Stable isotope analyses show that Ancient Beringian diets were seasonally structured and included substantial summer salmon.
Abstract
The earliest Native Americans have often been portrayed as either megafaunal specialists or generalist foragers, but this debate cannot be resolved by studying the faunal record alone. Stable isotope analysis directly reveals the foods consumed by individuals. We present multi-tissue isotope analyses of two Ancient Beringian infants from the Upward Sun River site (USR), Alaska (~11,500 years ago). Models of fetal bone turnover combined with seasonally-sensitive taxa show that the carbon and nitrogen isotope composition of USR infant bone collagen reflects maternal diets over the summer. Using comparative faunal isotope data, we demonstrate that although terrestrial sources dominated maternal diets, salmon was also important, supported by carbon isotope analysis of essential amino acids and bone bioapatite. Tooth enamel samples indicate increased salmon use between spring and summer. Our results do not support either strictly megafaunal specialists or generalized foragers but indicate that Ancient Beringian diets were complex and seasonally structured.
INTRODUCTION
Identifying the subsistence strategies of the earliest inhabitants of the Americas remains a contentious problem; these populations have been portrayed as megafaunal specialists or as broad-spectrum foragers (1, 2). Addressing this issue is a key in understanding the initial colonization, routes of dispersal, and settlement of the continent. Analyses of archaeological faunal assemblages are essential for elucidating diet breadth but cannot fully resolve it since the compositions of these assemblages may be biased by factors related to preservation, animal processing, and recovery (3). In contrast, stable isotope analysis of human remains provides a powerful tool for directly quantifying ancient diets and revealing the food sources actually consumed by individuals (4). The remains of two infants from the Upward Sun River (USR) site (Fig. 1), the earliest human remains in eastern Beringia, offer a rare opportunity to investigate the diet of ancient Native Americans using stable isotope analysis.
Fig. 1. Location of USR and related archaeological sites.
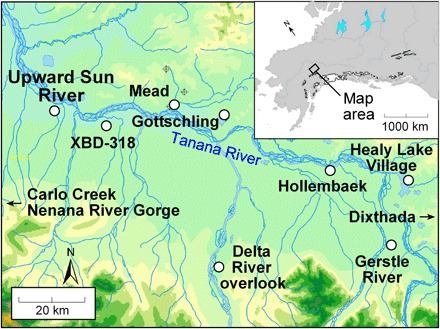
Two female infants were recovered in a single burial dating to ~11,500 calibrated years before present (cal yr. BP) at the USR site in interior Alaska (5). The older individual (Xach’itee’aanenh T’eede Gaay or USR1) was a newborn infant (~3 to 4 weeks), and the younger individual (Yełkaanenh T’eede Gaay or USR2) was a late-term prenate (5). Genomic analyses indicate that the two infants are the first known members of one of the two basal branches of Native Americans, termed Ancient Beringians (6). These infants belong to different maternal lineages (C1b and B2 haplogroups, respectively) (7) and provide two independent windows into the paleodiets of the mothers and, more broadly, of Ancient Beringians at the Pleistocene-Holocene transition. USR and the infants are assigned to the Denali Complex, a widespread cultural group present in eastern Beringia from 12,500 to 6000 cal yr. BP (5).
Here, we present multitissue stable isotope analyses of the USR infants to model the diets of their mothers based on a suite of comparative archaeological faunal data from the region, including large terrestrial herbivores (bison and wapiti), small game (hare, ground squirrel, and grouse/ptarmigan), salmon, and freshwater whitefish (tables S1 to S4 and fig. S1). Terminal Pleistocene human remains from North America are extremely rare, and to date, no isotope-based dietary analyses using regional archaeofaunal specimens have been conducted for this time period.
To gain a holistic understanding of the diet, we analyzed multiple tissues, as they reflect different dietary components: (i) carbon and nitrogen stable isotope values (δ13C and δ15N values, respectively) of bulk bone collagen, (ii) δ13C values of bone collagen single essential amino acids (EAAs) (compound specific isotope analysis), (iii) δ13C values of bone bioapatite, and (iv) δ13C values of tooth enamel. Dietary protein sources are reflected in (i) and (ii), and whole diet carbon sources (carbohydrates, lipids, and proteins) are reflected in (iii) and (iv) (4, 8, 9). Because USR2 died before birth, and USR1 likely died shortly after birth, the isotopic composition of their tissues will reflect maternal diets during pregnancy while the tissues were forming (10, 11), although we consider the possible effects of gestation and/or breastfeeding on isotope values.
Season of death for both infants is estimated as early August based on multiple proxies, including seasonally sensitive plant and animal taxa (berries, immature and mature ground squirrel, and salmon) (table S5). To determine the dietary window represented by the bone tissues of USR1 and USR2, we developed models of fetal bone collagen and mineral turnover rates.
RESULTS
Prenatal bone isotopic composition reflects recent maternal diet
Our models of fetal bone tissue turnover indicate rapid turnover rates for bone collagen and mineral during gestation (table S6 and fig. S2). For example, for an infant at age 40 gestational weeks, around 17% of existing bone collagen was formed in the previous week (Fig. 2). Applying the turnover models to the USR infants shows that the majority (~70%) of bone collagen was synthesized in the last 5 to 8 weeks of the infants’ lives (table S7). On the basis of the estimated time of death, the infants’ bone tissues would largely reflect maternal diets over the summer.
Fig. 2. Proportion of bone collagen formed in preceding gestational weeks at given ages.
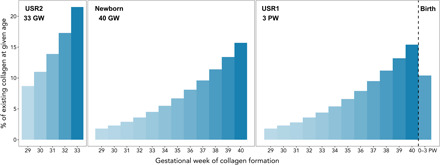
For a prenate at age 33 gestational weeks (GW), 64% of existing collagen was formed in the previous 4 weeks (30th to 33rd week), while for a newborn (40 GW), 55% of bone collagen was formed in the previous 4 weeks. For a neonate at 3 postnatal weeks (PW), 26% of bone collagen was formed in the previous 4 weeks, and 68% was formed in the previous 8 weeks. Estimates are based on a new model of fetal bone collagen turnover (tables S6 and S7); postnatal estimates are based on a previously published model (15).
We assume that bone collagen formed in utero reflects mother’s diet since the fetus derives its nutrition from the maternal bloodstream via the placenta. However, we recognize that the question of whether mother and fetal bone collagen isotope values are identical is unresolved (12). We addressed this uncertainty by incorporating variance parameters for trophic discrimination factors (TDFs; the change in isotope values between food sources and consumer tissues) into our mixing models, as well as by examining the sensitivity of our mixing models to varied TDFs.
Terrestrial resources dominated but salmon was also important in maternal diets
We analyzed the bulk bone collagen isotope data of the humans and ancient regional fauna both graphically (13) and using a Bayesian multisource mixing model (MixSIAR) (14) to estimate dietary protein sources for the mothers of USR1 and USR2 (Figs. 3 and 4 and tables S8 and S9). The USR infants have identical bone collagen δ13C values (−18.4‰), but USR1 has a slightly higher δ15N value (9.1‰) compared to USR2 (8.7‰) (table S2), possibly reflecting the onset of nursing, which leads to 15N enrichment in tissues (11). Using the δ15N value for USR2 as a prenursing baseline, we estimate that the slight bone collagen 15N enrichment of 0.4‰ would be achieved between 3 to 4 weeks of nursing, assuming normal bone growth and remodeling for USR1 and a maximum 15N enrichment of 3.0‰ through breastfeeding (11, 15).
Fig. 3. Bone collagen δ13C and δ15N values for USR infants and regional fauna.
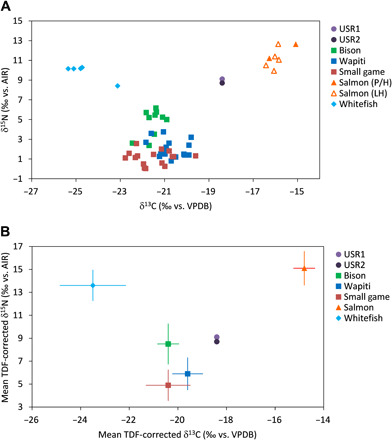
Small game is an average of hare, ground squirrel, and grouse/ptarmigan. (A) Raw bone collagen δ13C and δ15N values. Salmon (P/H) includes specimens dating to the terminal Pleistocene/early Holocene; Salmon (LH) includes specimens dating to the Late Holocene. (B) Isospace plot showing mean faunal δ13C and δ15N values corrected for TDFs is as follows: δ13CTDF-corrected = δ13Ccollagen + 1.1‰ (±0.2‰) and δ15NTDF-corrected = δ15Ncollagen + 3.8‰ (±1.1‰) (66). Source error bars show ±1 SD [combined source and TDF SD calculated following (71)]. Salmon includes P/H and LH specimens.
Fig. 4. Models of USR maternal diets based on infant bone collagen δ13C and δ15N values.
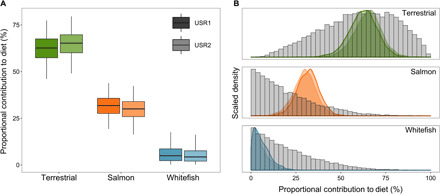
Estimated contributions (%) of food sources to maternal diets were determined using a five-source (bison, wapiti, small game, salmon, and whitefish), two-biotracer (δ13Ccollagen and δ15Ncollagen) mixing model in MixSIAR. Terrestrial is an a posteriori aggregation of bison, wapiti, and small game. (A) Box plots showing median (center line), 50% credible interval (CI) (box edges), and 95% CI (error bars) for estimated contributions. (B) Posterior distributions (colored) superimposed on prior distributions (gray) of source contributions for USR1 (darker color) and USR2 (lighter color). Prior is the uninformative Dirichlet (equal weights for all five original sources).
Biplots of infant and faunal δ15N and δ13C values (Fig. 3, A and B) show that the infant isotope values fall within the dietary mixing space bounded by the faunal isotope values (after correcting for TDFs), demonstrating that their isotopic compositions can be explained by a mixture of the selected food sources in maternal diets (13). The infant isotope values are located close to those of the TDF-corrected terrestrial fauna (bison, wapiti, and small game), indicating that one or more of these resources contributed substantially to maternal diets. However, the infant δ13C values are higher than those of the terrestrial sources (and the whitefish), indicating that a 13C-enriched source, salmon, must have contributed to the diets of both mothers. Furthermore, a plot that includes rarer secondary fauna shows that no other source exhibits the combination of 13C and 15N enrichment necessary to explain the infant isotopic signatures in the absence of salmon (fig. S3).
Bayesian mixing model results for both USR1 and USR2 indicate that although their mothers derived most of their dietary protein from terrestrial resources, salmon also contributed a substantial proportion, with negligible contributions from freshwater fish (Fig. 4A and table S9). Mean dietary contribution estimates for USR1 are 62 ± 8% terrestrial (an a posteriori aggregation of bison, wapiti, and small game), 32 ± 6% salmon, and 6 ± 5% freshwater whitefish. Results are very similar for USR2, with mean estimates of 65 ± 8% terrestrial, 30 ± 7% salmon, and 5 ± 4% freshwater whitefish. Bayesian 95% credible intervals (CIs) for each of the three sources indicate that there are no credible solutions composed of solely terrestrial sources (table S9). All solutions within the 95% CI also contain at least 16% salmon, although there are solutions in this interval that include no whitefish. Mixing model performance indicators are strong when considering the three major resource categories (terrestrial, salmon, and whitefish), with relatively constrained proportional contribution estimates and narrow, unimodal posterior distributions that diverge from the prior distributions, indicating that diet contribution estimates are robust and informed by the isotope data (Fig. 4, A and B) (14, 16). However, mixing model output cannot clearly resolve the relative importance of the discrete terrestrial resources, as there is a high degree of uncertainty for contribution estimates for bison, wapiti, and small game (table S9 and fig. S4).
Given the uncertainty in the estimate of the nitrogen stable isotope TDF (Δ15N), we examined the sensitivity of mixing model contribution estimates to different magnitudes of Δ15N. We varied Δ15N in increments of 1‰ around our main model choice of Δ15N = 3.8‰, spanning 2.8 to 5.8‰. We found that the mean contribution estimates for terrestrial, salmon, and whitefish sources are fairly robust to changes in Δ15N (table S10). For example, for each 1‰ increase in Δ15N, the estimated mean salmon contribution decreases by only about five percentage points.
Amino acid and bioapatite δ13C values support a mix of terrestrial and marine sources
We measured the stable carbon isotope composition of several bone collagen EAAs (table S11). Because EAAs cannot be synthesized by animals, their carbon is routed from dietary protein with minimal modification and reflects the stable carbon isotopic composition of primary production sources at the base of the food web (17). In addition, individual amino acid δ13C values can be normalized to control for baseline variation in isotope values, allowing comparisons of diets among human groups from different regions or time periods (18). Here, we use two normalized EAA dietary markers, Δ13CVal-Phe and Δ13CLys-Phe, calculated from published data, to discriminate between aquatic and terrestrial consumers (Fig. 5 and table S12) [modified from (18, 19)]. There is clear separation among consumers, with higher values of Δ13CVal-Phe and Δ13CLys-Phe for marine and freshwater consumers compared to terrestrial consumers, with no overlap in values. These differences parallel those found in algae and plants, which are major primary producers at the base of aquatic and terrestrial food webs, respectively. The USR infant values fall between those of the terrestrial and marine consumers, as expected if maternal diets were based on a combination of terrestrial mammals and salmon.
Fig. 5. Bone collagen EAA δ13C dietary markers for the USR infants compared to terrestrial, marine, and freshwater protein consumers.
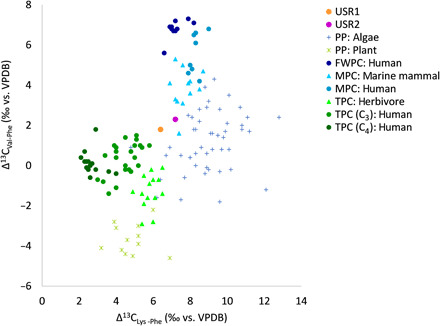
Comparative values were calculated from published data (table S12) (17–19, 72–74). Δ13CVal-Phe = δ13Cvaline − δ13Cphenylalanine and Δ13CLys-Phe = δ13Clysine − δ13Cphenylalanine. Values are also shown for primary producers. PP, primary producer; FWPC, freshwater protein consumer; MPC, marine protein consumer; TPC, terrestrial protein consumer.
We also measured the stable carbon isotope composition of USR1 bone bioapatite, which is −15.5‰. Bioapatite sample quality was evaluated using Fourier transform infrared spectroscopy (20, 21), and sample quality indictors including carbonate-to-phosphate ratio (0.28) and infrared splitting factor (2.6) are within the parameters of well-preserved archaeological samples (21). We used δ13Cbioapatite and δ13Ccollagen values in tandem to provide additional insights into maternal diet. Experimental evidence from animals fed controlled diets shows that δ13Ccollagen and δ13Cbioapatite distributions form two distinct regression lines depending on whether protein is derived from (i) C3 terrestrial sources or from (ii) marine or C4 terrestrial sources (22). Because C4 plants are extremely rare in the subarctic (23), we consider the second regression line as reflecting a marine diet. USR1 falls between the two protein lines but nearer to the C3 terrestrial protein line (fig. S5), in strong agreement with mixing model results based on bone collagen δ13C and δ15N values.
The spacing between δ13Cbioapatite and δ13Ccollagen values (Δ13Cbioapatite-collagen) for USR1 is 2.9‰, which indicates that the δ13C value of the whole diet is lower than that of dietary protein (9). For diets based on terrestrial animals, Δ13Cbioapatite-collagen is predicted to narrow as the 13C-depleted lipid fraction of the diet increases, with a predicted spacing of around 3‰ when lipid comprises 50% of macronutrients (24). Overall, the USR spacing suggests a high animal lipid content, which is consistent with diet inferences based on our other proxies, and further suggests substantial consumption of large mammals, which have a much higher percentage of whole body fat than small mammals (25).
Serial enamel samples suggest an increase in salmon consumption during summer
Tooth enamel forms incrementally and is not remodeled, so isotopic analysis of serial samples from this tissue can detect changes in diet over the course of tooth formation (26). For USR1, we sampled enamel from the upper second deciduous incisor (di2), which begins forming around the 17th gestational week (27). Serial samples of enamel from di2 produced δ13C values of −14.2‰ for the incisal section (formed earlier, ~February/March to May) and −13.4‰ from the cervical section (formed later, ~May to August). The increase in USR1 enamel δ13C values over time of nearly 1‰ suggests the incorporation of 13C-enriched salmon in maternal diet during later development, consistent with the timing of modern salmon runs in the Tanana basin (28).
Independent proxies support a predominantly terrestrial diet with some salmon
Zooarchaeological analyses of contemporaneous sites in the Tanana basin (29, 30) and USR-specific hearth sediment isotopic analyses (31) provide independent proxies for Ancient Beringian paleodiets. Very large mammals (primarily bison and wapiti) are found at all Denali Complex sites with fauna (n = 16; 100% ubiquity), while small mammals (e.g., ground squirrel and hare), birds (terrestrial and waterfowl), and fish are found at fewer sites (29, 44, and 25% ubiquity, respectively) (tables S13 and S14) (29, 30). Among ungulates, bison (56% occurrence) and wapiti (31%) are most common, followed by caribou and sheep (19%) and moose (6%). Some short-term Denali hunting camps, such as Gerstle River (32), are dominated by bison and wapiti and a narrow range of weapon technology, while Denali residential base camps, such as USR and Broken Mammoth, have a much wider range of faunal taxa (table S14) (5, 33).
Paleoindian use of salmon has only recently been confirmed in a Denali site, at USR component 3 (34). We report here the identification of chum salmon (Oncorhynchus keta) remains from a second site (XBD-318) in the Tanana basin. The salmon specimen was recovered from an archaeological component buried in loess (35) and directly dated to 12,680 to 12,770 cal yr. BP (UGAMS-26403; 10,830 ± 40 BP). The specimen has been confirmed as an anadromous chum salmon through DNA and isotopic analyses (tables S1 to S4).
There is also strong agreement between our estimated terrestrial and salmon contributions to diet based on bone collagen δ13C and δ15N values and those based on carbon stable isotope analyses of fatty acids in sediments from multiple hearths at USR component 3 (table S15 and fig. S6) (31). Collectively, the regional zooarchaeological record and hearth chemical profiling from the burial component support terrestrial subsistence dominated by bison and wapiti, with some use of salmon, freshwater resources, and small game.
DISCUSSION
These stable isotope-based paleodiet models provide unique windows into the diets of two Ancient Beringian women over a single spring and summer period 11,500 years ago, providing a link between the broad-scale subsistence patterns observed in the archaeological record over millennia and the short-term foraging decisions made by individuals. Our results indicate that over a summer season, the mothers of USR1 and USR2 obtained a majority of dietary protein from terrestrial food resources but salmon also contributed substantially, while freshwater fish were of minor importance. Salmon were probably only available in the last 4 to 6 weeks of the infants’ lives, corresponding to the time period during which the bulk of bone collagen was forming, and the tooth enamel data from USR1 indicate that there was increased consumption of a 13C-enriched source, likely salmon, between the spring and summer seasons.
Current models for terminal Pleistocene subsistence and mobility in eastern Beringia indicate that the availability of terrestrial resources, and particularly the availability of megafauna, was the principal conditioning factor for making economic and land use decisions (30). Our paleodiet models do not indicate either strictly megafaunal specialists or generalized broad-spectrum foragers. Collectively, these diet models combined with archaeological data from Denali Complex large mammal hunting stations (32) and residential base camps such as USR (5) provide a more holistic perspective on Ancient Beringian diets. We have shown that several independent proxies of diet, including isotopic analyses of multiple human tissues and hearth sediment fatty acids (31), along with regional zooarchaeological evidence (30), support the conclusion that Ancient Beringian diets were complex and seasonally structured. While the diet models produced here support the primacy of terrestrial resources to Ancient Beringian diets, the isotopic evidence for substantial inputs from salmon suggests that this resource too was important in shaping mobility and settlement systems.
Salmon were historically a critical resource for Athabascan populations in interior Alaska (36), and they continue to be important today. An extensive diet survey of modern indigenous Alaskan populations (37) found that Athabascan groups in the Tanana region derive 24% of their dietary protein from salmon, a figure very similar to our estimate for the salmon contribution to Ancient Beringians living in the area 11,500 years ago.
MATERIALS AND METHODS
Archaeological human and faunal specimens
All human and faunal bone samples analyzed in this study are presented and described in table S1. All specimens came from existing collections permanently curated at the University of Alaska Museum of the North or under active investigation at research laboratories.
USR infants
Excavation and analyses of the USR infant remains were conducted under a Memorandum of Agreement (MOA) signed by the State of Alaska (the land owner), the National Science Foundation (the lead federal agency), the Healy Lake Tribal Council (the federally recognized tribal authority), and the Tanana Chiefs Conference (the regional nonprofit tribal consortium). An amendment to the MOA was signed by all parties to allow for destructive analysis of skeletal remains for the purposes of diet reconstruction through stable isotope analyses and assessment of genetic relationships through DNA analyses. We are grateful for the support and cooperation of all parties.
Osteological and genetic studies of the USR infants have been published previously (5–7). Mitochondrial DNA sequences show that the infants belong to two separate mitochondrial lineages (B2 and C1b) and, thus, are not maternally related (7). Whole-genome sequence data indicate that relatedness between the USR infants is within the range of half siblings or first cousins, and they confirm that the two infants are female (6).
Osteometric estimates of age at death are around birth to 6 weeks for USR1 and around 33 weeks in utero for USR2 (5). The age estimate based on tooth development is slightly higher for USR1 (~12 postnatal weeks). The discrepancy between the osteometric and dental ages may have resulted from differences between Ancient Beringians and the modern European-derived individuals used to the develop the age schedules, or bone growth could have lagged behind chronological age for USR1, although no skeletal anomalies or pathologies are apparent (5). The uncertainty ranges surrounding the point age estimates are larger for the dental methods (±~4 to 13 weeks) than for the osteometric methods (±~2 weeks) (5, 38). Given that the one-sigma uncertainty ranges for the two methods overlap at around age 3 to 4 weeks, we suggest that this is a reasonable estimate of the age at death for USR1.
Single middle ribs from USR1 and USR2 were selected for bone collagen extraction. The rib from USR1 produced sufficient material to also allow bioapatite extraction. In addition, enamel from a single tooth (deciduous right maxillary lateral incisor) from USR1 was serially sampled for δ13C analysis. Serial enamel sampling was attempted for USR2, but the samples were too small for analysis.
Faunal remains
We selected faunal taxa based on their presence in the USR component 3 faunal assemblage (directly associated with the infants) and/or their ubiquity in regional terminal Pleistocene/early Holocene faunal assemblages. Faunal remains directly associated with the USR infants are generally too burned to be good candidates for collagen extraction and stable isotope analysis. Therefore, we selected faunal remains from other archaeological sites in the Tanana basin of central Alaska to encompass the suite of potential faunal resources used by USR inhabitants and to capture food source isotopic variation (table S1). We used molecular methods (including DNA and/or protein fingerprinting) to confirm or refine the taxonomic identifications of all salmon and nearly all ungulates (tables S1, S3, and S4 and data file S1). To control for interlaboratory and interstudy variation in measurements of δ13Ccollagen and δ15Ncollagen (39), all faunal specimens were analyzed specifically for this study using the same collagen extraction method and, except for a single specimen, using the same stable isotope laboratory (table S2).
We considered two sets of faunal food sources: (i) primary fauna, which includes only those fauna confirmed in USR component 3 (directly associated with the infants) and/or fauna that are ubiquitous in terminal Pleistocene/early Holocene faunal assemblages in the Tanana basin (bison, wapiti, small game, salmon, and whitefish); and (ii) secondary fauna, which includes taxa that are not confirmed in USR Component 3 but that occasionally occur in the faunal record for this time period (caribou, sheep, and waterfowl) (tables S13 and S14) (5, 30). Additional details on faunal sample composition are found in the Supplementary Materials.
DNA taxonomic identification of selected faunal remains
DNA extraction
All pre–polymerase chain reaction (PCR) activities were conducted in the ancient DNA facility at the Laboratories of Molecular Anthropology and Microbiome Research at the University of Oklahoma. This facility is a dedicated workspace for processing aged, degraded, and/or low copy number DNA samples. Precautions aimed to minimize and monitor the introduction of contamination are practiced in the laboratory.
Approximately 50 mg or less of bone material was subsampled from each specimen. The subsamples were submerged in 6% (w/v) sodium hypochlorite for 4 min (40). The sodium hypochlorite was poured off, and the samples were quickly submerged in DNA-free water twice.
The bone samples were transferred to 1.5-ml tubes, to which aliquots of 500 μl of 0.5 M EDTA were added, and the tubes gently rocked at room temperature for >48 hours. An extraction negative control, to which no bone material was added, accompanied each batch of extractions, typically in a ratio of 1:7 with the samples.
DNA was extracted following the method described by Kemp et al. (41). Ninety microliters of proteinase K (Bio Basic catalog no. 32181) at a concentration of 1 mg/30 μl (or >20 U/30 μl) was added to each sample, and the tubes were incubated at 64° to 65°C for 3 hours. Following proteinase K digestion, the tubes were centrifuged at 15,000 rpm for 1 min to pellet any undigested bone, dirt, and/or “sludge.” All centrifugation steps in this study were conducted with an Eppendorf centrifuge 5424. The liquid was carefully moved to a new 1.5-ml tube, to which 750 μl of 2.5% “resin” (i.e., 2.5% celite in 6 M guanidine HCl) and 250 μl of 6 M guanidine HCl were added. The tubes were vortexed multiple times over approximately a 2-min period.
Promega Wizard minicolumns were attached to 3 ml of Luer-Lok syringe barrels (minus the plunger) and placed on a vacuum manifold. Three milliliters of DNA-free water was first pulled across the columns with the intent to wash away potential contaminating DNA. The DNA/resin mixture was subsequently pulled across the columns. The silica pellet on the minicolumn was then rinsed by pulling 3 ml of 80% isopropanol across the columns.
The minicolumns were then placed in new 1.5-ml tubes and centrifuged at 10,000 rpm for 2 min to remove excess isopropanol. The minicolumns were transferred to new 1.5-ml tubes. Fifty microliters of DNA-free water heated to 64° to 65°C was added to the minicolumns and left for 3 min before centrifugation of the tubes for 30 s at 10,000 rpm. This step was repeated, amounting to 100 μl of extracted DNA. Ten microliters of the full concentration eluates and extraction negative controls were diluted 1:10 with water and used as described below [under standard PCR, rescue PCR, and PCR buffer enhancer P (PEC-P)].
Inhibition test and repeat silica extraction
The remaining volumes of the full concentration DNA eluates were tested for the presence of PCR inhibitors following the rationale of Kemp et al. (41) using a “turkey collective” as the ancient DNA positive control (see their figure 1). DNA recovered from seven or more archaeological turkey (Meleagris gallopavo) bones (42) was pooled together to make the turkey collective. The choice to pool these individual extractions was made with the intention to reduce variation between turkey DNA eluates in both endogenous mitochondrial DNA copy number and possible inhibitors coextracted with the turkey DNA. Before they are used in experiments, each turkey collective was demonstrated to PCR amplify consistently (in six or more amplifications), hence serving as a positive control.
Fifteen microliters of PCRs were conducted to amplify a 186–base pair portion of turkey displacement loop using the primers “T15709F” and “T15894R” described by Kemp et al. (42). The components of these PCRs were as follows: 1× Omni Klentaq Reaction Buffer (including a final concentration of 3.5 mM MgCl2), 0.32 mM deoxynucleotide triphosphates (dNTPs), 0.24 μM of each primer, 0.3 U of Omni Klentaq LA polymerase, and 1.5 μl of turkey collective DNA. These reactions were spiked with 1.5 μl of potentially inhibited, full concentration DNA eluates recovered from the samples under investigation here. The extraction negative controls were also tested for inhibitors in this manner. These PCRs were run in parallel with reactions that contained only turkey collective DNA (i.e., were not spiked). These reactions served as positive controls and allowed us to preclude PCR failure from contributing to our results. PCR negatives also accompanied each round of amplification, allowing us to monitor for possible contamination. Additional positive controls of modern turkey DNA were added in the post-PCR laboratory only before initiating amplification, serving as another check for possible PCR failure. Following denaturing at 94°C for 3 min, 60 cycles of PCR was conducted at 94°C for 15 s, 60°C for 15 s, and 68°C (note that this is the optimal extension temperature for Omni Klentaq LA polymerase) for 15 s. Last, a 3-min extension period at 68°C was conducted before bringing the reactions to 10°C.
If the turkey collective failed to amplify when spiked with any given ancient DNA eluate, then we considered the eluate to be inhibited. In the case that spiking the ancient DNA permitted amplification of the turkey collective DNA, we consider that DNA eluate to be inhibitor “free.”
Full concentration eluates deemed to be inhibited using the test outlined above were subjected to repeat silica extraction (41). To the remaining volume of the eluate, 750 μl of 2.5% resin and 250 μl of 6 M guanidine HCl were added. The samples were vortexed numerous times over a 2-min period. The extraction then followed procedures described above under “DNA extraction,” except that the volume used to elute the DNA from column matched the volume being repeat silica extracted. For example, if the starting volume was 87 μl, then 43.5 μl of DNA-free water heated to 65°C was added to the minicolumns and left for 3 min before centrifugation. This step was repeated twice for a total volume of 87 μl.
These repeat silica eluates were tested again for inhibition, as described above. Those still deemed to be inhibited were once again repeat silica extracted and tested again for inhibition. This was carried out until all full concentration eluates were deemed to be uninhibited.
Standard PCR, rescue PCR, and PEC-P
All of the full concentration, uninhibited eluates and the original 1:10 dilutions of the full concentration eluates were subject to three forms of PCR. First, “standard” PCRs contained 1× Omni Klentaq Reaction Buffer, 0.32 mM dNTPs, 0.24 μM of each primer, 0.3 U of Omni Klentaq LA polymerase, and 1.5 μl of template DNA. Second, rescue PCR at a 25% increase was carried out, as described by Johnson and Kemp (43). Rescue PCRs contained 1.25× Omni Klentaq Reaction Buffer (including a final concentration of 4.375 mM MgCl2), 0.4 mM dNTPs, 0.3 μM of each primer, 0.375 U of Omni Klentaq LA polymerase, and 1.5 μl of template DNA. Last, we used PEC-P. The composition of this enhancer cocktail is proprietary, and no safety data sheet is made available at the DNA Polymerase Technology website (www.klentaq.com/). However, Palmer et al. (44) used PEC-P to increase their success in species identification of archaeological remains of smelt and other small fishes. These PCR reactions contained 1× Omni Klentaq Reaction Buffer (including a final concentration of 3.5 mM MgCl2), 0.32 mM dNTPs, 0.24 μM of each primer, 0.3 U of Omni Klentaq LA polymerase, 20% (v/v) PEC-P, and 1.5 μl of template DNA. All three forms of PCR were conducted as follows: (i) 94°C for 3 min; (ii) 60 cycles of PCR at 94°C for 15 s, the annealing temperature for 15 s (table S3), and 68°C for 15 s; and (iii) a 3-min extension period at 68°C before bringing the reactions to 10°C.
Primers
Primers are listed in table S3. For salmonid species identification, we used previously described primers by Jordan et al. (45). Note that Jordan et al. (45) originally described their reverse primer in the wrong orientation. The corrected primers are OST12S-forward (5′-GCTTAAAACCCAAAGGACTTG-3′) and OST12S-reverse (5′-CTACACCTCGACCTGACGTT-3′). Other primers were designed in this study to differentiate moose (Alces alces), bison (Bison priscus), elk (Cervus canadensis), Dall sheep (Ovis dalli), and caribou (Rangifer tarandus).
Sequencing results
Sequencing results are shown in table S4, and species identification is noted in table S1. Samples 16.179, 16.180, 16.181, 16.182, 16.184, and 17.276 were identified as bison (B. priscus) with repeatable results. DNA from sample 16.185, identified as bison based on morphology and as Bison/Bos based on ZooMS results, failed to amplify despite repeated attempts.
Sample 17.284 was identified as a caribou (R. tarandus) from the amplicon produced with primers Art3F/Art3R. Despite repeated PCRs, this observation was not repeatable. However, note that this sample was believed to be a caribou based on morphology.
Samples 15.301, 16.109, and 17.106 were identified as chum salmon (O. keta), as described by Jordan et al. (45), exhibiting the following mutations relative to the rainbow trout (Oncorhynchus mykiss), reference sequence (DQ288271.1): 660T and 713T. Samples 17.109, 17.110, and 17.112 were also identified as chum salmon with an additional derived mutation of a thymine (T) deletion at nucleotide position 668. Replication was possible for these six salmonid specimens. A single amplicon from sample 17.111 revealed this specimen to be a chum salmon with a T deletion at nucleotide position 668. Sequences from two additional amplicons from this sample showed additional but different “mutations,” likely attributable to postmortem damage.
Samples 16.171, 16.175, and 16.176 were identified as elk (C. canadensis) with primers ElkCOIF/ElkCOIR, matching the reference sequence (JF443209.1). Only a single amplicon was produced for sample 16.171, despite repeated attempts at amplification. Three additional amplicons produced from sample 16.175 differed from the reference sequence by different mutations, likely as a product of postmortem damage. A second amplicon sequenced from sample 16.176 also showed signs of damage with mutations at nucleotide positions 230T and 234T. Samples 16.171, 16.175, and 16.176 were identified as elk by morphological assessment and as Cervidae by ZooMS analysis. DNA from samples 16.168, 16.169, 16.170, 16.172, 16.173, 16.174, and 16.177 failed to amplify.
Molecular taxonomic identification of selected faunal remains (ZooMS)
A subsample of approximately 1 to 2 mg of bone collagen (below) was resuspended with 50 mM ammonium bicarbonate and digested with 0.4 μg of sequencing grade trypsin (Promega, UK) overnight at 37°C. Digests were then acidified to 0.1% trifluoroacetic acid (TFA) and fractionated into 10 and 50% acetonitrile (in 0.1% TFA) before being dried down to completion by centrifugal evaporation and resuspension in 10 μl of 0.1% TFA. One microliter of resuspended peptide mixture was then spotted onto a stainless steel matrix-assisted laser desorption/ionization–time-of-flight (MALDI-TOF) plate along with an equal volume of α-cyano hydroxycinnamic acid matrix (10 mg/ml) in 50% ACN (acetonitrile)/0.1% TFA and allowed to air dry. MALDI-TOF mass spectrometry was carried out using a Bruker Ultraflex II with 2000 laser acquisitions over the range mass/charge ratio of 700 to 3700, and the resultant spectra were compared with reference spectra published previously (46). Results are shown in table S1, fig. S1, and data file S1.
Sample pretreatment and stable isotope measurements
All carbon and nitrogen stable isotope measurements are expressed in delta notation (as δ13C and δ15N, respectively) relative to internationally accepted standards as follows: δ = (Rsample – Rstandard)/Rstandard, where R is the ratio of the heavy to light isotope (e.g., 13C/12C) (47); by convention, this quantity is multiplied by 1000 to report the result in parts per thousand (‰). The international standard for carbon is Vienna Pee Dee belemnite (VPDB) and that for nitrogen is atmospheric N2 (AIR). The standard reference materials used to calibrate raw isotopic measurements to the internationally accepted scales are described for each laboratory below.
Bone collagen (δ13C and δ15N values)
All human and faunal bone collagen extractions were conducted in the same laboratory (Laboratory of Environmental Archaeology, University of Alaska Fairbanks) using the modified Longin (48) protocol described previously (34). Briefly, powdered bone was demineralized in HCl (0.5 M), followed by treatment in NaOH (0.1 M), and gelatinization at 70°C in dilute HCl (0.001 M, pH 3). Bone collagen samples were submitted to the Washington State University Stable Isotope Core Laboratory for carbon and nitrogen stable isotope measurement on a Thermo Finnigan Delta Plus XP continuous flow isotope ratio mass spectrometer, coupled to a Costech elemental analyzer (ECS 4010). The carbon and nitrogen stable isotope compositions were calibrated relative to VPDB and AIR, respectively, using at least two internal standards, which had been previously calibrated against internationally certified standards (table S16). In addition, all sample sequences included the same quality control check standard, casein (B2155 Elemental Microanalysis), as a check of the normalization. Precision was calculated as the pooled SD of all repeated measures of calibration and check standards over the relevant analytical runs following Szpak et al. (49) and was ±0.14‰ for both δ13C and δ15N. One extracted bone collagen sample (#17.260) was analyzed by the Center for Applied Isotope Studies at the University of Georgia. This laboratory calibrated relative to VPDB and AIR using two in-house standards, spinach (δ13C = −27.22‰ and δ15N = 0.19‰) and bovine (δ13C = −17.53‰ and δ15N = 8.14‰), which had been previously calibrated to National Institute of Standards and Technology standards.
All bone collagen samples used in this study met well-accepted quality standards: %N > 5%, %C > 13%, an atomic C/N ratio of 2.9 to 3.6, and an collagen yield of >1% (table S2) (50–53). Further, our δ13C and δ15N values for ungulates are generally comparable to those previously reported for archaeological specimens in the region and elsewhere in Alaska, although there is variation among studies (54–57). We further note that the stable carbon isotope composition of our salmon specimens, as in other archaeological and modern Pacific salmon samples, is relatively depleted in 13C compared to many marine fishes, likely due to the offshore feeding location of many salmon species (58–62).
Bone collagen EAAs (δ13C values)
We measured amino acid δ13C values using gas chromatography–combustion–stable isotope ratio mass spectrometry (GC-C-IRMS) in the Alaska Stable Isotope Facility (ASIF) at the niversity of Alaska Fairbanks. To prepare samples for analysis, we hydrolyzed dried collagen (~1 mg) using 1 ml of HCl (6 M) at 110°C for 20 hours and dried them under N2. We derivatized the amino acids to N-acetyl methyl esters (NACMEs) for analysis by GC-C-IRMS (63). First, we methylated the amino acids with acidified methanol, prepared by the addition of acetyl chloride to methanol (1:6, v/v) in an ice bath. The methylation was completed during 75°C incubation for 1 hour. Samples were then dried down under N2 and acetylated with the addition of acetic anhydride, triethylamine, and acetone (1:2:5, v/v/v) and an incubation at 60°C for 10 min. The NACMEs (i.e., the derivatized amino acids) were dried down and purified with a phosphate buffer wash [1 M potassium phosphate and 1 M sodium phosphate (pH 7)], extracted with chloroform, and, again, dried down under N2. Last, the NACMEs were dissolved in ethyl acetate. A mix of amino acid standards with known δ13C values was prepared alongside samples, and an internal standard (norleucine) was added to standards and samples. Amino acid NACMEs were injected onto a VF-35ms column (Agilent) in a TRACE 1310 GC interfaced with an Isolink II (Thermo Scientific), combusted into CO2 gas, and the carbon isotope ratios of individual NACME peaks were measured on a Delta V Plus IRMS (Thermo Scientific). The δ13C calculation was based on NACME peak integration, which was performed by the program Isodat (version 3.0, Thermo Scientific). Peak integration was visually assessed for correct identification, width and background assignment, and adequate baseline separation between peaks. We measured the δ13C in six EAAs: isoleucine (Ile), leucine (Leu), lysine (Lys), phenylalanine (Phe), threonine (Thr), and valine (Val). The δ13C values of individual amino acids are measured relative to the δ13C values of standard amino acids; these are internal (in-house ASIF) standards, and their δ13C, before derivatization, are given in table S17. Internal (rather than international) individual amino acids are used to account for the potential influence of slightly different fractionations that can occur between batches of sample derivatizations and to allow for the correction of carbon added during the derivatization process (17, 63). Analytical precision was <0.3‰ for each of the amino acids in both the standards (<0.2‰) and samples (with the exception of phenylalanine that was <0.8‰ in samples and <0.2‰ in standards).
Bone bioapatite (δ13C values)
The USR1 bone sample yielded sufficient bulk bone powder to allow both collagen and bioapatite extraction, but the USR2 bone sample was exhausted after collagen extraction. Bone bioapatite for USR1 was extracted following Garvie-Lok et al. (20) with minor modifications. Briefly, organics were removed from cleaned, powdered bone by soaking in 2% NaOCl for 48 hours (with a refresh at 24 hours), followed by rinsing and removal of contaminating carbonate by treatment in 0.1 M acetic acid (unbuffered) for 24 hours, and followed by a final rinsing and freeze drying for 48 hours. Bioapatite samples were submitted to the University of California Santa Cruz Stable Isotope Laboratory for carbon stable isotope measurement by acid digestion using a Thermo Scientific Kiel IV carbonate device interfaced to a Thermo Scientific MAT 253 isotope ratio mass spectrometer. Carbon stable isotope composition was calibrated relative to VPDB using NBS-18 and the internal standard CM12 (Carrara Marble; δ13C = 2.05‰). The internal standard was previously calibrated against NBS-19 and NBS-18. Precision for δ13C over the analytical run was calculated as defined above and was 0.11‰.
Tooth enamel bioapatite (δ13C values)
Enamel from a single tooth (deciduous right maxillary lateral incisor) from USR1 was serially sampled for δ13C analysis. Serial enamel sampling was attempted for USR2, but yields were too low for analysis. Because enamel forms incrementally and is not remodeled once formed, serial samples can be used to detect changes in diet over the course of tooth formation. Tooth enamel formation for the maxillary di2 begins at approximately 163 days before birth (~17 gestational weeks) (27). Assuming a time of death in early August, if USR1 died at birth (40 gestational weeks), then her di2 tooth enamel likely began forming in early March, whereas if USR1 lived for several weeks after birth, then the start of enamel formation would be pushed back accordingly (e.g., ~mid-February if USR1 lived for 3 weeks, ~mid-January if USR lived for 6 weeks, etc.). Enamel formation begins at the cusp of the developing tooth and proceeds toward the tooth cervix. The tooth was serially sampled in two locations, one (cuspal) reflecting earlier development (i.e., ~spring) and one (cervical) reflecting later development (i.e., ~summer).
To prepare enamel bioapatite, the tooth was mechanically cleaned to remove surface contaminants and then ultrasonically cleaned in deionized double-distilled water (DDH2O) for 30 min. Approximately 2.0 mg of cleaned enamel powder was drilled from the tooth. The samples were first treated in microcentrifuge tubes with 3% H2O2 for 15 min and then were rinsed three times with DDH2O water before being treated with 0.1 M CH3COOH for 15 min. Samples were rinsed three times with DDH2O water and then dried overnight in an oven at 60°C. δ13C values were measured by continuous-flow IRMS at IsoForensics in Salt Lake City, Utah using a GasBench (Thermo Scientific) interfaced to an isotope ratio mass spectrometer (Thermo Finnigan MAT 253). The carbon stable isotope composition was calibrated relative to VPDB using NBS-19 and LSVEC. Precision for δ13C over the analytical run was calculated as described above and was 0.10‰.
Stable isotope mixing models and diet estimation
To estimate proportional contributions of food sources to the diets of the USR infants’ mothers, we used MixSIAR (14), a Bayesian stable isotope mixing model framework available as an open-source package in R (64).
Food source selection and aggregation
For diet estimation using stable isotope mixing models, the number of diet sources should be kept as low as possible without excluding important sources (14, 16, 65). Bayesian mixing models assume that all sources included in the model contributed to diet, so it is important to include only sources that were actually consumed (65). For the MixSIAR analyses, we used the five sources (bison, salmon, small game, wapiti, and whitefish) described above as “primary fauna.”
Trophic discrimination factors
We used the following collagensource-to-collagenconsumer TDFs (with associated SDs) following Bocherens et al. (66): Δ13Cconsumer collagen−source collagen = 1.1 ± 0.2‰ and Δ15Nconsumer collagen−source collagen = 3.8 ± 1.1‰. We further assumed that fetal and neonate bone collagen isotope values reflect those of the mother’s diet (after adjusting for the standard TDFs) but with caveats. The assumption is supported by limited isotopic analyses of accessible proteinaceous hard tissues (fingernails) in living mother/infant pairs, which show that newborn and mother fingernail δ15N and δ13C values are similar (11, 67). A study of hair from newborn-mother pairs likewise found a negligible difference (+0.4‰) for δ13C values but a +0.9‰ enrichment for δ15N (68). Studies comparing archaeological adult and infant bone collagen isotope values are mixed for δ15N values, with a meta-analysis by Reynard and Tuross (12) showing neonate bone collagen δ15N values both above and below the adult female mean.
Addressing uncertainty in the δ15N TDF (Δ15N)
To address uncertainty in Δ15N between consumer and source collagen, as well as between mother’s diet and fetal/neonate bone collagen, (i) we include a variance parameter in our Δ15N our isotope mixing models, and (ii) we explore the sensitivity of model mean proportional contribution estimates to various magnitudes of Δ15N. We set Δ15N variance at 1.2‰, the squared SD of 1.1‰ found by Bocherens et al. (66) in their meta-analysis of prey-to-predator, collagen-to-collagen offsets in δ15N values. We varied Δ15N in increments of 1‰ around our main model choice of 3.8‰, from 2.8 to 5.8‰, to approximate the range of reported nitrogen isotope TDFs for humans and other mammals (69, 70).
Statistical analyses
We used the Bayesian multisource mixing model MixSIAR (14) to estimate dietary protein sources for the mothers of USR1 and USR2. MixSIAR models were run separately for USR1 and USR2. Table S8 provides the consumer and food source isotope values, TDFs, and associated uncertainties included in the models. Markov chain Monte Carlo parameters were as follows: (i) “normal” run: chain length = 100,000, burn-in = 50,000, thinning = 50, and chains = 3; (ii) alpha.prior: Dirichlet prior (default, 1; uninformative); and (iii) error structure: residual error: FALSE (not included) and process error: TRUE (included) (note that this is the only error structure appropriate for fitting a single consumer) (14).
Supplementary Material
Acknowledgments
We thank representatives from the Healy Lake Tribal Council and Tanana Chiefs Conference for support. We also thank S. Shirar, Collection Manager at UAMN, for aiding in collections selection; C. Monroe, R. Frome, M. Labonte, and E. Palmer for conducting genetic species identifications and other assistance in the laboratory (University of Oklahoma); and IsoForensics and L. Chesson for isotope analyses on the tooth enamel samples. Funding: This work was supported, in part, by NSF grant 1521501 to C.M.H., B.A.P., and J.D.R., with a subcontract to B.M.K.; NSF grants 1223119, 1439293, and 1558332 to B.A.P.; NSF grant 1521365 to B.P.F.; and M. J. Murdock Charitable Trust Grant SR-201811010 for instrumentation to M.J.W. Author contributions: Conceptualization: C.M.H., B.A.P., B.P.F., and J.D.R. Methodology: C.M.H., B.A.P., T.T., B.M.K., E.J.B., and M.B. Formal analysis: C.M.H., B.A.P., T.T., and C.T.C. Investigation: C.M.H., B.A.P., H.J.M., T.T., B.M.K., E.J.B., M.J.W., M.B., J.J.J., B.L.B., F.B.L., R.A.S., and J.D.R. Resources: C.M.H., B.A.P., T.T., B.M.K., E.J.B., M.J.W., M.B., F.B.L., and J.D.R. Writing (original draft): C.M.H. and B.A.P. Writing (review and editing): C.M.H., B.A.P., H.J.M., T.T., B.P.F., B.M.K., E.J.B., M.J.W., M.B., C.T.C, J.J.J., B.L.B., F.B.L., R.A.S., and J.D.R. Visualization: C.M.H., B.A.P., and C.T.C. Competing interests: The authors declare that they have no competing interests. Data and materials availability: DNA sequences have been deposited in GenBank (accession numbers MN931661-MN931683). All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/36/eabc1968/DC1
REFERENCES AND NOTES
- 1.T. A. Surovell, N. M. Waguespack, Human prey choice in the late Pleistocene and its relation to megafaunal extinctions, in American Megafaunal Extinctions at the End of the Pleistocene, G. Haynes, Ed. (Springer, 2009), pp. 77–105. [Google Scholar]
- 2.Cannon M. D., Meltzer D. J., Explaining variability in Early Paleoindian foraging. Quat. Int. 191, 5–17 (2008). [Google Scholar]
- 3.E. J. Reitz, E. S. Wing, Zooarchaeology (Cambridge Univ. Press, 1999). [Google Scholar]
- 4.M. P. Richards, Isotope analysis for diet studies, in Archaeological Science: An Introduction, M. P. Richards, K. Britton, Eds. (Cambridge Univ. Press, 2020), pp. 125–143. [Google Scholar]
- 5.Potter B. A., Irish J. D., Reuther J. D., McKinney H. J., New insights into Eastern Beringian mortuary behavior: A terminal Pleistocene double infant burial at Upward Sun River. Proc. Natl. Acad. Sci. U.S.A. 111, 17060–17065 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moreno-Mayar J. V., Potter B. A., Vinner L., Steinrücken M., Rasmussen S., Terhorst J., Kamm J. A., Albrechtsen A., Malaspinas A.-S., Sikora M., Reuther J. D., Irish J. D., Malhi R. S., Orlando L., Song Y. S., Nielsen R., Meltzer D. J., Willerslev E., Terminal Pleistocene Alaskan genome reveals first founding population of Native Americans. Nature 553, 203–207 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Tackney J. C., Potter B. A., Raff J., Powers M., Watkins W. S., Warner D., Reuther J. D., Irish J. D., O’Rourke D. H., Two contemporaneous mitogenomes from terminal Pleistocene burials in eastern Beringia. Proc. Natl. Acad. Sci. U.S.A. 112, 13833–13838 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Webb E. C., Lewis J., Shain A., Kastrisianaki-Guyton E., Honch N. V., Stewart A., Miller B., Tarlton J., Evershed R. P., The influence of varying proportions of terrestrial and marine dietary protein on the stable carbon-isotope compositions of pig tissues from a controlled feeding experiment. Sci. Technol. Archaeolog. Res. 3, 28–44 (2017). [Google Scholar]
- 9.S. H. Ambrose, L. Norr, Experimental evidence for the relationship of the carbon isotope ratios of whole diet and dietary protein to those of bone collagen and carbonate, in Prehistoric Human Bone: Archaeology at the Molecular Level, J. B. Lambert, G. Grupe, Eds. (Springer-Verlag, 1993), pp. 1–37. [Google Scholar]
- 10.Richards M. P., Mays S., Fuller B. T., Stable carbon and nitrogen isotope values of bone and teeth reflect weaning age at the medieval Wharram Percy site, Yorkshire, UK. Am. J. Phys. Anthropol. 119, 205–210 (2002). [DOI] [PubMed] [Google Scholar]
- 11.Fuller B. T., Fuller J. L., Harris D. A., Hedges R. E. M., Detection of breastfeeding and weaning in modern human infants with carbon and nitrogen stable isotope ratios. Am. J. Phys. Anthropol. 129, 279–293 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Reynard L. M., Tuross N., The known, the unknown and the unknowable: Weaning times from archaeological bones using nitrogen isotope ratios. J. Archaeol. Sci. 53, 618–625 (2015). [Google Scholar]
- 13.Phillips D. L., Gregg J. W., Source partitioning using stable isotopes: Coping with too many sources. Oecologia 136, 261–269 (2003). [DOI] [PubMed] [Google Scholar]
- 14.Stock B. C., Jackson A. L., Ward E. J., Parnell A. C., Phillips D. L., Semmens B. X., Analyzing mixing systems using a new generation of Bayesian tracer mixing models. PeerJ 6, e5096 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsutaya T., Yoneda M., Quantitative reconstruction of weaning ages in archaeological human populations using bone collagen nitrogen isotope ratios and approximate Bayesian computation. PLOS ONE 8, e72327 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Phillips D. L., Inger R., Bearhop S., Jackson A. L., Moore J. W., Parnell A. C., Semmens B. X., Ward E. J., Best practices for use of stable isotope mixing models in food-web studies. Can. J. Zool. 92, 823–835 (2014). [Google Scholar]
- 17.Larsen T., Ventura M., Andersen N., O’Brien D. M., Piatkowski U., McCarthy M. D., Tracing carbon sources through aquatic and terrestrial food webs using amino acid stable isotope fingerprinting. PLOS ONE 8, e73441 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Honch N. V., McCullagh J. S. O., Hedges R. E. M., Variation of bone collagen amino acid δ13C values in archaeological humans and fauna with different dietary regimes: Developing frameworks of dietary discrimination. Am. J. Phys. Anthropol. 148, 495–511 (2012). [DOI] [PubMed] [Google Scholar]
- 19.Webb E. C., Honch N. V., Dunn P. J. H., Eriksson G., Lidén K., Evershed R. P., Compound-specific amino acid isotopic proxies for detecting freshwater resource consumption. J. Archaeol. Sci. 63, 104–114 (2015). [Google Scholar]
- 20.Garvie-Lok S. J., Varney T. L., Katzenberg M. A., Preparation of bone carbonate for stable isotope analysis: The effects of treatment time and acid concentration. J. Archaeol. Sci. 31, 763–776 (2004). [Google Scholar]
- 21.Beasley M. M., Bartelink E. J., Taylor L., Miller R. M., Comparison of transmission FTIR, ATR, and DRIFT spectra: Implications for assessment of bone bioapatite diagenesis. J. Archaeol. Sci. 46, 16–22 (2014). [Google Scholar]
- 22.Froehle A. W., Kellner C. M., Schoeninger M. J., FOCUS: Effect of diet and protein source on carbon stable isotope ratios in collagen: Follow up to Warinner and Tuross (2009). J. Archaeol. Sci. 37, 2662–2670 (2010). [Google Scholar]
- 23.Wooller M. J., Zazula G. D., Edwards M., Froese D. G., Boone R. D., Parker C., Bennett B., Stable carbon isotope compositions of eastern Beringian grasses and sedges: Investigating their potential as paleoenvironmental indicators. Arct., Antarc., Alp. Res. 39, 318–331 (2007). [Google Scholar]
- 24.Fernandes R., Nadeau M.-J., Grootes P. M., Macronutrient-based model for dietary carbon routing in bone collagen and bioapatite. Archaeol. Anthropol. Sci. 4, 291–301 (2012). [Google Scholar]
- 25.Cordain L., Miller J. B., Eaton S. B., Mann N., Holt S. H., Speth J. D., Plant-animal subsistence ratios and macronutrient energy estimations in worldwide hunter-gatherer diets. Am. J. Clin. Nutr. 71, 682–692 (2000). [DOI] [PubMed] [Google Scholar]
- 26.Balasse M., Reconstructing dietary and environmental history from enamel isotopic analysis: Time resolution of intra-tooth sequential sampling. Int. J. Osteoarchaeol. 12, 155–165 (2002). [Google Scholar]
- 27.Mahoney P., Incremental enamel development in modern human deciduous anterior teeth. Am. J. Phys. Anthropol. 147, 637–651 (2012). [DOI] [PubMed] [Google Scholar]
- 28.Cappiello T. A., Bromaghin J. F., Mark-recapture abundance estimate of fall-run chum salmon in the Upper Tanana River, Alaska, 1995. Alaska Fishery Research Bulletin 4, 12–35 (1997). [Google Scholar]
- 29.Potter B. A., A first approximation of Holocene inter-assemblage variability in central Alaska. Arct. Anthropol. 45, 89–113 (2008). [Google Scholar]
- 30.B. A. Potter, C. E. Holmes, D. R. Yesner, Technology and economy among the earliest prehistoric foragers in interior eastern Beringia, in Paleoamerican Odyssey, K. E. Graf, C. V. Ketron, M. R. Waters, Eds. (Center for the Study of the First Americans, Texas A&M University, 2013), pp. 81–103. [Google Scholar]
- 31.Choy K., Potter B. A., McKinney H. J., Reuther J. D., Wang S. W., Wooller M. J., Chemical profiling of ancient hearths reveals recurrent salmon use in Ice Age Beringia. Proc. Natl. Acad. Sci. U.S.A. 113, 9757–9762 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Potter B. A., Models of faunal processing and economy in early Holocene interior Alaska. Environ. Archaeol. 12, 3–23 (2007). [Google Scholar]
- 33.Yesner D. R., Human dispersal into interior Alaska: Antecedent conditions, mode of colonization, and adaptation. Quat. Sci. Rev. 20, 315–327 (2001). [Google Scholar]
- 34.Halffman C. M., Potter B. A., McKinney H. J., Finney B. P., Rodrigues A. T., Yang D. Y., Kemp B. M., Early human use of anadromous salmon in North America at 11,500 y ago. Proc. Natl. Acad. Sci. U.S.A. 112, 12344–12348 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.B. A. Potter, E. P. Gaines, P. M. Bowers, M. Proue, “Results of the 2006 cultural resource survey of proposed Alaska Railroad northern rail extension routes and ancillary facilities, Alaska” (NLUR technical report #278b-c, Northern Land Use Research Inc., 2007).
- 36.H. T. Allen, Report of an Expedition to the Copper, Tananá, and Kóyukuk Rivers, in the Territory of Alaska, in the Year 1885 (Government Printing Office, 1887).
- 37.C. Ballew, A. Ross, R. S. Wells, V. Hiratsuka, Final Report on the Alaska Traditional Diet Survey (Alaska Native Health Board, 2004).
- 38.AlQahtani S. J., Hector M. P., Liversidge H. M., Accuracy of dental age estimation charts: Schour and Massler, Ubelaker and the London Atlas. Am. J. Phys. Anthropol. 154, 70–78 (2014). [DOI] [PubMed] [Google Scholar]
- 39.Pestle W. J., Crowley B. E., Weirauch M. T., Quantifying inter-laboratory variability in stable isotope analysis of ancient skeletal remains. PLOS ONE 9, e102844 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barta J. L., Monroe C., Kemp B. M., Further evaluation of the efficacy of contamination removal from bone surfaces. Forensic Sci. Int. 231, 340–348 (2013). [DOI] [PubMed] [Google Scholar]
- 41.Kemp B. M., Monroe C., Judd K. G., Reams E., Grier C., Evaluation of methods that subdue the effects of polymerase chain reaction inhibitors in the study of ancient and degraded DNA. J. Archaeol. Sci. 42, 373–380 (2014). [Google Scholar]
- 42.Kemp B. M., Judd K., Monroe C., Eerkens J. W., Hilldorfer L., Cordray C., Schad R., Reams E., Ortman S. G., Kohler T. A., Prehistoric mitochondrial DNA of domesticate animals supports a 13th century exodus from the northern US southwest. PLOS ONE 12, e0178882 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johnson B. M., Kemp B. M., Rescue PCR: Reagent-rich PCR recipe improves amplification of degraded DNA extracts. J. Archaeol. Sci. Rep. 11, 683–694 (2017). [Google Scholar]
- 44.Palmer E., Tushingham S., Kemp B. M., Human use of small forage fish: Improved ancient DNA species identification techniques reveal long term record of sustainable mass harvesting of smelt fishery in the northeast Pacific Rim. J. Archaeol. Sci. 99, 143–152 (2018). [Google Scholar]
- 45.Jordan L. G., Steele C. A., Thorgaard G. H., Universal mtDNA primers for species identification of degraded bony fish samples. Mol. Ecol. Resour. 10, 225–228 (2010). [DOI] [PubMed] [Google Scholar]
- 46.Buckley M., Harvey V. L., Chamberlain A. T., Species identification and decay assessment of Late Pleistocene fragmentary vertebrate remains from Pin Hole Cave (Creswell Crags, UK) using collagen fingerprinting. Boreas 46, 402–411 (2017). [Google Scholar]
- 47.Coplen T. B., Guidelines and recommended terms for expression of stable-isotope-ratio and gas-ratio measurement results. Rapid Commun. Mass Spectrom. 25, 2538–2560 (2011). [DOI] [PubMed] [Google Scholar]
- 48.Longin R., New method of collagen extraction for radiocarbon dating. Nature 230, 241–242 (1971). [DOI] [PubMed] [Google Scholar]
- 49.Szpak P., Metcalfe J. Z., Macdonald R. A., Best practices for calibrating and reporting stable isotope measurements in archaeology. J. Archaeol. Sci. Rep. 13, 609–616 (2017). [Google Scholar]
- 50.van Klinken G. J., Bone collagen quality indicators for palaeodietary and radiocarbon measurements. J. Archaeol. Sci. 26, 687–695 (1999). [Google Scholar]
- 51.Ambrose S. H., Preparation and characterization of bone and tooth collagen for isotopic analysis. J. Archaeol. Sci. 17, 431–451 (1990). [Google Scholar]
- 52.DeNiro M. J., Postmortem preservation and alteration of in vivo bone collagen isotope ratios in relation to palaeodietary reconstruction. Nature 317, 806–809 (1985). [Google Scholar]
- 53.Richards M. P., Pettitt P. B., Trinkaus E., Smith F. H., Paunović M., Karavanić I., Neanderthal diet at Vindija and Neanderthal predation: The evidence from stable isotopes. Proc. Natl. Acad. Sci. U.S.A. 97, 7663–7666 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guthrie D. R., New carbon dates link climatic change with human colonization and Pleistocene extinctions. Nature 441, 207–209 (2006). [DOI] [PubMed] [Google Scholar]
- 55.Potter B. A., Reuther J. D., Newbold B. A., Yoder D. T., High resolution radiocarbon dating at the Gerstle River Site, central Alaska. Am. Antiq. 77, 71–98 (2012). [Google Scholar]
- 56.Lanoë F. B., Reuther J. D., Holmes C. E., Hodgins G. W. L., Human paleoecological integration in subarctic eastern Beringia. Quat. Sci. Rev. 175, 85–96 (2017). [Google Scholar]
- 57.Leonard J. A., Vila C., Fox-Dobbs K., Koch P. L., Wayne R. K., Van Valkenburgh B., Megafaunal extinctions and the disappearance of a specialized wolf ecomorph. Curr. Biol. 17, 1146–1150 (2007). [DOI] [PubMed] [Google Scholar]
- 58.Szpak P., Orchard T. J., Gröcke D. R., A Late Holocene vertebrate food web from southern Haida Gwaii (Queen Charlotte Islands, British Columbia). J. Archaeol. Sci. 36, 2734–2741 (2009). [Google Scholar]
- 59.Johnson S. P., Schindler D. E., Trophic ecology of Pacific salmon (Oncorhynchus spp.) in the ocean: A synthesis of stable isotope research. Ecol. Res. 24, 855–863 (2009). [Google Scholar]
- 60.Misarti N., Finney B., Maschner H., Wooller M. J., Changes in northeast Pacific marine ecosystems over the last 4500 years: Evidence from stable isotope analysis of bone collagen from archeological middens. The Holocene 19, 1139–1151 (2009). [Google Scholar]
- 61.Britton K., McManus-Fry E., Nehlich O., Richards M., Ledger P. M., Knecht R., Stable carbon, nitrogen and sulphur isotope analysis of permafrost preserved human hair from rescue excavations (2009, 2010) at the precontact site of Nunalleq, Alaska. J. Archaeol. Sci. Rep. 17, 950–963 (2018). [Google Scholar]
- 62.Byers D. A., Yesner D. R., Broughton J. M., Coltrain J. B., Stable isotope chemistry, population histories and late prehistoric subsistence change in the Aleutian Islands. J. Archaeol. Sci. 38, 183–196 (2011). [Google Scholar]
- 63.Corr L. T., Berstan R., Evershed R. P., Optimisation of derivatisation procedures for the determination of δ13C values of amino acids by gas chromatography/combustion/isotope ratio mass spectrometry. Rapid Commun. Mass Spectrom. 21, 3759–3771 (2007). [DOI] [PubMed] [Google Scholar]
- 64.R Core Team, R: A language and environment for statistical computing (R Foundation for Statistical Computing, 2017); www.R-project.org/.
- 65.Brown C. J., Brett M. T., Adame M. F., Stewart-Koster B., Bunn S. E., Quantifying learning in biotracer studies. Oecologia 187, 597–608 (2018). [DOI] [PubMed] [Google Scholar]
- 66.Bocherens H., Drucker D. G., Germonpré M., Lázničková-Galetová M., Naito Y. I., Wissing C., Brůžek J., Oliva M., Reconstruction of the Gravettian food-web at Předmostí I using multi-isotopic tracking (13C, 15N, 34S) of bone collagen. Quat. Int. 359-360, 211–228 (2015). [Google Scholar]
- 67.M. L. Fogel, N. Tuross, D. W. Owsley, Nitrogen isotope tracers of human lactation in modern and archaeological populations, in Annual report of the Director of the Geophysical Laboratory, 1988–1989 (Carnegie Institution, 1989), pp. 111–117. [Google Scholar]
- 68.de Luca A., Boisseau N., Tea I., Louvet I., Robins R. J., Forhan A., Charles M. A., Hankard R., δ15N and δ13C in hair from newborn infants and their mothers: A cohort study. Pediatr. Res. 71, 598–604 (2012). [DOI] [PubMed] [Google Scholar]
- 69.O’Connell T. C., Kneale C. J., Tasevska N., Kuhnle G. G. C., The diet-body offset in human nitrogen isotopic values: A controlled dietary study. Am. J. Phys. Anthropol. 149, 426–434 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vanderklift M. A., Ponsard S., Sources of variation in consumer-diet δ15N enrichment: A meta-analysis. Oecologia 136, 169–182 (2003). [DOI] [PubMed] [Google Scholar]
- 71.B. C. Stock, B. X. Semmens, MixSIAR GUI user manual v3.1 (2016); https://github.com/brianstock/MixSIAR/.
- 72.Choy K., Smith C. I., Fuller B. T., Richards M. P., Investigation of amino acid δ13C signatures in bone collagen to reconstruct human palaeodiets using liquid chromatography–isotope ratio mass spectrometry. Geochim. Cosmochim. Acta 74, 6093–6111 (2010). [Google Scholar]
- 73.Raghavan M., McCullagh J. S. O., Lynnerup N., Hedges R. E. M., Amino acid δ13C analysis of hair proteins and bone collagen using liquid chromatography/isotope ratio mass spectrometry: Paleodietary implications from intra-individual comparisons. Rapid Commun. Mass Spectrom. 24, 541–548 (2010). [DOI] [PubMed] [Google Scholar]
- 74.Webb E. C., Honch N. V., Dunn P. J. H., Linderholm A., Eriksson G., Lidén K., Evershed R. P., Compound-specific amino acid isotopic proxies for distinguishing between terrestrial and aquatic resource consumption. Archaeol. Anthropol. Sci. 10, 1–18 (2018). [Google Scholar]
- 75.Hobson K. A., Clark R. G., Assessing avian diets using stable isotopes I: Turnover of 13C in tissues. The Condor 94, 181–188 (1992). [Google Scholar]
- 76.Hobson K. A., Atwell L., Wassenaar L. I., Yerkes T., Estimating endogenous nutrient allocations to reproduction in Redhead Ducks: A dual isotope approach using δD and δ13C measurements of female and egg tissues. Funct. Ecol. 18, 737–745 (2004). [Google Scholar]
- 77.D. R. Yesner, Faunal extinction, hunter-gatherer foraging strategies, and subsistence diversity among eastern Beringian Paleoindians, in Foragers of the Terminal Pleistocene in North America, R. B. Walker, B. N. Driskell, Eds. (University of Nebraska Press, 2007), pp. 15–31. [Google Scholar]
- 78.Yesner D. R., Moose hunters of the boreal forest? A re-examination of subsistence patterns in the western subarctic. Arctic 42, 97–108 (1989). [Google Scholar]
- 79.Lanoë F. B., Reuther J. D., Holmes C. E., Potter B. A., Small mammals and paleovironmental context of the terminal pleistocene and early holocene human occupation of central Alaska. Geoarchaeology 35, 164–176 (2019). [Google Scholar]
- 80.R. A. McKennan, The Upper Tanana Indians. Yale University Publications in Anthropology, No. 55 (Yale University, 1959).
- 81.Richards M. P., Hedges R. E. M., Variations in bone collagen δ13C and δ15N values of fauna from Northwest Europe over the last 40 000 years. Palaeogeogr. Palaeoclimatol. Palaeoecol. 193, 261–267 (2003). [Google Scholar]
- 82.Hedges R. E. M., Stevens R. E., Richards M. P., Bone as a stable isotope archive for local climatic information. Quat. Sci. Rev. 23, 959–965 (2004). [Google Scholar]
- 83.Mann D. H., Groves P., Kunz M. L., Reanier R. E., Gaglioti B. V., Ice-age megafauna in Arctic Alaska: Extinction, invasion, survival. Quat. Sci. Rev. 70, 91–108 (2013). [Google Scholar]
- 84.Espinasse B., Hunt B. P. V., Coll Y. D., Pakhomov E. A., Investigating high seas foraging conditions for salmon in the North Pacific: Insights from a 100-year scale archive for Rivers Inlet sockeye salmon. Can. J. Fish. Aquat. Sci. 76, 918–927 (2018). [Google Scholar]
- 85.Guiry E., Royle T. C. A., Matson R. G., Ward H., Weir T., Waber N., Brown T. J., Hunt B. P. V., Price M. H. H., Finney B. P., Kaeriyama M., Qin Y., Yang D. Y., Szpak P., Differentiating salmonid migratory ecotypes through stable isotope analysis of collagen: Archaeological and ecological applications. PLOS ONE 15, e0232180 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Potter B. A., Irish J. D., Reuther J. D., Gelvin-Reymiller C., Holliday V. T., A terminal Pleistocene child cremation and residential structure from eastern Beringia. Science 331, 1058–1062 (2011). [DOI] [PubMed] [Google Scholar]
- 87.C. R. Holloway, Paleoethnobotany in interior Alaska, thesis, University of Alaska Fairbanks (2016). [Google Scholar]
- 88.Sheriff M. J., Boonstra R., Palme R., Buck C. L., Barnes B. M., Coping with differences in snow cover: The impact on the condition, physiology and fitness of an arctic hibernator. Conserv. Physiol. 5, cox065 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Buck C. L., Barnes B. M., Annual cycle of body composition and hibernation in free-living arctic ground squirrels. J. Mammal. 80, 430–442 (1999). [Google Scholar]
- 90.Sheriff M. J., Buck C. L., Barnes B. M., Autumn conditions as a driver of spring phenology in a free-living arctic mammal. Clim. Change Resp. 2, 4 (2015). [Google Scholar]
- 91.L. Halpin, Living Off the Land: Contemporary Subsistence in Tetlin, Alaska (Alaska Department of Fish and Game, 1987). [Google Scholar]
- 92.Trotter M., Hixon B. B., Sequential changes in weight, density, and percentage ash weight of human skeletons from an early fetal period through old age. Anat. Rec. 179, 1–18 (1974). [DOI] [PubMed] [Google Scholar]
- 93.A. Rohatgi, WebPlotDigitizer Version 4.1 (2018); https://automeris.io/WebPlotDigitizer.
- 94.Nakano K., Iwamatsu T., Wang C. M., Tarasima M., Nakayama T., Sasaki K., Tachikawa E., Noda N., Mizoguchi E., Osawa M., High bone turnover of type I collagen depends on fetal growth. Bone 38, 249–256 (2006). [DOI] [PubMed] [Google Scholar]
- 95.Leggett R. W., Eckerman K. F., Williams L. R., Strontium-90 in bone: A case study in age-dependent dosimetric modeling. Health Phys. 43, 307–322 (1982). [DOI] [PubMed] [Google Scholar]
- 96.Rauch F., Schoenau E., Changes in bone density during childhood and adolescence: An approach based on bone’s biological organization. J. Bone Miner. Res. 16, 597–604 (2001). [DOI] [PubMed] [Google Scholar]
- 97.J. W. Wood, Dynamics of Human Reproduction: Biology, Biometry, Demography (Aldine De Gruyter, 1994). [Google Scholar]
- 98.Bowers P. M., Reuther J. D., AMS re-dating of the Carlo Creek Site, Nenana Valley, central Alaska. Curr. Res. Pleist. 25, 58–61 (2008). [Google Scholar]
- 99.B. A. Potter, J. D. Reuther, Site chronology, in Archaeological Investigations at Delta River Overlook, B. A. Potter, Ed. (Archaeology GIS Laboratory, University of Alaska Fairbanks, 2018), pp. 58–70. [Google Scholar]
- 100.Potter B. A., Radiocarbon chronology of central Alaska: Technological continuity and economic change. Radiocarbon 50, 181–204 (2008). [Google Scholar]
- 101.Wooller M. J., Kurek J., Gaglioti B. V., Cwynar L. C., Bigelow N., Reuther J. D., Gelvin-Reymiller C., Smol J. P., An ~11,200 year paleolimnological perspective for emerging archaeological findings at Quartz Lake, Alaska. J. Paleolimnol. 48, 83–99 (2012). [Google Scholar]
- 102.B. A. Potter, Site structure and organization in Central Alaska: Archaeological investigations at Gerstle River, thesis, University of Alaska Fairbanks (2005). [Google Scholar]
- 103.Shapiro B., Drummond A. J., Rambaut A., Wilson M. C., Matheus P. E., Sher A. V., Pybus O. G., Gilbert M. T. P., Barnes I., Binladen J., Willerslev E., Hansen A. J., Baryshnikov G. F., Burns J. A., Davydov S., Driver J. C., Froese D. G., Harington C. R., Keddie G., Kosintsev P., Kunz M. L., Martin L. D., Stephenson R. O., Storer J., Tedford R., Zimov S., Cooper A., Rise and fall of the Beringian steppe bison. Science 306, 1561–1565 (2004). [DOI] [PubMed] [Google Scholar]
- 104.Meiri M., Lister A. M., Collins M. J., Tuross N., Goebel T., Blockley S., Zazula G. D., van Doorn N., Dale Guthrie R., Boeskorov G. G., Baryshnikov G. F., Sher A., Barnes I., Faunal record identifies Bering isthmus conditions as constraint to end-Pleistocene migration to the New World. Proc. Biol. Sci. 281, 20132167 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cook J. P., Historic archaeology and ethnohistory at Healy Lake, Alaska. Arctic 42, 109–118 (1989). [Google Scholar]
- 106.Alter S. E., Newsome S. D., Palumbi S. R., Pre-whaling genetic diversity and population ecology in eastern Pacific gray whales: Insights from ancient DNA and stable isotopes. PLOS ONE 7, e35039 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.C. R. Harington, Ed., Annotated Bibliography of Quaternary Vertebrates of Northern North America (University of Toronto Press, 2003). [Google Scholar]
- 108.Harrison R. G., Katzenberg M. A., Paleodiet studies using stable carbon isotopes from bone apatite and collagen: Examples from Southern Ontario and San Nicolas Island, California. J. Anthropol. Archaeol. 22, 227–244 (2003). [Google Scholar]
- 109.E. J. Bartelink, Resource intensification in pre-contact central California: A bioarchaeological perspective on diet and health patterns among hunter-gatherers from the lower Sacramento Valley and San Francisco Bay, thesis, Texas A&M University (2006). [Google Scholar]
- 110.J. Tackney, J. Coltrain, J. Raff, D. O’Rourke, Ancient DNA and stable isotopes: Windows on Arctic prehistory, in The Oxford Handbook of the Prehistoric Arctic, T. M. Friesen, O. K. Mason, Eds. (Oxford Univ. Press, 2016), pp. 51–79. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/36/eabc1968/DC1


