Abstract
Tumor necrosis factor-inducible gene 6 protein (TSG-6) is a cytokine secreted by mesenchymal stem cells (MSCs) and regulates MSC stemness. We previously reported that TSG-6 changes primary human hepatic stellate cells (pHSCs) into stem-like cells by activating yes-associated protein-1 (YAP-1). However, the molecular mechanism behind the reprogramming action of TSG-6 in pHSCs remains unknown. Cluster of differentiation 44 (CD44) is a transmembrane protein that has multiple functions depending on the ligand it is binding, and it is involved in various signaling pathways, including the Wnt/β-catenin path-way. Given that β-catenin influences stemness and acts down-stream of CD44, we hypothesized that TSG-6 interacts with the CD44 receptor and stimulates β-catenin to activate YAP-1 during TSG-6-mediated transdifferentiation of HSCs. Immuno-precipitation assays showed the interaction of TSG-6 with CD44, and immunofluorescence staining analyses revealed the colocalization of TSG-6 and CD44 at the plasma membrane of TSG-6-treated pHSCs. In addition, TSG-6 treatment upregulated the inactive form of phosphorylated glycogen synthase kinase (GSK)-3β, which is a negative regulator of β-catenin, and promoted nuclear accumulation of active/nonphosphorylated β-catenin, eventually leading to the activation of YAP-1. However, CD44 suppression in pHSCs following CD44 siRNA treatment blocked the activation of β-catenin and YAP-1, which inhibited the transition of TSG-6-treated HSCs into stem-like cells. Therefore, these findings demonstrate that TSG-6 interacts with CD44 and activates β-catenin and YAP-1 during the conversion of TSG-6-treated pHSCs into stem-like cells, suggesting that this novel pathway is an effective therapeutic target for control-ling liver disease.
Keywords: β-catenin, CD44, Hepatic stellate cell, Reprogramming, TSG-6
INTRODUCTION
Tumor necrosis factor-inducible gene 6 protein (TSG-6), one of cytokines released from mesenchymal stem cells (MSCs), is also involved in maintaining the stem cell properties of MSCs (1, 2). TSG-6 suppression in bone marrow-derived MSCs results in them losing their morphology, proliferative ability and differentiation capacity (2). In addition, we previously reported that TSG-6 induces hepatic stellate cells (HSCs) to gain stemness by activating yes-associated protein 1 (YAP-1), which is a well-known factor associated with stemness, resulting in reprogramming of HSCs into stem cell-like cells (3). How-ever, it remains unknown what receptors and intercellular sig-naling pathways are involved in TSG-6-mediated YAP-1 acti-vation and fate-change of HSCs.
Cluster of differentiation 44 (CD44) is a multifunctional cell-surface receptor that participates in hematopoietic differen-tiation, lymphocyte activation, inflammation and response to bacterial infection (4-7). CD44 is mainly expressed by immune cells, such as leukocytes, neutrophils and macrophages, and it regulates their functions, such as cell adhesion, proliferation, survival, motility, and migration (5). In addition, CD44 is a well-known marker of stem cells and cancer stem cells, and it plays a pivotal role in maintaining the properties of stem cells (8-10). CD44, a major receptor for hyaluronic acid (HA), binds components of the extracellular matrix, such as laminin and fibronectin, in addition to cytokines and growth factors (11). Depending on what ligand is bound to CD44, CD44 can activate a series of key signaling pathways, including Rho GTPases, Ras-mitogen-activated protein kinase (MAPK), phos-phatidylinositol 3-kinase (PI3K)/protein kinase B (AKT) and Wnt/β-catenin pathways, which impact cell behavior and function (12-15). A previous study by Choi et al. showed that TSG-6 interacts with the CD44 receptor on macrophages to inhibit nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κb) signaling and to suppress the production of pro-inflammatory cytokines (16). However, the association between CD44 and TSG-6 in the liver is poorly understood.
β-catenin, a core component of Wnt/β-catenin signaling, plays an important role in embryonic development, adult and various human diseases, including cancer (17-19). In the activated Wnt signaling pathway, phosphorylated glycogen synthase kinase (p-GSK)-3β, an inactive form, allows an active form of nonphosphorylated β-catenin in the cytoplasm to enter into the nucleus where it regulates target gene expression (20, 21). In the nucleus, active β-catenin forms a complex with T-cell factor 4 (TCF4), which drives the transcription of target genes including YAP-1 (22, 23). Wnt/β-catenin signaling is essential for maintaining the pluripotency of stem cells and for somatic cell reprogramming (24). Disruption of β-catenin leads to the failure in maintaining the pluripotency of embryonic stem cells and reprogramming of somatic cells (24). Recently, it has been suggested that CD44 is a positive upstream regu-lator of β-catenin in developing brain and cancer cells, such as chronic myeloid leukemia and glioblastoma (15, 25, 26). However, the association of CD44 with β-catenin in the liver remains unclear.
Emerging evidence shows that liver resident nonparen-chymal cells, such as Kupffer cells, progenitor cells, and HSCs, express CD44, and CD44 is involved in the proliferation, migration and activation of these cells (27-29). Given that the CD44 receptor is expressed on the cell surface of activated HSCs and that TSG-6 promotes their transdifferentiation into stem-like cells, we hypothesized that CD44 is involved in TSG-6-mediated reprogramming of HSCs. To test this hypo-thesis, we investigated whether TSG-6 interacts with CD44 and whether CD44 stimulates signals to activate YAP-1 in human primary HSCs (pHSCs). We found that TSG-6 interacts with CD44 and that its interaction activates β-catenin and ulti-mately YAP-1 to induce the fate-change of pHSCs into stem cell-like cells. Therefore, these findings demonstrate that CD44 is a novel receptor for TSG-6 in inducing the cellular transition of HSCs, suggesting that the TSG-6 stimulated CD44 pathway is a potential therapeutic target for treating liver disease.
RESULTS
Extracellular TSG-6 interacts with the cell surface receptor CD44 in pHSCs
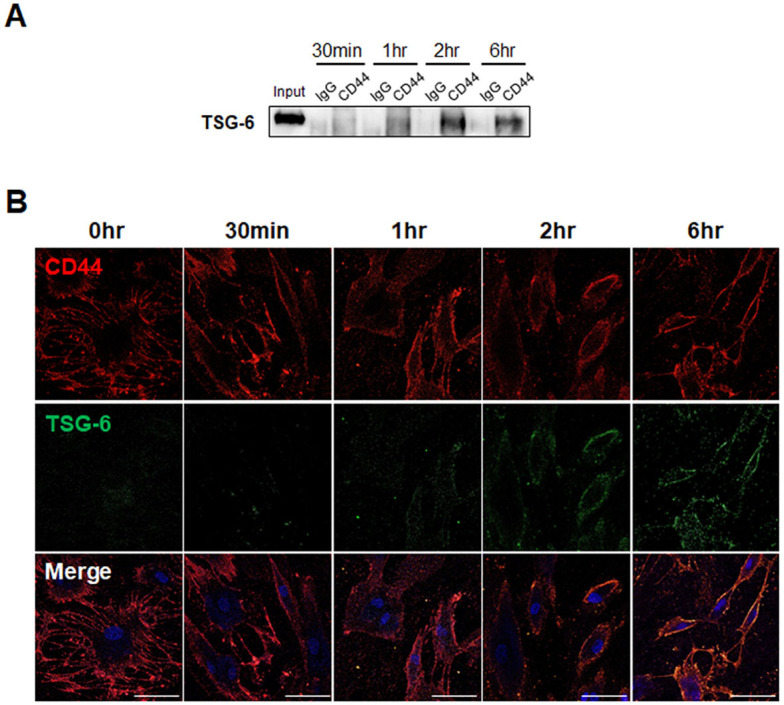
It was previously demonstrated that TSG-6 switches activated pHSCs to stem-like cells (3). Given that CD44 is involved in stemness and fate change and is expressed by HSCs (8, 29), we first examined whether the TSG-6 peptide interacts with CD44 to determine the association between CD44 and TSG-6 action in HSCs. We conducted an immunoprecipitation assay using pHSCs that were treated with TSG-6 for 30 min, 1, 2, and 6 hours. Cell lysates isolated from TSG-6-exposed pHSCs were immunoprecipitated with an anti-CD44 antibody or normal IgG and were then analyzed by immunoblot with an anti-TSG-6 antibody (Fig. 1A). In anti-CD44 immunoprecipitates, TSG-6 started to be observed at 30 min and showed higher expression at 2 and 6 hours after TSG-6 treatment, indicating that the greatest amount of interaction between TSG-6 and CD44 occurred at these time points. TSG-6 was detected in the input control, but no band for TSG-6 was detected in anti-IgG immunoprecipitates from TSG-6-treated pHSC lysates. To verify the interaction between TSG-6 and CD44, we performed immuno-fluorescence staining using fluorescein isothiocyanate (FITC)-conjugated TSG-6 peptides to visualize its localization with CD44 in pHSCs. Immunofluorescence staining showed that CD44 (red), which is a cell surface receptor, was mostly loca-lized at the plasma membrane of pHSCs treated with or without TSG-6 (Fig. 1B). Green fluorescence in pHSCs was not detected at 0 hours, and FITC-conjugated TSG-peptides were only faintly observed at 30 min. Their presence at the plasma membrane of pHSCs became clear, and colocalization of TSG-6 with CD44 was evident at 1, 2, and 6 hours after TSG-6 treatment. Consistent with the immunoprecipitation assay, CD44 and TSG-6 were mostly colocalized on the cell surface of pHSCs at 2 and 6 hours after TSG-6 treatment. These results demonstrate that extracellular TSG-6 peptides interact with CD44 on the cell surface of pHSCs, suggesting that CD44 is involved in TSG-6-mediated reprogramming of HSCs.
Fig. 1.
Interaction of TSG-6 with CD44 receptor in human pHSCs. (A) Immunoblots with TSG-6 in the immunoprecipitated pHSC lysates with CD44. pHSCs were treated with TSG-6 for 30 min, 1 hour, 2 hours, and 6 hours, respectively and then immunoprecipi-tated with CD44 antibody or IgG as negative control. Input sam-ples containing the same amount of TSG-6 protein used for in vitro treatment were used as positive controls. Data shown represent one of three experiments with similar results. (B) Immunofluo-rescence staining for FITC-conjugated TSG-6 (green) and CD44 (red) in pHSCs treated with TSG-6. DAPI was used in staining for nucleus. Representative images obtained from three experiments with similar results are shown (scale bar = 50 µm).
TSG-6 induces activation of β-catenin and subsequently YAP-1 in pHSCs
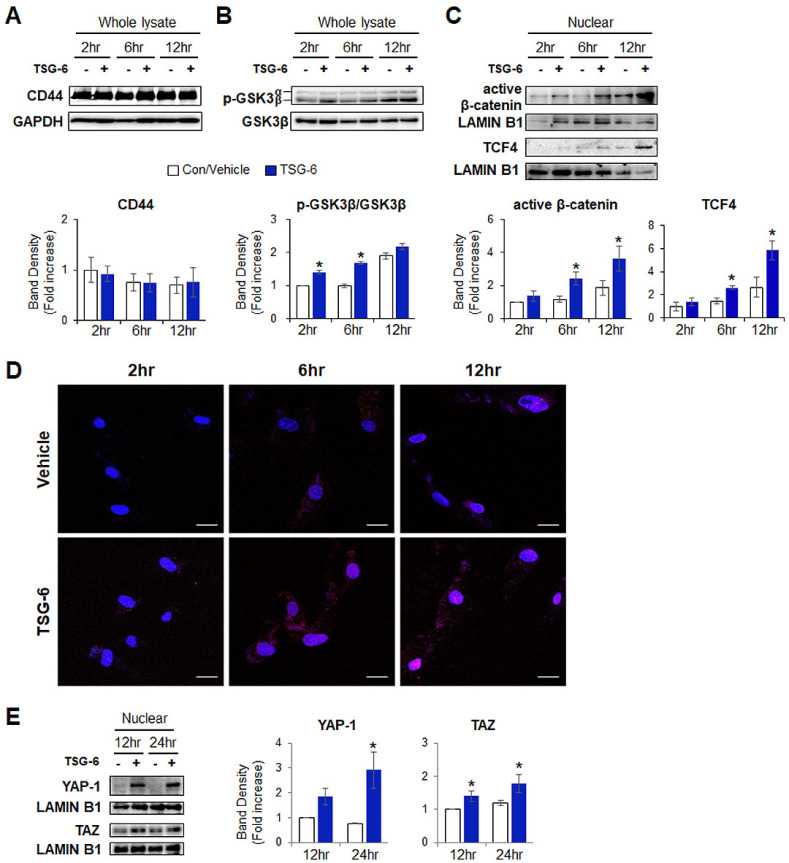
β-catenin, which is a main effector molecule in the Wnt signaling pathway, is a downstream effector of CD44 (25). In addition, Wnt/β-catenin signaling has been reported to directly regulate the expression of YAP-1 in colorectal cancer (23). Given that TSG-6 induced transdifferentiation of pHSCs into stem-like cells through YAP-1 (3) and that it could interaction with CD44, we assessed whether TSG-6 and CD44 influenced the activation of β-catenin in pHSCs. A strong interaction between TSG-6 and CD44 was observed at 2 and 6 hours in TSG-6-treated pHSCs, as shown by immunoprecipitation and immunofluorescence assays. Therefore, later time points (2, 6 and 12 hours) were analyzed to probe the downstream pathway of CD44 following activation by TSG-6. The protein level of CD44 was not significantly changed before and after TSG-6 treatment (Fig. 2A), as shown by immunofluorescent staining analysis for CD44 (Fig. 1B). GSK3β regulates β-catenin by phosphorylating it, and phosphorylated β-catenin undergoes ubiquitination-mediated degradation in the cytoplasm. The protein level of p-GSK3β, which is an inactive form, significantly increased at 2 and 6 hours in TSG-6-treated pHSCs compared to the vehicle-treated pHSCs (Fig. 2B). The amount of nonphos-phorylated nuclear β-catenin, which is the active form, showed a significant increase at 6 and 12 hours in TSG-6-treated pHSCs compared to vehicle-treated cells (Fig. 2C). TCF4, the transcrip-tional partner of nuclear β-catenin, was also upregulated in these cells. These results were also confirmed by immunofluorescence staining for active/nonphosphorylated β-catenin. Nuclear accu-mulation of active β-catenin (red) was first observed at 2 hours and was apparent in TSG-6-treated pHSCs at 6 and 12 hours, whereas nonphosphorylated β-catenins were rarely observed in the nucleus at 2 hours, and a small quantity of nonphos-phorylated β-catenins was detected in the nucleus at 6 and 12 hours in the vehicle-treated pHSCs, as observed under a con-focal microscope (Fig. 2D). YAP-1 activity was examined at 12 and 24 hours because nuclear β-catenin, a regulator of YAP, was accumulated at 6 and 12 hours in TSG-6-treated pHSCs. Com-pared to vehicle-treated pHSCs, the level of YAP-1 in the nucleus, where YAP-1 is active, was elevated at 12 and 24 hours and showed a significant increase at 24 hours (Fig. 2E). The level of transcriptional coactivator with PDZ-binding motif (TAZ), the co-activator of Hippo pathway, was also significantly elevated at 12 and 24 hours. These results demonstrate that TSG-6 promotes GSK3β phosphorylation and translocation of β-catenin into the nucleus and subsequently activates YAP-1, indicating that TSG-6-mediated translocation of β-catenin into the nucleus is closely associated with YAP-1 activity in TSG-6-treated pHSCs.
Fig. 2.
TSG-6 increases the levels of p-GSK3β (inactive form), nuclear β-catenin, and active YAP-1 in pHSCs. (A) Immunoblot and cumulative densitometric analyses of CD44, (B) inactive/phosphorylated (p-) GSK3β and GSK3β in whole lysate of pHSCs treated with TSG-6 for 2, 6, and 12 hours, respectively. GAPDH was used as an internal control. (C) Immunoblot and cumulative densitometric analyses of active/non-phosphorylated b-cate-nin and TCF4 in nuclear extracts of these cells. LAMIN B1 was used as an internal control for nuclear fraction. (D) Immunofluorescence staining for active/non-phosphorylated β-catenin (red) in these cells. DAPI was used for nuclear counterstaining. Representative images ob-tained from three experiments with similar results are shown (scale bar = 20 µm). (E) Immunoblot and cumulative densitometric analyses for nuclear YAP-1 and TAZ in these cells. All data shown represent one of three experiments with similar results. Mean ± SD results are graphed (*P < 0.05 vs. vehicle-treated pHSCs).
Suppression of CD44 abrogates the effect of TSG-6 in the fate-change of HSCs into stem-like cells
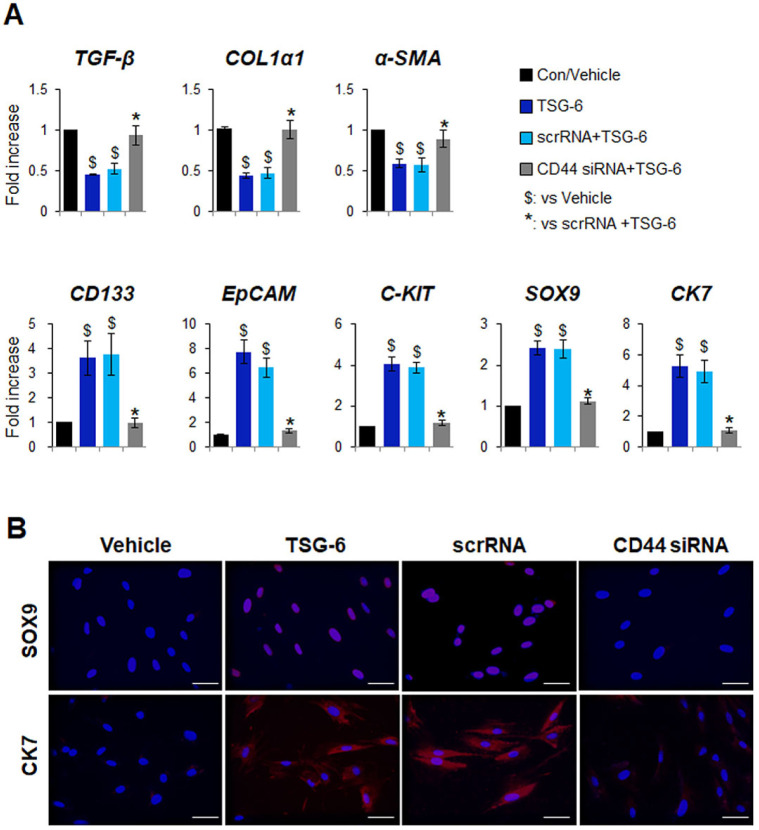
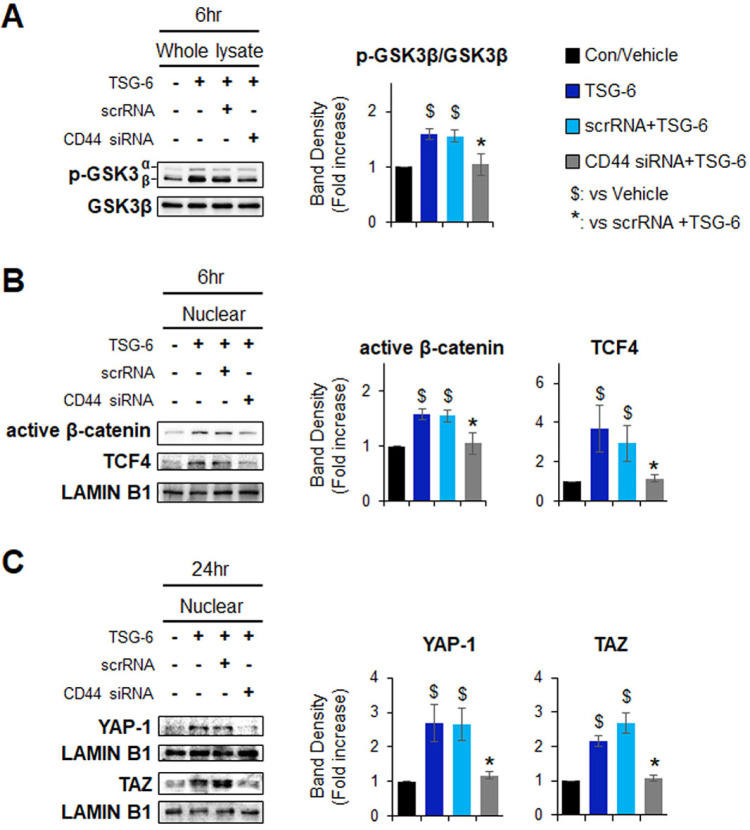
To probe whether the TSG-6-mediated HSC transition is dependent on CD44, CD44 expression in pHSCs was suppressed by CD44 siRNA. The knockdown efficiency for CD44 was confirmed at both the mRNA and protein levels after transfection. Scrambled siRNA (scrRNA) was used as a negative control (Supplementary Fig. S1). After siRNA transfection, these cells were treated with TSG-6 or a vehicle control for 24 hours. QRT-PCR analysis showed that the expression of markers of HSC activation, including transforming growth factor beta (TGF-β), collagen 1 alpha 1 (COL1α1), and alpha smooth muscle actin (α-SMA), significantly decreased in TSG-6-treated pHSCs with (scrRNA + TSG-6 group) or without scrRNA (TSG-6 group) compared with vehicle-treated cells. However, the downregu-lated expression of these markers was restored in CD44-sup-pressed pHSCs treated with TSG-6 (CD44 siRNA + TSG-6 group) (Fig. 3A). Furthermore, there was highly upregulated ex-pression of stem cell markers, such as CD133, epithelial cell adhesion molecule (EpCAM), C-KIT, SRY-box transcription factor 9 (SOX9), and cytokeratin 7 (CK7), in both the TSG-6 and scrRNA + TSG-6 groups, and the expression was significantly lower in the CD44 siRNA + TSG-6 group. Immunofluorescence staining for SOX9 (red) or CK7 (red) confirmed the qRT-PCR results by showing that accumulated SOX9- or CK7-positive cells in TSG-6-given pHSCs with or without scrRNA were rarely detected in CD44-inhibited HSCs that also had TSG-6 (Fig. 3B). In addition, the protein levels of p-GSK3β, active β-catenin, and TCF4 were significantly decreased in the CD44 siRNA + TSG-6 group compared with the TSG-6 and scrRNA + TSG-6 groups at 6 hours (Fig. 4A and 4B). The amount of YAP-1 and TAZ in the nucleus was lower in pHSCs with CD44 siRNA than pHSCs with or without scrRNA at 24 hours post TSG-6 treatment (Fig. 4C). These results suggest that TSG-6 interacts with CD44 and activates its downstream effectors β-catenin and YAP-1 to regulate the HSC transition into stem-like cells, indicating that TSG-6 influences HSC trans-differentiation through CD44.
Fig. 3.
Depletion of CD44 blocks TSG-6-mediated transition of HSCs into stem-like cells. (A) qRT-PCR analysis of fibrotic markers including TGF-β, COL1α1, α-SMA, and stem cell markers including CD133, EpCAM, C-KIT, SOX9, CK7, in vehicle-(vehicle) or TSG-6-treated pHSCs (TSG-6) transfected with or without scramble or siRNA targeting CD44 (scrRNA + TSG-6, CD44 siRNA + TSG-6). pHSCs were treated with scrRNA or CD44 siRNA for 24 hours before TSG-6 treatment and then exposed to vehicles or TSG-6 for 24 hours. Data shown represent one of three experiments with similar results. Mean ± SD results are graphed (one-way ANOVA with Tukey corrections, $P < 0.05 vs. vehicle, *P < 0.05 vs. scrRNA + TSG-6). (B) Immunofluorescent staining for SOX9 (red) or CK7 (red) in these cells. DAPI was used for nuclear counterstaining. Representative images obtained from three experiments with similar results are shown (scale bar = 20 µm).
Fig. 4.
Inhibition of CD44 decreases expressions of pGSK3β, nuclear β-catenin and active YAP-1 in TSG-6-treated pHSCs. (A) Immunoblot and cumulative densitometric analyses of inactive/p-GSK3β, GSK3β in whole lysate of vehicle-(vehicle) or TSG-6-treated pHSCs (TSG-6) trans-fected with scramble siRNA (scrRNA + TSG-6) or siRNA targeting CD44 (CD44 siRNA + TSG-6). pHSCs were treated with scrRNA or CD44 siRNA for 24 hours before TSG-6 treatment and then treated with vehicles or TSG-6 for 6 hours. (B) Immunoblot and cumulative densitometric analyses of active β-catenin and TCF4 in nuclear extracts of these cells. LAMIN B1 was used as an internal control for nuclear fraction. (C) Immunoblot and cumulative densi-tometric analyses of nuclear YAP-1 and TAZ in scrRNA or CD44 siRNA-transfected pHSCs treated with vehicle or TSG-6 for 24 hours. All data shown represent one of three experiments with si-milar results. Mean ± SD results are graphed (one-way ANOVA with Tukey corrections, $P < 0.05 vs. vehicle, *P < 0.05 vs. scrRNA + TSG-6).
DISCUSSION
CD44 is a transmembrane protein that has multiple functions depending on the ligand that it binds, although the main actions of CD44 are to regulate wound healing and to regulate the immune system (6). There are several well-known ligands for CD44, such as HA, osteopontin, fibronectin, collagens, and laminin (11). Among these ligands, HA is known as the main ligand for the CD44 receptor, and when HA binds to CD44, downstream signaling is activated, including the PI3K/Akt and extracellular signal-regulated kinase (Erk) 1/2 pathways, which modulate cell proliferation, differentiation, migration, survival and angiogenesis (14). CD44 is highly expressed in cancer cells, including cancer stem cells, and it has been shown to be important in tumor growth and cancer metastasis (9, 30). In the liver, CD44 is highly expressed in fibrotic liver compared with healthy liver, and it is also expressed by infiltrating lymphocytes, Kupffer cells and activated HSCs (28). Hence, the CD44 receptor has been regarded as a promising target of anti-fibrotic agents. In the current study, we demonstrate for the first time that CD44 is a novel receptor for TSG-6 in HSCs. Immunoprecipitation assays and immunofluorescence staining showed that TSG-6 interacts with the CD44 receptor on the cell surface of pHSCs after TSG-6 treatment (Fig. 1). We previously demonstrated that TSG-6 induces inactivation, senescence and fate-change of HSCs and that it efficiently induces the reprogramming of activated HSCs into functional stem cell-like cells by activating YAP-1 during these processes (3). However, we did not reveal the molecular mechanisms underlying the link between TSG-6 and YAP-1 activation. Herein, we reveal that extracellular TSG-6 binds to the CD44 receptor on the surface of activated HSCs, upregulates the level of inactive GSK3β, promotes nuclear translocation of active β-catenin and finally contributes to YAP-1 activation (Fig. 1 and 2). The subsequent signaling pathway triggered by TSG-6 was abrogated in CD44-depleted pHSCs, and TSG-6 treatment failed to induce the expression of stem cell markers in the absence of CD44 (Fig. 3 and 4), suggesting that TSG-6 binding to CD44 and the subsequent activation of β-catenin/YAP-1 is essential for the reprogramming action of TSG-6 in HSCs.
However, there are other possible mechanisms to support the actions of CD44 on HSCs, although our findings suggest that direct interaction of TSG-6 with CD44 drives the fate-change of HSCs. Structural homology analyses report that both CD44 and TSG-6 contain an HA-binding domain and that TSG-6 has a higher binding affinity for HA than it does for CD44 (31, 32). Because HA is known to promote HSC activation and liver fibrosis via CD44 (29), it is possible that TSG-6 inhibits the binding of HA to CD44 by capturing HA, and reduction of HA binding to CD44 promotes HSC inacti-vation and senescence, leading to the fate-change of HSCs. To prove this possibility, further studies are needed to investigate possible associations among TSG-6, CD44, and HA.
In conclusion, we demonstrate that TSG-6 interacts with the CD44 receptor, and activates β-catenin/YAP-1 to drive the TSG-6-mediated fate-change of HSCs, suggesting that the CD44 pathway could be an effective target of TSG-6-based therapy for treating liver fibrosis.
MATERIALS AND METHODS
Cell experiments and CD44 siRNA transfection
Human pHSCs (purchased from Zen-Bio Inc., NC, USA) were seeded on 10 cm2 plates at a density of 2 × 106 cells or on 6 well-plates at a density of 2 × 105 cells, and cultured in Dulbecco’s Modified Eagle’s Medium (DMEM; Gibco, Life Technologies, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS; Gibco) and 1% penicillin/streptomycin (P/S; Gibco) at 37°C in a humidified atmosphere containing 5% CO2. pHSCs at 70-80% confluence were serum-starved overnight in medium containing no FBS, and then, were treated with 20 ng/ml of TSG-6 (R&D systems, Minneapolis, MN, USA) or vehicle for 30 min, 1, 2, 6, 12, and 24 hours, respectively. The concentration of TSG-6 was determined based on our previous study (3). For visualizing the localization of TSG-6, FITC-conjugated TSG-6 peptides (CUSABIO, Houston, TX, USA) were used in the same manner as described above. To examine the effect of TSG-6 in CD44-suppressed pHSCs, pHSCs at 50-60% confluence were serum-starved overnight, cultured in antibiotics-free medium with 2% FBS for 24 hours, and then transfected with 25nM of CD44 (Dharmacon, Chicago, IL, USA) or scramble siRNA (Dharmacon) using Lipofectamine RNAiMAX (Invitrogen, Life Technologies, CA, USA) according to the manufacturer’s instructions for 24 hours. After washing, TSG-6 or Vehicle was given to CD44-depleted pHSCs for 1 day. These experiments were repeated at least three times.
Statistical analysis
Results are expressed as the mean ± SD. Statistical differences were determined by Student’s t-test or one-way analysis of variance (ANOVA) using SPSS statistics 20 software, followed by the post hoc Tukey’s test. P-values < 0.05 were considered statistically significant.
Supplemental Materials
ACKNOWLEDGEMENTS
This work was supported by 2-year grant of Pusan National University.
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicting interests.
References
- 1.Carelli S, Colli M, Vinci V, Caviggioli F, Klinger M, Gorio A. Mechanical Activation of Adipose Tissue and Derived Mesenchymal Stem Cells: Novel Anti-Inflam-matory Properties. Int J Mol Sci. 2018;19:pii: E267. doi: 10.3390/ijms19010267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Romano B, Elangovan S, Erreni M, et al. TNF-Sti-mulated Gene-6 Is a Key Regulator in Switching Stemness and Biological Properties of Mesenchymal Stem Cells. Stem Cells. 2019;37:973–987. doi: 10.1002/stem.3010. [DOI] [PubMed] [Google Scholar]
- 3.Wang S, Kim J, Lee C, et al. Tumor necrosis factor-inducible gene 6 reprograms hepatic stellate cells into stem-like cells, which ameliorates liver damage in mouse. Biomaterials. 2019;219:119375. doi: 10.1016/j.biomaterials.2019.119375. [DOI] [PubMed] [Google Scholar]
- 4.Cao H, Heazlewood SY, Williams B, et al. The role of CD44 in fetal and adult hematopoietic stem cell regulation. Haematologica. 2016;101:26–37. doi: 10.3324/haematol.2015.135921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baaten BJ, Li CR, Bradley LM. Multifaceted regulation of T cells by CD44. Commun Integr Biol. 2010;3:508–512. doi: 10.4161/cib.3.6.13495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson P, Ruffell B. CD44 and its role in inflammation and inflammatory diseases. Inflamm Allergy Drug Targets. 2009;8:208–220. doi: 10.2174/187152809788680994. [DOI] [PubMed] [Google Scholar]
- 7.van der Windt GJ, Florquin S, de Vos AF, et al. CD44 deficiency is associated with increased bacterial clearance but enhanced lung inflammation during Gram-negative pneumonia. Am J Pathol. 2010;177:2483–2494. doi: 10.2353/ajpath.2010.100562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thapa R, Wilson GD. The Importance of CD44 as a Stem Cell Biomarker and Therapeutic Target in Cancer. Stem Cells Int. 2016;2016:2087204. doi: 10.1155/2016/2087204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Du L, Wang H, He L, et al. CD44 is of Functional Importance for Colorectal Cancer Stem Cells. Clin Cancer Res. 2008;14:6751–6760. doi: 10.1158/1078-0432.CCR-08-1034. [DOI] [PubMed] [Google Scholar]
- 10.Zhu H, Mitsuhashi N, Klein A, et al. The role of the hyaluronan receptor CD44 in mesenchymal stem cell migration in the extracellular matrix. Stem Cells. 2006;24:928–935. doi: 10.1634/stemcells.2005-0186. [DOI] [PubMed] [Google Scholar]
- 11.Lesley J, Hyman R, Kincade PW. CD44 and its interaction with extracellular matrix. Adv Immunol. 1993;54:271–335. doi: 10.1016/S0065-2776(08)60537-4. [DOI] [PubMed] [Google Scholar]
- 12.Bourguignon LY. Hyaluronan-mediated CD44 acti-vation of RhoGTPase signaling and cytoskeleton function promotes tumor progression. Semin Cancer Biol. 2008;18:251–259. doi: 10.1016/j.semcancer.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Orian-Rousseau V, Morrison H, Matzke A, et al. Hepatocyte growth factor-induced Ras activation requires ERM proteins linked to both CD44v6 and F-actin. Mol Biol Cell. 2007;18:76–83. doi: 10.1091/mbc.e06-08-0674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herishanu Y, Gibellini F, Njuguna N, et al. Acti-vation of CD44, a receptor for extracellular matrix com-ponents, protects chronic lymphocytic leukemia cells from spontaneous and drug induced apoptosis through MCL-1. Leuk Lymphoma. 2011;52:1758–1769. doi: 10.3109/10428194.2011.569962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmitt M, Metzger M, Gradl D, Davidson G, Orian-Rousseau V. CD44 functions in Wnt signaling by regulating LRP6 localization and activation. Cell Death Differ. 2015;22:677–689. doi: 10.1038/cdd.2014.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choi H, Lee RH, Bazhanov N, Oh JY, Prockop DJ. Anti-inflammatory protein TSG-6 secreted by activated MSCs attenuates zymosan-induced mouse peritonitis by decrea-sing TLR2/NF-kappaB signaling in resident macro-phages. Blood. 2011;118:330–338. doi: 10.1182/blood-2010-12-327353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhan T, Rindtorff N, Boutros M. Wnt signaling in cancer. Oncogene. 2017;36:1461–1473. doi: 10.1038/onc.2016.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Steinhart Z, Angers S. Wnt signaling in develop-ment and tissue homeostasis. Development. 2018;145:pii: dev146589. doi: 10.1242/dev.146589. [DOI] [PubMed] [Google Scholar]
- 19.Shi J, Zheng H, Yuan L. High NDRG3 Expression Facilitates HCC Metastasis by Promoting Nuclear Translocation of b-Catenin. BMB Rep. 2019;52:451–456. doi: 10.5483/BMBRep.2019.52.7.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Behrens J, Jerchow BA, Würtele M, et al. Functional Interaction of an Axin Homolog, Conductin, with b-Cate-nin, APC, and GSK3β. Science. 1998;280:596–599. doi: 10.1126/science.280.5363.596. [DOI] [PubMed] [Google Scholar]
- 21.He TC, Sparks AB, Rago C, et al. Identification of c-MYC as a target of the APC pathway. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 22.Bottomly D, Kyler SL, McWeeney SK, Yochum GS. Identification of β-catenin binding regions in colon cancer cells using ChIP-Seq. Nucleic Acids Res. 2010;38:5735–5745. doi: 10.1093/nar/gkq363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Konsavage WM, Jr, Kyler SL, Rennoll SA, Jin G, Yochum GS. Wnt/beta-catenin signaling regulates Yes-associated protein (YAP) gene expression in colorectal carcinoma cells. J Biol Chem. 2012;287:11730–11739. doi: 10.1074/jbc.M111.327767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miki T, Yasuda SY, Kahn M. Wnt/beta-catenin signaling in embryonic stem cell self-renewal and somatic cell reprogramming. Stem Cell Rev Rep. 2011;7:836–846. doi: 10.1007/s12015-011-9275-1. [DOI] [PubMed] [Google Scholar]
- 25.Chang G, Zhang H, Wang J, et al. CD44 targets Wnt/beta-catenin pathway to mediate the proliferation of K562 cells. Cancer Cell Int. 2013;13:117. doi: 10.1186/1475-2867-13-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu X, Morales FC, Agarwal NK, Dogruluk T, Gagea M, Georgescu MM. Moesin is a glioma pro-gression marker that induces proliferation and Wnt/beta-catenin pathway activation via interaction with CD44. Cancer Res. 2013;73:1142–1155. doi: 10.1158/0008-5472.CAN-12-1040. [DOI] [PubMed] [Google Scholar]
- 27.Yovchev MI, Grozdanov PN, Joseph B, Gupta S, Dabeva MD. Novel hepatic progenitor cell surface markers in the adult rat liver. Hepatology. 2007;45:139–149. doi: 10.1002/hep.21448. [DOI] [PubMed] [Google Scholar]
- 28.Satoh T, Ichida T, Matsuda Y, et al. Interaction between hyaluronan and CD44 in the development of dimethylnitrosamine-induced liver cirrhosis. J Gastroenterol Hepatol. 2000;15:402–411. doi: 10.1046/j.1440-1746.2000.02164.x. [DOI] [PubMed] [Google Scholar]
- 29.Kikuchi S, Griffin CT, Wang SS, Bissell DM. Role of CD44 in epithelial wound repair: migration of rat hepatic stellate cells utilizes hyaluronic acid and CD44v6. J Biol Chem. 2005;280:15398–15404. doi: 10.1074/jbc.M414048200. [DOI] [PubMed] [Google Scholar]
- 30.Chen C, Zhao S, Karnad A, Freeman JW. The biology and role of CD44 in cancer progression: thera-peutic implications. J Hematol Oncol. 2018;11:64. doi: 10.1186/s13045-018-0605-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lesley J, English NM, Gal I, Mikecz K, Day AJ, Hyman R. Hyaluronan binding properties of a CD44 chimera containing the link module of TSG-6. J Biol Chem. 2002;277:26600–26608. doi: 10.1074/jbc.M201068200. [DOI] [PubMed] [Google Scholar]
- 32.Lesley J, Gal I, Mahoney DJ, et al. TSG-6 modulates the interaction between hyaluronan and cell surface CD44. J Biol Chem. 2004;279:25745–25754. doi: 10.1074/jbc.M313319200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.