ABSTRACT
Spinal cord pMN progenitors sequentially produce motor neurons and oligodendrocyte precursor cells (OPCs). Some OPCs differentiate rapidly as myelinating oligodendrocytes, whereas others remain into adulthood. How pMN progenitors switch from producing motor neurons to OPCs with distinct fates is poorly understood. pMN progenitors express prdm8, which encodes a transcriptional repressor, during motor neuron and OPC formation. To determine whether prdm8 controls pMN cell fate specification, we used zebrafish as a model system to investigate prdm8 function. Our analysis revealed that prdm8 mutant embryos have fewer motor neurons resulting from a premature switch from motor neuron to OPC production. Additionally, prdm8 mutant larvae have excess oligodendrocytes and a concomitant deficit of OPCs. Notably, pMN cells of mutant embryos have elevated Shh signaling, coincident with the motor neuron to OPC switch. Inhibition of Shh signaling restored the number of motor neurons to normal but did not rescue the proportion of oligodendrocytes. These data suggest that Prdm8 regulates the motor neuron-OPC switch by controlling the level of Shh activity in pMN progenitors, and also regulates the allocation of oligodendrocyte lineage cell fates.
This article has an associated ‘The people behind the papers’ interview.
KEY WORDS: Sonic hedgehog, Motor neurons, Oligodendrocytes, pMN progenitors, Spinal cord, Zebrafish
Highlighted Article: Expression and functional analyses revealed that Prdm8 regulates the timing of a motor neuron-oligodendrocyte switch and oligodendrocyte lineage cell identity in the zebrafish spinal cord by dampening Shh activity in pMN progenitors.
INTRODUCTION
Oligodendrocytes, one of the major glial cell types of the central nervous system (CNS) of vertebrate animals, increase the speed of axon electrical impulses and support neuron health by wrapping axons with a myelin membrane (Simons and Nave, 2016). In the spinal cord, most oligodendrocytes originate from ventral pMN progenitors (Noll and Miller, 1993; Warf et al., 1991), which express the basic helix loop helix (bHLH) transcription factor Olig2 (Lu et al., 2000; Novitch et al., 2001; Zhou and Anderson, 2002; Zhou et al., 2000). pMN progenitors first produce motor neurons followed by oligodendrocyte precursor cells (OPCs) (Richardson et al., 2000; Rowitch, 2004). After specification, some OPCs rapidly differentiate as myelinating oligodendrocytes, whereas other OPCs persist into adulthood (Dawson et al., 2003; Pringle et al., 1992; Wolswijk and Noble, 1989). The switch from motor neuron to OPC production and the subsequent regulation of oligodendrocyte differentiation require tight control of gene expression through a complex network of interacting transcription factors and extracellular cues. Although many factors that promote oligodendrocyte differentiation and myelination have been identified (Elbaz and Popko, 2019; Emery, 2010; He and Lu, 2013; Sock and Wegner, 2019; Zuchero and Barres, 2013), the mechanisms that regulate the onset of OPC specification and maintain some of them in a non-myelinating state are not well understood.
During early neural tube patterning, pMN progenitors are specified by the morphogen Sonic hedgehog (Shh). The Shh ligand, secreted by the notochord, a mesodermal structure below the ventral spinal cord, and by the floor plate, the ventralmost cells of the neural tube, signals neural progenitors to acquire ventral identities (Echelard et al., 1993; Martí et al., 1995; Roelink et al., 1994). The Shh ligand binds to its transmembrane receptor Patched (Ptch), which allows intercellular Shh signaling to be transduced by Smoothened (Smo). Upon Shh binding, Smo is internalized to promote GliA transcriptional activity by inhibiting its cleavage to GliR (Briscoe and Thérond, 2013; Danesin and Soula, 2017; Ribes and Briscoe, 2009). Graded Shh activity induces expression of genes that encode bHLH and homeodomain proteins at distinct positions on the dorsoventral axis (Briscoe and Thérond, 2013; Briscoe et al., 2000; Poh et al., 2002). The duration of Shh signaling also influences cell fate and gene expression. Initially, high ventral Shh signaling activates expression of Olig2, then sustained Shh activity promotes expression of Nkx2.2 adjacent to the floor plate, ventral to Olig2, thus forming two distinct ventral progenitor domains (Dessaud et al., 2007, 2010). The sequential induction of cross-repressive transcription factors by graded morphogen activity along the dorsoventral axis creates spatially restricted progenitor domains that sequentially give rise to specific neurons and glia (Briscoe et al., 2000; Lek et al., 2010; Nishi et al., 2015).
Shh activity is necessary for the establishment of the pMN domain and motor neuron formation, and subsequently, a transient increase in Shh activity coincides with, and is required for, timely OPC specification (Danesin and Soula, 2017). Pharmacological and genetic reduction of Shh signaling in chick, mouse and zebrafish spinal cords results in prolonged motor neuron formation and impaired OPC formation (Al Oustah et al., 2014; Danesin et al., 2006; Hashimoto et al., 2017; Jiang et al., 2017; Ravanelli and Appel, 2015; Touahri et al., 2012). Furthermore, in chick neural tube explants treated with exogenous Shh, premature formation of OPCs occurs at the expense of motor neurons (Danesin et al., 2006; Orentas et al., 1999). Thus, transient elevation of Shh activity is required to induce the transition from motor neuron to OPC production. The temporal change in Shh activity is in part due to the upregulation of Sulfatase 1/2 by p3 cells, which are ventral to pMN progenitors, before OPC specification (Al Oustah et al., 2014; Danesin et al., 2006; Jiang et al., 2017). Sulfatase expression increases the local Shh ligand concentration available to pMN progenitors (Al Oustah et al., 2014; Danesin et al., 2006), and loss of Sulfatase 1/2 functions delay the motor neuron-OPC switch (Jiang et al., 2017). Whether additional mechanisms contribute to the modulation of Shh signaling strength to regulate fate specification over time, is not well understood.
In addition to expressing distinct combinations of bHLH and homeodomain transcription factors, subsets of spinal cord progenitors express specific PRDI-BF1 and RIZ homology domain containing (Prdm) proteins (Zannino and Sagerström, 2015). This family of proteins contains an N-terminal SET domain followed by a varied number of C-terminal zinc-finger repeats and they act as transcriptional regulators or co-factors implicated in nervous system patterning, neural stem cell maintenance and differentiation (Baizabal et al., 2018; Chittka et al., 2012; Hanotel et al., 2014; Hernandez-Lagunas et al., 2011; Kinameri et al., 2008; Ross et al., 2012; Thélie et al., 2015; Yildiz et al., 2019). In the ventral mouse spinal cord, neural progenitors express Prdm8 from embryonic (E) day 9.5 to E13.5 (Kinameri et al., 2008; Komai et al., 2009), corresponding to the period of motor neuron and OPC formation. The function of Prdm8 in spinal cord development is not yet known, but in the mouse telencephalon Prdm8 forms a repressive complex with Bhlhb5, a bHLH transcription factor closely related to Olig2, to regulate axonal migration (Ross et al., 2012). Moreover, in the retina, Prdm8 promotes the formation of a subset of rod bipolar cells and regulates amacrine cell type identity (Jung et al., 2015). These data raise the possiblility that Prdm8 regulates pMN cell development.
To investigate prdm8 expression and function in pMN progenitors, we used the developing zebrafish spinal cord as a model. Our expression analysis showed that pMN progenitors express prdm8 before and during the switch from motor neuron to OPC production, and that, subsequently, differentiating oligodendrocytes downregulate prdm8 expression. Because Prdm8 can control cell fate, we therefore hypothesized that Prdm8 regulates motor neuron and OPC specification. To test this hypothesis, we performed a series of experiments to assess changes in pMN cell fates resulting from loss of prdm8 function. Our data reveal that prdm8 mutant embryos have a deficit of late-born motor neurons, excess differentiating oligodendrocytes and a deficit of OPCs. Birth-dating studies showed that the motor neuron deficit results from a premature switch from motor neuron to OPC production. prdm8 mutant embryos have abnormally high levels of Shh signaling and pharmacological suppression of Shh signaling rescued the motor neuron deficit but not the formation of excess oligodendrocytes. Taken together, our data suggest that Prdm8 dampens Shh signaling activity to modulate the timing of the motor neuron-OPC switch, and secondarily regulates the myelinating fate of oligodendrocyte lineage cells.
RESULTS
pMN progenitors and oligodendrocyte lineage cells express prdm8
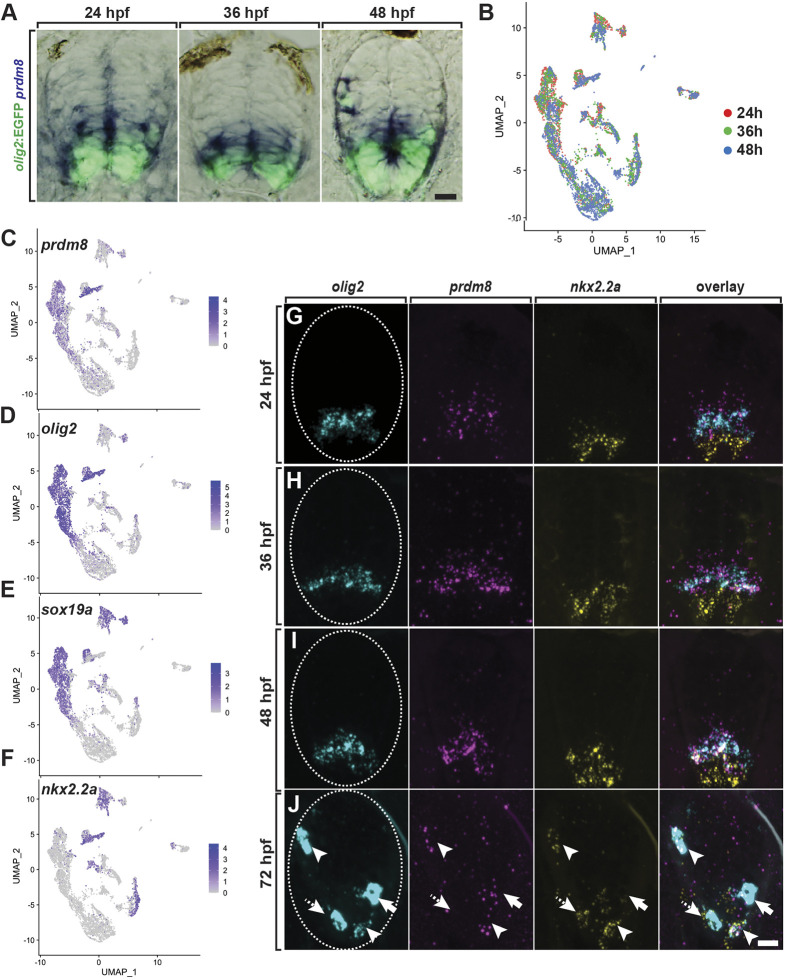
To begin our investigation of prdm8 function, we first assessed the temporal and spatial features of prdm8 expression in the zebrafish spinal cord during development. To do so, we performed in situ RNA hybridization (RNA ISH) using transverse sections obtained from Tg(olig2:EGFP) embryos and larvae. pMN cells in these fish express EGFP driven by olig2 regulatory DNA (Shin et al., 2003). pMN cells and cells dorsal to the pMN domain expressed prdm8 at 24, 36 and 48 h postfertilization (hpf) (Fig. 1A). This is consistent with previous data revealing that cells of the pMN, p2 and p1 domains in the developing mouse spinal cord express Prdm8 (Kinameri et al., 2008; Komai et al., 2009). Next we evaluated pMN cell expression of prdm8 through development using a single cell RNA-seq (scRNA-seq) dataset obtained from olig2:EGFP+ cells isolated from 24, 36 and 48 hpf embryos, a period spanning the formation of most motor neurons and OPCs. An aligned Harmony clustering analysis and uniform manifold approximation and projection (UMAP) of the data, revealed that gene expression profiles formed several distinct clusters (Fig. 1B). Plotting individual gene expression profiles revealed that many olig2+ sox19a+ cells, which likely represent pMN progenitors, also expressed prdm8 (Fig. 1C-E).
Fig. 1.
pMN progenitors express prdm8. (A) Representative transverse sections of trunk spinal cord (dorsal up), showing prdm8 RNA (blue) and olig2:EGFP (green) expression. Developmental stages noted at the top. (B) Harmony visualization of the scRNA-seq dataset from olig2:EGFP+ spinal cord cells obtained from 24, 36 and 48 hpf Tg(olig2:EGFP) embryos. Each point represents one cell (n=6489). Colors represent sample time points. (C-F) DimPlots of selected transcripts. Cells are colored by expression level (gray is low, purple is high). prdm8 expression overlaps extensively with olig2 and sox19a, and partially with nkx2.2a. (G-J) Representative transverse trunk spinal cord sections processed for fluorescent ISH to detect olig2, prdm8 and nkx2.2a mRNA at 24 hpf (G) 36 hpf (H) 48 hpf (I) and 72 hpf (J). Arrowheads indicate prdm8+/nkx2.2a+/olig2+ cells; solid arrows denote prdm8+/nkx2.2a–/olig2+ cells; dashed arrows indicate prdm8–/nkx2.2a+/olig2+ cells; and dashed ovals outline the spinal cord. Scale bars: 10 μm.
Following motor neuron formation, pMN progenitors begin to form OPCs. This is initiated by the reorganization of ventral progenitor domains, such that pMN cells that enter the oligodendrocyte lineage begin to co-express Nkx2.2 and Olig2 (Agius et al., 2004; Fu et al., 2002; Kessaris et al., 2001; Soula et al., 2001; Zhou et al., 2001). Our scRNA-seq data show that at 48 hpf, cells that expressed nkx2.2a and olig2 also expressed prdm8, signifying that nascent OPCs express prdm8 (Fig. 1F). We validated these observations using fluorescent RNA ISH. Consistent with our scRNA-seq data, prdm8 mRNA puncta were present in the pMN domain marked by olig2 expression at 24, 36 and 48 hpf (Fig. 1G-I). At 24 and 36 hpf, cells that expressed prdm8 and olig2 were adjacent to more ventral nkx2.2a+ p3 domain cells (Fig. 1G,H), but at 48 hpf some cells at the p3/pMN border expressed all three transcripts (Fig. 1I). By 72 hpf, the expression of olig2 mRNA was mostly depleted from the pMN domain but evident at a high level in oligodendrocyte lineage cells. At this stage, some olig2+ cells expressed both prdm8 and nkx2.2a, some expressed only nkx2.2a and others expressed only prdm8 (Fig. 1J). These data indicate that following pMN progenitor cell expression, prdm8 expression is differentially maintained by oligodendrocyte lineage cells.
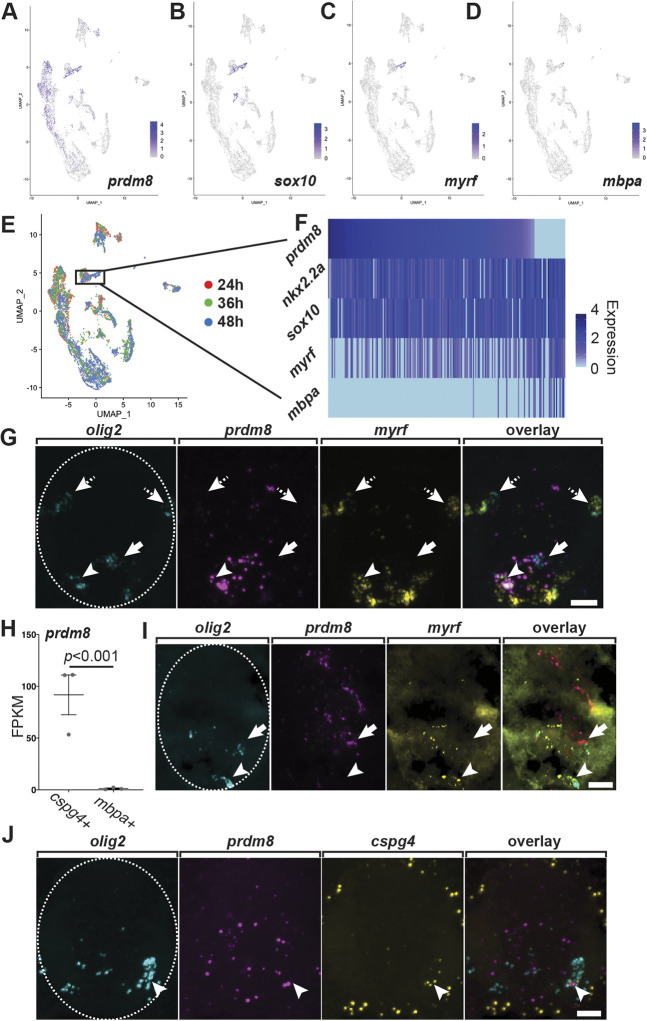
To determine the identity of prdm8+ cells, we compared prdm8 expression with the expression of genes characteristic of the oligodendrocyte lineage in the scRNA-seq dataset. At 48 hpf, a subset of cells that expressed prdm8 also expressed sox10, which encodes a transcription factor expressed by all oligodendrocyte lineage cells (Britsch et al., 2001; Kuhlbrodt et al., 1998; Park et al., 2002) (Fig. 2A,B). Some prdm8+ sox10+ cells also expressed myrf, which encodes Myelin regulatory factor, a transcription factor required for oligodendrocyte differentiation (Emery et al., 2009) (Fig. 2C). This dataset included only a few mbpa+ cells, and these appeared as a small subset of sox10+ myrf+ cells (Fig. 2D). Therefore, these cells represent pre-myelinating oligodendrocytes. A heatmap representation of these cells (Fig. 2E,F) showed that most sox10+ nkx2.2a+ cells expressed prdm8 at high levels. However, cells that also expressed myrf and mbpa at higher levels had little prdm8 expression. These data suggest that pre-myelinating oligodendrocytes downregulate prdm8 expression as they differentiate.
Fig. 2.
Differentiating oligodendrocytes downregulate prdm8 expression. (A-D) DimPlots of select transcripts. Cells are colored by expression level (gray is low, purple is high). prdm8 expression overlaps considerably with sox10 and myrf but not with mbpa. (E) Harmony visualization of clustered samples across all time points. Pre-myelinating oligodendrocytes at 48 hpf (blue; n=220) are indicated by the box. (F) Heatmap showing selected transcripts expressed by pre-myelinating oligodendrocytes at 48 hpf. (G) Representative transverse trunk spinal cord sections obtained from 72 hpf larvae processed for fluorescent ISH (dorsal is up). Arrowheads mark prdm8+/myrf+/olig2+ pre-myelinating oligodendrocytes that express prdm8; dashed arrows point to prdm8–/myrf+/olig2+ pre-myelinating oligodendrocytes that do not express prdm8; and solid arrows denote prdm8+/myrf–/olig2+ OPCs. (H) prdm8 expression (FPKM) in OPCs (n=3) and oligodendrocytes (n=3) isolated from batched 7 dpf larvae. Data are mean±s.e.m. with individual data points indicated. Statistical significance assessed by one-way ANOVA. (I,J) Representative transverse trunk spinal cord sections obtained from 7 dpf larvae processed for fluorescent ISH (dorsal on top). (I) Arrowheads denote a prdm8–/myrf+/olig2+ myelinating oligodendrocyte and solid arrow denotes a prdm8+/myrf–/olig2+ OPC. (J) Arrowheads indicate a prdm8+/cspg4+/olig2+ OPC and the dashed oval outlines the spinal cord boundary. Scale bars: 10 μm.
To corroborate these observations, we examined prdm8, myrf and olig2 mRNA expression using fluorescent RNA ISH in the trunk spinal cord of 72 hpf larvae. Consistent with our scRNA-seq findings, this revealed that a majority of olig2+ cells expressed either prdm8 or myrf, and that few olig2+ cells expressed both genes (Fig. 2G). To determine whether some oligodendrocyte lineage cells maintain prdm8 expression, we assessed RNA-seq data collected from cspg4+ OPCs and mbpa+ oligodendrocytes isolated from 7 days postfertilization (dpf) larvae (Ravanelli et al., 2018). We found that OPCs expressed prdm8 at 75-fold higher levels than oligodendrocytes (Fig. 2H). We validated these data using fluorescent RNA ISH to label prdm8 mRNA at 7 dpf in combination with myrf to mark oligodendrocytes (Fig. 2I), or cspg4 to mark OPCs (Fig. 2J). This revealed that OPCs, but not oligodendrocytes, expressed prdm8, thus confirming our RNA-seq data. We therefore conclude that pMN progenitors and OPCs express prdm8, and that prdm8 expression declines in oligodendrocytes undergoing differentiation.
Zebrafish larvae lacking prmd8 function have excess oligodendrocytes and a deficit of OPCs
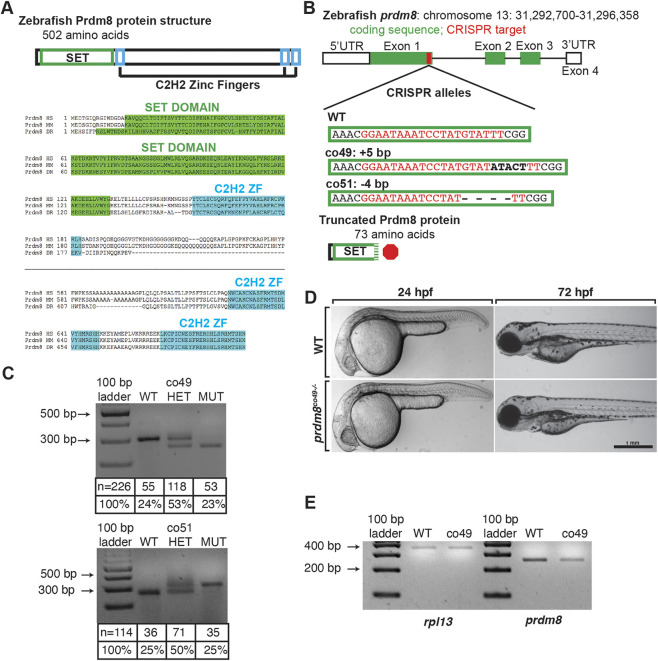
Zebrafish prdm8 encodes a 502 amino acid protein containing an N-terminal PR/SET domain and three zinc-finger domains, similar to its human and mouse orthologs (Fig. 3A). To investigate Prdm8 function, we used CRISPR/Cas9 to create gene-disrupting mutations within the first exon (Fig. 3B). We verified the efficiency of single guide RNA (sgRNA) targeting using diagnostic fluorescent PCR and subsequently raised injected embryos to adulthood. We identified F0 adults that transmitted mutations through the germ line and selected two, prdm8co49 and prdm8co51, for further analysis. DNA sequencing revealed that the co49 allele contained a 5 bp insertion, whereas the co51 allele had a 4 bp deletion. Both alleles were predicted to result in translation frameshifting leading to premature translation termination and proteins truncated within the PR/SET domain (Fig. 3B). Because the C-terminal zinc finger domains of mouse Prdm8 are necessary for nuclear localization (Eom et al., 2009), we predicted that the truncated proteins produced by the co49 and co51 alleles would be non-functional. Genotyping assays revealed that F1 heterozygous adults transmitted mutant alleles to progeny with Mendelian frequencies (Fig. 3C). Homozygous mutant embryos had no discernible morphological phenotype at 24 hpf or at early larval stages (Fig. 3D). To assess the effect of the co49 allele on prdm8 transcript levels, we performed RT-PCR. This revealed a partial reduction in the levels of mutant prdm8 mRNA relative to wild type (Fig. 3E).
Fig. 3.
Generation and characterization of prdm8 loss-of-function mutations. (A) Zebrafish Prdm8 protein structure is depicted as an empty black box with the SET domain highlighted in green and the C2H2 zinc-finger domains in blue. Alignment of Prdm8 amino acid sequences from human (HS), mouse (MM), and zebrafish (DR). Conserved SET domain and C2H2 zinc-finger domains are shown as green or blue boxes, respectively. (B) Schematic representing prdm8 gene structure. The sequence targeted for CRISPR/Cas9-mediated mutagenesis is marked by a red line in exon 1. The wild-type sequence CRISPR target sequence is shown below in red text and the co49 insertion and co51 deletion are shown in bold or dashes, respectively. Both mutations are predicted to produce 73 amino acid proteins truncated at the C-terminal end of the SET domain. (C) Images of agarose gels showing prdm8 DNA fragmentation following dCAPS genotyping of homozygous wild-type (WT), heterozygous and homozygous mutant embryos with sample genotype frequencies. (D) Representative images of living 24 and 72 hpf wild-type and prdm8co49−/− embryos. (E) Image of RT-PCR gel, showing decreased expression of prdm8 mRNA in prdm8 mutant embryos compared with control, with no difference in expression of the control transcript rpl13.
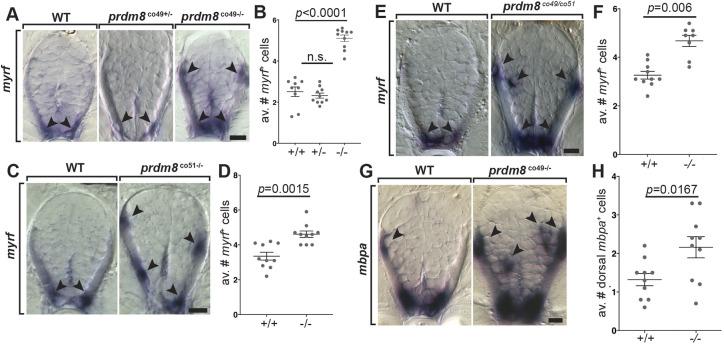
To determine whether prdm8 regulates the formation of oligodendrocytes, we performed RNA ISH to detect myrf expressed by wild-type and prdm8 mutant larvae. At 72 hpf, larvae homozygous for the co49 allele had almost twice as many spinal cord myrf+ oligodendrocytes compared with wild-type siblings (Fig. 4A,B). Heterozygous siblings were not different from wild type (Fig. 4A,B). Larvae homozygous for the co51 allele similarly had excess myrf+ oligodendrocytes relative to wild-type siblings (Fig. 4C,D). Larvae trans-heterozygous for the co49 and co51 alleles also had a greater number of myrf+ oligodendrocytes (Fig. 4E,F), indicating that this phenotype results specifically from loss of prdm8 function and not as a consequence of an off-target mutation produced by CRISPR/Cas9. We additionally examined the expression of mbpa. Consistent with our myrf data, co49 homozygous mutant larvae had ∼twofold more dorsal mbpa+ oligodendrocytes than wild-type siblings (Fig. 4G,H). These data indicate that Prdm8 limits oligodendrocyte formation.
Fig. 4.
prdm8 mutant larvae have excess oligodendrocytes. (A,C,E,G) Representative trunk spinal cord transverse sections obtained from 72 hpf larvae showing mRNA expression patterns detected by ISH. Images and quantification of myrf expression in wild-type (WT), heterozygous and homozygous co49 mutant larvae (A,B), wild-type and homozygous co51 mutant larvae (C,D), and wild-type and co49/co51 mutant larvae (E,F). Arrowheads mark myrf+ oligodendrocytes. (G,H) Images of mbpa expression and quantification of dorsal mbpa+ oligodendrocytes in wild-type and homozygous co49 mutant larvae. Arrowheads denote mbpa+ oligodendrocytes. n=10 larvae for each genotype except wild type in B (n=9) and co49/co51 mutant larvae in F (n=8). Data are mean±s.e.m. with individual data points indicated. Statistical significance was evaluated by Mann–Whitney U test (B,D,F) and an unpaired, two-tailed Student's t-test (H). n.s., not significant. Scale bars: 10 μm.
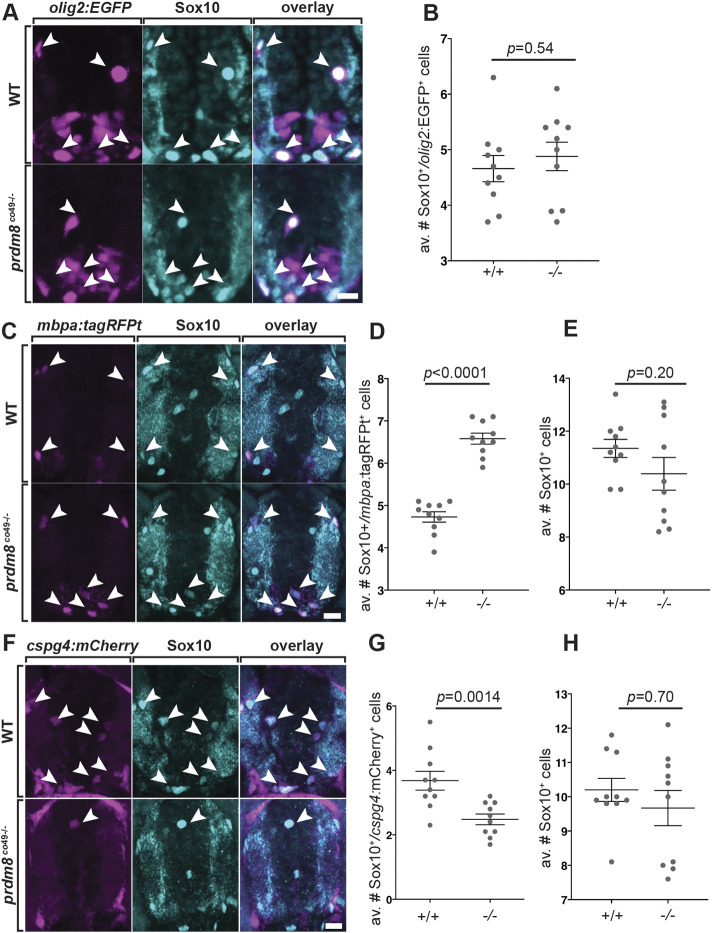
To identify the source of excess oligodendrocytes, we first counted the number of oligodendrocyte lineage cells using an antibody to detect expression of Sox10 in spinal cord sections of wild-type and mutant larvae carrying the Tg(olig2:EGFP) reporter. At 72 hpf, homozygous prdm8 mutant larvae had the same number of Sox10+ olig2:EGFP+ cells as control larvae (Fig. 5A,B). To determine the proportion of Sox10+ cells that differentiated as oligodendrocytes, we then performed immunohistochemistry to detect Sox10 on sections obtained from 5 dpf larvae carrying a Tg(mbpa:tagRFPt) transgenic reporter. This experiment showed that homozygous prdm8 mutant larvae had a significant increase in the number of Sox10+ mbpa:tagRFPt+ oligodendrocytes without a change in the total number of Sox10+ cells relative to sibling controls (Fig. 5C-E). To assess the OPC population, we labeled sections from 5 dpf larvae carrying a Tg(cspg4:mCherry) transgenic reporter (Ravanelli et al., 2018) with Sox10 antibody. Homozygous prdm8 mutant larvae had fewer OPCs than wild-type siblings but total oligodendrocyte lineage cells were unchanged (Fig. 5F-H). These data indicate that Prdm8 regulates the proportion of OPCs that differentiate as oligodendrocytes.
Fig. 5.
prdm8 mutant larvae have more myelinating oligodendrocytes and a deficit of OPCs. (A,C,F) Representative images of trunk spinal cord transverse sections processed to detect Sox10 expression (blue) in combination with transgenic reporter gene expression (pink). (A,B) The number of Sox10+ olig2:EGFP+ oligodendrocyte lineage cells is similar in 72 hpf wild-type (WT) and prdm8co49 mutant larvae. Arrowheads indicate oligodendrocyte lineage cells. n=10 for both genotypes. (C-E) prdm8co49 mutant larvae at 5 dpf have more Sox10+ mbpa:tagRFP+ oligodendrocytes (arrowheads) than wild-type larvae but there was no difference in total Sox10+ oligodendrocyte lineage cells. n=14 for both genotypes. (F-H) prdm8co49 mutant larvae at 5 dpf have fewer Sox10+ cspg4:mCherry+ OPCs (arrowheads) than wild-type larvae (G), but the number of total Sox10+ oligodendrocyte lineage cells was unchanged (H). n=14 for both genotypes. Data are mean±s.e.m. with individual data points indicated. Statistical significance was evaluated by an unpaired, two-tailed Student's t test (B,D,E) and Mann–Whitney U test (G,H). Scale bars: 10 μm.
prdm8 regulates the timing of a neuron-glia switch by repressing neural tube Shh signaling activity
We next investigated whether prdm8 regulates motor neuron formation from pMN progenitors, which precedes OPC specification. To do so, we used an antibody to detect Isl1/2 (Isl), which marks post-mitotic motor neurons (Ericson et al., 1992). At 24 hpf, homozygous prdm8 mutant embryos had the same number of Isl+ olig2:EGFP+ motor neurons as controls (Fig. 6A,B), signifying that prdm8 mutant embryos initiate motor neuron formation normally. By contrast, at 36 and 48 hpf, prdm8 mutant embryos had fewer motor neurons than control embryos (Fig. 6C-F), suggesting that Prdm8 is required to maintain motor neuron production from pMN progenitors.
Fig. 6.
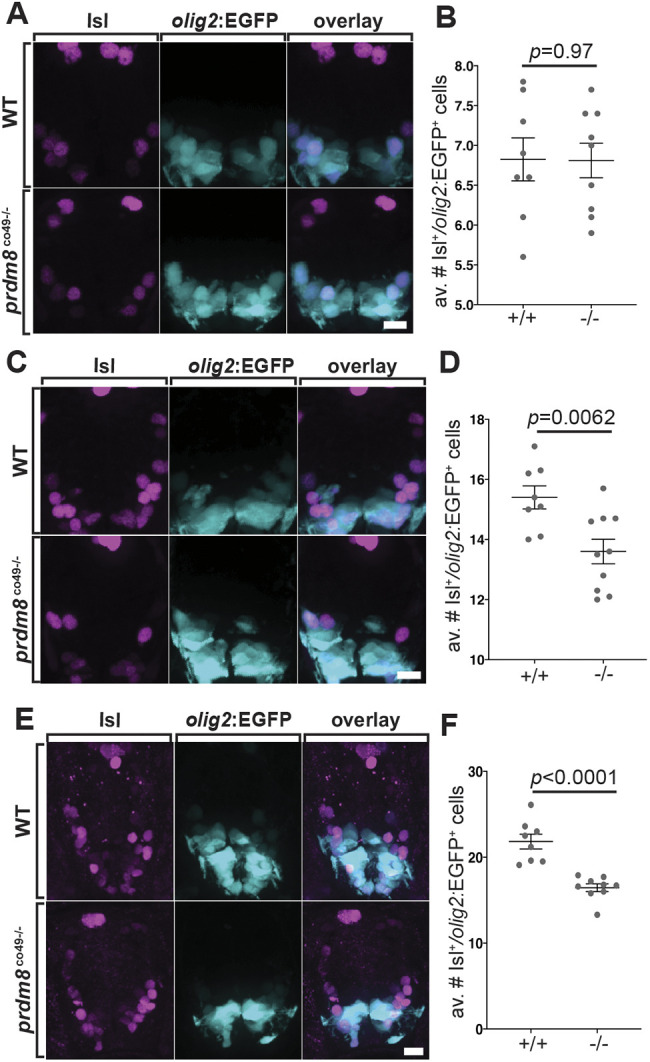
prdm8 mutant embryos have a deficit of motor neurons. (A,C,E) Representative images of trunk spinal cord transverse sections processed to detect Isl expression (pink) in combination with olig2:EGFP (blue). (A,B) The number of Isl+ olig2:EGFP+ motor neurons is similar in 24 hpf wild-type (WT) (n=8) and prdm8co49 mutant (n=9) embryos. (C,D) prdm8co49 mutant embryos at 36 hpf (n=10) have fewer Isl+ olig2:EGFP+ motor neurons than wild-type embryos (n=8). (E,F) prdm8co49 mutant embryos at 48 hpf (n=9, 16.4±0.46) have fewer Isl+ olig2:EGFP+ motor neurons than wild-type embryos (n=8, 21.8±0.86). Data are mean±s.e.m. with individual data points indicated. Statistical significance was evaluated by an unpaired, two-tailed Student's t-test (B,F) and Mann–Whitney U test (D). Scale bars: 10 μm.
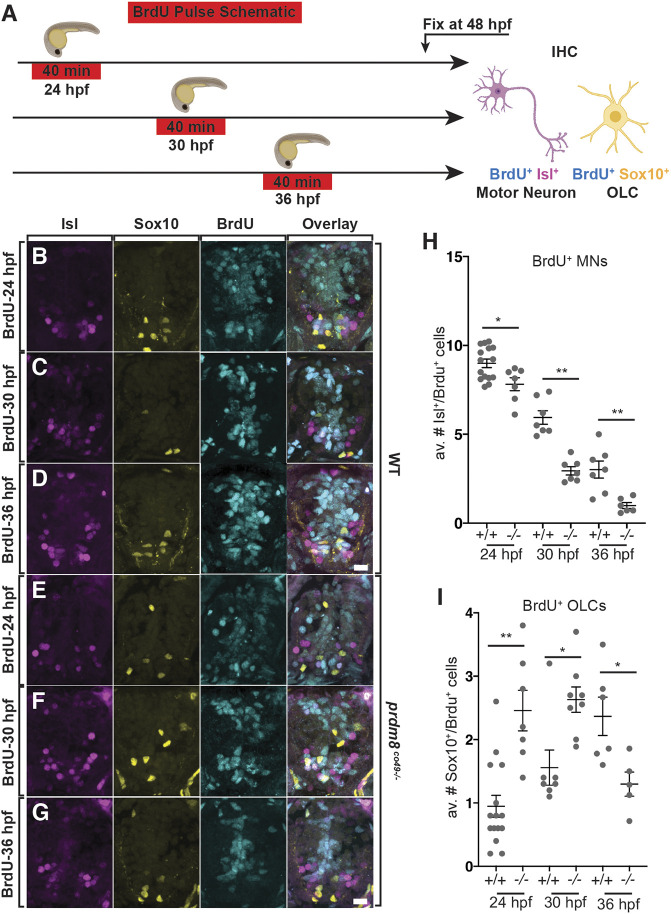
One possible explanation for these data is that in the absence of prdm8 function, pMN progenitors prematurely switch from motor neuron to OPC production, resulting in a deficit of motor neurons. To test this prediction, we exposed 24, 30 and 36 hpf embryos to a pulse of bromodeoxyuridine (BrdU) to label cells in S-phase and used immunohistochemistry at 48 hpf to identify the progeny of the labeled progenitors (Fig. 7A). Compared with stage-matched wild-type control embryos, homozygous prdm8 mutant embryos exposed to BrdU at each time point had a deficit of BrdU+ Isl+ motor neurons (Fig. 7B-H). By contrast, mutant embryos pulsed with BrdU at 24 and 30 hpf had more BrdU+ Sox10+ cells than controls (Fig. 7B,C,E,F,I), whereas those pulsed at 36 hpf had fewer (Fig. 7D,G,I). These data indicate that prdm8 mutant embryos prematurely terminate motor neuron formation and concomitantly produce OPCs earlier than normal. However, mutant embryos also prematurely terminate OPC production. Altogether, these data raise the possibility that Prdm8 prevents premature OPC specification, thus preserving pMN progenitors for motor neuron fate.
Fig. 7.
prdm8 mutant embryos prematurely switch from motor neuron to OPC production. (A) Schematic of BrdU pulses. (B-G) Representative images of trunk spinal cord sections from 48 hpf embryos treated with BrdU and processed to detect Isl (pink), Sox10 (yellow) and BrdU (blue). Wild-type (B) and prdm8co49−/− (E) embryos pulsed with BrdU at 24 hpf. Wild-type (C) and prdm8co49−/− (F) embryos pulsed with BrdU at 30 hpf. Wild-type (D) and prdm8co49−/− (G) embryos pulsed with BrdU at 36 hpf. (H) Quantification of Isl+/BrdU+ motor neurons pulsed with BrdU at 24 hpf in wild type (n=15) and prdm8co49−/− (n=7); 30 hpf in wild type (n=7) and prdm8co49−/− (n=7); and 36 hpf in wild type (n=7) and prdm8co49−/− (n=6). (I) Quantification of Sox10+/BrdU+ cells pulsed with BrdU at 24 hpf in wild type (n=15) and prdm8co49−/− (n=7); 30 hpf in wild type (n=7) and prdm8co49−/− (n=8); and 36 hpf in wild-type (n=6) and prdm8co49−/− (n=5). Data are mean±s.e.m. with individual data points indicated. Statistical significance was evaluated by Mann-Whitney U test. *P<0.05, **P<0.001. Analysis of embryos pulsed with BrdU at 24 hpf represent data collected from two laboratory replicates. Scale bars: 10 μm.
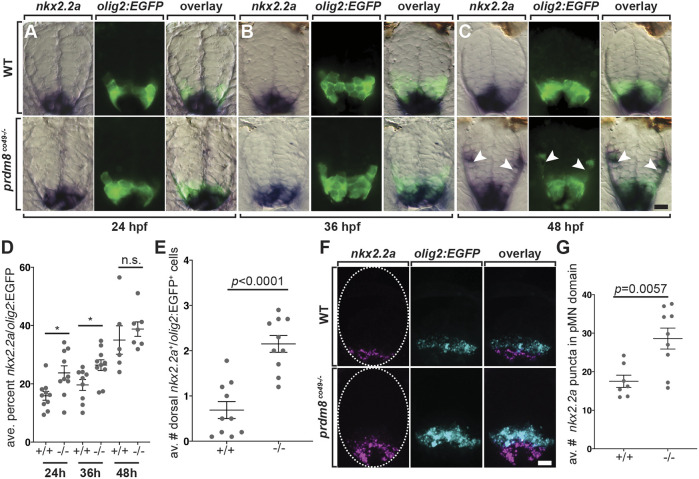
OPC specification coincides with and requires a dorsal expansion of Nkx2.2 expression from the p3 domain, resulting in co-expression of Nkx2.2 and Olig2 (Kessaris et al., 2001; Kucenas et al., 2008; Soula et al., 2001; Xu et al., 2000). Thus, one possible mechanism by which prdm8 prevents premature OPC specification is regulation of the time at which pMN progenitors express nkx2.2a. To examine this possibility, we first used RNA ISH to evaluate the area of nkx2.2a expression within the olig2:EGFP+ pMN domain. At 24 and 36 hpf, nkx2.2a expression appeared to be expanded dorsolaterally into the pMN domain of mutant embryos (Fig. 8A,B). Measurement of the area of overlap of nkx2.2a and olig2:EGFP expression confirmed this observation, revealing a significant increase in the area of coincident olig2:EGFP and nkx2.2a expression in mutant embryos (Fig. 8D). By 48 hpf, there was no difference in nkx2.2a expression in the pMN domain between wild-type and mutant embryos (Fig. 8C,F). However, the number of dorsal olig2:EGFP+ OPCs that expressed nkx2.2a at 48 hpf was increased almost threefold in prdm8 mutant embryos compared with controls (Fig. 8C,E). We also used fluorescent RNA ISH to label nkx2.2a and olig2 mRNA, and quantified nkx2.2a puncta in the pMN domain at 28 hpf, before OPC specification. prdm8 mutant embryos had more nkx2.2a mRNA puncta localized to pMN cells than controls (Fig. 8F,G). Altogether, these data suggest that prdm8 controls the timing of OPC specification by controlling the time at which pMN cells initiate nkx2.2a expression.
Fig. 8.
pMN cells prematurely express nkx2.2a in prdm8 mutant embryos. (A-C) Representative transverse sections of trunk spinal cord (dorsal up) showing prdm8 RNA (blue) and olig2:EGFP (green) expression. Developmental stages are noted at the bottom. Arrowheads indicate dorsally migrated oligodendrocyte lineage cells. (D) The area of nkx2.2a expression in the pMN domain is greater at 24 (n=10) and 36 hpf (co49, n=9; wild type, n=10) in prdm8 mutants embryos compared with controls and there is no difference at 48 hpf (co49 n=6; wild type, n=7). (E) prdm8co49−/− (n=10) have more dorsal OPCs (nkx2.2a+/olig2:EGFP+) than wild-type embryos (n=10) at 48 hpf. (F) Representative transverse trunk spinal cord sections obtained from 28 hpf embryos processed for fluorescent ISH to detect olig2 (blue) and nkx2.2a (pink) mRNA. Dashed ovals outline the spinal cord. (G) More nkx2.2a puncta are located within the olig2+ pMN domain of prdm8co49−/− embryos (n=9) compared with wild-type embryos (n=7). Data are mean±s.e.m. with individual data points indicated. Statistical significance was evaluated by an unpaired, two-tailed Student's t-test. n.s., not significant. *P<0.05. Scale bars: 10 μm.
At the end of neurogenesis, ventral spinal cord cells transiently elevate Shh signaling activity, which is necessary for OPC specification (Orentas et al., 1999; Soula et al., 2001; Touahri et al., 2012). Experimentally increasing Shh levels caused premature termination of motor neuron formation and precocious OPC formation (Danesin et al., 2006), similar to the loss of prdm8 function and thereby raising the possibility that Prdm8 suppresses Shh activity in the ventral spinal cord. To test this possibility, we probed for expression of ptch2, a transcriptional target of the Shh signaling pathway. At 24 hpf, prdm8 mutant embryos appeared to express more ptch2 than wild-type embryos (Fig. 9A). By 48 hpf, there was no visible difference in ptch2 expression between genotypes (Fig. 9A). Next, we used fluorescent RNA ISH to quantify ptch2 expression. At 24 and 36 hpf, prdm8 mutant embryos expressed more ptch2 mRNA relative to total spinal cord area compared with controls (Fig. 9B-F). Wild-type and prdm8 mutant embryos similarly expressed shha, which encodes a Shh ligand, suggesting that the elevated level of ptch2 expression results from increased Shh signaling activity and is independent of ligand expression (Fig. 9G). To further validate that loss of prdm8 leads to elevated Shh activity, we probed for expression of boc1, which encodes a Shh co-receptor that is negatively regulated by Shh signaling (Tenzen et al., 2006). At 24 hpf, prdm8 mutant embryos expressed less boc1 than controls, which is in line with the idea that loss of prdm8 function leads to increased Shh activity (Fig. 9H,I). These results are consistent with the possibility that Prdm8 suppresses Shh response in pMN cells to regulate the transition between motor neuron and OPC formation.
Fig. 9.
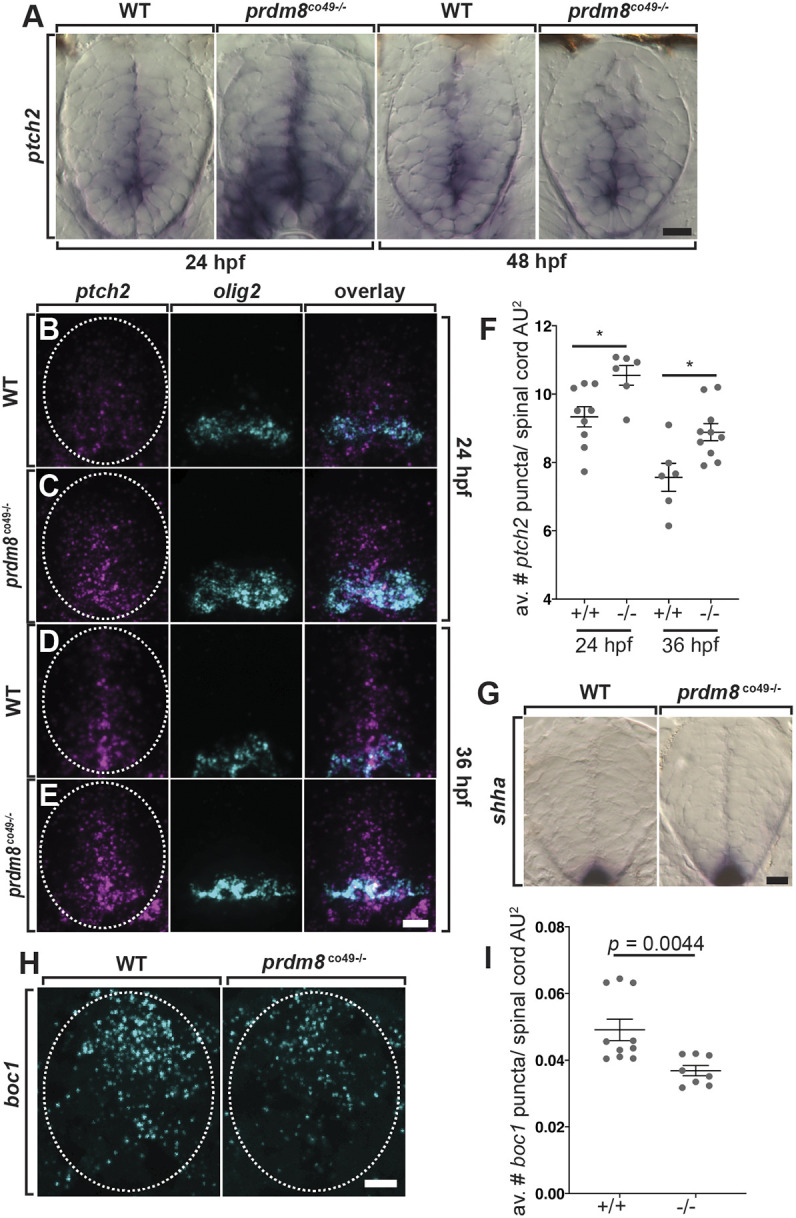
Spinal cord cells of prdm8 mutant embryos have elevated Shh signaling activity. (A) Representative transverse sections of trunk spinal cords obtained from 24 and 48 hpf wild-type (WT) and prdm8co49−/− embryos (dorsal up) showing ptch2 RNA expression. (B-E) Representative transverse trunk spinal cord sections processed for fluorescent ISH to detect olig2 (blue) and ptch2 (pink) mRNA at 24 hpf (B,C) and 36 hpf (D,E). (F) prdm8co49−/− embryos have more ptch2 puncta per AU2 of spinal cord at 24 hpf (n=6) and 36 hpf (n=10) than wild-type embryos at 24 hpf (n=9) and 36 hpf (n=6). (G) Representative transverse sections of trunk spinal cord (dorsal up) showing shha RNA expression in 24 hpf wild-type and prdm8co49−/− embryos. (H) Representative transverse trunk spinal cord sections processed for fluorescent ISH to detect boc1 (blue) mRNA at 24 hpf. (I) Wild-type embryos (n=6) have more boc1 puncta per AU2 of spinal cord at 24 hpf than prdm8co49−/− embryos (n=8). Data are mean±s.e.m. with individual data points indicated. Statistical significance was evaluated by an unpaired, two-tailed Student's t-test (F) and by Mann-Whitney U test (I). *P<0.001. Dashed oval outlines the spinal cord boundary. Scale bars: 10 μm.
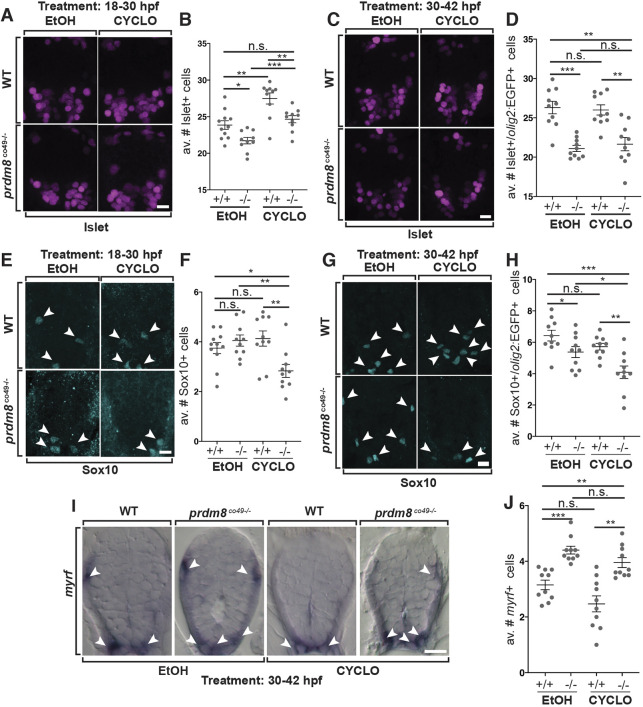
Because the premature transition between motor neuron and OPC production resulting from lack of prdm8 function resembled the effect of abnormally elevated Shh signaling, we predicted that the number of motor neurons in prdm8 mutant embryos could be rescued by inhibiting Shh activity. To test this prediction, we treated prdm8 mutant and wild-type embryos with a low concentration of cyclopamine to partially block Shh signal transduction from 18 to 30 hpf and assessed motor neuron number at 48 hpf. The number of motor neurons was similar in prdm8 mutant embryos treated with cyclopamine and wild-type embryos treated with vehicle control (Fig. 10A,B), consistent with our prediction. Furthermore, both wild-type and prdm8 mutant embryos treated with cyclopamine had more motor neurons than their genotype-matched controls (Fig. 10B), raising the possibility that suppression of Shh signaling delays the motor neuron to OPC switch, resulting in the formation of excess motor neurons. By contrast, treatment with cyclopamine from 30 to 42 hpf, after most spinal cord neurogenesis is normally completed, had no effect on motor neuron number in either wild-type or prdm8 mutant embryos (Fig. 10C,D). Consistent with our previous assessments, vehicle control-treated mutant embryos had fewer motor neurons than wild-type siblings (Fig. 10A-D). These data therefore support the possibility that Prdm8 suppresses Shh signaling within pMN cells to regulate the termination of motor neuron production and the timing of the neuron-glia switch.
Fig. 10.
Shh inhibition rescues the motor neuron but not oligodendrocyte phenotypes of prdm8 mutant embryos. (A,C,E,G) Representative images of trunk spinal cord sections from 48 hpf embryos treated with 0.5 μM cyclopamine (CYCLO) or ethanol (EtOH) from 18 to 30 hpf (A,E) or 30 to 42 hpf (C,G), and processed to detect Isl (A,C) or Sox10 (E,G) expression. (A,B) Wild-type (WT) embryos treated with EtOH control solution and prdm8 mutant embryos treated with cyclopamine have similar numbers of motor neurons. (C,D) There are fewer motor neurons (Islet+) in prdm8co49−/− embryos treated with EtOH and cyclopamine compared with wild-type embryos treated with EtOH. (E,F) There are fewer OPCs (Sox10+; arrowheads) in prdm8co49−/− embryos treated with cyclopamine and no difference in OPCs prdm8co49−/− embryos treated with EtOH compared with wild-type embryos treated with EtOH. (G,H) There are fewer OPCs (Sox10+; arrowheads) in prdm8co49−/− embryos treated with cyclopamine and slightly fewer OPCs in prdm8co49−/− embryos treated with EtOH compared with wild-type embryos treated with EtOH. (I) Representative trunk spinal cord transverse sections obtained from 72 hpf larvae treated with 0.5 μM cyclopamine or EtOH from 30 to 42 hpf showing myrf mRNA expression detected by in situ RNA hybridization. (I,J) prdm8co49−/− embryos treated with EtOH or cyclopamine have more oligodendrocytes (myrf+; arrowheads) than wild-type embryos treated with EtOH. n=10 for all genotypes and treatments except for wild-type embryos treated with EtOH (n=11) (A,E). Data are mean±s.e.m. with individual data points indicated. Statistical significance was evaluated by an unpaired, two-tailed Student's t-test. *P<0.05; **P<0.001; ***P<0.0001; n.s., not significant. Scale bars: 10 μm.
We next tested whether suppressing Shh signaling in prdm8 mutant embryos affected oligodendrocyte development. As above, there was no difference in the number of Sox10+ cells between wild-type and mutant larvae treated with a control solution between 18 and 30 hpf (Fig. 10E,F). Whereas wild-type embryos treated with control solution or 0.5 μM cyclopamine from 18 to 30 hpf had similar numbers of Sox10+ cells, prdm8 mutant embryos treated with cyclopamine had a significant deficit of Sox10+ cells compared with mutant embryos treated with control solution and wild-type embryos treated with cyclopamine (Fig. 10E,F). Treating embryos with cyclopamine from 30 to 42 hpf similarly caused prdm8 mutant embryos to have fewer Sox10+ cells than mutant embryos treated with control solution and wild-type embryos treated with cyclopamine (Fig. 10G,H). One possible explanation for these observations is that, although cyclopamine treatment delays the premature neuron-glia transition in prdm8 mutant embryos, it does not rescue the premature termination of OPC production, thereby resulting in a deficit of oligodendrocyte lineage cells.
We have shown above that prdm8 mutant larvae have excess oligodendrocytes and a deficit of cspg4+ OPCs. To determine whether this phenotype results from misregulated Shh signaling, we treated embryos with cyclopamine from 30 to 42 hpf and examined expression of myrf as a marker for myelinating oligodendrocytes. prdm8 mutant larvae treated with control solution and cyclopamine had similar numbers of myrf+ oligodendrocytes (Fig. 10I,J). Thus, suppressing Shh signaling did not rescue the excess oligodendrocyte phenotype of prdm8 mutant larvae, raising the possibility that Prdm8 regulates oligodendrocyte formation independently of its role in controlling the timing of a neuron-glia switch.
DISCUSSION
The neuron-glia switch, whereby neural progenitors produce neurons followed by glia, is a general feature of developing nervous systems (Rowitch and Kriegstein, 2010). Despite its important role in diversifying neural cell fate, the mechanisms that cause the switch and determine its timing remain poorly understood. In the ventral spinal cord, a temporally regulated rise in Shh signaling activity appears to trigger pMN progenitors to switch from motor neuron to OPC production (Danesin and Soula, 2017). Our results now indicate that Prdm8 suppresses Shh signaling activity within pMN progenitors to control the timing of the motor neuron-OPC switch.
Distinct types of neurons and glia arise from distinct subpopulations of progenitor cells aligned along the dorsoventral axis of the spinal cord. A large body of work conducted over the past 30 years has shown that the identities of these progenitor populations are determined by combinatorial expression of an extensive array of bHLHs and homeodomain transcription factors (Sagner and Briscoe, 2019). Additionally, specific subpopulations of spinal cord progenitors also express members of the Prdm protein family, although how these factors contribute to spinal cord development has received considerably less attention (Zannino and Sagerström, 2015). For example, dorsal progenitors express Prdm13, which regulates the balance between excitatory and inhibitory interneuron production by blocking the activity of bHLH transcription factors that drive expression of genes required for excitatory neuron differentiation (Chang et al., 2013; Mona et al., 2017). p1 progenitors express Prdm12, which is required for V1 interneuron formation (Thélie et al., 2015), and Prdm14 promotes Islet2 expression and axon outgrowth in motor neurons (Liu et al., 2012). Finally, pMN, p1 and p2 progenitors express Prdm8 (Kinameri et al., 2008; Komai et al., 2009). Although the spinal cord function of Prdm8 had not been previously investigated, evidence indicating that Prdm8 regulates the specification of retinal cells (Jung et al., 2015), and that Prdm8 can interact with bHLH transcription factors (Ross et al., 2012; Yildiz et al., 2019), raises the possibility that Prdm8 contributes to mechanisms that determine spinal cord progenitor fate.
The first main finding of our work is that zebrafish spinal cord cells express prdm8 similarly to mouse (Kinameri et al., 2008; Komai et al., 2009). Our data show that pMN progenitors express prdm8 throughout developmental neurogenesis and gliogenesis, but prdm8 expression subsides from the ventral spinal cord as pMN progenitors are depleted at the onset of the larval period. Furthermore, our analysis extends the mouse expression data by showing that oligodendrocyte lineage cells also express prdm8. Specifically, our data indicate that newly specified OPCs express prdm8 but then downregulate it as they initiate oligodendrocyte differentiation. By contrast, larval OPCs, marked by cspg4 expression, maintain prdm8 expression. Altogether, these observations raise the possibility that Prdm8 regulates pMN progenitor fate specification and, subsequently, OPC differentiation.
Our second main conclusion is that Prdm8 regulates the timing of the motor neuron to OPC switch by determining how strongly pMN progenitors respond to Shh signaling. Specifically, we found that prdm8 mutant embryos have a deficit of late-born motor neurons because of a premature neuron-glia switch, that mutant spinal cord cells express abnormally high levels of ptch2 and that the motor neuron deficit was rescued by treating mutant embryos with a low concentration of a Shh inhibitor. This finding supports previous evidence that a transient burst of Shh signaling activity initiates the switch from motor neuron to OPC production (Danesin and Soula, 2017). This burst is mediated, at least in part, by sulfatase function (Al Oustah et al., 2014; Danesin et al., 2006; Jiang et al., 2017). Sulfatases are secreted by ventral spinal cord cells and increase the range of Shh ligand in the extracellular matrix by regulating the sulfation state of heparan sulfate proteoglycans (Farzan et al., 2008; Yan and Lin, 2009). How cells receive and process extracellular signals also can influence signaling strength. In particular, Notch signaling increases the sensitivity of neural cells to Shh signaling (Kong et al., 2015; Ravanelli et al., 2018; Stasiulewicz et al., 2015). Currently, we do not know how Prdm8 suppresses Shh signaling activity within pMN progenitors. Because Prdm8 functions as a transcriptional inhibitor (Chen et al., 2018; Eom et al., 2009; Iwai et al., 2018; Ross et al., 2012), it might suppress expression of factors that transduce Shh signaling. For example, Prdm8 could suppress expression of the Shh co-receptors Cdon and Gas1, which enhance cell response to Shh (Allen et al., 2007, 2011). Alternatively, Prdm8 could limit expression of Notch signaling effectors that enhance Shh signaling. The identification of genes misregulated in prdm8 mutant embryos, combined with the determination of genomic loci targeted by Prdm8, should help uncover the regulatory function of Prdm8 in pMN progenitor specification.
Finally, we found that prdm8 mutant larvae have excess oligodendrocytes at the apparent expense of OPCs. There are at least two possible explanations for this phenotype. Because our expression data show that cells undergoing oligodendrocyte differentiation downregulate prdm8 expression, the first possibility is that Prdm8 inhibits OPC differentiation and, therefore, in its absence, OPCs that normally persist into the larval stage instead develop as myelinating oligodendrocytes. A second possibility is that Prdm8 regulates the allocation of pMN progenitors for distinct oligodendrocyte lineage cell fates. Previously, in a process we called progenitor recruitment, we showed that motor neurons, OPCs that rapidly differentiate and OPCs that persist into the larval stage arise from distinct pMN progenitors that sequentially initiate olig2 expression (Ravanelli and Appel, 2015; Ravanelli et al., 2018). We also found that slightly higher levels of Shh signaling favor the formation of oligodendrocytes over larval OPCs, which is similar to the oligodendrocyte phenotype of prdm8 mutant animals. However, inhibiting Shh with cyclopamine did not restore oligodendrocytes and OPCs to their normal numbers, raising the possibility that Prdm8 regulates oligodendrocyte lineage cell fate independently of Shh signaling. Identifying Prdm8 regulatory targets combined with detailed cell lineage analysis will help us to discriminate between these possibilities.
MATERIALS AND METHODS
Zebrafish lines and husbandry
All animal work was approved by the Institutional Animal Care and Use Committee at the University of Colorado School of Medicine. All non-transgenic embryos were obtained from pairwise crosses of males and females from the AB strain. Embryos were raised at 28.5°C in E3 media [5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2 and 0.33 mM MgSO4 (pH 7.4), with sodium bicarbonate], sorted for good health and staged according to developmental morphological features and hpf (Kimmel et al., 1995). Developmental stages are described in the Results section for individual experiments. Sex cannot be determined at embryonic and larval stages. Embryos were randomly assigned to control and experimental conditions for BrdU and pharmacological treatments. The transgenic lines used were Tg(olig2:EGFP)vu12 (Shin et al., 2003), Tg(mbpa:tagRFPt)co25 (Hines et al., 2015) and Tg(cspg4:mCherry)co28 (Ravanelli et al., 2018). All transgenic embryos were obtained from pairwise crosses of males or females from the AB strain to males or females of each transgenic line used.
Generation of CRISPR/Cas9 mutant zebrafish lines
We designed a sgRNA for the zebrafish prdm8 gene using the CRISPOR web tool (www.crispor.tefor.net) (Table S1). The sgRNA was constructed by annealing sense and anti-sense single-stranded oligonucleotides containing 5′ BsaI overhangs, and was inserted into BsaI-linearized pDR274 using the Quick Ligation Kit (New England BioLabs) (Table S1). The plasmid was transformed into chemically competent DH5a cells and purified from individual colony liquid cultures using a Qiagen Spin Miniprep Kit. To make the sgRNA, we linearized purified pDR274, containing the guide sequence, with DraI and used a T7 RNA polymerase for in vitro transcription (New England BioLabs). The pMLM3613 plasmid encoding cas9 was used for in vitro transcription using the SP6 mMessage mMachine Kit (Ambion) according to manufacturer's instructions. The sgRNA and cas9 mRNA were co-injected into single cell AB zebrafish embryos at the following concentrations: 200 ng/μl cas9 mRNA and 150 ng/μl prdm8 mRNA.
The following day, injected embryos were assayed for sgRNA activity by DNA extraction and two rounds of PCR amplification, first to amplify the prdm8 CRISPR target with gene-specific primers containing a M13F extension to the 5′ end of the forward primer (5′-TGTAAAACGACGGCCAGT-3′) and a second to add a fluorescein tag to the 5′ end of the amplified region (Table S2). The fluorescein-tagged PCR product was analyzed using capillary gel electrophoresis to detect product length. To detect F0 founders, we set up pairwise crosses of injected adults with ABs and screened their offspring for mutagenic events by fluorescent PCR and capillary gel electrophoresis. We used Sanger sequencing to determine the sequence of mutant alleles. We identified two mutant alleles, one with a 5 bp insertion (prdm8co49) and another with a 4 bp deletion (prdm8co51) (Fig. 3B). We were unable to recover homozygous mutant adults; therefore, these lines were maintained as heterozygotes through pairwise crosses with ABs or transgenic lines.
Derived cleaved amplified polymorphic sequencing (dCAPS) genotyping
To genotype embryos and adults, we designed a dCAPs assay to insert a restriction site into either the mutant or wild-type allele via PCR. Specific forward primers were designed for each allele, one that added a BsrGI restriction site into the prdm8co49 allele and another that added a NdeI restriction site into the wild-type allele for prdm8co51 identification (Table S2). PCR products were digested with the appropriate enzymes and samples were run on a 2.5% agarose gel; the prdm8co49 digest creates 267 and 37 bp digested mutant fragments and a 299 bp undigested wild-type fragment; the prdm8co51 digest creates two digested wild-type fragments of 260 and 55 bp, and a 315 bp undigested mutant fragment (Fig. 3C).
RNA extraction and reverse transcription PCR
We genotyped and decapitated euthanized 36 hpf embryos to isolate trunk tissue. We then added TRIzol to the trunks of each embryo and pooled trunks according to genotype. The pooled tissue was homogenized by trituration with a needle and phenol/chloroform was used to extract RNA. After precipitating RNA with isopropyl alcohol, cDNA was made using the SuperScript IV First-Strand Synthesis kit (Thermo Fisher Scientific) according to the manufacturer's instructions. To assess the expression of prdm8 mRNA, we designed primers overlaying the junctions between exons 1/2 and exons 2/3, resulting in the amplification of a 250 bp fragment. As a control, we used primers overlapping exons 1/2 and 3/4 to detect rpl13 mRNA. The same amount of cDNA was added to each PCR reaction and the products were detected on a 2.5% gel.
BrdU labeling
Embryos and larvae were dechorionated, incubated in a 20 μM BrdU solution for 40 min on ice at indicated time points (Fig. 7A) and subsequently washed four times (5 min each time) with embryo medium. Embryos and larvae were allowed to develop until 2 dpf in embryo E3 media. Samples were fixed in 4% paraformaldehyde (PFA) in 1× PBS, embedded (1.5% agar and 5% sucrose), sectioned and prepared for immunohistochemistry as described below.
Cyclopamine treatment
Cyclopamine (Cayman Chemical, 11321) was reconstituted in ethanol to make a 10 mM stock and stored at −20°C. Dechorionated embryos were treated with 0.5 μM cyclopamine or an equal concentration of ethanol alone in E3 media at indicated time points. Following treatment, embryos were washed three times with E3 media and grown to designated time points before fixation.
Whole-mount in situ RNA hybridization
In situ RNA hybridizations were performed as described previously (Hauptmann and Gerster, 2000). Probes included ptch2 (Concordet et al., 1996), nkx2.2a (Barth and Wilson, 1995), myrf, mbpa (Brösamle and Halpern, 2002) and prdm8 (Table S2). Plasmids were linearized with the appropriate restriction enzymes and cRNA preparation was carried out using Roche DIG-labeling reagents and T3, T7 or SP6 RNA polymerases (New England Biolabs). After staining, embryos were embedded in 1.5% agar/5% sucrose and frozen over dry ice. Transverse sections (20 μm) were cut using a Leica CM 1950 cryostat, collected on microscope slides and mounted with 75% glycerol.
Fluorescent in situ RNA hybridization
Fluorescent in situ RNA hybridization was performed using the RNAScope Multiplex Fluorescent V2 Assay Kit (Advanced Cell Diagnostics) on 12 μm PFA-fixed and agarose-embedded cryosections according to the manufacturer's instructions with the following modification: slides were covered with Parafilm for all 40°C incubations to maintain moisture and disperse reagents across the sections. The zebrafish olig2-C1, nkx2.2a-C2, ptch2-C2, myrf-C2, cspg4-C2, boc1-C3 and prdm8-C3 transcript probes were designed and synthesized by the manufacturer, and used at a dilution of 1:50. Transcripts were fluorescently labeled with Opal520 (1:1500), Opal570 (1:500) and Opal650 (1:1500) using the Opal 7 Kit (Perkin Elmer, NEL797001KT).
Cold-active protease cell dissociation and fluorescence-activated cells (FACs)
Tg(olig2:EGFP) euthanized embryos (at 24, 36 and 48 hpf) were collected in 1.7 ml microcentrifuge tubes and deyolked in 100 μl of prechilled Ca-free Ringer's solution [116 mM NaCl, 2.6 mM KCl and 5 mM HEPES (pH 7.0)] on ice. Embryos were pipetted intermittently with a p200 micropipette for 15 min and left for 5 min. Protease solution (500 μl; 10 mg/ml BI protease, 125 U/ml DNase, 2.5 mM EDTA and 1× PBS) was added to microcentrifuge tubes on ice for 15 min and embryos were homogenized every 3 min with a p100 micropipette for 15 min. Stop solution (200 μl; 30% FBS, 0.8 mM CaCl2 and 1× PBS) was then mixed into the tubes. Samples were then spun down at 400 g for 5 min at 4°C, and supernatant was removed. On ice, 1 ml of chilled suspension medium [1% fetal bovine serum (FBS), 0.8 mM CaCl2, 50 U/ml penicillin and 0.05 mg/ml streptomycin] was added to the samples, which were then spun down again at 400 g for 5 min at 4°C. The supernatant was removed, and 400 μl of chilled suspension medium was added and the solution was filtered through a 35 μm strainer into a collection tube. Cells were FAC sorted to distinguish EGFP+ cells using a MoFlo XDP100 cell sorter at the CU-SOM Cancer Center Flow Cytometry Shared Resource and collected in 1.7 ml FBS-coated microcentrifuge tubes in 200 μl of 1× PBS.
scRNA sequencing
The Chromium Box from 10x Genomics was used to capture cells using a Chromium Single Cell 3′ Reagent Kit (PN-1000075). Libraries were sequenced on the Illumina NovaSeq 6000 Instrument. FASTQ files were analyzed using Cell Ranger Software. We obtained 2174 (24 h), 2555 (36 h) and 3177 (48 h) cells yielding a mean of 118,014 (24 h), 65,182 (36 h) and 96,053 (48 h) reads per cell with a median of 1929 (24 h), 1229 (36 h) and 1699 (48 h) genes identified per cell.
Raw sequencing reads were demultiplexed, mapped to the zebrafish reference genome (build GRCz11/danRer11) and summarized into gene expression matrices using Cell Ranger (version 3.0.1). The resulting count matrices were further filtered in Seurat 3.1.0 (www.satijalab.org/seurat/) to remove cell barcodes with fewer than 250 detectable genes, more than 5% of unique molecular identifiers (UMIs) derived from mitochondrial genes, or more than 50,000 UMIs (to exclude putative doublets). This filtering resulted in 6489 single cells across all samples (1952 from 24 hpf, 2147 from 36 hpf and 2390 from 48 hpf). After standard Seurat normalization, principal component analysis was carried out using the 1291 most variable genes. Next, dimensionality reduction was performed using uniform manifold approximation and projection on the first 15 principal components. Differential expression and marker gene identification was performed using MAST (Finak et al., 2015).
Immunohistochemistry
Larvae were fixed using 4% PFA/1× PBS overnight at 4°C. Embryos were washed with 1× PBS, rocked at room temperature, embedded in 1.5% agar/5% sucrose, frozen over dry ice and sectioned in 20 or 15 µm transverse increments using a cryostat microtome. Slides were place in Sequenza racks (Thermo Scientific), washed three times (for 5 min each time) in 0.1% Triton X-100/1× PBS (PBSTx), blocked for 1 h in 2% goat serum/2% bovine serum albumin/PBSTx and then placed in primary antibody (in block) overnight at 4°C. The primary antibodies used included: rabbit anti-Sox10 (1:500; Park et al., 2005); mouse anti-Islet (1:500; Developmental Studies Hybridoma Bank, AB2314683); rat anti-BrdU (1:100; Abcam, AB6326); or mouse JL-8 Living Colors (1:500; Clontech, 632380) to restore Tg(olig2:EGFP) fluorescence after RNA ISH. Sections were washed for 1 h at room temperature with PBSTx and then incubated for 2 h at room temperature with secondary antibodies at a 1:200 dilution in block. The secondary antibodies used included: AlexaFluor 488 anti-rabbit (Invitrogen, A11008), AlexaFluor 588 anti-rabbit (Invitrogen, A11011), AlexaFluor 647 anti-rabbit (Jackson ImmunoResearch, 111606144), AlexaFluor 488 anti-mouse (Life Technologies, A11001), AlexaFluor 568 anti-mouse (Invitrogen, A11004) and AlexaFluor 568 anti-rat (Invitrogen, A11077). Sections were washed for 1 h with PBSTx and mounted in VectaShield (Vector Laboratories).
Imaging
Fixed sections of embryos and larvae were imaged on a Zeiss Cell Observer SD 25 spinning disk confocal system or a Zeiss Axiovert 200 microscope equipped with a PerkinElmer spinning disk confocal system. IHC cell counts were collected using a 20× objective (n.a. 0.8) and representative images were collected using a 40× oil immersion objective (n.a. 1.3). Wild-type 1 dpf larvae were positioned on top of a 2% agarose plate and imaged using a Leica M165FC dissection scope with a SPOT RT3 camera. RNA ISH sections were imaged using differential interference contrast optics and a Zeiss AxioObserver compound microscope. Cell counts and representative images were acquired at a 40× magnification (n.a. 0.75). Images are presented as extended z-projections or a single plan (RNA ISH) collected using Volocity (PerkinElmer) or Zen (Carl Zeiss) imaging software. Image brightness and contrast were adjusted in Photoshop (Adobe) or ImageJ (National Institutes of Health).
Data quantification and statistical analysis
Immunohistochemistry and RNA ISH
Evaluations of nkx2.2a and olig2:EGFP domain overlap were performed in transverse sections by collecting single plane wide-field images of each section. The area of overlap was quantified using ImageJ by outlining the area of expression of olig2:EGFP and the area of nkx2.2a expression within the total olig2:EGFP area. Quantifications of fluorescent cell numbers in transverse sections were performed by collecting confocal z stacks of the entire section. Quantifications of RNA ISH cell numbers in transverse sections were performed by viewing the entire z plane. Data for each embryo were collected from ten consecutive trunk spinal cord sections and n represents the average number of cells per section in one embryo. All cell counts on sections were performed by a researcher who was blinded to the identities of the slides except Fig 4A-D and Fig. 6E,F.
Fluorescent RNA ISH
Quantification of fluorescent RNA ISH hybridization was carried out on z projections collected at identical exposures. All quantifications were performed with ImageJ Fiji using a custom script created by Karlie Fedder, University of Colorado, Department of Pediatrics, https://github.com/rebeccaorourke-cu/Prdm8-regulates-pMN-progenitor-specification. First, ten 0.5 μm z intervals were maximum z projected and background was subtracted using a 2-rolling ball. The image was then thresholded by taking two standard deviations above the mean fluorescence intensity. A region of interest was drawn around the pMN domain or spinal cord, and puncta were analyzed using the ‘analyze particles’ feature with a size of 0.01 to infinity and circularity of 0.00 to 1.00. All thresholded puncta were inspected to ensure single molecules were selected. Puncta with an area of only one pixel were removed from the dataset. Data for each embryo were collected from five consecutive trunk spinal cord sections and n represents the average number of puncta in a region of interest per section in a single embryo.
Statistical analysis
We plotted all data and performed all the statistical analyses in GraphPad Prism. All data are expressed as mean±s.e.m. Normality was assessed with a D'Agostino and Pearson omnibus test. For statistical analysis, we used an unpaired Student's two-tailed t-test for all data with normal distributions, or Mann–Whitney tests for non-normal data. Unless otherwise stated, all graphs represent data collected from one laboratory replicate, sampling fish from multiple crosses with no inclusion or exclusion criteria. P values not provided in the graphs are indicated as follows: *P<0.05, **P<0.001, ***P<0.0001.
Supplementary Material
Acknowledgements
We thank Christina Kearns for isolating cells for scRNA-seq and members of the Appel lab and the Section of Developmental Biology for discussions and advice. Cell sorting was performed by the University of Colorado Cancer Center Flow Cytometry Shared Resource, supported by a Cancer Center Support grant (P30CA046934). scRNA-seq was performed by the University of Colorado Anschutz Medical Campus Genomics Shared Resource Core Facility, supported by a Cancer Center Support grant (P30CA046934). Single cell RNA-seq and bioinformatics analysis was supported by a pilot award from the University of Colorado RNA Bioscience Initiative. The University of Colorado Anschutz Medical Campus Zebrafish Core Facility was supported by a National Institutes of Health grant (P30 NS048154).
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: K.S., B.A.; Formal analysis: K.S., R.O., A.G.; Investigation: K.S.; Resources: B.A.; Writing - original draft: K.S.; Writing - review & editing: B.A.; Supervision: B.A.; Project administration: B.A.; Funding acquisition: B.A.
Funding
This work was supported by a National Institutes of Health grant (NS406668 to B.A.) and a gift from the Gates Frontiers Fund to B.A. Deposited in PMC for release after 12 months.
Data availability
The single cell RNA-seq data have been deposited in GEO under accession number GSE155988.
Supplementary information
Supplementary information available online at https://dmm.biologists.org/lookup/doi/10.1242/dev.191023.supplemental
Peer review history
The peer review history is available online at https://dev.biologists.org/lookup/doi/10.1242/dev.191023.reviewer-comments.pdf
References
- Agius E., Soukkarieh C., Danesin C., Kan P., Takebayashi H., Soula C. and Cochard P. (2004). Converse control of oligodendrocyte and astrocyte lineage development by Sonic hedgehog in the chick spinal cord. Dev. Biol. 270, 308-321. 10.1016/j.ydbio.2004.02.015 [DOI] [PubMed] [Google Scholar]
- Al Oustah A., Danesin C., Khouri-Farah N., Farreny M.-A., Escalas N., Cochard P., Glise B. and Soula C. (2014). Dynamics of sonic hedgehog signaling in the ventral spinal cord are controlled by intrinsic changes in source cells requiring sulfatase 1. Development 141, 1392-1403. 10.1242/dev.101717 [DOI] [PubMed] [Google Scholar]
- Allen B. L., Tenzen T. and McMahon A. P. (2007). The Hedgehog-binding proteins Gas1 and Cdo cooperate to positively regulate Shh signaling during mouse development. Genes Dev. 21, 1244-1257. 10.1101/gad.1543607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen B. L., Song J. Y., Izzi L., Althaus I. W., Kang J.-S., Charron F., Krauss R. S. and McMahon A. P. (2011). Overlapping roles and collective requirement for the coreceptors GAS1, CDO, and BOC in SHH pathway function. Dev. Cell 20, 775-787. 10.1016/j.devcel.2011.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baizabal J.-M., Mistry M., García M. T., Gómez N., Olukoya O., Tran D., Johnson M. B., Walsh C. A. and Harwell C. C. (2018). The epigenetic state of PRDM16-regulated enhancers in radial glia controls cortical neuron position. Neuron 98, 945-962.e8. 10.1016/j.neuron.2018.04.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth K. A. and Wilson S. W. (1995). Expression of zebrafish nk2.2 is influenced by sonic hedgehog/vertebrate hedgehog-1 and demarcates a zone of neuronal differentiation in the embryonic forebrain. Development 121, 1755-1768. [DOI] [PubMed] [Google Scholar]
- Briscoe J. and Thérond P. P. (2013). The mechanisms of Hedgehog signalling and its roles in development and disease. Nat. Rev. Mol. Cell Biol. 14, 416-429. 10.1038/nrm3598 [DOI] [PubMed] [Google Scholar]
- Briscoe J., Pierani A., Jessell T. M. and Ericson J. (2000). A homeodomain protein code specifies progenitor cell identity and neuronal fate in the ventral neural tube. Cell 101, 435-445. 10.1016/S0092-8674(00)80853-3 [DOI] [PubMed] [Google Scholar]
- Britsch S., Goerich D. E., Riethmacher D., Peirano R. I., Rossner M., Nave K. A., Birchmeier C. and Wegner M. (2001). The transcription factor Sox10 is a key regulator of peripheral glial development. Genes Dev. 15, 66-78. 10.1101/gad.186601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brösamle C. and Halpern M. E. (2002). Characterization of myelination in the developing zebrafish. Glia 39, 47-57. 10.1002/glia.10088 [DOI] [PubMed] [Google Scholar]
- Chang J. C., Meredith D. M., Mayer P. R., Borromeo M. D., Lai H. C., Ou Y.-H. and Johnson J. E. (2013). Prdm13 mediates the balance of inhibitory and excitatory neurons in somatosensory circuits. Dev. Cell 25, 182-195. 10.1016/j.devcel.2013.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Gao W., Pu L., Zhang L., Han G., Zuo X., Zhang Y., Li X., Shen H., Wang X. et al. (2018). PRDM8 exhibits anti-tumor activities toward hepatocellular carcinoma by targeting NAP1L1. Hepatology 68, 994-1009. 10.1002/hep.29890 [DOI] [PubMed] [Google Scholar]
- Chittka A., Nitarska J., Grazini U. and Richardson W. D. (2012). Transcription factor positive regulatory domain 4 (PRDM4) recruits protein arginine methyltransferase 5 (PRMT5) to mediate histone arginine methylation and control neural stem cell proliferation and differentiation. J. Biol. Chem. 287, 42995-43006. 10.1074/jbc.M112.392746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Concordet J. P., Lewis K. E., Moore J. W., Goodrich L. V., Johnson R. L., Scott M. P. and Ingham P. W. (1996). Spatial regulation of a zebrafish patched homologue reflects the roles of sonic hedgehog and protein kinase A in neural tube and somite patterning. Development 122, 2835-2846. [DOI] [PubMed] [Google Scholar]
- Danesin C. and Soula C. (2017). Moving the Shh source over time: what impact on neural cell diversification in the developing spinal cord? J. Dev. Biol. 5, 4 10.3390/jdb5020004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danesin C., Agius E., Escalas N., Ai X., Emerson C., Cochard P. and Soula C. (2006). Ventral neural progenitors switch toward an oligodendroglial fate in response to increased Sonic hedgehog (Shh) activity: involvement of Sulfatase 1 in modulating Shh signaling in the ventral spinal cord. J. Neurosci. 26, 5037-5048. 10.1523/JNEUROSCI.0715-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson M. R. L., Polito A., Levine J. M. and Reynolds R. (2003). NG2-expressing glial progenitor cells: an abundant and widespread population of cycling cells in the adult rat CNS. Mol. Cell. Neurosci. 24, 476-488. 10.1016/S1044-7431(03)00210-0 [DOI] [PubMed] [Google Scholar]
- Dessaud E., Yang L. L., Hill K., Cox B., Ulloa F., Ribeiro A., Mynett A., Novitch B. G. and Briscoe J. (2007). Interpretation of the sonic hedgehog morphogen gradient by a temporal adaptation mechanism. Nature 450, 717-720. 10.1038/nature06347 [DOI] [PubMed] [Google Scholar]
- Dessaud E., Ribes V., Balaskas N., Yang L. L., Pierani A., Kicheva A., Novitch B. G., Briscoe J. and Sasai N. (2010). Dynamic assignment and maintenance of positional identity in the ventral neural tube by the morphogen sonic hedgehog. PLoS Biol. 8, e1000382 10.1371/journal.pbio.1000382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echelard Y., Epstein D. J., St-Jacques B., Shen L., Mohler J., McMahon J. A. and McMahon A. P. (1993). Sonic hedgehog, a member of a family of putative signaling molecules, is implicated in the regulation of CNS polarity. Cell 75, 1417-1430. 10.1016/0092-8674(93)90627-3 [DOI] [PubMed] [Google Scholar]
- Elbaz B. and Popko B. (2019). Molecular control of oligodendrocyte development. Trends Neurosci. 42, 263-277. 10.1016/j.tins.2019.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery B. (2010). Regulation of oligodendrocyte differentiation and myelination. Science 330, 779-782. 10.1126/science.1190927 [DOI] [PubMed] [Google Scholar]
- Emery B., Agalliu D., Cahoy J. D., Watkins T. A., Dugas J. C., Mulinyawe S. B., Ibrahim A., Ligon K. L., Rowitch D. H. and Barres B. A. (2009). Myelin gene regulatory factor is a critical transcriptional regulator required for CNS myelination. Cell 138, 172-185. 10.1016/j.cell.2009.04.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eom G. H., Kim K., Kim S.-M., Kee H. J., Kim J.-Y., Jin H. M., Kim J.-R., Kim J. H., Choe N., Kim K.-B. et al. (2009). Histone methyltransferase PRDM8 regulates mouse testis steroidogenesis. Biochem. and Biophy. Research Comm. 388, 131-136. 10.1016/j.bbrc.2009.07.134 [DOI] [PubMed] [Google Scholar]
- Ericson J., Thor S., Edlund T., Jessell T. and Yamada T. (1992). Early stages of motor neuron differentiation revealed by expression of homeobox gene Islet-1. Science 256, 1555-1560. 10.1126/science.1350865 [DOI] [PubMed] [Google Scholar]
- Farzan S. F., Singh S., Schilling N. S. and Robbins D. J. (2008). Hedgehog processing and biological activity. Am. J. Physiol. Gastrointest. Liver Physiol. 294, G844-G849. 10.1152/ajpgi.00564.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finak G., McDavid A., Yajima M., Deng J., Gersuk V., Shalek A. K., Slichter C. K., Miller H. W., McElrath M. J., Prlic M. et al. (2015). MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 16, 278 10.1186/s13059-015-0844-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H., Qi Y., Tan M., Cai J., Takebayashi H., Nakafuku M., Richardson W. and Qiu M. (2002). Dual origin of spinal oligodendrocyte progenitors and evidence for the cooperative role of Olig2 and Nkx2.2 in the control of oligodendrocyte differentiation. Development 129, 681-693. [DOI] [PubMed] [Google Scholar]
- Hanotel J., Bessodes N., Thélie A., Hedderich M., Parain K., Van Driessche B., Brandão K. D. O., Kricha S., Jorgensen M. C., et al. (2014). The Prdm13 histone methyltransferase encoding gene is a Ptf1a–Rbpj downstream target that suppresses glutamatergic and promotes GABAergic neuronal fate in the dorsal neural tube. Dev. Biol. 386, 340-357. 10.1016/j.ydbio.2013.12.024 [DOI] [PubMed] [Google Scholar]
- Hashimoto H., Jiang W., Yoshimura T., Moon K.-H., Bok J. and Ikenaka K. (2017). Strong sonic hedgehog signaling in the mouse ventral spinal cord is not required for oligodendrocyte precursor cell (OPC) generation but is necessary for correct timing of its generation. Neurochem. Int. 119, 178-183. 10.1016/j.neuint.2017.11.003 [DOI] [PubMed] [Google Scholar]
- Hauptmann G. and Gerster T. (2000). Multicolor whole-mount in situ hybridization. Methods Mol. Biol. 137, 139-148. 10.1385/1-59259-066-7:139 [DOI] [PubMed] [Google Scholar]
- He L. and Lu Q. R. (2013). Coordinated control of oligodendrocyte development by extrinsic and intrinsic signaling cues. Neurosci. Bull. 29, 129-143. 10.1007/s12264-013-1318-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Lagunas L., Powell D. R., Law J., Grant K. A. and Artinger K. B. (2011). Prdm1a and olig4 act downstream of Notch signaling to regulate cell fate at the neural plate border. Dev. Biol. 356, 496-505. 10.1016/j.ydbio.2011.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hines J., Ravanelli A., Schwindt R., Scott E. and Appel B. (2015). Neuronal activity biases axon selection for myelination in vivo. Nat. Neurosci. 18, 683-689. 10.1038/nn.3992 [DOI] [PMC free article] [PubMed]
- Iwai R., Tabata H., Inoue M., Nomura K.-I., Okamoto T., Ichihashi M., Nagata K.-I. and Mizutani K.-I. (2018). A Prdm8 target gene Ebf3 regulates multipolar-to-bipolar transition in migrating neocortical cells. Biochem. Biophys. Res. Commun. 495, 388-394. 10.1016/j.bbrc.2017.11.021 [DOI] [PubMed] [Google Scholar]
- Jiang W., Ishino Y., Hashimoto H., Keino-Masu K., Masu M., Uchimura K., Kadomatsu K., Yoshimura T. and Ikenaka K. (2017). Sulfatase 2 modulates fate change from motor neurons to oligodendrocyte precursor cells through coordinated regulation of Shh signaling with sulfatase 1. Dev. Neurosci. 39, 361-374. 10.1159/000464284 [DOI] [PubMed] [Google Scholar]
- Jung C. C., Atan D., Ng D., Ploder L., Ross S. E., Klein M., Birch D. G., Diez E. and McInnes R. R. (2015). Transcription factor PRDM8 is required for rod bipolar and type 2 OFF-cone bipolar cell survival and amacrine subtype identity. Proc. Natl. Acad. Sci. USA 112, E3010-E3019. 10.1073/pnas.1505870112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessaris N., Pringle N. and Richardson W. D. (2001). Ventral neurogenesis and the neuron-glial switch. Neuron 31, 677-680. 10.1016/S0896-6273(01)00430-5 [DOI] [PubMed] [Google Scholar]
- Kimmel C., Ballard W., Kimmel S., Ullmann B. and Schilling T. (1995). Stages of embryonic development of the zebrafish. Dev. Dyn. 203, 253-310. 10.1002/aja.1002030302 [DOI] [PubMed]
- Kinameri E., Inoue T., Aruga J., Imayoshi I., Kageyama R., Shimogori T. and Moore A. W. (2008). Prdm proto-oncogene transcription factor family expression and interaction with the Notch-Hes pathway in mouse neurogenesis. PLoS ONE 3, e3859 10.1371/journal.pone.0003859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komai T., Iwanari H., Mochizuki Y., Hamakubo T. and Shinkai Y. (2009). Expression of the mouse PR domain protein Prdm8 in the developing central nervous system. Gene Expr. Patterns 9, 503-514. 10.1016/j.gep.2009.07.005 [DOI] [PubMed] [Google Scholar]
- Kong J. H., Yang L., Briscoe J., Novitch Correspondence B. G., Dessaud E., Chuang K., Moore D. M., Rohatgi R. and Novitch B. G. (2015). Notch activity modulates the responsiveness of neural progenitors to sonic hedgehog signaling. Dev. Cell 33, 373-387. 10.1016/j.devcel.2015.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucenas S., Snell H. and Appel B. (2008). nkx2.2a promotes specification and differentiation of a myelinating subset of oligodendrocyte lineage cells in zebrafish. Neuron Glia Biol. 4, 71-81. 10.1017/S1740925X09990123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlbrodt K., Herbarth B., Sock E., Hermans-Borgmeyer I. and Wegner M. (1998). Sox10, a novel transcriptional modulator in glial cells. J. Neurosci. 18, 237-250. 10.1523/JNEUROSCI.18-01-00237.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek M., Dias J. M., Marklund U., Uhde C. W., Kurdija S., Lei Q., Sussel L., Rubenstein J. L., Matise M. P., Arnold H.-H. et al. (2010). A homeodomain feedback circuit underlies step-function interpretation of a Shh morphogen gradient during ventral neural patterning. Development 137, 4051-4060. 10.1242/dev.054288 [DOI] [PubMed] [Google Scholar]
- Liu C., Ma W., Su W., Zhang J., Brand M., Furutani-Seiki M., Haffter P., Hammerschmidt M., Heisenberg C. P., Jiang Y. J. et al. (2012). Prdm14 acts upstream of islet2 transcription to regulate axon growth of primary motoneurons in zebrafish. Development 139, 4591-4600. 10.1242/dev.083055 [DOI] [PubMed] [Google Scholar]
- Lu Q. R., Yuk D.-I., Alberta J. A., Zhu Z., Pawlitzky I., Chan J., McMahon A. P., Stiles C. D. and Rowitch D. H. (2000). Sonic hedgehog-regulated oligodendrocyte lineage genes encoding bHLH proteins in the mammalian central nervous system. Neuron 25, 317-329. 10.1016/S0896-6273(00)80897-1 [DOI] [PubMed] [Google Scholar]
- Martí E., Takada R., Bumcrot D. A., Sasaki H. and McMahon A. P. (1995). Distribution of Sonic hedgehog peptides in the developing chick and mouse embryo. Development 121, 2537-2547. [DOI] [PubMed] [Google Scholar]
- Mona B., Uruena A., Kollipara R. K., Ma Z., Borromeo M. D., Chang J. C. and Johnson J. E. (2017). Repression by PRDM13 is critical for generating precision in neuronal identity. Elife 6, e25787 10.7554/eLife.25787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishi Y., Zhang X., Jeong J., Peterson K. A., Vedenko A., Bulyk M. L., Hide W. A. and McMahon A. P. (2015). A direct fate exclusion mechanism by Sonic hedgehog-regulated transcriptional repressors. Development 142, 3286-3293. 10.1242/dev.124636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noll E. and Miller R. H. (1993). Oligodendrocyte precursors originate at the ventral ventricular zone dorsal to the ventral midline region in the embryonic rat spinal cord. Development 118, 563-573. [DOI] [PubMed] [Google Scholar]
- Novitch B. G., Chen A. I. and Jessell T. M. (2001). Coordinate regulation of motor neuron subtype identity and pan-neuronal properties by the bHLH repressor Olig2. Neuron 31, 773-789. 10.1016/S0896-6273(01)00407-X [DOI] [PubMed] [Google Scholar]
- Orentas D. M., Hayes J. E., Dyer K. L. and Miller R. H. (1999). Sonic hedgehog signaling is required during the appearance of spinal cord oligodendrocyte precursors. Development 126, 2419-2429. [DOI] [PubMed] [Google Scholar]
- Park H.-C., Mehta A., Richardson J. S. and Appel B. (2002). olig2 is required for zebrafish primary motor neuron and oligodendrocyte development. Dev. Biol. 248, 356-368. 10.1006/dbio.2002.0738 [DOI] [PubMed] [Google Scholar]
- Park H. C., Boyce J., Shin J. and Appel B. (2005). Oligodendrocyte specification in zebrafish requires notch-regulated cyclin-dependent kinase inhibitor function. J. Neurosci. 25, 6836-6844. 10.1523/JNEUROSCI.0981-05.2005 [DOI]
- Poh A., Karunaratne A., Kolle G., Huang N., Smith E., Starkey J., Wen D., Wilson I., Yamada T. and Hargrave M. (2002). Patterning of the vertebrate ventral spinal cord. Int. J. Dev. Biol. 46, 597-608. [PubMed] [Google Scholar]
- Pringle N. P., Mudhar H. S., Collarini E. J. and Richardson W. D. (1992). PDGF receptors in the rat CNS: during late neurogenesis, PDGF alpha-receptor expression appears to be restricted to glial cells of the oligodendrocyte lineage. Development 115, 535-551. [DOI] [PubMed] [Google Scholar]
- Ravanelli A. M. and Appel B. (2015). Motor neurons and oligodendrocytes arise from distinct cell lineages by progenitor recruitment. Genes Dev. 29, 2504-2515. 10.1101/gad.271312.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravanelli A. M., Kearns C. A., Powers R. K., Wang Y., Hines J. H., Donaldson M. J. and Appel B. (2018). Sequential specification of oligodendrocyte lineage cells by distinct levels of Hedgehog and Notch signaling. Dev. Biol.444, 93-106. 10.1016/j.ydbio.2018.10.004 [DOI]
- Ribes V. and Briscoe J. (2009). Establishing and interpreting graded Sonic Hedgehog signaling during vertebrate neural tube patterning: the role of negative feedback. Cold Spring Harb. Perspect. Biol. 1, a002014 10.1101/cshperspect.a002014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson W. D., Smith H. K., Sun T., Pringle N. P., Hall A. and Woodruff R. (2000). Oligodendrocyte lineage and the motor neuron connection. Glia 29, 136-142. [DOI] [PubMed] [Google Scholar]
- Roelink H., Augsburger A., Heemskerk J., Korzh V., Norlin S., Ruiz i Altaba A., Tanabe Y., Placzek M., Edlund T., Jessell T. M. et al. (1994). Floor plate and motor neuron induction by vhh-1, a vertebrate homolog of hedgehog expressed by the notochord. Cell 76, 761-775. 10.1016/0092-8674(94)90514-2 [DOI] [PubMed] [Google Scholar]
- Ross S. E., Mccord A. E., Jung C., Atan D., Mok S. I., Hemberg M., Kim T.-K., Salogiannis J., Hu L., Cohen S. et al. (2012). Bhlhb5 and Prdm8 form a repressor complex involved in neuronal circuit assembly. Neuron. January 26, 292-303. 10.1016/j.neuron.2011.09.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowitch D. and Kriegstein A. (2010). Developmental genetics of vertebrate glial-cell specification. Nature468, 214-222. 10.1038/nature09611 [DOI]
- Sagner A. and Briscoe J. (2019). Establishing neuronal diversity in the spinal cord: a time and place. Development146, dev182154. 10.1242/dev.182154 [DOI]
- Shin J., Park H. C., Topczewska J. M., Madwsley D. J. and Appel B. (2003). Neural cell fate analysis in zebrafish using olig2 BAC transgenics. Methods Cell Sci.25, 7-14. 10.1023/B:MICS.0000006847.09037.3a [DOI]
- Simons M. and Nave K.-A. (2016). Oligodendrocytes: myelination and axonal support. Cold Spring Harb. Perspect. Biol. 8, a020479 10.1101/cshperspect.a020479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sock E. and Wegner M. (2019). Transcriptional control of myelination and remyelination. Glia 67, 2153-2165. 10.1002/glia.23636 [DOI] [PubMed] [Google Scholar]
- Soula C., Danesin C., Kan P., Grob M., Poncet C. and Cochard P. (2001). Distinct sites of origin of oligodendrocytes and somatic motoneurons in the chick spinal cord: oligodendrocytes arise from Nkx2.2-expressing progenitors by a Shh-dependent mechanism. Development 128, 1369-1379. [DOI] [PubMed] [Google Scholar]
- Stasiulewicz M., Gray S. D., Mastromina I., Silva J. C., Bjö Rklund M., Seymour P. A., Booth D., Thompson C., Green R. J., Hall E. A. et al. (2015). A conserved role for Notch signaling in priming the cellular response to Shh through ciliary localisation of the key Shh transducer Smo. Development 142, 2291-2303. 10.1242/dev.125237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenzen T., Allen B. L., Cole F., Kang J. S., Krauss R. S. and McMahon A. P. (2006). The Cell surface membrane proteins Cdo and Boc are components and targets of the hedgehog signaling pathway and feedback network in mice. Dev. Cell 10, 647-656. 10.1016/j.devcel.2006.04.004 [DOI] [PubMed] [Google Scholar]
- Thélie A., Desiderio S., Hanotel J., Quigley I., Van Driessche B., Rodari A., Borromeo M. D., Kricha S., Lahaye F., et al. (2015). Prdm12 specifies V1 interneurons through cross-repressive interactions with Dbx1 and Nkx6 genes in Xenopus. Development 142, 3416-3428. 10.1242/dev.121871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touahri Y., Escalas N., Benazeraf B., Cochard P., Danesin C. and Soula C. (2012). Sulfatase 1 promotes the motor neuron-to-oligodendrocyte fate switch by activating Shh signaling in Olig2 progenitors of the embryonic ventral spinal cord. J. Neurosci. 32, 18018-18034. 10.1523/JNEUROSCI.3553-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warf B. C., Fok-Seang J. and Miller R. H. (1991). Evidence for the ventral origin of oligodendrocyte precursors in the rat spinal cord. J. Neurosci. 11, 2477-2488. 10.1523/JNEUROSCI.11-08-02477.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolswijk G. and Noble M. (1989). Identification of an adult-specific glial progenitor cell. Development 105, 387-400. 10.1016/0922-3371(89)90618-7 [DOI] [PubMed] [Google Scholar]
- Xu X., Cai J., Fu H., Wu R., Qi Y., Modderman G., Liu R. and Qiu M. (2000). Selective expression of Nkx-2.2 transcription factor in chicken oligodendrocyte progenitors and implications for the embryonic origin of oligodendrocytes. Mol. Cell. Neurosci. 16, 740-753. 10.1006/mcne.2000.0916 [DOI] [PubMed] [Google Scholar]
- Yan D. and Lin X. (2009). Shaping morphogen gradients by proteoglycans. Cold Spring Harb. Perspect. Biol. 1, a002493 10.1101/cshperspect.a002493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz O., Downes G. B. and Sagerström C. G. (2019). Zebrafish prdm12b acts independently of nkx6.1 repression to promote eng1b expression in the neural tube p1 domain. Neural Dev. 14, 5 10.1186/s13064-019-0129-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zannino D. A. and Sagerström C. G. (2015). An emerging role for prdm family genes in dorsoventral patterning of the vertebrate nervous system. Neural Dev. 10, 24 10.1186/s13064-015-0052-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q. and Anderson D. J. (2002). The bHLH transcription factors OLIG2 and OLIG1 couple neuronal and glial subtype specification. Cell 109, 61-73. 10.1016/S0092-8674(02)00677-3 [DOI] [PubMed] [Google Scholar]
- Zhou Q., Wang S. and Anderson D. J. (2000). Identification of a novel family of oligodendrocyte lineage-specific basic helix-loop-helix transcription factors. Neuron 25, 331-343. 10.1016/S0896-6273(00)80898-3 [DOI] [PubMed] [Google Scholar]
- Zhou Q., Choi G. and Anderson D. J. (2001). The bHLH transcription factor Olig2 promotes oligodendrocyte differentiation in collaboration with Nkx2.2. Neuron 31, 791-807. 10.1016/S0896-6273(01)00414-7 [DOI] [PubMed] [Google Scholar]
- Zuchero J. B. and Barres B. A. (2013). Intrinsic and extrinsic control of oligodendrocyte development. Curr. Opin. Neurobiol. 23, 914-920. 10.1016/j.conb.2013.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.