Abstract
ANKHD1 (ankyrin repeat and KH domain containing 1) is a large protein characterized by the presence of multiple ankyrin repeats and a K-homology domain. Ankyrin repeat domains consist of widely existing protein motifs in nature, they mediate protein-protein interactions and regulate fundamental biological processes, while the KH domain binds to RNA or ssDNA and is associated with transcriptional and translational regulation. In recent years, studies containing relevant information on ANKHD1 in cancer biology and its clinical relevance, as well as the increasing complexity of signaling networks in which this protein acts, have been reported. Among the signaling pathways of interest in oncology regulated by ANKHD1 are Hippo signaling, JAK/STAT, and STMN1. The scope of the present review is to survey the current knowledge and high-light future perspectives for ANKHD1 in the malignant phenotype of cancer cells, exploring biological, functional, and clinical reports of this protein in cancer.
Keywords: ANKHD1, Ankyrin repeat and KH domain containing 1, Cancer, JAK/STAT, YAP1
BACKGROUND
Multiple ankyrin repeats single KH domain (mask) protein was initially identified in Drosophila melanogaster and is characterized as a large protein with 25 ankyrin repeats grouped into two blocks and a K-homology domain (KH) (1). In human beings, the mask has two orthologous proteins: ANKHD1 (ankyrin repeat and KH domain containing 1, also known as human MASK1) and ANKRD17 (ankyrin repeat domain 17, also known as human MASK2). Ankyrin repeat domains (ARD) comprise several stacked ankyrin repeats (ANK), which are widely existing protein motifs in nature, consisting of 30-34 amino acid residues that fold into a β-turn, followed by two antiparallel α-helices, and mediate protein-protein interactions (2, 3). Unlike other protein domains, such as the SH2 and SH3 domains, ARD does not recognize a specific amino acid sequence or protein structure, and it can interact with several other domains (4, 5). ARD-containing proteins are found in all species and are fundamental to biological processes, including cell cycle control, transcrip-tional regulation, differentiation, apoptosis, and cytoskeletal organization (3, 6). The KH domains bind to RNA or ssDNA, and are found in proteins associated with transcriptional and translational regulation (7-9).
In recent years, many studies containing relevant informa-tion on ANKHD1 in cancer biology and its clinical relevance, as well as the increasing complexity of signaling networks in which this protein acts, have been reported. Thus, understanding ANKHD1-related cellular and molecular functions may pave the avenues for new insights into this protein in the field of oncology. In the present review, we analyze the evidence of ANKHD1 in the malignant phenotype of cancer cells (i.e. cell proliferation, migration, invasion, and tumorigenesis), exploring biological, functional, and clinical descriptions of the involve-ment of this protein in neoplasias.
ANKHD1: GENE AND PROTEIN ORGANIZATION
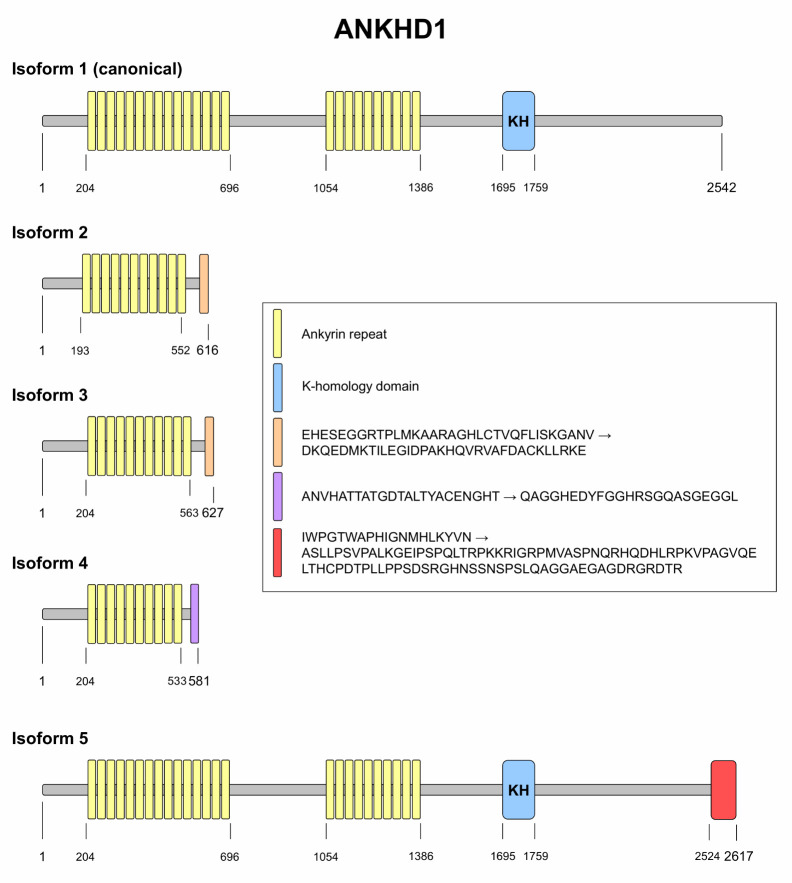
The entire human ANKHD1 (also named as MASK, MASK1, VBARP, and PP2500) gene is located at 5q31.3 and is about 147 Kb (start: 139781399 and end: 139929163 bp; orientation: plus strand). The complexity related to ANKHD1 transcripts and isoforms is increasing with the advancement of large-scale genetic sequencing and proteomic techniques. Currently, there are 21 different transcripts for the ANKHD1 gene deposited at Ensembl (www.ensembl.org), and a new ANKHD1 isoform unrelated to the transcripts previously reported at UniProt (www.uniprot.org). However, the present review will focus on the description and details of the 5 most consolidated transcript variants that are consistent in Ensembl, UniProt, and NCBI (https://www.ncbi.nlm.nih.gov/) databases.
The transcript variant 1 (start: 139781399 and end: 139919441 bp; orientation: plus strand; 34 exons; mRNA: 8233 bp, tran-script ID: ENST00000360839.7) is the longest transcript variant and encodes the ANKHD1 isoform 1 (canonical; 2542 amino acid [aa] protein). The transcript variant 2 (start: 139781399 and end: 139852062 bp; orientation: plus strand; 11 exons; mRNA: 2161 bp, transcript ID: ENST00000394722.7) presents an alternate in-frame splice site in the 5` coding region, lacks several exons, uses an alternate 3` terminal exon and encodes the ANKHD1 isoform 2, a shorter isoform that presents a distinct C-terminus compared to isoform 1 (616 aa protein). The transcript variant 3 (start: 139781399 and end: 139852062 bp; orientation: plus strand; 11 exons; mRNA: 2194 bp, tran-script ID: ENST00000394723.7) lacks several exons, uses an alternate 3` terminal exon, and encodes the ANKHD1 isoform 3 that also has a distinct C-terminus compared to isoform 1 (627 aa protein). The transcript variant 4 (start: 139781399 and end: 139852062 bp; orientation: plus strand; 10 exons, mRNA: 2084 bp, transcript ID: ENST00000616482.4) also lacks several exons, uses an alternate 3` terminal exon, and encodes the shortest ANKHD1 isoform (isoform 4; 581 aa protein), which also presents a distinct C-terminus compared to isoform 1.
Finally, the transcript variant 5 (also known as ANKHD1-EIF4EBP3) is a readthrough transcript involving ANKHD1 and EIF4EBP3 genes (start: 139781399 and end: 139929163 bp; orientation: plus strand; 36 exons, mRNA: 8349 bp; protein: 2617 aa, transcript ID: ENST00000532219.5). The comparative primary structure of ANKHD1 isoforms is illustrated in Fig. 1.
Fig. 1.
ANKHD1 isoforms. The primary structures of the main ANKHD1 isoforms are illustrated. The isoform 1 was considered the canonical isoform. The protein domains and amino acid positions are indicated. The data for figure construction was obtained from Ensembl (www. ensembl.org), UniProt (www.uniprot.org) and NCBI (https://www.ncbi.nlm.nih.gov/) databases.
Although ANKHD1 presents several isoforms, the specific functions of each isoform, as well as the subcellular localiza-tion pattern and their expression in different tissues and cell types, have been little explored so far. Smaller isoforms of ANKHD1 lack the KH domain, which has been implicated in the binding to microRNAs (miRNAs) and long noncoding RNAs (lncRNAs), modulating their availability and/or stability (10, 11). On the other hand, the repeats of ARD are quite preserved in all isoforms and appear to be necessary and sufficient for many of the protein interactions that we will discuss in the following lines. Larger ANKHD1 isoforms (1 and 5) present nuclear localization sequences (also known as NLS) that seem to play a key role in ANKHD1 nuclear translocation and in its function as a co-activator of YAP1 (12). Indeed, transfection assays using smaller ANKHD1 isoforms revealed a predominantly cytoplasmic distribution of these proteins (13).
ANKHD1 transcripts were widely expressed in normal tissues and cancer cell lines at different levels. For instance, the transcript variant 5 was highly expressed in the lymph node, while the high levels of the transcript variant 1 were found in the rectum. Similar levels of all transcript variants evaluated were reported in melanoma, breast, peripheral blood leukocytes, spleen, liver, and bone marrow (14). Thus, the data obtained so far suggest potential distinct functions for ANKHD1 isoforms, which is still a vast field to be explored.
SIGNALING NETWORK AND CELLULAR PROCESS REGULATED BY ANKHD1
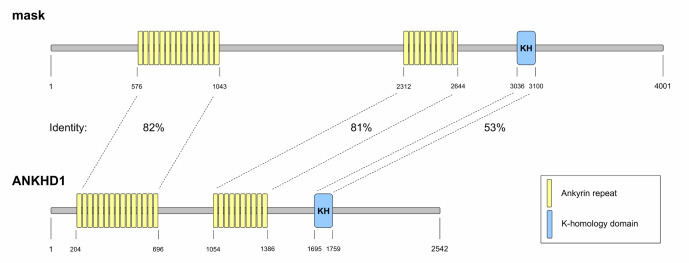
The human protein ANKHD1 is orthologous to the mask protein of Drosophila, so many cellular processes attributed to ANKHD1 were initially characterized in Drosophila models. A comparative analysis of the identity of the main domains between mask and ANKHD1 is illustrated in Fig. 2. Mask was initially identified playing a role in tyrosine kinase receptor-related signaling, including the EGFR pathway (1). This func-tion was associated with mask genetic interaction with the protein tyrosine phosphatase corkscrew (csw), which is ortho-logous to the human protein PTPN11 (also known as SHP2) (1). In Drosophila, loss of mask has been associated with the induction of apoptosis, reduction of cell proliferation and differentiation, and failure in signal transduction mediated by tyrosine kinase receptors (1). Later, the interaction between ANKHD1 and SHP2 was described in human tumor cell lines, LNCaP and K562, but their functional role remains largely unexplored (15). Downregulation of the small isoform of ANKHD1 that lacks the KH domain (also named as VBARP; ANKHD1 isoform 3) triggers caspase-3/7 activation and apop-tosis (13).
Fig. 2.
Comparative identity between mask and ANKHD1. The primary structure of mask (Drosophila melanogaster) and ANKHD1 (Homo sapiens; isoform 1) are illustrated. The protein domains, amino acid positions, and identity between regions are indicated. The data for figure construction was obtained from the UniProt database (www.uniprot.org), and the identities of indicated regions were compared using the Blastp database (https://blast.ncbi.nlm.nih.gov/).
In a screen for genetic modulators of the JAK/STAT signaling pathway in Drosophila models, mask was identified as a potential modulator for pathway activation (16). Recently, the role of ANKHD1 in regulating the JAK/STAT pathway has been the subject of research. In human cells, ANKHD1 modulates the levels of cytokine and growth factor receptors, including GP130, erythropoietin receptor, and thrombopoietin receptor (17). Furthermore, ANKHD1 silencing reduced STAT3 activation, which is an indicative of JAK/STAT pathway activity (17). In a murine model of autosomal dominant polycystic kidney disease, ANKHD1 increases STAT5 phosphorylation, induces cell proliferation, and contributes to the development and progression of the disease (18).
In 2013, it was described that mask acts as a yorkie (yki) cofactor, regulating the activity of the Hippo signaling pathway (19, 20). This discovery had a major impact, since the Hippo pathway has been related to morphogenesis and tissue homeostasis in normal development and is largely deregulated in cancer (21, 22). The interaction of ANKHD1/YAP1 (ortho-logizes mask/yki) has also been validated in human cells. Indeed, ANKHD1 has been shown to be important for nuclear translocation of YAP1, which is essential for its role as a transcriptional coactivator, as well as for the activation of YAP1-mediated genes (19, 20). In addition, implications of ANKHD1/YAP1 axis were highlighted for cell cycle progres-sion, migration, and invasion in vitro, and tumor growth in vivo (23-25). Recently, Sidor et al. (12) have reported mechanistic studies emphasizing that ANKHD1 is essential for the stability and nuclear localization of YAP1.
Despite strong indications that ANKHD1 regulates the JAK/STAT and Hippo pathway to exercise its role on cell prolife-ration, it has been shown that ANKHD1 may acts through other signaling repertoires to exert this cellular function. In leukemia cells that do not express significant levels of YAP1 and activation of the JAK/STAT pathway, the inhibition of ANKHD1 reduces proliferation and migration in vitro and tumorigenesis in vivo (26). In the molecular scenario, ANKHD1 physically interacts with SIVA1, contributing to STMN1 activity and microtubule dynamics (26). The activation of STMN1 is important for the proliferation and clonal growth of leukemia cells (27, 28).
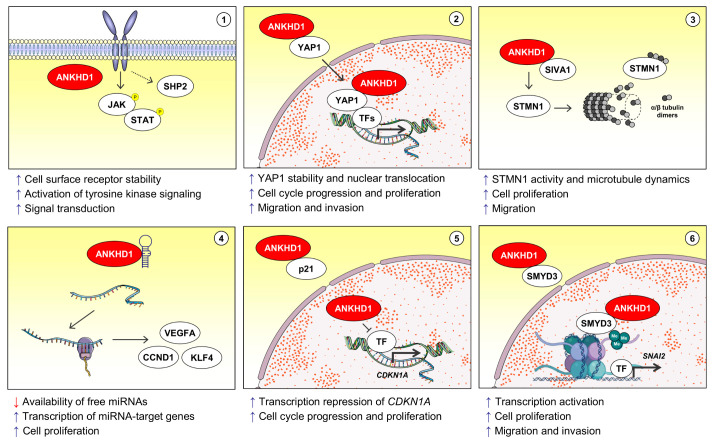
Accordingly, other studies have expanded the repertoire of signaling pathways and cellular processes mediated by ANKHD1. It has been reported that ANKHD1 interacts and represses CDKN1A (also known as p21) expression, promoting cell cycle progression (29, 30), and that ANKHD1 silencing induced DNA damage markers, and reduced core histones, PCNA expression and S-phase progression (31). In another context, ANKHD1 interacts with SMYD3, a lysine methyltransferase, and mediates the pro-migratory and pro-invasive phenotype through the activation of SMYD3-modulated genes, especially SNAI2 (also known as Slug) (32). It was also shown that ANKHD1 is able to bind to and sequester a subset of tumor suppressor miRNAs (i.e. miR-29a, miR-196a, and miR-205) through its K-homology domain, altering the expression of target genes of these miRNAs, including CCND1, VEGFA, and KLF4, and promoting cell cycle progression and proliferation (10). In Drosophila, mask genetically interacts with parkin to modulate mitochondrial morphology and homeostasis (33). Recently, it has been demonstrated that ANKHD1 is able to dimerize and deform lipid membranes, negatively regulating early endosome enlargement by its vesiculation ability (34). The role of ANKHD1 in the regulation of multiple signaling pathways and related cellular processes is illustrated in Fig. 3.
Fig. 3.
ANKHD1-mediated signaling pathways and cellular processes. (1) ANKHD1 stabilizes cell surface receptors, contributing to the activation of tyrosine kinase-mediated signaling pathways (e.g. JAK/STAT) and enhancing signal transduction. The SHP2 protein is an important regulator of tyrosine kinase and may be potentially involved in this process, as described by the crosstalk EGFR/mask/csw. (2) ANKHD1 stabilizes and participates in the nuclear translocation of YAP1, which contributes to the activation of the expression of genes related to increased cell cycle progression, proliferation, migration, and invasion. (3) ANKHD1 binds to SIVA1 preventing SIVA1-mediated STMN1 inhibition. The activity of STMN1 increases the microtubule dynamics, which contributes to cell proliferation and migration. (4) Experimental evidence indicates that ANKHD1, through its K-homology domain, reduces the availability of oncosuppressive miRNA, which allows expression of genes related to the malignant phenotype (e.g. CCND1, VEGFA and KLF4). (5) ANKHD1 interacts and inhibits p21, in addition to binding and repressing the promoter region of the gene CDKN1A. ANKHD1/p21 axis leads to progression in the cell cycle and increased pro-liferation. (6) ANKHD1 interacts with SMYD3 and assists in the tri-methylation at the 4th lysine residue of histone H3 (H3K4me3), this epigenetic alteration provides the expression of genes involved in cellular proliferation, migration, and invasion. Abbreviations: P, phosphorylation; Me, Methylation; TF, transcription factor.
ANKHD1 IMPLICATIONS IN CANCER
The aberrant expression of ANKHD1, as well as its potential prognostic value, has been reported in some types of cancer. In this setting, ANKHD1 is highly expressed in primary samples and cell lines derived from patients with acute leukemia and multiple myeloma (15, 29), which has been associated with cell proliferation, migration, and tumorigenesis (26, 30). Similarly, ANKHD1 expression is elevated in patients with renal cell carcinoma (10). In prostate cancer cells, ANKHD1 promotes cell cycle progression and increases tumor growth (23).
In breast cancer patients, high ANKHD1 expression was associated with reduced relapse-free survival in two independent data sets (20). In a similar way, high ANKHD1 was associated with reduced overall survival and recurrence-free survival in hepatocellular carcinoma patients (32). It was also demonstrated that ANKHD1 is widely expressed in both, normal and tumor colorectal tissues, but high ANKHD1 expression was associated with invasion of adjacent tissues of colorectal cancer patients (24). Functional assays confirmed that ANKHD1 promotes colorectal cancer cell migration and invasion in vitro and in vivo (24). ANKHD1 was highly expressed in non-small-cell lung tumors compared to normal adjacent tissue, and elevated ANKHD1 expression was associated with advanced pathological tumor-node-metastasis stage, lymph node metastasis, and poor survival outcomes in non-small-cell lung cancer patients (25). In lung cancer cell lines, ANKHD1 overexpression augmented cell proliferation, invasion, and tumor growth, while ANKHD1 silencing suppressed these malignant characteristics (25). Recently, it was reported that ANKHD1 was increased, enhanced the LINC00346 stability (a lncRNA), induced cell proliferation, and promoted angiogenesis in vitro and in vivo in glioma-associated endothelial cells (11). The main experimental and clinical findings of the participation of ANKHD1 in cancer are summarized in Table 1.
Table 1.
Summary of clinical and functional findings about ANKHD1 in cancer
| Cancer type | Expression | Notes and biological relevance | Ref. |
|---|---|---|---|
| Acute leukemia | ↑ ANKHD1 | ↑ Cell differentiation | (14) |
| It was found in primary leukemia blasts and cell lines | (15) | ||
| ↓ ANKHD1 | ↓ Proliferation and migration in vitro | (26) | |
| ↓Tumorigenesis in vivo | |||
| Multiple myeloma | ↑ ANKHD1 | It was found in primary CD138+ multiple myeloma cells and cell lines | (29) |
| ↓ ANKHD1 | ↓ Proliferation, migration, and cell cycle progression in vitro | (30, 31) | |
| ↓ Tumor growth in vivo | |||
| Breast cancer | ↑ ANKHD1 | ↓ Relapse-free survival | (20) |
| Prostate cancer | ↑ ANKHD1 | It was found in prostate cancer cells | (23) |
| ↓ ANKHD1 | ↓ Proliferation and cell cycle in vitro | ||
| ↓Tumorigenesis in vivo | |||
| Renal cancer cell | ↑ ANKHD1 | It was found in primary samples from renal cancer cell patients | (10) |
| ↑ Proliferation and cell cycle progression | |||
| Colorectal cancer | ↑ ANKHD1 | ↑ Tumor infiltration depth | (24) |
| ↓ ANKHD1 | ↓ Proliferation, migration, and invasion in vitro | ||
| ↓ Tumor growth and metastasis in vivo | |||
| Hepatocellular carcinoma | ↑ ANKHD1 | ↓ Overall survival and recurrence-free survival | (32) |
| Lung cancer | ↑ ANKHD1 | It was found in non-small-cell lung cancer tissues and cell lines | (25) |
| ↑ Metastasis | |||
| ↑ Proliferation and invasion | |||
| ↓ Prognosis | |||
| Glioma | ↑ ANKHD1 | It was found in glioma-associated endothelial cells | (11) |
| ↑ Proliferation and angiogenesis in vitro and in vivo |
FUTURE PERSPECTIVE FOR ANKHD1 IN ONCOLOGY AND CONCLUSION REMARKS
The complexity of the ANKHD1-mediated signaling network has grown in recent years. Due to the unique characteristics of this protein, current evidence indicates that ARD present in ANKHD1 serves as a signaling anchor providing stability for proteins and allowing or preventing interactions between different proteins (12, 26). Understanding the functions of the KH domain of ANKHD1 is still a topic to be explored and may considerably diversify the role of ANKHD1 in cellular processes, since the KH domain can bind, sequester or stabilize nucleic acids (e.g. miRNAs, lncRNAs) and modulate several relevant target-genes (10, 11). The identification of the relevant protein motifs for each protein interaction may help in the develop-ment of molecules capable of preventing these interactions and inhibiting the cellular processes of interest. In addition, recent evidence indicated that ANKHD1 may be relevant for aberrant angiogenesis in the tumor microenvironment (11), opening new perspectives of investigation.
Another point to be discussed is that the functional studies that silence ANKHD1 do not specify which isoforms are depleted. In this sense, only the study using the ANKHD1 isoform 3 showed that its silencing induced caspase activation and apoptosis (13). Of note, the expression of the different ANKHD1 isoforms may be tissue- and context-specific (14). Taken together, these data suggest that ANKHD1 may perform different functions according to the predominant isoform and the repertoire of ligands available in each cell type.
In drosophila, mask is the only protein form, but in humans, there are two orthologous proteins. Despite recent data, indi-cating ANKHD1 or ANKRD17 acted similarly in the nuclear translocation of YAP1 (12), results from knockout mice suggest additional non-redundant roles for ANKHD1 and ANKRD17, since the knockout of one of the proteins results in develop-mental failure and embryo lethality (18, 35). More in-depth studies on the impact of ANKHD1 knockout in murine models are still needed.
In summary, the data generated so far indicate that ANKHD1 has an important repertoire of interacting protein already related to the development and progression of cancer, high-lighting the positive regulation of YAP1, JAK/STAT, and STMN1. The clinical impact of ANKHD1 in different neoplasms is still scarce, but the results so far suggest that the high expression of ANKHD1 is related to worse clinical outcomes. Functional studies are consistent, even though by different signaling path-ways, that ANKHD1 promotes cell proliferation, migration, invasion, and tumor growth.
ACKNOWLEDGEMENTS
This study was supported by the grants #2017/24993-0, #2019/23864-7, and #2019/25421-5, São Paulo Research Foundation (FAPESP), and grant #402587/2016-2, Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq). The authors thanks Fernanda T. Udinal, from the Hemocentro Foundation of Ribeirão Preto, São Paulo, Brazil, for the English language review.
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicting interests.
References
- 1.Smith RK, Carroll PM, Allard JD, Simon MA. MASK, a large ankyrin repeat and KH domain-containing protein involved in Drosophila receptor tyrosine kinase signaling. Development. 2002;129:71–82. doi: 10.1242/dev.129.1.71. [DOI] [PubMed] [Google Scholar]
- 2.Li J, Mahajan A, Tsai MD. Ankyrin repeat: a unique motif mediating protein-protein interactions. Biochemistry. 2006;45:15168–15178. doi: 10.1021/bi062188q. [DOI] [PubMed] [Google Scholar]
- 3.Islam Z, Nagampalli RSK, Fatima MT, Ashraf GM. New paradigm in ankyrin repeats: Beyond protein-protein interaction module. Int J Biol Macromol. 2018;109:1164–1173. doi: 10.1016/j.ijbiomac.2017.11.101. [DOI] [PubMed] [Google Scholar]
- 4.Sedgwick SG, Smerdon SJ. The ankyrin repeat: a diversity of interactions on a common structural frame-work. Trends Biochem Sci. 1999;24:311–316. doi: 10.1016/S0968-0004(99)01426-7. [DOI] [PubMed] [Google Scholar]
- 5.Mosavi LK, Cammett TJ, Desrosiers DC, Peng ZY. The ankyrin repeat as molecular architecture for protein recognition. Protein Sci. 2004;13:1435–1448. doi: 10.1110/ps.03554604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hollenbeck JJ, Danner DJ, Landgren RM, Rainbolt TK, Roberts DS. Designed ankyrin repeat proteins as scaffolds for multivalent recognition. Biomacromolecules. 2012;13:1996–2002. doi: 10.1021/bm300455f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Valverde R, Edwards L, Regan L. Structure and function of KH domains. FEBS J. 2008;275:2712–2726. doi: 10.1111/j.1742-4658.2008.06411.x. [DOI] [PubMed] [Google Scholar]
- 8.Nazarov IB, Bakhmet EI, Tomilin AN. KH-Domain poly(C)-binding proteins as versatile regulators of multiple biological processes. Biochemistry (Mosc) 2019;84:205–219. doi: 10.1134/S0006297919030039. [DOI] [PubMed] [Google Scholar]
- 9.Nicastro G, Taylor IA, Ramos A. KH-RNA interactions: back in the groove. Curr Opin Struct Biol. 2015;30:63–70. doi: 10.1016/j.sbi.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 10.Fragiadaki M, Zeidler MP. Ankyrin repeat and single KH domain 1 (ANKHD1) drives renal cancer cell proliferation via binding to and altering a subset of miRNAs. J Biol Chem. 2018;293:9570–9579. doi: 10.1074/jbc.RA117.000975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang C, Zheng J, Liu X, et al. Role of ANKHD1/LINC00346/ZNF655 feedback loop in regulating the glio-ma angiogenesis via staufen1-mediated mRNA decay. Mol Ther Nucleic Acids. 2020;20:866–878. doi: 10.1016/j.omtn.2020.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sidor C, Borreguero-Munoz N, Fletcher GC, Elbediwy A, Guillermin O, Thompson BJ. Mask family proteins ANKHD1 and ANKRD17 regulate YAP nuclear import and stability. Elife. 2019;8:e48601. doi: 10.7554/eLife.48601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miles MC, Janket ML, Wheeler ED, et al. Molecular and functional characterization of a novel splice variant of ANKHD1 that lacks the KH domain and its role in cell survival and apoptosis. FEBS J. 2005;272:4091–4102. doi: 10.1111/j.1742-4658.2005.04821.x. [DOI] [PubMed] [Google Scholar]
- 14.Santos Duarte Ada S, Traina F, Favaro PM, et al. Characterisation of a new splice variant of MASK-BP3(ARF) and MASK human genes, and their expression patterns during haematopoietic cell differentiation. Gene. 2005;363:113–122. doi: 10.1016/j.gene.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 15.Traina F, Favaro PM, Medina Sde S, et al. ANKHD1, ankyrin repeat and KH domain containing 1, is over-expressed in acute leukemias and is associated with SHP2 in K562 cells. Biochim Biophys Acta. 2006;1762:828–834. doi: 10.1016/j.bbadis.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 16.Muller P, Kuttenkeuler D, Gesellchen V, Zeidler MP, Boutros M. Identification of JAK/STAT signalling components by genome-wide RNA interference. Nature. 2005;436:871–875. doi: 10.1038/nature03869. [DOI] [PubMed] [Google Scholar]
- 17.Fisher KH, Fragiadaki M, Pugazhendhi D, et al. A genome-wide RNAi screen identifies MASK as a positive regulator of cytokine receptor stability. J Cell Sci. 2018;131:jcs209551. doi: 10.1242/jcs.209551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patera F, Hautbergue GM, Wilson P, Evans PC, Ong AC, Fragiadaki M. Ankhd1 enhances polycystic kidney disease development via promoting proliferation and fibrosis. bioRxiv. 2020 doi: 10.1101/2020.03.04.977017. doi: 10.1101/2020.03.04.977017. [DOI] [Google Scholar]
- 19.Sidor CM, Brain R, Thompson BJ. Mask proteins are cofactors of Yorkie/YAP in the Hippo pathway. Curr Biol. 2013;23:223–228. doi: 10.1016/j.cub.2012.11.061. [DOI] [PubMed] [Google Scholar]
- 20.Sansores-Garcia L, Atkins M, Moya IM, et al. Mask is required for the activity of the Hippo pathway effector Yki/YAP. Curr Biol. 2013;23:229–235. doi: 10.1016/j.cub.2012.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Du Toit A. Cell signalling: A new Hippo pathway component. Nat Rev Mol Cell Biol. 2013;14:196. doi: 10.1038/nrm3534. [DOI] [PubMed] [Google Scholar]
- 22.Yu FX, Zhao B, Guan KL. Hippo pathway in organ size control, tissue homeostasis, and cancer. Cell. 2015;163:811–828. doi: 10.1016/j.cell.2015.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Machado-Neto JA, Lazarini M, Favaro P, et al. ANKHD1, a novel component of the Hippo signaling pathway, promotes YAP1 activation and cell cycle pro-gression in prostate cancer cells. Exp Cell Res. 2014;324:137–145. doi: 10.1016/j.yexcr.2014.04.004. [DOI] [PubMed] [Google Scholar]
- 24.Yao P, Li Y, Shen W, et al. ANKHD1 silencing suppresses the proliferation, migration and invasion of CRC cells by inhibiting YAP1-induced activation of EMT. Am J Cancer Res. 2018;8:2311–2324. [PMC free article] [PubMed] [Google Scholar]
- 25.Liu X-F, Han Q, Rong X-Z, et al. ANKHD1 pro-motes proliferation and invasion of non-small-cell lung cancer cells via regulating YAP oncoprotein expression and inactivating the Hippo pathway. Int J Oncol. 2020;56:1175–1185. doi: 10.3892/ijo.2020.4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Machado-Neto JA, Lazarini M, Favaro P, et al. ANKHD1 silencing inhibits Stathmin 1 activity, cell prolife-ration and migration of leukemia cells. Biochim Biophys Acta. 2015;1853:583–593. doi: 10.1016/j.bbamcr.2014.12.012. [DOI] [PubMed] [Google Scholar]
- 27.Machado-Neto JA, de Melo Campos P, Favaro P, et al. Stathmin 1 is involved in the highly proliferative phenotype of high-risk myelodysplastic syndromes and acute leukemia cells. Leuk Res. 2014;38:251–257. doi: 10.1016/j.leukres.2013.11.013. [DOI] [PubMed] [Google Scholar]
- 28.Machado-Neto JA, Saad ST, Traina F. Stathmin 1 in normal and malignant hematopoiesis. BMB Rep. 2014;47:660–665. doi: 10.5483/BMBRep.2014.47.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dhyani A, Duarte AS, Machado-Neto JA, Favaro P, Ortega MM, Olalla Saad ST. ANKHD1 regulates cell cycle progression and proliferation in multiple myeloma cells. FEBS Lett. 2012;586:4311–4318. doi: 10.1016/j.febslet.2012.10.037. [DOI] [PubMed] [Google Scholar]
- 30.Dhyani A, Machado-Neto JA, Favaro P, Saad ST. ANKHD1 represses p21 (WAF1/CIP1) promoter and promotes multiple myeloma cell growth. Eur J Cancer. 2015;51:252–259. doi: 10.1016/j.ejca.2014.11.012. [DOI] [PubMed] [Google Scholar]
- 31.Dhyani A, Favaro P, Saad STO. ANKHD1 is an S phase protein required for histone synthesis and DNA repair in multiple myeloma cells. Blood Cells Mol Dis. 2020;84:102460. doi: 10.1016/j.bcmd.2020.102460. [DOI] [PubMed] [Google Scholar]
- 32.Zhou Z, Jiang H, Tu K, et al. ANKHD1 is required for SMYD3 to promote tumor metastasis in hepatocellular carcinoma. J Exp Clin Cancer Res. 2019;38:18. doi: 10.1186/s13046-018-1011-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu M, Li X, Tian X, Wu C. Mask loss-of-function rescues mitochondrial impairment and muscle degeneration of Drosophila pink1 and parkin mutants. Hum Mol Genet. 2015;24:3272–3285. doi: 10.1093/hmg/ddv081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kitamata M, Hanawa-Suetsugu K, Maruyama K, Suetsugu S. Membrane-deformation ability of ANKHD1 is involved in the early endosome enlargement. iScience. 2019;17:101–118. doi: 10.1016/j.isci.2019.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hou SC, Chan LW, Chou YC, et al. Ankrd17, an ubiquitously expressed ankyrin factor, is essential for the vascular integrity during embryogenesis. FEBS Lett. 2009;583:2765–2771. doi: 10.1016/j.febslet.2009.07.025. [DOI] [PubMed] [Google Scholar]