Abstract
High levels of glycolysis have been associated with cancer metastasis, tumor recurrence and poor outcomes. In this light, single cells that exhibit high glycolysis are specific targets for therapy. However, the study of these cells requires efficient tools for their isolation. We use a droplet microfluidic technique developed in our lab, Sorting by Interfacial Tension (SIFT), to isolate cancer cell subpopulations based on glycolysis without the use of labels or active sorting components. By controlling the flow conditions on chip, the threshold of selection can be modified, enabling the isolation of cells with different levels of glycolysis. Hypoxia in tumors, that can be simulated with treatment with CoCl2, leads to an increase in glycolysis and more dangerous tumors. The device was used to enrich CoCl2 treated MDA-MB 231 breast cancer cells from an untreated population. It is also used to sort K562 human chronic myelogenous leukemia cells that have either been treated or untreated with 2-Deoxy-D-glucose (2DG), a pharmaceutical that targets cell metabolism. The technique provides a facile and robust way of separating cells based on elevated glycolytic activity; a biomarker associated with cancer cell malignancy.
Introduction
The measurement of behavior and activity at the level of single-cells is critical in understanding what drives key processes of cell biology and disease states. For example, disease progression and tumor growth in cancer are believed to result from minor populations of tumorigenic cells, whereas the majority of cancer cells have little impact on tumor growth and metastasis.1,2 A tumor is a complex and dynamic structure with each cell having a different biological activity, metabolic requirements and microenvironment. The cell to cell variability, masked in bulk measurements, causes the tumor to be affected not only by spatial but also by temporal conditions.3 This inherent heterogeneity highlights the need for tools capable of sorting or isolating cells and cell subpopulations based on criteria of particular relevance to the disease state.
Fluorescence-activated cell sorting (FACS) is the workhorse of cell sorting providing high throughput and selectivity. Cells are typically sorted based on the reading of a specific fluorescent marker.4 As an alternative, the encapsulation of cells in microfluidic droplets of picoliter to nanoliter volume enables the selection of individual cells based on their secretions.5–7 However, these cell sorting techniques rely on cell markers that lack generality and can be highly heterogeneous in cancer cells.8,9 In addition, they require a synchronization of observation (typically by fluorescence) and sorting of the cells.
This has led to the development of label-free microfluidic techniques that sort cells based on their physical characteristics, e.g. size, density, dielectric properties or deformability.10 Prominent techniques in this area are acoustophoresis,11,12 dielectrophoresis,13,14 deterministic lateral diffusion15,16 and inertial microfluidics,17–19 which are cost effective while providing high throughput. These techniques have the ability to efficiently sort cell types (e.g. separating circulating tumor cells from whole blood)19,20 and separating live and dead cells21–23. However, many applications would also demand selection based on cellular activity rather than physical characteristics. Moreover, within a cell population that share very similar physical properties, such as the same cell type, these techniques would have difficulty separating cells based on criteria directly correlated to cancer malignancy.
Aberrant metabolism is a hallmark of cancer,24 and is used as a general biomarker for cancer diagnostics and monitoring. A key observation of cancer metabolism was made almost one hundred years ago by Otto Warburg, that discovered that cancer cells uptake large amounts of glucose converting it to lactate via glycolysis even in the presence of oxygen (‘aerobic glycolysis’, or the so-called ‘Warburg effect’).25,26 This elevated metabolism of cancer cells is the basis of PET scans as radioactive glucose concentrates in a tumor and is routinely used for cancer detection, monitoring and staging.27 The secretion of large amounts of lactate is accompanied by secretion of protons, leading to an extracellular environment of tumors that can be in the pH range of 6.2–6.9.28,29 Cancer cells thrive in this acidic environment.30 There is increasing evidence that high extracellular acidity is not only a byproduct of the disease, but rather a disease driver.31 Tumor microenvironment, in particular hypoxia, can promote elevated cellular glycolysis.32 Elevated glycolysis and high levels of lactate release in turn are linked to cancer aggressivity, promoting metastasis and tumor recurrence.33–36 Therefore, single cells that show high lactate release rates and glycolysis are suspected to be more malignant and glycolysis is recognized as an emerging target in both cancer therapy and to counteract chemotherapy resistance.34,37,38 Given the correlation of glycolysis and malignancy, a label-free technique that selects cells based on glycolysis could have a major impact in oncology.
Acidification from glycolysis is already used as a way to detect cancer cells. Notably, a recent paper demonstrated a novel method using droplet microfluidics to identify cancer cells, including rare circulating tumor cells (CTCs) and several cancer cell lines (breast, colorectal and ovarian), within a large background of white blood cells.39 Single cells were encapsulated in picoliter droplets in oil. Within a few minutes, droplets containing cancer cells showed a marked change in droplet pH due to secretion of protons from elevated glycolysis. Likewise, extracellular acidification of bulk cells is the basis of the metabolic analyzer, Agilent’s Seahorse XF technology,40 used in over 2500 publications for both fundamental research and for the study of disease states. These approaches allow the measurement of glycolysis, but not the sorting based on glycolysis. The selection of cells is much more powerful as it enables the further study, such as genomic and transcriptomic analysis, of a selected cell subpopulation of interest.
We have recently developed a label-free and passive method to sort droplets based on pH dubbed Sorting by Interfacial Tension (SIFT).41,42 The technique utilizes an increase in droplet interfacial tension with decreasing pH in the presence of a specific surfactant. This change in interfacial tension with pH provides the crucial handle for droplet sorting. Droplets within the device are ‘pancake shaped’, confined by the top and bottom of the channel. These flattened droplets encounter a microfabricated rail of increased height that is oriented diagonally relative to the direction of flow. Droplets expand their height upon entry into the rail thus reducing their surface area and surface energy. Droplets of high pH (low interfacial tension) are immediately pushed off the rail by the oil entrainment flow. However, the entrainment flow is insufficient to push off droplets of low pH (high interfacial tension) and they follow the rail laterally towards a different chip exit. This strategy enables the efficient sorting of droplets as a function of pH. SIFT can be used to sort cells based on metabolism as glycolysis is accompanied with the secretion of protons leading to a decrease in droplet pH. The concurrent increase in droplet interfacial tension is then leveraged for cell sorting.
We have previously shown that SIFT can be used to sort encapsulated enzymes based on activity,41 empty vs. cell occupied droplets and live vs. dead cells42. Here, we expand its use and show how the platform can be used to isolate cancer cell subpopulations based on glycolysis without the use of markers or active sorting components. By controlling the flow conditions on chip, we show that the pH threshold of selection can be modified, enabling the selection of cells with different levels of glycolysis. The technique is used to enrich cells that have been exposed to hypoxia or therapeutics that target glycolysis. This demonstrates that the technique can be used to enrich cells that would have both lower and higher glycolysis as compared to untreated cells. The technique provides a facile and robust way of separating cells based on glycolytic activity, a biomarker that can track cancer cell malignancy.
Materials and Methods
Cells:
MDA-MB 231 human mammary gland/breast cancer cells were grown in Dulbecco’s Modified Eagle Medium (DMEM) and K562 human chronic myelogenous leukemia cells were grown in ATCC-formulated Iscove’s Modified Dulbecco’s Medium (IMDM). Both cell lines were purchased from ATCC. Media was supplemented with 10% fetal bovine serum (HyClone, GE Healthcare Life Sciences, Logan, UT) and 2% v/v penicillin-streptomycin (10,000 units/mL-10,000μg/mL) solution (Gibco, Life Technologies Corporation, Grand Island, NY). Growing conditions were 37 °C in a 5% CO2 atmosphere.
On the day of experiment, MDA-MB 231 cells were trypsinized and rinsed three times with 1X PBS. K562 cells were centrifuged and rinsed one time with 1X PBS. Cells were labeled with Calcein AM (Thermo Fischer, Waltham, MA), a viability fluorescent dye, for 30 min at 37 °C and 4% CO2 atmosphere. After incubation, the cells were centrifuged, the supernatant aspirated off and the cell pellet was resuspended in equal amounts of 2.5 mM PBS buffer and a DMEM media mix. The DMEM media mix used for the on-chip experiments did not contain fetal bovine serum (deproteinated media) or sodium bicarbonate, and was supplemented with Pluronic F-68 (1% w/w, Affymetrix Inc., Maumee, OH), Optiprep (15% v/v, Fresenius Kabi Norge AS for Axis-Shield PoCAS, Oslo, Norway) and 0.1 mg/mL pyranine (AAT Bioquest Inc., Sunnyvale, CA). Pluronic F-68 is added to the system to increase droplet stability and cell viability, whereas Optiprep limits cell sedimentation in tubing and droplets. Pyranine served as a fluorescent ratiometric pH probe. pH and osmolality (determined with Vapro Vapor Pressure Osmometer 5520, Wescor, ELITech Biomedical Systems, Logan, UT) of both solutions were verified and adjusted to physiological values (pH 7.4; 280–320 mOsmol) prior to experiment. Cell density was measured (Cellometer Auto T4 Bright Field Cell Counter, Nexcelcom Bioscience LLC, Lawrence, MA) and adjusted to about 5*105 cells/mL to ensure single-cell occupation of droplets.
Cell Treatment with CoCl2:
48 hours prior to harvesting MDA-MB 231 cells, the media was exchanged with fresh media containing 100 μM CoCl2 (Spectrum Chemical Mfg. Corp., Gardena, CA). Control cells were treated the same while excluding CoCl2. After 48 hours, treatment and control cells were harvested as described above. To ensure continual effects of the treatment, the wash solution also contained 100 μM CoCl2, however, the final on-chip solutions contained no CoCl2. To distinguish treated from non-treated cells on-chip, only treated cells were labeled with Calcein AM.
Cell Treatment with 2-Deoxy-D-glucose (2DG):
K562 suspension cells were harvested and resuspended in low glucose media supplemented with 100 mM 2DG (Sigma Aldrich) whereas a Calcein AM labeled control population was resuspended in the same media without 2DG. Samples were incubated for 3.5 h at 37 °C and 4% CO2 atmosphere. After incubation, the cells were prepared for on-chip experiments as described above.
Microfluidic Device:
The overall microfluidic chip design and dimensions in the rail region are shown in Figures S1 and S2, respectively. Chips with channel depth modulations were fabricated from polydimethylsiloxane (PDMS), utilizing the dry-film photoresist soft lithography technique previously reported by Stephan et al.,43 as this technique facilitates easy prototyping with multi-level designs. The PDMS chip was irreversibly bonded to a glass slide via plasma treatment. To render the internal surfaces of the channel hydrophobic, the channels were treated with Novec™ 1720 Electronic Grade Coating (3M, Maplewood, MN) for 30 min at 150 °C.
Measurements:
Fluid flow in the microfluidic device was regulated with a computer-controlled syringe pump system (Nemesys, Cetoni, Korbussen, Germany). Typical flow conditions can be found in Table S1 in the supplemental information. Chip temperature was maintained at 37 °C for all experiments using a heating stage with control module and temperature feedback (CHS-1 heating plate, TC-324C temperature controller, Warner Instruments, Hamden, CT). The chip consisted of three main functional regions (Figure S1): 1) encapsulation of cells in droplets, 2) incubation to enable glycolysis induced changes in pH and 3) sorting of the droplets based on differences in interfacial tension. The Aqueous Inlet injected the cellular solution onto the chip. Using a flow focuser, droplets in oil, 0.1 % w/w Picosurf-1 (Sphere Fluidics Limited, Cambridge, United Kingdom) in Novec™ 7500, were formed. The Oil Outlet was set to a flow in the opposite direction of the main flow to remove excess oil,44 allowing for the droplets to pack tightly in the incubation region. Incubation is an important aspect of the SIFT chip, and a tight packing allows for a longer, uniform incubation time for all encapsulated cells. Cells spend 6 – 8 minutes in the incubation region, before the channel narrows and droplets enter the sorting region. A flow in the opposite direction of the main flow at the Oil and Droplet Outlet removed approximately half the droplets. This reduced the number of droplets in the sorting region, reducing the risk of droplets getting bumped off the rail prematurely due to droplet interactions. An exchange to QX100 oil (QX100 Droplet Generation Oil for Probes, Biorad, Hercules, CA) using the QX100 Inlet and Oil Entrainment Inlet in the sorting region was necessary to enable separation on the rail due to QX100 induced changes in interfacial tension. QX100 is exchanged just prior to sorting as it impedes lactate production of cells and has low biocompatibility.42
Images and videos were taken on an inverted fluorescence microscope (Olympus IX-51) equipped with a 4X objective, equipped with a shuttered LED fluorescence excitation source (Spectra-X light engine, Lumencor, Beaverton, OR) and a high-speed camera (VEO-410, Vision Research, Wayne, NJ). The field of view for video and images allowed the simultaneous observation of both the end of the incubator and the sorting rail. The excitation source had individual addressable LEDs that were coupled to an Arduino (Arduino LLC, Scarmagno, Italy) to rapidly alternate between different colored LEDs using simple TTL triggering. Droplets were excited with alternating violet (395 nm BP 25 nm), blue (440 nm BP 20 nm) and green excitation (561 nm BP 14 nm) at a rate of 100 frames per second (33 fps for each color). The microscope filter cube contained a dual-edge dichroic mirror (Di03-R488/561-t1–25×36, Semrock, IDEX Health & Science LLC Rochester, NY) and dual-band emission filter (FF01–523/610–25, Semrock) that enabled transmission of both pyranine and Calcein AM fluorescence.
Data Analysis:
Images and videos were analyzed using ImageJ software.45 Green excitation was used to identify cells labeled with Calcein AM. The pH of droplets at the end of the incubator was determined from the ratio of fluorescence intensity from background-subtracted blue and violet excitation. A calibration curve (Figure S3) was produced from the fluorescence ratio of droplets of known pH (range of pH 6.2 – 8.0) measured on chip. Calibration solutions were prepared with identical media and pyranine concentrations as cellular solutions. The calibration plot was fit to a linear curve. It was observed that the slope of the calibration curve remained constant from day to day with small adjustments in the y-intercept. Calibration curves for experiments could thus be produced using the known pH of empty droplets in experiments to determine the y-intercept of the calibration curve.
Logistic regression was used to statistically estimate optimal pH thresholds to separate selected from non-selected cells. The pH threshold was defined at a 50% predicted probability that the cell would be selected. The standard error of the prediction was used to obtain a 95% confidence interval around that threshold. To assess how the flow rate influenced the pH selection threshold, a multiple logistic regression was fit, predicting selection from flow rate, pH and their interaction. This model predicted the probability that a particular cell will be selected based on the pH and flow rate, and allows the influence of one of these variables to depend on the other.
Results and Discussion
Droplet microfluidics allows the encapsulation and transport of cells in picoliter to nanoliter droplets for large scale biological analysis.5,46,47 Droplets are particularly well-suited for measuring cell secretions as they remain confined in the small droplet volume. This leads to both a fast and large change in analyte concentration enabling sensitive detection at the level of single cells.48,49
We recently reported that using a specific surfactant, QX100 Droplet Generation Oil for Probes, hereby called QX100, the interfacial tension of droplets can be very sensitive to pH and enables the sorting of droplets and cells based on droplet pH.41,42 Unlike other techniques that sort droplets based on interfacial tension,50,51 the droplets remain stabilized from coalescence with surfactants at all times. This technique was dubbed Sorting by Interfacial Tension (SIFT). The general chip geometry is presented in Figure S1. Briefly, cells are encapsulated in droplets with typical diameters of 55 to 90 μm. This droplet size ensures that the droplets are flattened, confined by the top and bottom of the channel, as the channel height is only 25 μm. Cell density is typically kept low to ensure that most droplets are either empty or singly occupied. Droplets are incubated by flowing through a long serpentine channel. Empty droplets remain at their initial pH after incubation. However, after incubation, droplets containing cells achieve a lower pH as the cells secrete protons from glycolysis. Although respiration can also lead to acidification via carbon dioxide dissolution,52 this would not be a contributor in this device as CO2 would not remain confined within a droplet. The length of the serpentine channel was optimized in prior work,42 to ensure a substantial change in droplet pH after incubation.
After incubation, droplets then enter a wider channel, the sorting region of the chip (Figure 1) containing a tapered rail oriented at 45 degrees relative to the direction of flow. Flattened droplets expand (reduce their surface energy) when they enter the rail that has increased height (40 μm).53,54 Droplets at high pH, thus low interfacial tension, enter the rail but the hydrodynamic drag pushes them off the rail (“Unselected”). However, when droplets at lower pH, and thus higher interfacial tension, enter the rail, the hydrodynamic drag is insufficient to push them off the rail. These droplets follow the rail upwards and exit the rail as it reaches a point (“Selected”). Two chip exits allow a separate collection of the unselected and selected droplets. As detailed in a previous paper,42 0.1% w/w Picosurf-1 (Sphere Fluidics Limited, Cambridge, United Kingdom) in Novec™ 7500 is used for droplet encapsulation and incubation while QX100 is used for the droplet sorting step. Compared to an earlier chip design,41,42 the sorting region here is narrower (1.5 mm compared to 3 mm), that was found to improve sorting robustness while reducing the use of oil. In prior work, droplets containing cells were separated from empty droplets or droplets containing dead cells.42 In that case, the pH difference between the selected and unselected droplets was relatively large, 0.3 pH units. We show here much finer sorting capacity, the selection of individual cells with elevated glycolytic activity.
Figure 1.
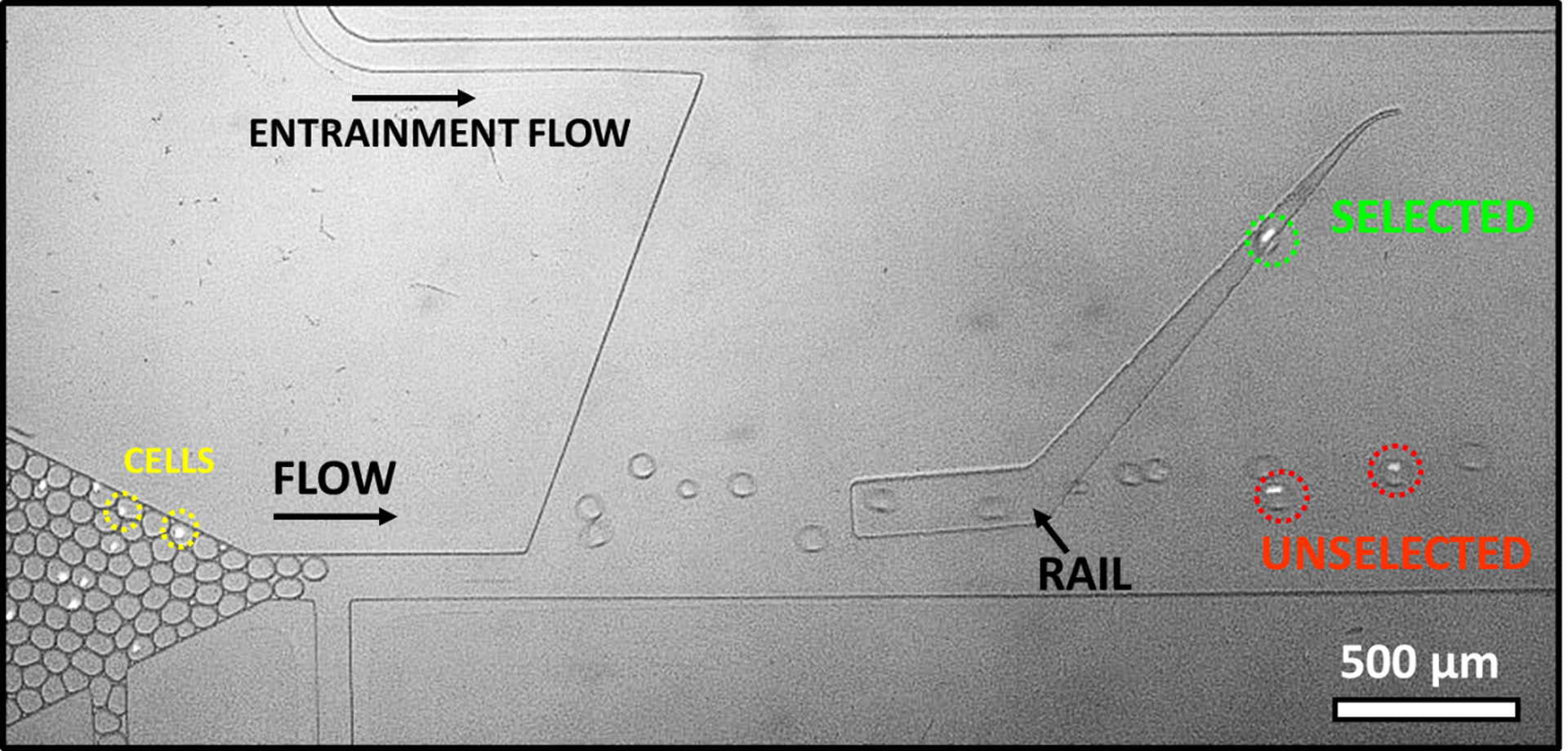
Selection of droplets of different pH using SIFT. Image shows the end of incubator and sorting region. K562 cells (a few circled in yellow) are labeled with Calcein AM for better visibility. Droplet pH is analyzed at the end of incubator before entering into the sorting region. Droplets containing a highly glycolytic cell (circled in green) ride the rail laterally up, leaving the rail at the top (selected). Droplets containing no cell or less active cells (circled in red) are at higher pH. They do not ride the rail or are only slightly deflected up the sorting rail (unselected).
The principle of sorting of droplets of different pH is shown in Figure 1 and supplemental video S1. The green excitation allowed a clear identification of cells using the label Calcein AM (a few cells are circled in yellow in Figure 1). The pH of droplets was determined using a ratiometric fluorescence pH probe, pyranine. The measurement was performed at the end of the incubator (bottom left of Figure 1) while the droplets were moving slowly and before contact with QX100 (QX100 leads to a further acidification of the droplets upon adsorption, likely indicating an acidic surfactant). This method allowed the pH determination of all droplets entering the sorting region. Due to the heterogeneous cellular metabolism rates, encapsulating cells in droplets provides a simple method to produce a distribution of droplets of varying pH. As the intent in this first experiment was to characterize sorting with respect to pH, a higher cell density (2*106 cells/ mL) was used leading to more droplets with cells and also droplets with multiple cells. After an incubation of 8 minutes, empty droplets were at pH 7.37 ± 0.02 (average ± error of the mean), unchanged from the initial pH of the media. Droplets with cells had a wide distribution of pH ranging from 6.41 to 7.42. The droplets in the lower range of pH typically contained multiple cells.
Droplets were then classified as unselected if they failed to ride the rail upwards or selected if they rode the rail upwards. The lateral midway point of the rail was used to distinguish between the two classifications as these led to different channel exits. In Figure 1, unselected droplets, circled in red, were at pH of 7.26 and 6.91. The selected droplet, circled in green, was at a lower pH of 6.74. Droplets were sorted at a rate of about 30 droplets per second.
Droplet selection using SIFT was found to be dependent on the interplay of droplet pH, size and entrainment flow.42 For droplets of constant size, the flow from the Oil Entrainment Inlet (top left inlet in Figure 1) provides an easily modified, user-defined parameter that sets the pH for droplet selection. Higher entrainment flow increases the hydrodynamic drag in the sorting region and thus selects for droplets with higher interfacial tension and thus lower pH.
The pH of selected and unselected droplets for specific entrainment flows is summarized in Figure 2a. At a given flow rate, higher pH droplets tend to be unselected while lower pH droplets are selected. There are some cases of overlap, especially at lower flow rates, where unselected droplets have a lower pH than some selected droplets. The general trend is that a higher entrainment flow leads to a decreasing pH that separates the population of selected and unselected droplets. At the lowest flow rate, 60% of droplets with cells are selected while at the highest flow rate 4% of droplets with cells are selected. At all flow rates studied, empty droplets not containing cells (at pH 7.37 in this experiment) are unselected and are not included in the plot.
Figure 2.
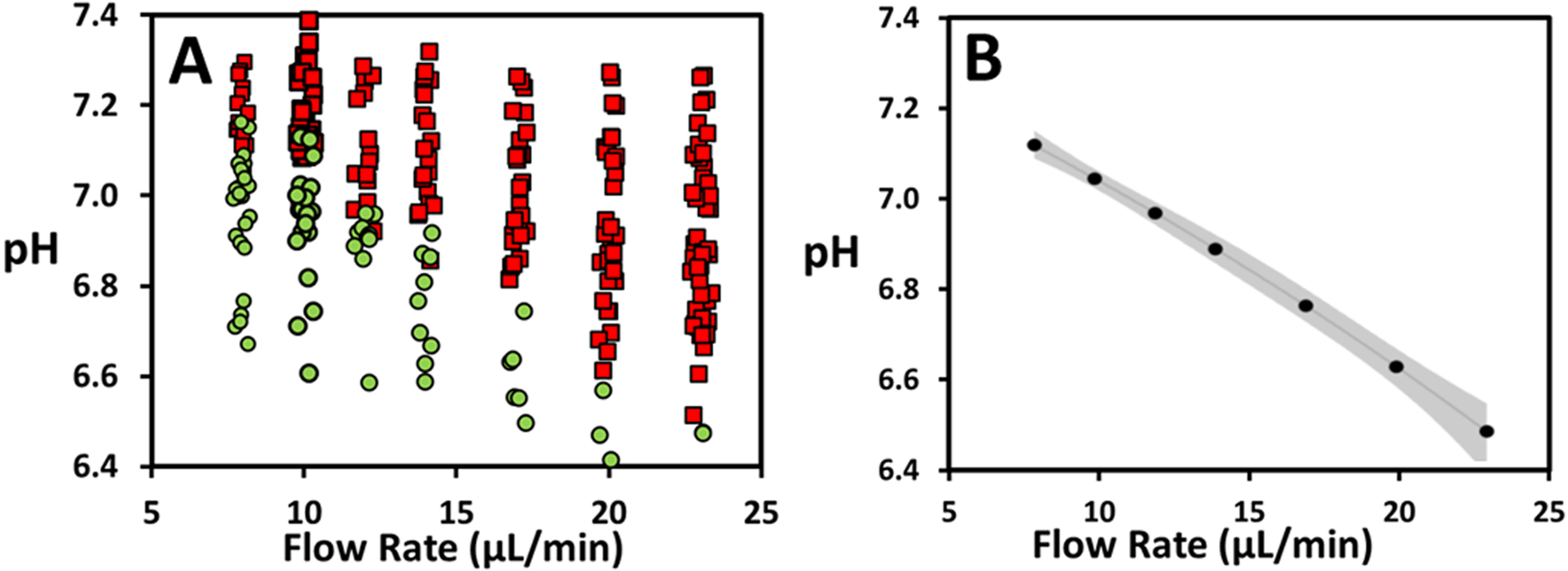
A) pH of droplets containing K562 cells plotted vs applied flow rate (μL/min) of Oil Entrainment Inlet. Square, red markers represent unselected; circular, green markers represent selected droplets. A small amount of noise (jitter) was added to the x-value to avoid overlap of data points. B) Estimated pH threshold as determined by logistic regression fits. 95% confidence interval is indicated in grey.
A pH selection threshold can be defined as the pH where there is an equal probability that a droplet is selected or unselected. This pH threshold was determined by fitting binary selected/unselected data vs. pH with a logistic regression (individual model fits shown in Supplemental Figure S4). A multiple logistic regression fit was performed that assessed the combined influence of pH and flow rate on selection probability, allowing an estimate of the pH threshold at any specified flow rate. The resultant pH threshold as a function of flow rate with 95% confidence limit is summarized in Figure 2b. pH threshold ranges from values of 7.12 to 6.48 from the lowest to highest flow rates measured. The confidence interval for each pH threshold is small. It ranges from 0.02 pH units for a flowrate of 10 μL/min to 0.07 for a flow rate of 23 μL/min, providing a sensitive and reliable separation. The thresholds overlap with the pH range of droplets with single cells under conditions studied in this paper. The lower threshold of 6.48 would be lower than the typical pH for droplets containing single cells under similar conditions. This indicates that the technique has the flexibility to isolate rare cells with a glycolytic activity that is significantly elevated compared to the average. It is worth noting that the pH range of droplets for a given cell type can be controlled with several user-defined experimental parameters. Specifically, a lower average droplet pH can be attained by increasing the incubation time, or by decreasing the droplet size or buffering capacity. The correlation between threshold and flow rate will vary slightly with droplet size. As droplet size may vary for individual experiments, adjustments in flow rate would need to be adapted to allow a consistent cell selection population.
Tumors tend to exhibit a large range of heterogeneity with fluctuating levels of hypoxia, acidification, glucose, lactate, and energy availability.32 These tumor microenvironment characteristics are due to remodeling of the tumor vasculature and extracellular matrix within the tumor that can result in heterogeneous circulation of oxygen resulting in hypoxia.55 Hypoxia has been observed to initiate transcription programs that alter cellular metabolism and induce vascularization.32 These alterations can result in upregulation of MCT and GLUT1 transporters resulting in increased glycolysis and have been correlated with poor outcomes.56 Hypoxic conditions are reported to influence lactate release and the glycolytic index, and hence to increase malignancy of cells.32,56,57
Subsequent experiments are designed to sort cancer cells grown in different oxygenation conditions, mimicking the microenvironment found in tumors. Treatment of MDA-MB 231 cells with CoCl2 in the growing media will induce hypoxic pathways including overexpression of HIF-1.58 Chemical treatment with CoCl2 serves as a common substitute to growth in low-oxygen environment as it provides a simple and consistent treatment that is not influenced by temporary fluctuations in oxygen concentration.59 MDA cells were grown for 48 hours while exposed to 100 μM of CoCl2. For simplicity, CoCl2 treated cells are deemed hypoxia treated in the discussion below. A control population of cells was treated identically with the exclusion of CoCl2. Prior to mixing the two cell populations and injection onto the chip, the hypoxic population was labeled with Calcein AM to enable differentiation between the hypoxic and the control population. The labeling of cells with Calcein AM was found to have no influence on the pH of the droplets (data not shown).
Figure 3 and supplemental video S2 show an example of the sorting of a hypoxia treated cell (labeled with Calcein AM, circled in orange) and a control cell (unlabeled, circled in white). After incubation, the droplet containing a hypoxia treated cell was determined to be at pH 6.82 while the droplet containing a control cell was measured to be 7.19. The droplet containing a hypoxic cell is selected as it rides the rail upwards while the droplet containing a control cell is pushed off the rail to the unselected exit.
Figure 3.
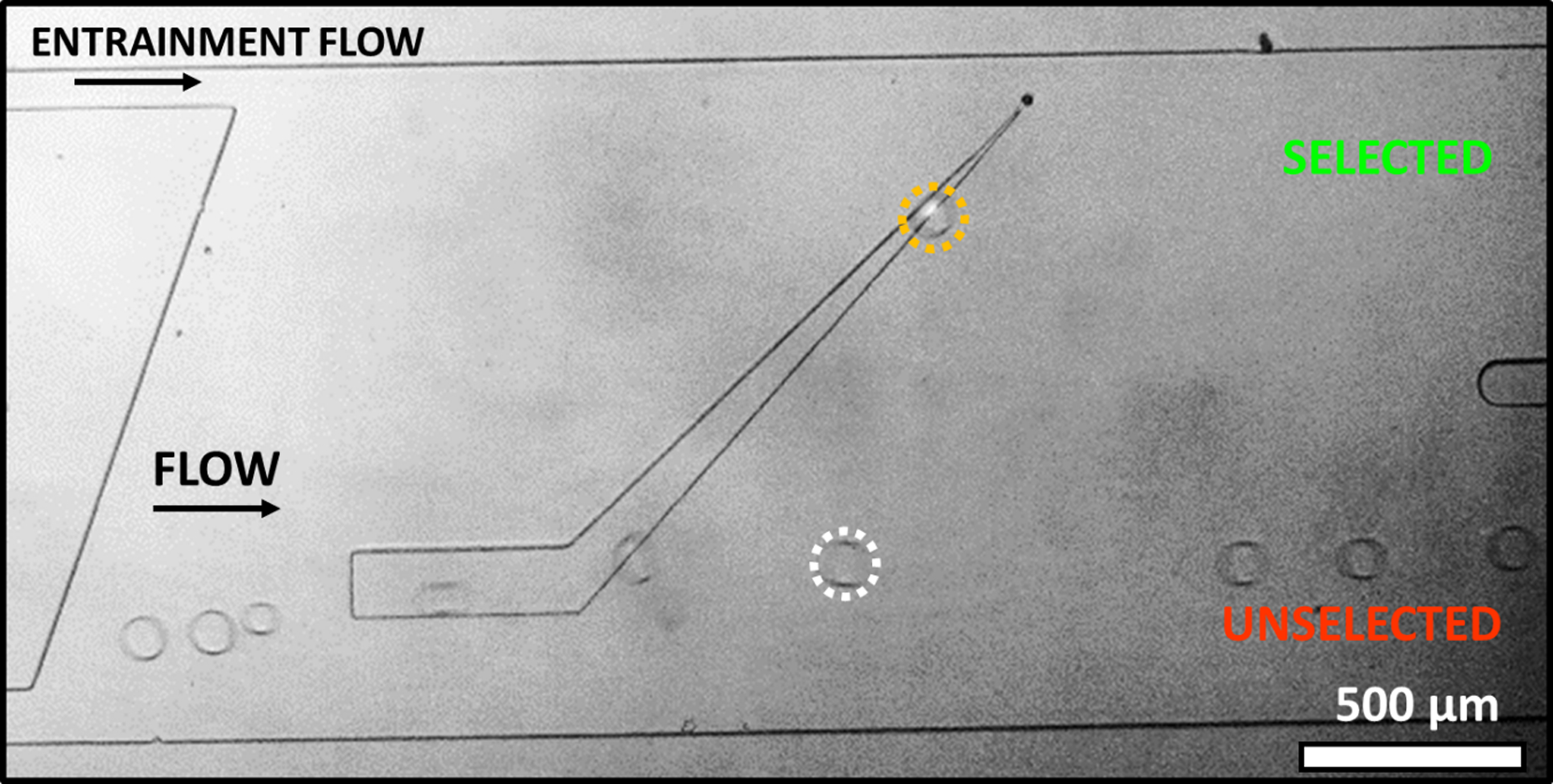
SIFT device sorting of hypoxia treated and untreated MDA-MB 231 cells. Hypoxic cells are labelled with Calcein AM to enable differentiation of cell populations. Cell grown under hypoxic conditions (circled in orange) with low pH, pH 6.82, rides the rail laterally up (selected). Control cell (circled in white) grown under normoxic conditions with higher pH (pH 7.19) is only slightly deflected by the rail (unselected).
Figure 4 summarizes the pH and sorting behavior over a population of hypoxia treated and control cells. The pH of the two populations of cells were measured at the end of the incubator with each point representing a droplet containing a single cell (Figure 4a). The control droplets showed an average pH value of 7.14 ± 0.02 (N = 66). The average pH of hypoxia treated cells was 6.95 ± 0.02 (N = 54). Although there is overlap between the two populations, hypoxia induces glycolysis and thus lowered average pH values. Furthermore, some hypoxic cells showed very high glycolysis, with droplet pH values as low as 6.64. The standard deviation of pH values of the hypoxia cells was 0.18 compared to 0.13 for the control cells. This broad heterogeneity in pH is indicative of a unique metabolism for each analyzed cell.
Figure 4.
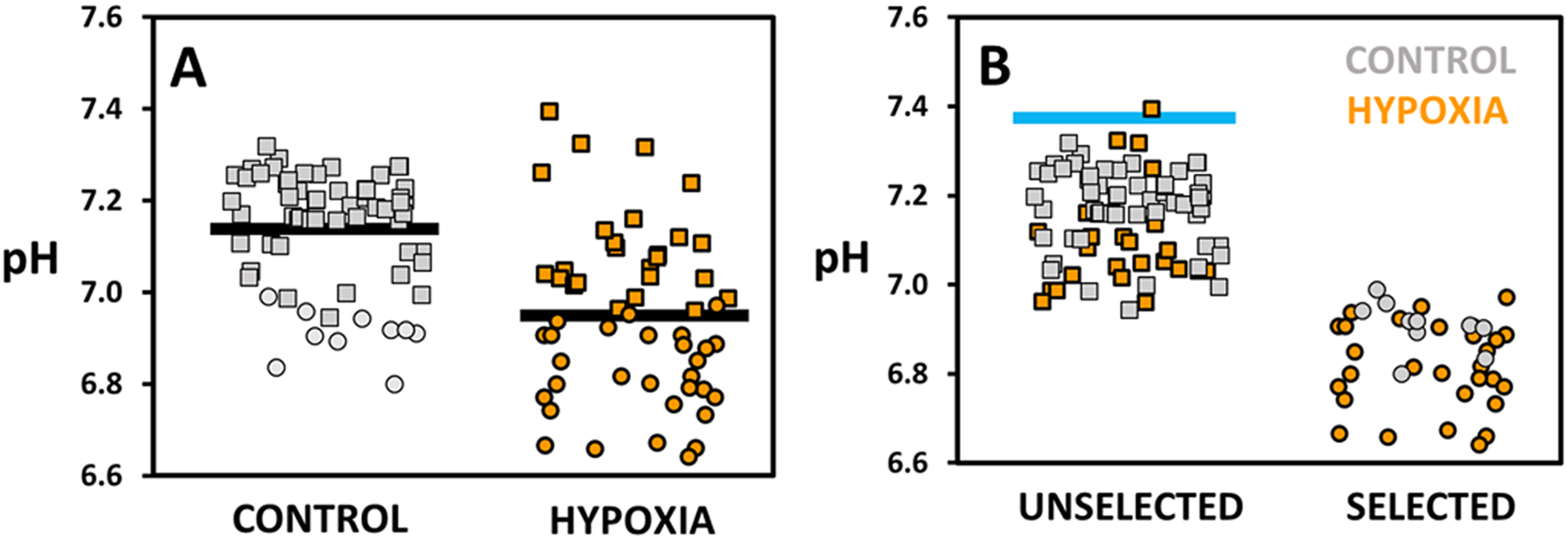
pH of droplets containing either control (grey) or hypoxia treated (orange) MDA-MB 231 cells. Square markers annotate unselected, circular markers selected droplets. pH values were determined via a fluorescence intensity ratio. A) Droplets presented as control and hypoxia cell populations. Average pH values, indicated by a black line, are 7.14 ± 0.02 (N = 66) for control and 6.95 ± 0.02 (N = 54) for hypoxia cells. B) Droplets presented as unselected and selected populations. Blue line indicates the pH of empty droplets (pH 7.37 ± 0.02). The pH threshold of selection was determined by a logistic regression fit and was found to be 6.96 ± 0.02 (threshold ± 95% confidence interval) (fit shown in Figure S5a). The sorting of droplets leads to an enrichment of hypoxia treated cells from 45 % (N = 54) before sorting to 75 % (N = 29) of selected cells.
Figure 4b shows the corresponding pH and identity, control or hypoxia treated, of selected and unselected droplets. The blue line annotates the pH value of empty droplets (pH 7.37 ± 0.02) that are unselected. The range of pH of the selected droplets (pH 6.64 – 6.99) is lower than those of unselected (pH 6.95 – 7.40). The pH threshold of selection was determined as presented earlier by a logistic regression fit and was found to be 6.96 ± 0.02 (threshold ± 95% confidence interval) (Figure S5a). As the pH of droplets containing hypoxia treated cells is lower, the sorting of droplets leads to an enrichment of hypoxia treated cells from 45 % (N = 54) before sorting to 75 % (N = 29) of selected cells. Control cells are slightly enriched in the unselected cells from 55 % (N = 66) to 69 % (N = 56). The method was also used for the enrichment of hypoxia treated cells in other cancer cell lines. The technique led to an enrichment from 54 % to 86 % of hypoxia treated cells in K562 cells (Figure S6a and b). The pH threshold was determined to be 6.93 ± 0.05 (Figure S6c).
The device selects cells that have lower pH, indicative of higher glycolytic rates. In this initial study using cell lines, cells deemed to have high glycolytic levels are relative to the rest of the population of cells. However further studies of more cell types and biologically relevant samples, (blood samples or liquid tumors) could better elucidate the pH values and differences (genomic, transcriptomic and proteomic) of cells displaying high levels of glycolysis. Within tumors, higher glycolytic rates are essential to cancer cell survival and tumor growth.60 SIFT enables the collection and further analysis of cells that would be deemed particularly malignant cells. Enrichment of these cells is important as a small amount of very malignant cancer cells can be the driving force behind disease progression. The sorting also preferentially selects cells that have been hypoxia treated. SIFT thus presents a way to select cells exposed to hypoxia that are known to have increased malignancy. Some control cells are among the selected cells. It is important to note that these selected control cells have droplet pH that are similar to the hypoxia treated cells. This shows that the determining factor of droplet selection is whether the droplet pH is below the pH threshold, regardless of whether the droplet is in the control or treated cell population. As the selected control cells also display high glycolysis, by this measure these cells would potentially share similar malignant traits.
The SIFT technique also allows enrichment of cells that have been treated or not treated with a pharmaceutic that inhibits metabolism and glycolysis. In this case, the non-treated cells serve as a model for a subpopulation of cells that may not respond to pharmaceutic treatment. Glycolysis is an emergent target in cancer therapy, especially for chemoresistant cells.34,37,38 2-Deoxy-D-glucose (2DG) hinders the glycolytic pathway by being transformed to phosphorylated 2DG through hexokinase, getting trapped inside a cell and in turn inhibiting hexokinase itself.38,61 This process induces intercellular ATP depletion62 and is linked to suppression of cell proliferation.63 It is currently suggested as a treatment for several cancers, such as lymphoma64 and leukemia65.
In contrast to hypoxia treated cells, pharmaceutical treatment with 2DG would lead to glycolysis that is on average below the basal rate.66 One population of K562 cells was suspended in low-glucose media supplemented with 100 mM 2DG for 3.5 hours, while the control population (labeled with Calcein AM) was held in the same media without 2DG. Cell populations were washed once with PBS and resuspended together in PBS buffer and media mix without 2DG prior to injection on chip.
Figure 5a displays the pH after incubation of droplets containing single cells for both control and 2DG treated cell populations. Control cells show an average pH of 6.97 ± 0.03 (N = 46) while 2DG treated cells show a value of 7.23 ± 0.02 (N = 41). The standard deviation of the control population is larger, 0.22 compared to 0.12 for the 2DG treated cells.
Figure 5.
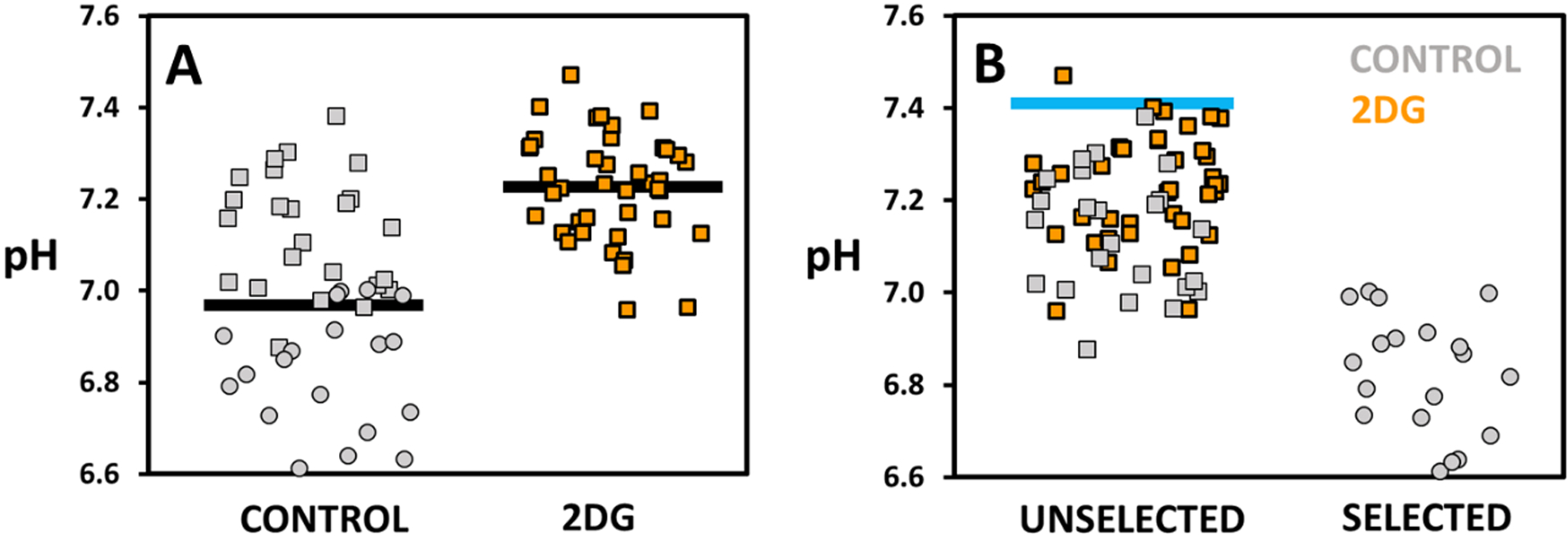
pH of droplets containing either control (grey) or 2DG treated (orange) K562 cells. Square markers annotate unselected, circular markers selected droplets. A) Droplets presented as control and 2DG treated cell populations. Average pH values, indicated by a black line, are 6.97 ± 0.03 (N = 46) for control and 7.23 ± 0.02 (N = 41) for 2DG treated cells. B) Droplets presented as unselected and selected populations. Blue line indicates the pH of empty droplets (pH 7.41 ± 0.04). The selection threshold was determined to be 6.96 ± 0.06 (fit shown in Figure S5b). While 2DG treated cells make up 47 % of cells prior to sorting, selected cells contain exclusively control cells (N = 22) excluding all the 2DG cells.
Figure 5b shows the pH and type, control or 2DG, of unselected and selected droplets. Here, the pH of empty droplets was 7.41 ± 0.04. The selection threshold was determined to be 6.96 ± 0.06 (Figure S5b). While 2DG treated cells make up 47 % of cells prior to sorting, selected cells contain exclusively control cells (N = 22) excluding all the 2DG cells. 2DG treated cells are enriched from 47 % (N = 41) to 63 % (N = 41) within the unselected cells. Thus, SIFT can be used to remove cells that have lower glycolytic activity. More importantly, this demonstrates that the technique can be used to isolate and then study a subpopulation of cells that may not be responsive to pharmaceutic treatment targeting glycolysis.
Conclusions
We show here how SIFT provides a way to sort cells based on single-cell glycolytic activity. This is a significant difference to other label-free microfluidic techniques that select cells based on their physical characteristics, such as size, density, deformability and dielectric properties. By selection based on differences in metabolism, cells can be selected that share similar physical features. To demonstrate this capacity, we enriched cells of similar type that have been exposed to hypoxia or pharmaceuticals that target metabolism. This has relevance to tumor cells that, exposed to different oxygen and nutrient environments, would have differences in glycolysis. There is substantial evidence indicating that glycolysis activity tracks with malignancy.33–36 The device can thus be used to isolate the most malignant cells. It also has potential in separating cancer cells from normal cells in a liquid biopsy or cell suspension of a solid tumor. The criterion of selection can be controlled to be more or less stringent by altering the flow conditions in the channel, thus setting the bar for the population of isolated cells. The sorting is robust, enabling the accurate sorting of droplets separated by 0.10 pH units or less for most flow rates. As a passive technique, it is easy and inexpensive to implement as it requires no markers, excitation sources, detectors, or active sorting components.
Although the focus here was on cancer where aberrant metabolism is a defining trait,24 the cell sorting platform described here could have an impact in many fields of research. Glycolysis plays a fundamental role in cell signaling, activation, proliferation and disease progression.67 Changes in metabolism from oxidative phosphorylation to glycolysis have been linked to such processes as the activation of T cells68,69 and the reprograming and proliferation of stem cells70,71. Moreover, many disease states, including diabetes, cancer, obesity and metabolic disorders, involve altered glycolysis.72 The device enables the separation, based on glycolytic profile, for a wide range of cell types for further analysis or culture. Furthermore, the device can also be used to sort cells based on the glycolytic and metabolic response to drug treatment, stimuli or stresses (nutrient, temperature, toxins, hypoxia).
Supplementary Material
Acknowledgments
AJGR acknowledges generous funding from the Clare Boothe Luce Scholar program. CZ also acknowledges support from the Inclusive Excellence Postdoctoral Program at Santa Clara University. We would also like to thank International Electronic Components Inc. for their generous donation of dry photoresist. Paul Abbyad is supported for this project by a National Science Foundation Career Award, Grant Number 1751861, the National Institutes of Health under grant 1R15GM129674-01 work and the Henry Dreyfus Teacher-Scholar Awards Program.
Footnotes
Supporting Information Available: This material is available free of charge via the Internet at http://pubs.acs.org.
Supporting Information: Videos of selection of droplets containing cells, SIFT device channel geometry, sorting rail dimensions and position, calibration curve for the pH sensitive ratiometric fluorescent probe pyranine, logistic regression fits of data, additional experiment of sorting under hypoxic conditions using K562 cells, and a table with typical flow parameters used in experiments.
References
- (1).Bonnet D; Dick JE Human Acute Myeloid Leukemia Is Organized as a Hierarchy That Originates from a Primitive Hematopoietic Cell. Nat. Med 1997, 3 (7), 730–737. [DOI] [PubMed] [Google Scholar]
- (2).Meacham CE; Morrison SJ Tumour Heterogeneity and Cancer Cell Plasticity. Nature 2013, 501 (7467), 328–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Danhier P; Bański P; Payen VL; Grasso D; Ippolito L; Sonveaux P; Porporato PE Cancer Metabolism in Space and Time: Beyond the Warburg Effect. Biochim. Biophys. Acta - Bioenerg 2017, 1858 (8), 556–572. [DOI] [PubMed] [Google Scholar]
- (4).Herzenberg LA; Parks D; Sahaf B; Perez O; Roederer M; Herzenberg LA The History and Future of the Fluorescence Activated Cell Sorter and Flow Cytometry: A View from Stanford. Clin. Chem, 2002, 48, 1819–1827. [PubMed] [Google Scholar]
- (5).Agresti JJ; Antipov E; Abate AR; Ahn K; Rowat AC; Baret J-C; Marquez M; Klibanov AM; Griffiths AD; Weitz DA Ultrahigh-Throughput Screening in Drop-Based Microfluidics for Directed Evolution. Proc. Natl. Acad. Sci. U.S.A 2010, 107 (9), 4004–4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Wang BL; Ghaderi A; Zhou H; Agresti J; Weitz DA; Fink GR; Stephanopoulos G Microfluidic High-Throughput Culturing of Single Cells for Selection Based on Extracellular Metabolite Production or Consumption. Nat. Biotech 2014, 32 (5), 473–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Matuła K; Rivello F; Huck WT S. Single-Cell Analysis Using Droplet Microfluidics. Adv. Biosyst 2019, 1900188. [DOI] [PubMed] [Google Scholar]
- (8).Magee JA; Piskounova E; Morrison SJ Cancer Stem Cells: Impact, Heterogeneity, and Uncertainty. Cancer Cell 2012, 21 (3), 283–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Ferreira MM; Ramani VC; Jeffrey SS Circulating Tumor Cell Technologies. Mol. Oncol 2016, 10 (3), 374–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Wyatt Shields Iv C; Reyes CD; López GP Microfluidic Cell Sorting: A Review of the Advances in the Separation of Cells from Debulking to Rare Cell Isolation. Lab Chip 2015, 15 (5), 1230–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Petersson F; Åberg L; Swärd-Nilsson AM; Laurell T Free Flow Acoustophoresis: Microfluidic-Based Mode of Particle and Cell Separation. Anal. Chem 2007, 79 (14), 5117–5123. [DOI] [PubMed] [Google Scholar]
- (12).Zalis MC; Reyes JF; Augustsson P; Holmqvist S; Roybon L; Laurell T; Deierborg T Label-Free Concentration of Viable Neurons, HESC and Cancer Cells by Means of Acoustophoresis. Integr. Biol 2016, 8, 332–340. [DOI] [PubMed] [Google Scholar]
- (13).Huang Y; Joo S; Duhon M; Heller M; Wallace B; Xu X Dielectrophoretic Cell Separation and Gene Expression Profiling on Microelectronic Chip Arrays. Anal. Chem 2002, 74 (14), 3362–3371. [DOI] [PubMed] [Google Scholar]
- (14).Vahey MD; Voldman J An Equilibrium Method for Continuous-Flow Cell Sorting Using Dielectrophoresis. Anal. Chem 2008, 80 (9), 3135–3143. [DOI] [PubMed] [Google Scholar]
- (15).Huang LR; Cox EC; Austin RH; Sturm JC Continuous Particle Separation through Deterministic Lateral Displacement. Science 2004, 304 (5673), 987–991. [DOI] [PubMed] [Google Scholar]
- (16).Beech JP; Holm SH; Adolfsson K; Tegenfeldt JO Sorting Cells by Size, Shape and Deformability. Lab Chip 2012, 12 (6), 1048–1051. [DOI] [PubMed] [Google Scholar]
- (17).Hur SC; Tse HTK; di Carlo D Sheathless Inertial Cell Ordering for Extreme Throughput Flow Cytometry. Lab Chip 2010, 10 (3), 274–280. [DOI] [PubMed] [Google Scholar]
- (18).Hur SC; Henderson-MacLennan NK; McCabe ERB; di Carlo D Deformability-Based Cell Classification and Enrichment Using Inertial Microfluidics. Lab Chip 2011, 11 (5), 912–920. [DOI] [PubMed] [Google Scholar]
- (19).Rao J; Dhar M; Pao E; Renier C; Go DE; Che J; Montoya R; Conrad R; Matsumoto M; Heirich K; Triboulet M; Jeffrey SS; Garon E; Goldman J; Rao NP; Kulkarni R; Sollier E; di Carlo D Label-Free Enumeration, Collection and Downstream Cytological and Cytogenetic Analysis of Circulating Tumor Cells. Sci. Rep 2016, 96 (October 2015), 117A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Antfolk M; Magnusson C; Augustsson P; Lilja H; Laurell T Acoustofluidic, Label-Free Separation and Simultaneous Concentration of Rare Tumor Cells from White Blood Cells. Anal. Chem 2015, 87 (18), 9322–9328. [DOI] [PubMed] [Google Scholar]
- (21).Shafiee H; Sano MB; Henslee EA; Caldwell JL; Davalos RV Selective Isolation of Live/Dead Cells Using Contactless Dielectrophoresis (CDEP). Lab Chip 2010, 10 (4), 438–445. [DOI] [PubMed] [Google Scholar]
- (22).Yang AHJ; Soh HT Acoustophoretic Sorting of Viable Mammalian Cells in a Microfluidic Device. Anal. Chem 2012, 84, 10756–10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Kwon T; Yao R; Hamel JFP; Han J Continuous Removal of Small Nonviable Suspended Mammalian Cells and Debris from Bioreactors Using Inertial Microfluidics. Lab Chip 2018, 18 (18), 2826–2837. [DOI] [PubMed] [Google Scholar]
- (24).Hanahan D; Weinberg RA Hallmarks of Cancer: The next Generation. Cell 2011, 144 (5), 646–674. [DOI] [PubMed] [Google Scholar]
- (25).Warburg OH Über Den Stoffwechsel Der Carcinomzelle. Naturwissenschaften 1924, 12 (50), 1131–1137. [Google Scholar]
- (26).Warburg O On the Origin of Cancer Cells. Science 1956, 123 (3191), 309–314. [DOI] [PubMed] [Google Scholar]
- (27).Surasi DS; Bhambhvani P; Baldwin JA; Almodovar SE; O’Malley JP 18F-FDG PET and PET/CT Patient Preparation: A Review of the Literature. J. Nuc. Med. Technol 2014, 42 (1), 5–13. [DOI] [PubMed] [Google Scholar]
- (28).Wike-Hooley JL; Haveman J; Reinhold HS The Relevance of Tumour pH to the Treatment of Malignant Disease. Radiother. Oncol. 1984, 2 (4), 343–366. [DOI] [PubMed] [Google Scholar]
- (29).Justus CR; Dong L; Yang L v. Acidic Tumor Microenvironment and pH-Sensing G Protein-Coupled Receptors. Front. Physiol December 5, 2013, p 354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Liberti M.v; Locasale JW The Warburg Effect: How Does It Benefit Cancer Cells? Trends Biochem. Sci 2016, 41 (3), 211–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Corbet C; Feron O Tumour Acidosis: From the Passenger to the Driver’s Seat. Nat. Rev. Cancer 2017, 17 (10), 577–593. [DOI] [PubMed] [Google Scholar]
- (32).Hsu PP; Sabatini DM Cancer Cell Metabolism: Warburg and Beyond. Cell 2008, 134 (5), 703–707. [DOI] [PubMed] [Google Scholar]
- (33).Walenta S; Wetterling M; Lehrke M; Schwickert G; Sundfør K; Rofstad EK; Mueller-Klieser W High Lactate Levels Predict Likelihood of Metastases, Tumor Recurrence, and Restricted Patient Survival in Human Cervical Cancers. Cancer Res. 2000, 60 (4), 916–21. [PubMed] [Google Scholar]
- (34).Doherty J; Cleveland J Targeting Lactate Metabolism for Cancer Therapeutics. J. Clin. Invest 2013, 123 (9), 3685–3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Martinez-Outschoorn UE; Prisco M; Ertel A; Tsirigos A; Lin Z; Pavlides S; Wang C; Flomenberg N; Knudsen ES; Howell A; Pestell RG; Sotgia F; Lisanti MP Ketones and Lactate Increase Cancer Cell “Stemness,” Driving Recurrence, Metastasis and Poor Clinical Outcome in Breast Cancer: Achieving Personalized Medicine via Metabolo-Genomics. Cell Cycle 2011, 10 (8), 1271–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Cairns RA; Harris IS; Mak TW Regulation of Cancer Cell Metabolism. Nature Rev. Cancer 2011, 11 (2), 85–95. [DOI] [PubMed] [Google Scholar]
- (37).Zhao Y; Butler EB; Tan M Targeting Cellular Metabolism to Improve Cancer Therapeutics. Cell Death Dis. 2013, 4 (3), e532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Pelicano H; Martin DS; Xu RH; Huang P Glycolysis Inhibition for Anticancer Treatment. Oncogene 2006, 25 (34), 4633–4646. [DOI] [PubMed] [Google Scholar]
- (39).del Ben F; Turetta M; Celetti G; Piruska A; Bulfoni M; Cesselli D; Huck WTS; Scoles G A Method for Detecting Circulating Tumor Cells Based on the Measurement of Single-Cell Metabolism in Droplet-Based Microfluidics. Angew. Chem. – Int. Ed 2016, 55 (30), 8581–8584. [DOI] [PubMed] [Google Scholar]
- (40).Wu M; Neilson A; Swift AL; Moran R; Tamagnine J; Parslow D; Armistead S; Lemire K; Orrell J; Teich J; Chomicz S; Ferrick DA Multiparameter Metabolic Analysis Reveals a Close Link between Attenuated Mitochondrial Bioenergetic Function and Enhanced Glycolysis Dependency in Human Tumor Cells. Am. J. Physiol. Cell Physiol 2007, 292 (1), C125–36. [DOI] [PubMed] [Google Scholar]
- (41).Horvath DG; Braza S; Moore T; Pan CW; Zhu L; Pak OS; Abbyad P Sorting by Interfacial Tension (SIFT): Label-Free Enzyme Sorting Using Droplet Microfluidics. Anal. Chim. Acta 2019, 1089, 108–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Pan CW; Horvath DG; Braza S; Moore T; Lynch A; Feit C; Abbyad P Sorting by Interfacial Tension (SIFT): Label-Free Selection of Live Cells Based on Single-Cell Metabolism. Lab Chip 2019, 19 (8), 1344–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Stephan K; Pittet P; Renaud L; Kleimann P; Morin P; Ouaini N; Ferrigno R Fast Prototyping Using a Dry Film Photoresist: Microfabrication of Soft-Lithography Masters for Microfluidic Structures. J. Micromech. Microeng 2007, 17, N69. [Google Scholar]
- (44).Frenz L; Blank K; Brouzes E; Griffiths AD Reliable Microfluidic On-Chip Incubation of Droplets in Delay-Lines. Lab Chip 2009, 9 (10), 1344–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Abramoff MD; Magalhaes PJ; Ram SJ Image Processing with ImageJ. Biophotonics Int. 2004, 11 (7), 36–43. [Google Scholar]
- (46).el Debs B; Utharala R; Balyasnikova I v; Griffiths, A. D.; Merten, C. a. Functional Single-Cell Hybridoma Screening Using Droplet-Based Microfluidics. Proc. Natl. Acad. Sci. U.S.A 2012, 109 (29), 11570–11575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Huebner A; Bratton D; Whyte G; Yang M; Demello AJ; Abell C; Hollfelder F Static Microdroplet Arrays: A Microfluidic Device for Droplet Trapping, Incubation and Release for Enzymatic and Cell-Based Assays. Lab Chip 2009, 9 (5), 692–698. [DOI] [PubMed] [Google Scholar]
- (48).Mazutis L; Gilbert J; Ung WL; Weitz DA; Griffiths AD; Heyman JA Single-Cell Analysis and Sorting Using Droplet-Based Microfluidics. Nat. Protoc 2013, 8 (5), 870–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Mongersun A; Smeenk I; Pratx G; Asuri P; Abbyad P Droplet Microfluidic Platform for the Determination of Single-Cell Lactate Release. Anal. Chem 2016, 88 (6), 3257–3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Kurup GK; Basu AS Passive, Label-Free Droplet Sorting by Chemical Composition Using Tensiophoresis. Micro Total Analysis Systems 2012. 2012, 76–78. [Google Scholar]
- (51).Kurup GK; Basu AS Deterministic Protein Extraction from Droplets Using Interfacial Drag and Tensiophoresis. Micro Total Analysis Systems 2013. 2013, No. October, 1344–1346. [Google Scholar]
- (52).Mookerjee SA; Goncalves RLS; Gerencser AA; Nicholls DG; Brand MD The Contributions of Respiration and Glycolysis to Extracellular Acid Production. Biochim. Biophys. Acta - Bioenerg 2015, 1847 (2), 171–181. [DOI] [PubMed] [Google Scholar]
- (53).Abbyad P; Dangla R; Alexandrou A; Baroud CN Rails and Anchors: Guiding and Trapping Droplet Microreactors in Two Dimensions. Lab Chip 2011, 11, 813–821. [DOI] [PubMed] [Google Scholar]
- (54).Dangla R; Lee S; Baroud CN Trapping Microfluidic Drops in Wells of Surface Energy. Phys. Rev. Let 2011, 107 (12), 124501. [DOI] [PubMed] [Google Scholar]
- (55).Vaupel P Tumor Microenvironmental Physiology and Its Implications for Radiation Oncology. Sem. Radiat. Oncol 2004, 14 (3), 198–206. [DOI] [PubMed] [Google Scholar]
- (56).Macheda ML; Rogers S; Best JD Molecular and Cellular Regulation of Glucose Transporter (GLUT) Proteins in Cancer. J. Cell. Physiol 2005, 202 (3), 654–662. [DOI] [PubMed] [Google Scholar]
- (57).Park JS; Lee S; Jeong AL; Han S; Ka HI; Lim JS; Lee MS; Yoon DY; Lee JH; Yang Y Hypoxia-Induced IL-32β Increases Glycolysis in Breast Cancer Cells. Cancer Let 2015, 356 (2), 800–808. [DOI] [PubMed] [Google Scholar]
- (58).Piret J-P; Mottet D; Raes M; Michiels C CoCl2, a Chemical Inducer of Hypoxia-Inducible Factor-1, and Hypoxia Reduce Apoptotic Cell Death in Hepatoma Cell Line HepG2. Ann. N. Y. Acad. Sci 2002, 973, 443–447. [DOI] [PubMed] [Google Scholar]
- (59).Guo M; Song LP; Jiang Y; Liu W; Yu Y; Chen GQ Hypoxia-Mimetic Agents Desferrioxamine and Cobalt Chloride Induce Leukemic Cell Apoptosis through Different Hypoxia-Inducible Factor-1α Independent Mechanisms. Apoptosis 2006, 11 (1), 67–77. [DOI] [PubMed] [Google Scholar]
- (60).Yu L; Chen X; Sun X; Wang L; Chen S The Glycolytic Switch in Tumors: How Many Players Are Involved? J. Cancer 2017, 3430–3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Wick AN; Drury DR; Nakada HI; Wolfe JB Localization of the Primary Metabolic Block Produced by 2-Deoxyglucose. J. Biol. Chem 1957, 224 (2), 963–969. [PubMed] [Google Scholar]
- (62).Karczmar GS; Arbeit JM; Toy BJ; Speder A; Weiner MW Selective Depletion of Tumor ATP by 2-Deoxyglucose and Insulin, Detected by 31P Magnetic Resonance Spectroscopy. Cancer Res. 1992, 52 (1), 71–76. [PubMed] [Google Scholar]
- (63).Kang HT; Hwang ES 2-Deoxyglucose: An Anticancer and Antiviral Therapeutic, but Not Any More a Low Glucose Mimetic. Life Sci. 2006, 78 (12), 1392–1399. [DOI] [PubMed] [Google Scholar]
- (64).Zagorodna O; Martin SM; Rutkowski DT; Kuwana T; Spitz DR; Knudson CM 2-Deoxyglucose-Induced Toxicity Is Regulated by Bcl-2 Family Members and Is Enhanced by Antagonizing Bcl-2 in Lymphoma Cell Lines. Oncogene 2012, 31 (22), 2738–2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Gu L; Yi Z; Zhang Y; Ma Z; Zhu Y; Gao J Low Dose of 2-Deoxy-D-Glucose Kills Acute Lymphoblastic Leukemia Cells and Reverses Glucocorticoid Resistance via N-Linked Glycosylation Inhibition under Normoxia. Oncotarget 2017, 8 (19), 30978–30991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Teslaa T; Teitell MA Techniques to Monitor Glycolysis. Methods Enzymol 2014, 542, 91–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Lunt SY; vander Heiden MG Aerobic Glycolysis: Meeting the Metabolic Requirements of Cell Proliferation. Annu. Rev. Cell Dev. Biol 2011, 27 (1), 441–464. [DOI] [PubMed] [Google Scholar]
- (68).Buck MD; O’Sullivan D; Pearce EL T Cell Metabolism Drives Immunity. J. Exp. Med 2015, 212 (9), 1345–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Gubser PM; Bantug GR; Razik L; Fischer M; Dimeloe S; Hoenger G; Durovic B; Jauch A; Hess C Rapid Effector Function of Memory CD8+ T Cells Requires an Immediate-Early Glycolytic Switch. Nat. Immunol 2013, 14 (10), 1064–1072. [DOI] [PubMed] [Google Scholar]
- (70).Mathieu J; Zhou W; Xing Y; Sperber H; Ferreccio A; Agoston Z; Kuppusamy KT; Moon RT; Ruohola-Baker H Hypoxia-Inducible Factors Have Distinct and Stage-Specific Roles during Reprogramming of Human Cells to Pluripotency. Cell Stem Cell 2014, 14 (5), 592–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Prigione A; Rohwer N; Hoffmann S; Mlody B; Drews K; Bukowiecki R; Blümlein K; Wanker EE; Ralser M; Cramer T; Adjaye J HIF1α Modulates Cell Fate Reprogramming Through Early Glycolytic Shift and Upregulation of PDK1–3 and PKM2. Stem Cells 2014, 32 (2), 364–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).DeBerardinis RJ; Thompson CB Cellular Metabolism and Disease: What Do Metabolic Outliers Teach Us? Cell 2012, 148 (6), 1132–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


