Abstract
CD97, a member of the adhesion G-protein coupled receptor family, is normally expressed on leukocytes and smooth muscles. CD97 is also expressed in a variety of solid cancers, particularly those with aggressive metastatic phenotypes. Here we characterize the clinical significance of CD97 in acute myeloid leukemia (AML). We analyzed 173 patients from the TCGA AML data set and found that CD97 was higher in cytogenetically normal patients compared with cytogenetically abnormal patients (p = 0.023). High CD97 was also associated with NPM1 mutations (p = 0.0033). Patients with high CD97 expression had shorter overall (median: 7.35 months vs. 24.1 months, p = 0.0015) and disease-free (median DFS: 8.2 months vs. 18.2 months, p = 0.017) survival. Importantly, we identified pathways involved in the leukemia stem cell interaction with the bone marrow niche, such as integrin, CXCR4, and interleukin-8, among the most upregulated signaling pathways in patients with high CD97 expression. Our results suggest that high CD97 expression is associated with poor clinical outcome and indicate a need for future functional and mechanistic studies to investigate the role of CD97 in AML.
Acute myeloid leukemia (AML) is a heterogeneous, hematologic malignancy characterized by clonal proliferation of myeloid precursors [1]. It is the most common acute leukemia in adults. Overall survival of patients with AML remains dismal (<50% for younger patients and <10% for older patients) because of the high rate of relapse [2]. Cytogenetic and molecular genetic alterations provide significant prognostic information for determining the response to chemotherapy and survival outcome.
Adhesion G protein-coupled receptors (aGPCRs) were recently reported to be widely deregulated in AML [3,4]. Among aGPCRs, CD97 is particularly interesting; it is expressed predominantly in hematopoietic cells [5] and has been recently identified as a marker for leukemia stem cells [6]. Several ligands have been reported to bind CD97: CD55, which is a negative regulator of the complement cascade [7]; chondroitin sulfate, a component of the extracellular matrix (ECM) [8]; and the integrin α5β1 [8]. The association with integrins is relevant to AML, as integrin and ILK signaling pathways play crucial roles in leukemia stem cell (LSC) survival and interaction with the bone marrow (BM) niche [9,10].
In solid cancers, CD97 association with integrin was found to regulate invasion, migration, and angiogenesis [11]. In undifferentiated anaplastic thyroid carcinoma, CD97 overexpression was associated with metastatic lesions [12,13]. Additionally, high CD97 expression was also reported to be associated with increased lymph vessel invasion and poor clinical outcome in colorectal carcinomas [14]. In pancreatic cancer, CD97 expression significantly correlated with tumor aggressiveness [15]. More recently, CD97 was also shown to be the first member of the EGF-TM7 family to be directly involved in cell signaling. Ward et al. [16] demonstrated functional synergy and heterodimerization between CD97 and lysophosphatidic acid receptor 1 (LPAR1), resulting in increased RHO-GTP levels in a Gα12/13-dependent signaling pathway in thyroid and prostate cancers, as well as correlating with increased tumor invasion.
The role of CD97 in invasion and signaling suggests that it is an important therapeutic target. In this study, we characterized CD97 expression in patients with AML using publicly available data sets to determine its association with patient’s clinical and molecular characteristics and clinical outcome. We also identified signaling pathways that are deregulated in patients with high CD97.
Methods
Patient data sets
Available data on 173 of 200 patients with previously untreated AML with complete clinical and RNA expression data were studied from the TCGA data set [17]. This data set includes patients with previously untreated AML, all of whom had been diagnosed and received treatment according to the National Comprehensive Cancer Network (NCCN) guidelines between November 2001 and March 2010. The risk group stratification of the patients was done according to NCCN guidelines. The patients were assigned subtype classifications according to the French–American–British (FAB) classifications. Patients included in the study were assessed for the most frequently found somatic mutations in AML, such as FLT3, NPM1, IDH1/2, and TET2. Patient’s clinical, gene expression (Z score), mutation, gene methylation, and survival data were downloaded from the TCGA database available at cBioPortal on September 9, 2018. Metzeler-163 [18], Metzeler-79 [18], and Bullinger [19] data sets were downloaded from Oncomine.
Ethics approval and consent to participate were not necessary, as we studied public de-identified data.
Gene expression analyses
We downloaded publicly available and analyzed RNA sequencing data of 173 patients with acute myeloid leukemia and complete clinical and molecular data [20,21]. We dichotomized patients into two groups based on Z scores ≥1 (high) and <1 (low). We also validated the associations using a Z score of 2 as the cutoff as well as the median of the mRNA log2 transformed data.
Statistical analyses
Overall survival (OS) was defined as the time between diagnosis and death from any reason, and disease-free survival (DFS) was defined as the time between diagnosis and removal from study because of relapse or death (two of the patients with missing information regarding their DFS were not included in the DFS analysis). We generated Kaplan–Meier survival curves for comparison of OS and DFS between patients with Z ≥1 and Z <1 CD97 expression. Additionally, Kaplan–Meier survival curves were generated for patients with Z ≥1 and Z <1 CD97 expression after stratification by age, cytogenetic status, transplant status, and NPM1 mutation status. To determine the association between CD97 expression and patients’ clinical and molecular characteristics, we employed T test and Fisher’s exact test for continuous and categorical variables, respectively, using the GraphPad Prism software package (Version 5.0, GraphPad Software Inc., La Jolla, CA). We used Stata SE 12.0 to perform a Cox proportional hazards model to conduct survival analysis of the effect of CD97 expression on OS and DFS after adjusting for other clinical/molecular factors. A statistical cutoff of p < 0.05 was used for inclusion of variables into the multivariate survival analysis from univariate analysis.
Ingenuity pathway analysis (IPA)
Patients were grouped as CD97 high (Z >2) and CD97 low (Z <–1). Pearson correlation scores of CD97 versus all the available gene expression data in TCGA were calculated, and upregulated genes with a score >0.5 and downregulated genes with a score <–0.5 were exported into IPA. Signaling pathways were evaluated for CD97 high and CD97 low patients.
Results
CD97 expression in AML
CD97 mRNA (RNA Seq V2 RSEM) expression data were downloaded from the TCGA data set on cbioportal [20,21]. Histograms representing the distribution of CD97 mRNA log2-transformed data and CD97 Z-scores are provided in Supplementary Figure E1A and B (online only, available at www.exphem.org). The scatterplot of CD97 log2-transformed mRNA expression against CD97 Z score is shown in Figure E1C (Pearson’s r = 0.944, p ≤ 0.001). CD97 mRNA levels were compared among patients classified according to the FAB. Although patients in the M5 subgroup had relatively higher CD97 expression, there was no significant difference in CD97 expression among the subgroups (Supplementary Figure E1D). When patients were classified based on cytogenetic status into cytogenetically normal AML (CN-AML) (N = 80) and cytogenetically abnormal AML (CA-AML) (N = 90), we found that patients with CN-AML had significantly higher CD97 expression compared with patients with CA-AML (1.31-fold, p = 0.023) (Supplementary Figure E1E).
We also analyzed CD97 expression according to the NCCN AML classification in which patients are grouped based on their molecular and cytogenetic risk status into favorable, intermediate, and poor risk groups and found no significant difference between these groups (Supplementary Figure E1F).
Additionally, we used the Vizome data analysis tool, which contains data from the BEAT AML cohort [22], and examined the level of CD97 expression in relapsed (N = 46) versus de novo AML (N = 288) samples and found no significant difference between the two groups (Supplementary Figure E1G).
We dichotomized the patients in the TCGA data set based on their CD97 mRNA expression Z score (RNA Seq V2 RSEM) into CD97 high (Z score ≥1, N = 26) and CD97 low (Z score <1, N = 147). Patients with high CD97 expression had significantly higher white blood cell counts (WBCs) (median: 56.1 vs. 13.1 [× 109/L], p = 0.004) and higher percentages of bone marrow blasts (median: 83% vs. 71%, p = 0.019) (Table 1).
Table 1.
Clinical characteristics of the AML cohort in the TCGA data set with respect to CD97 expression
| Total | CD97 low (Z <1) | CD97 high (Z ≥1) |
p Value |
||
|---|---|---|---|---|---|
| T test | Fisher exact test | ||||
| Sex, No. (%) | 1 | ||||
| Female | 81 (46.8) | 69 (85.2) | 12 (14.8) | ||
| Male | 92 (53.1) | 78 (84.8) | 14 (15.2) | ||
| Age, y (range) | 0.385 | ||||
| Median | 58 | 57 | 62.5 | ||
| Mean | 55.2 ± 1.22 | 54.7 ± 1.32 | 57.7 ± 3.38 | ||
| WBC count x109/L | 0.004 | ||||
| Median | 17 | 13.1 | 56.1 | ||
| Mean | 36.63 ± 3.50 | 32.44 ± 3.81 | 60.30 ± 7.48 | ||
| PB blasts, % | 0.25 | ||||
| Median | 39 | 34.5 | 49 | ||
| Mean | 39.59 ± 2.44 | 38.38 ± 2.68 | 46.2 ± 5.85 | ||
| BM blasts, % | 0.019 | ||||
| Median | 72 | 71 | 83 | ||
| Mean | 69.08 ± 1.45 | 67.65 ± 1.59 | 77.15 ± 3.17 | ||
| NCCN subtype, No. | vs. favorable | ||||
| Favorable | 33 | 29 | 4 | ||
| Intermediate | 92 | 76 | 16 | 0.5 | |
| Poor | 45 | 40 | 5 | 0.6 | |
| FAB subtype, No. | vs. M5 | ||||
| M0 | 16 | 15 | 1 | 0.042 | |
| M1 | 44 | 39 | 5 | 0.028 | |
| M2 | 38 | 34 | 4 | 0.026 | |
| M3 | 16 | 12 | 4 | 0.476 | |
| M4 | 34 | 29 | 5 | 0.081 | |
| M5 | 18 | 11 | 7 | ||
| M6 | 2 | 2 | |||
| M7 | 3 | 3 | |||
High CD97 expression is associated with NPM1 mutations
To understand potential molecular genetic aberrations that may lead to or be associated with high CD97 in AML, we analyzed its expression with respect to the mutational status of patients with AML. CD97 was significantly higher in patients with NPM1 mutation (n = 48) than in patients with NPM1 wild type (n = 125, 1.56-fold, p < 0.0001; Figure 1A). CD97 was also significantly higher in patients with FLT3 mutations (ITD and point mutations) (n = 49) than in patients carrying FLT3 wild type (n = 124, 1.4-fold, p = 0.0008; Figure 1B). Additionally, CD97 was significantly lower in the patients with RUNX1 mutations (n = 17) than in patients with the wild-type RUNX1 (n = 156) (42.1% lower, p = 0.0002; Figure 1C). No significant association was observed between CD97 expression and mutations in DNMT3A, IDH1, IDH2, TET2, TP53, CEBPA, WT1, and KIT.
Figure 1.
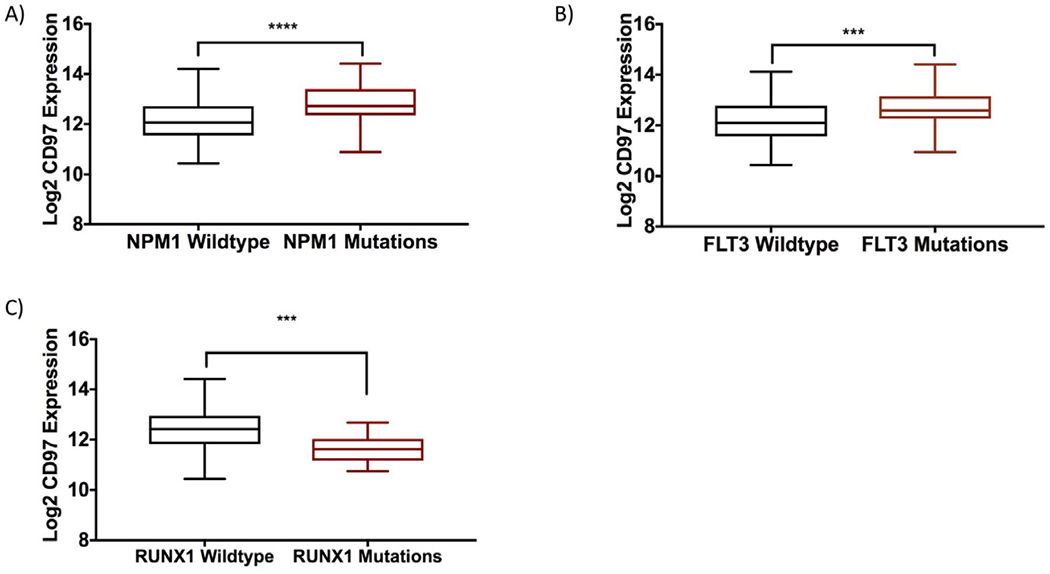
Association of CD97 mRNA expression with patient mutational status. Relative CD97 log2 mRNA expression in (A) patients with NPM1 mutations versus wild type; (B) patients with FLT3 mutations versus wild type; and (C) patients with RUNX1 mutations versus wild type. ***p < 0.001. ****p < 0.0001.
When we dichotomized patients according to CD97 Z scores, we found a higher frequency of NPM1 mutations in high CD97 (Z ≥1) patients than in low CD97 (Z < 1) patients (52% vs. 30.6%, Fisher exact p = 0.0065). No association was found between CD97 upregulation and mutations in other genes in the dichotomized analysis (Table 2).
Table 2.
Association of CD97 expression with patient mutational status in the TCGA data set
| Total | CD97 low (Z <1) | CD97 high (Z ≥1) |
p Value |
||
|---|---|---|---|---|---|
| T test | Fischer exact test | ||||
| FLT3, No. (%) | 0.1005 | ||||
| Present | 49 (28.3) | 38 (77.5) | 11 (22.4) | 0.0008 | |
| Absent | 124 (71.67) | 109(87.9) | 15 (12.1) | ||
| IDH1, No. (%) | 0.09 | 0.4718 | |||
| Mutated | 16 (9.2) | 15 (93.7) | 1 (6.2) | ||
| Wild type | 157 (92.3) | 132 (84.0) | 25 (15.9) | ||
| IDH2, No. (%) | 0.77 | 0.7238 | |||
| Mutated | 17 (9.8) | 14 (82.3) | 3 (17.6) | ||
| Wild type | 156 (90.1) | 133 (85.2) | 23 (14.7) | ||
| RUNX1, No. (%) | 0.0002 | 0.070 | |||
| Mutated | 17 (9.8) | 17 (100) | 0 (0) | ||
| Wild type | 156 (90.1) | 130 (83.3) | 26 (16.6) | ||
| TET2, No. (%) | 0.10 | 0.1311 | |||
| Mutated | 15 (8.6) | 15 (100) | 0 (0) | ||
| Wild type | 158 (91.3) | 132 (83.5) | 26 (16.4) | ||
| NRAS, No. (%) | 0.57 | 1 | |||
| Mutated | 12 (6.9) | 10 (83.3) | 2 (16.6) | ||
| Wild type | 161 (93.0) | 137 (85.0) | 24 (14.9) | ||
| CEBPA, No. (%) | 0.54 | 0.6944 | |||
| Mutated | 13 (7.5) | 12 (92.3) | 1 (7.6) | ||
| Wild type | 160 (92.5) | 135 (84.3) | 25 (15.6) | ||
| WT1, No. (%) | 0.42 | 0.3624 | |||
| Mutated | 10 (5.7) | 10 (100) | 0 (0) | ||
| Wild type | 163 (94.2) | 137 (84.0) | 26 (15.9) | ||
| DNMT3A, No. (%) | 0.122 | 0.1453 | |||
| Mutated | 45 (26.0) | 35 (77.7) | 10 (22.2) | ||
| Wild type | 128 (73.9) | 112 (87.5) | 16 (12.5) | ||
| NPM1, No. (%) | <0.0001 | 0.0033 | |||
| Mutated | 48 (27.7) | 34 (70.8) | 14 (29.1) | ||
| Wild type | 125 (72.3) | 113 (90.4) | 12 (9.6) | ||
| TP53 | 0.34 | ||||
| Mutated | 14 (8.09) | 13 (92.8) | 1 (7.1) | 0.6970 | |
| Wild type | 159 (91.9) | 134 (84.2) | 25 (15.7) | ||
High CD97 expression is associated with poor clinical outcome
The overall survival of patients in the CD97 high group (Z ≥1) was significantly shorter than that of patients in the CD97 low group (median OS: 7.35 vs. 24.1 months, p = 0.0015; Figure 2A). Patients with high CD97 expression had significantly shorter DFS than patients with low CD97 (median DFS: 8.2 months vs. 18.2 months, p = 0.017; Figure 2B). To further validate the association between high CD97 and poor clinical outcome, we stratified patients into Z ≥ 2 and Z < 2 for survival analysis. Patients with high CD97 expression had significantly shorter OS and DFS than patients with low CD97 (OS—median: 7.5 months vs. 20.5 months, p = 0.04; DFS—median: 7.3 months vs. 17 months, p = 0.03; Supplementary Figure E2A and B, online only, available at www.exphem.org). Because patients with APL or t(15, 17) are treated differently (with ATRA) and have better clinical outcome than the rest of patients with AML, we excluded these patients from the survival analysis. Yet, the OS of the CD97 high group (Z ≥1) remained significantly shorter than that of the CD97 low patients (median: 7.35 months vs. 19 months, p= 0.003; Figure 2C). A similar trend was observed in DFS (median: 8.2 months vs. 16.1 months, p= 0.08; Figure 2D). Among patients with APL, the OS of patients in the CD97 high group (Z ≥1) was significantly shorter than that of the CD97 low patients (p= 0.03; Supplementary Figure E3A, online only, available at www.exphem.org). APL patients with high CD97 expression had significantly shorter DFS than patients with low CD97 (p = 0.003; Supplementary Figure E3B).
Figure 2.
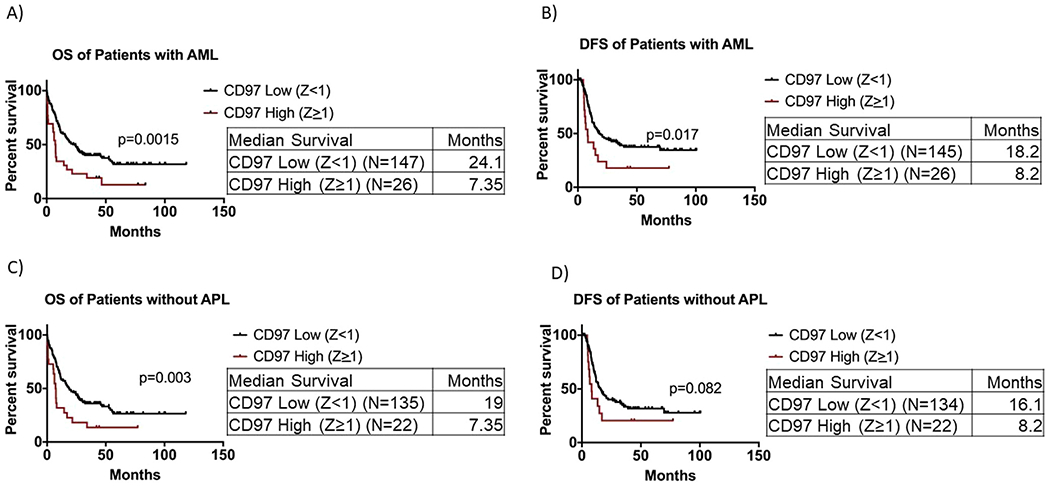
Survival analysis of AML patients with respect to CD97 expression. (A) Overall survival of 173 AML patients with CD97 Z score ≥1 and CD97 Z score <1. (B) Disease-free survival of 171 AML patients with CD97 Z score ≥1 and CD97 Z score <1. (C) Overall survival of patients without APL with CD97 Z score ≥1 and CD97 Z score <1. (D) Disease-free survival of patients without APL with CD97 Z score ≥1 and CD97 Z score <1.
When patients were stratified according to their cytogenetic status, we found that among CA-AML patients, those with CD97 high (Z ≥1) had shorter overall survival than those with CD97 low; however, this was not statistically significant (median OS: 7.2 months vs. 19.2 months, p = 0.159; Figure 3A). Yet the difference in OS between the two groups was statistically significant among the CN-AML patients (median OS: 7.5 months vs. 24.6 months, p = 0.0045; Figure 3B). Also, there was a significant decrease in the DFS of CD97 high patients (Z ≥1) compared with CD97 low patients (median: 10.8 months vs. 39 months, p = 0.043; Figure 3C) in CA-AML but not in CN-AML patients (median: 8.2 months vs. 12.1 months, p = 0.25; Figure 3D).
Figure 3.
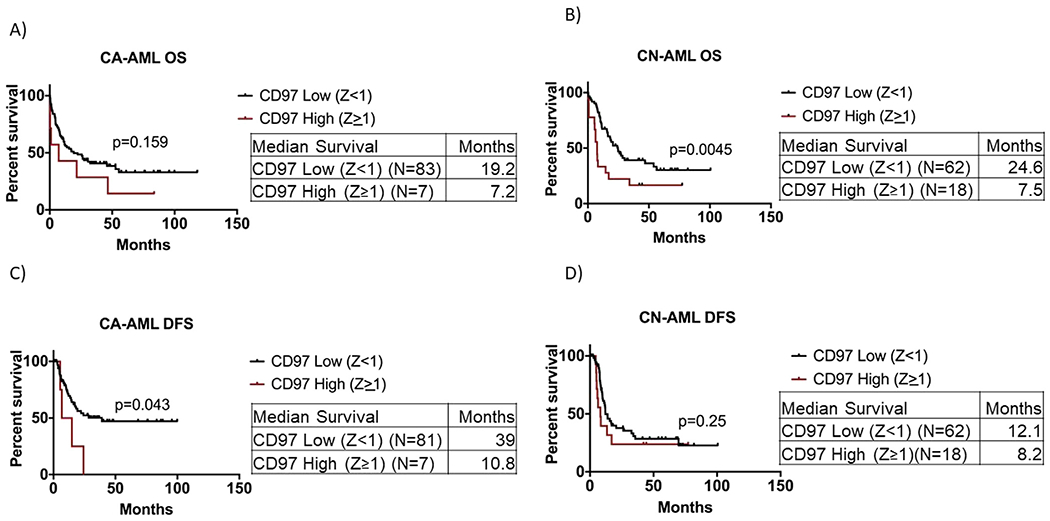
Survival analysis of AML patients with respect to CD97 expression after stratification based on cytogenetic status. (A) Overall survival of AML patients with CD97 high (Z score ≥1) versus CD97 low (Z score <1) in cytogenetically abnormal (CA-AML) patients. (B) Overall survival of AML patients with CD97 high (Z score ≥1) versus CD97 low (Z score <1) in cytogenetically normal (CN-AML) patients. (C) Disease-free survival of AML patients with CD97 high (Z score >1) versus CD97 low (Z score <1) in CA-AML patients. (D) Disease-free survival of AML patients with CD97 high (Z score ≥1) versus CD97 low (Z score <1) in CN-AML patients.
In multivariate survival analysis, CD97 high expression (Z ≥1) was associated with shorter overall survival when adjusted for age, cytogenetic risk, and transplant status (HR = 1.96, 95% CI: 1.19–3.24, p = 0.009; Table 3). A similar trend was observed when patients with t(15,17) were excluded from the analysis: CD97 expression (Z ≥1) was associated with shorter overall survival (HR = 1.56, 95% Cl: 0.97–2.83, p = 0.066).
Table 3.
Cox proportional hazards model for overall survival in patients with AML comparing patients with high (Z ≥1) CD97 expression and patients with low (Z <1) CD97 expression.
| Variable | Hazard ratio | 95% Confidence interval | p Value | |
|---|---|---|---|---|
| Age | 1.03 | 1.01 | 1.04 | 0.002 |
| Molecular risk | 0.002 | |||
| Intermediate | 3.03 | 1.51 | 6.07 | 0.002 |
| Poor | 6.57 | 3.10 | 13.9 | <0.001 |
| Transplant status (Y/N) | 0.47 | 0.29 | 0.74 | 0.001 |
| CD97 (Z ≥1) | 1.96 | 1.19 | 3.24 | 0.009 |
When patients were stratified according to whether they received a transplant or not, we found that only in patients who did not receive a transplant was CD97 high expression (Z ≥1) associated with significantly shorter OS and DFS (median OS: 5.5 months vs. 11.4 months, p = 0.0078; median DFS: 5.9 months vs. 32.7 months, p = 0.007) (Figure 4A, C).
Figure 4.
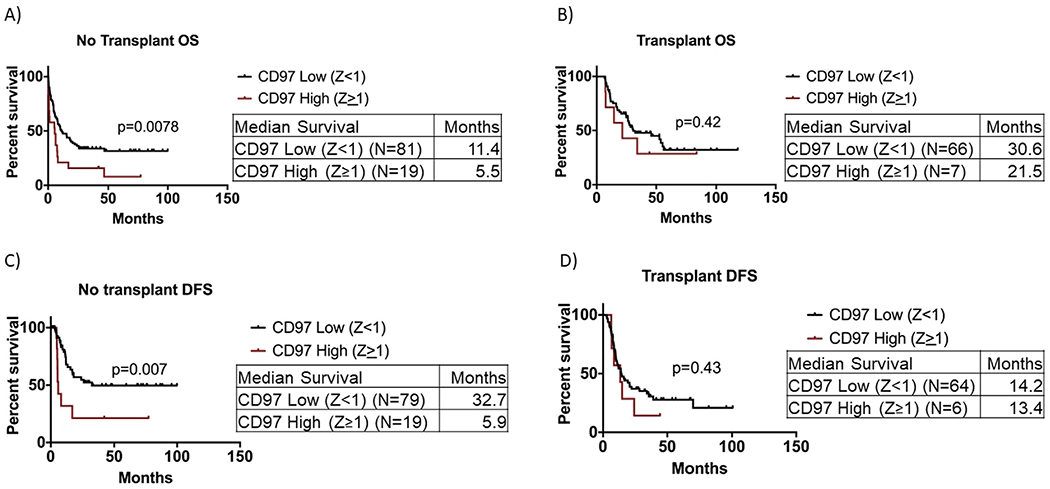
Survival analysis of AML patients with respect to CD97 expression after stratification based on patient transplant status. (A) Overall survival of AML patients with CD97 high (Z score ≥1) versus CD97 low (Z score <1) in patients who did not receive a transplant. (B) Overall survival of AML patients with CD97 high (Z score ≥1) versus CD97 low (Z score <1) in patients who received a transplant. (C) Disease-free survival of AML patients with CD97 high (Z score ≥1) versus CD97 low (Z score <1) in patients who did not receive a transplant. (D) Disease-free survival of AML patients with CD97 high (Z score ≥1) versus CD97 low (Z score <1) in patients who received a transplant.
No significant difference was observed in OS (Figure 4B) and DFS (Figure 4D) between the CD97 high and low in patients who received a transplant.
We also analyzed the association of CD97 expression with clinical outcome in additional data sets: Metzeler-163 [18] (N = 163 CN-AML patients), Metzeler-79 [18] (N = 79 CN-AML patients) and Bullinger (N = 105 CN-AML and CA-AML patients) [19] data sets (histograms representing the distribution of CD97 mRNA expression are illustrated in Supplementary Figure E4A–C [online only, available at www.exphem.org]). Patients were dichotomized based on median CD97 expression into CD97 high (above median) and CD97 low (below median). In the Metzeler-79 data set (which includes CN-AML only), patients with CD97 high expression had shorter overall survival (median OS: 8.9 months vs. 42.01 months, p = 0.0027; Figure 5A). Similarly, in the Metzeler-163 data set (which includes CN-AML only), patients with high CD97 expression survived for shorter times than patients with low CD97 expression; however, the difference in survival was not statistically significant (median OS: 8.6 months vs. 14.2 months, p = 0.23; Figure 5B). On the contrary, in the Bullinger data set, which includes both CN and CA-AML, there was no significant association between CD97 expression and clinical outcome (median survival: 13.15 months vs. 18.74 months, p = 0.6; Figure 5C). This discrepancy between the Bullinger data set survival analysis and those of other data sets is possibly due to the difference in the microarray gene probe used to measure CD97. We also analyzed the TCGA data set using CD97 median expression to dichotomize patients into high and low CD97 expression groups. We found that patients with high CD97 expression had shorter overall survival (median OS: months 11 vs. 27.4 months, p = 0.008; Figure 5D).
Figure 5.
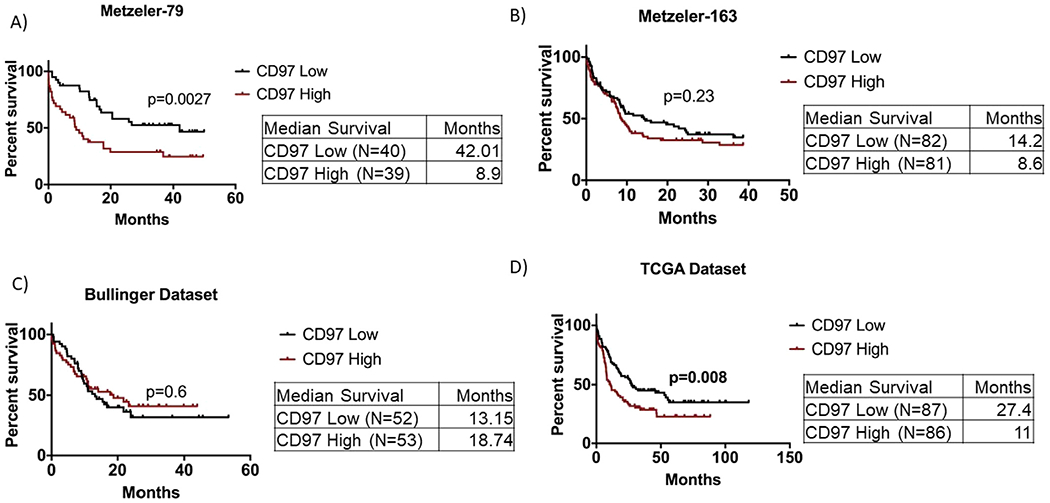
Survival analysis of AML patients with respect to CD97 expression in different data sets. Overall survival of patients with AML in four data sets, in which patients were dichotomized based on CD97 median mRNA expression into CD97 high and CD97 low according to the log2 median-centered expression. (A) Metzeler-79 data set. (B) Metzeler-163 data set. (C) Bullinger data set. (D) TCGA data set.
CD97 expression is associated with survival status of patients with NPM1 mutation
Because CD97 expression was associated with NPM1 mutation, which is a favorable prognostic marker in AML, we examined the association between CD97 expression and clinical outcome in patients with NPM1 mutation. We stratified patients according to NPM1 mutational status and performed survival analysis in each group. We found that in patients with wild-type NPM1, high CD97 expression was associated with a significantly shorter OS (median survival: 6.35 months vs. 22.3 months, p = 0.0081; Figure 6A) and DFS (median survival: 6.7 months vs. 22.2 months, p = 0.0015; Supplementary Figure E5A, online only, available at www.exphem.org). In patients with NPM1 mutation, a similar trend but not statistically significant association was observed between high CD97 expression and shorter OS (median survival: 7.5 months vs. 24.1 months, p = 0.11; Figure 6B) but not DFS (median survival: 10.8 months vs. 12.1 months, p = 0.75; Supplementary Figure E5B).
Figure 6.
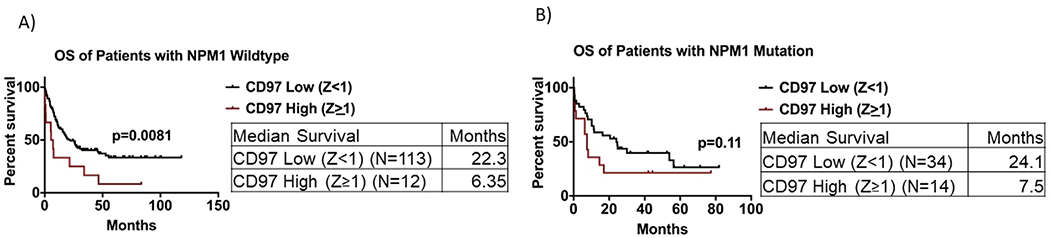
Survival analysis of AML patients with respect to CD97 expression after stratification based on NPM1 mutation status. (A) Overall survival of patients with CD97 high (Z score ≥1) versus CD97 low (Z score <1) among patients with NPM1 wild-type gene. (B) Overall survival of patients with CD97 high (Z score ≥1) versus CD97 low (Z score <1) among patients with NPM1 mutated gene.
As patients with NPM1 mutation have a better prognosis and patients with FLT3 mutation have poor prognosis, we further analyzed the association between CD97 expression and patients harboring FLT3 and NPM1 mutations to gain better insight. We identified 8 of 173 patients in the TCGA data that have both FLT3 mutation, NPM1 mutation, and CD97 Z ≥1. When we compared overall survival between patients with CD97 Z >1 (N = 8) and those with CD97 Z < 1 (N = 18) among those with FLT3 and NMP1 mutations, we found that patients with CD97 Z ≥1 had significantly worse survival compared with the Z ≤1 group (p = 0.04, median survival: 7.5 months vs. 24.6 months; Supplementary Figure E5C).
High CD97 expression is associated with poor survival in older patients
Because older patients with AML have significantly worse overall survival, we performed survival analysis in patients with AML stratified by their age into younger patients (<60) and older patients (≥60). In patients ≥60, high CD97 (Z ≥1) expression was associated with shorter OS compared with low CD97 (Z <1) (median OS: 3.3 months vs. 11 months, p = 0.0019; Figure 7A) and a decrease in DFS though not significant (median DFS: 5.9 months vs. 17 months, p = 0.092; Supplementary Figure E6A, online only, available at www.exphem.org). A similar trend but not statistically significant association was observed in younger patients (median OS: 27.75 months vs. 53.9 months; p = 0.2; Figure 7B) and (median DFS: 11.7 months vs. 20.8 months, p = 0.1; Supplementary Figure E6B).
Figure 7.
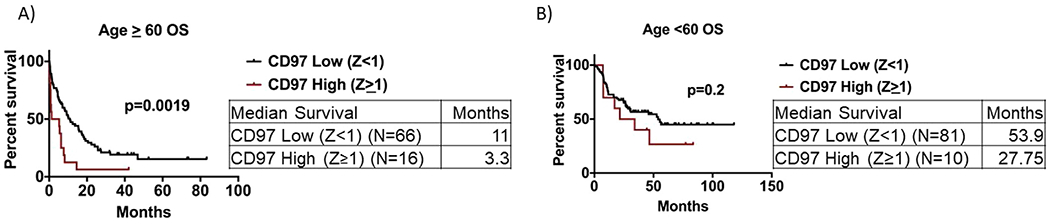
Survival analysis of AML patients with respect to CD97 expression after stratification based on age. (A) Overall survival of patients ≥60 years of age with CD97 high (Z score ≥1) versus CD97 low (Z score <1). (B) Overall survival of patients <60 years of age with CD97 high (Z score ≥1) versus CD97 low (Z score <1).
Pathways involved in the leukemia stem cell interaction with the bone marrow niche are activated in patients with high CD97 expression
We further evaluated the CD97-associated cell signaling pathways through ingenuity pathway analysis (Supplementary Figure E7, online only, available at www.exphem.org). Patients were divided into CD97 high (Z >2, N = 11) and CD97 low (Z <–1, N = 10). Pearson correlation scores of CD97 versus all the available gene expression data in TCGA were calculated and analyzed in IPA. Our analysis revealed that the integrin signaling, IL-8 signaling, CXCR4 signaling, and ILK signaling pathways were all activated in patients with high CD97 expression (Z >2). Importantly, these signaling pathways were also downregulated in patients with low CD97 expression (Z <–1). This further validates the association between CD97 levels and activation of pathways involved in the LSC–BM niche interaction.
Discussion
CD97 has been reported to play a role in disease progression of a variety of malignancies by regulating cell invasion and metastasis. In glioma, high CD97 expression is associated with worse survival [23]. In colorectal cancer, high CD97 expression correlated with the cells’ dedifferentiation ability [14]. With use of several cancer models including AML cells, CD97 knockdown was found to decrease cell migration and adhesion [16,23,24].
Here we report that high CD97 mRNA expression is significantly associated with poorer overall survival in AML patients. This is particularly relevant in older patients: median overall survival of patients >60 years with high CD97 was less than 4 months. This is significantly lower than the already dismal survival of these patients. In AML, the median age at diagnosis is 67 years, and more than 60% of newly diagnosed patients are older than 60 years [25]. Because of several factors, including comorbidities and disease complications, older patients are faced with challenging and not optimal therapeutic approaches compared with younger patients. Therefore, the identification of outcome predictors and possible viable targets in this subset of patients will greatly affect disease understanding and treatment outcome.
Interestingly, we found that patients with FLT3 mutations had high CD97 expression. Previous in vitro studies found that AML cell lines harboring a FLT3- ITD mutation had higher CD97 expression than cell lines with FLT3-WT [26]. Similarly, it was also reported that inhibiting FLT3-ITD in AML cell lines with PKC412, a FLT3-ITD inhibitor, resulted in reduced CD97 expression [26]. The mechanism by which FLT3-ITD and CD97 pathways interact remains unclear. Additionally, our analysis also reveals a positive association between CD97 and NPM1 mutations. NPM1 mutations are associated with a favorable outcome in AML [27]. Contrastingly, there is a trend for patients with NPM1 mutation and high CD97 expression (Z ≥1) to have a worse survival outcome compared with patients with NPM1 mutation and low CD97 expression. Additionally, patients with NPM1 wild type and high CD97 expression (Z ≥1) have significantly worse survival outcome compared with patients with NPM1 wild type and low CD97 expression. This suggests a benefit to including CD97 expression in prediction of survival in patients with NPM1 mutation. The mechanistic interplay between CD97 and NPM1 is yet to be determined.
The identification of integrin and ILK signaling pathway to be activated in CD97 high groups is of particular interest, as CD97 was previously reported to act as a chemoattractant for migration and invasion of human umbilical vein endothelial cells (HUVECs) in an integrindependent manner [11]. Importantly, integrin and ILK signaling pathways have been shown to contribute to leukemia stem cell survival [10]. Additionally, IL-8 and CXCR4 signaling pathways are among the most significant pathways in the high CD97 group. IL-8 was previously reported to be regulated by hypoxia, as well as to influence the niche formation in the bone marrow by inducing the migration of mesenchymal stromal cells (MSC) [28]. Furthermore, high IL-8 expression also correlated with a worse clinical outcome in non-APL AML [28]. Interestingly, mAbCD97 inhibits IL-8-induced HSC/HPC mobilization [29], suggesting that CD97 may potentially regulate the IL-8 signaling pathway. Similarly, CXCR4 expression is associated with poor prognosis of patients with AML [30]. CXCR4 plays an important role in the crosstalk between leukemic cells and the bone marrow niche [31,32]. The association between CXCR4 and CD97 is yet to be determined. Yet, knockdown of CD97 in MV4-11 AML cells impaired cell adhesion on a MSC monolayer [26]. Additionally, the extracellular domain of CD97 binds to integrins α5β1 and αvβ3, stimulating endothelial cell migration and invasion [11]. Furthermore, the interaction of CD97 with Thy-1 plays a role in the regulation of leukocyte trafficking to inflammatory sites [33]. Because CD97 binding partners (integrins and Thy-1) are also present in the MSCs, it is plausible that CD97 upregulation may contribute to the interaction between LSCs and the bone marrow niche.
Conclusions
Patients with high CD97 expression had shorter overall and disease-free survival. CD97 was higher in cytogenetically normal patients compared with cytogenetically abnormal patients. Additionally, high CD97 expression was also associated with NPM1 mutation. Our findings demonstrate that CD97 expression contributes to the clinical outcome of patients with AML, particularly older patients. This study provides a rationale for further functional and mechanistic studies aiming to understand the role of CD97 in AML.
Supplementary Material
Acknowledgments
We thank the Bioinformatics Core at the Norris Medical Library, the University of Southern California, and The Cancer Genome Atlas (TCGA) program. We acknowledge the University of Southern California School of Pharmacy Seed Fund. This study was funded by CTSI KL-2 funding for HA.
Footnotes
Conflict of interest disclosure
The authors declare no competing interests.
References
- 1.Lowenberg B Acute myeloid leukemia: the challenge of capturing disease variety. Hematology Am Soc Hematol Educ Program. 2008;1:1–11. 10.1182/asheducation-2008.1.1. [DOI] [PubMed] [Google Scholar]
- 2.Dombret H, Gardin C. An update of current treatments for adult acute myeloid leukemia. Blood. 2016;127:53–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maiga A, Lemieux S, Pabst C, et al. Transcriptome analysis of G protein-coupled receptors in distinct genetic subgroups of acute myeloid leukemia: identification of potential disease-specific targets. Blood Cancer J. 2016;6:e431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang J, Wu S, Alachkar H. Characterization of upregulated adhesion GPCRs in acute myeloid leukemia. Transl Res. In press. doi: 10.1016/j.trsl.2019.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jaspars LH, Vos W, Aust G, Van Lier RA, Hamann J. Tissue distribution of the human CD97 EGF-TM7 receptor. Tissue Antigens. 2001;57:325–331. [DOI] [PubMed] [Google Scholar]
- 6.Bonardi F, Fusetti F, Deelen P, van Gosliga D, Vellenga E, Schuringa JJ. A proteomics and transcriptomics approach to identify leukemic stem cell (LSC) markers. Mol Cell Proteomics. 2013;12:626–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hamann J, Vogel B, van Schijndel GM, van Lier RA. The sevenspan transmembrane receptor CD97 has a cellular ligand (CD55, DAF). J Exp Med. 1996;184:1185–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qian YM, Haino M, Kelly K, Song WC. Structural characterization of mouse CD97 and study of its specific interaction with the murine decay-accelerating factor (DAF, CD55). Immunology. 1999;98:303–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matsunaga T, Takemoto N, Sato T, et al. Interaction between leukemic-cell VLA-4 and stromal fibronectin is a decisive factor for minimal residual disease of acute myelogenous leukemia. Nat Med. 2003;9:1158–1165. [DOI] [PubMed] [Google Scholar]
- 10.Alachkar H, Santhanam R, Maharry K, et al. SPARC promotes leukemic cell growth and predicts acute myeloid leukemia outcome. J Clin Invest. 2014;124:1512–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang T, Ward Y, Tian L, et al. CD97, an adhesion receptor on inflammatory cells, stimulates angiogenesis through binding integrin counterreceptors on endothelial cells. Blood. 2005;105:2836–2844. [DOI] [PubMed] [Google Scholar]
- 12.Aust G, Eichler W, Laue S, et al. CD97: A dedifferentiation marker in human thyroid carcinomas. Cancer Res. 1997;57:1798–1806. [PubMed] [Google Scholar]
- 13.Ward Y, Lake R, Martin PL, et al. CD97 amplifies LPA receptor signaling and promotes thyroid cancer progression in a mouse model. Oncogene. 2012;32:2726–2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Steinert M, Wobus M, Boltze C, et al. Expression and regulation of CD97 in colorectal carcinoma cell lines and tumor tissues. Am J Pathol. 2002;161:1657–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He Z, Wu H, Jiao Y, Zheng J. Expression and prognostic value of CD97 and its ligand CD55 in pancreatic cancer. Oncol Lett. 2015;9:793–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ward Y, Lake R, Yin JJ, et al. LPA receptor heterodimerizes with CD97 to amplify LPA-initiated RHO-dependent signaling and invasion in prostate cancer cells. Cancer Res. 2011;71:7301–7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.The Cancer Genome Atlas Research Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Metzeler KH, Hummel M, Bloomfield CD, et al. An 86-probe-set gene-expression signature predicts survival in cytogenetically normal acute myeloid leukemia. Blood. 2008;112:4193–4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bullinger L, Dohner K, Bair E, et al. Use of gene-expression profiling to identify prognostic subclasses in adult acute myeloid leukemia. N Engl J Med. 2004;350:1605–1616. [DOI] [PubMed] [Google Scholar]
- 20.Cerami E, Gao J, Dogrusoz U, et al. The cBio Cancer Genomics Portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao J, Aksov BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBio-Portal. Sci Signal. 2013;6:pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tyner JW, Tognon CE, Bottomly D, et al. Functional genomic land-scape of acute myeloid leukaemia. Nature. 2018;562:526–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Safaee M, Clark AJ, Oh MC, et al. Overexpression of CD97 confers an invasive phenotype in glioblastoma cells and is associated with decreased survival of glioblastoma patients. PLoS One. 2013;8:e62765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yin Y, Xu X, Tang J, et al. CD97 promotes tumor aggressiveness through the traditional g protein-coupled receptor-mediated signaling in hepatocellular carcinoma. Hepatology. 2018;68:1865–1878. [DOI] [PubMed] [Google Scholar]
- 25.Medeiros BC, Satram-Hoang S, Hurst D, Hoang KQ, Momin F, Reyes C. Big data analysis of treatment patterns and outcomes among elderly acute myeloid leukemia patients in the United States. Ann Hematol. 2015;94:1127–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wobus M, Bornhäuser M, Jacobi A, et al. Association of the EGF-TM7 receptor CD97 expression with FLT3-ITD in acute myeloid leukemia. Oncotarget. 2015;6:38804–38815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Y, He P, Liu F, et al. Prognostic significance of NPM1 mutations in acute myeloid leukemia: A meta-analysis. Mol Clin Oncol. 2014;2:275–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuett A, Rieger C, Perathoner D, et al. IL-8 as mediator in the microenvironment-leukaemia network in acute myeloid leukaemia. Sci Rep. 2015;5:18411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Pel M, Hagoort H, Kwakkenbos MJ, Hamann J, Fibbe WE. Differential role of CD97 in interleukin-8-induced and granulocyte-colony stimulating factor-induced hematopoietic stem and progenitor cell mobilization. Haematologica. 2008;93:601–604. [DOI] [PubMed] [Google Scholar]
- 30.Spoo AC, Lubbert M, Wierda WG, Burger JA. CXCR4 is a prognostic marker in acute myelogenous leukemia. Blood. 2007;109: 786–791. [DOI] [PubMed] [Google Scholar]
- 31.Monaco G, Belmont JW, Konopleva M, Andreeff M. Correlation between CXCR4 and homing or engraftment of acute myelogenous leukemia. Cancer Res. 2004;64:6832. author reply 6832–6833. [DOI] [PubMed] [Google Scholar]
- 32.Kremer KN, Peterson KL, Schneider PA, et al. SDF-1/CXCR4 signaling induces apoptosis in acute myeloid leukemia cells via regulation of the Bcl-2 family members Bcl-XL, Noxa, and Bak. J Biol Chem. 2013;288:22899–22914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wandel E, Saalbach A, Sittig D, Gebhardt C, Aust G. Thy-1 (CD90) is an interacting partner for CD97 on activated endothelial cells. J Immunol. 2013;188:1442–1450. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


