Abstract
Background:
Carbapenem-resistant Enterobacteriaceae (CRE) are a global threat. Here, we describe the clinical and molecular characteristics of Centers for Disease Control and Prevention (CDC)-defined CRE in the US.
Methods:
The second Consortium on Resistance Against Carbapenems in Klebsiella and other Enterobacteriaceae (CRACKLE-2, ClinicalTrials.gov: NCT03646227) is a prospective, multicenter, cohort study. Patients hospitalized in 49 US hospitals, with clinical cultures positive for CDC-defined CRE between 30 April 2016 and 31 August 2017 were included. Primary outcome was desirability of outcome ranking (DOOR) at 30 days. Clinical data and bacteria were collected, and whole genome sequencing (WGS) was performed.
Findings:
1,040 patients with unique isolates were included; 449 (43%) with infection and 591 (57%) with colonization. CDC-defined CRE admission rate was 57 CDC-defined CRE admissions/100,000 admissions (95% CI: 45–71). Three subsets of CDC-defined CRE were identified: carbapenemase-producing Enterobacteriaceae (618/1,040, 59%); non-carbapenemase-producing CRE (194/1,040, 19%); and unconfirmed CRE (228/1,040, 22%; initially reported as CRE, but susceptible to carbapenems in two central laboratories). Klebsiella pneumoniae carbapenemase (KPC)-producing clonal group 258 K. pneumoniae was the most common carbapenemase-producing Enterobacteriaceae. In 449 patients with CDC-defined CRE infections, DOOR outcomes were not significantly different in patients with carbapenemase-producing Enterobacteriaceae, non-carbapenemase-producing CRE, and unconfirmed CRE. At 30 days 107/449 (24%, 95% CI 20–28%) patients had died.
Interpretation:
Among patients with CDC-defined CRE, similar outcomes were observed among three subgroups, including the novel unconfirmed CRE group. CDC-defined CRE represent diverse bacteria, whose spread may not respond to interventions directed to carbapenemase-producing Enterobacteriaceae.
Funding:
National Institutes of Health
INTRODUCTION
Antimicrobial resistance is an important threat to global public health.1,2 Carbapenem-resistant Enterobacteriaceae (CRE) rank among the top three critical multi-drug resistant pathogens on the priority list of the World Health Organization (WHO).3 The subset of CRE that produce carbapenemases – carbapenemase-producing Enterobacteriaceae (CPE) – are of high clinical and public health concern, as they may spread quickly in healthcare systems.4
Globally, common carbapenemases in Enterobacteriaceae include the Klebsiella pneumoniae carbapenemases (KPC), oxacillinase (OXA)-48-like β-lactamases, and metallo-β-lactamases, such as New-Delhi-metallo-β-lactamases (NDM), the “active in imipenem” (IMP) family of carbapenemases, and Verona integron–encoded metallo-β-lactamases (VIM).1 When expressed in enteric bacteria, KPC are resistant to inactivation by clavulanic acid, sulbactam, and tazobactam.5 In the retrospective INCREMENT cohort, a 43% all-cause 30-day mortality in 437 patients with CPE bloodstream infection was observed.6 Patients in INCREMENT originated from 12 countries including the US.6 KPC-producing K. pneumoniae was the predominant species of CPE in the INCREMENT study.6 In hospitalized patients in low-income and middle-income countries (LMIC), bloodstream infection due to CRE is associated with an adjusted hazard ratio of 1·75 (95% CI 1·04–2·94) for in-hospital mortality.7 Of note, in LMICs, only a minority of CR K.pneumoniae were part of clonal group 258, and blaNDM was the most commonly identified carbapenemase-encoding gene.7 In a microbiologically survey of 1,801 CRE – defined as in vitro resistance to any carbapenem – isolates from China, 86% of isolates were CPE. In CPE, KPC-producing ST11 K. pneumoniae were the most common.8
In 2012, the Centers for Disease Control and Prevention (CDC) defined CRE as Enterobacteriaceae with non-susceptibility to imipenem, meropenem and/or doripenem, and resistance to extended-spectrum cephalosporins (ceftriaxone, ceftazidime, ceftizoxime, and cefotaxime).9 In 2015, the CDC definition was updated to the current definition of CRE, which includes in vitro resistance to one or more carbapenems – including ertapenem – without any requirement for cephalosporin resistance.10 In the US, a more detailed understanding of outcomes, and the impact of bacterial characteristics on those outcomes, in patients with CRE is needed. This improved understanding will help guide future interventional trials. Therefore, the aim of this study is to describe in detail the clinical spectrum of patients diagnosed with CDC-defined CRE infection or colonization in the US, their outcomes, and the phenotypic and genotypic characterization of their isolates. Our research question is whether carbapenemase production in CRE is associated with adverse clinical outcomes. In that context, we report the clinical and molecular epidemiology of 1,040 patients with CDC-defined CRE, hospitalized in 49 participating US hospitals.
METHODS
Patients
The second Consortium on Resistance Against Carbapenems in Klebsiella and other Enterobacteriaceae (CRACKLE-2) is a prospective, observational, multicenter study with consecutive enrollment of hospitalized patients.11,12 Patients were eligible for inclusion in the study if CDC-defined CRE was isolated in a clinical culture from any anatomic site during hospitalization; surveillance cultures were not eligible. There was no age exclusion. The first qualifying culture episode during the first admission for each unique patient enrolled during the study period (30 April 2016 – 31 August 2017) with an available CDC-defined CRE isolate was included. Twenty-six study sites with 49 U.S. hospitals in 15 states and the District of Columbia contributed patients. The 49 study hospitals are compared to 6,282 US hospitals in Supplementary Table 1. The final study size was derived by inclusion of all eligible patients within the study period. The study was approved by the Institutional Review Boards of all the health systems involved with a waiver of consent.
Clinical data Clinical data, including race/ethnicity (which were included to facilitate comparisons with non-study populations) were obtained from the electronic health record (EHR). Infections were defined by standard criteria (Supplementary Materials), otherwise, positive cultures were considered colonization.12 At 90 days after discharge, data on post-hospitalization death and readmission were collected from the EHR. Treatment was divided into empirical antibiotics (those given prior to the date of the antibacterial susceptibility report) and definitive treatment (antibiotics given after susceptibility results were available).
Clinical outcomes
Outcomes were evaluated at 30 days after the index culture. The primary outcome was a desirability of outcome ranking (DOOR) analysis assessing three deleterious events (lack of clinical response, unsuccessful discharge, and adverse events; see Supplementary Materials for definitions) in addition to survival at 30 days after the index culture.13 The best outcome was defined as being alive without deleterious events. The worst outcome was death. Three levels in between these two extremes were: alive with 1, 2, and 3 deleterious events, respectively. As only 1 out of 450 patients with CRE infection fell into the “alive with 3 events” level, that level was grouped post hoc with the “alive with 2 events” level for analysis, with 4 total levels of outcomes. DOOR is a method to compare groups using a single ordinal patient-centric outcome. This ordinal outcome represents a global assessment of patient well-being including efficacy and safety components. Analyses consist of estimating the probability of a more desirable result in one group relative to another with a probability of 50% implying equality of groups.11,13 A probability of greater than 50% – with a 95% significance interval that excludes 50% – implies superiority of one group compared to the other. Similarly, a probability of less than 50% – with a 95% significance interval that excludes 50% – implies inferiority of one group compared to the other. Secondary outcomes included 30-day all-cause mortality, 90-day all-cause mortality, clinical response, and 90-day readmissions in those who were discharged alive.
Microbiology
CRE were defined according to current CDC guidelines, applied in local clinical microbiology laboratories.10 Briefly, CDC-defined CRE were Enterobacteriaceae that tested resistant to any of the carbapenems (i.e., minimum inhibitory concentrations [MIC] of ≥ 4 μg/mL for doripenem, meropenem, or imipenem, OR ≥ 2 μg/mL for ertapenem) or were documented to harbor a gene encoding a carbapenemase or were positive for carbapenemase production. For Enterobacteriaceae that exhibit intrinsic imipenem non-susceptibility (i.e. Morganella morganii, Proteus species, Providencia species), resistance to carbapenems other than imipenem was required. Eligibility was based on antimicrobial susceptibility testing performed in local contributing clinical microbiology laboratories. Bacterial identification and carbapenem susceptibility testing were performed in these laboratories using MicroScan (Beckman Coulter, Atlanta, GA), Vitek 2, Etest (both bioMérieux, Durham, NC), BD Phoenix, BBL disks (both Becton Dickinson, Durham, NC), Sensititre (Thermo Fisher, Waltham, MA), disc diffusion, or in-house agar dilution. Central carbapenem susceptibility testing was performed in two independent central research laboratories using Etest and Microscan (Beckman Coulter, Atlanta, GA).
Whole genome sequencing and genome analysis
Sequencing was performed at three locations: Molecular Resource Facility, Rutgers (Rutgers; Illumina NextSeq500), UTHealth (Illumina MiSeq), and Baylor College of Medicine (Illumina HiSeq X). Sequence type (ST) was defined as an allele combination of housekeeping genes (n=7) resulting in a number that identify the genetic background of a bacterial isolate based on multilocus sequence typing. Clonal groups (CG) were defined as related STs differing only in one or two alleles. The CGs are named according to the central (main) ST. Due to the genetic heterogeneity of the Enterobacter spp., genomic clades were used to show the population structure of Enterobacter isolates. Genomic clades in Enterobacter spp. were defined by pairwise average nucleotide identity-based distance matrix and core single nucleotide polymorphism-based phylogeny analysis. Mean average nucleotide identity values within a clade were usually ≥ 95%, while the values between clades were mainly <95%. The average nucleotide identity and single nucleotide polymorphism phylogeny were concordant in clustering the genomes into phylogenetic clades. A genomic cluster within highly related isolates was defined as a <20 single nucleotide polymorphisms in the core genome by phylogenetic analyses. Details of sequencing, bioinformatics, and phylogenetic analyses are available in the Supplementary Materials.
Statistical analysis
Distributions across groups for continuous variables were compared using the Kruskal-Wallis test. Pearson chi-squared testing across three groups was used for categorical variables. CDC-defined CRE admission rates and robust 95% CIs were estimated using a generalized linear mixed effects model (glimmix) with hospital as a random effect (Supplementary Materials).
To compare outcomes between CPE, non-carbapenemase-producing CRE, and unconfirmed CRE, pairwise DOOR analyses were performed (Supplementary Materials).13 A weight was calculated for each patient using the following variables based on their clinical relevance: origin (home vs. other), Charlson comorbidity index (CCI >3 vs. 3), and age at culture, resulting in a pseudo-population of weights where the three CDC-defined CRE groups were similar at baseline based on the IPW variables. Desirability of outcome ranking probabilities and 95% bootstrap confidence intervals were then calculated using the weighted population. Less than 1% of outcome data were missing (Supplementary Materials). Because of the potential for type 1 error due to multiple comparisons, findings for analyses of secondary endpoints should be interpreted as exploratory. P-values ≤0·05 were considered statistically significant, and all tests were 2-sided. All analyses were performed using SAS software version 9·4 (SAS Institute).
Role of the funding source
The sponsor of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report. The corresponding author had full access to all the data in the study and had final responsibility for the decision to submit for publication.
RESULTS
Subsets of CDC-defined CRE
Three mutually exclusive subsets were identified in 1,040 CDC-defined CRE isolates (Table 1). Carbapenemase genes were present in 618/1,040 (59%, 95% CI 56%–62%) CRE (“carbapenemase-producing Enterobacteriaceae”). In 194/1,040 (19%, 95% CI 16%–21%) of CDC-defined CRE in vitro resistance to at least one carbapenem was confirmed in absence of any carbapenemase gene. These, except for five imipenem-resistant Proteus species, were defined as non-carbapenemase-producing CRE (non-CP-CRE). Carbapenemase genes were not found in an additional 228/1,040 (22%, 95% CI 19%–24%) of CDC-defined CRE. These CDC-defined CRE – while they were identified as carbapenem-resistant by local laboratories - were found to be susceptible or intermediate to all tested carbapenems in both central laboratories (Supplementary Table 2). These isolates were defined as unconfirmed CRE (U-CRE).
Table 1.
Baseline characteristics.
| Characteristic | CPE | Non-CP-CRE | U-CRE | All | P Valuef |
|---|---|---|---|---|---|
| n | 618 (59) | 194 (19) | 228 (22) | 1,040 | |
| region | <0·001 | ||||
| Midwest | 119 (19) | 31 (16) | 24 (11) | 174 (17) | |
| Northeast | 316 (51) | 67 (35) | 70 (31) | 453 (44) | |
| South | 136 (22) | 67 (35) | 105 (46) | 308 (30) | |
| West | 47 (8) | 29 (15) | 29 (13) | 105 (10) | |
| age, median (IQR) | 64 (54, 75) | 64 (53, 75) | 63 (51, 74) | 64 (53, 75) | 0·51 |
| sex, male | 329 (53) | 128 (66) | 127 (56) | 584 (56) | 0·008 |
| race | 0·73 | ||||
| white | 292 (47) | 96 (49) | 103 (45) | 491 (47) | |
| black | 201 (33) | 55 (28) | 79 (35) | 335 (32) | |
| othera | 125 (20) | 43 (22) | 46 (20) | 214 (21) | |
| hispanic ethnicity | 74 (12) | 26 (13) | 25 (11) | 125 (12) | 0·74 |
| CCI, median (IQR)b | 3 (1, 5) | 3 (1, 5) | 2 (1, 4) | 3 (1, 5) | 0·01 |
| PBS, median (IQR)c | 3 (2, 6) | 3 (1, 6) | 2 (0, 5) | 3 (2, 6) | 0·01 |
| Time to positive culture, days, median (IQR)d | 2 (0, 16) | 11 (1, 30) | 3 (0, 13) | 3 (0, 18) | <0·001 |
| Community onset | 147 (24) | 42 (22) | 63 (28) | 253 (24) | 0·38 |
| admitted frome | <0·001 | ||||
| home | 323 (52) | 127 (65) | 151 (66) | 601 (58) | |
| long-term chronic care | 172 (28) | 23 (12) | 35 (15) | 230 (22) | |
| hospital transfer | 79 (13) | 39 (20) | 34 (15) | 152 (15) | |
| long term acute care | 41 (7) | 3 (2) | 7 (3) | 51 (5) | |
| transferred from foreign country | 3 (<1) | 2 (1) | 0 (0) | 5 (<1) | |
| hospice | 0 (0) | 0 (0) | 1 (<1) | 1 (<1) | |
| hospital | |||||
| tertiary care center | 471 (76) | 162 (84) | 175 (77) | 808 (78) | 0·10 |
| bed number | 0·22 | ||||
| 0–499 | 147 (24) | 54 (28) | 67 (29) | 268 (26) | |
| 500–999 | 189 (31) | 64 (33) | 74 (32) | 327 (31) | |
| 1,000+ | 282 (46) | 76 (39) | 87 (38) | 445 (43) | |
| culture | <0·001 | ||||
| blood: infection | 75 (12) | 25 (13) | 30 (13) | 130 (13) | |
| urine: infection | 84 (14) | 20 (10) | 25 (11) | 129 (12) | |
| urine: colonization | 175 (28) | 39 (20) | 61 (27) | 275 (26) | |
| respiratory: infection | 41 (7) | 15 (8) | 11 (5) | 67 (6) | |
| respiratory: colonization | 129 (21) | 29 (15) | 43 (19) | 201 (19) | |
| wound: infection | 32 (5) | 10 (5) | 17 (7) | 59 (6) | |
| wound: colonization | 41 (7) | 12 (6) | 18 (8) | 71 (7) | |
| intra-abdominal: infection | 19 (3) | 29 (15) | 10 (4) | 58 (6) | |
| other: infection | 2 (<1) | 3 (2) | 1 (<1) | 6 (1) | |
| other: colonization | 20 (3) | 12 (6) | 12 (5) | 44 (4) |
All data is shown as n (%) unless otherwise specified.
Other races included asian (n=40), native Hawaiian/pacific islander (n=3), multi-racial (n=5), and those patients for whom race was not specified in the medical record (n=166) Community onset defined as home origin with first positive culture date less than 3 days from date of admission.
CCI; Charlson comorbidity index. CCI is a chronic comorbidity score with a range from 0 to 37, with higher scores indicating more comorbid conditions present. A patient with a score of 3 could have three level 1 comorbid conditions (e.g. dementia, chronic pulmonary disease, and congestive heart failure), or one level 1 (e.g. dementia) and one level 2 comorbid condition (e.g. leukemia), or one level 3 condition (moderate or severe liver disease).29
PBS; Pitt bacteremia score. PBS is an acute severity of illness score. Higher scores indicate more severe illness. A patient with a score of 3 would have one level 1 marker (e.g. disoriented mental status) and one level 2 marker of acute illness (e.g. hypotension).30
Time to first positive culture indicates the number of days from admission to the collection date of the index culture, with 0 indicating that the index culture was obtained on the day of admission.
For analysis purposes grouped as home/transferred from foreign country, long term acute care/hospital transfer, and long term chronic care/hospice.
P value comparing distributions where applicable. CPE: carbapenemase-producing Enterobacteriaceae. Non-CP-CRE: non-carbapenemase-producing carbapenem-resistant Enterobacteriaceae (CRE). U-CRE: unconfirmed CRE. IQR Interquartile range.
Clinical epidemiology
The CDC-defined CRE admission rates in participating hospitals during the study period are shown in Figure 1. The mean CDC-defined CRE admission rate was 57 CDC-defined CRE admissions per 100,000 admissions (95% CI: 45–71). From the 49 participating hospitals, 1,040 patients met criteria for inclusion (Table 1). In participating hospitals in the Northeast 316/453 (70%) CDC-defined CRE were CPE as compared to 136/308 (44%) in the South (difference 26%, 95% CI 19%–33%, p<0·001). Fifty-eight (601/1,040) percent of patients were admitted from home. Patients with CPE (213/618, 34%) were more likely to be admitted from long-term care settings as compared to patients with non-CP-CRE (26/194,13%, difference 21%, 95% CI 15%–27%) and U-CRE (42/228, 18%, difference 16% 95% CI 10%–22%), p<0·001. Compared to patients with U-CRE, patients with CPE had more chronic comorbid conditions (median CCI 3 [IQR=1–3] vs. 2 [IQR=1–4]) and were more acutely ill (median Pitt bacteremia score 3 [IQR=2–6] vs. 2 [IQR=0–5]).
Figure 1. CDC-defined CRE admission rates at participating hospitals.
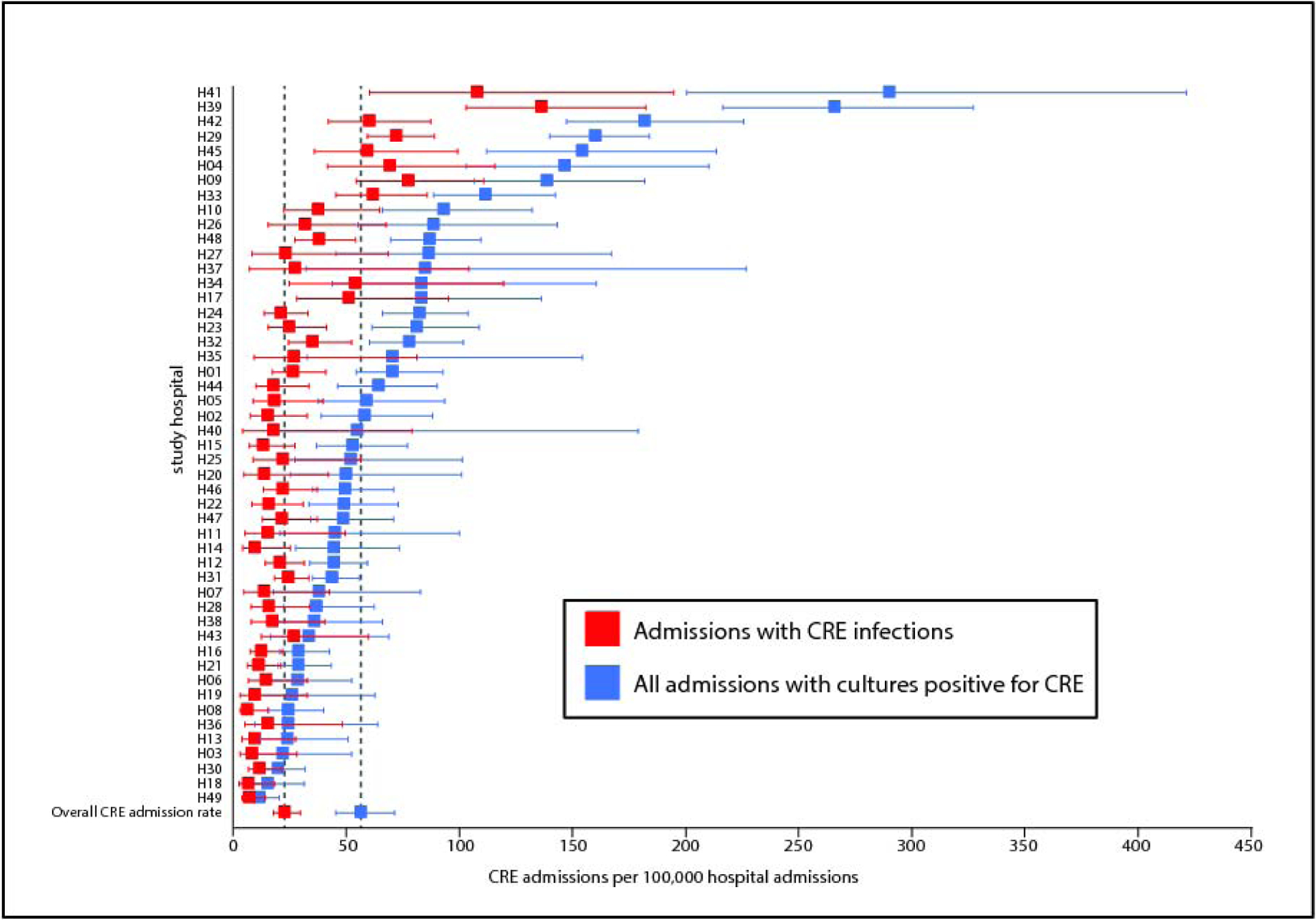
The rates of all admissions during which CRE were identified (blue), and of admissions during which at least one CRE infection was diagnosed (red) are shown. CDC-defined CRE admission rates and robust 95% CIs were estimated using a generalized linear mixed effects model with hospital as a random effect. Error bars indicate 95% confidence intervals.
Microbiologic characteristics
The most common source of CRE isolates was urine (404/1,040, 39%), followed by respiratory (268/1,040, 26%), blood (130/1,040, 13%), and wound (130/1,040, 13%). CRE infection was present in 449 (43%) patients, with the remaining 591 (57%) classified as CRE-colonization. Within the group of CDC-defined CRE-infected patients, those with non-CP-CRE were less likely to have urinary tract infections (20/102, 20%, difference 14%; 95% CI 4%–23%) and more likely to have abdominal infections (29/102, 28%, difference 21%; 95% CI 12%−30%) compared to patients with CPE (urine: 84/253, 33%; abdominal: 19/253, 8%). (p<0·001 for overall distribution). Species distribution and susceptibility to antibiotics as reported by the local microbiology laboratories are summarized in Table 2 and Supplementary Tables 3 and 4. Eighty-three percent (493/593) of K. pneumoniae identified as CDC-defined CRE by local laboratories were carbapenemase-producing Enterobacteriaceae, compared to 24% (46/192, difference 59%; 95% CI 52%–66%) of Enterobacter species and 31% (38/122, difference 52%; 95% CI 43%–61%) of Escherichia coli (p<0·001). For 105/228 (46%) of U-CRE, ertapenem was the only carbapenem with in vitro resistance as reported by the local microbiology laboratory, as compared to 4% (27/618, difference 42%; 95% CI 35%–48%) in carbapenemase-producing Enterobacteriaceae. MIC distribution, as determined in one of the central laboratories, is shown in Supplementary Figure 1. U-CRE and non-CP-CRE were more susceptible to non-carbapenem antibiotics, as compared to CPE (Supplementary Table 4). The 2012 CDC criteria for CRE would have defined 520/618 (84%) of carbapenemase-producing Enterobacteriaceae, but only 97/194 (50%, difference 34%, 95% CI 27%–42%) of non-CP-CRE, and 65/228 (29%, difference 56%, 95% CI 49%−62%) of U-CRE as CDC-defined CRE (p<0·001).
Table 2.
Bacterial characteristics including distribution of common β-lactamase genes.
| CPE (n=618) | Non-CP-CRE (n=194) | U-CRE (n=228) | All (n=1,040) | P value | |
|---|---|---|---|---|---|
| Species | <0·001 | ||||
| K. pneumoniae | 493 (80) | 52 (27) | 48 (21) | 593 (57) | |
| ST258 K. pneumoniae | 334 (54) | 6 (3) | 4 (2) | 344 (33) | |
| Enterobacter spp. | 46 (7) | 73 (38) | 73 (32) | 192 (18) | |
| E. coli | 38 (6) | 33 (17) | 51 (22) | 122 (12) | |
| ST131 E. coli | 22 (4) | 13 (7) | 24 (11) | 59 (6) | |
| non-K. pneumoniae Klebsiella spp. | 14 (2) | 26 (13) | 16 (7) | 56 (5) | |
| other | 27 (4) | 10 (5) | 40 (18) | 77 (7) | |
| Meets 2012 CDC criteria for CRE | 520 (84) | 97 (50) | 65 (29) | 682 (66) | <0·001 |
| Carbapenemasesa | |||||
| blaKPC-2 | 313 (51) | 313 (30) | |||
| blaKPC-3 | 253 (41) | 253 (24) | |||
| other blaKPCb | 7 (1) | 7 (1) | |||
| blaNDM-1 | 15 (2) | 15 (1) | |||
| other blaNDMc | 7 (1) | 7 (1) | |||
| blaOXA-48 | 6 (1) | 6 (1) | |||
| Other blaOXA-48-liked | 15 (2) | 15 (1) | |||
| Othere | 10 (2) | 10 (1) | |||
| Extended spectrum β-lactamase | |||||
| blaCTX-M | 121 (20) | 59 (30) | 45 (20) | 225 (22) | 0·004 |
| blaSHVf | 217 (35) | 19 (10) | 14 (6) | 250 (24) | <0·001 |
| blaTEMg | 0 (0) | 0 (0) | 1 (<1) | 1 (<1) | 0·41 |
| blaAmpC | 116 (19) | 112 (58) | 141 (62) | 369 (35) | <0·001 |
Totals exceed 100%, as 8 isolates carried more than one carbapenemase gene.
Other blaKPC included blaKPC-4 (3), blaKPC-6 (1), blaKPC-8 (1), and blaKPC-18 (2).
Other blaNDM included blaNDM-5 (6), blaNDM-7 (1).
Other blaoxa-48-like included blaoxa-181 (2), blaoxa-232 (13).
other carbapenemases included blaVIM (4), blaIMI (2), blaSME (3), blaNMC-A (1).
Those blaSHV that are considered extended spectrum β-lactamase genes, including blaSHV-12 (217), blaSHV-7 (12), blaSHV-30 (11), blaSHV-2 (5), blaSHV-5 (4), and blaSHV-105 (1).
blaTEM-10. CPE: carbapenemase-producing Enterobacteriaceae. Non-CP-CRE: non-carbapenemase-producing carbapenem-resistant Enterobacteriaceae (CRE). U-CRE: unconfirmed CRE.
Molecular epidemiology and bacterial population structure
An interactive version of Figure 2 is available online (http://arlg.med.unc.edu/crackle/password:B@ct3r1@). Observed carbapenemase genes (Table 2, Supplementary Tables 3 and 5, and Figure 2) included blaKPC-2 (313/618, 51%), blaKPC-3 (253/618, 41%), blaNDM (22/618, 3%), and blaOXA-48-like (21/618, 3%). Extended spectrum β-lactamase genes found in CPE included extended spectrum β-lactamase blaSHV (217/618, 35%) and blaCTX-M (121/618, 20%). Non-CP-CRE were more likely to carry blaCTX-M (59/194, 30%, difference 11%; 95% CI 4%–18%), whereas blaAmpC carriage was associated with both non-CP-CRE (112/194, 58%) and U-CRE (141/228, 62%). Mutations in either or both genes encoding outer membrane porins OmpK35 and OmpK36 were present in 120/194 (62%) of non-CP-CRE and 49/228 (21%) of U-CRE.
Figure 2. Phylogenetic population structures.
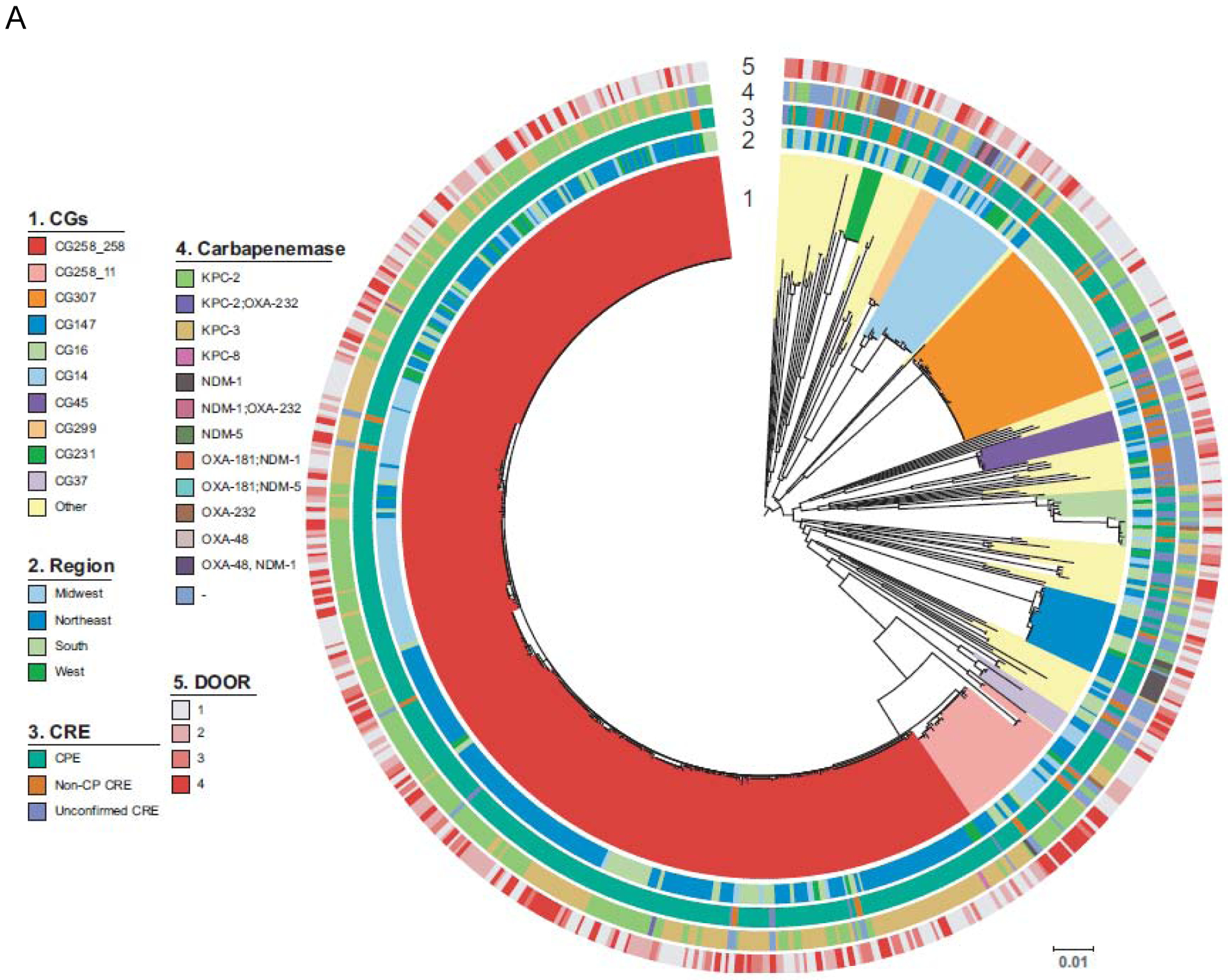
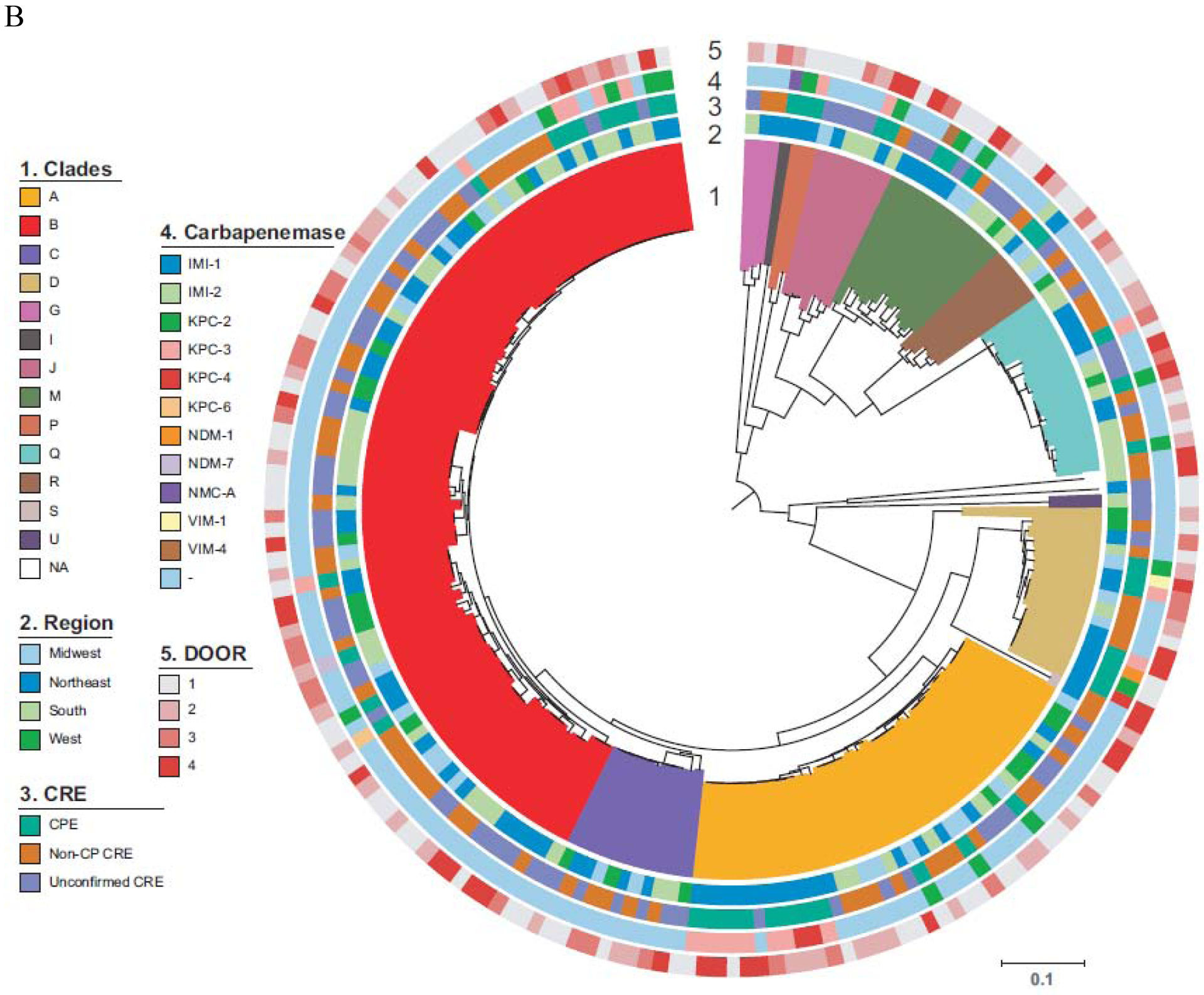

Circle 1 shows clonal groups (CG, 2.A.), Genomic clusters (2.B.), and Sequence Types (ST, 2.C.), Circle 2. indicates region of origin, Circle 3 CPE vs. non-CP-CRE vs. U-CRE, Circle 4 indicates carbapenemase present where applicable and Circle 5 indicates the DOOR level (Level 1; alive without deleterious events. Level 2; alive with 1 deleterious event. Level 3; alive with 2 or 3 deleterious events. Level 4; death). A. K. pneumoniae B. Enterobacter spp. C. E. coli. Of note, an interactive version of this Figure is available online (http://arlg.med.unc.edu/crackle/ password: B@ct3r1@).
The most common clonal group (CG) of K. pneumoniae was CG258 (382/593, 64%), representing 364/493 (74%) of carbapenemase-producing K. pneumoniae (Figure 2.A.). Of 382 CG258 K. pneumoniae, 364 (95%) were carbapenemase-producing, harboring primarily blaKPC-2 (200, 55%) and blaKPC-3 (161, 44%). Among carbapenemase producing CG258 K. pneumoniae isolates, ST258 encompassed 92% of isolates (334/364). After CG258, the most frequent clonal group was CG307 (44/593, 7%), concentrated in the participating hospitals of Houston, TX. Similar to CG258, 37 (84%) of 44 CG307 isolates were carbapenemase-producing, with blaKPC-2 detected in 35 (95%) of these 37 isolates. All CG307 harbored blaCTX-M, a common group of extended-spectrum β-lactamases. Geographic distribution of ST307 K. pneumoniae is shown in Supplementary Figure 2. Enterobacter isolates were the second most frequent group of CDC-defined CRE (Figure 2.B.). Due to the observed genetic heterogeneity of the Enterobacter cloacae complex, genetic clades were used to show the population structure of Enterobacter isolates.14 In 146/192 (76%) of Enterobacter species no carbapenemase genes were present. In the remaining 46/192 (24%), blaKPC-2 (n=16), blaKPC-3 (n=19), and typical carbapenemase genes previously described in Enterobacter species were found (blaIMI-1, blaIMI-2, and blaNMC-A; one of each). Additionally, various metallo-β-lactamases genes were identified, including blaNDM-1, blaNDM-7, blaVIM-1, and blaVIM-4 (one of each).
In E. coli, diverse genetic lineages were observed (Figure 2.C.). In 84/122 (69%) E. coli, no carbapenemase genes were present. ST131 accounted for 37/122 (30%), and was present in hospitals belonging to all geographical areas studied. The most common carbapenemases in E. coli were blaKPC-2 (n=15), and blaKPC-3 (n=14), with sporadic isolates containing blaNDM-5 (n=4), blaOXA-232 (n=2), and blaOXA-48 (n=3).
Phylogenetic reconstructions of K. pneumoniae, E. coli and Enterobacter spp. comparing infecting vs colonizing isolates (Supplementary Figure 3) indicate that both groups of isolates are highly related suggesting that infecting isolates are likely originating from initial colonization events.
Clinical outcomes
Of the total 1,040 patients, 449 (43%) met criteria for CDC-defined CRE infection. Using DOOR outcomes at 30 days after index culture, 183 (41%) patients with infection were alive without events, 97 (22%) alive with one event, 62 (14%) alive with 2 or 3 events, and 107 (24%) were dead (Table 3). Outcomes were not significantly different between groups after adjusting for possible confounding factors. Inverse probability weighted (IPW) DOOR analyses indicated no significant differences between groups. In DOOR analysis, 50% likelihood of a better outcome is equal to no difference between groups, whereas a greater than 50% probability combined with a 95% confidence interval that does not cross 50%, indicates a significantly higher likelihood of a better outcome in one group versus the other. IPW-adjusted probabilities of a patient with CPE vs. non-CP-CRE, CPE vs. U-CRE, and non-CP-CRE vs U-CRE having a better outcome are 52% (95% CI 45%–58%), 52% (95% CI 44%–61%), and 51% (95% CI 41%–60%), respectively. In the subset of patients with invasive infections (pneumonia, bacteremia, or intra-abdominal infection, n=256), there was also no significant difference in DOOR outcome (IPW-adjusted probability of a better outcome of 54%, 95% CI=42%–66%). Similarly, in DOOR analyses stratified based on Pitt Bacteremia score, and when time from admission to first positive culture was included as an additional IPW-confounder, differences between CRE groups were not observed (Supplementary Tables 6 and 7). Likewise, when limiting IPW-adjusted DOOR analysis to 238 patients infected with K. pneumoniae, no significant difference between groups was observed (data not shown). All-cause 30-day and 90-day mortality in patients with CDC-defined CRE infections was 24% (107/449, 95% CI 20%–28%), and 31% (137/449, 95% CI 26%–35%), respectively (Table 3). Mortality rates were not significantly different between patients infected with carbapenemase-producing Enterobacteriaceae, non-CP-CRE, and U-CRE. Among 325 patients discharged alive after CDC-defined CRE infection, 150 (46%, 95% CI 41%–52%) were readmitted within 90 days, with a median time to readmission of 21 days (IQR 8–44 days). Antibiotic treatment is outlined in Supplementary Table 8. In patients with U-CRE, 204/449 (38%) and 155/414 (37%) received a carbapenem as part of their empiric or definitive treatment regimen, respectively. In 591 patients with CDC-defined CRE colonization, 30-day mortality was 111/591 (19%, 95% CI 16%–22%). The 90-day readmission rate in 469 patients with CDC-defined CRE colonization who were discharged alive was 186/469 (40%, 95% CI 35%–44%).
Table 3.
Outcomes in patients with CRE infections.
| CPE (n=253) | Non-CP-CRE (n=102) | U-CRE (n=94) | All (n=449) | P value | |
|---|---|---|---|---|---|
| DOOR at 30 days | n/ad | ||||
| alive without events | 106 (42) | 37 (36) | 40 (43) | 183 (41) | |
| alive with 1 event | 53 (21) | 26 (25) | 18 (19) | 97 (22) | |
| alive with 2 or 3 events | 31 (12) | 17 (17) | 14 (15) | 62 (14) | |
| dead | 63 (25) | 22 (22) | 22 (23) | 107 (24) | |
| DOOR components at 30 daysa | |||||
| not discharged | 103 (41) | 45 (44) | 36 (38) | 184 (41) | 0·70 |
| readmitted | 32 (13) | 12 (12) | 14 (15) | 58 (13) | 0·79 |
| lack of clinical response | 86 (34) | 35 (34) | 32 (34) | 153 (34) | >0·99 |
| lack of symptomatic response | 74 (29%) | 29 (28) | 28 (30) | 131 (29) | 0·98 |
| relapse | 12 (5) | 3 (3) | 3 (3) | 18 (4) | 0·66 |
| remains on anti-CRE antibiotic | 8 (3) | 9 (9) | 7 (7) | 24 (5) | 0·06 |
| renal failure | 13 (5) | 5 (5) | 5 (5) | 23 (5) | 0·99 |
| C. difficile infection | 3 (1) | 2 (2) | 0 (0) | 5 (1) | 0·42 |
| LOS, days, median (IQR) | 19 (9, 38) | 29 (12, 60) | 15 (6, 35) | 20 (8, 45) | 0·002 |
| Post-culture LOS, days, median (IQR) | 11 (5, 22) | 16 (6, 26) | 10 (4, 19) | 12 (5, 23) | 0·02 |
| 30-day mortality | 63 (25) | 22 (22) | 22 (23) | 107 (24) | 0·80 |
| 90-day mortality | 79 (31) | 33 (32) | 25 (27) | 137 (31) | 0·64 |
| 90-day readmissionsb | 81/183 (44%) | 37/69 (54%) | 32/73 (44%) | 150/325 (46%) | 0·37 |
| clinical response | 167 (66%) | 67 (66%) | 62 (66%) | 296 (66%) | >0·99 |
| Dispositionc | 0·03 | ||||
| death | 63 (25) | 27 (26) | 19 (20) | 109 (24) | |
| home | 72 (28) | 38 (37) | 40 (43) | 150 (33) | |
| hospice | 7 (3) | 6 (6) | 2 (2) | 15 (3) | |
| long term acute care | 29 (11) | 7 (7) | 2 (2) | 38 (8) | |
| long term care | 71 (28) | 21 (21) | 24 (26) | 116 (26) | |
| transfer other hospital | 11 (4) | 3 (3) | 6 (6) | 20 (4) | |
| transferred to a foreign country | 0 (0) | 0 (0) | 1 (1) | 1 (<1) |
Desirability of outcome ranking (DOOR) analysis components as defined in Supplementary Materials.
in patients discharged alive. LOS length of hospital stay.
Grouped for analysis purposes as death/hospice, home/transferred to a foreign country, LTAC/transfer other hospital, long-term care.
Inverse probability weighted (IPW) DOOR analyses indicated no significant differences between groups. IPW-adjusted probabilities of a patient with CPE vs. non-CP-CRE, CPE vs. U-CRE, non-CP-CRE vs U-CRE having a better outcome are 52% (95% CI 45%–58%), 52% (95% CI 44%–61%), 51% (95% CI 41%–60%). CPE: carbapenemase-producing Enterobacteriaceae. Non-CP-CRE: non-carbapenemase-producing carbapenem-resistant Enterobacteriaceae (CRE). U-CRE: unconfirmed CRE.
DISCUSSION
In this contemporary analysis of CRE in hospitalized US patients, three clinically and molecularly distinct subsets of CRE were identified. CPE are generally considered of the greatest epidemiologic interest for their association with poor outcomes and ability to spread rapidly throughout healthcare systems. However, in this cohort, 41% of isolates meeting the current CDC guidelines did not carry carbapenemase genes, and 22% were not carbapenem-resistant upon centralized laboratory re-testing. Thus, resources dedicated to halting the spread of CPE may therefore be directed at bacteria of lesser public health concern. Correct identification of carbapenemase production at the patient, hospital, regional and national levels is important for treatment selection, infection control, and prevention of spread.
Clinical outcomes were not significantly different regardless of infection with carbapenemase-producing Enterobacteriaceae, non-CP-CRE, or U-CRE. As most CPE are KPC-producing ST258 K. pneumoniae, this comparison is primarily between these strains and a genetically much more diverse group of Enterobacteriaceae of various species. Three non-mutually exclusive explanations for this finding may be considered. First, current CDC criteria may identify patients with infections that – regardless of the underlying mechanism of carbapenem resistance and/or in vitro reproducibility of the phenotype – are associated with high risk of mortality and readmissions. Second, improved treatment options for CPE infections that were available during the study period may have decreased the difference in patient outcomes predicted based on earlier studies. Third, the label of “CRE” may lead to unnecessary treatment with more toxic and/or less effective antibiotics. A retrospective, single-center study evaluated 83 patients with CDC-defined CRE bacteremia diagnosed between 2013 and 2016. In that cohort, infection with CPE was compared to non-CP-CRE. While limited by a small sample size, and inclusion of only bacteremia cases, infection with CPE was marginally associated with both increased 14-day (aOR 4·92, 95% CI 1·01–24·81, p=0·05) and 30-day mortality (aOR 3·19, 95% CI 0·99–10·25; p=0·05).15 Ceftazidime-avibactam – superior to polymyxins in the treatment of CRE infections – was not available during that study.11,15,16 Thus, it is possible that treatment of high-risk patients with CPE infections with ceftazidime-avibactam resulted in improved outcomes.11 However, only a subset of patients with CPE received ceftazidime-avibactam as empiric (17%), and definitive therapy (23%), respectively. Furthermore, 95% of patients with non-CP-CRE received a carbapenem in that single-center study, as compared to less than 40% in the current study. Carbapenems are superior to piperacillin/tazobactam and are considered by many to be the preferred treatment for patients with severe infections with ceftriaxone-resistant, carbapenem-susceptible Enterobacteriaceae.15,17
The high rate of non-CPE amongst CDC-defined CRE appears to be a direct consequence of the change in definition implemented in 2015. When we applied the 2012 definition in this dataset, the percentage of CDC-defined CRE without carbapenemases decreased to 24% (162/682). Based on similar outcomes between patients infected with different subgroups of CDC-defined CRE, this broad definition may well be justified. However, control of infections with bacteria in these various subgroups may not respond to the same infection prevention and control strategies. In a 2017 CDC surveillance study, 68% of CRE were non-carbapenemase producers.18 In addition, 22% of CDC-defined CRE were not carbapenem resistant upon central testing. The simplest explanation for these U-CRE is that they reflect major errors of automated carbapenem susceptibility testing, specifically when using ertapenem. In addition, U-CRE may in part represent isolates with ertapenem MICs close to breakpoints, in which a single dilution difference might change the interpretation of susceptibility from resistant to intermediate. However, when tested in the central laboratory, ertapenem MICs ranged widely in these isolates. Another explanation is loss of resistance genes during transport and passage. However, loss of a carbapenemase-containing plasmid would not explain the high rates of meropenem and imipenem susceptibility observed at the contributing local microbiology laboratories. Also, as U-CRE are found in patients, who are clinically different from patients with carbapenemase-producing Enterobacteriaceae, and display a species distribution and non-carbapenem susceptibility pattern distinct from carbapenemase-producing Enterobacteriaceae, a stochastic random event is unlikely to explain the observed U-CRE. Regardless, infection with U-CRE seems to be an indicator of increased risk of mortality to the same extent as infection with carbapenemase-producing Enterobacteriaceae.
In carbapenemase-producing Enterobacteriaceae, CG 258 K. pneumoniae containing blaKPC remains the most common.19 In addition, ST307 is now the most common K. pneumoniae lineage containing blaCTX-M and blaKPC in the Houston area, supporting the introduction of this novel high-risk clone. Previous reports suggest that ST307 is likely to follow a similar pattern of spread to CG258.20,21
Treatment-emergent resistance to ceftazidime-avibactam has been reported in ST307 K. pneumoniae, similar to CG258 strains.22,23 Additional mechanisms of resistance were also seen, such as CRE containing blaOXA-48-like and genes encoding metallo-β-lactamases. Horizontal gene transfer may cause spread of blaNDM, blaVIM, and blaOXA in a comparable manner to blaKPC.
The most common genetic lineage of E. coli in the CRE isolates was a highly related clade of ST131. Given that ST131 is the most common E. coli lineage among pathogenic isolates causing extra-intestinal disease worldwide, acquisition of carbapenem resistance among these isolates is concerning.24,25 Most disturbing are those ST131 E. coli, which have acquired a carbapenemase gene, as these clones have high potential for causing severe invasive disease.26,27
Limitations.
This study has several limitations. First, hospitals were selected based on interest of site investigators, rather than as a random selection. Small hospitals were underrepresented in our study hospitals and large teaching hospitals were overrepresented. Therefore, these findings should not be extrapolated to hospitals with fewer than 100 beds. However, the study hospitals represented a wide range of sizes, ownership models, community vs. tertiary care, as well as a wide variety in CRE admission rates. Second, this was a consent-waived study, and only EHR data were included. This approach allows for unbiased, sequential inclusion, regardless of ability to provide consent. Third, patients and isolates were compared in three groups for several variables, which may introduce problems with multiple comparisons. However, the primary outcome variable – the DOOR analysis – was a priori defined, and no significant difference was observed between groups. Fourth, our sampling was limited to a single country. The epidemiology of CRE in other parts of the world may be substantially different. Ongoing studies in the MDRO Network are evaluating the international epidemiology of CRE.
Conclusions.
In the US, Klebsiella pneumoniae carbapenemase (KPC)-producing clonal group 258 K. pneumoniae is the most common carbapenemase-producing Enterobacteriaceae. Among US patients with CDC-defined CRE in 49 hospitals in 26 sites, there were similar clinical outcomes among three subgroups, including the novel U-CRE group. These data provide guidance for clinical practice and public health policy; CDC-defined CRE represent a diverse group of bacteria, whose spread may not respond to interventions directed solely to carbapenemase-producing Enterobacteriaceae. Regardless of CRE subgroup, CDC-defined CRE infections are associated with poor outcomes.
Supplementary Material
Research in context.
Evidence before this study
Resistance to carbapenems in Enterobacteriaceae is a threat to global public health. We searched MEDLINE (1966 to July 1, 2019) and Google Scholar (1966 to July 1, 2019), using the terms “carbapenem resistant Enterobacteriaceae”, “carbapenemase”, and “mortality”. Our search identified reports of surveillance studies conducted by the US Centers for Disease Control and Prevention (CDC). These reports indicate that a subset of CDC-defined CRE in the US do not produce carbapenemases. Data from large retro spective cohorts of patients with carbapenemase–producing Enterobacteriaceae (CPE) from Europe, US and China indicate a predominance of KPC-producing K. pneumoniae. Retrospective multi-center data on CPE bloodstream infections show a 30-day all-cause mortality of 43%. Furthermore, in low-income and middle-income countries, carbapenem resistance is associated with a 15% absolute increase in in-hospital mortality among inpatients with a bloodstream infection due to Enterobacteriaceae. A single-center, retrospective study from the US suggested that infection with carbapenemase-producing CRE (CPE) is associated with increased mortality as compared to non-carbapenemase-producing CRE (non-CP-CRE).
Added value of this study
In this study, we provide comprehensive clinical and whole genome sequencing (WGS) data for a cohort of 1,040 unique patients with CDC-defined CRE. In addition to CPE, and non-CP-CRE, we identified a novel subset of CDC-defined CRE. These unconfirmed CRE met criteria for CRE at the clinical lab, but were found to be carbapenem-susceptible in two central laboratories. Clinical outcomes in patients infected with these three subsets were similar. Analyses of WGS data reveal that clonal group 258 Klebsiella pneumoniae remains the most common CPE. However, clonal group 307 is also on the rise.
Implications of all the available evidence
Among US patients with CDC-defined CRE, there were similar clinical outcomes among three subgroups, including the novel unconfirmed CRE group. CDC-defined CRE represent a diverse group of bacteria, whose spread may not respond to interventions directed solely to carbapenemase-producing Enterobacteriaceae.
Acknowledgments:
Dr van Duin had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Role of Funder: This study is supported by the National Institute of Allergy And Infectious Diseases of the National Institutes of Health (NIAID) under Award Number UM1AI104681. NIAID had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript nor the decision to submit the manuscript for publication, or to veto publication, or to control which journal the paper was submitted to.
The investigators would like to thank all the patients and their families. This publication made use of the PubMLST website (https://pubmlst.org/) developed by Keith Jolley, at the University of Oxford.28 The development of that website was funded by the Wellcome Trust. This work was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under Award Number UM1AI104681, and number R21AI114508. VGF was supported by a Mid-Career Mentoring Award 2K24-AI093969 from the National Institutes of Health. In addition, research reported in this publication was supported in part by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under Award Numbers R01AI143910 (DvD), R01AI090155 (BNK), R21AI135250 (BNK), R21AI117338 (LC), K08-AI113317 (TTT), R01AI100560 (RAB), R01AI063517 (RAB), R01AI072219 (RAB), K24AI121296 (CAA), R01AI134637 (CAA), R21AI143229 (CAA). This study was supported in part by funds and/or facilities provided by the Cleveland Department of Veterans Affairs, Award Number 1I01BX001974 to RAB from the Biomedical Laboratory Research & Development Service of the VA Office of Research and Development and the Geriatric Research Education and Clinical Center VISN 10 (RAB) and UTHealth Searle Award (BH) and UTHealth Presidential Collaborative Award (CAA). YD was supported by research awards R01AI104895, R21AI123747, and R21AI135522 from the NIH. KSK is supported by the National Institute of Allergy and Infectious Diseases (Division of Microbiology and Infectious Diseases protocol 10-0065 and R01AI119446).
Conflict of Interest:
D.v.D.: Allergan, Achaogen, Qpex, Shionogi, Tetraphase, Sanofi-Pasteur, T2 Biosystems, NeuMedicine, Roche, MedImmune, Astellas, Merck Advisory Board. Travel reimbursement from IDSA, ASM and ESCMID. C.A.A. Grant support from Merck, MeMed Diagnostics and Entasis Therapeutics. Royalties from UptoDate, Harrison’s Principles of Internal Medicine and Mandell’s Principles and Practice of Infectious Diseases and reimbursement for Travel from IDSA and ASM. G.W.: Research support from Allergan. S.S.R: Research support from bioMerieux, BD Diagnostics, BioFire, OpGen, Hologic, Diasorin, Accelerate and Roche. M.J.S.: Advisory Board: Achaogen, Shionogi; Grant support from Merck and Allergan. Y.D.: Grant support: The Medicines Company, Accelerate Diagnostics, NIH. Advisory board: Meiji, Tetraphase, Roche, Geom. K.S.K: Allergan, Consultant, Grant Investigator and Speaker’s Bureau, Consulting fee, Grant recipient and Speaker honorarium. Merck – Grant recipient, consultant. Xellia – consultant. Achaogen – consultant. J.C.G.: Speaker: Allergan, Melinta, Merck. Consultant: Achaogen, Tetraphase, Melinta, Merck, grant: Merck L.M.A.: Honoraria: Pfizer, MSD, Merck; Paid Advisory Board: Achaogen, Nabriva, Roche Diagnostics. R.P. Research support from CD Diagnostics, bioMerieux, BioFire, Curetis, Merck, Contrafect, Hutchison Biofilm Medical Solutions, Accelerate Diagnostics, Allergan, EnBiotix, Contrafect and The Medicines Company, is or has been a consultant to Curetis, Specific Technologies, Selux Dx, GenMark Diagnostics, Roche, PathoQuest, Heraeus Medical, and Qvella (monies are paid to Mayo Clinic), has a patent on Bordetella pertussis/parapertussis PCR issued, a patent on a device/method for sonication with royalties paid by Samsung to Mayo Clinic, and a patent on an anti-biofilm substance issued, and received travel reimbursement from ASM and IDSA, an editor’s stipend from ASM and IDSA, and honoraria from the NBME, Up-to-Date and the Infectious Diseases Board Review Course. F.P.: Grants from Accelerate, Merck, Pfizer. J.J.L.: consultancy: Merck. D.L.P.: Board membership: Merck, Pfizer, Shionogi, Achaogen, AstraZeneca, Leo Pharmaceuticals, Bayer, GlaxoSmithKline, Cubist, Venatorx, Accelerate, grants from Shionogi, Merck (MSD), speaker bureau: Pfizer. S.E.: consultancy: Takeda / Millennium, Pfizer, Roche, Novartis, Achaogen, ACTTION, Genentech, Amgen, GSK, AstraZeneca, Teva, Zeiss, Dexcom, Claret Medical, Vir, Arrevus, Five Prime, Shire, Alexion, Gilead, Spark, Nuvelution, Tracon, WAVE, Advantagene, Braeburn, Cardinal Health, Lipocine, Microbiotix, Stryker. V.G.F.: Grant/Research Support: MedImmune, Cerexa/Forest/Actavis/Allergan, Pfizer, Advanced Liquid Logics, Theravance, Novartis, Cubist/Merck; Medical Biosurfaces; Locus; Affinergy; Contrafect; Karius; Genentech, Regeneron, BasileaPaid Consultant: Pfizer, Novartis, Galderma, Novadigm, Durata, Debiopharm, Genentech, Achaogen, Affinium, Medicines Co., Cerexa, Tetraphase, Trius, MedImmune, Bayer, Theravance, Cubist, Basilea, Affinergy, Janssen, xBiotech, Contrafect, Regeneron, Basilea, Destiny. Membership: Merck Co-Chair V710 Vaccine. Educational fees: Green Cross, Cubist, Cerexa, Durata, Theravance; Debiopharm. Royalties: UpToDate. R.A.B.: Grant/Research Support: Achaogen, Allecra, Entasis, Merck, Roche, Shionogi, Wockhardt. All remaining authors have nothing to disclose.
MDRO Network Investigators (listed by center alphabetically): Albert Einstein College of Medicine, Bronx, New York, United States of America: Gregory Weston, Belinda Ostrowsky; Boston University, Boston, Massachusetts, United States of America: Judith J. Lok; Case Western Reserve University, Cleveland, Ohio, United States of America: Robert A. Bonomo, T. Nicholas Domitrovic, Kristine M. Hujer, Andrea M. Hujer, Susan D. Rudin, Steven H. Marshall, Robert A. Salata, Federico Perez; Cleveland Clinic, Cleveland, Ohio, United States of America: Eric Cober, Sandra S. Richter; Duke Clinical Research Institute: Rebekka Arias, Carol Hill; Duke University, Durham, North Carolina, United States of America: Vance G. Fowler Jr., Deverick J. Anderson; Emory University, Atlanta, Georgia, United States of America: Jesse T. Jacob; Fudan University, Shanghai, China: Minggui Wang; Hackensack Meridian Health, Nutley, New Jersey, United States of America: Liang Chen, Samit Desai, Barry N. Kreiswirth, Claudia Manca, Jose R. Mediavilla; Icahn School of Medicine at Mount Sinai, New York, New York, United States of America: Gopi Patel; Mayo Clinic, Rochester, Minnesota, United States of America: Robin Patel; Medical College of Wisconsin and Froedtert Memorial Lutheran Hospital, Milwaukee, Wisconsin, United States of America: Sara Revolinski; MedStar Washington Hospital Center, Washington, District of Columbia, United States of America: Glenn Wortmann; MetroHealth Medical Center, Cleveland, Ohio, United States of America: Robert C. Kalayjian; North Shore University Hospital, Manhasset, New York, United States of America: Angela Kim; Ochsner Clinic Foundation, New Orleans, Louisiana, United States of America: Julia Garcia-Diaz; Stony Brook University, Stony Brook, New York, United States of America: Bettina C. Fries; SUNY Downstate Medical Center, New York, New York, United States of America: Brandon Eilertson; Temple University School of Pharmacy, Philadelphia, Pennsylvania, United States of America: Jason C. Gallagher; The George Washington University, Rockville, Maryland, United States of America: Michelle Earley, Scott Evans, Lauren Komarow; University of California, Los Angeles, California, United States of America: Omai B. Garner; University of California San Francisco, San Francisco, California, United States of America: Henry F. Chambers; University of Illinois College of Medicine at Peoria, Peoria, Illinois, United States of America: John J. Farrell; University of Miami Miller School of Medicine and Jackson Health System, Miami, Florida, United States of America: Lilian M. Abbo; University of Michigan, Ann Arbor, Michigan, United States of America: Keith S. Kaye; University of North Carolina, Chapel Hill, North Carolina, United States of America: Courtney Luterbach, David van Duin; University of Pennsylvania, Philadelphia, Pennsylvania, United States of America: Jennifer H. Han; University of Pittsburgh, Pittsburgh, Pennsylvania, United States of America: Yohei Doi; University of Queensland, Queensland, Australia: David L. Paterson; University of Southern California, Los Angeles, California, United States of America: Darren Wong; University of Texas Health Science Center at Houston, Houston, Texas, United States of America: Cesar A. Arias, Blake Hanson, An Dinh, Diana Panesso, William Shropshire, Truc T. Tran; Vanderbilt University, Nashville, Tennessee, United States of America: Ritu Banerjee; Wayne State University, Detroit, Michigan, United States of America: Sorabh Dhar; Weill Cornell Medicine, New York, New York, United States of America: Michael J. Satlin; Yale School of Medicine, New Haven, Connecticut, United States of America: Matthew Grant.
| MDRO Investigators (alphabetical by surname) | ||
|---|---|---|
| First name | Middle initial | surname |
| Lilian | M. | Abbo |
| Deverick | J. | Anderson |
| Rebekka | Arias | |
| Cesar | A. | Arias |
| Ritu | Banerjee | |
| Robert | A. | Bonomo |
| Henry | F. | Chambers |
| Liang | Chen | |
| Eric | Cober | |
| Samit | Desai | |
| Sorabh | Dhar | |
| An | Dinh | |
| Yohei | Doi | |
| T. Nicholas | Domitrovic | |
| Michelle | Earley | |
| Brandon | Eilertson | |
| Scott | Evans | |
| John | J. | Farrell |
| Vance | G. | Fowler |
| Bettina | C. | Fries |
| Jason | C. | Gallagher |
| Julia | Garcia-Diaz | |
| Omai | B. | Garner |
| Matthew | Grant | |
| Jennifer | H. | Han |
| Blake | Hanson | |
| Carol | Hill | |
| Kristine | M. | Hujer |
| Andrea | M. | Hujer |
| Jesse | T. | Jacob |
| Robert | C. | Kalayjian |
| Keith | S. | Kaye |
| Angela | Kim | |
| Lauren | Komarow | |
| Barry | N. | Kreiswirth |
| Judith | J. | Lok |
| Courtney | Luterbach | |
| Claudia | Manca | |
| Steven | H. | Marshall |
| Jose | R. | Mediavilla |
| Belinda | Ostrowsky | |
| Diana | Panesso | |
| Gopi | Patel | |
| Robin | Patel | |
| David | L. | Paterson |
| Federico | Perez | |
| Sara | Revolinski | |
| Sandra | S. | Richter |
| Susan | D. | Rudin |
| Robert | A. | Salata |
| Michael | J. | Satlin |
| William | Shropshire | |
| Truc | T. | Tran |
| David | van Duin | |
| Minggui | Wang | |
| Gregory | Weston | |
| Darren | Wong | |
| Glenn | Wortmann | |
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Publisher's Disclaimer: Disclaimer. The contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health or the Department of Veterans Affairs.
REFERENCES
- 1.van Duin D, Doi Y. The global epidemiology of carbapenemase-producing Enterobacteriaceae. Virulence 2017; 8(4): 460–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Logan LK, Weinstein RA. The Epidemiology of Carbapenem-Resistant Enterobacteriaceae: The Impact and Evolution of a Global Menace. J Infect Dis 2017; 215(suppl_1): S28–S36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organization. WHO priority pathogens list for R&D of new antibiotics. 2017; http://www.who.int/mediacentre/news/releases/2017/bacteria-antibiotics-needed/en/.
- 4.Haverkate MR, Bootsma MC, Weiner S, et al. Modeling spread of KPC-producing bacteria in long-term acute care hospitals in the Chicago region, USA. Infect Control Hosp Epidemiol 2015; 36(10): 1148–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bush K. Carbapenemases: Partners in crime. J Glob Antimicrob Resist 2013; 1(1): 7–16. [DOI] [PubMed] [Google Scholar]
- 6.Gutierrez-Gutierrez B, Salamanca E, de Cueto M, et al. Effect of appropriate combination therapy on mortality of patients with bloodstream infections due to carbapenemase-producing Enterobacteriaceae (INCREMENT): a retrospective cohort study. Lancet Infect Dis 2017; 17(7): 726–34. [DOI] [PubMed] [Google Scholar]
- 7.Stewardson AJ, Marimuthu K, Sengupta S, et al. Effect of carbapenem resistance on outcomes of bloodstream infection caused by Enterobacteriaceae in low-income and middle-income countries (PANORAMA): a multinational prospective cohort study. Lancet Infect Dis 2019; 19(6): 601–10. [DOI] [PubMed] [Google Scholar]
- 8.Wang Q, Wang X, Wang J, et al. Phenotypic and Genotypic Characterization of Carbapenem-resistant Enterobacteriaceae: Data From a Longitudinal Large-scale CRE Study in China (2012–2016). Clin Infect Dis 2018; 67(suppl_2): S196–S205. [DOI] [PubMed] [Google Scholar]
- 9.Guh AY, Bulens SN, Mu Y, et al. Epidemiology of Carbapenem-Resistant Enterobacteriaceae in 7 US Communities, 2012–2013. JAMA 2015; 314(14): 1479–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Centers for Disease Control and Prevention. Facility Guidance for Control of Carbapenem-resistant Enterobacteriaceae (CRE) November 2015 Update - CRE Toolkit https://wwwcdcgov/hai/pdfs/cre/CRE-guidance-508pdf 2015; last accessed 4/1/2019.
- 11.van Duin D, Lok JJ, Earley M, et al. Colistin Versus Ceftazidime-Avibactam in the Treatment of Infections Due to Carbapenem-Resistant Enterobacteriaceae. Clin Infect Dis 2018; 66(2): 163–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Duin D, Perez F, Rudin SD, et al. Surveillance of Carbapenem-Resistant Klebsiella pneumoniae: Tracking Molecular Epidemiology and Outcomes through a Regional Network. Antimicrob Agents Chemother 2014; 58(7): 4035–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Evans SR, Rubin D, Follmann D, et al. Desirability of Outcome Ranking (DOOR) and Response Adjusted for Duration of Antibiotic Risk (RADAR). Clin Infect Dis 2015; 61(5): 800–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoffmann H, Roggenkamp A. Population genetics of the nomenspecies Enterobacter cloacae. Appl Environ Microbiol 2003; 69(9): 5306–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tamma PD, Goodman KE, Harris AD, et al. Comparing the Outcomes of Patients With Carbapenemase-Producing and Non-Carbapenemase-Producing Carbapenem-Resistant Enterobacteriaceae Bacteremia. Clin Infect Dis 2017; 64(3): 257–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shields RK, Nguyen MH, Chen L, et al. Ceftazidime-Avibactam Is Superior to Other Treatment Regimens against Carbapenem-Resistant Klebsiella pneumoniae Bacteremia. Antimicrob Agents Chemother 2017; 61(8): e00883–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harris PNA, Tambyah PA, Lye DC, et al. Effect of Piperacillin-Tazobactam vs Meropenem on 30-Day Mortality for Patients With E coli or Klebsiella pneumoniae Bloodstream Infection and Ceftriaxone Resistance: A Randomized Clinical Trial. JAMA 2018; 320(10): 984–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Woodworth KR, Walters MS, Weiner LM, et al. Vital Signs: Containment of Novel Multidrug-Resistant Organisms and Resistance Mechanisms - United States, 2006–2017. MMWR Morb Mortal Wkly Rep 2018; 67(13): 396–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kitchel B, Rasheed JK, Patel JB, et al. Molecular epidemiology of KPC-producing Klebsiella pneumoniae isolates in the United States: clonal expansion of multilocus sequence type 258. Antimicrob Agents Chemother 2009; 53(8): 3365–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Castanheira M, Farrell SE, Wanger A, Rolston KV, Jones RN, Mendes RE. Rapid expansion of KPC-2-producing Klebsiella pneumoniae isolates in two Texas hospitals due to clonal spread of ST258 and ST307 lineages. Microbial drug resistance 2013; 19(4): 295–7. [DOI] [PubMed] [Google Scholar]
- 21.Ocampo AM, Chen L, Cienfuegos AV, et al. A Two-Year Surveillance in Five Colombian Tertiary Care Hospitals Reveals High Frequency of Non-CG258 Clones of Carbapenem-Resistant Klebsiella pneumoniae with Distinct Clinical Characteristics. Antimicrob Agents Chemother 2016; 60(1): 332–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giddins MJ, Macesic N, Annavajhala MK, et al. Successive Emergence of Ceftazidime-Avibactam Resistance through Distinct Genomic Adaptations in blaKPC-2-harboring Klebsiella pneumoniae Sequence Type 307 Isolates. Antimicrob Agents Chemother 2018; 62(3): e02101–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shields RK, Chen L, Cheng S, et al. Emergence of ceftazidime-avibactam resistance due to plasmid-borne blaKPC-3 mutations during treatment of carbapenem-resistant Klebsiella pneumoniae infections. Antimicrob Agents Chemother 2016; 61(3): e02097–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han JH, Johnston B, Nachamkin I, et al. Clinical and molecular epidemiology of Escherichia coli sequence type 131 among hospitalized patients colonized intestinally with fluoroquinolone-resistant E. coli. Antimicrob Agents Chemother 2014; 58(11): 7003–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nicolas-Chanoine MH, Bertrand X, Madec JY. Escherichia coli ST131, an intriguing clonal group. Clin Microbiol Rev 2014; 27(3): 543–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Duin D, Paterson DL. Multidrug-Resistant Bacteria in the Community: Trends and Lessons Learned. Infect Dis Clin North Am 2016; 30(2): 377–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pitout JD, DeVinney R. Escherichia coli ST131: a multidrug-resistant clone primed for global domination. F1000Res 2017; 6: 195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jolley KA, Maiden MC. BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 2010; 11: 595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Charlson ME, Pompei P, Ales KL, MacKenzie CR. A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. J Chronic Dis 1987; 40(5): 373–83. [DOI] [PubMed] [Google Scholar]
- 30.Henderson H, Luterbach CL, Cober E, et al. The Pitt Bacteremia Score Predicts Mortality in Non-Bacteremic Infections. Clin Infect Dis 2019; In press: doi: 10.1093/cid/ciz528. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


