ABSTRACT
The Hippo-Yap pathway regulates multiple cellular processes in response to mechanical and other stimuli. In Drosophila, the polarity protein Lethal (2) giant larvae [L(2)gl], negatively regulates Hippo-mediated transcriptional output. However, in vertebrates, little is known about its homolog Llgl1. Here, we define a novel role for vertebrate Llgl1 in regulating Yap stability in cardiomyocytes, which impacts heart development. In contrast to the role of Drosophila L(2)gl, Llgl1 depletion in cultured rat cardiomyocytes decreased Yap protein levels and blunted target gene transcription without affecting Yap transcript abundance. Llgl1 depletion in zebrafish resulted in larger and dysmorphic cardiomyocytes, pericardial effusion, impaired blood flow and aberrant valvulogenesis. Cardiomyocyte Yap protein levels were decreased in llgl1 morphants, whereas Notch, which is regulated by hemodynamic forces and participates in valvulogenesis, was more broadly activated. Consistent with the role of Llgl1 in regulating Yap stability, cardiomyocyte-specific overexpression of Yap in Llgl1-depleted embryos ameliorated pericardial effusion and restored blood flow velocity. Altogether, our data reveal that vertebrate Llgl1 is crucial for Yap stability in cardiomyocytes and its absence impairs cardiac development.
KEY WORDS: Hippo-Yap pathway, Valvulogenesis, Cardiac development, Zebrafish
Summary: Mutagenic analyses in rat and zebrafish reveal a new role for Llgl1 promoting Yap protein stability, which facilitates more precise modulation of Hippo-Yap signaling in cardiomyocytes and zebrafish cardiac development.
INTRODUCTION
Lethal giant larvae (Lgl) proteins are regulators of cell polarity, cell junction stability and composition, as well as endomembrane activities, including vesicle trafficking and acidification (Jossin et al., 2017; Greenwood et al., 2016; Wang et al., 2011; Yamanaka et al., 2003). Furthermore, within vertebrate cancer cells, the two paralogs Llgl1 and Llgl2 affect cell migration (Greenwood et al., 2016; Kashyap et al., 2013). In zebrafish neuroepithelia, Llgl1 controls apical domain size and loss of function results in increased Notch activation (Clark et al., 2012). Although the focus of that research was retinal neurogenesis, it was observed that knockdown of Llgl1 also affected cardiac development, which is analyzed more deeply in this study.
In Drosophila, L(2)gl has been shown to regulate the Hippo-Yap pathway (Grzeschik et al., 2010; Parsons et al., 2014a). Hippo-Yap signaling is essential for the development and maintenance of many organs. This pathway consists of several kinases and their associated activator proteins that form a core kinase complex that governs the activity of the transcriptional co-activators Yap and Wwtr1 (also known as Taz) (Hao et al., 2008; Liu et al., 2011). The regulatory inputs that influence the activity of the kinase complex are still poorly defined and likely vary among different organs and tissues. Regulation of the Hippo-Yap pathway was first characterized in Drosophila, and it was revealed that L(2)gl, along with other polarity factors, functions upstream of the core kinase complex and negatively regulates Yap/Wwtr1 transcriptional output (Parsons et al., 2014a). In Drosophila, l(2)gl mutations disrupt the membrane localization of both components of the core kinase complex and Yorkie (the Drosophila Yap homolog), resulting in increased nuclear enrichment of Yorkie and excessive target gene activation (Grzeschik et al., 2010). Exactly how l(2)gl mutations result in increased nuclear Yorkie activity is unclear (Parsons et al., 2014a). However, the organization and composition of Hippo-Yap pathway components at cell junctions regulates Yorkie activity in other contexts, such as promoting activation of the core kinase complex (Sun et al., 2015). Whether the loss of Lgl1 in vertebrates impacts Hippo-Yap signaling is currently unknown.
The role of the Hippo-Yap pathway in cardiac development is well documented (Wang et al., 2018). In cardiomyocytes, Hippo-Yap signaling regulates proliferation and aspects of differentiation (Wang et al., 2018). Mutations that negatively affect Hippo core kinase activity or transgenic over-expression of Yap result in cardiomegaly (Flinn et al., 2019; Heallen et al., 2011; von Gise et al., 2012; Xin et al., 2013). Conversely, the deletion of Yap or Tead1 in embryonic cardiomyocytes results in decreased cardiomyocyte proliferation and a thinned myocardial wall (Chen et al., 1994; von Gise et al., 2012; Xin et al., 2013) or cardiobifida (Miesfeld and Link, 2014). In the heart, Hippo-Yap is crucial for cardiomyocytes, as epicardial Hippo-Yap signaling is essential for the proper differentiation of epicardium-derived cells, including fibroblasts and vascular smooth muscle cells (Xiao et al., 2018). Hippo-Yap signaling, like that of Notch, is also important for endocardial expression of neuregulin (Artap et al., 2018), and has been implicated in regulating atrioventricular valve development (Zhang et al., 2014). In humans, mutations in the upstream Hippo-Yap pathway modulator dachsous (DCHS1) cause mitral valve prolapse, a common cardiac valve disease (Durst et al., 2015). Considering the crucial role of Hippo-Yap signaling in cardiac development and the link between L(2)gl and Hippo-Yap signaling in Drosophila, we assessed the role of Lgl1 on Yap activity in vertebrate cardiomyocytes and investigated its effect on cardiac development and function in the zebrafish model. We found that depletion of Llgl1 in vertebrate cardiomyocytes results in decreased levels of Yap protein and reduced transcription of Yap target genes. Our data indicate that Llgl1 is required for normal zebrafish cardiac development and function, as assessed by cardiomyocyte morphology, hemodynamics and valvulogenesis. Interestingly, although our study focuses on the systemic depletion of Llgl1, the aberrant cardiac phenotypes associated with Llgl1 were ameliorated with exogenous expression of Yap, specifically in cardiomyocytes. Collectively, this study is the first to define the role of Llgl1 in Hippo-Yap regulation in cardiomyocytes and offers a potential mechanism for modulating Hippo-Yap signaling more precisely in cardiomyocytes, an area of increasing interest in the field of cardiac regeneration (Heallen et al., 2011; Morikawa et al., 2017; von Gise et al., 2012; Xin et al., 2013).
RESULTS
Llgl1 depletion in neonatal rat cardiomyocytes decreases Yap protein and Yap transcriptional target expression
In Drosophila, L(2)gl protein modulates both the Notch and Hippo-Yap pathways (Grzeschik et al., 2010; Parsons et al., 2014b). In a previous study, we found that knockdown of zebrafish Llgl1, one of the two vertebrate homologs of Drosophila L(2)gl, resulted in increased Notch activity and reduced retinal neurogenesis (Clark et al., 2012). In those studies, we also noted severe cardiac effusion and dysmorphic hearts. Although the role of vertebrate Llgl1 on Notch activity has been characterized (Clark et al., 2012), less is known about its potential impact on Hippo-Yap signaling. Because Yap signaling is known to modulate cardiomyocyte development (von Gise et al., 2012), and given the strong Llgl1 depletion phenotype with heart morphogenesis (Clark et al., 2012), we investigated the relationship between Llgl1 function and Hippo-Yap signaling within cardiomyocytes. We initially depleted Llgl1, as well as Llgl2, in neonatal rat cardiomyocytes using siRNAs. siRNA knockdown of Llgl1 or Llgl2, or both paralogs, did not alter Yap mRNA abundance, as measured by qRT-PCR (Fig. 1A). However, knockdown of either Llgl1 or Llgl2 did result in compensatory upregulation of the paralog transcript. Knockdown of Yap did not affect mRNA abundance of either Llgl1 or Llgl2. Interestingly, in contrast to Drosophila, in which mutation of l(2)gl results in elevated Yorkie nuclear localization and transcriptional activity (Grzeschik et al., 2010; Parsons et al., 2014a), knockdown of Llgl1 or Llgl2 in primary rat cardiomyocytes resulted in decreased levels of YAP protein, as assessed by western blot (Fig. 1B, Table S4). Consistent with lower YAP protein levels, the expression of YAP-TEAD target genes decreased in cells treated with a combination of Llgl1 and Llgl2 siRNA. Gene Set Enrichment Analysis (GSEA) on RNA-seq data from cardiomyocytes simultaneously depleted of Llgl1 and Llgl2 revealed a decrease in enrichment for YAP/WWTR1-TEAD direct transcriptional targets (Fig. 1C). This reference gene set was defined by YAP ChIP-Seq and YAP knockdown experiments performed on human MDA-MB-231 cells (Zanconato et al., 2015). We independently verified this GSEA by running the enrichment analysis with RNA-seq data generated from cultured rat cardiomyocytes treated with either Yap siRNA or negative control siRNA (Flinn et al., 2019, Fig. S1). Thus, although depletion of Llgl1 and Llgl2 does not alter the transcription of Yap, Llgl function is required to maintain appropriate Yap protein levels and Yap-TEAD transcriptional activity in vertebrate cardiomyocytes.
Fig. 1.
Knockdown of Llgl1 in cultured neonatal rat cardiomyocytes decreases YAP protein levels. (A) qRT-PCR results denoting RNA transcript levels of Llgl1, Llgl2 or Yap in isolated neonatal rat cardiomyocytes following siRNA treatments. n=3. One-way ANOVA, Tukey's multiple comparisons test showing difference compared with negative control. Error bars indicate s.e.m. (B) Western blot analysis of anti-YAP and anti-γ-tubulin staining in siRNA treated rat cardiomyocytes. Quantification of anti-YAP band signal normalized to anti-γ-tubulin signal. n=6 for negative sequence, n=3 for untreated, n=9 for Llgl1, n=6 for Llgl2 and n=6 for Yap siRNA-treated cells. One-way ANOVA, Tukey's multiple comparisons test. (C) GSEA for YAP/WWTR1/TEAD target genes using RNAseq data from siRNA-treated rat cardiomyocytes targeting Llgl1 and Llgl2 or a negative control sequence. n=3. Error bars indicate s.e.m. *P<0.05, **P<0.01, ***P<0.001.
Developmental defects and lethality of llgl1 mutant zebrafish
To understand how loss of Llgl1 influences Yap protein levels in vivo and to test whether Llgl1 also has a function during cardiac development, we generated an llgl1 mutant zebrafish line. Transcription activator-like effector nucleases (TALENs) were used to generate a ten (llgl1mw83) nucleotide deletion in exon 1 of llgl1, resulting in a frameshift mutation and generation of an early stop codon (Fig. 2A, Fig. S2). Homozygous llgl1 mutant (llgl1−/−) embryos closely resembled the morpholino knockdown phenotype characterize previously in Clark et al. (2012). (Fig. 2B, Figs S3,S4). Pericardial effusion, which can result from cardiac dysfunction, was apparent in all llgl1−/− embryos by 3 days postfertilization (dpf) but varied in expressivity, which was similarly observed in morpholino-treated embryos (Fig. 2B, Fig. S3). Interestingly, a subset of llgl1−/− embryos recovered from pericardial effusion to resemble wild-type siblings by 5 dpf and survived to adulthood. Variable expressivity was not the result of differential maternal contribution of Llgl1 protein or llgl1 mRNA, as either maternal or paternal llgl1−/− fish outcrossed to an llgl1+/− heterozygote yielded the same spectrum of expressivity among their offspring (Fig. 2C). Although a subset of llgl1−/− fish recovered by 5 dpf, those displaying more overt pericardial effusion at 3 dpf progressed to exhibit edema throughout the body and died by 6 dpf. Approximately two-thirds of the llgl1−/− fish survived to adulthood based on a comparison of measured survival versus Mendelian-predicted proportions of homozygous mutants from a cross of llgl1−/− and llgl1+/− parents (Fig. 2D, Table S5). Altogether, these observations of llgl1−/− mutant fish demonstrate overt morphological phenotypes and impaired survival in a subset of mutants. We next used this line to characterize zebrafish cardiac development and function in the absence of llgl1.
Fig. 2.
Assessment of llgl1 mutant gross morphology. (A) Diagram depicting the 10 bp deletion generated by TALENs in exon 1 of llgl1. (B) Embryos (3 dpf) from an llgl1+/−×llgl1−/− cross categorized into three grades of severity: class 0, wild-type phenotype; class 1 (mild), subtle signs of pericardial effusion at 3 dpf; class 2, pronounced pericardial effusion with a small subset displaying smaller eyes; and class 3 (severe), pronounced pericardial effusion and/or body edema, small eyes and hearts fail to undergo cardiac looping (Movie 5). At 5 dpf, llgl1−/− embryos are undistinguishable from llgl1+/− siblings. Scale bars: 1 mm. (C) Quantification of phenotype classes from llgl1+/−×llgl1−/− crosses comparing maternal contribution. Three clutches were used for each condition. n=109 for maternal llgl1−/− and n=160 for paternal llgl1−/−. χ2 test. (D) Adult genotyping from six pooled llgl1+/−×llgl1−/− crosses, greater than 4 months of age. The dashed line represents the predicted Mendelian number for each genotype. n=58 for llgl1+/− and n=31 for llgl1−/−. Two-tailed binomial test. Error bars indicate s.e.m. *P<0.05.
Llgl1 depletion impairs cardiac development and function in zebrafish
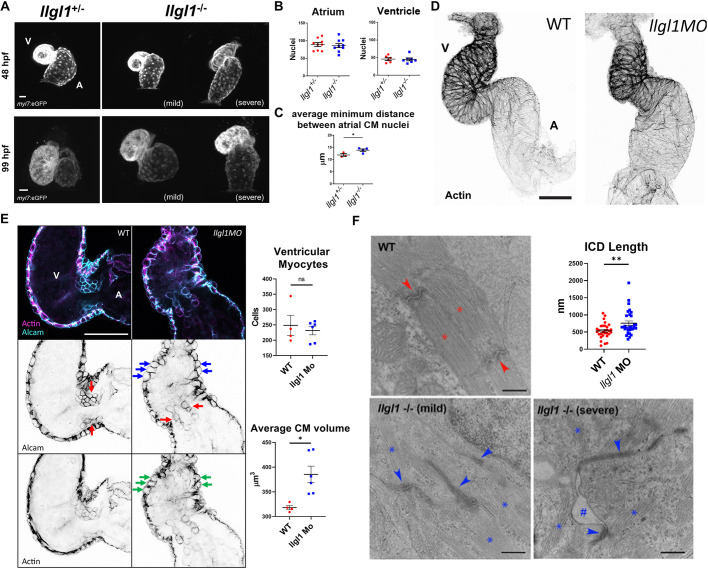
To assess how the loss of Llgl1 affects cardiac development, we crossed the llgl1mw83 line with a cardiomyocyte-reporter transgenic line Tg(myl7:eGFP)mw45 to visualize cardiomyocytes during development. At 48 hours postfertilization (hpf), a subset of llgl1−/− embryos failed to undergo cardiac looping, whereas llgl1−/− embryos with slight pericardial effusion resembled llgl1+/− siblings (Fig. 3A). This phenotype was not due to changes in atrial or ventricular cardiomyocyte cell numbers, as this did not change between groups (Fig. 3B). However, by 99 hpf, llgl1−/− hearts appeared larger than llgl1+/− siblings (Fig. 3A). Analysis using Imaris software (Bitplane) revealed that a loss of Llgl1 resulted in larger atrial cardiomyocytes, as indicated by increased internuclear distances, which is indicative of cardiomyocyte hypertrophy (Fig. 3C). As llgl1−/− mutant embryos do not withstand microinjection, potentially because of epidermal adhesion/resealing defects, a previously validated llgl1 morpholino was used to mediate the knockdown of Llgl1 for subsequent co-injection experiments (Clark et al., 2012). We first assessed cardiac phenotypes of llgl1 morphant embryos that present a strong similarity in gross morphology to llgl1−/− mutants (Fig. S3). Embryos injected with the llgl1 morpholino were examined for the pericardial effusion phenotype before analysis and the observed phenotype varied in severity (Fig. S3). Additionally, as with llgl1−/− mutant embryos, llgl1 morphants failed to undergo cardiac looping at 48 hpf, resulting in dysmorphic ventricles (Fig. 3D, Fig. S5). Confocal analysis of 48 hpf llgl1 morphant hearts revealed larger rounded ventricular cardiomyocytes with an abnormal distribution of actin (Fig. 3E, Fig. S6). Before trabeculation, ventricular cardiomyocytes develop dense cortically localized myofibrils in the basal/luminal region of the cell (Reischauer et al., 2014). To test whether this atypical distribution of myofibrils might affect the contractile structures of the heart, we performed transmission electron microscopy, which revealed disorganized and reduced sarcomere bundles in llgl1 morphant cardiomyocytes at 48 hpf (Fig. S7). Similar phenotypes were observed in llgl1−/− mutant embryos at 4 dpf, which presented dysgenic sarcomeres and enlarged dysmorphic intercalated discs (Fig. 3F, Fig. S7). In embryos with a severe pericardial effusion phenotype, adhesion between cardiomyocytes and the surrounding cells was affected. The intercalated disc phenotype is reminiscent to the enlarged and dysmorphic adherens junctions observed in llgl1 morphant retinal neuroepithelia (Clark et al., 2012). Collectively, these results show that Llgl1 regulates the development of cardiomyocyte morphology and contraction in the first few days of life in zebrafish.
Fig. 3.
Loss of llgl1 disrupts normal heart development. (A) Representative confocal z-stack images of myl7:eGFP expression in hearts at 48 hpf and 99 hpf in llgl1+/− and llgl1−/− siblings. A, atrium; V, ventricle. (B) Quantification of atrial and ventricular cardiomyocyte nuclei in 48 hpf llgl1 morphants versus uninjected controls. n=9 for atrial nuclei analysis, n=6 for uninjected control and n=7 for llgl1 morpholino microinjection for ventricular nuclei analysis. (C) Quantification of average minimum distance between atrial cardiomyocyte nuclei in llgl1+/− and llgl1−/− siblings at 48 hpf. n=3 for llgl1+/− hearts and n=4 for llgl1−/− hearts. (D) Representative confocal z-stack images of actin-stained 48 hpf hearts. (E) Representative single-section confocal micrograph of alcam or actin-stained 48 hpf hearts, focusing on the AVC. Red arrows depict valve leaflets, blue arrows depict large rounded ventricular cardiomyocytes and green arrows depict abnormal actin staining in the myocardium. Quantification of ventricular cardiomyocyte number and volume. n=4 for untreated embryos and n=6 for llgl1 morpholino-treated embryos. (F) Representative electron micrographs of 4 dpf wild-type and llgl1−/− cardiomyocytes. Red asterisks denote normal sarcomeres of wild type, whereas blue asterisks denote thin and disorganized sarcomeres of llgl1−/− cardiomyocytes. Red arrows illustrate normal intercalated discs of wild type, whereas blue arrows indicate elongated dysmorphic intercalated discs of llgl1−/− cardiomyocytes. The hashtag indicates loss of adhesion between cardiomyocytes. Quantification of cardiomyocyte intercalated disc (ICD) length (top right). n=30 intercalated discs per group compiled from three animals per group, and ten micrographs per animal. Two-tailed unpaired Student's t-test. Error bars indicate s.e.m. ns, not significant, *P<0.05, **P<0.01. Scale bars: 25 μm (A); 50 μm (D,E); 500 nm (F).
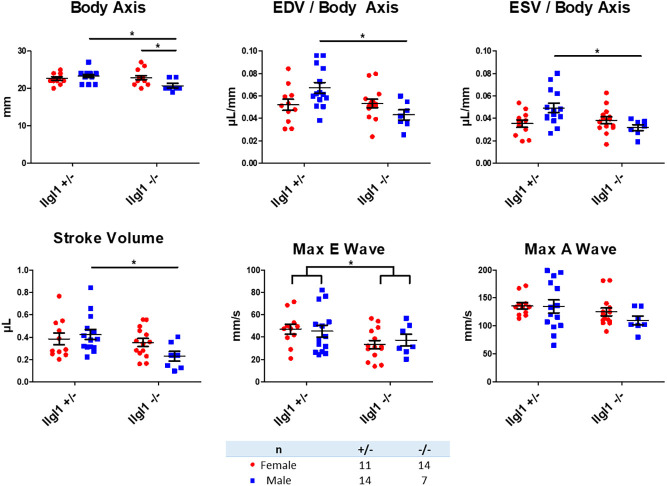
Although ∼30% of llgl1−/− fish die, the others recover and survive to adulthood. We analyzed three clutches of siblings raised from llgl1−/−×llgl1+/− crosses using echocardiography to assess cardiac function between genotypes. Echocardiograms revealed variations in cardiac physiology among adult llgl1−/− males, as indicated by statistically smaller end-diastolic and end-systolic volume (EDV and ESV, respectively) measurements when normalized to body length (Fig. 4). Stroke volume was also significantly reduced in males. Although there was no difference in the A wave peak velocity between groups, we observed a significant decrease in maximum E wave velocity in llgl1−/− fish compared with llgl1+/− siblings, with no observable differences between sexes of the same genotype. These results illustrated a dysfunction in passive filling of the ventricle during early diastole in llgl1−/− fish and a decrease in cardiac function in llgl1−/− male fish compared with llgl1+/− male siblings. Altogether, our observations of llgl1 mutant and morphant fish illustrate that a loss of Llgl1 in zebrafish results in dysmorphic heart development and impaired cardiac function in adult males.
Fig. 4.
Adult male llgl1 mutant zebrafish display smaller hearts. Quantification of echocardiogram data from 5-month-old zebrafish. n=11 for llgl1+/− females, n=14 for llgl1−/− females, n=14 for llgl1+/− males and n=7 llgl1−/− males. Two-way ANOVA, Tukey's multiple comparisons test. Error bars indicate s.e.m. *P<0.05.
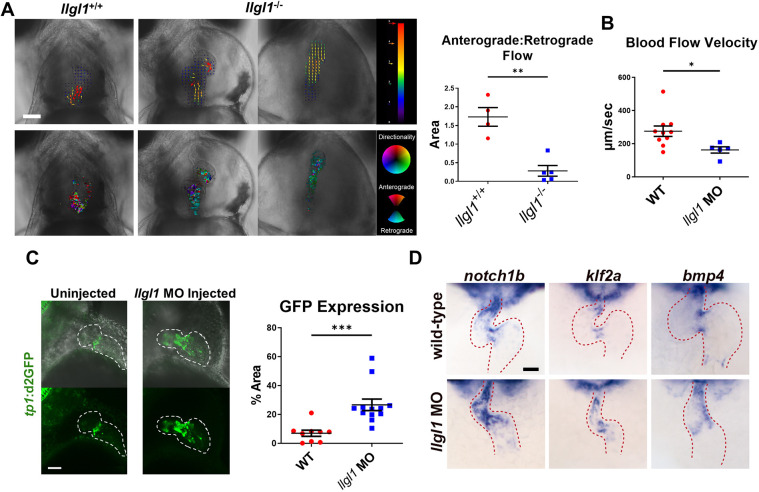
Intracardiac fluid forces are essential for embryonic cardiogenesis (Andrés-Delgado and Mercader, 2016; Haack and Abdelilah-Seyfried, 2016; Steed et al., 2016; Paolini and Abdelilah-Seyfried, 2018). In the developing zebrafish heart, valve leaflets are normally formed by 96 hpf, ensuring unidirectional blood flow (Steed et al., 2016). Given the abnormal morphology of cardiomyocytes and pericardial effusion phenotype in Llgl1-depleted embryos, we hypothesized that hemodynamics and valvulogenesis were also affected. Both llgl1−/− and llgl1+/− embryo hearts displayed oscillatory flow between the heart chambers at 78 hpf (Movies 1, 2). However, at 99 hpf, we observed unidirectional flow within llgl1+/− hearts, whereas llgl1−/− hearts continued to exhibit substantial oscillatory flow (Movies 3-5). To assess ventricular regurgitation, the degree of anterograde or retrograde flow during diastasis was quantified, revealing a significant increase in retrograde flow in llgl1−/− embryos (Fig. 5A). Similarly, we observed a significant decrease in blood flow velocity in llgl1 morphants (Fig. 5B, Movies 6, 7). As impaired blood flow can suppress valvulogenesis, we analyzed the developing valve using a Notch1 reporter transgenic line, Tg(tp1-MmHbb:d2GFP)mw43, which was used to identify the developing valve (Samsa et al., 2015). Quantification of the short-lived d2GFP protein by confocal microscopy revealed a significant increase in the area encompassed by Notch signaling within 2 dpf llgl1 morphants (Fig. 5C). Notch activity was apparent throughout the ventricle, rather than localized to the atrioventricular canal (AVC) as seen in control embryos. This result is similar to the findings in llgl1 morphant retinal progenitor cells, which also showed increased Notch activity (Clark et al., 2012). Finally, in situ hybridization of notch1b, bmp4 and klf2a mRNAs, which mark valve cells, revealed disrupted valvulogenesis in llgl1 morphants (Fig. 5D, Figs S7-S9). Collectively, our data indicate that deletion of Lgl1 disrupts unidirectional blood flow that is accompanied by impaired valvulogenesis. Loss of hemodynamic forces likely compounds valve dysgenesis and heart chamber defects.
Fig. 5.
Valve morphology and hemodynamics are compromised in llgl1 mutants. (A) Representative images from videos acquired using light microscopy of 99 hpf hearts. A heart from an llgl1−/− embryo with slight pericardial effusion is displayed in the middle, whereas on the right, a heart with severe pericardial effusion is depicted. Above: blood flow was analyzed during diastasis using the particle image velocimetry in Fiji. Arrow vectors show directionality distance moved in pixels. Below: particle image velocimetry (PIV) data were analyzed using the PIV analyzer function in Fiji and qualified for anterograde or retrograde flow. Quantification of anterograde versus retrograde area within the heart chambers during diastasis is depicted in the graph (right). n=4 for llgl1+/+ embryos and n=5 for llgl1−/− embryos. (B) Blood flow velocity measurements analyzed from videos. n=10 for untreated embryos and n=5 for llgl1 morpholino-treated embryos. (C) Representative images from confocal z-stack micrographs of hearts of 2 dpf embryos carrying the tp1 transgenic Notch reporter. The area of cardiac GFP expression is depicted in the graph. n=9 for untreated embryos and n=12 for llgl1 morpholino-treated embryos. (D) Representative images of in situ hybridization of valve markers in 48 hpf fish. Ventral view focusing on the heart (red dashed line). Color model depicts the ventricle in red, atrium in blue, AVC in yellow and venous pole in purple. Two-tailed unpaired Student's t-test. Error bars indicate s.e.m. *P<0.05, **P<0.01, ***P<0.001. Scale bars: 100 µm.
Exogenous expression of Yap in zebrafish cardiomyocytes ameliorates the abnormal cardiac physiological function associated with loss of Llgl1
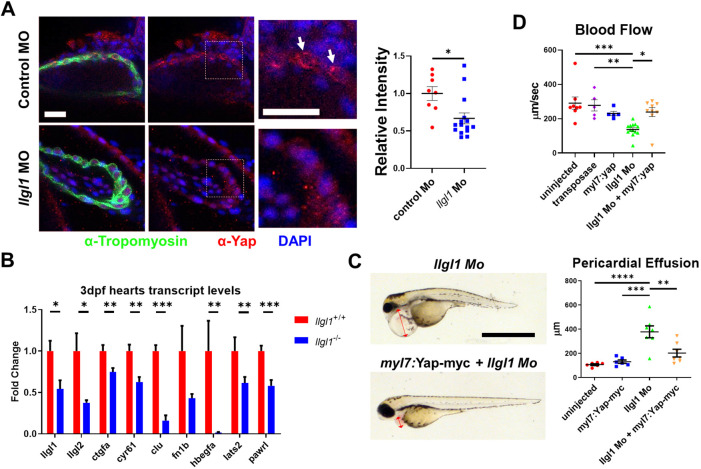
As we found that depletion of Llgl1 reduced YAP protein levels in rat cardiomyocytes in vitro, we next tested whether, in contrast to Drosophila, in vivo depletion of Llgl1 also reduced Yap protein levels in zebrafish cardiomyocytes. Anti-Yap staining in 2 dpf zebrafish showed a significant decrease in Yap protein signature in the tropomyosin-expressing ventricular cardiomyocytes of llgl1 morphants compared with controls (Fig. 6A). Ventricular size was measured using confocal microscopy and quantified with Imaris software. Furthermore, qRT-PCR analysis revealed a decrease in Yap-regulated transcripts in hearts isolated from 3 dpf llgl1−/− embryos compared with wild type (Fig. 6B). The expression of both llgl1 and llgl2 transcripts was also significantly decreased. Taken together, these data indicate that Llgl1 regulates Yap protein levels and associated transcriptional activity in zebrafish cardiomyocytes.
Fig. 6.
Llgl1 promotes appropriate Yap protein levels in cardiomyocytes, and exogenous expression of Yap in cardiomyocytes ameliorates deleterious cardiac physiology associated with loss of Llgl1. (A) Representative images of 2 dpf zebrafish ventricles from single plane confocal micrographs (left). Boxes indicate area of enlargement. Arrows depict Yap within the cardiomyocytes. Quantification of the intensity of anti-Yap signature in tropomyosin+ cells normalized to the control morpholino group (right). n=8 for untreated embryos and n=15 for llgl1 morpholino-treated embryos. (B) qRT-PCR transcript analysis for llgl1, llgl2 and genes regulated by Yap activity in llgl1+/+ or llgl1−/− 3 dpf zebrafish hearts. n=10 for each group. (C) Representative images of 3 dpf embryos microinjected with llgl1 morpholino with or without Tol2-mediated integration of myl7:yap-myc (left). Arrows depict the region measured for pericardial effusion. Quantification of pericardial effusion in 3 dpf embryos (right). n=6 for uninjected control group, n=6 for the myl7:yap-myc control group, n=7 for llgl1 morphants and n=7 for llgl1 morphants with myl7:yap-myc expression. (D) Quantification of blood flow velocity in 2 dpf embryos. n=8 for the uninjected control group, n=5 for the transposase only control group, n=5 for the myl7:yap-myc control group, n=13 for llgl1 morphants and n=9 for llgl1 morphants with myl7:yap-myc expression. Two-tailed unpaired Student's t test (A,B) and One-way ANOVA, Tukey's multiple comparisons test (C,D). Error bars indicate s.e.m. *P<0.05, **P<0.01, ***P<0.001,****P<0.0001. Scale bars: 50 µm (A); 1 mm (C).
To assess the extent to which the loss of Llgl1 cardiac phenotype was caused by decreased levels of Yap protein, we addressed whether exogenous expression of Yap would rescue heart development in llgl1 morphants. Specifically, we microinjected Tol2-based plasmid DNA encoding myc-tagged Yap driven by a cardiomyocyte-specific promoter (myl7:Yap-myc). This plasmid has been used previously and the expressed protein has been verified at 2 dpf in cardiomyocytes using western blot (Flinn et al., 2019). Interestingly, llgl1−/− embryos were sensitive to microinjection, failing to develop after perturbance at the one-cell stage, thus necessitating the use of the llgl1 morpholino. Exogenous expression of Yap in cardiomyocytes ameliorated the pericardial effusion phenotype of llgl1 morphants at 3 dpf (Fig. 6C). Furthermore, analysis of hemodynamic forces showed a statistically significant increase in blood flow velocity with co-injection of the myl7:Yap-myc construct in llgl1 morphants compared with llgl1 morphants alone (Fig. 6D). No statistical difference in blood flow velocity was observed between the myl7:Yap-myc-treated llgl1 morphants or the myl7:Yap-myc-treated wild-type embryos compared with the control groups. This rescue of cardiac function in llgl1 morphants by Yap overexpression in cardiomyocytes is consistent with a role for Llgl1 in controlling Yap protein levels.
Loss of Llgl1 affects Yap localization and levels
In vertebrates, the levels of Yap protein are partially controlled by regulated degradation. In addition to promoting 14-3-3 binding and cytoplasmic retention, Lats kinases also affect Yap activity via phospho-mediated degradation (Zhao et al., 2010). We suspected that Lats activity might underlie the reduction of Yap protein levels within Llgl1-depleted cardiomyocytes and could explain the observed differences between Drosophila and vertebrates, as the phosphodegron sequence is not conserved in Yorkie. Studies in human cell lines revealed that Ser 381 of YAP can be phosphorylated by LATS, resulting in subsequent phosphorylation by CK1δ/ε at S384 and S387, and E3ubiquitin-mediated degradation via β-TRCP (Zhao et al., 2010). This triple serine region is highly conserved across vertebrates (Zhao et al., 2010). Indeed, in zebrafish Yap, replacement of the Lats target residue serine S335 with non-phosphorylatable alanine results in increased Yap stability and activity in cardiomyocytes (Flinn et al., 2019). To test Yap localization and phospho-degradation, we generated two constructs expressing eGFP-Yap fusion proteins either with or without the serine 355 to alanine mutation. In addition, we generated a non-phosphorylatable serine 54 to alanine mutant that disrupts Tead-binding and moderates potential overexpression phenotypes (Miesfeld et al., 2015). Comparisons between the two transgenes revealed changes in the localization and degradation of Yap caused by loss of Llgl1.
Membrane localization of Yap protein at both the lateral surface and adherens junction-like intercalated discs of cardiomyocytes has been shown to regulate the activity of Yap (Morikawa et al., 2017; Flinn et al., 2020). Given its role in establishing apical-basal polarity, we hypothesized that loss of Llgl1 would disrupt membrane localization of Yap, thus making it accessible for Lats-mediated phosphodegradation. To assess the function of Llgl1 on Yap stability, we used the epidermis as a model that shows strong membrane localization of Yap and can be clearly and easily imaged by whole-embryo confocal microscopy (Fig. S10). In an attempt to rescue embryos exhibiting severe pericardial effusion, we conducted cold rearing and staged embryos to 5 dpf as described by Kimmel et al. (1995). We have previously shown that loss of retinal pigmented epithelium in yap1−/− mutant fish can be rescued by slowing development (Miesfeld et al., 2015). When llgl1−/− embryos were reared at low temperature (20.5°C), the severity of edema was diminished and all mutants survived longer; this revealed an epidermal dysgenesis along their fins (Fig. S11A,B). In comparison, cold rearing of wild-type fish did not result in abnormal phenotypes. Electron micrographs of the llgl1−/− mutant fin epidermis showed an array of irregularities. Specifically, the density of desmosomes was reduced between the outer enveloping layer and basal epidermal layer, and the basement membrane of llgl1−/− mutant fin epidermis was disordered compared with the compact composition seen in llgl1+/− fins. Actinotrichia, collagen-rich skeletal components appeared dysmorphic and were trapped within the basal epidermal layer (Fig. S11C). Additionally, apical junctions of the enveloping layer were elongated and more electron dense (Fig. 7A), resembling phenotypes observed in the intercalated discs of cardiomyocytes (Fig. 3F, Fig. S7) and apical junctions of retinal progenitor cells in llgl1 morphants (Clark et al., 2012). As Yap localizes to cell junctions in the epidermis, endothelium and cardiomyocytes (Morikawa et al., 2017; Neto et al., 2018; Schlegelmilch et al., 2011), we next assayed whether loss of Llgl1 resulted in a displacement of Yap from the cell cortex. To visualize Yap localization in the zebrafish epidermis, we expressed eGFP-YapS54A and eGFP-YapS54A;S335A under the epidermal krt18 promoter via the Tol2 transposable constructs. Expression of plasmid DNA revealed that eGFP-YapS54A localized to epidermal cell membranes in 30 hpf control embryos (Fig. 7B). Compared with controls, the llgl1 morphant epidermis lacked membrane localization and, although the eGFP-YapS54A signal varied from cell to cell, the eGFP-YapS54A signal was significantly decreased in intensity. In contrast, expression of the eGFP-YapS54A;S335A variant, which is resistant to phospho-degradation, showed no change in abundance compared with control embryos but was also delocalized from the cell cortex. Similarly, an antibody that recognizes Yap revealed a subcellular displacement and reduction in expression levels in 48 hpf llgl1−/− mutants (Fig. 7C, Fig. S10). Collectively, our results suggest that loss of Llgl1 affects Yap localization and consequently turnover, potentially through the remodeling of apical junctions in epithelia and intercalated discs of cardiomyocytes (Fig. 8). Furthermore, these data suggest that the loss of Yap protein associated with depletion of Llgl1 is ultimately the result of Lats kinase activity.
Fig. 7.
Loss of Llgl1 in zebrafish epidermis results in dysmorphic junctions and increased Yap phospho-degradation. (A) Representative transmission electron micrographs of fin epidermis from 5 dpf stage cold-reared embryos (left). Arrows indicate apical junctions. Quantification of apical junction length of the outer epidermal layer and the abundance of desmosomes between the epidermal layers (right). n=11 for llgl1+/− and 14 for llgl1−/− embryos. (B) Representative confocal z-stack micrographs of 30 hpf zebrafish epidermis expressing either eGFP-YapS54A or eGFP-YapS54,355A. Arrowheads indicate membrane localization of the transgene (left). Quantification of the intensity of the transgene signal in positively expressed cells is depicted (right). n=13 for eGFP-YapS54A intensity quantification. n=10 for untreated and n=12 llgl1 morpholino-treated embryos for eGFP-YapS54,335A intensity. (C) Representative confocal z-stack micrographs of 2 dpf embryos immunostained with an antibody against Yap. Two-tailed unpaired Student's t-test. Error bars indicate s.e.m. ns, not significant, *P<0.05, **P<0.01, ***P<0.001. Scale bars: 500 nm (A); 50 µm (B,C).
Fig. 8.
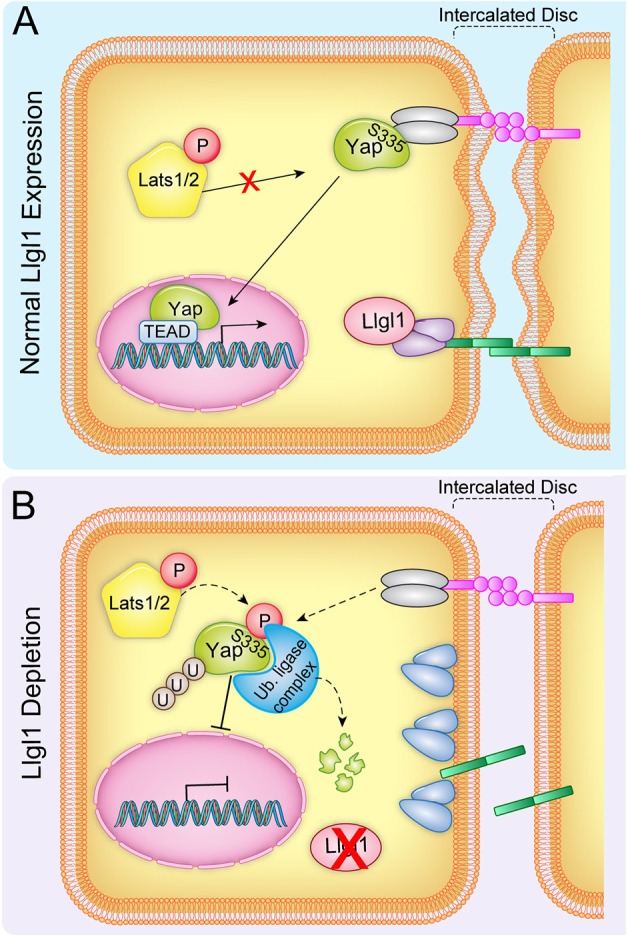
A model for the role of Llgl1 in modulating Yap stability. Cartoon depicting the proposed mechanism by which Llgl1 prevents Yap phosphodegradation. Llgl1 is necessary to maintain appropriate protein composition of cell junctions, such as intercalated discs in cardiomyocytes or adherens junctions of epithelia in which Yap colocalizes with cell-junctional components such as α-catenin. Loss of Llgl1 disrupts the normal composition of intercalated discs between cardiomyocytes, resulting in the displacement of Yap and subsequent phosphorylation by Lats kinases, leading to targeted degradation via ubiquitin ligation.
DISCUSSION
The Hippo pathway regulates tissue growth in the response to soluble cues, cell density, mechanical force and other stimuli (Deng et al., 2015; Dupont et al., 2011; Fletcher et al., 2018). However, a great challenge remains in identifying those signals upstream that sense, mediate and modify the physiological inputs into this pathway. We found that Llgl1 regulates Yap protein levels and transcriptional activity. However, as we could not find a Llgl1 antibody specific for zebrafish, we cannot rule out splicing variants or alternative start sites accounting for some diminished form of Llgl1 expression in llgl1mw83 mutant fish. Nonetheless, we show that depletion of Llgl1 in cultured rat and zebrafish cardiomyocytes, and epidermal cells in vivo, resulted in lower levels of total Yap protein. Furthermore, depletion of Llgl1 also resulted in diminished expression of Yap-TEAD target genes in both llgl1−/− mutant zebrafish and cultured rat cardiomyocytes. Our in vivo data show that loss of Llgl1 resulted in the delocalization of Yap from the membrane in epidermal cells, rendering Yap susceptible to degradation. The effect of Llgl1 on Yap levels, but not cell junction localization, depended on serine 335 of Yap, which is a known target of Lats kinases that prime Yap for degradation. As opposed to the function of L(2)gl in Drosophila, our collective observations illustrate a role for Llgl1 as a positive mediator of Yap activity.
Several observations suggest that Llgl1 affects Yap stability through cell junction remodeling. Similar to the llgl1 morphant retinal neuroepithelium (Clark et al., 2012), we found that apical junctions in the epidermis of the llgl1−/− mutant and intercalated discs in cardiomyocytes of llgl1 mutants were elongated and more electron dense compared with wild-type cells. Multiple junction-associated proteins have been implicated in regulating Yap activity. These include α-catenin in the intercalated discs of cardiomyocytes (Li et al., 2015; Vite et al., 2018) and apical junctions of skin keratinocytes (Schlegelmilch et al., 2011). In addition, Yap is associated at the lateral cell cortex in cardiomyocytes by the dystrophin-glycoprotein complex (Morikawa et al., 2017). Although disruption of the dystrophin-glycoprotein complex in neonatal Mdx (Dmd) null mice, which are deficient in dystrophin, displaces Yap from the lateral membrane regions, nuclear localization of Yap is unaltered (Morikawa et al., 2017). However, a role for Lats kinases in modulating the stability of delocalized Yap in Mdx mice was shown by cardiomyocyte-specific knockout of Sav, a positive mediator of Lats activity. These double mutant mice showed a striking increase in Yap localization and transcriptional activity compared with wild-type littermates. This observation is in line with the role of Lats kinases in modulating the levels of Yap following delocalization from the cell membrane. Other cell junction-associated factors have been implicated in modulating the activity of Lats kinases, including those of the Ajuba protein family (Sun and Irvine, 2013). Work with Drosophila and cultured mammalian cells, demonstrated that both activators and inhibitors of Hippo pathway activity localize to apical cell junctions, and that junction reorganization is associated with changes in Yorkie/Yap activity (Sun et al., 2015). Furthermore, in cardiomyocytes Amotl1 has been implicated in shuttling Yap from the intercalated disc junction to the nucleus in a manner that depends on the activity of Fat4, an atypical cadherin that localizes to cell junctions and is essential for intercalated disc integrity (Ragni et al., 2017). Altogether, these published data, coupled with our observations, indicate that Llgl1 promotes the protein composition of cell junction complexes that facilitate Yap localization and protection from phospho-mediated proteasome targeting and degradation.
We further identified a novel role for Llgl1 as a physiological modifier of zebrafish cardiac development. Phenotypic consequences of Llgl1 disruption on heart morphogenesis included cardiac looping defects, abnormal cardiomyocyte morphology and atrioventricular valve dysgenesis. Our global Llgl1 depletion model precludes us from determining whether abnormal myocardial development subsequently impaired valve development, or whether Llgl1 contributes to these developmental processes independently. Evidence from the literature demonstrates that altered hemodynamics can impair valvulogenesis (Donat et al., 2018; Haack and Abdelilah-Seyfried, 2016; Steed et al., 2016; Paolini and Abdelilah-Seyfried, 2018). Thus, a primary input to myocardial contractility and blood flow velocity could secondarily contribute to impaired valvulogenesis, and therefore compound hemodynamic-dependent phenotypes through a feed-forward loop. Our data demonstrate that cardiac hemodynamics is at least partially mediated by Llgl1 and Yap activity in cardiomyocytes themselves, as cardiomyocyte-specific rescue of Yap in llgl1 morphants restored blood flow velocity to baseline levels. We have yet to determine whether Llgl1 has a direct role in regulating valvulogenesis. However, because Llgl1 can control signaling factors such as Yap and Notch, both of which are known to contribute to atrioventricular valve development, this is a possibility. Future studies will address this outstanding issue.
The role for an apico-basal cell polarity factor in regulating cardiac morphogenesis is not unprecedented. Scribble (Scrib), which complexes with Llgl1 and Discs Large family members to regulate basolateral cell identity in many cell types, results in cardiac dysgenesis when deleted from mouse cardiomyocytes (Boczonadi et al., 2014). In these Scrib-conditional depletion mice, differentiation of cardiomyocytes was compromised, resulting in structural abnormalities of the ventricular myocardium, such as ventricular septal defects. Interestingly, like llgl1−/− mutant zebrafish, most Scrib-deficient embryos recovered and survived to adulthood, although the mature hearts showed elevated fibrosis. Whether loss of Scrib affects Yap activity was not evaluated in that study. Additionally, loss of Llgl2, the paralog of Llgl1, in zebrafish causes a lack of hemidesmosome formation between the basal epidermis and the underlying basement membrane (Sonawane et al., 2005). This results in a catastrophic loss of epidermal integrity. Similarly, llgl1−/− mutants with severe pericardial effusion show a variety of abnormalities within the developing epidermis. Collectively, our data show that Llgl1 mediates cardiomyocyte development and valvulogenesis, and thus influences heart development in zebrafish, at least in part, through Yap availability in cardiomyocytes. Although variants in human Llgl1 have not been found to associate with cardiovascular anomalies, our studies provide additional evidence for the role of Hippo-Yap pathway regulation in congenital heart disease.
MATERIALS AND METHODS
Animals
All protocols used in these studies were approved by the local Animal Care and Use Committee, and conform to the Guide for the Care and Use of Laboratory Animals published by the National Institutes of Health. The llgl1mw83 knockout mutant was generated using a pair of TALENs (Table S1), producing a 10 bp deletion (c. 55_64del) that resulted in frameshift mutations and the generation of an early stop codon (Fig. S2). A transgenic zebrafish line expressing enhanced GFP under the myl7 cardiomyocyte-specific promoter Tg(myl7:eGFP)mw45 was used to visualize cardiomyocytes (Miesfeld and Link, 2014). A Notch reporter transgenic zebrafish line, Tg(tp1-MmHbb:d2GFP)mw43, was used to visualize valve development (Clark et al., 2012). Sprague Dawley rat neonates (Charles River Laboratories) were used for cardiac cell isolation. Zebrafish were euthanized by anesthetization with tricaine followed by an ice bath, and rat pups were euthanized by decapitation.
Genotyping
The presence or absence of the llgl1mw83 allele was verified by PCR amplification using the following primers: F, 5′-ACCGACTGACGCCTAACGGAA-3′; and R, 5′-CATGCGGTGGTCGTTGATGCCT-3′. Genomic DNA was extracted using the Puregene Core Kit A (Qiagen) according to the manufacturer's protocol. Primers flanking the mutation were used to amplify a 375 bp product. Digestion with AlwNI (New England BioLabs) yielded 140 and 240 bp fragments for the wild-type allele, whereas the llgl1mw83 amplicon remained undigested.
Microinjections
All plasmids were generated using Gateway (Invitrogen) entry clones in conjunction with the Tol2kit (Kwan et al., 2007). Terminally myc-tagged Yap was inserted downstream of the cardiomyocyte-specific promoter myl7, and eGFP-Yap fusion proteins containing mutations at S54A or S54A;S335A were inserted downstream of the krt18 promoter for expression in epithelia cells. ZDR embryos were microinjected at the one-cell stage with 4.6 nl of a solution containing 8 ng/µl plasmid and 15 ng/µl transposase mRNA. The llgl1 ATG (5′-CCGTCTGAACCTAAACTTCATCATC-3′) and control (5′-CCTCTTACCTCAGTTACAATTTATA-3′) morpholinos were microinjected as described by Clark et al. (2012).
Zebrafish immunohistochemistry
Epidermal anti-Yap staining was performed on 48 hpf embryos using anti-Yap antibody (Cell Signaling Technology, 4912S) as described by Miesfeld et al. (2015). To image hearts, zebrafish embryos were fixed at 48 hpf and stained for actin using rhodamine phalloidin (Sigma-Aldrich, 658740; dilution) at 1:100 stock concentration, and anti-alcam [Developmental Studies Hybridoma Bank (DHSB)] before heart extraction for confocal imaging on a LSM 710 microscope (Zeiss) as described by Renz et al. (2015). Anti-Yap staining in the heart was accomplished using anti-Yap (Abcam, ab81183) and anti-tropomyosin (DSHB, CH1) antibodies. Antibody dilutions are provided in Table S4. Similarly, whole-mount in situ hybridization was performed on 48 hpf zebrafish embryos as described by Renz et al. (2015) using the following probes: klf2a (Renz et al., 2015); notch1b (Walsh and Stainier, 2001); and bmp4 (Walsh and Stainier, 2001). Both notch1b and bmp4 plasmids were gifts from Didier Stainier (Max Planck Institute for Heart and Lung Research, Bad Nauheim, Germany).
Confocal microscopy
Zebrafish embryos were fixed in a solution of 4.0% paraformaldehyde at 4°C overnight. Embryos were then embedded in 1.0% low-melt agarose and imaged using a D-Eclipse C1 Microscope System (Nikon) with EZ-C1 software (version 4.11). The resulting images were analyzed using Fiji ImageJ to determine the signal intensity of compiled z-stack images, or Imaris (Bitplane) for nearest neighbor nuclear distance, cell number and cell size.
Echocardiography
Echocardiographs were obtained from 5-month-old zebrafish using a Vevo 770 with an RMV 708 probe (VisualSonics). Zebrafish were anesthetized in 150 ppm tricane and 45 ppm isoflurane before imaging. Scans were taken in triplicate and measured in a genotype-masked manner using the Vevo 770 software package (ver 3.0.0 build 55.00) as demonstrated by Lee et al. (2016).
Embryonic blood flow imagining
Blood flow was analyzed using Fiji ImageJ from captured video. Particle image velocimetry was used to show the direction of blood flow during diastasis, whereas blood velocity was determined by the distance erythrocytes traveled in the dorsal aorta during a single pulse.
Electron microscopy
Zebrafish embryos were fixed in a solution containing 1.0% paraformaldehyde and 2.5% glutaraldehyde overnight at 4°C. The embryos were then treated with 1.0% osmium tetroxide in phosphate buffer, dehydrated through a series of methanol solutions, and infused with EMbed 812 resin (Electron Microscopy Sciences). Plastic sections were stained with uranyl acetate (Electron Microscopy Sciences). The Hitachi H-600 transmission electron microscope with an ORCA-100 digital camera (Hamamatsu) was used for transmission electron microscopy.
siRNA knockdown in cultured rat cardiac cells
Primary cardiac myocytes and fibroblasts were obtained from 2-day-old Sprague Dawley rat hearts. Hearts were excised and digested with the Neonatal Heart Dissociation Kit (Miltenyi Biotec) according to the manufacturer's protocol. Cardiomyocytes were isolated through an isotonic Percoll gradient centrifugation. Isolated cardiomyocytes were plated in Dulbecco's modified eagle medium (DMEM, Life Technologies) containing 1 g/l glucose, supplemented with 15% fetal bovine serum (FBS) and penicillin-streptomycin (Life Technologies). After 16 h, the cells were transfected with two different siRNAs designed against the same target gene (to achieve robust knockdown), each at a concentration of 25 nM. siRNAs designed against Yap, Llgl1, Llgl2, a combination of Llgl1 and Llgl2, or negative control siRNA (Table S2) were transfected with Mission siRNA Transfection Reagent (Sigma-Aldrich). Untreated and transfected cells were cultured in DMEM supplemented with 7.5% FBS for 48 hours. Cells were then collected in TRIzol Reagent for RNA extraction for qRT-PCR and RNAseq, or collected in lysis buffer for western blotting as described by O'Meara et al. (2015).
RNAseq
RNA was extracted from neonatal rat primary cardiomyocytes transfected with siRNA designed against a combination of Llgl1 and Llgl2 or negative control siRNA (Table S2), using the TRIzol (Ambion) extraction protocol recommended by the manufacturer. RNA quality was determined using an Agilent BioAnalyzer. RNA libraries were prepared by BGI Americas using the low-input poly-A BGISEQ-500RSRNASeq protocol. Sequencing results were analyzed by BGI Americas. Primary sequencing data were generated with a BGISEQ-500 50SE at 20 million reads per sample. Bowtie2 was used to map reads to a reference genome. GSEA (Broad Institute) was performed on RNAseq results (fragments per kilobase million).
qRT-PCR
RNA was extracted using TRIzol (Thermo Fisher Scientific) phenol chloroform extraction. cDNA was generated using a Superscript III First-Strand Synthesis System RT-PCR Kit (Invitrogen) according to the manufacturer's instructions. qRT-PCR was performed using a CFX96 and CFX Connect Real-Time System (Bio-Rad) using SsoAdvanced SYBR Green Supermix (Bio-Rad) or TaqMan Gene Expression Master Mix (Thermo Fisher Scientific). Primers used are listed in Table S3. Quantifications were normalized to TATA-binding protein (Tbp) for in vitro rat cell cultures and eef1a1l1 for zebrafish hearts. Product size and uniformity was assessed via melt curve analysis.
Western blotting
Cell lysates were acquired from siRNA-treated primary rat cardiomyocytes as described by O'Meara et al. (2015). Gel electrophoresis was performed using a 4-15% Mini-PROTEAN TGX precast gel (Bio-Rad) followed by blotting onto Immobilon-FL PVDF membranes (Millipore). Antibodies and concentrations used for immunochemistry are described in Table S4. Protein detection was performed using an Odyssey infrared imager (LI-COR Biosciences).
Statistical analysis
Data were analyzed using Prism 7 (GraphPad). Two-way ANOVA followed by Tukey's multiple comparisons tests were performed on samples with two experimental factors. Statistical comparisons between two groups were analyzed by Student's t-test, and for three or more groups by one-way ANOVA followed by Tukey's multiple comparisons test. A χ2 test of independence was used to compare the degree of embryonic pericardial effusion between maternal lineages, whereas a two-tail binomial test was used to compare genotypes in adult populations with expected Mendelian ratios.
Supplementary Material
Acknowledgements
We acknowledge Shelby Hader, Dr Dawid Chabowski and Dr David Gutterman (Department of Physiology, The Medical College of Wisconsin) for their assistance with imaging zebrafish vasculature.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: C.C.O., B.A.L.; Methodology: M.A.F., C.O., Z.J.B.; Formal analysis: M.A.F., C.O., Z.J.B., T.C.W., J.A.A., S.A.-S, C.C.O., B.A.L.; Investigation: M.A.F., C.O., Z.J.B., J.R.B., A.K., T.C.W., J.A.A., S.A.-S, C.C.O., B.A.L.; Writing - original draft: M.A.F., C.C.O., B.A.L.; Writing - review & editing: M.A.F., C.O., Z.J.B., J.R.B., A.K., T.C.W., J.A.A., S.A.-S, C.C.O., B.A.L.; Supervision: C.C.O., B.A.L.; Project administration: B.A.L.; Funding acquisition: J.A.A., S.A.-S, C.C.O., B.A.L.
Funding
This work was supported by the Cardiovascular Center and Research and Education Program Fund at the Medical College of Wisconsin (C.C.O., J.A.A. and B.A.L.); by Advancing a Healthier Wisconsin Co-Investigator Grant (B.A.L. and C.C.O.); by the National Institutes of Health (R01 HL141159 to C.C.O., 5R01HL131788 and 1S10 OD025038 to J.A.A., and T32 HL134643 to M.A.F.); by the Cardiovascular Center's A.O. Smith Fellowship Scholars Program (M.A.F.); by the Medical College of Wisconsin Cardiovascular Center [FP00012308 to J.A.A.]; by Excellence cluster REBIRTH (SFB958 to S.A.-S.); by Deutsche Forschungsgemeinschaft (SE2016/7-2 and SE2016/10-1 to S.A.-S.); and by the Deutsches Zentrum für Herz-Kreislaufforschung (S.A.-S.). Deposited in PMC for release after 12 months.
Data availability
The RNAseq data generated in this study are available in the GEO under accession number GSE121929.
Supplementary information
Supplementary information available online at https://dev.biologists.org/lookup/doi/10.1242/dev.193581.supplemental
Peer review history
The peer review history is available online at https://dev.biologists.org/lookup/doi/10.1242/dev.193581.reviewer-comments.pdf
References
- Andrés-Delgado L. and Mercader N. (2016). Interplay between cardiac function and heart development. Biochim. Biophys. Acta Mol. Cell Res. 1863, 1707-1716. 10.1016/j.bbamcr.2016.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artap S., Manderfield L. J., Smith C. L., Poleshko A., Aghajanian H., See K., Li L., Jain R. and Epstein J. A. (2018). Endocardial Hippo signaling regulates myocardial growth and cardiogenesis. Dev. Biol. 440, 22-30. 10.1016/j.ydbio.2018.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boczonadi V., Gillespie R., Keenan I., Ramsbottom S. A., Donald-Wilson C., Al Nazer M., Humbert P., Schwarz R. J., Chaudhry B. and Henderson D. J. (2014). Scrib:Rac1 interactions are required for the morphogenesis of the ventricular myocardium. Cardiovasc. Res. 104, 103-115. 10.1093/cvr/cvu193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Friedrich G. A. and Soriano P. (1994). Transcriptional enhancer factor 1 disruption by a retroviral gene trap leads to heart defects and embryonic lethality in mice. Genes Dev. 8, 2293-2301. 10.1101/gad.8.19.2293 [DOI] [PubMed] [Google Scholar]
- Clark B. S., Cui S., Miesfeld J. B., Klezovitch O., Vasioukhin V. and Link B. A. (2012). Loss of Llgl1 in retinal neuroepithelia reveals links between apical domain size, Notch activity and neurogenesis. Development 139, 1599-1610. 10.1242/dev.078097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng H., Wang W., Yu J., Zheng Y., Qing Y. and Pan D. (2015). Spectrin regulates Hippo signaling by modulating cortical actomyosin activity. eLife 4, e06567 10.7554/eLife.06567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donat S., Lourenço M., Paolini A., Otten C., Renz M. and Abdelilah-Seyfried S. (2018). Heg1 and Ccm1/2 proteins control endocardial mechanosensitivity during zebrafish valvulogenesis. eLife 7, e28939 10.7554/eLife.28939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont S., Morsut L., Aragona M., Enzo E., Giulitti S., Cordenonsi M., Zanconato F., Le Digabel J., Forcato M., Bicciato S. et al. (2011). Role of YAP/TAZ in mechanotransduction. Nature 474, 179-183. 10.1038/nature10137 [DOI] [PubMed] [Google Scholar]
- Durst R., Sauls K., Peal D. S., deVlaming A., Toomer K., Leyne M., Salani M., Talkowski M. E., Brand H., Perrocheau M. et al. (2015). Mutations in DCHS1 cause mitral valve prolapse. Nature 525, 109-113. 10.1038/nature14670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher G. C., Diaz-de-la-Loza M.-C., Borreguero-Muñoz N., Holder M., Aguilar-Aragon M. and Thompson B. J. (2018). Mechanical strain regulates the hippo pathway in Drosophila. Development 145, dev159467 10.1242/dev.159467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flinn M. A., Jeffery B. E., O'Meara C. C. and Link B. A. (2019). Yap is required for scar formation but not myocyte proliferation during heart regeneration in zebrafish. Cardiovasc. Res. 115, 570-577. 10.1093/cvr/cvy243 [DOI] [PubMed] [Google Scholar]
- Flinn M. A., Link B. A. and O'Meara C. C. (2020). Upstream regulation of the Hippo-Yap pathway in cardiomyocyte regeneration. Semin. Cell Dev. Biol. 100, 11-19. 10.1016/j.semcdb.2019.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood E., Maisel S., Ebertz D., Russ A., Pandey R., Schroeder J., Greenwood E., Maisel S., Ebertz D., Russ A. et al. (2016). Llgl1 prevents metaplastic survival driven by epidermal growth factor dependent migration. Oncotarget 7, 60776-60792. 10.18632/oncotarget.11320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzeschik N. A., Parsons L. M., Allott M. L., Harvey K. F. and Richardson H. E. (2010). Lgl, aPKC, and Crumbs regulate the Salvador/Warts/Hippo pathway through two distinct mechanisms. Curr. Biol. 20, 573-581. 10.1016/j.cub.2010.01.055 [DOI] [PubMed] [Google Scholar]
- Haack T. and Abdelilah-Seyfried S. (2016). The force within: endocardial development, mechanotransduction and signalling during cardiac morphogenesis. Development 143, 373-386. 10.1242/dev.131425 [DOI] [PubMed] [Google Scholar]
- Hao Y., Chun A., Cheung K., Rashidi B. and Yang X. (2008). Tumor suppressor LATS1 Is a negative regulator of oncogene YAP. J. Biol. Chem. 283, 5496-5509. 10.1074/jbc.M709037200 [DOI] [PubMed] [Google Scholar]
- Heallen T., Zhang M., Wang J., Bonilla-Claudio M., Klysik E., Johnson R. L. and Martin J. F. (2011). Hippo pathway inhibits wnt signaling to restrain cardiomyocyte proliferation and heart size. Science 332, 458-461. 10.1126/science.1199010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jossin Y., Lee M., Klezovitch O., Kon E., Cossard A., Lien W.-H., Fernandez T. E., Cooper J. A. and Vasioukhin V. (2017). Llgl1 connects cell polarity with cell-cell adhesion in embryonic neural stem cells. Dev. Cell 41, 481-495.e5. 10.1016/j.devcel.2017.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashyap A., Zimmerman T., Ergül N., Bosserhoff A., Hartman U., Alla V., Bataille F., Galle P. R., Strand S. and Strand D. (2013). The human Lgl polarity gene, Hugl-2, induces MET and suppresses snail tumorigenesis. Oncogene 32, 1396-1407. 10.1038/onc.2012.162 [DOI] [PubMed] [Google Scholar]
- Kimmel C. B., Ballard W. W., Kimmel S. R., Ullmann B. and Schilling T. F. (1995). Stages of embryonic development of the zebrafish. Dev. Dyn. 203, 253-310. 10.1002/aja.1002030302 [DOI] [PubMed] [Google Scholar]
- Kwan K. M., Fujimoto E., Grabher C., Mangum B. D., Hardy M. E., Campbell D. S., Parant J. M., Yost H. J., Kanki J. P. and Chien C.-B. (2007). The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev. Dyn. 236, 3088-3099. 10.1002/dvdy.21343 [DOI] [PubMed] [Google Scholar]
- Lee L., Genge C. E., Cua M., Sheng X., Rayani K., Beg M. F., Sarunic M. V. and Tibbits G. F. (2016). Functional assessment of cardiac responses of adult zebrafish (Danio rerio) to acute and chronic temperature change using high-resolution echocardiography. PLoS ONE 11, e0145163 10.1371/journal.pone.0145163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Gao E., Vite A., Yi R., Gomez L., Goossens S., van Roy F. and Radice G. L. (2015). Alpha-catenins control cardiomyocyte proliferation by regulating Yap activity. Circ. Res. 116, 70-79. 10.1161/CIRCRESAHA.116.304472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Huang W. and Lei Q. (2011). Regulation and function of the TAZ transcription co-activator. Int. J. Biochem. Mol. Biol. 2, 247-256. [PMC free article] [PubMed] [Google Scholar]
- Miesfeld J. B. and Link B. A. (2014). Establishment of transgenic lines to monitor and manipulate Yap/Taz-Tead activity in zebrafish reveals both evolutionarily conserved and divergent functions of the Hippo pathway. Mech. Dev. 133, 177-188. 10.1016/j.mod.2014.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miesfeld J. B., Gestri G., Clark B. S., Flinn M. A., Poole R. J., Bader J. R., Besharse J. C., Wilson S. W. and Link B. A. (2015). Yap and Taz regulate retinal pigment epithelial cell fate. Development 142, 3021-3032. 10.1242/dev.119008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morikawa Y., Heallen T., Leach J., Xiao Y. and Martin J. F. (2017). Dystrophin–glycoprotein complex sequesters Yap to inhibit cardiomyocyte proliferation. Nature 547, 227 10.1038/nature22979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neto F., Klaus-Bergmann A., Ong Y. T., Alt S., Vion A.-C., Szymborska A., Carvalho J. R., Hollfinger I., Bartels-Klein E., Franco C. A. et al. (2018). YAP and TAZ regulate adherens junction dynamics and endothelial cell distribution during vascular development. eLife 7, e31037 10.7554/eLife.31037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Meara C. C., Wamstad J. A., Gladstone R. A., Fomovsky G. M., Butty V. L., Shrikumar A., Gannon J. B., Boyer L. A. and Lee R. T. (2015). Transcriptional reversion of cardiac myocyte fate during mammalian cardiac regeneration. Circ. Res. 116, 804-815. 10.1161/CIRCRESAHA.116.304269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paolini A. and Abdelilah-Seyfried S. (2018). The mechanobiology of zebrafish cardiac valve leaflet formation. Curr. Opin. Cell Biol. 55, 52-58. 10.1016/j.ceb.2018.05.007 [DOI] [PubMed] [Google Scholar]
- Parsons L. M., Grzeschik N. A. and Richardson H. E. (2014a). lgl Regulates the Hippo Pathway independently of Fat/Dachs, Kibra/Expanded/Merlin and dRASSF/dSTRIPAK. Cancers (Basel) 6, 879-896. 10.3390/cancers6020879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons L. M., Portela M., Grzeschik N. A. and Richardson H. E. (2014b). Lgl regulates Notch signaling via endocytosis, independently of the apical aPKC-Par6-Baz polarity complex. Curr. Biol. 24, 2073-2084. 10.1016/j.cub.2014.07.075 [DOI] [PubMed] [Google Scholar]
- Ragni C. V., Diguet N., Le Garrec J.-F., Novotova M., Resende T. P., Pop S., Charon N., Guillemot L., Kitasato L., Badouel C. et al. (2017). Amotl1 mediates sequestration of the Hippo effector Yap1 downstream of Fat4 to restrict heart growth. Nat. Commun. 8, 14582 10.1038/ncomms14582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reischauer S., Arnaout R., Ramadass R. and Stainier D. Y. R. (2014). Actin binding GFP allows 4D in vivo imaging of myofilament dynamics in the zebrafish heart and the identification of Erbb2 signaling as a remodeling factor of myofibril architecture. Circ. Res. 115, 845-856. 10.1161/CIRCRESAHA.115.304356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renz M., Otten C., Faurobert E., Rudolph F., Zhu Y., Boulday G., Duchene J., Mickoleit M., Dietrich A.-C., Ramspacher C. et al. (2015). Regulation of β1 integrin-Klf2-mediated angiogenesis by CCM proteins. Dev. Cell 32, 181-190. 10.1016/j.devcel.2014.12.016 [DOI] [PubMed] [Google Scholar]
- Samsa L. A., Givens C., Tzima E., Stainier D. Y. R., Qian L. and Liu J. (2015). Cardiac contraction activates endocardial notch signaling to modulate chamber maturation in zebrafish. Development 142, 4080-4091. 10.1242/dev.125724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlegelmilch K., Mohseni M., Kirak O., Pruszak J., Rodriguez J. R., Zhou D., Kreger B. T., Vasioukhin V., Avruch J., Brummelkamp T. R. et al. (2011). Yap1 acts downstream of α-catenin to control epidermal proliferation. Cell 144, 782-795. 10.1016/j.cell.2011.02.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonawane M., Carpio Y., Geisler R., Schwarz H., Maischein H.-M. and Nuesslein-Volhard C. (2005). Zebrafish penner/lethal giant larvae 2 functions in hemidesmosome formation, maintenance of cellular morphology and growth regulation in the developing basal epidermis. Development 132, 3255-3265. 10.1242/dev.01904 [DOI] [PubMed] [Google Scholar]
- Steed E., Boselli F. and Vermot J. (2016). Hemodynamics driven cardiac valve morphogenesis. Biochim. Biophys. Acta Mol. Cell Res. 1863, 1760-1766. 10.1016/j.bbamcr.2015.11.014 [DOI] [PubMed] [Google Scholar]
- Sun G. and Irvine K. D. (2013). Ajuba family proteins link JNK to Hippo signaling. Sci. Signal. 6, ra81 10.1126/scisignal.2004324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S., Reddy B. V. V. G. and Irvine K. D. (2015). Localization of Hippo signalling complexes and Warts activation in vivo. Nat. Commun. 6, 8402 10.1038/ncomms9402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vite A., Zhang C., Yi R., Emms S. and Radice G. L. (2018). Alpha-catenin-dependent cytoskeletal tension controls Yap activity in the heart. Development 145, dev149823 10.1242/dev.149823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Gise A., Lin Z., Schlegelmilch K., Honor L. B., Pan G. M., Buck J. N., Ma Q., Ishiwata T., Zhou B., Camargo F. D. et al. (2012). YAP1, the nuclear target of Hippo signaling, stimulates heart growth through cardiomyocyte proliferation but not hypertrophy. Proc. Natl. Acad. Sci. USA 109, 2394-2399. 10.1073/pnas.1116136109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh E. C. and Stainier D. Y. R. (2001). UDP-glucose dehydrogenase required for cardiac valve formation in zebrafish. Science 293, 1670-1673. 10.1126/science.293.5535.1670 [DOI] [PubMed] [Google Scholar]
- Wang T., Liu Y., Xu X.-H., Deng C.-Y., Wu K.-Y., Zhu J., Fu X.-Q., He M. and Luo Z.-G. (2011). Lgl1 activation of Rab10 promotes axonal membrane trafficking underlying neuronal polarization. Dev. Cell 21, 431-444. 10.1016/j.devcel.2011.07.007 [DOI] [PubMed] [Google Scholar]
- Wang J., Liu S., Heallen T. and Martin J. F. (2018). The Hippo pathway in the heart: pivotal roles in development, disease, and regeneration. Nat. Rev. Cardiol. 15, 672 10.1038/s41569-018-0063-3 [DOI] [PubMed] [Google Scholar]
- Xiao Y., Hill M. C., Zhang M., Martin T. J., Morikawa Y., Wang S., Moise A. R., Wythe J. D. and Martin J. F. (2018). Hippo signaling plays an essential role in cell state transitions during cardiac fibroblast development. Dev. Cell 45, 153-169.e6. 10.1016/j.devcel.2018.03.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin M., Kim Y., Sutherland L. B., Murakami M., Qi X., McAnally J., Porrello E. R., Mahmoud A. I., Tan W., Shelton J. M. et al. (2013). Hippo pathway effector Yap promotes cardiac regeneration. Proc. Natl. Acad. Sci. USA 110, 13839-13844. 10.1073/pnas.1313192110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka T., Horikoshi Y., Sugiyama Y., Ishiyama C., Suzuki A., Hirose T., Iwamatsu A., Shinohara A. and Ohno S. (2003). Mammalian Lgl forms a protein complex with PAR-6 and aPKC independently of PAR-3 to regulate epithelial cell polarity. Curr. Biol. 13, 734-743. 10.1016/S0960-9822(03)00244-6 [DOI] [PubMed] [Google Scholar]
- Zanconato F., Forcato M., Battilana G., Azzolin L., Quaranta E., Bodega B., Rosato A., Bicciato S., Cordenonsi M. and Piccolo S. (2015). Genome-wide association between YAP/TAZ/TEAD and AP-1 at enhancers drives oncogenic growth. Nat. Cell Biol. 17, 1218-1227. 10.1038/ncb3216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., von Gise A., Liu Q., Hu T., Tian X., He L., Pu W., Huang X., He L., Cai C.-L. et al. (2014). Yap1 is required for endothelial to mesenchymal transition of the atrioventricular cushion. J. Biol. Chem. 289, 18681-18692. 10.1074/jbc.M114.554584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Li L., Tumaneng K., Wang C.-Y. and Guan K.-L. (2010). A coordinated phosphorylation by Lats and CK1 regulates YAP stability through SCFbeta-TRCP. Genes Dev. 24, 72-85. 10.1101/gad.1843810 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.