Abstract
Extracellular vesicles (EVs), are important for intercellular communication in both physiological and pathological processes. To explore the potential of cancer derived EVs as disease biomarkers for diagnosis, monitoring, and treatment decision, it is necessary to thoroughly characterize their biomolecular content. The aim of the study was to characterize and compare the protein content of EVs derived from three different cancer cell lines in search of a specific molecular signature, with emphasis on proteins related to the carcinogenic process. Oral squamous cell carcinoma (OSCC), pancreatic ductal adenocarcinoma (PDAC) and melanoma brain metastasis cell lines were cultured in CELLine AD1000 flasks. EVs were isolated by ultrafiltration and size-exclusion chromatography and characterized. Next, the isolated EVs underwent liquid chromatography-mass spectrometry (LC-MS) analysis for protein identification. Functional enrichment analysis was performed for a more general overview of the biological processes involved. More than 600 different proteins were identified in EVs from each particular cell line. Here, 14%, 10%, and 24% of the identified proteins were unique in OSCC, PDAC, and melanoma vesicles, respectively. A specific protein profile was discovered for each cell line, e.g., EGFR in OSCC, Muc5AC in PDAC, and FN1 in melanoma vesicles. Nevertheless, 25% of all the identified proteins were common to all cell lines. Functional enrichment analysis linked the proteins in each data set to biological processes such as “biological adhesion”, “cell motility”, and “cellular component biogenesis”. EV proteomics discovered cancer-specific protein profiles, with proteins involved in processes promoting tumor progression. In addition, the biological processes associated to the melanoma-derived EVs were distinct from the ones linked to the EVs isolated from OSCC and PDAC. The malignancy specific biomolecular cues in EVs may have potential applications as diagnostic biomarkers and in therapy.
1. Introduction
Extracellular vesicles (EVs) are released by cells into the extracellular space and are classified according to their size and biogenesis [1, 2]. Accordingly, EVs with diameters of 30–100 nm and of endosomal origin are defined as exosomes [2]. The EVs which originate by direct outward budding of the cell membrane are named microvesicles (100–1000 nm) and apoptotic bodies (>1000 nm) [2]. EVs are important players in cell-cell communication in health and disease [3] due to their diverse content of biomolecules, such as lipids, nucleic acids, and proteins [4].
EVs are quite abundant in biofluids as they are continuously released by cells [2]. In some diseases, e.g., cancer, the amount of EVs in the biofluids increases [5]. The EVs in the blood of cancer patients are released both by normal and cancer cells, and their number is estimated to be twice of that found in the blood of healthy individuals [6–8]. Oncogenes in cancer-derived EVs can modulate normal host cells, e.g. fibroblasts and macrophages, as well as local cancer cells and metastatic cells [9–11]. In this manner, tumor-derived EVs can contribute to and maintain the Hallmarks of cancer, a panel of acquired abilities of malignant tumors such as cancer cell proliferation, evasion of growth suppressors, resistance to cell death, migration, and invasion as well as modulating normal cells to favor tumor progression by transforming the microenvironment into a more permissive one [12–15].
Additionally, since EVs contain signaling molecules, they are considered be a potential source of diagnostic biomarkers for the prediction of disease, as well as in disease monitoring and treatment decision making [10, 16]. To explore the potential of cancer derived EVs as possible diagnostic and prognostic markers and to expand our understanding of their influence in cell signaling in disease progression, there is a need to isolate EVs from the other components in cell culture supernatant or biofluid of interest (e.g. blood, saliva, urine) [17]. One method for EV separation is combining two size-based separation techniques (ultrafiltration (UF) and size exclusion chromatography (SEC)). These use the size of EVs to separate them from other components that are present in the biofluid or cell culture media. Once isolated, it is of great value to characterize the EV content. Mass spectrometry (MS)-analysis allows for the identification and characterization of proteins in EV samples. However, the enormous amount of data produced by this technique can be quite extensive [18]. Therefore, to extract meaningful information from the extensive list of proteins, Gene Ontology (GO) has become a useful resource, by associating a GO term to each protein or group of proteins in the data set [18]. Overall, GO is a standardized language, or ontology, that describes the function of a gene or a gene product (RNAs or proteins) in three key domains: biological processes, molecular functions, and cellular components [18–20].
In this study, we characterized the protein content of small EVs (diameters under 200 nm), most likely exosomes and small microvesicles. These were isolated by the combination of UF and SEC from cell culture supernatant of three distinct cancer cell lines: oral squamous cell carcinoma (OSCC), pancreatic ductal adenocarcinoma (PDAC), and melanoma, all with poor prognosis [21, 22]. We aimed to provide information on and compare the protein content carried by EVs from these different malignancies. Overall, we attempted to identify an EV specific molecular signature characteristic of each cancer which could be explored for diagnostic work and/or in the prediction of prognosis of these malignancies. In addition, we studied the protein content of the EVs in relation to the carcinogenic processes known as Hallmarks of cancer.
2 Methods
2.1 Cell culture
Small EVs were isolated from the culture media of human oral squamous cell carcinoma (OSCC) (PE/CA-PJ49/E10; ECACC, Salisbury, UK), pancreatic ductal adenocarcinoma (PDAC) (BxPC3, ATCC, Manassas, USA), and human melanoma brain metastasis (H3; a kind gift from Prof. F. Thonsen, University of Bergen, Norway). The cells were cultured as previously described [23]. In order to reduce the amount of EVs from the FBS, all cell lines where progressively adapted to Advanced media (Gibco, Life Techologies) supplemented with 1% exosome depleted FBS (Gibco, Life Technologies). Briefly, E10, BxPC3, and H3 cells were cultured in complete, conventional media (respectively: Iscove’s Modified Dulbecco’s medium (IMDM; SIGMA), Roswell Park Memorial Institute (RPMI; Gibco, Life Technologies), and Dulbecco’s Modified Eagle Medium (DMEM; Gibco, Life Technologies), all supplemented with 10% FBS). Thereafter, we adapted the cells by sub-culturing them in increasing ratios of Advanced DMEM (E10 and H3) and Advanced RPMI (BxPC3) all supplemented with 1% exosome depleted FBS, 2 mM L-glutamine (Thermo Scientific), and 1X Antibiotic Antimycotic Solution (penicillin, streptomycin, and amphotericin B (PSA), SIGMA). Sub-culturing was carried out during 8 weeks for the E10 and BxPC3 cell lines. Due to a lower proliferation rate of the H3 cells, sub-culturing of this cell line was carried out for a period of 10 weeks. At this point the conventional media had been completely replaced with the Advanced media. Adapted cells were grown in CELLine AD1000 reactor flasks (Argos Technologies), designed for mass production of adherent cells. Cells were kept at 37°C in a 5% CO2 atmosphere incubator.
2.2 EV isolation
Cell culture supernatants were collected once a week. After collection of the cell culture supernatant, the CELLine AD1000 flasks were washed with warm PBS. Cell culture supernatant and PBS were then pooled. Samples were centrifuged at 4000g for 5 min at room temperature, to remove cell debris and the supernatant was then centrifuged for another 45 min at 15000g, 20°C. Next, 15 ml of the 15000g supernatant was concentrated by UF using Amicon-Ultra 15 Centrifugal Filter Units (Merck Milipore) with a 50 kDa molecular weight cut-off (MWCO), to a final volume of 4 ml. Concentrates were loaded into 30 ml sepharose CL-2B (GE Healthcare Bio-Sciences AB) size-exclusion chromatography (SEC) columns. The concentrated cell culture supernatants were eluted through the columns with the continuous addition of filtered PBS. Eluates were collected by gravity in 20 fractions of 1 ml. For each fraction, protein quantification was determined by spectrophotometry (Absorbance 280nm) in a Nanodrop spectrophotometer (Thermofisher).
Following protein quantification, the EV enriched fractions were identified and pooled. Samples were characterized by nanoparticle tracking analysis (NTA), immunoaffinity capture (Exosome Human CD9 Flow Detection Kit (Dynal®, Thermo-Fisher Scientific)), western blot (WB) (anti-CD9 antibody (10626D, Invitrogen, Carlsbad, USA)) and transmission electron microscopy (TEM), as described elsewhere [23]. Briefly, using NTA, the particle concentration and their size were estimated. Samples were analyzed for the presence of classical exosome markers by western blot Isolated EVs were also characterized by immunoaffinity capture for the presence of CD9. Lastly, visualization of the isolated EVs was carried out by TEM.
2.3 Proteomics
Following characterization, proteomic analysis was carried out using a total of 20 μg of protein for each of the preparations (n = 3 for each of the cell lines), as described in detail in (S1 File). Briefly, each sample was precipitated and the EV proteins were digested by trypsin. The resulting peptides were analyzed on an Ultimate 3000 RSLCnano-UHPLC system connected to a Q Exactive mass spectrometer (Thermo Fisher Scientific, Bremen, Germany) equipped with a nano-electrospray ion source. The peptide mass/spectra were compared to a reference database (SwissProt database, Human) for peptide identification and the multiple identified peptides are assembled for protein identification. Protein identification and validation, including statistical analysis were carried out using Scaffold_4.8.7 (Proteome Software Inc.) (See S1 File).
2.4 Data analysis
Validation of liquid chromatography-mass spectrometry (LC-MS)-based peptide and protein identifications was carried out using Scaffold_4.9.0 (Proteome Software Inc., Portland, OR, USA). Here protein threshold was set at 99.0% to ensure that only proteins with 99% of probability of being present were produced, and the minimum number of peptides was set to 2. Furthermore, the peptide threshold was set at 95%, the minimum probability to determine if a given spectrum identifies a peptide.
Data from the vesicles isolated from E10, BxPC3, and H3 cell culture supernatants were analyzed and extracted individually from Scaffold. All datasets were screened and proteins that were not present in all 3 replicates within a cell line were not included for further analysis.
Comparison of the proteins from the different cell lines was carried out using the Functional Enrichment Analysis Tool (FunRich) (http://www.funrich.org). In addition, the proteins identified in the preparations from E10, BxPC3, and H3 were compared with the Vesiclepedia database (http://microvesicles.org), a web-based open source compendium of biomolecules (proteins, RNAs, lipids, and metabolites) found across all classes of EVs from published and unpublished studies [24]. Proteins from each dataset were ranked according to their average spectral count (SC), a semi-quantitative measure of protein abundance [25]. The top 20 proteins were extracted for a more detailed analysis. Furthermore, proteomes of the EVs from E10, BxPC3 and H3 cell lines were screened for the presence of tumor related proteins previously described in the literature, and their role in the acquisition of cancer characteristics known as Hallmarks of cancer [26–33].
Analysis of the proteomics datasets of the isolated EVs from the different cell lines was carried out using the Database for Annotation, Visualization and Integrated Discovery (DAVID) Bioinformatics Resources 6.8. The proteins were classified according to their gene ontology (GO) term and categorized into functional groups under the term “Biological processes” (GOTERM_BP_FAT). Next, the GO terms were grouped into “Functional annotation clusters”, where the terms are organized into groups based on the premise that genes with similar annotation are functionally related to each other [34]. Functional annotation clusters were ranked according to their enrichment score (ES), indicating the relative importance of each cluster [34, 35]. GO terms, ES and statistical determinants (p-values and False Discovery Rate, FDR) were calculated by the DAVID software.
3. Results
3.1 EV characterization
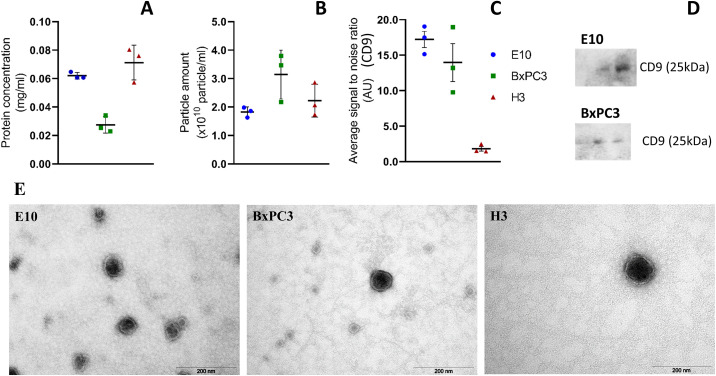
EVs derived from OSCC, PDAC, and melanoma brain metastasis cell lines were isolated from the cell culture supernatant combining UF and SEC, and were characterized by NTA, WB, immunoaffinity capture, and TEM (Fig 1). Average particle concentration, as determined by NTA, was 1.82x1010, 3.14x1010, and 2.22x1010 particles/ml for E10, BxPC3, and H3, respectively (Fig 1B). Immunoaffinity capture was also used to analyze the presence of CD9 in the small EVs (Fig 1C). E10 and BxPC3 were found to be positive for this marker, while its expression was very low in the vesicles from H3. The EVs were examined for the detection of the markers CD9, CD81, and CD63 by WB. Here only CD9 was detected (Fig 1D), and only in the vesicles derived from the E10 and BxPC3 cell lines. Analysis by TEM showed the presence of EVs with diameters in the range of 60–140 nm, presenting a dark central area surrounded by a lighter peripheral zone corresponding to the membrane of the small EVs (Fig 1E).
Fig 1. Characterization of the small EVs isolated from the cell culture supernatant of the E10, BxPC3, and H3 cell lines.
EVs were studied by protein concentration measurements (A), NTA for particle quantification (B), immunoaffinity capture targeting CD9 (C), and Western blot for the detection of CD9 in three sequential SEC fractions (20 μl per well) (D). Samples were negatively stained with 4% uranyl acetate (aqueous) for analysis by TEM (E). Data republished from Guerreiro et al [23].
The EVs underwent liquid-chromatography mass-spectrometry (LC-MS) analysis for identification and characterization of their protein content.
3.2 Overview of EV protein content
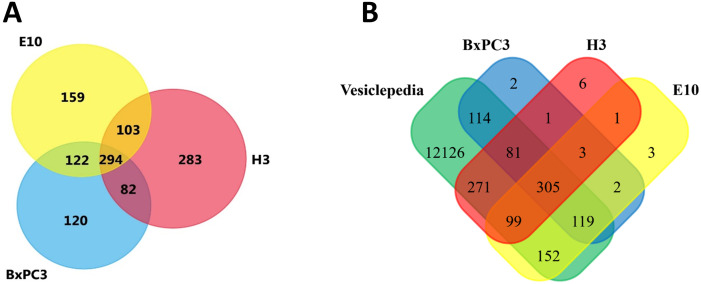
Proteomic analysis of the vesicles isolated from cell culture supernatant from the three cell lines identified 678 different proteins from the E10 derived EVs, 618 proteins from the BxPC3 derived EVs, and 762 proteins from H3 derived EVs (Fig 1A). Interestingly, 25% of all the proteins identified were common to all cell lines. Additionally, 7–10% of the total proteins identified were shared by two cell lines. In contrast, 14%, 10%, and 24% proteins were unique in E10, BxPC3, and H3 vesicles, respectively. Furthermore, most of the proteins in the vesicles from the cell lines were already identified and registered in the Vesiclepedia database [36] (Fig 1B), with only a few exceptions (9 proteins in E10, 8 in BxPC3, and 11 in H3; S1 Table). In addition, the 30 proteins most often identified in EVs according to Vesiclepedia, including the vesicle markers CD9, HSPA8, and ENO1, were also found to be present in our preparations. However, CD63 was detected only in the H3 vesicles (S2 Table).
3.3 Top 20 proteins of each cell line
The top 20 most highly expressed proteins in the EVs from each of the cell lines are listed in Table 1 together with their known functions. Here, EVs from E10 were found to be enriched in EGFR, ITGB4, NT5E, MYO1C, ATP1A1, and CTNNA1, while enriched proteins in the BxPC3 vesicles were mucins (MUC5AC, MUC5B, MUC16, and MUC2), MVP, COL7A1, PGK1, PLEC, and EEF2. In addition, the most enriched proteins in the EVs from the H3 cell line were HMCN1, HSP8A, APOE, LAMA1, HSP90AB1, CSPG4, PYGB, MYH10, and LAMB1. All the vesicles were also found to be enriched in HSPG2, GAPDH, CLTC, ACTG1, PKM, AGRN, and PTGFRN.
Table 1. The 20 most highly enriched proteins in the vesicles isolated from the E10, BxPC3, and H3 cell lines, ranked according to the average spectral count (SC).
Functions associated are as described in Uniprot Knowledgebase [37].
| E10 | BxPC3 | H3 | |||
|---|---|---|---|---|---|
| Gene name (SC) | Function | Gene name (SC) | Function | Gene name (SC) | Function |
| HSPG2 (323) | Integral component of basement membranes; promotes angiogenesis | HSPG2 (495) | Integral component of basement membranes; promotes angiogenesis | HSPG2 (291) | Integral component of basement membranes; promotes angiogenesis |
| AGRN (188) | Cellular proliferation, migration and oncogenic signaling | MUC5AC (407) | Escape of cancer cells from immunesurveillance/immunosuppressive agent | HMCN1 (276) | Cancer cell invasion and metastasis |
| EGFR (158) | Promotes tumorigenesis and angiogenesis | AGRN (160) | Cellular proliferation, migration and oncogenic signaling | FASN (162) | Contributor to transformation and tumorigenic potential; cellular growth and proliferation |
| MYH9 (142) | Cytoskeleton reorganization | MUC5B (135) | Promotes proliferation, migration and invasion of tumor cells | GAPDH (156) | Extracellular vesicle marker |
| ITGB4 (141) | Tumor cell proliferation, migration and survival; Regulation of keratinocyte polarity and motility | PKM (135) | Tumor cell proliferation and survival | FN1 (137) | Cell adhesion, cell motility, wound healing, and maintenance of cell shape |
| ACTG1 (128) | Cell motility | LAMA5 (110) | Thought to mediate the attachment, migration and organization of cells | DYNC1H1 (133) | Intracellular trafficking |
| FN1 (99) | Cell adhesion, cell motility, wound healing, and maintenance of cell shape | MVP (108) | Drug resistance; cell survival and malignant progression | CLTC (129) | Endocytosis and mitosis |
| MSN (93) | Cell adhesion and mobility | ACTG1 (99) | Cell motility | ACTG1 (115) | Cell motility |
| LAMA5 (92) | May mediate attachment, migration and organization of cells | CLTC (98) | Endocytosis and mitosis | HSP8A (104) | Tumor growth; antiapoptotic effects |
| ANXA2 (90) | Intercellular transport, cell division and migration | GAPDH (93) | Extracellular vesicle marker | PKM (104) | Tumor cell proliferation and survival |
| MUC5AC (90) | Escape of cancer cells from immunesurveillance/immunosuppressive agent | COL7A1 (88) | Promotes cancer progression | APOE (102) | Lipid transport; promotes cancer cell proliferation and survival |
| CLTC (89) | Endocytosis and mitosis | PTGFRN (84) | Promotion of metastasis | MYH10 (101) | Cell migration |
| NT5E (89) | Promoter tumor growth | LGALS3BP (82) | Promotes cell proliferation and inhibits apoptosis in cancer cells; promotes angiogenesis | LAMA1 (99) | May mediate attachment, migration and organization of cells |
| PTGFRN (87) | Promotion of metastasis | MUC16 (80) | Promotes cell growth, tumorigenesis and metastasis | CSPG4 (96) | Migration; epithelial to mesenchymal transition |
| PKM (83) | Tumor cell proliferation and survival | PGK1 (80) | Promotes invasion and metastasis | AGRN (95) | Cellular proliferation, migration and oncogenic signaling |
| GAPDH (80) | Extracellular vesicle marker | FASN (72) | Contributor to transformation and tumorogenic potential; cellular growth and proliferation | MYH9 (90) | Cytoskeleton reorganization |
| MYO1C (76) | Angiogenesis, glucose uptake and progression of cell cycle | PLEC (71) | Involved in tumor growth | PYGB (89) | Drives glycogenolysis in brain |
| LGALS3BP (73) | Promotes cell proliferation and angiogenesis; inhibits apoptosis in cancer cells | EEF2 (69) | Promotion of cancer cell growth | HSP90AB1 (88) | Molecular chaperone |
| ATPA1 (69) | Creates the electrochemical gradient of sodium and potassium ions, providing the energy for active transport of various nutrients | ITGB4 (65) | Tumor cell proliferation, migration and survival Regulation of keratinocyte polarity and motility | PTGFRN (85) | Promotion of metastasis |
| CTNNA1 (68) | Cell-adhesion. May be involved in cell differentiation; Invasion suppressor | MUC2 (63) | Tumor suppressive gene | LAMB1 (82) | Proliferation, adhesion, migration, invasion and angiogenesis |
Basement membrane-specific heparan sulfate proteoglycan core protein (HSPG2); Agrin (AGRN); Epidermal growth factor receptor (EGFR); Myosin-9 (MYH9); Integrin beta-4 (ITGB4); Actin, cytoplasmic 2 (ACTG1); Fibronectin (FN1); Moesin (MSN); Laminin subunit alpha-5 (LAMA5); Annexin A2 (ANXA2); Mucin-5AC (MUC5AC); Clathrin heavy chain 1 (CLTC); 5'-nucleotidase (NT5E); Prostaglandin F2 receptor negative regulator (PTGFRN); Pyruvate kinase PKM (PKM); Glyceraldehyde-3-phosphate dehydrogenase (GAPDH); Unconventional myosin-Ic (MYO1C); Galectin-3-binding protein (LGALS3BP); Na+/K+ transporting ATPase subunit alpha-1 (ATP1A1); Catenin alpha-1 (CTNNA1); Mucin-5B (MUC5B); Major vault protein (MVP); Collagen alpha-1(VII) chain (COL7A1); Mucin-16 (MUC16); Phosphoglycerate kinase 1 (PGK1); Fatty acid synthase (FASN); Plectin (PLEC); Elongation factor 2 (EEF2); Mucin-2 (MUC2); Hemicentin-1 (HMCN1); Cytoplasmic dynein 1 heavy chain 1 (DYNC1H1); Heat shock cognate 71 kDa protein (HSP8A); Apolipoprotein E (APOE); Laminin subunit alpha-1 (LAMA1); Heat shock protein HSP 90-beta (HSP90AB1); Chondroitin sulfate proteoglycan 4 (CSPG4); Glycogen phosphorylase, brain form (PYGH); Myosin-10 (MYH10); Laminin subunit beta-1 (LAMB1)
Thus, despite the presence of common proteins, the EVs isolated from the different cell lines showed an individual profile.
3.4 EV protein content related to the Hallmarks of cancer
When the proteins in our datasets were analyzed in view of the Hallmarks of cancer, we identified proteins with roles in the processes of angiogenesis, inflammation, cell proliferation, migration, immunity, and cell adhesion (Table 2).
Table 2. EVs isolated from cell culture supernatants of E10, BxPC3, and H3 contain proteins shown to modulate tumor cells and metastatic environment [26–33] (for more detailed information on protein function and average spectral counts (SC), see S3 Table).
| Role \ Cell line | E10 | (SC) | BxPC3 | (SC) | H3 | (SC) |
|---|---|---|---|---|---|---|
| Angiogenesis | ITGB1 | (68) | MFEG8 | (47) | MFEG8 | (61) |
| TNC | (63) | ITGB1 | (19) | TNC | (51) | |
| ICAM | (55) | ICAM | (9) | ITGB1 | (25) | |
| MFEG8 | (47) | ICAM | (2) | |||
| Inflammation | S100A9 | (15) | S100A9 | (9) | MIF | (8) |
| MIF | (14) | MIF | (7) | |||
| Proliferation | EGFR | (158) | ITGB4 | (65) | CSPG4 | (96) |
| ITGB4 | (141) | Muc4 | (43) | ITGA3 | (18) | |
| ITGA6 | (66) | Muc1 | (35) | CDC42 | (8) | |
| ITGA3 | (59) | ITGA6 | (32) | TIMP1 | (4) | |
| ITGA2 | (57) | EGFR | (26) | ITGA2 | (2) | |
| CSPG4 | (34) | ITGA3 | (16) | EGFR | (2) | |
| ITGA5 | (31) | ITGA2 | (15) | |||
| CDC42 | (15) | CDC42 | (14) | |||
| ADAM10 | (13) | TIMP1 | (12) | |||
| Muc1 | (11) | S100A14 | (10) | |||
| S100A14 | (9) | ADAM10 | (6) | |||
| TIMP1 | (9) | ITGA5 | (6) | |||
| Muc4 | (5) | CSPG4 | (4) | |||
| Migration | ITGB4 | (141) | ITGB4 | (65) | FN1 | (137) |
| FN1 | (99) | ITGA6 | (32) | CSPG4 | (96) | |
| ITGB1 | (68) | ITGB1 | (19) | TNC | (51) | |
| ITGA6 | (66) | ITGA3 | (16) | VIM | (47) | |
| TNC | (63) | ITGA2 | (15) | LOXL3 | (40) | |
| ITGA3 | (59) | CDC42 | (14) | ITGB1 | (25) | |
| ITGA2 | (57) | FN1 | (12) | ITGA3 | (18) | |
| CSPG4 | (34) | LOXL2 | (7) | CDC42 | (8) | |
| ITGA5 | (31) | ITGA5 | (6) | ITGA2 | (2) | |
| LOXL2 | (20) | ADAM10 | (6) | |||
| CDC42 | (15) | CSPG4 | (4) | |||
| ADAM10 | (13) | |||||
| Immunity | Muc5AC | (90) | Muc5AC | (407) | PMEL | (26) |
| LEG1 | (23) | LEG1 | (8) | LEG1 | (17) | |
| Muc5AC | (14) | |||||
| Adhesion | ITGB4 | (141) | ITGB4 | (65) | FN1 | (137) |
| FN1 | (99) | TINAGL1 | (58) | ITGB1 | (25) | |
| ITGB1 | (68) | THBS1 | (53) | ITGA3 | (18) | |
| ITGA6 | (66) | ITGA6 | (32) | Muc18 | (14) | |
| ITGA3 | (59) | ITGB1 | (19) | TINAGL1 | (2) | |
| ITGA2 | (57) | CD166 | (17) | ITGA2 | (2) | |
| TINAGL1 | (53) | ITGA3 | (16) | |||
| ITGA5 | (31) | ITGA2 | (15) | |||
| ALCAM | (22) | FN1 | (12) | |||
| THBS1 | (15) | ITGA5 | (6) | |||
| Muc18 | (4) | FN1 | (12) | |||
Integrin beta-1 (ITGB1), Tenascin C (TNC), S-100 calcium binding protein A9 (S100A9), Integrin, alpha 2 (ITGA2), Integrin, alpha 3 (ITGA3), Integrin, alpha 5 (ITGA5), Integrin, alpha 6 (ITGA6), Integrin, beta 4 (ITGB4), Mucin 1 (Muc1), Mucin 4 (Muc4), S-100 calcium binding protein A14 (S100A14), S-100 calcium binding protein A10 (ADAM10), Epidermal growth factor receptor (EGFR), Cell division control protein 42 homolog (CDC42), Chondroitin sulfate proteoglycan 4 (CSPG4), Metalloproteinase inhibitor 1 (TIMP1), Fibronectin (FN1), Lysyl oxidase homolog 2 (LOXL2), Lysyl oxidase homolog 3 (LOXL3), Vimentin (VIM), Mucin 5AC (Muc5AC), Melanocyte protein (PMEL), Mucin 18 (Muc18), Macrophage migration inhibitory factor (MIF), Intercellular adhesion molecule (ICAM), Thrombospondin-1 (THBS1), CD166 antigen (ALCAM), Galectin 1 (LEG1), Tubulointerstitial nephritis antigen-like (TINAGL1), Tissue factor (F3), Lactadhering (MFGE8)
The EVs from each of the cell lines demonstrated varied protein profiles related to these processes. For example, the panels of proteins associated with cell proliferation, migration, and adhesion were fairly extensive for both E10 and BxPC3. Of note is the presence of several members of the integrin family (IT), associated with cell proliferation, migration, and adhesion, such as ITGA2, ITGA3, ITGA6, ITGA5, ITGB1, and ITGB4 in the EVs derived from E10 and BxPC3. In contrast, only half of the cell proliferation-related proteins were found in EVs from the H3 cell line, most of them poorly expressed (with SC below 10). The number of proteins related to adhesion were also reduced in the H3 derived EVs. Here ITGB1, ITGA3, and ITGA2 were found in the vesicles from H3, the latter with fairly low SC.
Overall, the protein content of the EVs isolated from the different cell lines carry signals that promote cancer traits. However, the EVs from the H3 melanoma cell line have a more distinct protein signature than the other two, with respect to both the type of proteins as well as their amount present.
3.5 EV protein content in relation to biological processes
To better understand the roles and relationships of the proteins in the isolated EVs, Gene Ontology (GO) analysis was performed as described in the Materials and Methods section.
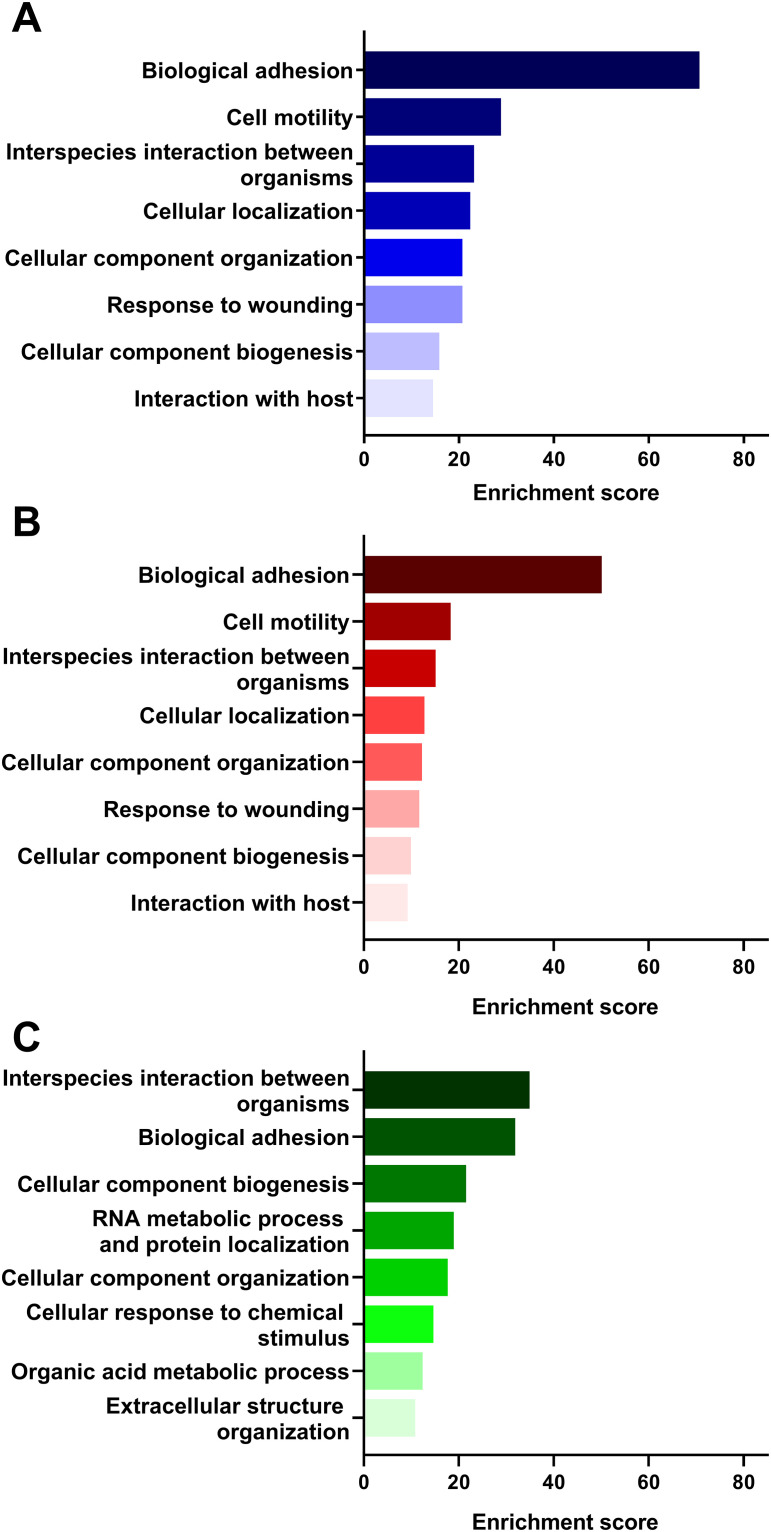
In the dataset of proteins identified in the E10 vesicles, the 8 most enriched biological processes in the functional annotation cluster analysis (Fig 3A) included biological adhesion, cell motility, interspecies interaction between organisms, cellular localization, cellular component organization, response to wounding, cellular component biogenesis, and interaction with host. Interestingly, the same biological processes were identified in the vesicles isolated from BxPC3 (Fig 3B). However, these presented lower enrichment scores than the E10 vesicles. The protein datasets from the EVs isolated from the H3 cell culture supernatant were distinct from the ones of E10 and BxPC3. In H3 (Fig 3C), enriched biological processes included interspecies interaction between organisms, biological adhesion, cellular component biogenesis, RNA metabolic process and protein localization, cellular component organization, cellular response to chemical stimulus, organic acid metabolic process, and extracellular structure organization.
Fig 3. Gene Ontology (GO) functional enrichment analysis of the biological processes (GOTERM_BP_FAT) of the identified proteins in the EVs isolated from the cell culture supernatant of E10 (A), BxPC3 (B), and H3 (C).
Biological processes for each cell line were ranked according to their enrichment score, revealing which processes were more relevant within each data set.
4. Discussion
Extracellular vesicles carry complex biomolecular cues and thus, play an important role in the communication between cells. In this study we analyzed and categorized the protein content from EVs derived from oral squamous cell carcinoma (OSCC), pancreatic ductal adenocarcinoma (PDAC), and melanoma brain metastasis cell lines (E10, BxPC3, and H3, respectively), all cancers with poor prognosis for the patient. Isolation of the EVs from the cell culture supernatants, prior to proteomic analysis, was carried out combining UF and SEC. The EVs were characterized by NTA, WB, immunoaffinity capture, and TEM.
4.1 Overview of protein content
Characterization of the protein content of the isolated vesicles by LC-MS identified a similar number of proteins for all preparations: 678 proteins for E10, 618 for BxPC3, and 762 for H3 (Fig 2A). When comparing the contents of proteins in the different preparations, a number of proteins were found to be unique for each of the cell lines (14%, 10%, and 24% of identified proteins in E10, BxPC3, and melanoma brain metastasis, respectively), suggesting that they have distinct EV profiles. Analysis of the protein datasets produced from the EVs isolated from E10, BxPC3, and H3 was carried out by comparing them with the Vesiclepedia database, a web-based directory of biomolecules (proteins, lipids, RNAs, and metabolites) identified in different classes of EVs [24]. Vesiclepedia is maintained by continuous contribution of the research community as well as manually curated data from published research [24, 36]. Here we found that most of the proteins in the EVs from the different cell lines had previously been reported (Fig 2B). However, the proteins in our samples not yet described in the Vesiclepedia database (S1 Table) were found to be involved in processes such as cell adhesion, apoptosis, regulation of exocytosis, and regulation of vesicular trafficking process. We further investigated the abundance of these proteins in our samples using spectral count (SC), a semi-quantitative measure of protein abundance in proteomic studies [25]. Here we found that the panel of proteins not described previously presented low SCs, most of them SC<5. The work from Lundgren et al., indicates that SC of less than five is unrealiable in studies with a small number of replicates [25, 38, 39]. Thus, it is not clear if the absence of these proteins from the database is due to no previous detection or to a low expression level. The 30 most frequently identified proteins in EVs, as reported in Vesiclepedia, were then scrutinized in our dataset (S2 Table). The proteins indexed in the Vesiclepedia list were found to be present in the EV samples from E10, BxPC3 and H3 with the exception of CD63. The reason for the abscence of CD63 in our samples is not clear and should be evaluated in future studies. This is a common extracellular vesicle marker, and its presence has been previously reported in the EVs derived from BxPC3, a cell line used in this study [40]. Nevertheless, CD63 positive vesicles have been described to increase in response to stress [41]. The gradual modification of culture media used in the present study, from the conventional media, supplemented with 10% FBS to the Advanced media, might have lowered the cellular stress, and, therefore, reduced the amount of CD63. Furthermore, the culture conditions may also influence the CD63 levels. Indeed, it has been reported that the presence of CD63 in vesicles derived from different colorectal cancer cell lines depended on whether the cell growth occurred in a 2D versus 3D culture system [42].
Fig 2.
A: Venn diagrams comparing the lists of all the proteins identified in the small EVs derived from OSCC (yellow), PDAC (blue), and melanoma cells (red). B: All proteins from the different datasets were compared with the proteins listed in the Vesiclepedia database (green).
4.2 The most abundant proteins in EVs isolated from each cell line
The proteins identified in the EVs of each cell line were ranked according to their average SC for further analysis. Due to the large datasets for each of the cell lines, the top 20 most abundant proteins were extracted (Table 1). A characteristic profile began to emerge for each of the cell lines. In the vesicles isolated from E10, EGFR and ITGB4 were found among the top 20. These proteins are known to have relevant roles in the development of OSCC [43]. EGFR is involved in the acquisition of self-sufficient growth-stimulatory signaling, while ITGB4 promotes the tumor ability for invasion and metastasis [43]. In BxPC3, the presence of several types of mucins (MUC5AC, MUC5B, and MUC16) reflected what is described for pancreatic tumors, which are known to overexpress these proteins [44]. For example, while absent in healthy pancreatic tissue, expression levels of MUC16 increase in moderately to poorly differentiated cancers, as is the case of the BxPC3 cell line used [44, 45]. The roles mucins hold in the progression of pancreatic cancer include regulation of the detachment of the cells from the primary tumor, invasion, and enhancement of the survival of detached cells [46–50]. In addition, these molecules also promote adhesion at metastatic sites [50–52]. Interestingly, MUC2 was found in our material. This is a tumor suppressor protein that is down regulated in pancreatic cancer [53]. Its presence among the proteins identified in the EVs derived from the BxPC3 cell line can be explained by the fact that the cells themselves of this particular cell line expresses MUC2 [54, 55]. For the top 20 highly expressed proteins in the EVs isolated from the H3 cell line, LAMA1 and LAMB1, as well as CSPG4, stand out. Here, LAMA1 and LAMB1 have been identified as potential markers for cutaneous melanoma metastasis as these proteins are involved in the modulation of cell adhesion and migration [37, 56, 57]. Also implicated in those latter processes is CSPG4 [37]. Furthermore, CSPG4 is involved in the activation of survival and growth pathways [58–60]. In addition to the proteins that are characteristic for each particular cell line, the proteomic results also identified proteins shared by all the cell lines. Those proteins were HSPG2, GAPDH, CLTC, ACTG1, PKM, AGRN, and PTGFRN. The specific role of HSPG2 and AGRN in EVs is not yet clear although they are known to be components of the extracellular matrix (ECM), the molecular scaffold for tissue organization [37, 61]. Together, this could support the hypothesis that EVs play a functional and structural role in ECM by participating in matrix organization by regulating cell migration or establishing molecular networks within the ECM [62]. In addition, GAPDH, a so-called housekeeping protein that is often identified in the proteome of EVs, does not have a clear role in EVs. It has been hypothesized that the presence of this protein, as well as other glycolytic enzymes, is related to the maintenance of the EVs’ own energy that may be required for the uptake of cancer derived EVs by cells in the tumor microenvironment [63]. The remaining proteins are involved in the maintenance of the membrane structure (ACTG1 and PTGFRN), the uptake of EVs by endocytosis (CLTC), and the release of exosomes (PKM), in particular from cancer cells [64–67].
In summary, our results highlight that the EVs isolated from each of the different cancer cell lines have distinct protein profiles. Cell line-specific profiles already begins to emerge when comparing the 20 most adundant proteins. Here, proteins that seem to provide tumors with signals that promote its progression are found among EV-related proteins. These EV related proteins are present in the datasets from all three cell lines.
4.3 Survey of EV protein contribution to cancer characteristics
The panel of proteins detected in the EVs from OSCC, PDAC, and melanoma cell lines (E10, BxPC3, and H3, respectively) indicates that these vesicles can play roles in angiogenesis, inflammation, proliferation, migration, as well as in modulation of immunity, all representing Hallmarks of cancer. Within this selection of proteins (Table 2), there were noticeable differences among the cell lines. For example, there were more proteins associated with cell proliferation in E10 and BxPC3 compared to H3. This can be due to the proliferative characteristics of each of the cell lines, with E10 presenting the highest growth rate and H3 the lowest. In addition, although EVs from the three cell lines expressed many of the same proteins, their amounts differed, as demonstrated by EGFR, Muc5AC, and FN1. EGFR was quite abundant in the EVs from the E10 cell line, but its expression levels were at a much reduced level in BxPC3. EGFR levels in H3 were barely detectable. The expression levels of EGFR in the E10 derived EVs may reflect the high expression of EGFR in OSCC tissue, as shown by immunohistochemistry (IHC), and its abundance in the saliva of OSCC patients, compared to controls [68, 69]. EGFR has been detected by IHC in PDAC tissues as well as in the EVs derived from PDAC cell lines, including BxPC3 [70, 71]. Studies on the transport of EGFR by tumor derived EVs revealed that the transported EGFR will localize in the membrane of the recipient cells, thus preparing distant sites for metastasis [72]. The presence of EGFR in the E10 and BxPC3 derived EVs may suggest that the same mechanism is at play with the exchange of growth factor receptors between the cells of the primary tumors, as well as between the tumor cells and other cells in the tumor microenvironment and even cells in distant sites. The significance of the difference we found in EGFR expression levels in the E10 and BxPC3 derived EVs needs to be further investigated. The very low expression levels of EGFR in the H3 derived EVs could be in line with what has been described in other studies where the presence of EGFR appears to only be associated with a subset of melanomas [73]. It has also been observed that EGFR expression in different melanoma cell lines and their EVs may vary [74].
In contrast, Muc5AC was found to be highly enriched in the BxPC3 derived EVs, while present in much lower amounts in the EVs from the other two cell lines. Muc5AC is a protein that stimulates angiogenesis in PDAC and might also suppress the immune response towards the tumor [75]. In patients, Muc5AC is known to be up-regulated in PDAC tissues as well as in blood [76]. In this manner, our findings are in line with these previous studies. We could not find published work describing the expression of this protein in OSCC and melanoma. However, other members of the Muc family, Muc1 and Muc4, are associated with high aggressiveness of OSCC [77, 78].
Lastly, FN1 was abundant in the H3 derived EVs but detected in a smaller amount in the E10 derived EVs. Furthermore, a dramatic drop in the FN1 content was found in the BxPC3 derived EVs. FN1 plays a role in cell adhesion, migration, and differentiation [32]. Furthermore, FN1 is highly expressed both in metastatic melanoma and OSCC cells [79, 80]. The expression levels of FN1 in H3 and E10 derived EVs reflect those results. H3 is a cell line established from a melanoma brain metastasis and E10 from a primary OSCC. In contrast to our results, analysis of FN1 by IHC in PDAC tissues has demonstrated that this protein is abundant in the tumor microenvironment [81]. Whether the very low amount of FN1 in our BxPC3 derived EVs could be due to a selective protein loading into the EVs requires further investigation.
Differences among the cell lines were also noticeable regarding members of the integrin family. The members of this family of cell adhesion receptors are relevant in cancer processes such as cell migration and invasion [82, 83]. In this study, a higher number of different integrins was detected in the EVs derived from E10 and BxPC3 cell lines, in comparison to H3. The low number of integrin members in the H3 derived EVs may be due to the cell line being generated from a melanoma brain metastasis. According to Hoshino et al the levels of integrin expression in exosomes derived from melanoma brain metastasis are down regulated [84]. Moreover, proteins identified in our datasets were also detected in studies on biofluids from OSCC, PDAC and melanoma patients. Some proteins present in the OSCC cell line derived EVs in the present study were identified by Winck et al in saliva from OSCC patients (e.g., complement C3) [85]. Similarly, EVs derived from the PDAC and melanoma cell lines included proteins (e.g., ALCAM and galectin-3, respectively) previously reported to be up-regulated in the serum of patients with PDAC and melanoma, respectively [86, 87]. This is an interesting finding and suggests a benefit of parallel studies on cancer cell lines and saliva/serum from cancer patients. This approach might contribute to the identification of possible prognostic markers in saliva/serum of patients.
Overall, the EV content reflects the characteristics of each cell line. Additionally, the cell line specific EV protein combinations point to the cellular phenotypical characteristics described as Hallmarks of cancer. Furthermore, the proteins identified have been detected in clinical cancer tissues in other studies, indicating that these may be relevant as diagnostic biomarkers for these cancers.
4.4 Potential roles of EV proteins in relation to biological processes
Functional enrichment analysis was performed using DAVID annotation clustering. Here, we identified the most enriched biological processes regulated by the hundreds of proteins identified in the EVs from each cell line (Fig 3). We found that the biological processes linked to the H3 derived EVs were quite different from the ones associated with either E10 or BxPC3. The most enriched biological processes unique for the H3 derived EVs were “RNA metabolic processes and protein localization”, “cellular response to chemical stimulus”, “organic acid metabolic process”, and “extracellular structure organization”. In contrast, “cell motility”, “cellular localization”, “response to wounding”, and “interaction with host” were the processes enriched in E10 and BxPC3. Nevertheless, several of these biological processes are found to be associated with cancer. For example, in H3 the “RNA metabolic processes and protein localization” and “cellular response to stimulus” highlight a possible dysregulation of RNA processing which may drive tumor progression [88]. “Cell motility” and “response to wounding” in E10 and BxPC3 are important for cancer cell invasion of surrounding tissues and in establishing metastasis [89, 90]. Whether the differences observed between H3 and E10/BxPC3 are a consequence of the first being a cell line derived from a metastasized melanoma, while both E10 and BxPC3 are derived from primary tumors, is not clear. However, there is evidence that a metastasis possesses a gene expression program distinct from the primary tumor that have allowed for the cancer cells to disseminate [91], which in turn may be reflected in the EV signaling.
The EVs derived from the three cancer cell lines also had some biological processes in common. “Interspecies interaction between organisms”, related to viral reproduction is one example. This process reflects the similarity of EVs and viruses even though EV biogenesis is a naturally occurring biological process while viruses hijack the cellular machinery for its propagation [92]. In addition, all three cell lines had in common the biological processes of “cellular component organization” and “cellular component biogenesis”, which are related to production, assembly, and localization of elements of a cell.
Together, these findings reveal that the EVs analyzed in our study contain proteins that can regulate biological processes towards a cancer phenotype, in a manner that is characteristic for each of the cell lines.
5. Concluding remarks
In conclusion, in the present study we used LC-MS to characterize the protein content of small EVs derived from OSCC, PDAC, and melanoma brain metastasis cell lines. More than 600 proteins were identified in the EVs of each cell line. Analysis of the datasets revealed cell-specific protein panels. In short, EVs derived from E10 had a high expression of EGFR, ITGB4, and NT5E, the EVs isolated from BxPC3 were enriched in mucins (MUC5A, MUC5B, MUC16, and MUC2), and the most abundant proteins in the EVs from the H3 cell line included HMCN1, HSP8A, and APOE. Furthermore, screening of the protein datasets for proteins involved in the Hallmarks of cancer also exposed cell-specific protein panels. Here, proteins associated with the promotion of processes such as angiogenesis, proliferation, migration, or adhesion were found to be present in the EVs derived from all three cell lines. This is relevant due to the role of EVs in the modulation of the cells in the tumor microenvironment (e.g. fibroblasts, macrophages and the cancer cells themselves) into a more permissive setting for tumor progression. The list of proteins involved, for example, in cell proliferation was quite extensive in E10 and BxPC3 in comparison to H3 where only half of the proteins were found and most of them poorly expressed. Functional enrichment analysis by gene ontology (GO) was carried out to extract a broader biological significance. Here, processes such as “biological adhesion”, “cell motility”, and “cellular component biogenesis”, all associated with aspects of cancer, were found. However, the processes identified from the proteins from the H3 derived EVs were distinct from the ones in the datasets from E10 and BxPC3, revealing an underlying difference between the cell lines. This study focuses on the protein content of small EVs, and does not explore other classes of biomolecules such as RNAs. Nevertheless, the protein lists generated and analyzed here may provide important information regarding the complex biomolecular cues EVs carry. In a clinical setting, biofluids from cancer patients will include a higher number of cancer-derived EVs in addition to non-cancer-related EVs. This will complicate the collection and examination of spacific caner-derived EVs. Hence, we believe it is important to start with a broad analytical approach that may assist in pinpointing the cancer-derived EVs in biofluids.
Supporting information
(PDF)
(PDF)
(PDF)
(PDF)
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This research has been funded by “Stiftelsen til tannlegevitenskapens fremme”, www.unifor.no (EMG, HKG, TMS), Faculty of Dentistry, University of Oslo (HKG, TMS), Nansenfondet, www.unifor.no (HG, TMS), and Research Council of Norway through its Centers of Excellence funding scheme, www.nfr.no; DEC (Grant No. 22325) and Helse Vest (Grant No. 911902/2013 and 912260/2019) The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Yang Y, Hong Y, Cho E, Kim GB, Kim IS. Extracellular vesicles as a platform for membrane-associated therapeutic protein delivery. Journal of extracellular vesicles. 2018;7(1):1440131 10.1080/20013078.2018.1440131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yanez-Mo M, Siljander PR, Andreu Z, Zavec AB, Borras FE, Buzas EI, et al. Biological properties of extracellular vesicles and their physiological functions. Journal of extracellular vesicles. 2015;4:27066 10.3402/jev.v4.27066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maas SLN, Breakefield XO, Weaver AM. Extracellular Vesicles: Unique Intercellular Delivery Vehicles. Trends in cell biology. 2017;27(3):172–88. 10.1016/j.tcb.2016.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abels ER, Breakefield XO. Introduction to Extracellular Vesicles: Biogenesis, RNA Cargo Selection, Content, Release, and Uptake. Cellular and molecular neurobiology. 2016;36(3):301–12. 10.1007/s10571-016-0366-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sadovska L, Eglitis J, Line A. Extracellular Vesicles as Biomarkers and Therapeutic Targets in Breast Cancer. Anticancer research. 2015;35(12):6379–90. [PubMed] [Google Scholar]
- 6.Willms E, Johansson HJ, Mäger I, Lee Y, Blomberg KEM, Sadik M, et al. Cells release subpopulations of exosomes with distinct molecular and biological properties. Scientific Reports. 2016;6(1):22519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kalluri R. The biology and function of exosomes in cancer. The Journal of clinical investigation. 2016;126(4):1208–15. 10.1172/JCI81135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Melo SA, Luecke LB, Kahlert C, Fernandez AF, Gammon ST, Kaye J, et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature. 2015;523(7559):177–82. 10.1038/nature14581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tai YL, Chu PY, Lee BH, Chen KC, Yang CY, Kuo WH, et al. Basics and applications of tumor-derived extracellular vesicles. Journal of biomedical science. 2019;26(1):35 10.1186/s12929-019-0533-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu S-L, Sun P, Li Y, Liu S-S, Lu YJTCR. Exosomes as critical mediators of cell-to-cell communication in cancer pathogenesis and their potential clinical application. 2019. 2019;8(1):298–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen F, Zhuang X, Lin L, Yu P, Wang Y, Shi Y, et al. New horizons in tumor microenvironment biology: challenges and opportunities. BMC Med. 2015;13:45-. 10.1186/s12916-015-0278-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kanada M, Bachmann MH, Contag CH. Signaling by Extracellular Vesicles Advances Cancer Hallmarks. Trends in cancer. 2016;2(2):84–94. 10.1016/j.trecan.2015.12.005 [DOI] [PubMed] [Google Scholar]
- 13.Bishop JM. Molecular themes in oncogenesis. Cell. 1991;64(2):235–48. 10.1016/0092-8674(91)90636-d [DOI] [PubMed] [Google Scholar]
- 14.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100(1):57–70. 10.1016/s0092-8674(00)81683-9 [DOI] [PubMed] [Google Scholar]
- 15.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–74. 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- 16.Rahbarghazi R, Jabbari N, Sani NA, Asghari R, Salimi L, Kalashani SA, et al. Tumor-derived extracellular vesicles: reliable tools for Cancer diagnosis and clinical applications. Cell communication and signaling: CCS. 2019;17(1):73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rosa-Fernandes L, Rocha VB, Carregari VC, Urbani A, Palmisano G. A Perspective on Extracellular Vesicles Proteomics. Frontiers in chemistry. 2017;5:102 10.3389/fchem.2017.00102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmidt A, Forne I, Imhof A. Bioinformatic analysis of proteomics data. BMC Systems Biology. 2014;8(2):S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature genetics. 2000;25(1):25–9. 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rhee SY, Wood V, Dolinski K, Draghici S. Use and misuse of the gene ontology annotations. Nature reviews Genetics. 2008;9(7):509–15. 10.1038/nrg2363 [DOI] [PubMed] [Google Scholar]
- 21.Nordin JZ, Lee Y, Vader P, Mager I, Johansson HJ, Heusermann W, et al. Ultrafiltration with size-exclusion liquid chromatography for high yield isolation of extracellular vesicles preserving intact biophysical and functional properties. Nanomedicine. 2015;11(4):879–83. 10.1016/j.nano.2015.01.003 [DOI] [PubMed] [Google Scholar]
- 22.Yu LL, Zhu J, Liu JX, Jiang F, Ni WK, Qu LS, et al. A Comparison of Traditional and Novel Methods for the Separation of Exosomes from Human Samples. Biomed Res Int. 2018;2018:3634563 10.1155/2018/3634563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guerreiro EM, Vestad B, Steffensen LA, Aass HCD, Saeed M, Ovstebo R, et al. Efficient extracellular vesicle isolation by combining cell media modifications, ultrafiltration, and size-exclusion chromatography. PloS one. 2018;13(9):e0204276 10.1371/journal.pone.0204276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pathan M, Fonseka P, Chitti SV, Kang T, Sanwlani R, Van Deun J, et al. Vesiclepedia 2019: a compendium of RNA, proteins, lipids and metabolites in extracellular vesicles. Nucleic acids research. 2019;47(D1):D516–d9. 10.1093/nar/gky1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lundgren DH, Hwang SI, Wu L, Han DK. Role of spectral counting in quantitative proteomics. Expert review of proteomics. 2010;7(1):39–53. 10.1586/epr.09.69 [DOI] [PubMed] [Google Scholar]
- 26.Ji H, Greening DW, Barnes TW, Lim JW, Tauro BJ, Rai A, et al. Proteome profiling of exosomes derived from human primary and metastatic colorectal cancer cells reveal differential expression of key metastatic factors and signal transduction components. Proteomics. 2013;13(10–11):1672–86. 10.1002/pmic.201200562 [DOI] [PubMed] [Google Scholar]
- 27.Herraiz C, Jimenez-Cervantes C, Sanchez-Laorden B, Garcia-Borron JC. Functional interplay between secreted ligands and receptors in melanoma. Seminars in cell & developmental biology. 2018;78:73–84. [DOI] [PubMed] [Google Scholar]
- 28.Dalla Pozza E, Forciniti S, Palmieri M, Dando I. Secreted molecules inducing epithelial-to-mesenchymal transition in cancer development. Seminars in cell & developmental biology. 2018;78:62–72. [DOI] [PubMed] [Google Scholar]
- 29.Yuan Y, Jiang YC, Sun CK, Chen QM. Role of the tumor microenvironment in tumor progression and the clinical applications (Review). Oncology reports. 2016;35(5):2499–515. 10.3892/or.2016.4660 [DOI] [PubMed] [Google Scholar]
- 30.Westgaard A, Schjolberg AR, Cvancarova M, Eide TJ, Clausen OP, Gladhaug IP. Differentiation markers in pancreatic head adenocarcinomas: MUC1 and MUC4 expression indicates poor prognosis in pancreatobiliary differentiated tumours. Histopathology. 2009;54(3):337–47. 10.1111/j.1365-2559.2009.03227.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park SY, Kim BH, Kim JH, Lee S, Kang GH. Panels of immunohistochemical markers help determine primary sites of metastatic adenocarcinoma. Archives of pathology & laboratory medicine. 2007;131(10):1561–7. [DOI] [PubMed] [Google Scholar]
- 32.Weinstein D, Leininger J, Hamby C, Safai B. Diagnostic and prognostic biomarkers in melanoma. The Journal of clinical and aesthetic dermatology. 2014;7(6):13–24. [PMC free article] [PubMed] [Google Scholar]
- 33.Park JE, Tan HS, Datta A, Lai RC, Zhang H, Meng W, et al. Hypoxic tumor cell modulates its microenvironment to enhance angiogenic and metastatic potential by secretion of proteins and exosomes. Molecular & cellular proteomics: MCP. 2010;9(6):1085–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang DW, Sherman BT, Tan Q, Collins JR, Alvord WG, Roayaei J, et al. The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biology. 2007;8(9):R183 10.1186/gb-2007-8-9-r183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4(1):44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- 36.Kalra H, Simpson RJ, Ji H, Aikawa E, Altevogt P, Askenase P, et al. Vesiclepedia: a compendium for extracellular vesicles with continuous community annotation. PLoS biology. 2012;10(12):e1001450 10.1371/journal.pbio.1001450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.UniProt Knowledgebase [https://www.uniprot.org/.
- 38.Old WM, Meyer-Arendt K, Aveline-Wolf L, Pierce KG, Mendoza A, Sevinsky JR, et al. Comparison of label-free methods for quantifying human proteins by shotgun proteomics. Molecular & cellular proteomics: MCP. 2005;4(10):1487–502. [DOI] [PubMed] [Google Scholar]
- 39.Wong JW, Sullivan MJ, Cagney G. Computational methods for the comparative quantification of proteins in label-free LCn-MS experiments. Briefings in bioinformatics. 2008;9(2):156–65. 10.1093/bib/bbm046 [DOI] [PubMed] [Google Scholar]
- 40.Shin SJ, Smith JA, Rezniczek GA, Pan S, Chen R, Brentnall TA, et al. Unexpected gain of function for the scaffolding protein plectin due to mislocalization in pancreatic cancer. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(48):19414–9. 10.1073/pnas.1309720110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kanemoto S, Nitani R, Murakami T, Kaneko M, Asada R, Matsuhisa K, et al. Multivesicular body formation enhancement and exosome release during endoplasmic reticulum stress. Biochemical and biophysical research communications. 2016;480(2):166–72. 10.1016/j.bbrc.2016.10.019 [DOI] [PubMed] [Google Scholar]
- 42.Szvicsek Z, Oszvald A, Szabo L, Sandor GO, Kelemen A, Soos AA, et al. Extracellular vesicle release from intestinal organoids is modulated by Apc mutation and other colorectal cancer progression factors. Cellular and molecular life sciences: CMLS. 2019;76(12):2463–76. 10.1007/s00018-019-03052-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Choi S, Myers JN. Molecular pathogenesis of oral squamous cell carcinoma: implications for therapy. Journal of dental research. 2008;87(1):14–32. 10.1177/154405910808700104 [DOI] [PubMed] [Google Scholar]
- 44.Moschovis D, Bamias G, Delladetsima I. Mucins in neoplasms of pancreas, ampulla of Vater and biliary system. World journal of gastrointestinal oncology. 2016;8(10):725–34. 10.4251/wjgo.v8.i10.725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Deer EL, Gonzalez-Hernandez J, Coursen JD, Shea JE, Ngatia J, Scaife CL, et al. Phenotype and genotype of pancreatic cancer cell lines. Pancreas. 2010;39(4):425–35. 10.1097/MPA.0b013e3181c15963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saitou M, Goto M, Horinouchi M, Tamada S, Nagata K, Hamada T, et al. MUC4 expression is a novel prognostic factor in patients with invasive ductal carcinoma of the pancreas. Journal of clinical pathology. 2005;58(8):845–52. 10.1136/jcp.2004.023572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Komatsu M, Yee L, Carraway KL. Overexpression of sialomucin complex, a rat homologue of MUC4, inhibits tumor killing by lymphokine-activated killer cells. Cancer research. 1999;59(9):2229–36. [PubMed] [Google Scholar]
- 48.Komatsu M, Tatum L, Altman NH, Carothers Carraway CA, Carraway KL. Potentiation of metastasis by cell surface sialomucin complex (rat MUC4), a multifunctional anti-adhesive glycoprotein. International journal of cancer. 2000;87(4):480–6. [DOI] [PubMed] [Google Scholar]
- 49.Tinder TL, Subramani DB, Basu GD, Bradley JM, Schettini J, Million A, et al. MUC1 enhances tumor progression and contributes toward immunosuppression in a mouse model of spontaneous pancreatic adenocarcinoma. Journal of immunology (Baltimore, Md: 1950). 2008;181(5):3116–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kaur S, Kumar S, Momi N, Sasson AR, Batra SK. Mucins in pancreatic cancer and its microenvironment. Nature reviews Gastroenterology & hepatology. 2013;10(10):607–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Geng Y, Marshall JR, King MR. Glycomechanics of the metastatic cascade: tumor cell-endothelial cell interactions in the circulation. Annals of biomedical engineering. 2012;40(4):790–805. 10.1007/s10439-011-0463-6 [DOI] [PubMed] [Google Scholar]
- 52.Zhao Q, Barclay M, Hilkens J, Guo X, Barrow H, Rhodes JM, et al. Interaction between circulating galectin-3 and cancer-associated MUC1 enhances tumour cell homotypic aggregation and prevents anoikis. Molecular cancer. 2010;9:154 10.1186/1476-4598-9-154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jonckheere N, Skrypek N, Van Seuningen I. Mucins and pancreatic cancer. Cancers. 2010;2(4):1794–812. 10.3390/cancers2041794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Levi E, Klimstra DS, Andea A, Basturk O, Adsay NV. MUC1 and MUC2 in pancreatic neoplasia. Journal of clinical pathology. 2004;57(5):456–62. 10.1136/jcp.2003.013292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yamada N, Hamada T, Goto M, Tsutsumida H, Higashi M, Nomoto M, et al. MUC2 expression is regulated by histone H3 modification and DNA methylation in pancreatic cancer. International journal of cancer. 2006;119(8):1850–7. 10.1002/ijc.22047 [DOI] [PubMed] [Google Scholar]
- 56.Chen J, Wu F, Shi Y, Yang D, Xu M, Lai Y, et al. Identification of key candidate genes involved in melanoma metastasis. Molecular medicine reports. 2019;20(2):903–14. 10.3892/mmr.2019.10314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Meves A, Nikolova E, Heim JB, Squirewell EJ, Cappel MA, Pittelkow MR, et al. Tumor Cell Adhesion As a Risk Factor for Sentinel Lymph Node Metastasis in Primary Cutaneous Melanoma. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2015;33(23):2509–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang J, Price MA, Neudauer CL, Wilson C, Ferrone S, Xia H, et al. Melanoma chondroitin sulfate proteoglycan enhances FAK and ERK activation by distinct mechanisms. The Journal of cell biology. 2004;165(6):881–91. 10.1083/jcb.200403174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang J, Price MA, Li GY, Bar-Eli M, Salgia R, Jagedeeswaran R, et al. Melanoma proteoglycan modifies gene expression to stimulate tumor cell motility, growth, and epithelial-to-mesenchymal transition. Cancer research. 2009;69(19):7538–47. 10.1158/0008-5472.CAN-08-4626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chekenya M, Krakstad C, Svendsen A, Netland IA, Staalesen V, Tysnes BB, et al. The progenitor cell marker NG2/MPG promotes chemoresistance by activation of integrin-dependent PI3K/Akt signaling. Oncogene. 2008;27(39):5182–94. 10.1038/onc.2008.157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jethwa SA, Leah EJ, Zhang Q, Bright NA, Oxley D, Bootman MD, et al. Exosomes bind to autotaxin and act as a physiological delivery mechanism to stimulate LPA receptor signalling in cells. Journal of cell science. 2016;129(20):3948–57. 10.1242/jcs.184424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rilla K, Mustonen AM, Arasu UT, Harkonen K, Matilainen J, Nieminen P. Extracellular vesicles are integral and functional components of the extracellular matrix. Matrix biology: journal of the International Society for Matrix Biology. 2019;75–76:201–19. [DOI] [PubMed] [Google Scholar]
- 63.Goran Ronquist K. Extracellular vesicles and energy metabolism. Clinica chimica acta; international journal of clinical chemistry. 2019;488:116–21. [DOI] [PubMed] [Google Scholar]
- 64.Adamo A, Brandi J, Caligola S, Delfino P, Bazzoni R, Carusone R, et al. Extracellular Vesicles Mediate Mesenchymal Stromal Cell-Dependent Regulation of B Cell PI3K-AKT Signaling Pathway and Actin Cytoskeleton. Frontiers in immunology. 2019;10:446 10.3389/fimmu.2019.00446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Merchant ML, Rood IM, Deegens JKJ, Klein JB. Isolation and characterization of urinary extracellular vesicles: implications for biomarker discovery. Nature reviews Nephrology. 2017;13(12):731–49. 10.1038/nrneph.2017.148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Costa Verdera H, Gitz-Francois JJ, Schiffelers RM, Vader P. Cellular uptake of extracellular vesicles is mediated by clathrin-independent endocytosis and macropinocytosis. Journal of controlled release: official journal of the Controlled Release Society. 2017;266:100–8. [DOI] [PubMed] [Google Scholar]
- 67.Wei Y, Wang D, Jin F, Bian Z, Li L, Liang H, et al. Pyruvate kinase type M2 promotes tumour cell exosome release via phosphorylating synaptosome-associated protein 23. Nature communications. 2017;8:14041 10.1038/ncomms14041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ang KK, Berkey BA, Tu X, Zhang HZ, Katz R, Hammond EH, et al. Impact of epidermal growth factor receptor expression on survival and pattern of relapse in patients with advanced head and neck carcinoma. Cancer research. 2002;62(24):7350–6. [PubMed] [Google Scholar]
- 69.Zanotti L, Paderno A, Piazza C, Pagan E, Bignotti E, Romani C, et al. Epidermal growth factor receptor detection in serum and saliva as a diagnostic and prognostic tool in oral cancer. The Laryngoscope. 2017;127(11):E408–e14. 10.1002/lary.26797 [DOI] [PubMed] [Google Scholar]
- 70.Yamanaka Y. The immunohistochemical expressions of epidermal growth factors, epidermal growth factor receptors and c-erbB-2 oncoprotein in human pancreatic cancer. Nihon Ika Daigaku zasshi. 1992;59(1):51–61. 10.1272/jnms1923.59.51 [DOI] [PubMed] [Google Scholar]
- 71.Adamczyk KA, Klein-Scory S, Tehrani MM, Warnken U, Schmiegel W, Schnolzer M, et al. Characterization of soluble and exosomal forms of the EGFR released from pancreatic cancer cells. Life sciences. 2011;89(9–10):304–12. 10.1016/j.lfs.2011.06.020 [DOI] [PubMed] [Google Scholar]
- 72.Zhang H, Deng T, Liu R, Bai M, Zhou L, Wang X, et al. Exosome-delivered EGFR regulates liver microenvironment to promote gastric cancer liver metastasis. Nature communications. 2017;8:15016 10.1038/ncomms15016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Boone B, Jacobs K, Ferdinande L, Taildeman J, Lambert J, Peeters M, et al. EGFR in melanoma: clinical significance and potential therapeutic target. Journal of cutaneous pathology. 2011;38(6):492–502. 10.1111/j.1600-0560.2011.01673.x [DOI] [PubMed] [Google Scholar]
- 74.Lazar I, Clement E, Ducoux-Petit M, Denat L, Soldan V, Dauvillier S, et al. Proteome characterization of melanoma exosomes reveals a specific signature for metastatic cell lines. Pigment cell & melanoma research. 2015;28(4):464–75. [DOI] [PubMed] [Google Scholar]
- 75.Hoshi H, Sawada T, Uchida M, Iijima H, Kimura K, Hirakawa K, et al. MUC5AC protects pancreatic cancer cells from TRAIL-induced death pathways. International journal of oncology. 2013;42(3):887–93. 10.3892/ijo.2013.1760 [DOI] [PubMed] [Google Scholar]
- 76.Kaur S, Smith LM, Patel A, Menning M, Watley DC, Malik SS, et al. A Combination of MUC5AC and CA19-9 Improves the Diagnosis of Pancreatic Cancer: A Multicenter Study. The American journal of gastroenterology. 2017;112(1):172–83. 10.1038/ajg.2016.482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hamada T, Nomura M, Kamikawa Y, Yamada N, Batra SK, Yonezawa S, et al. DF3 epitope expression on MUC1 mucin is associated with tumor aggressiveness, subsequent lymph node metastasis, and poor prognosis in patients with oral squamous cell carcinoma. Cancer. 2012;118(21):5251–64. 10.1002/cncr.27542 [DOI] [PubMed] [Google Scholar]
- 78.Hamada T, Wakamatsu T, Miyahara M, Nagata S, Nomura M, Kamikawa Y, et al. MUC4: a novel prognostic factor of oral squamous cell carcinoma. International journal of cancer. 2012;130(8):1768–76. 10.1002/ijc.26187 [DOI] [PubMed] [Google Scholar]
- 79.Li B, Shen W, Peng H, Li Y, Chen F, Zheng L, et al. Fibronectin 1 promotes melanoma proliferation and metastasis by inhibiting apoptosis and regulating EMT. OncoTargets and therapy. 2019;12:3207–21. 10.2147/OTT.S195703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Morita Y, Hata K, Nakanishi M, Omata T, Morita N, Yura Y, et al. Cellular fibronectin 1 promotes VEGF-C expression, lymphangiogenesis and lymph node metastasis associated with human oral squamous cell carcinoma. Clinical & experimental metastasis. 2015;32(7):739–53. [DOI] [PubMed] [Google Scholar]
- 81.Hu D, Ansari D, Zhou Q, Sasor A, Said Hilmersson K, Andersson R. Stromal fibronectin expression in patients with resected pancreatic ductal adenocarcinoma. World journal of surgical oncology. 2019;17(1):29 10.1186/s12957-019-1574-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bianconi D, Unseld M, Prager GW. Integrins in the Spotlight of Cancer. International journal of molecular sciences. 2016;17(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Desgrosellier JS, Cheresh DA. Integrins in cancer: biological implications and therapeutic opportunities. Nat Rev Cancer. 2010;10(1):9–22. 10.1038/nrc2748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hoshino A, Costa-Silva B, Shen T-L, Rodrigues G, Hashimoto A, Tesic Mark M, et al. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527(7578):329–35. 10.1038/nature15756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Winck FV, Prado Ribeiro AC, Ramos Domingues R, Ling LY, Riano-Pachon DM, Rivera C, et al. Insights into immune responses in oral cancer through proteomic analysis of saliva and salivary extracellular vesicles. Sci Rep. 2015;5:16305 10.1038/srep16305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pan S, Brentnall TA, Chen R. Proteomics analysis of bodily fluids in pancreatic cancer. Proteomics. 2015;15(15):2705–15. 10.1002/pmic.201400476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vereecken P, Cornelis F, Van Baren N, Vandersleyen V, Baurain JF. A synopsis of serum biomarkers in cutaneous melanoma patients. Dermatology research and practice. 2012;2012:260643 10.1155/2012/260643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yu LH, Huang QW, Zhou XH. Identification of Cancer Hallmarks Based on the Gene Co-expression Networks of Seven Cancers. Frontiers in genetics. 2019;10:99 10.3389/fgene.2019.00099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Liotta LA, Stracke ML, Aznavoorian SA, Beckner ME, Schiffmann E. Tumor cell motility. Seminars in cancer biology. 1991;2(2):111–4. [PubMed] [Google Scholar]
- 90.Sundaram GM, Ismail HM, Bashir M, Muhuri M, Vaz C, Nama S, et al. EGF hijacks miR-198/FSTL1 wound-healing switch and steers a two-pronged pathway toward metastasis. The Journal of experimental medicine. 2017;214(10):2889–900. 10.1084/jem.20170354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hartung F, Wang Y, Aronow B, Weber GF. A core program of gene expression characterizes cancer metastases. Oncotarget. 2017;8(60):102161–75. 10.18632/oncotarget.22240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nolte-'t Hoen E, Cremer T, Gallo RC, Margolis LB. Extracellular vesicles and viruses: Are they close relatives? Proceedings of the National Academy of Sciences of the United States of America. 2016;113(33):9155–61. 10.1073/pnas.1605146113 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(PDF)
(PDF)
(PDF)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.