and the KRT16 gene are part of the TFAP2A gene regulatory network.77 The protein expressed by the KRT16 gene has been shown to be a target for autoantibody responses in a preclinical CRPS model.61
Our results identified numerous PPIs related to the differentially methylated genes observed between groups. These findings of known functional interactions between proteins coded for by differentially methylated genes linked to CRPS strengthen arguments for the biological importance of these genes in CRPS. The 2 most prominent nodes reflected hormonal regulation processes and immune function.
The current findings further support the potential of immune-focused therapies for CRPS.29 To the extent that immune differences may play a mechanistic role in only a subset of patients with CRPS, profiles of differential methylation in CRPS-related immune genes could potentially help target immune-focused treatments to patients most likely to respond. The epigenetic changes indicated by CRPS-related methylation profiles might also help identify novel CRPS risk mechanisms that could point towards new treatments. Finally, from the methodological perspective, findings in the validation sample demonstrate the promise of using electronic health records linked to DNA for genomic studies of CRPS.
Several study limitations are acknowledged. First, although our sample of patients with CRPS was more than twice as large as any prior epigenetic work in patients with CRPS, the sample was nonetheless small and there were a large number of statistical tests. Risk of type I error was mitigated in part through the LIMMA analysis used, which accounts for intercorrelations among measures and thereby helps minimize type I error. We note that one CpG site in the COL11A1 gene was significant even after Bonferroni correction. In addition, the fact that 7 of the genes displaying differential methylation in the primary study were validated in the informatics-based clinical sample also supports at least some of the observed methylation differences as being real effects. Nonetheless, the ICD-9-based CRPS phenotype in this validation sample might limit interpretation of these validation results. Another potential limitation is that we cannot address the possibility that pain management medications, which may differ in patients with CRPS, or other clinical factors may have impacted on methylation patterns after development of CRPS. Finally, generalizability of study results must be considered in light of the relatively low mean pain intensity observed (4.7/10), the unusual initiating event (amputation), and atypical sample demographics, although all patients with CRPS did meet the Budapest criteria. Patients with CRPS seeking clinical care often report severe pain and are more likely to be female and older,8 whereas the current sample was primarily younger and male, due to the military nature of the sample. Replication of these findings in more traditional clinical CRPS samples would be desirable. The fact that 1 of the top 2 CpG sites exhibiting significant differential methylation in this study also displayed differential gene expression between patients with CRPS and nonpain controls in a prior nonmilitary sample37 suggests that current results might generalize to more clinically representative samples.
In summary, this study for the first time suggests a potential role for epigenetic mechanisms (differential DNA methylation) in risk of CRPS after injury. The pattern of results suggests that immune and inflammatory mechanisms in particular may be involved in influencing CRPS risk. Although our results require replication, these epigenetic findings build on prior studies increasingly pointing towards the potential value of interventions targeting immune and inflammatory pathways for enhancing care of patients with CRPS.
Supplementary Material
Acknowledgments
The authors acknowledge the assistance of Melissa Chont in this project.
This research was supported by Congressionally Directed Medical Research Programs and Department of Defense awards MR130082, W81XWH-15-2-0046, W81XWH-12-2-0129, W81XWH-11-2-0003, and NIH grant T32GM008600. Support was also provided by a research grant from the Reflex Sympathetic Dystrophy Syndrome Association. The data set used for validation analyses was obtained from Vanderbilt University Medical Center’s BioVU which is supported by institutional funding and CTSA award No. UL1 TR002243 from the NIH/National Center for Advancing Translational Sciences. The contents of this manuscript are solely the responsibility of the authors and do not necessarily represent official views of the National Center for Advancing Translational Sciences, the Department of Defense, the Department of Veterans Affairs, or the U.S. Government.
Footnotes
Conflict of interest statement
The authors have no conflicts of interest to declare.
Appendix A. Supplemental digital content
Supplemental digital content associated with this article can be found online at http://links.lww.com/PAIN/A818, and http://links.lww.com/PAIN/A819, and http://links.lww.com/PAIN/A820.
Sponsorships or competing interests that may be relevant to content are disclosed at the end of this article.
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal’s Web site (www.painjournalonline.com).
References
- [1].Aïm F, Klouche S, Frison A, Bauer T, Hardy P. Efficacy of vitamin C in preventing complex regional pain syndrome after wrist fracture: a systematic review and meta-analysis. Orthop Traumatol Surg Res 2017;103:465–70. [DOI] [PubMed] [Google Scholar]
- [2].Alexander GM, van Rijn MA, van Hilten JJ, Perreault MJ, Schwartzman RJ. Changes in cerebrospinal fluid levels of pro-inflammatory cytokines in CRPS. PAIN 2005;116:213–219. [DOI] [PubMed] [Google Scholar]
- [3].Basu S Bioactive eicosanoids: role of prostaglandin F(2α) and F2-isoprostanes in inflammation and oxidative stress related pathology. Mol Cells 2010;30:383–91. [DOI] [PubMed] [Google Scholar]
- [4].Bennett MI, Smith BH, Torrance N, Potter J. The S-LANSS score for identifying pain of predominantly neuropathic origin: validation for use in clinical and postal research. J Pain 2005;6:149–58. [DOI] [PubMed] [Google Scholar]
- [5].Birklein F, Drummond PD, Li W, Schlereth T, Albrecht N, Finch PM, Dawson LF, Clark JD, Kingery WS. Activation of cutaneous immune responses in complex regional pain syndrome. J Pain 2014;15:485–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Blaes F, Schmitz K, Tschernatsch M, Kaps M, Krasenbrink I, Hempelmann G, Bräu ME. Autoimmune etiology of complex regional pain syndrome (M. Sudeck). Neurology 2004;63:1734–6. [DOI] [PubMed] [Google Scholar]
- [7].Bruehl S An update on the pathophysiology of complex regional pain syndrome. Anesthesiol 2010;113:713–25. [DOI] [PubMed] [Google Scholar]
- [8].Bruehl S Complex regional pain syndrome. BMJ 2015;351:h2730. [DOI] [PubMed] [Google Scholar]
- [9].Bruehl S, Harden RN, Galer BS, Saltz S, Backonja M, Stanton-Hicks M. Complex regional pain syndrome: are there distinct subtypes and sequential stages of the syndrome? PAIN 2002;95:119–24. [DOI] [PubMed] [Google Scholar]
- [10].Bruehl S, Maihöfner C, Stanton-Hicks M, Perez RS, Vatine JJ, Brunner F, Birklein F, Schlereth T, Mackey S, Mailis-Gagnon A, Livshitz A, Harden RN. Complex regional pain syndrome: evidence for warm and cold subtypes in a large prospective clinical sample. PAIN 2016;157:1674–81. [DOI] [PubMed] [Google Scholar]
- [11].Buchheit T, Van de Ven T, Hsia HL, McDuffie M, MacLeod DB, White W, Chamessian A, Keefe FJ, Buckenmaier CT, Shaw AD. Pain phenotypes and associated clinical risk factors following traumatic amputation: results from veterans integrated pain evaluation research (VIPER). Pain Med 2016;17:149–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Carapito R, Gottenberg JE, Kotova I, Untrau M, Michel S, Naegely L, Aouadi I, Kwemou M, Paul N, Pichot A, Locke J, Bowman SJ, Griffiths B, Sivils KL, Sibilia J, Inoko H, Micelli-Richard C, Nocturne G, Ota M, Ng WF, Mariette X, Bahram S. A new MHC-linked susceptibility locus for primary Sjögren’s syndrome: MICA. Hum Mol Genet 2017;26:2565–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Coderre TJ, Xanthos DN, Francis L, Bennett GJ. Chronic post-ischemia pain (CPIP): a novel animal model of complex regional pain syndrome-type I (CRPS-I; reflex sympathetic dystrophy) produced by prolonged hindpaw ischemia and reperfusion in the rat. PAIN 2004;112:94–105. [DOI] [PubMed] [Google Scholar]
- [14].Deichmann U Epigenetics: the origins and evolution of a fashionable topic. Dev Biol 2016;416:249–54. [DOI] [PubMed] [Google Scholar]
- [15].de Mos M, Huygen FJ, van der Hoeven-Borgman M, Dieleman JP, Ch Stricker BH, Sturkenboom MC. Outcome of the complex regional pain syndrome. Clin J Pain 2009;25:590–7. [DOI] [PubMed] [Google Scholar]
- [16].de Mos M, Laferrière A, Millecamps M, Pilkington M, Sturkenboom MC, Huygen FJ, Coderre TJ. Role of NFkappaB in an animal model of complex regional pain syndrome-type I (CRPS-I). J Pain 2009;10:1161–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Dennis G Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol 2003;4:P3. [PubMed] [Google Scholar]
- [18].Denny JC, Bastarache L, Ritchie MD, Carroll RJ, Zink R, Mosley JD, Field JR, Pulley JM, Ramirez AH, Bowton E, Basford MA, Carrell DS, Peissig PL, Kho AN, Pacheco JA, Rasmussen LV, Crosslin DR, Crane PK, Pathak J, Bielinski SJ, Pendergrass SA, Xu H, Hindorff LA, Li R, Manolio TA, Chute CG, Chisholm RL, Larson EB, Jarvik GP, Brilliant MH, McCarty CA, KulloI J, Haines JL, Crawford DC, Masys DR, Roden DM. Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat Biotechnol 2013;31:1102–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Denny JC, Ritchie MD, Basford MA, Pulley JM, Bastarache L, Brown-Gentry K, Wang D, Masys DR, Roden DM, Crawford DC. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover genedisease associations. Bioinformatics 2010;26:1205–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].de Rooij AM, de Mos M, Sturkenboom MC, Marinus J, van den Maagdenberg AM, van Hilten JJ. Familial occurrence of complex regional pain syndrome. Eur J Pain 2009;13:171–7. [DOI] [PubMed] [Google Scholar]
- [21].de Rooij AM, de Mos M, van Hilten JJ, Sturkenboom MC, Gosso MF, van den Maagdenberg AM, Marinus J. Increased risk of complex regional pain syndrome in siblings of patients? J Pain 2009;10:1250–5. [DOI] [PubMed] [Google Scholar]
- [22].de Rooij AM, Florencia Gosso M, Haasnoot GW, Marinus J, Verduijn W, Claas FH, van den Maagdenberg AM, van Hilten JJ. HLA-B62 and HLA- DQ8 are associated with Complex Regional Pain Syndrome with fixed dystonia. PAIN 2009;145:82–5. [DOI] [PubMed] [Google Scholar]
- [23].Dirckx M, Schreurs MW, de Mos M, Stronks DL, Huygen FJ. The prevalence of autoantibodies in complex regional pain syndrome type I. Mediators Inflamm 2015;2015:718201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Dubuis E, Thompson V, Leite MI, Blaes F, Maihöfner C, Greensmith D, Vincent A, Shenker N, Kuttikat A, Leuwer M, Goebel A. Longstanding complex regional pain syndrome is associated with activating autoantibodies against alpha-1a adrenoceptors. PAIN 2014;155: 2408–17. [DOI] [PubMed] [Google Scholar]
- [25].Gamazon ER, Segrè AV, van de Bunt M, Wen X, Xi HS, Hormozdiari F, Ongen H, Konkashbaev A, Derks EM, Aguet F, Quan J; GTEx Consortium, Nicolae DL, Eskin E, Kellis M, Getz G, McCarthy MI, Dermitzakis ET, Cox NJ, Ardlie KG. Using an atlas of gene regulation across 44 human tissues to inform complex disease- and trait-associated variation. Nat Genet 2018;50:956–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Gamazon ER, Wheeler HE, Shah KP, Mozaffari SV, Aquino-Michaels K, Carroll RJ, Eyler AE, Denny JC; GTEx Consortium, Nicolae DL, Cox NJ, Im HK. A gene-based association method for mapping traits using reference transcriptome data. Nat Genet 2015;47:1091–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Gene Ontology Consortium. Creating the gene ontology resource: design and implementation. Genome Res 2001;11:1425–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Goebel A, Blaes F. Complex regional pain syndrome, prototype of a novel kind of autoimmune disease. Autoimmun Rev 2013;12:682–6. [DOI] [PubMed] [Google Scholar]
- [29].Goebel A, Jacob A, Frank B, Sacco P, Alexander G, Philips C, Bassett P, Moots R. Mycophenolate for persistent complex regional pain syndrome, a parallel, open, randomised, proof of concept trial. Scand J Pain 2018; 18:29–37. [DOI] [PubMed] [Google Scholar]
- [30].Goebel A, Leite MI, Yang L, Deacon R, Cendan CM, Fox-Lewis A, Vincent A. The passive transfer of immunoglobulin G serum antibodies from patients with longstanding complex regional pain syndrome. Eur J Pain 2011;15:504.e1–6. [DOI] [PubMed] [Google Scholar]
- [31].GTEx Consortium; Laboratory, Data Analysis &Coordinating Center (LDACC)—Analysis Working Group; Statistical Methods groups—Analysis Working Group; Enhancing GTEx (eGTEx) groups; NIH Common Fund; NIH/NCI; NIH/NHGRI; NIH/NIMH; NIH/NIDA; Biospecimen Collection Source Site—NDRI; Biospecimen Collection Source Site—RPCI; Biospecimen Core Resource—VARI; Brain Bank Repository—University of Miami Brain Endowment Bank; Leidos Biomedical—Project Management; ELSI Study; Genome Browser Data Integration &Visualization—EBI; Genome Browser Data Integration &Visualization—UCSC Genomics Institute, University of California Santa Cruz; Lead analysts; Laboratory, Data Analysis &Coordinating Center (LDACC); NIH program management; Biospecimen collection; Pathology; eQTL manuscript working group, Battle A, Brown CD, Engelhardt BE, Montgomery SB. Genetic effects on gene expression across human tissues. Nature 2017;550:204–13.29022597 [Google Scholar]
- [32].Guo S, Zhu Q, Jiang T, Wang R, Shen Y, Zhu X, Wang Y, Bai F, Ding Q, Zhou X, Chen G, He DY. Genome-wide DNA methylation patterns in CD4+ T cells from Chinese Han patients with rheumatoid arthritis. Mod Rheumatol 2017;27:441–7. [DOI] [PubMed] [Google Scholar]
- [33].Harden RN, Bruehl S, Perez RS, Birklein F, Marinus J, Maihofner C, Lubenow T, Buvanendran A, Mackey S, Graciosa J, Mogilevski M, Ramsden C, Chont M, Vatine JJ. Validation of proposed diagnostic criteria (the “Budapest criteria”) for complex regional pain syndrome. PAIN 2010;150:268–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Hettne KM, de Mos M, de Bruijn AG, Weeber M, Boyer S, van Mulligen EM, Cases M, Mestres J, van der Lei J. Applied information retrieval and multidisciplinary research: new mechanistic hypotheses in complex regional pain syndrome. J Biomed Discov Collab 2007;2:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Higashimoto T, Baldwin EE, Gold JI, Boles RG. Reflex sympathetic dystrophy: complex regional pain syndrome type I in children with mitochondrial disease and maternal inheritance. Arch Dis Child 2008;93: 390–7. [DOI] [PubMed] [Google Scholar]
- [36].Ignatov A, Robert J, Gregory-Evans C, Schaller HC. RANTES stimulates Ca2+ mobilization and inositol trisphosphate (IP3) formation in cells transfected with G protein-coupled receptor 75. Br J Pharmacol 2006; 149:490–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Jin EH, Zhang E, Ko Y, Sim WS, Moon DE, Yoon KJ, Hong JH, Lee WH. Genome-wide expression profiling of complex regional pain syndrome. PLoS One 2013;8:e79435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet 2012;13:484–92. [DOI] [PubMed] [Google Scholar]
- [39].Joyce AR, Palsson BØ. Predicting gene essentiality using genome-scale in silico models. Methods Mol Biol 2008;416:433–57. [DOI] [PubMed] [Google Scholar]
- [40].Kemler MA, van de Vusse AC, van den Berg-Loonen EM, Barendse GA, van Kleef M, Weber WE. HLA-DQ1 associated with reflex sympathetic dystrophy. Neurology 1999;53:1350–1. [DOI] [PubMed] [Google Scholar]
- [41].Kim JH, Kim YC, Nahm FS, Lee PB. The therapeutic effect of vitamin C in an animal model of complex regional pain syndrome produced by prolonged hindpaw ischemia-reperfusion in rats. Int J Med Sci 2017;14:97–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Klein J, Sato A. The HLA system. First of two parts. N Engl J Med 2000; 343:702–9. [DOI] [PubMed] [Google Scholar]
- [43].Kohr D, Singh P, Tschernatsch M, Kaps M, Pouokam E, Diener M, Kummer W, Birklein F, Vincent A, Goebel A, Wallukat G, Blaes F. Autoimmunity against the β2 adrenergic receptor and muscarinic-2 receptor in complex regional pain syndrome. PAIN 2011;152:2690–700. [DOI] [PubMed] [Google Scholar]
- [44].Kohr D, Tschernatsch M, Schmitz K, Singh P, Kaps M, Schäfer KH, Diener M, Mathies J, Matz O, Kummer W, Maihöfner C, Fritz T, Birklein F, Blaes F. Autoantibodies in complex regional pain syndrome bind to a differentiation-dependent neuronal surface autoantigen. PAIN 2009;143:246–51. [DOI] [PubMed] [Google Scholar]
- [45].Kunz M, König IR, Schillert A, Kruppa J, Ziegler A, Grallert H, Müller-Nurasyid M, Lieb W, Franke A, Ranki A, Panelius J, Koskenmies S, Hasan T, Kere J, Rönn AC, Simon JC, Schmidt E, Wenzel J, Tüting T, Landsberg J, Zeller T, Blankenberg S, Gläser R, Patsinakidis N, Kuhn A, Ibrahim SM. Genome-wide association study identifies new susceptibility loci for cutaneous lupus erythematosus. Exp Dermatol 2015;24:510–15. [DOI] [PubMed] [Google Scholar]
- [46].Lage K, Karlberg EO, StØrling ZM, Olason PI, Pedersen AG, Rigina O, Hinsby AM, Tümer Z, Pociot F, Tommerup N, Moreau Y, Brunak S. A human phenome-interactome network of protein complexes implicated in genetic disorders. Nat Biotechnol 2007;25:309–16. [DOI] [PubMed] [Google Scholar]
- [47].Lenz M, Uçeyler N, Frettlöh J, Höffken O, Krumova EK, Lissek S, Reinersmann A, Sommer C, Stude P, Waaga-Gasser AM, Tegenthoff M, Maier C. Local cytokine changes in complex regional pain syndrome type I (CRPS I) resolve after 6 months. PAIN 2013;154:2142–9. [DOI] [PubMed] [Google Scholar]
- [48].Li Y, Wilson HL, Kiss-Toth E. Regulating STING in health and disease. J Inflamm (Lond) 2017;14:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Maihöfner C, Handwerker HO, Neundörfer B, Birklein F. Mechanical hyperalgesia in complex regional pain syndrome: a role for TNF-alpha? Neurology 2005;65:311–13. [DOI] [PubMed] [Google Scholar]
- [50].McCaffary D STING signalling: an emerging common pathway in autoimmunity and cancer. Immunopharmacol Immunotoxicol 2017;39: 253–8. [DOI] [PubMed] [Google Scholar]
- [51].Nguyen T, Nioi P, Pickett CB. The Nrf2-antioxidant response element signaling pathway and its activation by oxidative stress. J Biol Chem 2009;284:13291–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Orlova IA, Alexander GM, Qureshi RA, Sacan A, Graziano A, Barrett JE, Schwartzman RJ, Ajit SK. MicroRNA modulation in complex regional pain syndrome. J Transl Med 2011;9:195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Orozco G, Eerligh P, Sánchez E, Zhernakova S, Roep BO, González-Gay MA, López-Nevot MA, Callejas JL, Hidalgo C, Pascual-Salcedo D, Balsa A, González-Escribano MF, Koeleman BP, Martín J. Analysis of a functional BTNL2 polymorphism in type 1 diabetes, rheumatoid arthritis, and systemic lupus erythematosus. Hum Immunol 2005;66:1235–41. [DOI] [PubMed] [Google Scholar]
- [54].Philips N, Auler S, Hugo R, Gonzalez S. Beneficial regulation of matrix metalloproteinases for skin health. Enzyme Res 2011;2011:427285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Ritchie MD, Denny JC, Crawford DC, Ramirez AH, Weiner JB, Pulley JM, Basford MA, Brown-Gentry K, Balser JR, Masys DR, Haines JL, Roden DM. Robust replication of genotype-phenotype associations across multiple diseases in an electronic medical record. Am J Hum Genet 2010; 86:560–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Roden DM, Pulley JM, Basford MA, Bernard GR, Clayton EW, Balser JR, Masys DR. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clin Pharmacol Ther 2008;84:362–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Rossin EJ, Lage K, Raychaudhuri S, Xavier RJ, Tatar D, Benita Y. International Inflammatory Bowel Disease Genetics Constortium, Cotsapas C, Daly MJ. Proteins encoded in genomic regions associated with immune-mediated disease physically interact and suggest underlying biology. PLoS Genet 2011;7:e1001273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Sánchez-Rodríguez R, Torres-Mena JE, Quintanar-Jurado V, Chagoya-Hazas V, Rojas Del Castillo E, Del Pozo Yauner L, Villa-Treviño S, Pérez-Carreón JI. Ptgr1 expression is regulated by NRF2 in rat hepatocarcinogenesis and promotes cell proliferation and resistance to oxidative stress. Free Radic Biol Med 2017;102:87–99. [DOI] [PubMed] [Google Scholar]
- [60].Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A 2003;100:9440–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Tajerian M, Hung V, Khan H, Lahey LJ, Sun Y, Birklein F, Krämer HH, Robinson WH, Kingery WS, Clark JD. Identification of KRT16 as a target of an autoantibody response in complex regional pain syndrome. Exp Neurol 2017;287:14–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Tan W, Song Y, Mo C, Jiang S, Wang Z. Analysis of gene expression profile microarray data in complex regional pain syndrome. Mol Med Rep 2017;16:3371–8. [DOI] [PubMed] [Google Scholar]
- [63].Tékus V, Hajna Z, Borbély É, Markovics A, Bagoly T, Szolcsányi J, Thompson V, Kemény ρ, Helyes Z, Goebel A. A CRPS-IgG-transfer-trauma model reproducing inflammatory and positive sensory signs associated with complex regional pain syndrome. PAIN 2014;155: 299–308. [DOI] [PubMed] [Google Scholar]
- [64].Tian B, Brasier AR. Identification of a nuclear factor kappa B-dependent gene network. Recent Prog Horm Res 2003;58:95–130. [DOI] [PubMed] [Google Scholar]
- [65].Tost J DNA methylation: an introduction to the biology and the disease-associated changes of a promising biomarker. Methods Mol Biol 2009; 507:3–20. [DOI] [PubMed] [Google Scholar]
- [66].Uçeyler N, Eberle T, Rolke R, Birklein F, Sommer C. Differential expression patterns of cytokines in complex regional pain syndrome. PAIN 2007;132:14–15. [DOI] [PubMed] [Google Scholar]
- [67].van de Beek WJ, Roep BO, van der Slik AR, Giphart MJ, van Hilten BJ. Susceptibility loci for complex regional pain syndrome. PAIN 2003;103: 93–7. [DOI] [PubMed] [Google Scholar]
- [68].Vaneker M, van der Laan L, Allebes WA, Goris J. Genetic factors associated with Complex Regional Pain Syndrome I: HLA DRB and TNF alpha promoter gene polymorphism. Disabil Med 2002;2: 69–74. [Google Scholar]
- [69].van Hilten JJ, van de Beek WJ, Roep BO. Multifocal or generalized tonic dystonia of complex regional pain syndrome: a distinct clinical entity associated with HLA-DR13. Ann Neurol 2000;48:113–6. [DOI] [PubMed] [Google Scholar]
- [70].van Rooijen DE, Roelen DL, Verduijn W, Haasnoot GW, Huygen FJ, Perez RS, Claas FH, Marinus J, van Hilten JJ, van den Maagdenberg AM. Genetic HLA associations in complex regional pain syndrome with and without dystonia. J Pain 2012;13:784–9. [DOI] [PubMed] [Google Scholar]
- [71].Wang F, Stefano GB, Kream RM. Epigenetic modification of DRG neuronal gene expression subsequent to nerve injury: etiological contribution to complex regional pain syndromes (Part I). Med Sci Monit 2014;20:1067–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Wang F, Stefano GB, Kream RM. Epigenetic modification of DRG neuronal gene expression subsequent to nerve injury: etiological contribution to complex regional pain syndromes (Part II). Med Sci Monit 2014;20:1188–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Wen H, Lei Y, Eun SY, Ting JP. Plexin-A4-semaphorin 3A signaling is required for Toll-like receptor- and sepsis-induced cytokine storm. J Exp Med 2010;207:2943–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Wesseldijk F, Huygen FJ, Heijmans-Antonissen C, Niehof SP, Zijlstra FJ. Six years follow- up of the levels of TNF-alpha and IL-6 in patients with complex regional pain syndrome type 1. Mediators Inflamm 2008;2008: 469439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Wesseldijk F, Huygen FJ, Heijmans-Antonissen C, Niehof SP, Zijlstra FJ. Tumor necrosis factor-alpha and interleukin-6 are not correlated with the characteristics of Complex Regional Pain Syndrome type 1 in 66 patients. Eur J Pain 2008;12:716–21. [DOI] [PubMed] [Google Scholar]
- [76].Yang T, Wang S, Yang X, Zheng Q, Wang L, Li Q, Wei M, Du Z, Fan Y. Upregulation of Bcl-2 and its promoter signals in CD4+ T cells during neuromyelitis optica remission. Front Neurosci 2017;11:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Zhang M Transcriptional regulatory element database. Available at: http://rulai.cshl.edu/cgi-bin/TRED/tred.cgi?process=home. Accessed March 5, 2019. [Google Scholar]
- [78].Zhao Q, Wei Y, Pandol SJ, Li L, Habtezion A. STING signaling promotes inflammation in experimental acute pancreatitis. Gastroenterology 2018; 154:1822–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.