Abstract
We report the development of a new strategy for the chemical analysis of live cells based on protein spherical nucleic acids (ProSNAs). The ProSNA architecture enables analyte detection via the highly programmable nucleic acid shell or a functional protein core. As a proof-of-concept, we use an i-motif as the nucleic acid recognition element to probe pH in living cells. By interfacing the i-motif with a forced-intercalation readout, we introduce a quencher-free approach that is resistant to false-positive signals, overcoming limitations associated with conventional fluorophore/quencher-based gold NanoFlares. Using glucose oxidase as a functional protein core, we show activity-based, amplified sensing of glucose. This enzymatic system affords greater than 100-fold fluorescence turn on in buffer, is selective for glucose in the presence of close analogs (i.e., glucose-6-phosphate), and can detect glucose above a threshold concentration of ~5 μM, which enables the study of relative changes in intracellular glucose concentrations.
The chemical analysis of live cells at the molecular level provides fundamental insight into dynamic cellular processes, informs about the role of intracellular analytes in disease progression, and has guided the development of new medical diagnostic tools.1–6 Although fluorescent probes based on both molecular recognition (binding-based sensing)3,7 and molecular reactivity (activity-based sensing)8 have led to significant new capabilities, the majority of techniques necessitate the fixing or lysis of the cells, the use of cytotoxic transfection reagents, or the genetic encoding of the cells. Specifically, protein- and nucleic acid-based approaches, such as enzyme-linked immunosorbent assays,9 genetically encoded-fluorescent proteins10 and RNA sensors,11 polymerase chain reaction,12 and fluorescence in situ hybridization,13 are routinely used to detect a wide variety of biological analytes. However, exogenous proteins and nucleic acids are not efficiently internalized by cells; thus, their development into live-cell intracellular probes is challenging.
To overcome these limitations, we have developed a powerful new class of intracellular probes based on protein spherical nucleic acids (ProSNAs).14,15 This design allows analyte detection via a quencher-free approach using either the nucleic acid or protein component. Additionally, this platform allows for the detection of intracellular analytes through binding or activity-based sensing. ProSNAs are based on the SNA architecture and consist of a protein core functionalized with a dense shell of radially oriented oligonucleotides. The SNA architecture is ideally suited for making intracellular measurements, as it is nontoxic to cells, elicits minimal immune response, can be taken up by cells without the need for transfection reagents, and is more resistant to nuclease degradation compared to traditionally used linear oligonucleotide probes.16 Additionally, it enables the intracellular delivery of functional proteins and confers stability against protease degradation.14,15
The first examples of SNA-based intracellular probes were NanoFlares (NFs).17,18 The NF construct consists of a gold nanoparticle core that acts as a quencher. Oligonucleotide duplexes comprising a recognition strand and a shorter fluorophore-labeled reporter strand are immobilized onto the gold nanoparticle through a gold–thiol linkage. Inside the cell, the target of interest displaces the reporter strand, as it binds to the recognition sequence and results in fluorescence turn on due to separation of the fluorophore and quencher. By designing the recognition strand to be complementary to nucleic acids in cells, genetic content can be measured.17–22 On the other hand, using aptamer and DNAzyme sequences, ions, small molecules, and proteins can be detected.23–25 NFs allow for live-cell genetic and metabolic analyses,17,23 the sorting and isolation of circulating tumor cells based on variations in genetic profiles,26 and the identification of diseased tissue in vivo.27–29
However, NFs suffer from several limitations. Since their fluorescence is solely dependent on the fluorophore’s distance from the gold core, NFs are susceptible to false-positive signals arising from nuclease degradation of the oligonucleotides, dehybridization of the reporter strands, or cleavage at the gold–thiol linkage.30 In addition, NFs rely on a displacement event for signal generation which retards probe–target binding kinetics.31 Finally, NFs can only be designed for targets with known nucleic acid-based recognition sequences.3
We hypothesized that the use of a quencher-free strategy coupled to an SNA architecture could overcome many of these challenges. To test this hypothesis, we first designed SNAs in which the nucleic acid sequences act as the recognition element. We used a duplex-sensitive dye, thiazole orange (TO), as a base surrogate in an oligonucleotide recognition sequence that is designed to bind to the target analyte (Figure 1, Figure S3). This class of dyes, derived from intercalators, has low fluorescence in a single-stranded oligonucleotide due to unrestricted rotation about the methine bridge in the molecules.32 In contrast, upon binding to the target analyte, the dye undergoes forced intercalation (FIT) between the oligonucleotide base pairs, thereby restricting its motion, leading to enhanced fluorescence.32–34 The use of a duplex-sensitive dye no longer necessitates the presence of a “chemically inert” gold core and allows us to expand the core selection. Here, we use a protein core as a model system due to its well-established biocompatibility, biodegradability, and known chemical topology, which allows for site-specific oligonucleotide attachment.35,36
Figure 1.
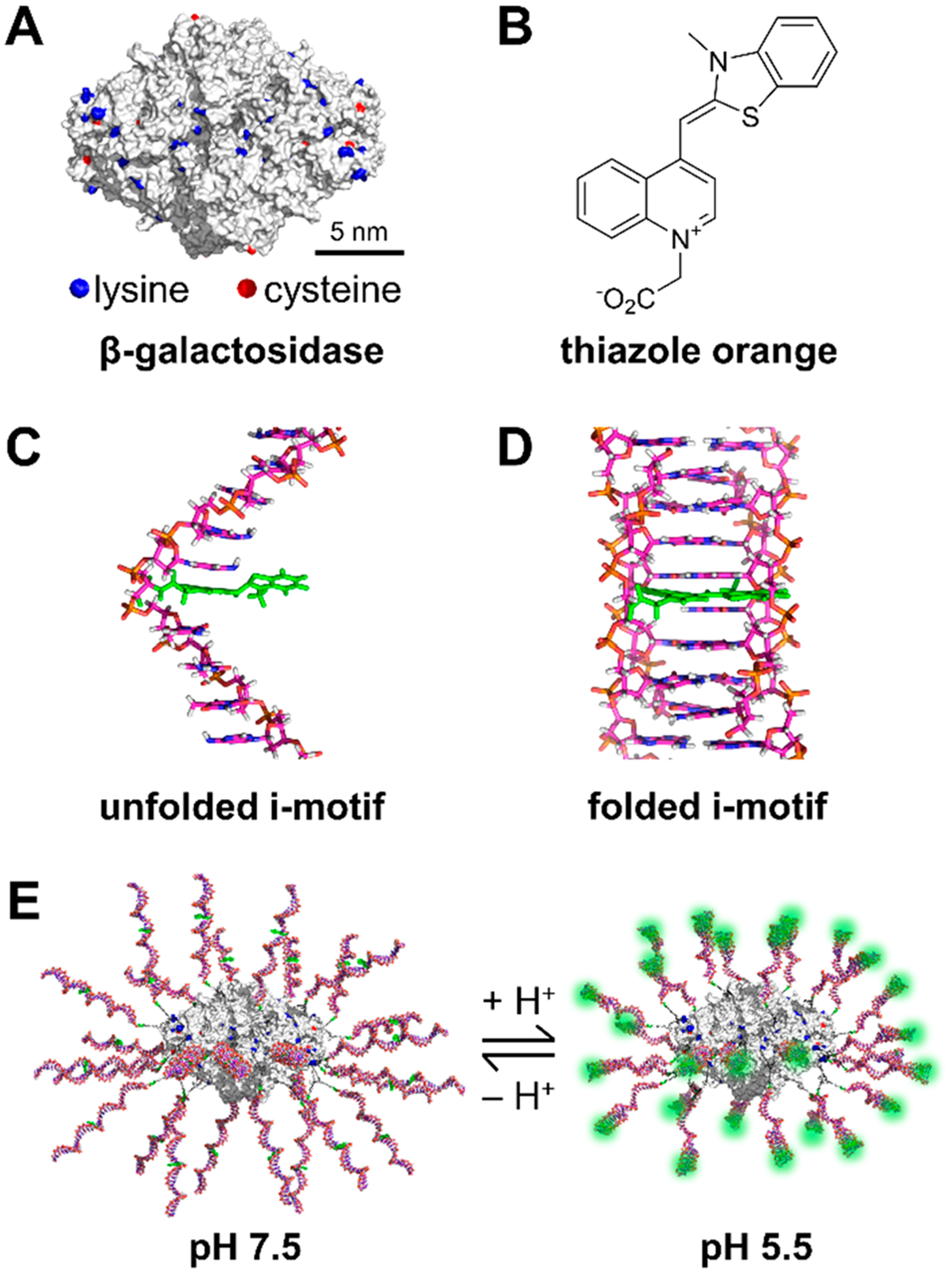
(A) Structure of β-galactosidase with lysine and cysteine residues highlighted. (B) Structure of the forced intercalation dye thiazole orange, TO (carboxymethylated derivative). (C) Unfolded i-motif sequence with a single base replaced with TO. (D) Folded i-motif with TO intercalated between base pairs. (E) Structure of ProTOn at pH 7.5 and pH 5.5. The formation of the i-motif structure leads to fluorescence turn on of TO.
For a proof-of-concept, we chose an i-motif sequence as the recognition strand that undergoes pH-dependent structural changes between an unfolded and a tetraplex form.37,38 The i-motif is an aptamer for protons, and we converted the i-motif into an FIT-aptamer using a strategy previously reported by our group.34 β-Galactosidase (β-gal) was chosen as the protein core, as it has been used in previous ProSNA studies.14,15 A fluorescent dye, Alexa Fluor 647 (AF-647), was covalently conjugated to the cysteine residues of the protein through maleimide–thiol coupling to enable the monitoring of the cellular uptake of the probe. Dibenzocyclooctyne terminated DNA was attached orthogonally to the lysine residues modified with PEG-azides (Figure S4). These modifications gave rise to a TO containing i-motif-β-gal ProSNA termed ProTOn (Figure 1).
We first established that ProTOn is capable of detecting pH changes in vitro (Figure 2A) by adding it to buffered solutions at different pH values. Our results show a gradual increase in the fluorescence enhancement of TO, saturating at 9-fold, when the pH of the solution changes from 7.5 to 5.0. No change in fluorescence signal is observed from AF-647 in this pH range. Similarly, ProSNAs containing a control sequence that does not form the i-motif structure results in no fluorescence enhancement in either the TO channel or the AF-647 channel in this pH range.
Figure 2.
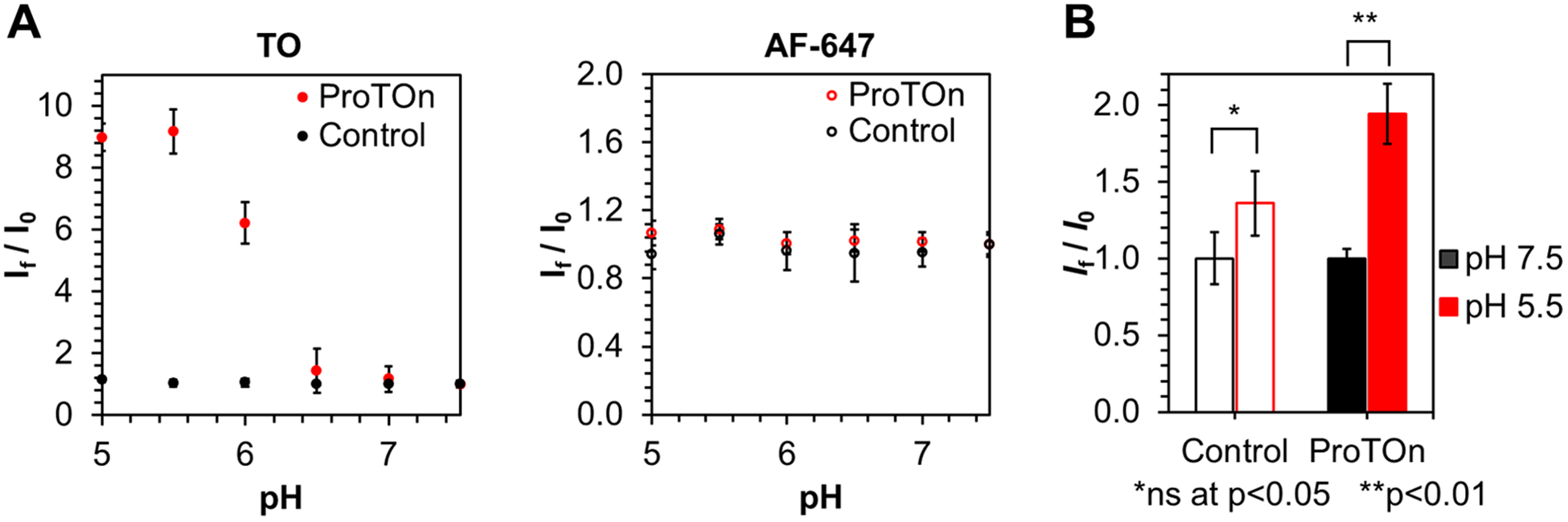
(A) In vitro fluorescence response of ProTOn and a control probe as a function of pH. The fluorescence of TO increases as pH decreases due to the formation of i-motifs in ProTOn. The control probe does not form an i-motif and, therefore, shows no change in fluorescence. The fluorescence of AF-647 remains unchanged for both ProTOn and the control probe. (B) TO channel fluorescence response of MDA-MB-231 cells treated with ProTOn and a control probe. ProTOn-treated cells clamped at pH 5.5 are almost twice as fluorescent as those clamped at pH 7.5. Cells treated with the control probe show no significant difference in fluorescence.
Next, we determined the ability of ProTOn to report changes in intracellular pH (Figure 2B). The MDA-MB-231 cell line, a human epithelial breast cancer cell line, was chosen as a model. These cells were incubated with 500 nM ProTOn (by DNA) in serum-free media for 3 h. The cells were then clamped at pH 5.5 and pH 7.5 and analyzed by flow cytometry. The mean fluorescence of cells at pH 5.5 is almost double that of cells at pH 7.5, consistent with the results obtained when pHrodo Red AM, a commercially available intracellular pH probe, is used as a benchmark (Figure S11). As before, control ProSNAs do not elicit a significant fluorescence change, demonstrating the specificity of ProTOn.
Importantly, unlike gold NFs, this new quencher-free ProSNA-based design is not prone to false-positive signals, as evidenced by in vitro nuclease degradation experiments (Figures S7, S9) and pulse-chase experiments in MDA-MB-231 cells (Figures S8, S10). Moreover, the use of an FIT-strategy does not require strand displacement, overcoming limitations associated with nonspecific dehybridization of the reporter strand and allowing faster binding to the target analyte.34 Taken together, these results show that ProSNA-based quencher-free probes constitute a next-generation platform for monitoring intracellular analytes in live cells compared to gold-based NFs.
Importantly, the use of a ProSNA allows one to not only detect analytes through the nucleic acid shell but also vastly expands the range of analytes that can be detected by taking advantage of the functional protein core. We hypothesized that using an enzyme, we can detect analytes for which nucleic acid-based recognition sequences are not known. To test this hypothesis, we designed a ProSNA for intracellular glucose detection using glucose oxidase (GOx) as the core (Figure 3). We chose glucose as a model analyte because of its fundamental importance to maintaining cellular functions, its high intracellular abundance (~0.1–2 mM),39,40 and the lack of a glucose aptamer with a biologically relevant binding affinity.41 Remarkably, the activity of GOx-SNAs remains unchanged compared to the native protein (Figure S14).
Figure 3.
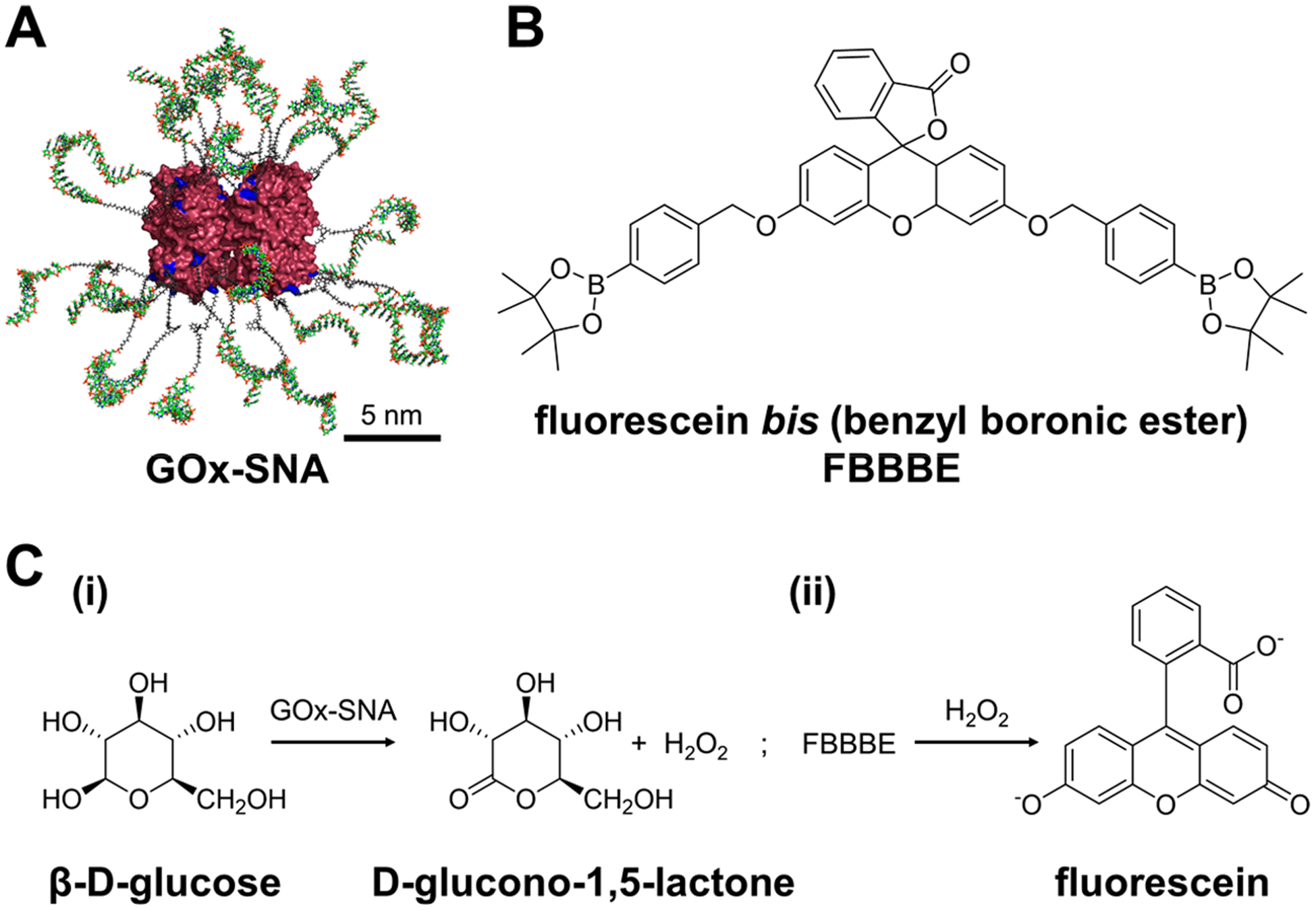
(A) Structure of glucose oxidase SNAs (GOx-SNAs). (B) Structure of fluorescein bis (benzyl boronic ester), FBBBE. (C) A two-step assay developed for glucose detection. (i) First, GOx-SNAs catalyze the conversion of β-d-glucose to d-glucono-1,5-lactone with the formation of H2O2. (ii) The H2O2 formed reacts with nonfluorescent FBBBE and yields highly fluorescent fluorescein.
To detect glucose intracellularly, we developed a new two-step assay. In the first step, cells are treated with GOx-SNAs in glucose-free media. Upon entering the cells, GOx-SNAs catalyze the conversion of glucose to d-glucono-1,5-lactone and produce hydrogen peroxide. In the second step, the cells are washed thoroughly and treated with fluorescein bis (benzyl boronic ester), FBBBE, a cell-permeable, nonfluorescent fluorescein derivative.42 In the presence of hydrogen peroxide, the boronate groups are cleaved and highly fluorescent fluorescein is formed and retained intracellularly. Therefore, the fluorescence is directly proportional to the amount of glucose in the cell. This assay results in a 120-fold fluorescence enhancement in the presence of glucose in vitro (Figure 4A). Due to the high specificity of GOx, the probe is selective against other sugars including sucrose, xylose, mannose, fructose, maltose, lactose, galactose, as well as glucose-6-phosphate which results from rapid phosphorylation of glucose upon cellular entry (Figure 4B, Figure S17).43
Figure 4.
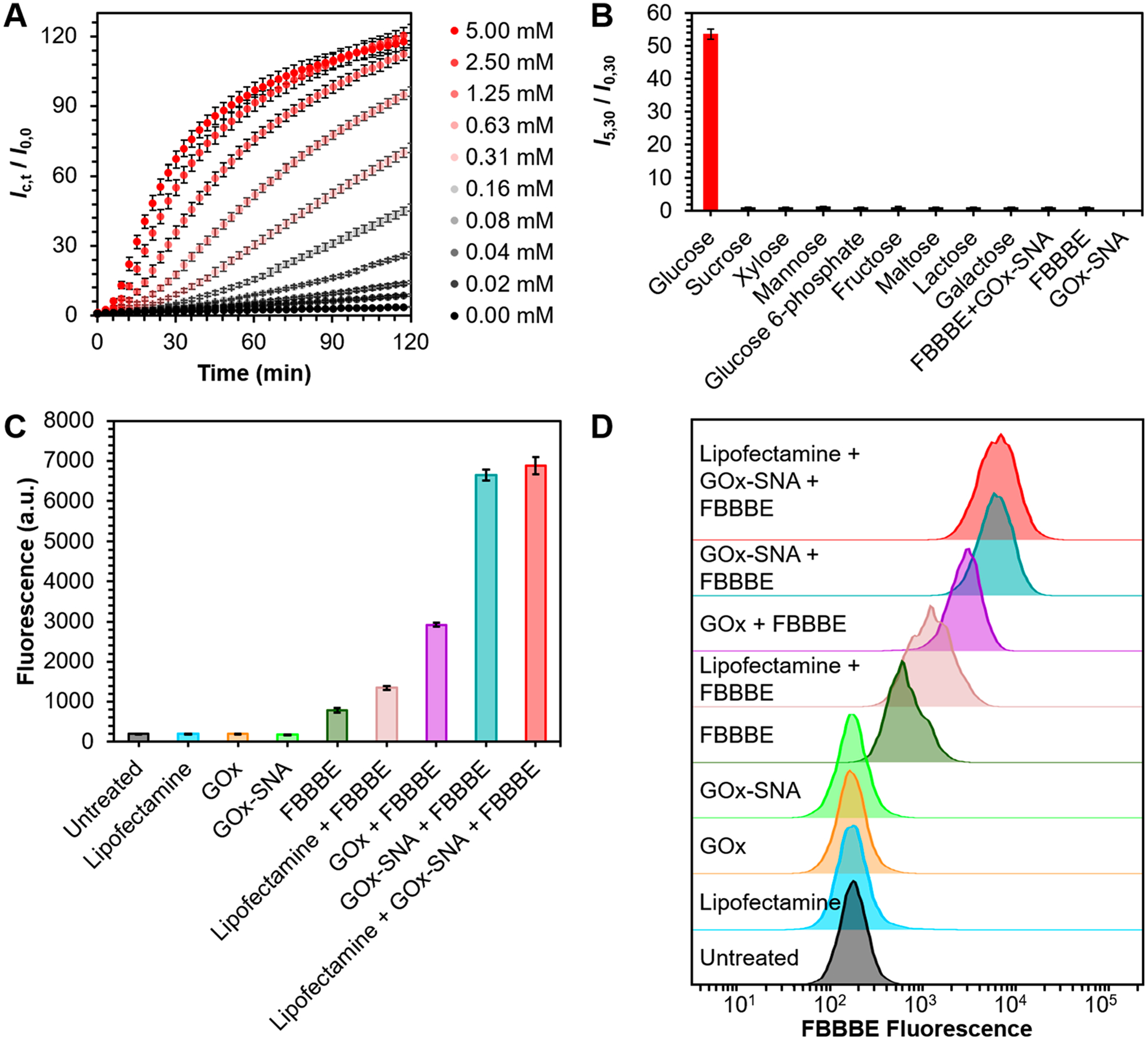
(A) In vitro fluorescence response of GOx-SNAs to increasing concentrations of glucose. The y-axis shows the observed fluorescence (Ic,t) for a particular concentration, c, of glucose at time, t, relative to the fluorescence (I0,0) of GOx-SNAs at the initial time point in the absence of glucose. Over 120-fold fluorescence enhancement is observed in the presence of glucose. (B) In vitro selectivity of GOx-SNAs against other sugars. (C) Fluorescence of EL4 cells under different treatment conditions. Cells treated with GOx-SNAs and FBBBE fluoresce ~12-fold more compared to cells treated with FBBBE alone. (D) Representative fluorescence histograms corresponding to data in (C).
To ensure the validity of this new assay in cells, we tested nine different cells lines (MDA-MB-231, MC38, U87, SKOV3, HDF, EL4, EG7-OVA, 4T1, and NIH/3T3), representing a mix of cancer and normal cells, adherent and suspension cells, and human and murine-derived cells (Figures S19–S27). Flow cytometry experiments show that cells treated with FBBBE alone have increased fluorescence compared to untreated cells. These results are expected because some fluorescein is formed due to the basal level of H2O2 produced in the cells. Cells pretreated with 40 nM GOx-SNAs (by protein) show up to an ~12-fold increase in fluorescence due to the elevated levels of H2O2 produced (Figure 4C). Modulating intracellular glucose levels impacts the fluorescence detected. GOx-SNA-treated cells exposed to media containing 25 mM glucose show an increase in fluorescence compared to those exposed to 0 mM glucose (Figure S28). Similarly, increasing glucose uptake by treating cells with 100 nM insulin results in higher fluorescence (Figure S29). In contrast, decreasing glucose uptake by using 10 μM cytochalasin B, a well-known glucose transport inhibitor,44 results in decreased fluorescence (Figure S30). Taken together, these experiments show that GOx-SNAs can be used as intracellular glucose probes, which, going forward, may aid in the high throughput screening of antihyperglycemic drugs.
In conclusion, we have developed a powerful new class of intracellular probes based on ProSNAs. Their modular structure allows one to change the protein core and the nucleic acid shell independently and detect analytes using either component as the sensing moiety through binding- or activity-based sensing. The programmable nature of nucleic acids, in principle, will allow for the detection of targets using aptamers, DNAzymes, or hybridization-based probes. The unparalleled specificity of proteins will give rise to probes that are highly selective for their targets. When one takes into account the many proteins that have been developed for sensing analytes in cell lysates or fluorescent proteins that are genetically encoded, the scope of possibilities is enormous, considering that these proteins could be repurposed as exogenous probes for detecting intracellular analytes in live cells and clinical samples.
Supplementary Material
ACKNOWLEDGMENTS
This material is based upon work supported by the Air Force Office of Scientific Research Award FA9550-17-1-0348 (oligonucleotide and SNA synthesis and characterization) and the Center for Bio-Inspired Energy Science, an Energy Frontier Research Center funded by the U.S. Department of Energy, Office of Science, Basic Energy Sciences Award DE-SC0000989 (i-motif and catalytic activity). It was also supported by the National Cancer Institute of the National Institutes of Health award U54CA199091 (cell studies and flow cytometry). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. This research was also sponsored by Air Force Research Laboratory under Agreement FA8650-15-2-5518 (gold NanoFlares and aptamer). This work made use of the IMSERC at Northwestern University, which has received support from the Soft and Hybrid Nanotechnology Experimental (SHyNE) Resource (NSF ECCS-1542205), the State of Illinois, and the International Institute for Nanotechnology (IIN). This work was supported by the Northwestern University – Flow Cytometry Core Facility supported by Cancer Center Support Grant (NCI CA060553). S.B.E. was supported in part by the Chicago Cancer Baseball Charities and the H Foundation at the Lurie Cancer Center of Northwestern University. We are grateful to Professor J. Annes and Z. Miao for helpful discussions.
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jacs.0c06866.
Experimental methods, additional discussion, and figures (PDF)
The authors declare no competing financial interest.
Contributor Information
Devleena Samanta, Department of Chemistry and International Institute for Nanotechnology, Northwestern University, Evanston, Illinois 60208, United States;.
Sasha B. Ebrahimi, Department of Chemical and Biological Engineering and International Institute for Nanotechnology, Northwestern University, Evanston, Illinois 60208, United States;.
Chad A. Mirkin, Department of Chemistry, Department of Chemical and Biological Engineering, and International Institute for Nanotechnology, Northwestern University, Evanston, Illinois 60208, United States;.
REFERENCES
- (1).Dean KM; Palmer AE Advances in Fluorescence Labeling Strategies for Dynamic Cellular Imaging. Nat. Chem. Biol 2014, 10 (7), 512–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Leung K; Chakraborty K; Saminathan A; Krishnan Y A DNA Nanomachine Chemically Resolves Lysosomes in Live Cells. Nat. Nanotechnol 2019, 14 (2), 176–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Samanta D; Ebrahimi SB; Mirkin CA Nucleic-Acid Structures as Intracellular Probes for Live Cells. Adv. Mater 2020, 32 (13), 1901743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Valm AM; Cohen S; Legant WR; Melunis J; Hershberg U; Wait E; Cohen AR; Davidson MW; Betzig E; Lippincott-Schwartz J Applying Systems-Level Spectral Imaging and Analysis to Reveal the Organelle Interactome. Nature 2017, 546 (7656), 162–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Park S; Aalipour A; Vermesh O; Yu JH; Gambhir SS Towards Clinically Translatable in Vivo Nanodiagnostics. Nat. Rev. Mater 2017, 2 (5), 17014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Ebrahimi SB; Samanta D; Mirkin CA DNA-Based Nanostructures for Live-Cell Analysis. J. Am. Chem. Soc 2020, 142 (26), 11343–11356. [DOI] [PubMed] [Google Scholar]
- (7).Schäferling M The Art of Fluorescence Imaging with Chemical Sensors. Angew. Chem., Int. Ed 2012, 51 (15), 3532–3554. [DOI] [PubMed] [Google Scholar]
- (8).Bruemmer KJ; Crossley SWM; Chang CJ Activity-Based Sensing: A Synthetic Methods Approach for Selective Molecular Imaging and Beyond. Angew. Chem. Int. Ed 2020, DOI: 10.1002/anie.201909690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Konstantinou GN Enzyme-Linked Immunosorbent Assay (ELISA). Methods Mol. Biol 2017, 1592, 79–94. [DOI] [PubMed] [Google Scholar]
- (10).Greenwald EC; Mehta S; Zhang J Genetically Encoded Fluorescent Biosensors Illuminate the Spatiotemporal Regulation of Signaling Networks. Chem. Rev 2018, 118 (24), 11707–11794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Paige JS; Nguyen-Duc T; Song W; Jaffrey SR Fluorescence Imaging of Cellular Metabolites with RNA. Science 2012. 335 (6073), 1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Nolan T; Hands RE; Bustin SA Quantification of MRNA Using Real-Time RT-PCR. Nat. Protoc 2006, 1 (3), 1559–1582. [DOI] [PubMed] [Google Scholar]
- (13).Trask BJ Human Cytogenetics: 46 Chromosomes, 46 Years and Counting. Nat. Rev. Genet 2002, 3 (10), 769–778. [DOI] [PubMed] [Google Scholar]
- (14).Brodin JD; Sprangers AJ; McMillan JR; Mirkin CA DNA-Mediated Cellular Delivery of Functional Enzymes. J. Am. Chem. Soc 2015, 137 (47), 14838–14841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Kusmierz CD; Bujold KE; Callmann CE; Mirkin CA Defining the Design Parameters for in Vivo Enzyme Delivery Through Protein Spherical Nucleic Acids. ACS Cent. Sci 2020, 6 (5), 815–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Cutler JI; Auyeung E; Mirkin CA Spherical Nucleic Acids. J. Am. Chem. Soc 2012, 134 (3), 1376–1391. [DOI] [PubMed] [Google Scholar]
- (17).Seferos DS; Giljohann DA; Hill HD; Prigodich AE; Mirkin CA Nano-Flares: Probes for Transfection and MRNA Detection in Living Cells. J. Am. Chem. Soc 2007, 129 (50), 15477–15479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Chinen AB; Guan CM; Ferrer JR; Barnaby SN; Merkel TJ; Mirkin CA Nanoparticle Probes for the Detection of Cancer Biomarkers, Cells, and Tissues by Fluorescence. Chem. Rev 2015, 115 (19), 10530–10574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Chen T; Wu CS; Jimenez E; Zhu Z; Dajac JG; You M; Han D; Zhang X; Tan W DNA Micelle Flares for Intracellular MRNA Imaging and Gene Therapy. Angew. Chem. Int. Ed 2013, 52 (7), 2012–2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Vilela P; Heuer-Jungemann A; El-Sagheer A; Brown T; Muskens OL; Smyth NR; Kanaras AG Sensing of Vimentin MRNA in 2D and 3D Models of Wounded Skin Using DNA-Coated Gold Nanoparticles. Small 2018, 14 (12), 1703489. [DOI] [PubMed] [Google Scholar]
- (21).Jayagopal A; Halfpenny KC; Perez JW; Wright DW Hairpin DNA-Functionalized Gold Colloids for the Imaging of MRNA in Live Cells. J. Am. Chem. Soc 2010, 132 (28), 9789–9796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Li N; Chang C; Pan W; Tang B A Multicolor Nanoprobe for Detection and Imaging of Tumor-Related Mrnas in Living Cells. Angew. Chem., Int. Ed 2012, 51 (30), 7426–7430. [DOI] [PubMed] [Google Scholar]
- (23).Zheng D; Seferos DS; Giljohann DA; Patel PC; Mirkin CA Aptamer Nano-Flares for Molecular Detection in Living Cells. Nano Lett. 2009, 9 (9), 3258–3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Wu P; Hwang K; Lan T; Lu Y A DNAzyme-Gold Nanoparticle Probe for Uranyl Ion in Living Cells. J. Am. Chem. Soc 2013, 135 (14), 5254–5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Chen T-T; Tian X; Liu C-L; Ge J; Chu X; Li Y Fluorescence Activation Imaging of Cytochrome c Released from Mitochondria Using Aptameric Nanosensor. J. Am. Chem. Soc 2015, 137 (2), 982–989. [DOI] [PubMed] [Google Scholar]
- (26).Halo TL; McMahon KM; Angeloni NL; Xu Y; Wang W; Chinen AB; Malin D; Strekalova E; Cryns VL; Cheng C; Mirkin CA; Thaxton CS NanoFlares for the Detection, Isolation, and Culture of Live Tumor Cells from Human Blood. Proc. Natl. Acad. Sci. U. S. A 2014, 111 (48), 17104–17109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Yeo DC; Wiraja C; Paller AS; Mirkin CA; Xu C Abnormal Scar Identification with Spherical-Nucleic-Acid Technology. Nat. Biomed. Eng 2018, 2, 227–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Liu Z; Zhao J; Zhang R; Han G; Zhang C; Liu B; Zhang Z; Han MY; Gao X Cross-Platform Cancer Cell Identification Using Telomerase-Specific Spherical Nucleic Acids. ACS Nano 2018, 12 (4), 3629–3637. [DOI] [PubMed] [Google Scholar]
- (29).Conde J; Oliva N; Artzi N Implantable Hydrogel Embedded Dark-Gold Nanoswitch as a Theranostic Probe to Sense and Overcome Cancer Multidrug Resistance. Proc. Natl. Acad. Sci. U. S. A 2015, 112 (11), E1278–E1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Yang Y; Huang J; Yang X; Quan K; Wang H; Ying L; Xie N; Ou M; Wang K FRET Nanoflares for Intracellular MRNA Detection: Avoiding False Positive Signals and Minimizing Effects of System Fluctuations. J. Am. Chem. Soc 2015, 137 (26), 8340–8343. [DOI] [PubMed] [Google Scholar]
- (31).Wilson BD; Hariri AA; Thompson IAP; Eisenstein M; Soh HT Independent Control of the Thermodynamic and Kinetic Properties of Aptamer Switches. Nat. Commun 2019, 10 (1), 5079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Köhler O; Jarikote DV; Seitz O Forced Intercalation Probes (FIT Probes): Thiazole Orange as a Fluorescent Base in Peptide Nucleic Acids for Homogeneous Single-Nucleotide-Polymorphism Detection. ChemBioChem 2005, 6 (1), 69–77. [DOI] [PubMed] [Google Scholar]
- (33).Hövelmann F; Gaspar I; Chamiolo J; Kasper M; Steffen J; Ephrussi A; Seitz O LNA-Enhanced DNA FIT-Probes for Multicolour RNA Imaging. Chem. Sci 2016, 7 (1), 128–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Ebrahimi SB; Samanta D; Cheng HF; Nathan LI; Mirkin CA Forced Intercalation (FIT)-Aptamers. J. Am. Chem. Soc 2019, 141 (35), 13744–13748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Abascal NC; Regan L The Past, Present and Future of Protein-Based Materials. Open Biol. 2018, 8 (10), 180113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Krall N; da Cruz FP; Boutureira O; Bernardes GJL Site-Selective Protein-Modification Chemistry for Basic Biology and Drug Development. Nat. Chem 2016, 8 (2), 103–113. [DOI] [PubMed] [Google Scholar]
- (37).Chakraborty K; Veetil A; Jaffrey SR; Krishnan Y Nucleic Acid–Based Nanodevices in Biological Imaging. Annu. Rev. Biochem 2016, 85, 349–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Choi J; Kim S; Tachikawa T; Fujitsuka M; Majima T PH-Induced Intramolecular Folding Dynamics of i-Motif DNA. J. Am. Chem. Soc 2011, 133 (40), 16146–16153. [DOI] [PubMed] [Google Scholar]
- (39).Cline GW; Petersen KF; Krssak M; Shen J; Hundal RS; Trajanoski Z; Inzucchi S; Dresner A; Rothman DL; Shulman GI Impaired Glucose Transport as a Cause of Decreased Insulin-Stimulated Muscle Glycogen Synthesis in Type 2 Diabetes. N. Engl. J. Med 1999, 341 (4), 240–246. [DOI] [PubMed] [Google Scholar]
- (40).Nascimento RAS; Özel RE; Mak WH; Mulato M; Singaram B; Pourmand N Single Cell “Glucose Nanosensor” Verifies Elevated Glucose Levels in Individual Cancer Cells. Nano Lett. 2016, 16 (2), 1194–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Nakatsuka N; Yang K-A; Abendroth JM; Cheung KM; Xu X; Yang H; Zhao C; Zhu B; Rim YS; Yang Y; Weiss PS; Stojanovic MN; Andrews AM Aptamer–Field-Effect Transistors Overcome Debye Length Limitations for Small-Molecule Sensing. Science 2018, 362 (6412), 319–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Daniel KB; Agrawal A; Manchester M; Cohen SM Readily Accessible Fluorescent Probes for Sensitive Biological Imaging of Hydrogen Peroxide. ChemBioChem 2013, 14 (5), 593–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Hers HG; Hue L Gluconeogenesis and Related Aspects of Glycolysis. Annu. Rev. Biochem 1983, 52 (1), 617–653. [DOI] [PubMed] [Google Scholar]
- (44).Estensen RD; Plagemann PGW Cytochalasin B: Inhibition of Glucose and Glucosamine Transport. Proc. Natl. Acad. Sci. U. S. A 1972, 69 (6), 1430–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


